Abstract
The promyelocytic leukemia protein PML is organized into nuclear bodies which mediate suppression of oncogenic transformation and of growth. The biochemical functions of PML bodies are unknown, despite their involvement in several human disorders. We demonstrate that eukaryotic initiation factor 4E (eIF4E) directly binds the PML RING, a domain required for association with bodies and for suppression of transformation. Nuclear eIF4E functions in nucleocytoplasmic transport of a subset of transcripts including Cyclin D1. Present studies indicate that some PML requires the evolutionarily older eIF4E protein for association with nuclear bodies. Further more, PML RING modulates eIF4E activity by drastically reducing its affinity for its substrate, 5′ m7G cap of mRNA. We demonstrate that eIF4E requires cap binding for transport of Cyclin D1 mRNA and subsequent transformation activity. Additionally, PML reduces the affinity of eIF4E for m7G mRNA cap, causing a reduction in Cyclin D1 protein levels and consequent transformation inhibition. PML is the first factor shown to modulate nuclear eIF4E function. These findings provide the first biochemical framework for understanding the transformation suppression activity of PML.
Keywords: eukaryotic initiation factor 4E/nuclear domain 10/promyelocytic leukemia/PML oncogenic domains/RING
Introduction
The promyelocytic leukemia protein (PML) is an important regulator of mammalian cell growth and apoptosis, and when overexpressed it suppresses oncogenic transformation (Melnick and Licht, 1999). These physiological functions of PML are correlated with its organization into nuclear matrix-associated multiprotein complexes (Melnick and Licht, 1999). These complexes are known as PML nuclear bodies, nuclear domain 10 (ND10) or PML oncogenic domains (POD). PML nuclear bodies are characteristically disrupted in acute promyelocytic leukemia (APL), spinocerebellar ataxia and by a variety of viruses (Melnick and Licht, 1999). In 98% of APL cases, a single PML allele is fused to the retinoic acid receptor α (RARα). The resulting PML–RARα fusion protein disrupts PML bodies in a dominant-negative fashion (Grimwade and Solomon, 1997). Remission in these patients is correlated with re-formation of bodies after treatment with all-trans retinoic acid (ATRA; Melnick and Licht, 1999). Despite their clear biomedical relevance, the molecular function of PML nuclear bodies has remained an enigma. The PML gene is absent from organisms such as the budding yeast Saccharomyces cerevisae (http:// genome-www.stanford.edu/Saccharomyces) and the fruit fly Drosophila melanogaster (http://www.fruitfly.org). This apparent lack of phylogenetic conservation of PML bodies is intriguing, since they are thought to underlie basic cellular processes in mammals.
The integrity of PML and its nuclear bodies is essential for its physiological functions (Melnick and Licht, 1999). Its N-terminal RING, double B-box, coiled-coil (RBCC) motif (Goddard et al., 1991) is required for association of PML with nuclear bodies (Borden et al., 1995, 1996). The RING and B-box are cysteine-rich zinc-binding domains that mediate protein–protein interactions (Borden, 2000). The three-dimensional structure of the PML RING reveals two zinc binding sites (I and II), and zinc binding is required for its native fold (Figure 1C; Borden et al., 1995). The transformation suppressive and pro-apoptotic functions of PML require intact RING and B-box domains. For example, single Cys-Ala substitutions that ablate zinc-binding in either domain disrupt body formation and lead to aberrant cell growth and loss of transformation suppression (Mu et al., 1994; Borden et al., 1995, 1996).
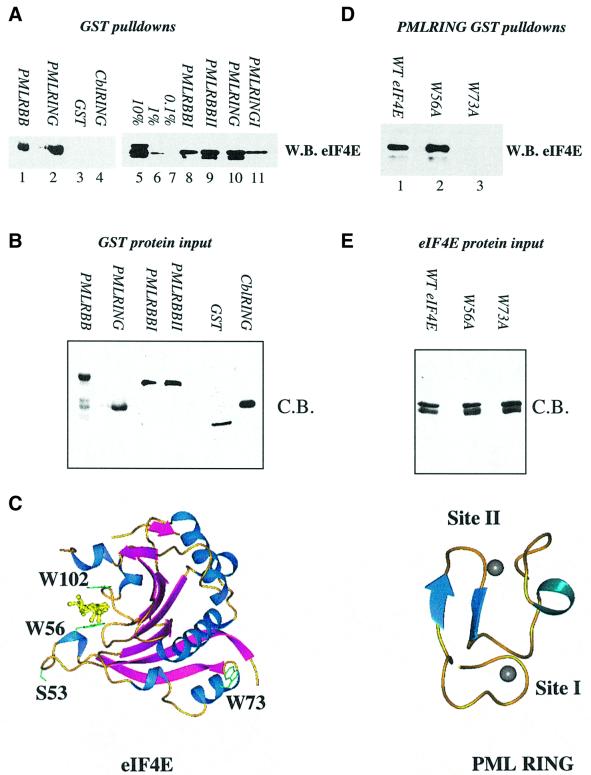
Fig. 1. Purified PML and eIF4E directly interact. (A and D) GST pull-down analysis of PML and eIF4E. Wild-type eIF4E (A) or mutant eIF4E (D) were incubated with GST fusion proteins as indicated. (B and E) Coomassie Blue-stained SDS–PAGE demonstrating production and purity of the mutant proteins used in (A) and (D). In all cases, proteins of the predicted molecular weight were produced. Further, identical quantities of each protein were used in the pull-down assays shown in (A) and (D). For each pull-down, fusion proteins attached to glutathione beads were resolved in the absence of bait proteins to demonstrate that there was no cross-reaction of these proteins with the antibodies used (data not shown). W.B., western blot; C.B., Coomassie Blue-stained SDS–PAGE. RBB indicates that constructs contained the RING and double B-box of PML; I and II indicate targeted mutation of either zinc site I or zinc site II. 10, 1 and 0.1% represent eIF4E input for (A). The lower of the two bands present in lanes 5, 9 and 10 of (A) and in (D) represents a minor degradation product of eIF4E, which is present in some eIF4E protein preparations and is still able to bind PML RING. (C) The three-dimensional structures of eIF4E and PML are shown (1ej1 and 1bor; Borden et al., 1995; Marcotrigiano et al., 1997). Arrows indicate β-sheets; coils, α-helix; balls, zinc atoms. The m7GpppG dinucleotide substrate of eIF4E is shown in yellow. Structures were rendered in PREPI (S.Islam and M.Sternberg).
Despite the progress in defining the physiological roles of PML in transformation suppression and growth, the biochemical basis for these actions remains unclear. Efforts to understand PML body function have centered on identifying its components and using this information to infer the function of the nuclear body itself. This type of analysis has limitations, since PML bodies are heterogeneous in composition and have constituents possessing a variety of seemingly unconnected functions. PML bodies contain a number of proteins involved in transcriptional regulation such as Sp100 (Sternsdorf et al., 1999). However, neither DNA nor RNA polymerase II associates with PML bodies under normal conditions (Maul et al., 2000; von Mikecz et al., 2000), suggesting that they are not sites of active transcription. Moreover, PML nuclear bodies contain proteins involved in nuclear RNA metabolism, translation and ribosome assembly such as eIF3/int-6 (Asano et al., 1997), the ribosomal P-proteins (Borden et al., 1998a) and especially interesting, the eukaryotic translation initiation factor eIF4E (Lai and Borden, 2000).
In contrast to PML, eIF4E promotes growth and oncogenic transformation (Sonenberg and Gingras, 1998). In the cytoplasm, eIF4E plays well defined biochemical roles acting as the limiting component in translation initiation (Sonenberg and Gingras, 1998). eIF4E, also known as the cap-binding protein, binds the 5′ m7G cap of mRNA where the cap intercalates between two tryptophans of eIF4E, W56 and W102 (Figure 1C; Marcotrigiano et al., 1997). Cap-binding by eIF4E is essential for initiation of cap-dependent translation (Keiper and Rhoads, 1997) and for regulation of mRNA stability (Dehlin et al., 2000). eIF4E directly binds several regulatory proteins through its dorsal surface. Mutation of W73 to alanine on this surface leads to loss of association of eIF4E with the eIF4G, eIF4E-BP1 and BP2 regulatory proteins (Ptushkina et al., 1998, 1999).
In the nucleus the majority of eIF4E is localized to bodies, and appears to function in nucleocytoplasmic mRNA transport of a subset of specific mRNAs (Lejbkowicz et al., 1992; Rosenwald et al., 1995; Rousseau et al., 1996; Lai and Borden, 2000). Overexpression of eIF4E is correlated with increased nucleocytoplasmic transport of Cyclin D1 mRNA, without affecting transport of other mRNAs such as GAPDH and actin (Rousseau et al., 1996; Lai and Borden, 2000). eIF4E overexpression does not lead to increased transcription of Cyclin D1 or to more efficient loading of these mRNAs onto polysomes, but to an increase in the cytoplasmic levels of Cyclin D1 mRNA (Rosenwald et al., 1995; Rousseau et al., 1996). Thus, the effective increase in Cyclin D1 protein results largely from increased mRNA transport. If serine 53 is mutated to alanine (Figure 1C), the resulting mutant eIF4E is unable to transport Cyclin D1 mRNA to the cytoplasm, unable to increase Cyclin D1 protein levels and subsequently unable to form foci in NIH 3T3 cells (Lazaris-Karatzas et al., 1990; Rousseau et al., 1996). The S53A mutant does not affect translation (Rousseau et al., 1996). Thus, eIF4E-dependent transport of Cyclin D1 mRNA is linked to its transformation activity.
