Abstract
Hepadnaviruses do not infect cultured cells, therefore our knowledge of the mechanism of the early stages of virus–cell interaction is rather poor. In this study, we show that dimethylsulfoxide (DMSO)-treated HepG2 hepatoblastoma cells are infected efficiently by serum-derived hepatitis B virus (HBV) as monitored by viral gene expression and replication markers. To measure virus attachment, a variety of HBV surface proteins (HBsAgs) were conjugated to polystyrene beads and their capacity to attach cells was visualized and quantified by light microscopy at a single-cell resolution. Remarkably, DMSO increases the attachment efficiency by >200-fold. We further identify the QLDPAF sequence within preS1 as the receptor-binding viral domain epitope. Interestingly, a similar sequence is shared by several cellular, bacterial and viral proteins involved in cell adhesion, attachment and fusion. We also found that the small HBsAg contains a secondary attachment site that recognizes a distinct receptor on the cell membrane. Furthermore, we provide evidence in support of multivalent HBV attachment with synergistic interplay. Our data depict a mechanistic view of virus attachment and ingestion.
Keywords: endocytosis/HBsAg proteins/HBV infection/QLDPAF motif/virus attachment
Introduction
Hepadnaviruses replicate almost exclusively in the liver. This liver tropism is believed to be determined, at least in part, by hepatocyte-specific receptors and co-receptors. Several cellular proteins were identified as putative viral receptors (reviewed in De Meyer et al., 1997). The gp180 protein was identified as the putative duck hepatitis B virus (DHBV) receptor (Kuroki et al., 1995; Breiner et al., 1998; Urban et al., 1998). The involvement of additional cellular proteins, e.g. gp120 and p55, in DHBV infection was proposed recently (Li et al., 1996; Guo and Pugh, 1997). A point of concern is that DHBV displays clear liver tropism yet the putative receptor candidates are not strictly liver specific. Our knowledge of hepatitis B virus (HBV) liver-specific receptors is poorer and is still in its infancy. Certainly, progress in understanding the early stages of virus–cell interaction will facilitate the research into resolving this issue.
Parallel studies and identification of the viral regions that bind the specific receptors on the target cell membrane have been more rewarding. The viral envelope is composed of a membrane originating from the cell in which several virus-encoded surface proteins are anchored. These include the small HBsAgs (p24 and gp28), the middle HBsAgs (gp33 and gp36) and the large HBsAgs (p39 and gp42). These proteins are presented on the surface of both the infectious viral particle (the 42 nm Dane particle) and the rod-shaped or spherical 22 nm subviral particles (SVPs). The latter are composed of the viral surface proteins and the cellular membrane. SVPs comprise most (90–99%) of the circulating viral particles in infectious serum as well as in culture media of infected or transfected cells. Of all the viral surface proteins, the large HBsAg of both avian and the mammalian hepadnaviruses is believed to play a major role in mediating virus attachment and entry into hepatocytes (Neurath et al., 1986; De Meyer et al., 1997; Le Seyec et al., 1999). Within the large HBsAg, the 21–47 amino acid epitope of preS1 was shown previously to mediate binding to the cell surface of HepG2 cells (Neurath et al., 1986). Antibodies directed against this epitope were shown to block infection (Neurath et al., 1989). Most of the preS1-interacting proteins that have been identified so far interact with this epitope (reviewed in De Meyer et al., 1997; Ryu et al., 2000). Thus, it appears that a single viral epitope is responsible for cell recognition and ingestion. However, as learned from other viruses, the notion of simple recognition and attachment to a single cell surface molecule is inaccurate (for a review see Haywood, 1994).
The study of HBV attachment and infection is hampered by the fact that the established human hepatocyte cell lines that retain the hepatocyte markers, such as HepG2 and Huh-7, are refractory to HBV infection. As the transfected HBV genome is transcribed efficiently in these cell lines, attachment and entry are believed to be the restrictive steps. Human primary hepatocytes, on the other hand, are variably susceptible to infection and for only a short period after culturing (Gripon et al., 1988; Pugh et al., 1995). However, upon their exposure to 1.5–2% dimethylsulfoxide (DMSO), enhanced and prolonged HBV (Gripon et al., 1988) and DHBV infection (Pugh and Summers, 1989) was obtained. We show here that DMSO-treated HepG2 cells manifest efficient HBV infection. It is therefore possible that DMSO improves HBV infection by inducing expression and presentation of differentiation-specific viral receptors, although other possibilities were not ruled out. The main obstacle to defining the underlying mechanisms is the lack of reliable tools to evaluate virus attachment to the target cells quantitatively.
