Abstract
UAF, a yeast RNA polymerase I transcription factor, contains Rrn5p, Rrn9p, Rrn10p, histones H3 and H4, and uncharacterized protein p30. Mutants defective in RRN5, RRN9 or RRN10 are unable to transcribe rDNA by polymerase I and grow extremely slowly, but give rise to variants able to grow by transcribing chromosomal rDNA by polymerase II. Thus, UAF functions as both an activator of polymerase I and a silencer of polymerase II for rDNA transcription. We have now identified the gene for subunit p30. This gene, UAF30, is not essential for growth, but its deletion decreases the cellular growth rate. Remarkably, the deletion mutants use both polymerase I and II for rDNA transcription, indicating that the silencer function of UAF is impaired, even though rDNA transcription by polymerase I is still occurring. A UAF complex isolated from the uaf30 deletion mutant was found to retain the in vitro polymerase I activator function to a large extent. Thus, Uaf30p plays only a minor role in its activator function. Possible reasons for slow growth caused by uaf30 mutations are discussed.
Keywords: RNA polymerase I/RNA polymerase switch/Saccharomyces cerevisiae/transcription factor UAF/UAF30
Introduction
Transcription of the 35S rRNA gene (rDNA) by RNA polymerase I (Pol I) in the yeast Saccharomyces cerevisiae requires transcription factors called upstream activation factor (UAF), core factor (CF), TATA binding protein (TBP) and Rrn3p in addition to Pol I (Keener et al., 1998; for review see Nomura, 1998). Like rRNA genes in other eukaryotic organisms studied, the promoter for rDNA in S.cerevisiae consists of two elements: a core element and an upstream element. For in vitro basal transcription observed with template lacking the upstream element, three components, Pol I, Rrn3p and CF, are sufficient, but high level transcription requires template containing both core and upstream elements and two additional transcription factors, UAF and TBP (Steffan et al., 1996; Keener et al., 1998). UAF is a multisubunit complex containing three proteins (Rrn5p, Rrn9p and Rrn10p) encoded by genes that are uniquely required for Pol I transcription in vivo: RRN5, RRN9 and RRN10 (Keys et al., 1996). Purified UAF complex contains three additional protein components, two of which were previously identified as histones H3 and H4, but the remaining protein (previously called p30) with an apparent molecular mass of 30 kDa has remained uncharacterized (Keys et al., 1996; Keener et al., 1997).
In addition to allowing a high level of Pol I transcription both in vitro and in vivo, UAF also functions to prevent, or ‘silence’, transcription of rDNA by RNA polymerase II (Pol II). Mutants with a defect in RRN5, RRN9 or RRN10 were originally isolated as galactose-dependent mutants from a strain carrying the 35S rRNA gene fused to the GAL7 promoter on a multicopy plasmid (‘helper plasmid’) (Nogi et al., 1991a; Keys et al., 1996). In the absence of such a helper plasmid, these UAF-defective mutants grow extremely slowly and are mostly unable to form colonies. However, we found that these UAF-defective mutants give rise to, with a low frequency, variants able to grow by transcribing endogenous rDNA by Pol II (Oakes et al., 1999; Vu et al., 1999; Siddiqi et al., 2001). The switch to this state (called PSW for polymerase switch for growth) consists of two steps: a mutational alteration in UAF and an expansion of chromosomal rDNA repeats. The first step, a mutation in UAF, is sufficient to eliminate all Pol I transcription and to allow Pol II transcription of chromosomal rDNA, but without the second step cells are unable to form a colony (and hence, the cells are in the state defined as N-PSW for no growth, but polymerase switched). In contrast, mutations in Pol I or other Pol I transcription factors, e.g. CF or rDNA chromatin-associated proteins, e.g. Sir2p, cannot independently lead to Pol II transcription of rDNA. Thus, UAF appears to have a specific role in silencing Pol II transcription of rDNA (Oakes et al., 1999).
In this paper, we report the identification of the gene encoding the previously uncharacterized UAF subunit protein, p30. We found that this gene, which we now call UAF30, is not essential for growth, but its deletion causes a slow growth phenotype. We also found that strains carrying a deletion of this gene use both Pol I and Pol II for rDNA transcription, indicating that the function of UAF to silence Pol II transcription of rDNA is impaired, even though Pol I transcription is still taking place. This observation demonstrates that the protein p30, or Uaf30p as we now call it, is indeed a subunit of UAF, participating in its Pol II silencing function. We also carried out experiments designed to examine the role of this subunit in the Pol I activating function of UAF. Our findings indicate that the latter function is at least partially retained in the absence of Uaf30p both in vivo and in vitro, thus explaining the finding that UAF30 is not essential. We discuss roles of Uaf30p in the two known functions of UAF, activation of Pol I transcription and silencing of Pol II transcription of rDNA.
Results
Identification of the gene for the 30 kDa subunit of UAF
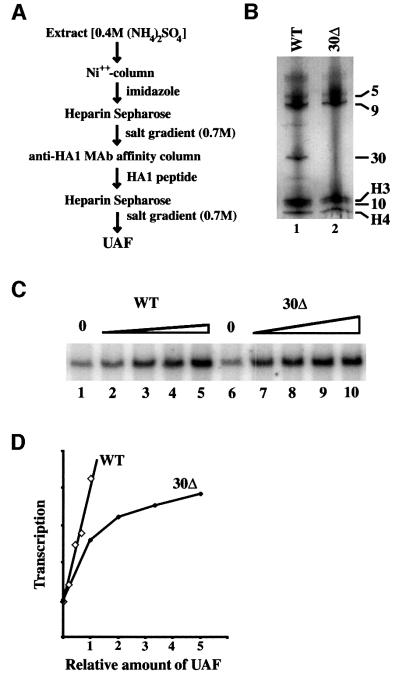
Transcription factor UAF was purified from strain NOY798 as described previously (Keener et al., 1997; see also below and Figure 6A) and subjected to SDS–PAGE (Figure 1, lane 1). The band corresponding to p30 was analyzed by tryptic digestion followed by HPLC/tandem mass spectrometry. The data obtained were compared with the OWL non-redundant protein sequence database, and an uncharacterized open reading frame (ORF), YOR295W, on chromosome XV was found to be the candidate gene (Figure 2A). This ORF encodes a 228 amino acid protein with a calculated molecular mass of 25 970 Da and a calculated isoelectric point of 9.01.
Fig. 6. Comparison of in vitro Pol I stimulatory activities of UAF and mutant UAF without Uaf30p. (A) Scheme for purification of UAF. (B) A silver-stained gel of purified UAF preparations from control strain NOY798 (lane 1) and uaf30 deletion strain NOY1047 (lane 2). Proteins in the preparations were precipitated with 20% TCA and then subjected to 10–15% SDS/pHast gel electrophoresis. (C and D) Comparison of stimulatory activities of purified UAF preparations from the mutant (‘30Δ’) and the control (‘WT’) strains. The mutant preparation was first concentrated by microcon (Millipore) and relative concentrations of UAF in the two preparations were determined by measuring the amounts of the tagged Rrn5p by western immunoblot analysis (data not shown). The results of transcription experiments shown in (C) were quantified and are shown in (D) based on the relative amounts found in the western analysis.