Previous studies have demonstrated that endogenous PML localizes with and can be immunoprecipitated by eIF4E (Lai and Borden, 2000). This interaction is functional, since PML antagonizes eIF4E-dependent mRNA transport and the RING domain of PML is required for this activity (Lai and Borden, 2000). However, it was not clear whether the antagonism of eIF4E function by PML was a direct consequence of a PML–eIF4E interaction. In addition, the biochemical basis for the antagonism and the physiological effects of this antagonism were unknown.
In order to determine biochemical functions for the PML protein and subsequently the nuclear body, we sought to identify proteins that interact directly with its functionally important RING domain. A biochemical analysis of these interactions should reveal molecular functions of the PML protein and a biochemical basis for its physiological effects. This report demonstrates that eIF4E is a RING-binding protein and that eIF4E itself forms both a functional and structural basis for some PML nuclear bodies. We show that the inhibition of eIF4E function by PML is a direct consequence of the interaction between these proteins. These findings have allowed us to develop a biochemical model for PML function as it pertains to eIF4E and suppression of transformation.
Results
The RING of PML directly binds eIF4E
In order to determine whether PML could directly alter eIF4E function, we first examined whether PML interacts directly with eIF4E and whether this interaction is through the RING domain of PML. We used bacterially expressed proteins purified to homogeneity. Constructs contained either the RING double B-box (PML RBB) or only the RING (PML RING). As can be seen from Figure 1A, both PML RING–GST (lane 2) and PML RBB–GST (lane 1) bind directly to eIF4E, demonstrating that the RING domain is sufficient for this interaction. These constructs bound ∼10% of the eIF4E input (lane 5) as indicated by western analysis. eIF4E does not bind GST alone (lane 3). Importantly, the PML RING–eIF4E interaction is specific, since the RING domain from an unrelated protein, Cbl, does not bind eIF4E (lane 4). The corresponding Coomassie Blue-stained gel demonstrates similar loading of all the GST constructs (Figure 1B). Thus, eIF4E interacts directly and specifically with the functionally important RING domain of PML.
Previous studies have indicated that other RING proteins, e.g. Msl2 (Copps et al., 1998), bind distinct partner proteins near zinc site I or II. We examined whether this would be the case for PML and eIF4E. Cysteines in either site I or II were mutated to alanine in PML RING or PML RBB and their ability to bind eIF4E was tested. As shown in Figure 1A, mutation of site I reduced eIF4E binding (lanes 8 and 11), whereas mutation of site II in either PML RBB (lane 9) or PML RING (data not shown) constructs was less detrimental to eIF4E binding (compare lanes 8 and 9). The faster migrating band is a degradation product observed in some protein preparations of eIF4E. Thus, our conclusions are based on the full-length eIF4E (the slower migrating band). Notably, although the site I mutation reduces binding, it does not abolish association with eIF4E. These data suggest that eIF4E binds to the RING near site I, but that unfolding of site I (by the cysteine to alanine mutation) does not completely disrupt the conformation of the eIF4E-binding site, suggesting that a short stretch of amino acids could be responsible for eIF4E recognition. In other eIF4E-binding proteins, the YXXXLΦ motif is used for recognition (Sonenberg and Gingras, 1998). PML, however, does not contain this motif, suggesting that another short motif in the RING, involving site I, is responsible for the PML–eIF4E association. The precise region is being defined currently by limited proteolysis and mass spectrometry (our unpublished observations).
To gain an insight into the molecular interaction, we examined the effects of m7G cap-binding by eIF4E and of specific mutations in eIF4E on its affinity for the PML RING. Mutation of tryptophan 56 to alanine did not alter the affinity for PML RING, relative to wild-type eIF4E (Figure 1D, compare lanes 1 and 2). However, this mutation substantially reduced retention of eIF4E on m7GTP-Sepharose beads (Figure 6C). Thus, the ability of eIF4E to bind the cap is not required for association with PML, and nor is the presence of W56. Conversely, we investigated whether the presence of m7G cap could inhibit the PML eIF4E interaction. Bacteria do not synthesize an m7G cap, thus all eIF4E produced in these assays is ‘cap free’. Therefore, the effect of adding the cap analog m7GpppG was examined. In agreement with the W56 mutation data above, pre-incubation of more than a 10-fold excess of m7GpppG with eIF4E did not alter the affinity of eIF4E for either PML RBB or PML RING (data not shown). W73 is located on the dorsal surface of eIF4E (Figure 1C) and is required for association with several regulatory proteins. We show that the W73A mutant protein has a substantially lower affinity for PML RING (Figure 1D, compare lanes 1 and 3). Thus, PML and eIF4E interact through functionally important parts of each protein.
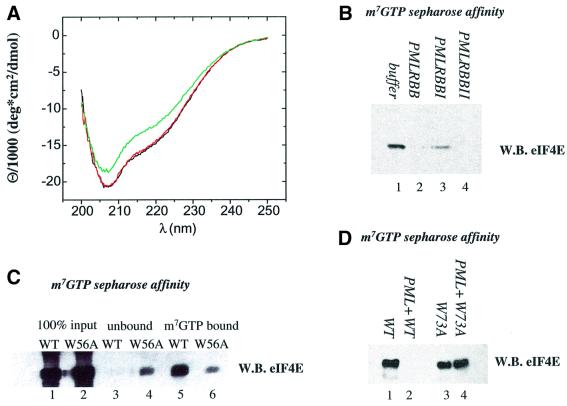
Fig. 6. (A) m7GpppG-binding dramatically affects the conformation of the eIF4E protein. Far-UV CD spectra of bacterially purified eIF4E at 0.24 µM (black), in complex with 3 µM GpppG (red) and with 3 µM m7GpppG (green). (B) PML dramatically reduces the affinity of eIF4E for m7GTP-Sepharose. Bacterially expressed and purified PML RBB or mutants in site I (PMLRBBI) or site II (PMLRBBII) of the RING were mixed with purified eIF4E and applied to m7GTP-Sepharose beads. (C) The W56A mutation reduces the affinity of eIF4E for m7GTP-Sepharose. The affinity of wild-type and W56A mutant of eIF4E was monitored by m7GTP-Sepharose affinity chromatography. The input for each protein preparation is shown, as is the material that bound m7GTP-Sepharose (lanes 5 and 6) and the unbound fraction (lanes 3 and 4). (D) PML does not reduce the affinity of the W73A eIF4E mutant for m7GTP-Sepharose. Results were monitored by western analysis using eIF4E mAb where WB indicates western blot.
PML and eIF4E co-localize in several leukemic cell lines
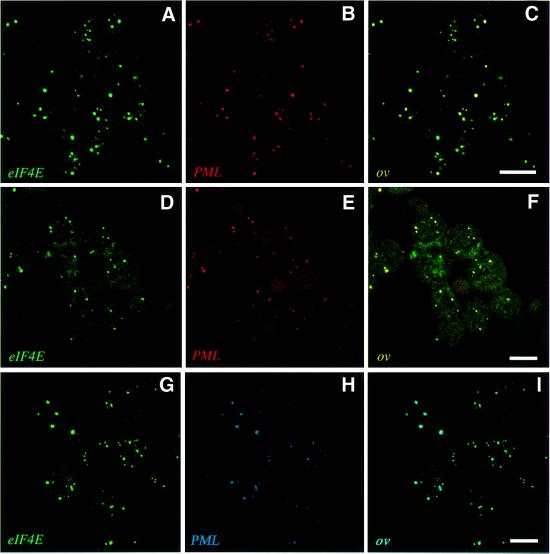
Previous studies have demonstrated that endogenous eIF4E co-localizes and co-immunoprecipitates with endogenous PML in NIH 3T3 cells (Lai and Borden, 2000). Using immunofluorescence methods in conjunction with confocal microscopy, we examined the localization of endogenous eIF4E and endogenous PML in some leukemic cell lines. In U937 cells, nearly all PML bodies associate with eIF4E bodies (Figure 2A–C, PML in red and eIF4E in green; G–I, PML in blue and eIF4E in green). Similar results were observed in K562 cells (Figure 2D–F). In addition, these proteins can be co-immunoprecipitated in these cell lines (data not shown). Identical results were obtained using either a combination of the PML polyclonal antibody (Borden et al., 1995) and a monoclonal antibody (mAb) to eIF4E (Campbell Dwyer et al., 2000; Dostie et al., 2000a,b; Lai and Borden, 2000) (Figure 2A–F), or using the combination of a PML mAb to 5E10 (Lai and Borden, 2000; Stuurman et al., 1992) and eIF4E mAb directly conjugated to fluorescein isothiocyanate (FITC) (Figure 2G–I). Both the PML and eIF4E antibodies have been characterized extensively elsewhere (Stuurman et al., 1992; Campbell Dwyer et al., 2000; Lai and Borden, 2000). There appears to be some variability amongst cell types in terms of the frequency of PML and eIF4E co-localization, since a higher frequency of co-localization is observed in myeloid cells as compared with fibroblasts such as NIH 3T3 (Lai and Borden, 2000), or PML+/+ mouse embryo fibroblasts (MEFs; Figure 3J). It is clear from these studies that the majority of PML nuclear bodies co-localize with eIF4E bodies in myeloid cells and that there are some eIF4E bodies present that do not localize with PML, consistent with the fact that in many cell types there are more eIF4E bodies than PML bodies. However, these results indicate that co-localization of PML and eIF4E in nuclear bodies is likely to be a general characteristic of mammalian cell lines.