Studies on characterization of the factors that mediate HBV and DHBV cell attachment using either labeled viral particles, anti-HBsAg antibodies (Klingmuller and Schaller, 1993; Pontisso et al., 1989) or coated beads (Neurath et al., 1986) have been reported. However, these methods neither demonstrate nor quantify attachment at the single-cell level. As an alternative approach, we developed a bead-mediated attachment (BMA) assay, where the attachment of a variety of HBV surface proteins, covalently conjugated to synthetic 6 µm beads, was visualized and quantified by light and electron microscopy. We show here that beads coated with subviral recombinant particles attach, albeit poorly, to HepG2 cells, but not to non-liver cells. The obtained attachment was increased by >200-fold upon DMSO treatment, a treatment that sensitized the cells to HBV infection. Furthermore, our quantitative assay permitted fine mapping of the major viral epitope and detection of a second minor region that is involved in attachment but via a distinct cellular receptor. Thus, our quantitative assessment of virus–host attachment permitted us to study the molecular mechanism of virus–host attachment, a step in infection that is inherently transient and, therefore, so far, has been difficult to study.
Results
DMSO improves HBV infection of HepG2 cells
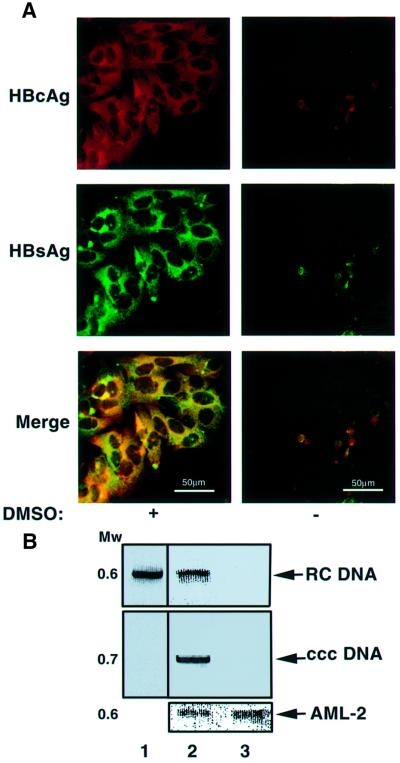
Treatment of primary liver culture cells with 1.5–2% DMSO results in enhanced and prolonged HBV (Gripon et al., 1988) and DHBV infection (Pugh and Summers, 1989). To apply this protocol to the established cell lines, HepG2 cells were treated with DMSO or left untreated, and incubated with HBV-positive human serum. Four days later, cells were analyzed by indirect immunoflourescent staining for the presence of viral proteins. Interestingly, HBcAg and HBsAg were both detected in the DMSO-treated cells, whereas in untreated plates a few cells were barely reactive (Figure 1A). These data suggest that DMSO improved HBV infection of the HepG2 cells. Next we checked whether this infection gives rise to viral replication. To this end, total DNA was extracted from the infected cells and from the HBV-positive sera virus stock. Equal amounts of DNA were analyzed by PCR for the presence of HBV relaxed circular (RC) and covalently closed circular (ccc) DNA, markers of productive infection and genome replication. RC DNA was detected both in the infected cells and in the virions, whereas cccDNA was detected only in the cells (Figure 1B). No viral DNA of any form was detected in cells not treated with DMSO. Both samples contain equal amounts of DNA as quantified by amplifying a genomic single gene (AML-2, Figure 1B, lower panel). To determine HBV replication directly, both untreated and DMSO-treated cells were harvested 5, 9 and 14 days after infection, and DNA from the cells was analyzed for RC, single-stranded (ss) and cccDNA by agarose gel electrophoresis and Southern blot hybridization. Remarkably, already 5 days after infection significant amounts of the DNA replication markers were detected only in DMSO-treated cells. HBV DNA accumulated further at 9 and 14 days (Figure 1C). Furthermore, accumulation of these replicative intermediate DNA forms is dependent on reverse transcription activity since they were not detected in the presence of lamivudine (3TC), a potent HBV reverse transcriptase inhibitor (Figure 1C, lane 8). Antibody raised against the 21–47 amino acid region of preS1 neutralizes infection (Neurath et al., 1989). This antibody (MA 18/7) displays similar activity in our system and blocked infection (Figure 1C, lane 9). These data strongly argue that DMSO sensitized HepG2 to efficient HBV infection and replication.