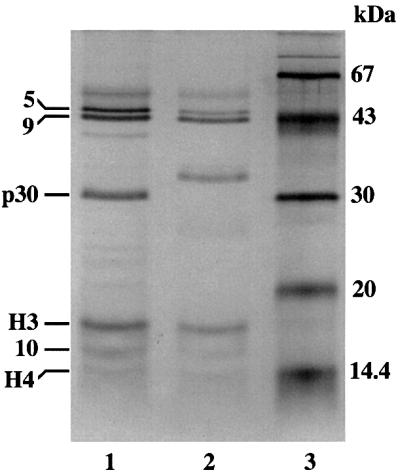
Fig. 1. Silver-stained gels of highly purified UAF (prepared from NOY798) and UAF containing the myc-tagged 30 kDa subunit (prepared from NOY1048). These two preparations are shown in lanes 1 and 2. Lane 3 contains molecular weight markers. The components of UAF are indicated as follows: 5, Rrn5p with the (HA1)3-(His)6 tag; 9, Rrn9p; 10, Rrn10p; H3 and H4, histones H3 and H4, respectively; p30, Uaf30p.
Fig. 2. The predicted amino acid sequence of Uaf30p (YOR295W ORF) (A) and SWIB domains found in Uaf30p and some other proteins (B). In (A), four tryptic peptides identified by mass spectrometry are underlined. In (B), SWIB domains in Uaf30p, YMR233p, S.pombe ORF SPCC285.17, human BAF60a and Chlamydia trachomatis topoisomerase I (cfTopI) (TrEMBL 084649) are aligned, and a consensus sequence (defined as residues identical or similar through the first four sequences) is shown. In addition to these proteins, a portion of Snf12p (also called Swp73p) is aligned according to Wang et al. (1996). It should be noted that Snf12p has an extra 30 amino acids inserted within the SWIB domain and its similarity to BAF60a (and other proteins shown) is not high in this region even though this protein is considered to be an S.cerevisiae homolog of human BAF60a (Wang et al., 1996). Residues that agree with the consensus sequence are indicated as shaded. In addition, residues showing identity or similarity in three out of the first four sequences are shaded.
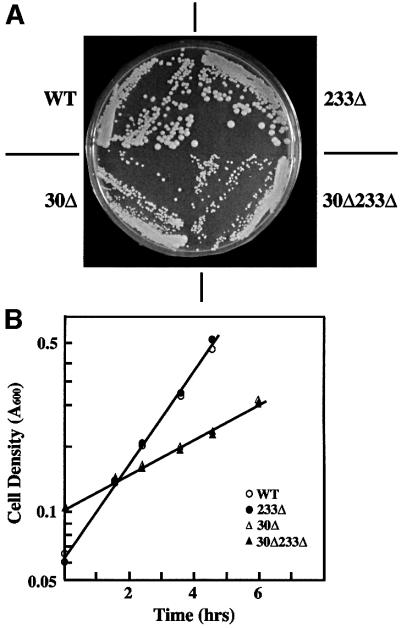
To confirm that the YOR295W ORF is the gene for p30, we cloned this ORF from yeast genomic DNA by PCR and constructed a null allele by replacing the coding region with the HIS3 gene. We then deleted the YOR295W ORF in a ‘wild-type’ yeast strain (NOY388 or NOY556) by a standard gene replacement technique and analyzed the phenotype of deletion strains. As shown in Figure 3 (deletion of YMR233W, also shown in Figure 3, will be discussed below), deletion of this ORF (‘uaf30Δ’; NOY1034 analyzed) was found to lead to a slow growth phenotype. This phenotype is clearly different from deletions of the RRN5, RRN9 and RRN10 genes, which encode other components of UAF. Deletions of the latter three genes cause much stronger growth defects, and allow only a very small fraction of cells to form colonies after switching to the PSW state (Vu et al., 1999; Siddiqi et al., 2001; see Introduction). To prevent switching to the PSW state, strains carrying deletion of either RRN5, RRN9 or RRN10 are maintained by introducing a helper plasmid carrying a fusion gene, GAL7-35S rDNA, on a multi-copy plasmid. In contrast, cells with the YOR295W ORF deletion grow fairly well without such a helper plasmid (∼3.8 to ∼2.5 h doubling time in the YEPD medium at 30°C; variability of the growth rate will be commented on below), but they still grow more slowly than wild-type cells (∼1.5 h doubling time in the same medium; see Figure 3B).
Fig. 3. Effects of deletion of UAF30, deletion of YMR233W ORF and deletion of both on cell growth. Strains NOY388 (WT), NOY1034 (uaf30Δ or ‘30Δ’), NOY1016 (ymr233wΔ or ‘233Δ’) and NOY1044 (uaf30Δ ymr233wΔ or 30Δ 233Δ) were streaked on YEPD and incubated at 30°C for 4 days (A), and also grown in YEPD liquid medium to measure growth rates (B).
We then constructed a CEN plasmid carrying the YOR295W ORF with a myc tag attached to the N-terminus of the ORF. This plasmid (pNOY512) was introduced into a strain carrying the chromosomal YOR295W ORF deletion [NOY1047; a YOR295W deletion strain derived from a strain carrying the (HA1)3-(His)6-tagged RRN5 for the purpose of UAF purification]. The resulting strain was found to have a growth rate identical to the control strain without the YOR295W deletion (data not shown), indicating that the myc epitope tag did not affect the function of the protein. UAF was then purified from this strain and the sizes of its protein components were compared with those of the corresponding components of a standard UAF preparation by SDS–PAGE. As shown in Figure 1 (compare lane 2 with lane 1), the mobility of p30 was slower relative to p30 of the standard UAF. The result demonstrates that the YOR295W ORF indeed encodes protein p30. As we described previously (Keys et al., 1996; see also Keener et al., 1997 and Figure 6A), p30 co-immunoprecipitates with the other UAF components and copurifies with these components through further purification steps, including several ionic exchange column chromatographic steps and a sizing column chromatography. There is no doubt that p30 is tightly associated with the other UAF components previously identified, forming a UAF complex with a size of ∼200 kDa (Keys et al., 1996). In addition, as we describe below, there is evidence indicating that this protein plays a role in the function of UAF to silence Pol II transcription of rDNA and possibly in stabilizing the structure of UAF for in vitro stimulation of Pol I transcription, and hence, can be considered a subunit of UAF. For these reasons, we now call this protein Uaf30p and the gene (YOR295W) UAF30.