Fig. 2. PML and eIF4E co-localize in several cell types. U937 (A–C) and K562 (D–F) cells were stained for eIF4E (in green) and PML (in red) with the overlay in yellow. PML and eIF4E were detected using polyclonal antiPML and eIF4E mAb, respectively. Identical results were obtained using anti-PML 5E10 mAb and eIF4E mAb conjugated directly to FITC when U937 cells (G–I) were stained for eIF4E (in green) and PML (in blue) with the overlay in aqua. The objective is 100×. Scale bars = 10 µM. Confocal micrographs represent single sections through the plane of the cells.
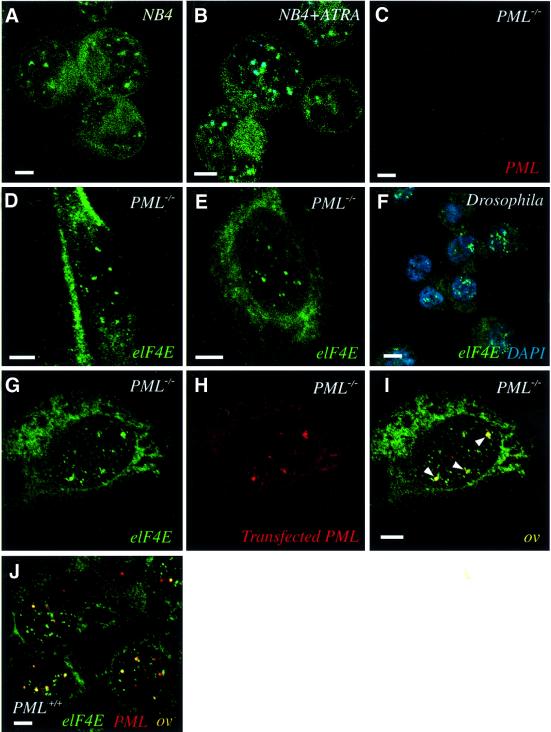
Fig. 3. eIF4E forms nuclear bodies in the absence of PML. (A) APL patient NB4 cells were stained with eIF4E mAb conjugated directly to FITC (eIF4E mAb-FITC, in green). After ATRA treatment (B), NB4 cells were stained for both eIF4E (eIF4E mAb-FITC, in green) and PML (5E10 mAb, in dark blue), with the overlay in aqua. PML–/– MEFs were stained separately with the polyclonal anti-PML (C) or eIF4E mAb (not conjugated to FITC) (D). (E) A nuclear matrix preparation of PML–/– MEFs was stained with eIF4E mAb. (F) D.melanogaster S2 cells were stained with eIF4E mAb (in green) and with DAPI (in blue) to define the nucleus. (G–I) PML–/– MEFs were transfected with the PML gene, and the localization of endogenous eIF4E (green) and transfected PML (red) were observed with the overlay in yellow. Arrows indicate co-localization. (J) PML+/+ MEFs stained for PML (red) and eIF4E (green), with the overlay in yellow. The objective is 100×. Scale bars = 5 µM. Confocal micrographs represent single optical sections through the plane of the cells.
eIF4E forms nuclear structures in the absence of PML bodies
We investigated whether eIF4E required PML to form nuclear bodies. In the NB4 cell line developed from an APL patient, PML bodies are disrupted by the PML– RARα fusion protein. Therefore, we monitored the localization of eIF4E as a function of ATRA treatment, which restores normal PML body morphology in NB4 cells. Importantly, eIF4E has body-like morphology in NB4 cells prior to ATRA treatment (Figure 3A, eIF4E in green). After ATRA treatment, PML bodies re-form and co-localize with eIF4E bodies, where co-localization is shown in aqua (Figure 3B, PML in dark blue). Results are similar to those observed in other cell types (Figure 2). Thus, in contrast to other PML partner proteins such as Sp100 (Zhong et al., 2000), eIF4E forms nuclear bodies independently of the distribution of PML.
Intriguingly, these data strongly suggest that eIF4E does not require PML for localization into nuclear bodies. To examine whether eIF4E nuclear bodies form in cells lacking PML altogether, we examined the localization of eIF4E in PML–/– MEFs that contain a targeted homozygous deletion of the PML gene (Wang et al., 1998). As expected, no PML is observed in these cells (Figure 3C). Cells stained for eIF4E contain a punctate nuclear pattern of eIF4E bodies that is indistinguishable from those observed in PML-containing cell types (Figure 3D and E). In addition, nuclear eIF4E bodies are detected by immunoelectron microscopy in PML–/– MEFs and U937 cells (data not shown). Therefore, eIF4E does not require PML for assembly into nuclear bodies. If PML assembles onto existing eIF4E nuclear bodies, expression of exogenous PML should reconstitute the PML–eIF4E interaction in PML–/– cells. When transfected, PML associates with endogenous eIF4E bodies (Figure 3G–I), suggesting that eIF4E bodies are sites of PML nuclear body assembly. As observed previously in other fibroblasts, there is an excess of eIF4E bodies relative to PML bodies in both the transfected PML–/– cells and the endogenous material in the PML+/+ cells (Figure 3J), suggesting that eIF4E bodies have a heterogeneous composition and diverse biochemical roles. For these fibroblasts, the frequency of PML bodies associated with eIF4E bodies is similar to results observed in NIH 3T3 cells (Lai and Borden, 2000).
PML nuclear bodies are characterized by their association with the nuclear matrix (Melnick and Licht, 1999). Therefore, we examined whether eIF4E bodies are also matrix associated in PML–/– cells. eIF4E bodies are morphologically unaltered by nuclear matrix preparation in PML–/– cells, indicating that eIF4E bodies are nuclear matrix-associated (Figure 3, compare D and E) and that PML is not required for this property.
We investigated whether cells from phylogenetically diverse organisms, such as D.melanogaster, which do not possess the PML gene, contain eIF4E bodies. Figure 3F shows that in D.melanogaster S2 cells, eIF4E localizes to nuclear bodies and is present diffusely throughout the cytoplasm. Similarly, eIF4E localizes to nuclear bodies in budding S.cerevisiae, as observed by immunoelectron microscopy (Lang et al., 1994). The nuclear organization of eIF4E in cells from phylogenetically diverse organisms is remarkably similar to that observed in mammalian cells, suggesting that eIF4E body assembly is independent of PML and that eIF4E nuclear bodies are phylogenetically conserved in eukaryotic cells, and may be ancestral to PML.
PML is dispersed by the m7G cap analog in cell culture
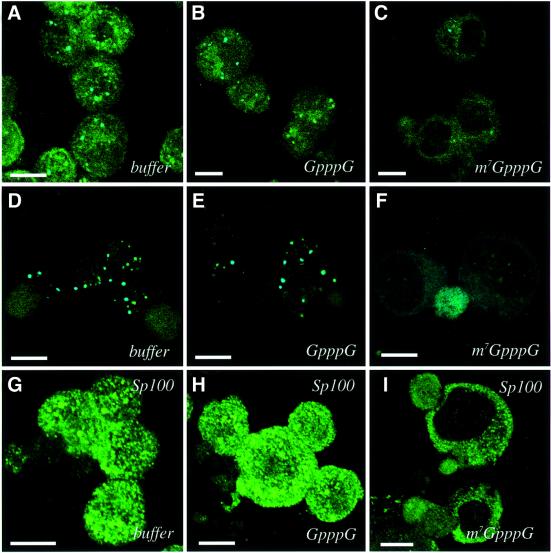
The above results indicate that the integrity of eIF4E bodies is independent of PML. Therefore, we examined the possibility that PML requires eIF4E for association with nuclear bodies. If this hypothesis is true, then disruption of eIF4E bodies should affect the distribution of PML. Previous studies with HeLa cells showed that eIF4E nuclear bodies are specifically disrupted by treatment with an analog of its mRNA substrate, m7GpppG (Dostie et al., 2000b). Thus, the effect of treatment with this analog on the morphology of PML nuclear bodies was examined (Figure 4). U937 cells were permeabilized and incubated with either m7GpppG cap analog or GpppG, which does not bind eIF4E efficiently (Dostie et al., 2000b). Following incubation, cells were washed to remove proteins released from the nucleus upon treatment and then fixed. Incubation with either GpppG or buffer alone does not affect the localization of PML, eIF4E or their co-localization, as seen by comparing Figure 4A and B with Figure 2 (PML in blue, eIF4E in green, co-localization in aqua). Strikingly, treatment with m7GpppG completely disperses these proteins from the bodies (Figure 4C). The same results were observed in K562 cells (Figure 4D–F). Similarly, m7GpppG treatment disassociates PML and eIF4E from bodies in NIH 3T3 cells and eIF4E bodies in PML–/– cells (data not shown). Experiments where each component was stained individually yielded the same results as the double staining experiments. We examined the effect of m7GpppG treatment on another PML body component, Sp100. Sp100, like several PML partner proteins, requires PML for association with bodies (Zhong et al., 2000). We found that Sp100 bodies were completely dispersed by m7GpppG while remaining intact in cells treated with GpppG or buffer (Figure 4G–I). Thus, treatment with the cap analog disrupts assembly of eIF4E, PML and Sp100 bodies in myeloid and non-myeloid cells. The ability of PML to associate with bodies appears to rely on the integrity of eIF4E nuclear bodies.