Fig. 1. Efficient infection of DMSO-treated HepG2 cells by HBV. (A) HepG2 cells were seeded on 18 mm coverslips and either DMSO treated or left untreated. After 6 days, cells were incubated for 14 h with HBV-positive sera containing 109 particles per ml, diluted in culture media. Subsequently, the unbound viruses were discarded and the cells were incubated further with (left panel) or without (right panel) DMSO for an additional 4 days. Viral infection was monitored by indirect immunoflourescent staining with αHBcAg (red) and αHBsAg (green) polyclonal sera, and by either RRX- or FITC-conjugated antibodies, respectively. The stained cells were visualized by scanning laser confocal microscopy. Yellow represents co-localization of HBcAg and αHBsAg. (B) Viral DNA in the duplicate samples was isolated from the infected cells and from the virions and subjected to PCR to detect the presence of HBV RC and cccDNA. The PCR results of the viral sample (lane 1), and extracts of infected cells that were DMSO treated (lane 2) or left untreated (lane 3) are shown. Control PCRs were performed for the endogenous AML-2. The migration position of the DNA is shown as the molecular weight in kb. (C) Southern blot analysis of total (lanes 2–9) and extrachromosomal DNA (lanes 10–11), extracted from HBV- and mock-infected (lane M) cells at the indicated days post-infection (dpi). Cells were either DMSO treated (lanes 2, 5–9 and 11) or left untreated (lanes 3, 4 and 10). The gel migration positions of relaxed circular (RC), covalently closed circular (ccc) and single-stranded (ss) forms are indicated. In lane 8, 100 µM lamivudine (3TC) was added to the culture medium 14 h after infection. Fresh medium containing 3TC was added every 3 days. To block infection by MA 18/7 neutralizing monoclonal anti-preS1 antibody, 200 µl of HBV-positive serum was pre-incubated with 0.5 µg of IgG for 4 h before infection. MW = DNA molecular weight in kb.
Preparation of HBV subviral particles (SVPs)
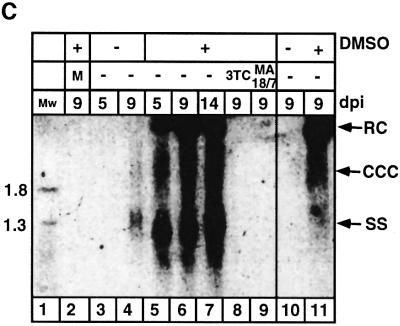
Our data suggest that DMSO might improve HBV infection by inducing expression and presentation of differentiation-specific viral receptors (receptor activation). To substantiate this, we developed the BMA assay that measures viral attachment at a single cell resolution. Recombinant HBV SVPs were produced in animal cells by utilizing the AL26 plasmid that contains an integrated form of HBV DNA (Faktor et al., 1988). Chinese hamster ovary (CHO) cells were transfected with AL26 plasmid, and high HBsAg producer lines were established that secrete HBsAg SVPs with the expected 1.22 g/ml density (Figure 2A). Electron microscopy revealed that the particles are spherical and homogeneous in size (22 nm in diameter). For protein composition analysis, metabolically 35S-labeled particles were prepared and analyzed by SDS–PAGE. The small HBsAgs p24 and p28 are the major components and the middle preS2 proteins, p33 and p36, are the minor components (Figure 2B and C), whereas the large preS1 proteins are barely detectable. However, the fact that these particles contain all three components was confirmed by immunoblotting utilizing different specific antibodies. Collectively, our analysis shows that the structure, density and composition of the purified recombinant SVPs are similar to those reported for serum-derived 22 nm SVPs.