Deletion of UAF30 allows transcription of rDNA repeats by both Pol I and Pol II
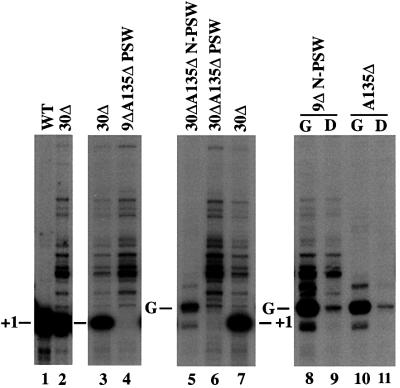
In contrast to strains carrying rrn5, rrn9 or rrn10 deletions, which are initially unable to grow unless they carry a helper plasmid and use a galactose medium, strains carrying the uaf30 deletion (uaf30Δ) grow fairly well, as described above. In strains carrying rrn5Δ, rrn9Δ or rrn10Δ mutations (and carrying a helper plasmid for growth), there is no transcription of chromosomal rDNA by Pol I. Instead, there is a low level of transcription by Pol II (recognized after transfer from galactose to glucose medium; see Figure 4, lane 9, for a rrn9Δ strain; Oakes et al., 1999). Pol II transcription reaches a significant level after rDNA repeat expansion, which allows cells to grow without a helper plasmid and thus achieve the PSW state (Oakes et al., 1999). For this reason, we first asked whether uaf30Δ strains synthesize rRNA using Pol I or Pol II. We analyzed RNA isolated from a uaf30Δ strain (NOY1035) by primer extension using a primer specific for the 35S precursor rRNA. Pol I transcription in wild-type strains results in a distinct start site for rRNA transcription, but Pol II transcription in PSW strains uses multiple upstream start sites (Oakes et al., 1999; Vu et al., 1999). This difference in start sites is evident in Figure 4 when comparing a wild-type strain (lane 1) with strain NOY897 (lane 4), which is rrn9Δ, rpa135Δ and grows without a helper plasmid.
Fig. 4. Primer extension analysis of precursor rRNA transcripts for detection of Pol II transcription of chromosomal rDNA. RNA was prepared from the following strains: NOY556 (WT), NOY1035 (uaf30Δ or ‘30Δ’) and NOY897 (rrn9Δ rpa135Δ PSW or ‘9Δ A135Δ PSW’) grown on YEPD (lanes 1–4). In one experiment, primer extension reactions were performed with 0.75 µg of total RNA for NOY556 (WT; lane 1) and 1.5 µg for NOY1035 (‘30Δ’; lane 2). In another experiment, 5 µg of total RNA for NOY1035 (‘30Δ’; lane 3) and 2.5 µg for NOY897 (lane 4) were analyzed. In the third experiment, NOY1037 (‘30Δ A135Δ N-PSW’; carries helper plasmid), NOY1050 (‘30Δ A135Δ PSW’) and NOY1034 (‘30Δ’) were grown on YEP-galactose (for NOY1037) or on YEPD (for NOY1050 and NOY1034) (lanes 5–7). RNAs used were 2.5 µg for NOY1037 and NOY1050, and 5.0 µg for NOY1034. In the fourth experiment, NOY921 (‘9Δ N-PSW’; carries helper plasmid) and NOY408–1a (‘A135Δ’; carries helper plasmid) were grown in supplemented synthetic galactose medium (G; lanes 8 and 10, respectively). Portions were used to isolate RNA, and the remaining portions were shifted to supplemented synthetic glucose medium and incubated for 1 h (D; lanes 9 and 11, respectively). RNAs analyzed were 2.5 µg for all these samples (lanes 8–11). The ‘+1’ refers to the published Pol I start site. Bands seen in lanes 4 and 6 represent start sites used by Pol II transcription of chromosomal rDNA. The bands marked as ‘G’ represent the rRNA start site originating from the GAL7-35SrDNA fusion gene on plasmids pNOY102 and pNOY103. Bands seen in lane 11 represent residual transcripts from the fusion gene on the plasmid that escaped from repression by glucose. The same residual bands together with the Pol II transcripts of chromosomal rDNA can be seen in lane 9. We note that the uaf30Δ strain analyzed here (lanes 2, 3 and 7) grew in YEPD media, and rRNA transcription by Pol II was 11.4, 13.3 and 14.1%, respectively, of total rRNA transcription, whereas the value obtained for the same strain grown in synthetic glucose medium (see experiments summarized in Table I) gave a corresponding value of 5.3%.
Interestingly, we have found that the uaf30Δ strain uses both Pol I and Pol II for rRNA transcription, as shown by the presence of both types of start site (Figure 4, lanes 2 and 3). Most transcripts are Pol I derived, but ∼10% of total rRNA transcription is clearly carried out by Pol II. (Values for Pol II transcription ranged from 5.3 to 14.1% and may vary depending on media used for cell growth; see the legend to Figure 4.) We conclude that, like a rrn5Δ, rrn9Δ or rrn10Δ mutation, the uaf30Δ mutation allows Pol II transcription of chromosomal rDNA; i.e. the Uaf30p subunit also participates in the UAF function to silence rDNA transcription by Pol II. However, in contrast to a rrn5Δ, rrn9Δ or rrn10Δ mutation, the uaf30Δ mutation does not abolish Pol I transcription (compare Figure 4, lanes 2, 3 and 7 with lane 9), but leaves it efficient enough so that the uaf30Δ mutant strain can attain a growth rate as high as ∼60% of the control UAF30 strain. In several experiments using a supplemented synthetic glucose medium, growth rates and rRNA synthesis rates of a uaf30Δ strain and a control UAF30 strain were compared and the results are summarized in Table I. The mutant strain showed an rRNA synthesis rate that is ∼30% of the control strain, as judged by both measurements of synthesis rates estimated from primer extension experiments and measurements of rRNA accumulation rates. Whether the decrease in rRNA synthesis rate (and that in growth rate) is a consequence of a defect in the function of UAF as a transcription factor of Pol I is discussed below.
Table I. Comparison of rRNA synthesis rates in uaf30Δ and UAF30 strains growing in a synthetic glucose medium.
| Strain | Growth rate |
[14C]uracil incorporation (ΔRNA/ΔA600) (3) | Accumulation rate |
rRNA synthesis rate (primer extension) (6) | ||
|---|---|---|---|---|---|---|
| DT (h) (1) | relative rate (2) | Total RNA (4) | rRNA (5) | |||
| UAF30 | 1.5 | (1.0) | 1.0 | 1.0 | 1.0 | 1.0 |
| uaf30Δ | 2.5 | (0.6) | 0.55 | 0.33 | 0.31 | 0.33 |
Two methods were used to compare rRNA synthesis between the wild type and uaf30 deletion mutant. One was the use of radioactive uracil to compare the rate of rRNA accumulation (column 5) and the second was to use primer extension to estimate rRNA synthesis rate (column 6). Strains NOY556 (UAF30) and NOY1035 (uaf30Δ::HIS3) were grown in supplemented SD medium containing [14C]uracil (5 µg/ml, 0.4 µCi/ml). Increases in cell density (A600) and total TCA-insoluble 14C-labeled RNA were determined during exponential phase of growth, and cellular RNA content was calculated as ΔRNA/ΔA600. Rates of accumulation of total RNA (4) were calculated by multiplying cellular RNA content (3) by growth rate (2). Rates of accumulation of rRNA (5) were calculated by multiplying this value (4) by a correction factor (to correct the small difference in the ratio, rRNA/total RNA, between the two strains, which was 0.93 as analyzed by gel electrophoretic analysis of 14C-labeled total RNA). Rates of rRNA synthesis per unit amount of cell mass (6) were calculated from the amounts of the 5′-ends of unstable 35S precursor rRNA per unit amount of RNA (determined by primer extension) combined with the values for cellular RNA content (3). The primer extension method depends on the fact that the 5′-ends of 35S precursor RNA are processed away from mature rRNA and have a very short half-life, and hence, the amounts of these unstable 5′-ends reflect rRNA synthesis rates. The fact that relative synthesis rates (1.0 versus 0.33) and relative accumulation rates (1.0 versus 0.31) are similar indicates that there is no significant difference in the efficiency of processing between the two strains. We also note that the primary extension method used to estimate rRNA synthesis rate has the advantage of avoiding the problem of cellular RNA precursor pools inherent in pulse labeling methods using radioactive RNA precursors. All the values are normalized to those for the control UAF30 strain except for doubling time (DT). It should also be noted that the uaf30Δ strain used showed a doubling time of 2.5 h in this medium throughout several experiments described here without any variability.