Fig. 4. Treatment with the m7GpppG cap analog disrupts the subnuclear distribution of eIF4E, PML and SP100. Cells were treated with buffer, GpppG or m7GpppG as indicated. U937 cells (A–C) and K562 cells (D–F) were stained for PML (dark blue) and eIF4E (green). The overlay is aqua. (G–I) K562 cells were stained for the PML partner protein Sp100. The objective is 100×. Scale bars = 10 µM. Confocal micrographs represent single sections through the plane of the cells.
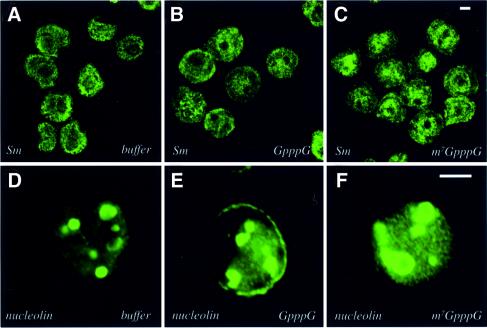
To examine the specificity of these treatments, we investigated the integrity of other nuclear structures. As can be seen from Figure 5A–C, the distribution of Sm proteins, which localize to nuclear speckles and are associated with mRNA splicing, is not altered by cap treatment, in agreement with Dostie et al. (2000b). Furthermore, the distribution of nucleolar and coiled body antigens, nucleolin (Figure 5D–F) and Nopp140 (data not shown) is unchanged by cap treatment. Furthermore, nucleoli appear intact after treatment when examined by phase contrast microscopy. Thus, the m7G cap treatment specifically disperses eIF4E, PML and Sp100 bodies.
Fig. 5. Treatment with the m7GpppG cap analog does not alter the subnuclear distribution of Sm speckles or nucleoli. Cells were treated with buffer, GpppG or m7GpppG as indicated. U937 and K562 cells were stained for Sm and nucleolin, respectively. The objective is 100×. Scale bars = 5 µM. Confocal micrographs represent single sections through the plane of the cells.
eIF4E undergoes a major conformational change upon m7GpppG binding
Large-scale conformational changes of eIF4E could alter its association with bodies and provide a mechanism for the dispersal of nuclear bodies upon m7G cap treatment. Therefore, we investigated whether eIF4E undergoes changes in conformation upon m7G cap binding. To monitor the conformation of the polypeptide backbone of eIF4E, we utilized circular dichroism (CD) difference spectroscopy (Fasman, 1996). In agreement with published structures, eIF4E has ∼60% α-helical character (Figure 6A). eIF4E was mixed with a 12.5-fold molar excess of m7GpppG so that all eIF4E molecules were complexed with m7GpppG. The resulting CD spectrum (Figure 6A) indicates that roughly 40 residues, ∼20% of the eIF4E protein, undergoes a large conformational change upon m7GpppG binding. Unmethylated GpppG, which does not bind eIF4E efficiently (Cai et al., 1999), does not alter its conformation (Figure 6A). The cap-induced conformational change is similar to that reported for yeast eIF4E (McCubbin et al., 1988; von Der Haar et al., 2000). Since 20% of the backbone residues in eIF4E undergo a specific conformational change upon m7GpppG cap binding in vitro, the dispersal of eIF4E and subsequent dispersal of PML bodies in cell culture may result from a structural rearrangement in eIF4E.
PML decreases affinity of eIF4E for its substrate
The above results raised the possibility that PML may regulate eIF4E in mammalian cells. To address this issue, we examined the ability of the RING of PML, which directly interacts with eIF4E (Figure 1), to modulate eIF4E function. Since the m7G cap is central to eIF4E function (Sonenberg and Gingras, 1998), we examined the possibility that PML affects cap binding (Figure 6B). Proteins were purified to homogeneity as shown in Figure 1. The eIF4E protein binds to m7GTP-Sepharose as expected (Figure 6B, lane 1). However, incubation with equimolar amounts of PML RBB–GST (lane 2) completely disrupts the ability of eIF4E to bind, whereas incubation with equimolar amounts of GST or with buffer shows no effect (data not shown). Similarly, PML RBB–GST site II-mutated protein (lane 4) or PML RING–GST (data not shown) drastically reduced the amount of eIF4E retained on the m7GTP-Sepharose. However, there is some eIF4E present on m7GTP-Sepharose after incubation with the PML RBB–GST site I-mutated protein (lane 3). These results are consistent with the fact that the site I mutation retained some, albeit significantly weaker, affinity for eIF4E, whereas the site II mutation bound eIF4E nearly as well as wild-type protein (Figure 1A). The W73A mutation, on the dorsal surface of eIF4E, did not alter the affinity of eIF4E for m7GTP-Sepharose (Figure 6D, compare lanes 1 and 3). Significantly, addition of PML to this mutant does not alter its affinity for m7GTP-Sepharose (compare lanes 2 and 4), consistent with the observation that PML does not associate with the W73A mutant (Figure 1B). Finally, none of the PML constructs bound to m7GTP-Sepharose in the absence of eIF4E (data not shown). Thus, PML substantially and directly reduces the affinity of eIF4E for its substrate, m7G cap, in a manner dependent on the region around site I of the PML RING and W73 on the dorsal surface of eIF4E.
Effects of PML on eIF4E-mediated transformation
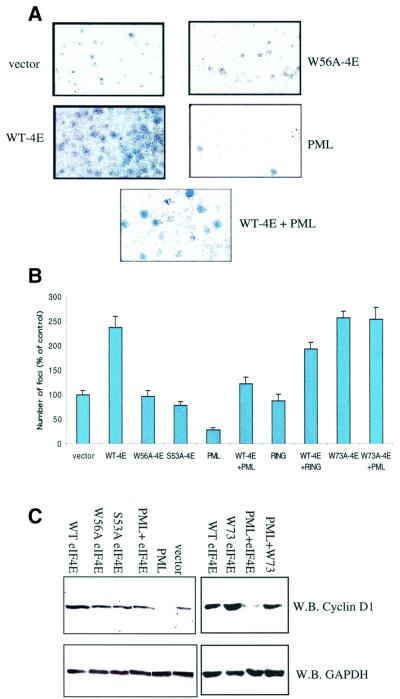
eIF4E overexpression results in oncogenic transformation whereas PML overexpression suppresses transformation (Lazaris-Karatzas et al., 1990; Melnick and Licht, 1999). Therefore, we determined the consequence of the PML– eIF4E interaction on cellular transformation. We initially examined whether eIF4E overexpression could transform PML–/– MEFs and whether exogenous PML could oppose eIF4E-mediated transformation (Figure 7A and B). Foci formation assays were carried out in PML–/– cells using stable transfection. It is clear from Figure 7A that wild-type eIF4E-overexpressing cells produce so many foci that it becomes difficult to quantitate them as the foci coalesce, as observed previously for eIF4E-overexpressing NIH 3T3 cells (Lazaris-Karatzas et al., 1990). For these experiments, only distinct foci of a minimal size and above were counted (Figure 7B). Thus, the number of foci in eIF4E-overexpressing cells may be underestimated.
Fig. 7. Growth suppression and Cyclin D1 levels in PML–/– cells. (A) Anchorage-dependent foci formation. Foci stained with Giemsa formed as a result of different transfections. Full-length PML and eIF4E were used in these assays. Cells were transfected as indicated: wild-type eIF4E (WT-4E), PML, PML site I RING mutant (RING), cap-binding mutant of eIF4E (W56A-4E), PML-binding mutant of eIF4E (W73A-4E), PML and WT-4E (WT-4E + PML) and vector. Identically sized areas were taken from representative regions of each petri dish. These results were quantitated in (B). Foci were counted in seven dishes per treatment, and the values represent the mean ± SD. Results are the average of at least two independent experiments. (C) Cyclin D1 levels. Western analysis of the indicated experiments 24 h after transfection. Fifty micrograms of total protein were loaded into each lane. GAPDH and Cyclin D1 proteins were detected using western analysis.
In vector-transfected control cells, a substantial number of foci were visible, consistent with the inherent transformed phenotype reported for PML–/– cells (Wang et al., 1998). However, substantially more foci were observed in cells overexpressing eIF4E than in vector controls (Figure 7A and B). Notably, expression of full-length PML in PML–/– cells reduced the number of foci relative to both the vector, presumably by acting on the endogenous eIF4E, and the eIF4E-overexpressing cells. Furthermore, PML reduced the number of foci observed in PML–/– cells transfected with both PML and eIF4E, relative to cells transfected with eIF4E alone or vector controls. Experiments using full-length PML mutated in site I of the RING revealed more foci as compared with wild-type PML, indicating that diminished affinity of PML for eIF4E reduces the ability of PML to suppress transformation. Co-expression of PML site I mutant and wild-type eIF4E consistently resulted in nearly the same number of foci formed as when eIF4E alone was overexpressed. Finally, we examined the effect of PML on the W73A eIF4E mutant. The W73A eIF4E mutant forms a similar number of foci as wild-type eIF4E. PML could not reduce the number of foci in the W73A mutant-overexpressing cells down to levels observed for the PML wild-type eIF4E co-expression experiments (Figure 7B), consistent with the reduced ability of PML to associate with the W73A mutant (Figure 1D). Interestingly, these results suggest that eIF4E-mediated transformation is independent of its translation activity, since the W73A mutant can not act in translation (Sonenberg and Gingras, 1998). Therefore, PML is able to suppress, at least in part, eIF4E-mediated transformation in a RING-dependent manner.