Fig. 2. Recombinant HBV SVPs containing all the HBV surface proteins. CHO cells stably transfected with the AL26 plasmid express 22 nm HBV subparticles. (A) Purified particles were fractionated on a CsCl gradient and the resulting fractions were assayed for HBsAg by radioimmunoassay using 125I-labeled anti-HBsAg antibodies. The density of HBsAg SVPs was calculated to be 1.22 g/ml, characteristic of the 22 nm spherical HBsAg SVPs. The insert shows 22 nm particles that were negatively stained by uranyl acetate followed by TEM. (B) The composition of the various surface proteins was determined by [35S]methionine labeling on SDS–PAGE and by western analysis with MA 18/7, the antibody specific for preS1 (αS1), MA Q19/10 (for preS2), and with polyclonal anti-S antibodies (αS). The various HBV surface proteins are indicated. (C) Schematic presentation of the different (S, L and M) HBsAg proteins with the corresponding antibody epitopes. The gray boxes represent the transmembrane regions.
DMSO improves SVP–bead attachment
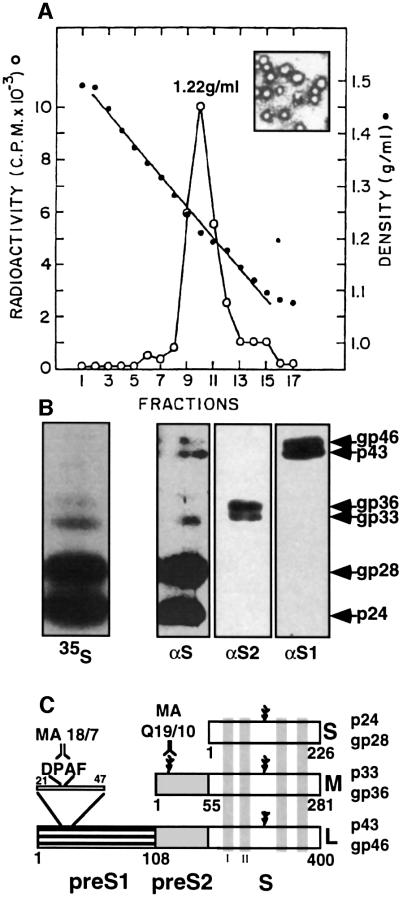
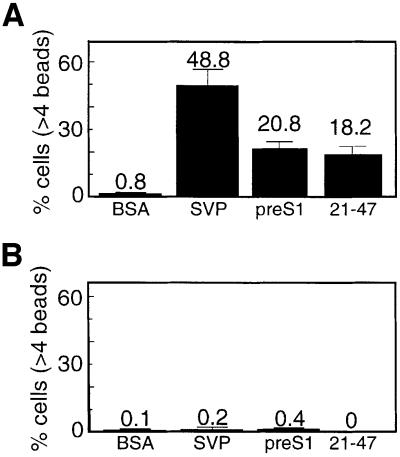
Recombinant SVPs containing all three HBsAg components were conjugated to synthetic beads to obtain SVP–beads. Control bovine serum albumin (BSA)-conjugated beads were also prepared and both were incubated for 16 h with untreated HepG2 cells. The unbound beads were removed, the cells were fixed and the number of attached beads per cell was determined by light microscopy. The percentage of cells that bind SVP–beads was significantly higher than for the control BSA–beads (Figure 3A and B), normally 50 and 18.7%, respectively. Remarkably, DMSO significantly improved SVP–bead attachment and ∼93% of the cells bind beads as compared with 50% of untreated cells (Figure 3C and D). Moreover, ∼49% of the cells bind more than four SVP–beads per cell (Figure 3D). This is 244-fold higher than that observed in untreated cells (0.2%, Figure 3B). The improvement in SVP–bead attachment is specific, as the majority (56%) of the cells do not bind BSA–beads at all and only 0.8% bind more than four beads per cell. Also the SVP–beads attached non-liver Cos-1 and HeLa cells poorly (data not shown), suggesting that SVP–bead attachment is cell type specific. We therefore concluded that DMSO increases not only the number of cells that bind HBsAg SVPs but also the binding capacity of a single cell. Similar results were obtained when purified serum-derived HBV particles were conjugated to the synthetic beads (data not shown). The fact that DMSO improved both HBV infection and SVP attachment suggests that the HBV receptors on the cell membrane became activated.