Comparison of deletion of UAF30 and RRN9 in establishing the PSW state in Pol I defective strains
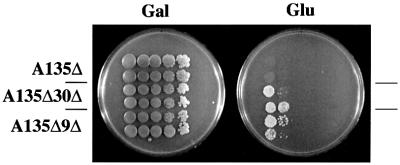
Strains that carry mutations in essential CF or Pol I subunit genes (e.g. rrn6 or rpa135 deletion, respectively) and grow by transcribing the GAL7-35SrDNA hybrid gene on a helper plasmid do not exhibit Pol II transcription of rDNA and cannot undergo polymerase switch; i.e. these mutant strains can grow on galactose, but fail to produce PSW variants on glucose plates. However, if a mutation in UAF subunit genes RRN5 or RRN9 (RRN10 not tested) was introduced into these strains carrying the rrn6 or rpa135 deletion, the resultant double mutants were able to produce variants that could grow on glucose (Oakes et al., 1999). We examined whether deletion of UAF30 also allows polymerase switching in Pol I mutant strains. We constructed a uaf30 deletion derivative of a Pol I mutant strain (NOY408-1a) carrying the rpa135 deletion. Colonies of the rpa135Δ strain and the rpa135Δ uaf30Δ strain (NOY1037) were analyzed by a galactose-glucose spot test for their ability to produce PSW variants. A positive control strain, a rrn9Δ derivative of the rpa135Δ strain (NOY896), was also analyzed. As shown in Figure 5, the strain carrying the uaf30Δ mutation in addition to rpa135Δ produced, upon a prolonged incubation, variants able to grow on glucose, as did the strain carrying the rrn9Δ mutation, whereas the rpa135Δ strain without the uaf30Δ mutation did not. This result is consistent with the conclusion that Uaf30p is important in silencing Pol II transcription of rDNA. We have also observed that, as in the case of switch from the N-PSW to the PSW state in rrn9Δ strains (Vu et al., 1999), when cells in the colonies formed on glucose plates shown in Figure 5 were analyzed once more by spot test, a majority of them failed to show growth on glucose, i.e. they were not stable PSW variants in both rrn9Δ rpa135Δ and uaf30Δ rpa135Δ strains. However, by repeated streaking on YEP glucose plates or streaking on glucose plus 5-fluoroorotic acid plates (5-fluoroorotic acid selecting cells that have lost the helper plasmid), it was easy to isolate stable rpa135Δ uaf30Δ PSW variants as well as stable rpa135Δ rrn9Δ PSW variants that are able to grow in the absence of a helper plasmid. One such rpa135Δ uaf30Δ PSW variant (NOY1050), which lost the helper plasmid, was analyzed for rDNA transcription by primer extension. As shown in Figure 4, the pattern of 5′-ends of precursor rRNA (lane 6) observed for this strain was almost identical to that observed for the rrn9Δ rpa135Δ PSW strain (NOY897, lane 4). As expected, no transcription of rDNA by Pol I or from the GAL7 promoter by Pol II was present (compare lane 6 with 5 and 7). In summary, we did not find a significant difference between UAF30 deletion and RRN9 deletion in establishing the PSW state in Pol I defective strains.
Fig. 5. Comparison of deletion of UAF30 and that of RRN9 in allowing Pol I deletion strain, NOY408-1a, to undergo polymerase switch. Two independent colonies of NOY408-1a (A135Δ), a uaf30Δ derivative (NOY1037) of NOY408–1a (A135Δ 30Δ) and a rrn9Δ derivative (NOY896) of NOY408-1a (A135Δ 9Δ) were suspended in water and analyzed by spotting aliquots of 10-fold serial dilutions on YEP-galactose (Gal) and YEPD (Glu). Plates incubated at 30°C for 11 days are shown.
Purification of UAF from a uaf30Δ strain
Deletion of UAF30 causes a decrease in growth rate as well as in rRNA synthesis rate. To study whether the decrease in rRNA synthesis rate observed in vivo is a consequence of a direct impairment of the UAF function as a Pol I transcription factor, we examined whether a stable UAF complex lacking Uaf30p is present in extracts of uaf30Δ strain NOY1047, and, if a complex is present, how its function as a transcription factor is affected by the absence of the Uaf30p subunit. By following the same purification procedure as used previously for the UAF30 strain (Figure 6A), we found that a UAF complex exists in the uaf30 deletion strain in an amount comparable to the wild type (examined by western immunoblot analysis; data not shown) and can be further purified. Figure 6B shows a silver-stained gel of a purified preparation analyzed side by side with a standard UAF preparation obtained from the UAF30 strain. The preparation from the uaf30Δ strain contained all the other UAF subunit proteins except for Uaf30p (compare lane 2 with 1). The results demonstrate that Uaf30p does not play a significant role in the assembly of the other subunits to form a complex.
The activity of the purified complex without the Uaf30p subunit was analyzed and compared with a control-purified UAF preparation (Figure 6C). Comparison of the activities after normalization to the amount of tagged Rrn5p indicated that the mutant UAF was ∼2-fold weaker than the control UAF (Figure 6D). It is not clear whether this difference indicates a functional role for Uaf30p in the activator function of UAF to stimulate rDNA transcription by Pol I. It is possible that the mutant UAF without the Uaf30p subunit might be more unstable and the differences in the activity between the mutant and control preparations observed after all purification steps might be due to an instability of the mutant UAF structure. In fact, comparison of the stimulatory activity at an earlier stage of purification (after the first heparin–Sepharose step shown in Figure 6A) showed a smaller decrease in the activity of the mutant UAF relative to the control UAF preparation (data not shown).
Protein encoded by YMR233W with a sequence similarity to Uaf30p is apparently not related to Uaf30p in its function(s) to support cell growth
The amino acid sequence of Uaf30p was analyzed for its similarity to other proteins in the protein sequence database. Among yeast genes, one uncharacterized ORF, YMR233W, was found to encode a protein that is similar to Uaf30p. The two proteins are similar in size (YMR233W ORF encoding a protein of 226 amino acids compared with Uaf30p, which is 228 amino acids) and in amino acid sequence (38% identity and 58% similarity). A portion of Uaf30p (51 amino acids from position 126 to 176) was also found to have a similarity to a portion of the 60 kDa subunit of human SWI/SNF chromatin-remodeling complexes, BAF60a (Wang et al., 1996; Xue et al., 2000), showing 43% identity and 61% similarity in this region (Figure 2B). We have also found in the database a hypothetical protein of Schizosaccharomyces pombe, SPCC285.17, which is a 234 amino acid protein and shows, in a 221 amino acid region (allowing three gaps), 34% identity and 54% similarity to Uaf30p. However, this S.pombe protein also shows the same degree of similarity to the S.cerevisiae YMR233W ORF and it is not clear whether the protein is an S.pombe ortholog of Uaf30p or of YMR233W protein. All three of these proteins show the same degree of strong similarity to human BAF60a in the region mentioned above (Figure 2B), suggesting that this region may represent a functionally significant domain.