We extended these studies to determine whether the cap-binding activity of eIF4E contributed to its ability to transform cells. The same number of foci were observed in cells overexpressing the W56A cap-binding mutant as in vector controls (Figure 7A and B). Thus, cap-binding activity is required for the transformation function of eIF4E. It is noteworthy that PML reduces the affinity of eIF4E for m7GTP-Sepharose by approximately the same fold as the W56A mutation does (Figure 6, compare B and C). These results suggest that the ability of PML to suppress eIF4E-mediated transformation is due at least in part to its ability to reduce the affinity of eIF4E for the cap.
As a control, the effects of the S53A eIF4E mutant proteins were monitored. Similarly to the results reported in NIH 3T3 cells (Lazaris-Karatzas et al., 1990), this mutation forms foci at the same levels as vector controls and the W56A cap-binding mutant in PML–/– cells. Thus, cap binding alone is necessary but not sufficient for eIF4E-mediated transformation.
Importance of cap binding to nuclear functions of eIF4E
Previous reports indicate that eIF4E-dependent nuclear mRNA transport of Cyclin D1 mRNA contributes to its ability to transform cells (Lazaris-Karatzas et al., 1990; Rousseau et al., 1996). We investigated the possibility that cap-binding activity was required for the nuclear transport function of eIF4E. We examined the effects of overexpression of eIF4E and the W56A cap-binding mutant on the expression of Cyclin D1 protein. eIF4E-overexpressing PML–/– cells expressed more Cyclin D1 protein than vector-transfected controls, whereas GAPDH protein levels did not change. Expression of the W56A eIF4E cap-binding mutant resulted in Cyclin D1 protein levels similar to the S53A mutant and much lower than those observed for wild-type eIF4E. In contrast, the W73A mutant produced levels of Cyclin D1 protein similar to wild-type eIF4E. Expression of PML in the PML–/– cells resulted in substantially reduced levels of Cyclin D1 protein (Figure 7C). In agreement with previous observations in NIH 3T3 cells (Lai and Borden, 2000), co-expression of PML and eIF4E resulted in lower Cyclin D1 protein levels than those observed with eIF4E alone. However, PML does not reduce Cyclin D1 protein levels in cells expressing the W73A mutant to the same levels observed for cells expressing both PML and wild-type eIF4E, consistent with the inability of PML to associate with this mutant. The smaller reduction observed for PML with the W73A mutant likely reflects the effect of PML on endogenous eIF4E. Thus, either direct reduction of the affinity of eIF4E for the m7G cap by the W56A mutation or reduction of affinity by direct interaction with PML results in decreased levels of Cyclin D1 protein. These effects are specific, as GAPDH (Figure 7C) or actin (data not shown; Lai and Borden, 2000) protein levels are not changed by overexpression of wild-type or mutant eIF4E constructs or by expression of PML. The specificity of these effects also suggests that changes in Cyclin D1 protein levels are not due to alterations of translation in the cytoplasm by overexpressing eIF4E mutants. Furthermore, the W73A mutant still increases Cyclin D1 protein levels relative to vector controls, but this mutant can not bind eIF4G to form an active translation initiation complex, again suggesting that alterations in Cyclin D1 levels are not a result of altered translation. As expected, northern analysis shows that Cyclin D1 mRNA levels are not altered in these experiments (data not shown), demonstrating that the effects are not transcriptional. Expression levels for wild-type and mutant PML proteins and wild-type and mutant eIF4E proteins were similar, as observed by western analysis and confocal microscopy. These data suggest that PML represses eIF4E-mediated transformation by reducing the affinity of eIF4E for the m7G cap of mRNA. One consequence of this effect is decreased transport of Cyclin D1 mRNA and suppression of transformation.
Discussion
In this report both the biochemical and physiological consequences of the interaction of PML with eIF4E are examined. Previous studies showed that PML reduced eIF4E-dependent transport of Cyclin D1 mRNA, leading to decreased levels of Cyclin D1 protein (Rousseau et al., 1996), but it was unknown if this regulatory pathway was mediated by a direct association between PML and eIF4E. In addition, the biochemical basis for this mRNA transport mechanism and its resulting physiological effects were unknown. We determined that PML directly binds eIF4E through the PML RING domain. The RING is required for association of PML with bodies and for its physiological functions, which include transformation suppression (Melnick and Licht, 1999). Through a direct interaction with the dorsal surface of eIF4E, the PML RING drastically reduces the affinity of eIF4E for its substrate, the 5′ m7G cap of mRNA. Other regulatory proteins that bind this surface either increase the affinity of eIF4E for the cap, e.g. hBP and full-length eIF4G, or do not alter cap affinity, e.g. a peptide corresponding to the eIF4G binding site (Ptushkina et al., 1998, 1999; von Der Haar et al., 2000). PML is the first example of a protein that directly reduces the affinity of eIF4E for the 5′ m7G cap.
We demonstrate that PML exerts, at least in part, its transformation suppressing functions by modulating the defining feature of eIF4E, i.e. cap binding. The importance of cap-binding to the nuclear function of eIF4E was not known. Our studies suggest that nuclear eIF4E requires cap-binding activity for its role in modulating the nucleocytoplasmic transport of Cyclin D1 mRNA (Rousseau et al., 1996; Lai and Borden, 2000), and strongly suggest that cap-binding activity is required for the transformation activity of eIF4E (Figure 7). The W56A eIF4E cap-binding mutant is unable to bind the m7G cap with high affinity and is consequently unable to increase Cyclin D1 protein levels, and therefore unable to transform cells (Figures 6 and 7). Similarly, PML reduces the cap affinity of wild-type eIF4E (Figure 6), which results in decreased levels of Cyclin D1 protein (Figure 7; Lai and Borden, 2000) and suppression of eIF4E-mediated transformation through interaction with W73 on the dorsal surface of eIF4E (Figure 7). These results suggest that the ability of PML to reduce the affinity of eIF4E for the m7G cap is a biochemical property that contributes to PML-mediated suppression of transformation. Our results suggest that PML functions in this capacity by reducing the cap affinity of eIF4E in the nucleus, therefore reducing its ability to preferentially transport Cyclin D1 mRNA to the cytoplasm. eIF4E modulates the transport of other transcripts, including ornithine decarboxylase and Pim-1 (Rousseau et al., 1996; Hoover et al., 1997) and it is therefore likely that these, and other yet to be identified mRNAs will also be modulated by PML. While it is tempting to speculate that modulation of Cyclin D1 levels is directly linked to the respective growth-promoting and growth-suppressing properties of eIF4E and PML, transport of other mRNAs may contribute significantly to these physiological effects. Several questions remain, including whether modulation of Cyclin D1 levels is directly responsible for these effects and whether specificity (i.e. which mRNAs are modulated in this way), will differ between cell types. However, monitoring Cyclin D1 allows us to monitor eIF4E-dependent transport in general, and thus to link this process to the physiological effects of PML and eIF4E.
Our findings indicate that the structural and therefore functional integrity of PML nuclear bodies does not rely only on the PML protein itself, but also on other proteins. Here, we show that eIF4E, a directly interacting partner of PML, can form nuclear bodies in PML–/– cells. PML appears to require intact eIF4E bodies in order to associate with nuclear bodies, since exogenous PML migrates to eIF4E bodies in PML–/– MEFs (Figure 4G–H). In addition, dispersal of PML bodies upon m7G cap treatment (Figure 4) is likely to be a direct consequence of reorganization within the nuclear body, resulting from alterations in the secondary structure of the eIF4E protein upon m7G cap binding (Figure 6A). It is unlikely that cap treatment disrupts the localization of PML through a direct competition with capped mRNA, since electon microscopy studies indicate that PML nuclear bodies do not contain RNA or DNA inside the body, but that mRNA is found at the periphery of the body (Boisvert et al., 2000). Furthermore, both PML and eIF4E nuclear distribution is unaltered either by RNAse or DNAse treatment, again suggesting that the integrity of these bodies does not rely on RNA or DNA (Stuurman et al., 1992; Melnick and Licht, 1999; Dostie et al., 2000b). Thus, the structural rearrangement of the eIF4E protein upon cap binding is the most likely explanation for the dispersal of eIF4E and PML upon m7G cap treatment. In myeloid cells, the majority of PML localizes to eIF4E bodies (Figure 2); however, a fraction of PML bodies does not associate with eIF4E. Since all PML bodies are disrupted by cap treatment, the possibility arises that PML associates with other nuclear cap-binding proteins, such as the cap-binding complex CBC found in organisms as divergent as yeast and mammals (Visa et al., 1996). Our preliminary data indicate that a small fraction of PML co-localizes and immunoprecipitates with the CBC in both myeloid and non-myeloid cells (N.Cohen and K.L.B.Borden, in preparation). Furthermore, although we have shown that the RING is required for nuclear body formation, presumably through interactions with eIF4E, previous studies by our group and others indicate that PML also requires its B-boxes and leucine coiled-coil domains for this function (Melnick and Licht, 1999). These observations suggest that although the PML RING–eIF4E interaction is required, other interactions through the B-boxes and leucine coiled-coil are also necessary for body formation and subsequent function.
Our observation that PML modulates the functions of a eukaryotic conserved protein, eIF4E, which plays a critical architectural role in the PML nuclear body organization, is consistent with several lines of reasoning. First, PML–/– mice are morphologically normal and acquire spontaneous cancers at rates only marginally higher than littermate controls (Wang et al., 1998), indicating that PML is not essential for survival. In contrast, deletion of the eIF4E gene appears impossible due to its essential function in cellular metabolism. Together these observations suggest that in addition to PML, more basic elements underlie PML body formation and function. Secondly, essential nuclear organelles such as nucleoli are largely conserved throughout eukaryotic evolution. However, the PML gene appears to be limited to mammals (Goddard et al., 1991), with no PML in D.melanogaster or S.cerevisiae. eIF4E, on the other hand, is conserved among eukaryotes from unicellular yeast to mammals (Sonenberg and Gingras, 1998), with eIF4E nuclear bodies detected in D.melano gaster (Figure 3) and yeast (Lang et al., 1994). These observations suggest that acquisition of PML in the course of mammalian evolution serves to modify the function of a more ancestral nuclear organelle constituted by eIF4E. Thirdly, the effect of virus infection on PML nuclear bodies is consistent with the presence of other more basic components. For example, upon arenavirus infection, the subcellular distribution of eIF4E is unaltered, whereas PML is redistributed to the cytoplasm (Borden et al., 1998b; Campbell Dwyer et al., 2000). Arenaviruses are able to establish chronic infection in tolerant hosts, suggesting that re-location of PML to the cytoplasm does not adversely affect cell survival (Borden et al., 1997), which is presumably maintained by preservation of eIF4E-containing bodies in the nucleus. Consistent with the idea that PML negatively regulates growth through interactions with eIF4E, the arenavirus-infected cells are more resistant to apoptotic stimuli than uninfected control cells and grow faster, presumably because regulation of eIF4E by PML has been uncoupled by infection (Borden et al., 1997).
In summary, we show that PML modulates eIF4E function by reducing its affinity for the m7G cap substrate, thereby allowing PML to directly modulate the mRNA transport function of eIF4E and to suppress its transformation activity. The ability of PML to decrease the affinity of eIF4E for the cap is the first biochemical function reported for this protein and provides the first mechanism for the transformation suppressive functions of PML. To date, PML is the only protein known to bind eIF4E in the nucleus and to modulate its nuclear function. Moreover, it appears that PML relies, at least in part, on the evolutionarily conserved eIF4E protein for PML nuclear body formation and function. It appears likely that in vivo, nuclear eIF4E could be subject to regulation by PML in response to changes in cell growth or cellular conditions.
Materials and methods
Reagents
The DNA construct of the RBB domain of human PML in pGEX corresponding to bases 158–840 in full-length PML has been described previously (Borden et al., 1998b). The RING domain of PML plus additional sequence corresponding to bases 158–240 and 802–840 was created by restriction of PML RBB with EagI. For cell culture studies full-length or mutant SV40-PML was used as described previously (Borden et al., 1998b; Lai and Borden, 2000). Full-length WT-eIF4E and the mutant S53A-eIF4E constructs in the mammalian expression vector pMV were described (Lazaris-Karatzas et al., 1990). This gene was subcloned into pGEX for protein interaction assays. The cbl construct contained nucleotides 1268–1441 encompassing the RING inserted into the BamHI site of pGEX. After thrombin cleavage of cbl–GST, cbl was N-terminally sequenced to confirm accurate cleavage. Laser desorption mass spectrometry confirmed that the cbl protein produced was the correct molecular weight, and NMR confirmed it was structured (Kentsis and Borden, 2000). Point mutations were engineered using the Strategene Quickchange kit. DNA sequencing was used to verify the integrity of all constructs.
Antibodies used against PML include 5E10 mAb, which recognizes human PML (Stuurman et al., 1992), and a polyclonal antibody that recognizes both human and mouse PML (Borden et al., 1995), which has been additionally characterized (Wang et al., 1998; Lai and Borden, 2000). The specificity of eIF4E mAb (Transduction Laboratories) has been verified previously (Lejbkowicz et al., 1992; Campbell Dwyer et al., 2000; Dostie et al., 2000a,b; Lai and Borden, 2000). Monoclonal antibodies against Sp100, Sm, nopp140 and nucleolin have been described previously (Lerner et al., 1981; Meier, 1996; Pinol-Roma, 1999; Ferbeyre et al., 2000).
All chemicals, unless otherwise stated, were of ACS grade from Sigma. Cap dinucleotide analogs m7GpppG and GpppG were obtained from New England Biolabs.
GST pull-down assays
GST fusion proteins (RBB–GST, RING–GST, eIF4E–GST and mutants) were expressed in Escherichia coli BL21 (DE-3) cells by induction with isopropyl-β-d-thiogalactopyranoside. Cell pellets were suspended in buffer A (0.5 M NaCl, 50 mM Na2PO4 and 1 µM ZnCl2 pH 7.5), supplemented with protease inhibitors (Complete, Boehringer Mannheim), lysed by sonication, solubilized with 0.1% Triton X-100 and cleared by centrifugation. Cleared lysates were adsorbed to G-Sepharose (Amersham) and extensively washed with buffer A. Proteins were cleaved with thrombin (Amersham) to release them from GST. The purity of all isolated proteins was verified by SDS–PAGE; UV spectroscopy and dynamic light scattering were used to confirm that free proteins were monomeric (Protein Solutions). Protein concentrations were determined using calculated extinction coefficients. GST fusion proteins bound to G-Sepharose were incubated with purified eIF4E in 0.5 ml of binding buffer [phosphate-buffered saline (PBS) supplemented with 0.3 M KCl and 1% NP-40] for 1 h at room temperature (RT) while tumbling. Beads were washed three times with binding buffer and sedimented by centrifugation. Sedimented proteins were identified using SDS–PAGE and western analysis. Our earlier studies with transfected material in NIH 3T3 cells suggested that eIF4E utilized site II, not site I, for interaction with PML. However, these experiments were carried out in the presence of endogenous PML, which may have mediated interactions between mutant PML and wild-type eIF4E. Since there is no endogenous PML or eIF4E in bacteria, the GST pull-down analysis described here gives more accurate results.
CD difference spectroscopy
Far-UV CD spectra were collected using a Jasco-810 spectropolarimeter with 8 mm tandem quartz cuvette (Hellma) in 50 mM NaCl, 10 mM Na2PO4, 1 µM ZnCl2 pH 7.5. eIF4E (0.24 µM) and substrates were placed into separate chambers of the tandem cuvette, and allowed to equilibrate to solvent conditions for 1 h at 23°C. Five spectra for each condition before and after mixing were collected using a 2 nm bandwidth and 1 nm resolution, and averaged. m7GpppG and GpppG were used at a saturating concentration of 3 µM. Relative ellipticity was converted to molar ellipticity as described previously (Fasman, 1996).
Cap-binding assays
m7GTP-Sepharose binding assays were performed as described previously (Ptushkina et al., 1999). Briefly, 15 ng each of eIF4E and GST fusion proteins were incubated with 40 µl of m7GTP-Sepharose (Pharmacia) in a total volume of 300 µl of buffer (10 mM sodium phosphate pH 7.5, 100 mM KCl, 1 mM dithiothreitol and 2 mM MgCl2). The mixture was incubated on a shaker at 4°C for 2 h. The resin was washed three times with 1 ml of buffer and in some cases the bound proteins were eluted with 80 µl of 0.1 mM m7GTP in buffer. Aliquots (20 µl) of the eluted fraction, or alternatively of beads, were analyzed on 15% SDS gel followed by western blotting using eIF4E mAb, yielding similar results.
Cell culture
U937 promonocytic cells, K562 chronic myelogenous leukemia cells and NB4 acute promyelocytic leukemia cells were grown in RPMI 1640 (Gibco) supplemented with 10% fetal bovine serum (FBS) (Gibco). When indicated, NB4 cells were treated with 1 µM ATRA (Sigma) for 2 days and harvested for examination. PML–/– mouse embryonic fibroblasts were grown in Dulbecco’s modified Eagle’s medium (Gibco) supplemented with 20% FBS (Wang et al., 1998), and D.melanogaster S2 cells in DES expression medium (Invitrogen) at 25°C. For Cyclin D1 and GAPDH detection, cells were lysed in lysis buffer (20 mM Tris–HCl, 150 mM NaCl, 1% NP-40 pH 7.4, supplemented with protease inhibitors) for 1 h on ice, and protein concentration was determined using Bio-Rad DC Protein Assay. Equal amounts of protein (50 µg) were loaded on each lane.