Because of its high degree of similarity to Uaf30p, we considered the possibility that the YMR233W protein might have a function related to that of Uaf30p. For example, the reason for the partial effect of the uaf30 deletion on growth (and on rRNA synthesis) might be the presence of the YMR233W protein carrying out a function overlapping that of the Uaf30p. We cloned a DNA region containing the YMR233W ORF from genomic DNA by PCR and then constructed strains in which this ORF region was deleted. We found that deletion of YMR233W from a standard wild-type strain (NOY556) or from a uaf30Δ strain did not cause any alteration in growth phenotypes, as judged either by colony size (Figure 3A) or by measuring doubling time in YEPD liquid medium (Figure 3B). We conclude that the (hypothetical) protein encoded by YMR233W does not play a functional role overlapping that of UAF30 and is unimportant for cellular growth under the conditions studied.
Discussion
Roles of the Uaf30p subunit in UAF functions to activate rDNA transcription by Pol I and to silence rDNA transcription by Pol II
We have identified the gene, now called UAF30, for the previously uncharacterized 30 kDa subunit of Pol I-specific transcription factor UAF and examined functional roles of the encoded protein, Uaf30p. In contrast to RRN5, RRN9 and RRN10, the (nearly) essential genes encoding three other specific subunits, we found that UAF30 is not essential, but its deletion causes a partial decrease in growth rate. In fact, we have found that a purified UAF complex missing the Uaf30p subunit largely retains an in vitro Pol I stimulatory activity, showing a decrease of activity 2-fold or less relative to the intact UAF. Thus, Uaf30p does not appear to play a significant role in the activation function of UAF. In contrast, Uaf30p plays a definite role in silencing Pol II transcription of chromosomal rDNA repeats, a role shared by the other three UAF subunits, Rrn5p, Rrn9p and Rrn10p, but not by other proteins in the Pol I transcriptional machinery or some known rDNA chromatin proteins tested (Oakes et al., 1999).
Mutant uaf30Δ strains show a decrease in rRNA synthesis rate. Under the particular conditions analyzed, the decrease was ∼70%. It is possible that Uaf30p plays a direct role in Pol I transcription of rDNA in vivo, and the decrease in growth rate observed in uaf30Δ strains is caused by a defect in the UAF function as an activator of Pol I transcription. A partial decrease of the in vitro activity observed with a purified mutant UAF preparation may reflect the in vivo decrease in rRNA synthesis rate. It should be noted, however, that the decrease in rRNA synthesis rate does not necessarily prove a defect in the Pol I machinery. It is known that the cellular content of ribosomes decreases with decreasing growth rate (see Kief and Warner, 1981; Warner, 1989). Therefore, the possibility cannot be excluded that cellular growth is inhibited by a defect in a function of Uaf30p separate from its possible role in the activator function of UAF, and that the rRNA synthesis rate is simply adjusted to that appropriate for decreased growth rate by a still uncharacterized regulatory mechanism. For example, it is possible that Pol II transcription of rDNA, i.e. a defect in the silencing function of UAF caused by the uaf30Δ deletion, leads to inhibition of rRNA synthesis indirectly, perhaps through a competition between the inefficient Pol II transcription machinery and the efficient Pol I transcription process. According to this model, silencing of Pol II transcription of rDNA by UAF is a physiologically important function. Silencing is presumably achieved by a specific rDNA chromatin structure, in which UAF plays a crucial role, and is important to ensure an efficient operation of the Pol I machinery.
Regarding the question discussed above, we compared growth rates of the Pol I deletion (rpa135Δ) strain carrying a GAL7-35SrDNA helper plasmid with its uaf30Δ mutant derivative (rpa135Δ uaf30Δ; N-PSW; containing helper plasmid), both of which are galactose dependent for growth. We reasoned that if the slow growth of uaf30Δ strains is due entirely to reduced efficiency of rDNA transcription by Pol I, either directly or indirectly, we would not expect any effect of the uaf30Δ mutation on growth when growth of cells is achieved by rRNA synthesis from the fusion gene on a helper plasmid using Pol II instead of Pol I. Unexpectedly, the uaf30Δ derivative of the Pol I deletion strain carrying a helper plasmid showed a significantly slower growth rate (doubling time ∼7 h) relative to the control Pol I deletion strain (doubling time ∼4.5 h) in galactose media. The result suggests that a decrease in growth rate observed for the original uaf30Δ strain (without the Pol I subunit deletion) in YEPD medium (Figure 3) may not be due entirely to a defect in the Pol I activation function of UAF, but rather at least partly to a defect(s) in some other function(s) of UAF. The possible defects in other functions may involve loss of Pol II silencing or other unidentified change(s) caused by the mutation, leading to harmful effects on growth separate from effects on Pol I transcription. Further studies are required to settle this question.
It was previously observed that rrn9Δ (or rrn5Δ or rrn10Δ) mutants growing in galactose media by transcribing the GAL7-35SrDNA fusion gene on a plasmid have a reduced rDNA repeat number (about half of the normal repeat number, i.e. ∼80), and that switch to the PSW state requires an expansion of repeats to ∼400 (Oakes et al., 1999). We observed that the uaf30Δ strains growing mostly by Pol I transcription of rDNA, but showing weak Pol II transcription of rDNA, carried rDNA with a repeat number expanded to a considerable extent, resembling PSW strains derived from rrn9Δ (or rrn5Δ or rrn10Δ) mutants (M.L.Oakes, unpublished observations). By freshly preparing a uaf30Δ disruption strain and measuring its growth rate and average rDNA repeat number of cell populations, we found that there is an increase in growth rate upon repeated subculture that appears to be accompanied by an increase in rDNA repeat number. The experiments shown in Figure 3 were carried out using freshly prepared uaf30Δ strains, which showed a doubling time of 3.8 h. After subsequent repeated subculturing, a clone of the uaf30Δ strain analyzed in the same YEPD medium showed a doubling time of 2.5 h and remained in the same state upon further subculturing (experiments shown in Table I were carried out using such a uaf30Δ strain). It is known that the FOB1 gene is required for rDNA repeat expansion and contraction (Kobayashi et al., 1998). Preliminary analysis of several fob1 deletion clones obtained from a uaf30Δ strain, which have stabilized rDNA repeat numbers, showed a correlation between increases in copy numbers and improvements in growth rate (L.Vu, M.L.Oakes and M.Nomura, unpublished observations). The significance of these observations is under current study.
Transcription of chromosomal rDNA by both Pol I and Pol II
In addition to the phenomenon of polymerase switch discovered for mutants of RRN5, RRN9 or RRN10 already discussed, there have been several reports describing transcription of rDNA by Pol II in different experimental systems (Smale and Tjian, 1985; Dhar et al., 1987; Conrad-Webb and Butow, 1995; Doelling and Pikaard, 1996). However, in none of these previous instances is the same chromosomal rDNA template transcribed by both Pol I and Pol II under a defined condition. Mutant uaf30Δ strains are unique in that cells use both Pol I and Pol II to transcribe chromosomal rDNA repeats. Two possibilities for this observation can be considered. The first possibility is that a given cell can take only one of the two reversible epigenetic states, one allowing transcription by Pol I and the other transcription by Pol II, and that the cell population is heterogeneous. This possibility may be less likely, but cannot be excluded. The second possibility is that both polymerases are engaged in rDNA transcription simultaneously within a single cell. In this case, certain rDNA repeats may be dedicated exclusively to Pol II transcription and the remaining repeats to Pol I transcription. Alternatively, the same rDNA repeats might be transcribed by both Pol I and Pol II. In either case, it would be interesting to ask whether rDNA repeats transcribed by Pol I and those transcribed by Pol II are localized within the same location (perhaps in the nucleolus at the nuclear periphery) or in separate regions (Pol I at the nuclear periphery as in normal strains, and Pol II at more interior locations as in PSW strains; see Oakes et al., 1999) in the nucleus. We have not examined these questions.
SWIB domains found in Uaf30p and YMR233W protein
The structural similarity of a portion of Uaf30p to a region of BAF60a subunit of human (and mouse) SWI/SNF complexes raises an interesting possibility of the presence of a domain with a common functional/structural role shared by Uaf30p and BAF60a (and BAF60b, BAF60c and their mouse homologs, all very similar to BAF60a, as well as S.cerevisiae YMR233Wp and its S.pombe homolog, SPCC285.17). Although a considerable amount of work has been done on SWI/SNF chromatin remodeling complexes (for recent reviews see Aalfs and Kingston, 2000; Sudarsanam and Winston, 2000), the role of the BAF60a subunit in these large multiprotein SWI/SNF complexes is unknown. [The yeast homolog of mammalian BAF60a is apparently Snf12p (also called Swp73p) (Cairns et al., 1996; Xue et al., 2000). Curiously, the common domain in question does not appear to be present in its intact form in Snf12p (see Figure 2B).] Nevertheless, both mammalian (or yeast) SWI/SNF chromatin remodeling complexes and the UAF complex play important roles as a transcriptional regulator influencing chromatin structures, and, therefore, the common structural feature of a domain in a subunit of these complexes may be related to such a function. Alternatively, this domain might simply be involved in protein–protein interactions. It is also interesting to note that the domains discussed here (called SWIB domains) are unique to eukaryotes, but a copy of the domain was found fused to the C-terminus of topoisomerase I in an obligate intracellular pathogen of humans, Chlamydia trachomatis (Stephens et al., 1998). It was speculated that this unique topoisomerase I may have evolved as a result of horizontal transfer of a eukaryotic gene and may function in the chromatin condensation–decondensation that is characteristic of the developmental cycle of this organism (Stephens et al., 1998).
In conclusion, we have identified the gene encoding the hitherto uncharacterized 30 kDa subunit of UAF. This subunit, Uaf30p, plays a significant role in silencing transcription of rDNA by Pol II, but its role in rDNA transcription by Pol I is apparently minor, at least in vitro. In this connection, there is a possibility that Uaf30p (and perhaps other subunits Rrn5p, Rrn9p and Rrn10p) plays a role in cell growth that is independent of the role of UAF as a Pol I-specific transcription factor. A unique finding in the present study is that mutant uaf30Δ cells use both Pol I and Pol II to transcribe chromosomal rDNA repeats. This finding poses an interesting question as to how cells can maintain and regulate two separate polymerase systems capable of carrying out apparently the same function, transcription of chromosomal rRNA genes. Studying such a question may be related to the general question of epigenetic control of rRNA gene functions. It might also lead to some insights regarding the evolution of the Pol I machinery and nucleolar structures, features unique to eukaryotic cells (see discussion in Oakes et al., 1999; Vu et al., 1999). Finally, the present study concludes identification of genes for all the known components of UAF, and should encourage more detailed analysis of the structure and functions of UAF and its individual components, especially histones H3 and H4, whose functional roles have yet to be studied experimentally.
Materials and methods
Yeast strains, plasmids and media
Yeast strains and plasmids used are listed in Table II. The UAF30 gene (YOR295W) was amplified from yeast genomic DNA by PCR using primers 1, 5′-AAAACTCGAGCAAGAGAAAAGCCACTGCCAAAGGC-3′ (containing an added XhoI site, which is underlined), and 2, 5′-AAAAGGATCCGCTGCTTCTTTAAATCGCGGTTCGT-3′ (containing an added BamHI site, which is underlined). The 1.7 kb PCR product contains the entire 700 bp UAF30 ORF, as well as 570 bp of upstream sequence and 477 bp of downstream sequence. The PCR product was digested with BamHI and XhoI and cloned into the same sites of pBluescript KS(–) to construct pNOY509 or into pRS316 (Sikorski and Hieter, 1989) to construct pNOY511.
Table II. Yeast strains and plasmids used.
| Strain or plasmid | Description |
|---|---|
| Strains | |
| NOY388 | MATa ade2-1 ura3-1 trp 1-1 leu2, 3–112 his3-11 can1-100 |
| NOY408–1a | MATα ade2-1 ura3-1 trp 1-1 leu2, 3-112 his3-11 can1-100 rpa135Δ::LEU2 pNOY102 |
| NOY556 | MATα ade2-1 ura3-1 trp1-1, leu2, 3-112 his3-11 can1-100 pNOY103 |
| NOY699 | MATα ade2-1 ura3-1 trp 1-1 leu2, 3-112 his3-11 can1-100 rrn5Δ::LEU2 pNOY103 |
| NOY798 | MATα ade2 ade3 leu2 ura3 trp1 his can1 rrn5::TRP1 (contains TRP1 insertion at the ClaI site, which is within the codon Ile161 of RRN5; Keys et al., 1996) and carries pNOY402 |
| NOY896 | same as NOY388, but rrn9Δ::HIS3 rpa135Δ::LEU2 pNOY103; N-PSW |
| NOY897 | same as NOY896, but PSW and lost pNOY103 |
| NOY921 | same as NOY388, but rrn9Δ::HIS3 fob1Δ::LEU2 pNOY103; N-PSW |
| NOY1016 | same as NOY556, but ymr233wΔ::TRP1 and does not carry pNOY103 |
| NOY1034 | same as NOY556, but uaf30Δ::HIS3 and does not carry pNOY103 |
| NOY1035 | same as NOY556, but uaf30Δ::HIS3 |
| NOY1037 | same as NOY408-1a, but uaf30Δ::HIS3; N-PSW |
| NOY1044 | same as NOY1016, but uaf30Δ::HIS3 |
| NOY1046 | same as NOY699, but carrying pNOY513 |
| NOY1047 | same as NOY1046, but without pNOY103 and uaf30Δ::HIS3 |
| NOY1048 | same as NOY1047, but carrying pNOY512 |
| NOY1050 | same as NOY1037, but PSW and without pNOY102 |
| Plasmids | |
| pRS314 | Escherichia coli–yeast shuttle vector carrying CEN6 ARSH4 TRP1 (Sikorski and Hieter, 1989) |
| pRS316 | Escherichia coli–yeast shuttle vector carrying CEN6 ARSH4 URA3 (Sikoroski and Hieter, 1989) |
| pNOY102 | high-copy-number plasmid carrying GAL7-35S rDNA, URA3, 2 µm, amp (Nogi et al., 1991b) |
| pNOY103 | high-copy-number plasmid carrying GAL7-35S rDNA, ADE3, URA3, 2 µm, amp (Nogi et al., 1991a) |
| pNOY402 | LEU2, CEN6, RRN5-(HA1)3-(His)6 (Keener et al., 1997) |
| pNOY509 | 1.7 kb genomic fragment carrying UAF30 gene and its promoter cloned into pBluescript KS(–) |
| pNOY510 | a derivative of pNOY509 containing uaf30Δ::HIS3 instead of UAF30 |
| pNOY511 | 1.7 kb genomic fragment carrying UAF30 gene and its promoter cloned into pRS316 (CEN6, URA3) |
| pNOY512 | N-terminal myc-tagged UAF30 in pRS316 (CEN6, URA3) |
| pNOY513 | RRN5-(HA1)3-(His)6 in pRS314 (CEN6, TRP1) |
To construct the uaf30Δ::HIS3 disruption plasmid (pNOY510), pNOY509 was first digested with NcoI and EcoRV to remove a 700 bp NcoI–EcoRV fragment containing the entire UAF30 ORF, blunt-ended with T4 DNA polymerase, and a blunt-ended 900 bp BamHI fragment of the HIS3 gene was then ligated, replacing the UAF30 ORF. The 1.2 kb XhoI–BamHI fragment carrying a uaf30Δ::HIS3 from this plasmid was used for disruption of UAF30. Disruption of UAF30 was confirmed by PCR analysis of genomic DNA from these strains, using primer 1 and an internal HIS3-specific primer.