Foci growth assay
PML–/– cells were plated at 1 × 105 cells in 100 mm2 dishes, and two plates per treatment were transfected with 10–15 µg of SV40-PML, SV40-PMLRING mutant, pMV-eIF4E WT and mutants (S53A, W73A, W56A), using SuperFect transfection reagent (Qiagen) according to the manufacturer’s instructions. Anchorage-dependent foci formation assays were conducted as previously described (Melnick et al., 2000). In all experiments 1 µg (or other amount in 1:10 ratio) of pIND containing Neomycin resistance gene (Invitrogen) was added. Single transfections were supplemented with an appropriate amount of empty vector to equalize the total amount of transfected DNA relative to the double transfections. Cells were selected in 1 mg/ml G418-containing medium. Forty-eight hours after transfection, cells were collected, diluted at 1:175, plated in 100 mm2 dishes and maintained for 7 days with addition of fresh medium containing 1 mg/ml G418 every 3 days. Cells were dried, stained with Giemsa 1:20 for 30 min at RT and foci counted. The numbers represent an average of seven plates per treatment. All experiments were repeated independently at least three times. In parallel, total protein concentrations were measured from identically treated plates as used in the foci assay. Total protein concentrations varied by treatment in the same way as the number of foci, indicating that focus-counting artifacts were not significant. Western analysis and confocal microscopy confirmed that mutant and wild-type constructs were expressed in similar levels in these assays. Furthermore, transfection efficiency was similar for the mutant and wild-type proteins as observed by confocal microscopy.
Cap treatment
Treatments were performed as previously described (Dostie et al., 2000b) with the following modifications: suspension cells (2–4 × 106) were collected and washed with PBS and then with transport buffer (as described previously by Dostie et al., 2000b), supplemented with protease inhibitors and incubated for 4 min at RT in transport buffer containing 0.4% Tween-20. Subsequently, cells were washed and resuspended in transport buffer containing 2 µg/ml bovine serum albumin (BSA) and 50 mM m7GpppG or GpppG. Adherent cells grown on coverslips were rinsed in transport buffer and incubated for 4 min at RT in transport buffer containing 40 µg/ml digitonin. Coverslips were rinsed and inverted over a drop of transport buffer containing 2 µg/ml BSA and 50 mM m7GpppG or GpppG. The reaction was carried out at 30°C for 25 min. Cells were rinsed, fixed in transport buffer containing 3.7% formaldehyde for 10 min at RT, and processed for immunofluorescence as described below.
Nuclear matrix preparation
Preparation of nuclear matrices was carried out as described previously (McNeil et al., 1998). Briefly, cells were washed twice for 15 min at 4°C with CSK buffer supplemented with 2 mM vanadyl ribonucleoside complex (VRC) and 2 mM phenylmethylsulfonyl fluoride (PMSF). DNA was digested by incubating cells at RT for 1 h in DB buffer supplemented with 100 µg/ml DNAse I, 2 mM VRC and 2 mM PMSF. Chromatin was then extracted by incubating for 10 min with DB buffer at 4°C, supplemented with 2 mM VRC and 0.25 M (NH4)2SO4. Cells were fixed with 3.7% formaldehyde at 4°C for 20 min and processed for immunofluorescence as described below.
Indirect immunofluorescence and laser scanning confocal microscopy
For indirect immunofluorescence analysis several fixation protocols were used. For PML, eIF4E and SP100 staining, cells were fixed in either 3.7% formaldehyde in PBS for 10 min at RT, or in MeOH for 10 min at –20°C. No significant difference in staining patterns was observed with either protocol; for Sm staining, cells were fixed in 3.7% formaldehyde in PBS for 10 min at RT; for nucleolin staining, cells were fixed in 2% formaldehyde in PBS for 20 min at RT followed by acetone at –20°C for 5 min; for cells treated with the cap, see cap treatment section for details. Cells were then washed and blocked as described previously (Lai and Borden, 2000). Antigens were detected by incubation with primary antibodies in blocking buffer (10% newborn calf serum, 0.1% Tween-20 in PBS) for 2 h at RT followed by secondary antibodies labeled with fluorochromes. For double labeling, slides were incubated with PML polyclonal antibody and eIF4E mAb in blocking buffer for 2 h at RT followed by FITC-conjugated goat anti-mouse IgG and Texas Red-conjugated donkey anti-rabbit IgG (Jackson Immunoresearch) secondary antibodies in blocking buffer for 1 h. Alternatively, slides were incubated with PML 5E10 mAb in blocking buffer for 2 h followed by Alexa Fluor (ultraviolet)-conjugated goat anti-mouse IgG (Molecular Probes), then slides were washed with PBS and additionally fixed in 3.7% formaldehyde in PBS for 10 min at RT followed by incubation with FITC-conjugated eIF4E mAb. Fluorescence was observed using a Leica inverted laser scanning confocal microscope, exciting at 568 nm (red), 488 nm (green) or 351/364 nm (blue). Micrographs represent single sections with a thickness of 300 nm. Under these conditions, no breakthrough among the blue, green and red channels was observed. Experiments were repeated at least twice with >500 cells in each sample.
Acknowledgments
Acknowledgements
We are grateful for the kind gift of antibodies from Paul Freemont, Gerd Maul, L.de Jong, Serafin Piñol-Roma and Thomas Meier. We thank Nahum Sonenberg for the pMV eIF4E and pMV S53A constructs, Pier Paolo Pandolfi for PML–/– cells and Serafin Piñol-Roma, Liliana Ossowski, Pauline McIntosh, Ari Melnick and Allan Capili for critical reading of the manuscript. Confocal laser scanning microscopy was performed at MSSM-LCSM core facility, supported with funding from NIH (1 S10 RR0 9145–01) and NSF (DBI-9724504). A.K. is supported by the NIH Medical Scientist Training Program and S.S. by the Samuel Waxman Cancer Research Foundation. K.L.B.B. is a scholar of the Leukemia and Lymphoma Society. Financial support was provided by the NIH (CA 80728 and CA 88991).
References
- Asano K., Merrick,W.C. and Hershey,J.W. (1997) The translation initiation factor eIF3-p48 subunit is encoded by int-6, a site of frequent integration by the mouse mammary tumor virus genome. J. Biol. Chem., 272, 23477–23480. [DOI] [PubMed] [Google Scholar]
- Boisvert F.M., Hendzel,M.J. and Bazett-Jones,D.P. (2000) Promyelo cytic leukemia (PML) nuclear bodies are protein structures that do not accumulate RNA. J. Cell Biol., 148, 283–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borden K.L. (2000) RING domains: master builders of molecular scaffolds? J. Mol. Biol., 295, 1103–1112. [DOI] [PubMed] [Google Scholar]
- Borden K.L.B., Boddy,M.N., Lally,J., O’Reilly,N.J., Martin,S., Howe,K., Solomon,E. and Freemont,P.S. (1995) The solution structure of the RING finger domain from the acute promyelocytic leukaemia proto-oncoprotein PML. EMBO. J, 14, 1532–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borden K.L.B., Lally,J.M., Martin,S.R., O’Reilly,N.J., Solomon,E. and Freemont,P.S. (1996) In vivo and in vitro characterization of the B1 and B2 zinc-binding domains from the acute promyelocytic leukemia protooncoprotein PML. Proc. Natl Acad. Sci. USA, 93, 1601–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borden K.L.B., Campbell Dwyer,E.J. and Salvato,M.S. (1997) The promyelocytic leukemia protein PML has a pro-apoptotic activity mediated through its RING domain. FEBS Lett., 418, 30–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borden K.L.B., Campbell Dwyer,E.J., Carlile,G.W., Djavani,M. and Salvato,M.S. (1998a) Two RING finger proteins, the oncoprotein PML and the arenavirus Z protein, colocalize with the nuclear fraction of the ribosomal P proteins. J. Virol., 72, 3819–3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borden K.L.B., Campbell Dwyer,E.J. and Salvato,M.S. (1998b) An arenavirus RING (zinc-binding) protein binds the oncoprotein promyelocyte leukemia protein (PML) and relocates PML nuclear bodies to the cytoplasm. J. Virol., 72, 758–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai A., Jankowska-Anyszka,M., Centers,A., Chlebicka,L., Stepinski,J., Stolarski,R., Darzynkiewicz,E. and Rhoads,R.E. (1999) Quantitative assessment of mRNA cap analogues as inhibitors of in vitro translation. Biochemistry, 38, 8538–8547. [DOI] [PubMed] [Google Scholar]
- Campbell Dwyer E.J., Lai,H., MacDonald,R.C., Salvato,M.S. and Borden,K.L. (2000) The lymphocytic choriomeningitis virus RING protein Z associates with eukaryotic initiation factor 4E and selectively represses translation in a RING-dependent manner. J. Virol., 74, 3293–3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copps K., Richman,R., Lyman,L.M., Chang,K.A., Rampersad-Ammons,J. and Kuroda,M.I. (1998) Complex formation by the Drosophila MSL proteins: role of the MSL2 RING finger in protein complex assembly. EMBO J., 17, 5409–5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehlin E., Wormington,M., Korner,C.G. and Wahle,E. (2000) Cap-dependent deadenylation of mRNA. EMBO J., 19, 1079–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostie J., Ferraiuolo,M., Pause,A., Adam,S.A. and Sonenberg,N. (2000a) A novel shuttling protein, 4E-T, mediates the nuclear import of the mRNA 5′ cap-binding protein, eIF4E. EMBO J., 19, 3142–3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostie J., Lejbkowicz,F. and Sonenberg,N. (2000b) Nuclear eukaryotic initiation factor 4E (eIF4E) colocalizes with splicing factors in speckles. J. Cell. Biol., 148, 239–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasman G.D. (ed.) (1996) Circular dichroism and the conformational analysis of biomolecules. Plenum Press, New York, NY.