To myc-tag UAF30 at the N-terminus, the UAF30 gene was re-amplified from pNOY511 using primers 3 and 2. Primer 3 contains a NcoI site followed by the myc tag sequence, then the UAF30 coding sequence beginning with the second codon (5′-AAAACCATGGCATCAGAGCAGAAGCTGATCTCAGAGGAGGACCTGAGCAGCGCTGAATTAAACGATTATAGTACG-3′; NcoI site underlined and UAF30 coding sequence in bold). The PCR product was digested with NcoI and BamHI, and was then used to replace the original NcoI–BamHI fragment in pNOY511. This results in a plasmid (pNOY512) carrying myc-tagged UAF30, which was confirmed by DNA sequencing analysis. The N-terminal amino acid sequence of the protein from this plasmid is MASEQKLISEEDLSSAELNDYST (myc tag sequence is in bold, Uaf30p sequence is underlined).
The construction of NOY1016 carrying a deletion of YMR233W ORF (ymr233wΔ::TRP1) was carried out in the following way. Two PCR primers, 5 and 6, were designed to have upstream or downstream sequences of YMR233W as well as upstream or downstream sequences from pRS314 flanking the TRP1: primer 5, 5′-ATTCCGATGGTTGACGCTATTTTGAGCGTATCCAACCCCGAGATTGTACTGAGAGTGCAC-3′ (the sequence in bold is pRS314 sequence upstream of the TRP1 gene); and primer 6, 5′-CGGGTGTTTGCTTTTGTTCCTCTTCATGTTTCACAATTTCCTGTCCGGTATTTCACACGG-3′ (the sequence in bold is pRS314 sequence downstream of the TRP1 sequence). PCR amplification of pRS314 resulted in a fragment 1117 bp long that contained the TRP1 gene flanked by YMR233W ORF N-terminus (+22 to +53) and C-terminus (+592 to +631) coding sequences. This PCR product was transformed into NOY556 and Trp+ transformants were selected. Disruption of YMR233W in one of the transformants (NOY1016) was confirmed by PCR analysis of genomic DNA using two primers flanking the YMR233W ORF.
Strain NOY1044, which carries the uaf30Δ::HIS3 and the ymr233wΔ:: TRP1 deletions, was constructed by replacing UAF30 in strain NOY1016 with a XhoI–BamHI digestion product of pNOY510 carrying uaf30Δ:: HIS3. Deletion of UAF30 was confirmed by PCR analysis of genomic DNA from this strain using primers 1 and 2 described above.
YEP-galactose, YEP-glucose (YEPD), synthetic galactose and synthetic glucose media were described previously (Nogi et al., 1991a). The following supplements were added to the synthetic media as appropriate to satisfy nutritional requirements: leucine (70 µg/ml), histidine (20 µg/ml), tryptophan (20 µg/ml), adenine (20 µg/ml) and uracil (20 µg/ml). Cells were grown at 30°C.
Identification of the gene for the 30 kDa subunit of UAF by mass spectrometry
UAF containing Rrn5p with (HA1)3-(His)6 attached at the C-terminus was isolated from strain NOY798 and purified as previously described (Keener et al., 1997; see Figure 6A for outline). Purified UAF was concentrated by ultrafiltration, subjected to SDS–PAGE, and stained by Coomassie Blue. The 30 kDa subunit band was excised and analyzed by mass spectrometry. The band was destained, reduced, alkylated, and digested with trypsin. Extracted tryptic peptides were analyzed by HPLC/tandem mass spectrometry using a custom built HPLC system and a ThermoFinnigan LCQ mass spectrometer (Falany et al., 2001). High quality spectra were searched against the OWL non-redundant database using SEQUEST (Eng et al., 1994). Matches to proteins of interest were validated by manually comparing experimental and predicted spectra.
Analysis of RNA by primer extension and [14C]uracil labeling
Analysis of 5′-ends of precursor rRNA by primer extension was carried out as described previously (Keener et al., 1998; Vu et al., 1999), using the 32P-5′-end-labeled primer 5′-ACACGCTGTATAGAGACTAGGC-3′, which hybridizes to 35S precursor rRNA 130 nucleotides downstream of the Pol I start site. Quantification was done with a PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
Cellular amounts of total RNA in cells growing in synthetic medium were determined by growing cells in the presence of [14C]uracil (5 µg/ml, 0.4 µCi/ml) and measuring increases in TCA-insoluble 14C-labeled RNA with increases in cell density (A600) at several time points. From these values, relative amounts of RNA per unit amount of cell mass during exponential growth were calculated. The amounts of 5′-ends of precursor rRNA per unit amount of cell mass were then calculated using these values and values obtained by primer extension. Rates of accumulation of RNA were also calculated from the values obtained for relative amount of RNA per cell mass and growth rates (see the legend to Table I).
In vitro Pol I transcription experiments
In vitro experiments examining UAF activity to stimulate Pol I transcription were carried out using purified components as described previously (Keener et al., 1998). Present in all reaction mixtures were: 0.2–0.4 nM wild-type linear template containing both upstream and core promoter elements (extending 210 bp upstream of the +1 start site), Rrn3p Pol I complex (prepared by incubation of Rrn3p, 2–6 nM, with Pol I, 10–15 nM), and much smaller amounts of TBP and CF. Concentrations of UAF preparations from the uaf30Δ strain were estimated by SDS–PAGE followed by western blot analysis using anti-HA1 antibodies and by comparison with control UAF preparations at the same purification steps from the UAF30 strain. Transcripts were labeled with [α-32P]GTP and quantified using a PhosphorImager. Degrees of stimulation by UAF were estimated by comparing with control reaction mixtures that did not contain UAF.
Acknowledgments
Acknowledgements
We thank Dr C.Greer for critical reading of the manuscript and S.VanAmburg for help in preparation of the manuscript. This work was supported by Public Health Service Grant GM35949 (to M.N.) and CA33752 (to M.K.Y.) from the National Institutes of Health.
References
- Aalfs J.D. and Kingston,R.E. (2000) What does ‘chromatin remodeling’ mean? Trends Biochem. Sci., 25, 548–555. [DOI] [PubMed] [Google Scholar]
- Cairns B.R., Levinson,R.S., Yamamoto,K.R. and Kornberg,R.D. (1996) Essential role of Swp73p in the function of yeast Swi/Snf complex. Genes Dev., 10, 2131–2144. [DOI] [PubMed] [Google Scholar]
- Conrad-Webb H. and Butow,R.A. (1995) A polymerase switch in the synthesis of rRNA in Saccharomyces cerevisiae. Mol. Cell. Biol., 15, 2420–2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar V.N., Miller,D.A., Kulkarni,A.B. and Miller,O.J. (1987) Human ribosomal DNA fragments amplified in hamster cells are transcribed only by RNA polymerase II and are not silver stained. Mol. Cell. Biol., 7, 1289–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doelling J.H. and Pikaard,C.S. (1996) Species-specificity of rRNA gene transcription in plants manifested as a switch in RNA polymerase specificity. Nucleic Acids Res., 24, 4725–4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng J.K., McCormak,A.L. and Yates,J.R.,III (1994) An approach to correlate tandem mass spectral data of peptides with amino acid sequences in protein database. J. Am. Soc. Mass Spectrom., 5, 976–989. [DOI] [PubMed] [Google Scholar]
- Falany M.L., Thames,A.M.,III, McDonald,J.M., Blair,H.C., McKenna,M.A., Moore,R.E., Young,M.K. and Williams,J.P. (2001) Osteoclasts secrete the chemotactic cytokine mim-1. Biochem. Biophys. Res. Commun., 281, 180–185. [DOI] [PubMed] [Google Scholar]
- Keener J., Dodd,J.A., Lalo,D. and Nomura,M. (1997) Histones H3 and H4 are components of upstream activation factor required for the high-level transcription of yeast rDNA by RNA polymerase I. Proc. Natl Acad. Sci. USA, 94, 13458–13462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keener J., Josaitis,C.A., Dodd,J.A. and Nomura,M. (1998) Reconstitu tion of yeast RNA polymerase I transcription in vitro from purified components. J. Biol. Chem., 273, 33795–33802. [DOI] [PubMed] [Google Scholar]
- Keys D.A., Lee,B.-S., Dodd,J., Nguyen,T.T., Vu,L., Fantino,E., Burson,L.M., Nogi,Y. and Nomura,M. (1996) Multiprotein transcription factor UAF interacts with the upstream element of the yeast RNA polymerase I promoter and forms a stable preinitiation complex. Genes Dev., 10, 887–903. [DOI] [PubMed] [Google Scholar]
- Kief D.R., and Warner,J.R. (1981) Coordinate control of syntheses of ribosomal ribonucleic acid and ribosomal proteins during nutritional shift-up in Saccharomyces cerevisiae. Mol. Cell. Biol., 1, 1007–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Heck,D.J., Nomura,M. and Horiuchi,T. (1998) Expansion and contractions of ribosomal DNA repeats in Saccharomyces cerevisiae: requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes Dev., 12, 3821–3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogi Y., Vu,L. and Nomura,M. (1991a) An approach for isolation of mutants defective in 35S ribosomal RNA synthesis in Saccharomyces cerevisiae.Proc. Natl Acad. Sci. USA, 88, 7026–7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogi Y., Yano,R. and Nomura,M. (1991b) Synthesis of large rRNAs by RNA polymerase II in mutants of Saccharomyces cerevisiae defective in RNA polymerase I. Proc. Natl Acad. Sci. USA, 88, 3962–3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura M. (1998) Transcriptional factors used by Saccharomyces cerevisiae RNA polymerase I and the mechanism of initiation. In Paule,M.R. (ed.), Transcription of Ribosomal RNA Genes by Eukaryotic RNA Polymerase I. Springer-Verlag and R.G. Landes Co., Austin, TX, pp. 155–172.
- Oakes M., Siddiqi,I., Vu.L., Aris,J. and Nomura,M. (1999) Transcriptional factor UAF, expansion and contraction of ribosomal DNA (rDNA) repeats, and RNA polymerase switch in transcription of yeast rDNA. Mol. Cell. Biol., 19, 8559–8569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqi I., Keener,J., Vu,L. and Nomura,M. (2001) Role of TATA binding protein (TBP) in yeast ribosomal DNA transcription by RNA polymerase I: defects in the dual functions of transcription factor UAF cannot be suppressed by TBP. Mol. Cell. Biol., 21, 2292–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smale S.T. and Tjian,R. (1985) Transcription of herpes simplex virus tk sequences under the control of wild-type and mutant human RNA polymerase I promoters. Mol. Cell. Biol., 5, 352–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffan J.S., Keys,D.A., Dodd,J.A. and Nomura,M. (1996) The role of TBP in rDNA transcription by RNA polymerase I in Saccharomyces cerevisiae: TBP is required for upstream activation factor-dependent recruitment of core factor. Genes Dev., 10, 2551–2563. [DOI] [PubMed] [Google Scholar]
- Stephens R.S. et al. (1998) Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science, 282, 754–759. [DOI] [PubMed] [Google Scholar]
- Sudarsanam P. and Winston,F. (2000) The Swi/Snf family nucleosome-remodeling complexes and transcriptional control. Trends Genet., 16, 345–351. [DOI] [PubMed] [Google Scholar]
- Vu L., Siddiqi,I., Lee,B.-S., Josaitis,C.A. and Nomura,M. (1999) RNA polymerase switch in transcription of yeast rDNA: role of transcription factor UAF (upstream activation factor) in silencing rDNA transcription by RNA polymerase II. Proc. Natl Acad. Sci. USA, 96, 4390–4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Xue,Y., Zhou,S., Kuo,A., Cairns,B.R. and Crabtree,G.R. (1996) Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev., 10, 2117–2130. [DOI] [PubMed] [Google Scholar]
- Warner J.R. (1989) Synthesis of ribosomes in Saccharomyces cerevisiae. Microbiol. Rev., 53, 256–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y., Canman,J.C., Lee,C.S., Nie,Z., Yang,D., Moreno,G.T., Young,M.K., Salmon,E.D. and Wang,W. (2000) The human SWI/SNF-B chromatin-remodeling complex is related to yeast Rsc and localizes at kinetochores of mitotic chromosomes. Proc. Natl Acad. Sci. USA, 97, 13015–13020. [DOI] [PMC free article] [PubMed] [Google Scholar]