- Ferbeyre G., de Stanchina,E., Querido,E., Baptiste,N., Prives,C. and Lowe,S.W. (2000) PML is induced by oncogenic ras and promotes premature senescence. Genes Dev., 14, 2015–2027. [PMC free article] [PubMed] [Google Scholar]
- Goddard A.D., Borrow,J., Freemont,P.S. and Solomon,E. (1991) Characterization of a zinc finger gene disrupted by the t(15;17) in acute promyelocytic leukemia. Science, 254, 1371–1374. [DOI] [PubMed] [Google Scholar]
- Grimwade D. and Solomon,E. (1997) Characterisation of the PML/RAR α rearrangement associated with t(15;17) acute promyelocytic leukaemia. Curr. Top. Microbiol. Immunol., 220, 81–112. [DOI] [PubMed] [Google Scholar]
- Hoover D.S., Wingett,D.G., Zhang,J., Reeves,R. and Magnuson,N.S. (1997) Pim-1 protein expression is regulated by its 5′-untranslated region and translation initiation factor elF-4E. Cell Growth Differ., 8, 1371–1380. [PubMed] [Google Scholar]
- Keiper B.D. and Rhoads,R.E. (1997) Cap-independent translation initiation in Xenopus oocytes. Nucleic Acids Res., 25, 395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kentsis A. and Borden,K. (2000) Construction of macromolecular assemblages in eukaryotic processes and their role in human disease: linking RINGs together. Current Peptide and Protein Sci., 1, 43–71. [DOI] [PubMed] [Google Scholar]
- Lai H.K. and Borden,K.L. (2000) The promyelocytic leukemia (PML) protein suppresses Cyclin D1 protein production by altering the nuclear cytoplasmic distribution of Cyclin D1 mRNA. Oncogene, 19, 1623–1634. [DOI] [PubMed] [Google Scholar]
- Lang V., Zanchin,N.I., Lunsdorf,H., Tuite,M. and McCarthy,J.E. (1994) Initiation factor eIF-4E of Saccharomyces cerevisiae. Distribution within the cell, binding to mRNA, and consequences of its overproduction. J. Biol. Chem., 269, 6117–6123. [PubMed] [Google Scholar]
- Lazaris-Karatzas A., Montine,K.S. and Sonenberg,N. (1990) Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature, 345, 544–547. [DOI] [PubMed] [Google Scholar]
- Lejbkowicz F., Goyer,C., Darveau,A., Neron,S., Lemieux,R. and Sonenberg,N. (1992) A fraction of the mRNA 5′ cap-binding protein, eukaryotic initiation factor 4E, localizes to the nucleus. Proc. Natl Acad. Sci. USA, 89, 9612–9616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner M.R., Boyle,J.A., Hardin,J.A. and Steitz,J.A. (1981) Two novel classes of small ribonucleoproteins detected by antibodies associated with lupus erythematosus. Science, 211, 400–402. [DOI] [PubMed] [Google Scholar]
- Marcotrigiano J., Gingras,A.C., Sonenberg,N. and Burley,S.K. (1997) Cocrystal structure of the messenger RNA 5′ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell, 89, 951–961. [DOI] [PubMed] [Google Scholar]
- Maul G.G., Negorev,D., Bell,P. and Ishov,A.M. (2000) Review: properties and assembly mechanisms of ND10, PML bodies, or PODs. J. Struct. Biol., 129, 278–287. [DOI] [PubMed] [Google Scholar]
- McCubbin W.D., Edery,I., Altmann,M., Sonenberg,N. and Kay,C.M. (1988) Circular dichroism and fluorescence studies on protein synthesis initiation factor eIF-4E and two mutant forms from the yeast Saccharomyces cerevisiae. J. Biol. Chem., 263, 17663–17671. [PubMed] [Google Scholar]
- McNeil S. et al. (1998) Targeting of the YY1 transcription factor to the nucleolus and the nuclear matrix in situ: the C-terminus is a principal determinant for nuclear trafficking. J. Cell. Biochem., 68, 500–510. [PubMed] [Google Scholar]
- Meier U.T. (1996) Comparison of the rat nucleolar protein nopp140 with its yeast homolog SRP40. Differential phosphorylation in vertebrates and yeast. J. Biol. Chem., 271, 19376–19384. [PubMed] [Google Scholar]
- Melnick A. and Licht,J.D. (1999) Deconstructing a disease: RARα, its fusion partners, and their roles in the pathogenesis of acute promyelocytic leukemia. Blood, 93, 3167–3215. [PubMed] [Google Scholar]
- Melnick A., Ahmad,K.F., Arai,S., Polinger,A., Ball,H., Borden,K.L., Carlile,G.W., Prive,G.G. and Licht,J.D. (2000) In-depth mutational analysis of the promyelocytic leukemia zinc finger BTB/POZ domain reveals motifs and residues required for biological and transcriptional functions. Mol. Cell. Biol., 20, 6550–6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu Z.M., Chin,K.V., Liu,J.H., Lozano,G. and Chang,K.S. (1994) PML, a growth suppressor disrupted in acute promyelocytic leukemia. Mol. Cell. Biol., 14, 6858–6867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinol-Roma S. (1999) Association of nonribosomal nucleolar proteins in ribonucleoprotein complexes during interphase and mitosis. Mol. Biol. Cell., 10, 77–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptushkina M., von der Haar,T., Vasilescu,S., Frank,R., Birkenhager,R. and McCarthy,J.E. (1998) Cooperative modulation by eIF4G of eIF4E-binding to the mRNA 5′ cap in yeast involves a site partially shared by p20. EMBO J., 17, 4798–4808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptushkina M., von der Haar,T., Karim,M.M., Hughes,J.M. and McCarthy,J.E. (1999) Repressor binding to a dorsal regulatory site traps human eIF4E in a high cap-affinity state. EMBO J., 18, 4068–4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenwald I.B., Kaspar,R., Rousseau,D., Gehrke,L., Leboulch,P., Chen,J.J., Schmidt,E.V., Sonenberg,N. and London,I.M. (1995) Eukaryotic translation initiation factor 4E regulates expression of Cyclin D1 at transcriptional and post-transcriptional levels. J. Biol. Chem., 270, 21176–21180. [DOI] [PubMed] [Google Scholar]
- Rousseau D., Kaspar,R., Rosenwald,I., Gehrke,L. and Sonenberg,N. (1996) Translation initiation of ornithine decarboxylase and nucleocytoplasmic transport of Cyclin D1 mRNA are increased in cells overexpressing eukaryotic initiation factor 4E. Proc. Natl Acad. Sci. USA, 93, 1065–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N. and Gingras,A.C. (1998) The mRNA 5′ cap-binding protein eIF4E and control of cell growth. Curr. Opin. Cell Biol., 10, 268–275. [DOI] [PubMed] [Google Scholar]
- Sternsdorf T., Jensen,K., Reich,B. and Will,H. (1999) The nuclear dot protein sp100, characterization of domains necessary for dimerization, subcellular localization, and modification by small ubiquitin-like modifiers. J. Biol. Chem., 274, 12555–12566. [DOI] [PubMed] [Google Scholar]
- Stuurman N., de Graaf,A., Floore,A., Josso,A., Humbel,B., de Jong,L. and van Driel,R. (1992) A monoclonal antibody recognizing nuclear matrix-associated nuclear bodies. J. Cell Sci., 101, 773–784. [DOI] [PubMed] [Google Scholar]
- Visa N., Izaurralde,E., Ferreira,J., Daneholt,B. and Mattaj,I.W. (1996) A nuclear cap-binding complex binds Balbiani ring pre-mRNA cotranscriptionally and accompanies the ribonucleoprotein particle during nuclear export. J. Cell Biol., 133, 5–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Der Haar T., Ball,P.D. and McCarthy,J.E. (2000) Stabilization of eukaryotic initiation factor 4E binding to the mRNA 5′-cap by domains of eIF4G. J. Biol. Chem., 275, 30551–30555. [DOI] [PubMed] [Google Scholar]
- von Mikecz A., Zhang,S., Montminy,M., Tan,E.M. and Hemmerich,P. (2000) CREB-binding protein (CBP)/p300 and RNA polymerase II colocalize in transcriptionally active domains in the nucleus. J. Cell Biol., 150, 265–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.G., Delva,L., Gaboli,M., Rivi,R., Giorgio,M., Cordon-Cardo,C., Grosveld,F. and Pandolfi,P.P. (1998) Role of PML in cell growth and the retinoic acid pathway. Science, 279, 1547–1551. [DOI] [PubMed] [Google Scholar]
- Zhong S., Muller,S., Ronchetti,S., Freemont,P.S., Dejean,A. and Pandolfi,P.P. (2000) Role of SUMO-1-modified PML in nuclear body formation. Blood, 95, 2748–2752. [PubMed] [Google Scholar]