Fig. 3. DMSO improves SVPs attachment. (A) HepG2 cells were seeded on coverslips and incubated with beads coated with either recombinant SVPs or BSA. Non-attached beads were removed and the cells were fixed and visualized by DIC light microscopy at 66× magnification. Attachment of beads was quantified for each representative microscopic field and the average attachment for a given number of beads per cell was determined. (B) The percentage of cells that attached beads out of the overall cell population (total) and the percentage of cells out of the total population that attached more than four beads per cell (high) for either BSA– or SVP–beads are presented together with their respective standard deviation bars. (C and D) As in (A) and (B), but cells were treated with 2% DMSO.
Bead attachment and endocytosis
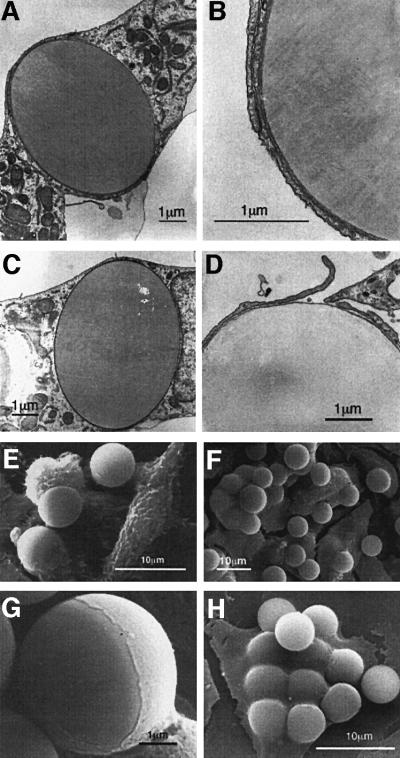
The effect of bead attachment on the cell surface was examined further by transmission electron microscopy (TEM, Figure 4A–D). Cells that bound SVP–beads show extensive membrane protrusions around the attached beads, that upon long incubation are internalized efficiently into DMSO-treated HepG2 cells (Figure 4C). This was hardly observed at all when either HepG2 cells not treated with DMSO or BSA-conjugated beads were used (not shown). Scanning electron microscopy (SEM) revealed a tight association of the beads with the cell surface (Figure 4E–H). In agreement with the TEM results, SVP-conjugated beads were partially (Figure 4G) or fully (Figure 4H) engulfed by the cells. The bead internalization, which is very probably the outcome of endocytosis, is rather an efficient process, and up to eight beads were found inside any given cell (Figure 4H). Interestingly, 21–47 peptide-conjugated beads (see below) were internalized efficiently whereas, under the same conditions, concanavalin A-conjugated beads, although able to attach the cells efficiently, did not internalize (data not shown). Together, these results demonstrate that the attachment of SVP–beads to HepG2 cells is followed by endocytosis.
Fig. 4. Attachment of SVP-conjugated beads is accompanied by endocytosis. (A–D) Late stages (A, B and D) and completion of SVP–bead engulfment (C) by the cell membrane are visualized by TEM. (B) is a higher magnification of (A). (E–H) Poor (E) and efficient (F) endocytosis of SVP–beads by untreated and DMSO-treated HepG2 cells, respectively, as visualized by SEM. Various stages of bead internalization are demonstrated (G and H).
Recombinant preS1 and cell attachment
To localize the region within HBsAg that plays a role in cell attachment, we prepared a recombinant preS1 protein and conjugated it to beads (preS1–beads). This region of the large HBsAg protein was shown previously to play an important role in receptor recognition (Neurath et al., 1986; De Meyer et al., 1997; Le Seyec et al., 1999). Untreated cells did not bind preS1–beads efficiently. However, DMSO treatment improved this step >50-fold (Figure 5A and B). Although the number of recombinant preS1 molecules per bead exceeds by far that of SVPs per bead (data not shown), the maximal attachment efficiency is ∼2-fold lower than that obtained with SVP–beads.
Fig. 5. The preS1 epitope (amino acids 21–47) mediates cell attachment. (A) Beads conjugated with either BSA, SVPs, recombinant preS1 or synthetic peptide encompassing amino acids 21–47 of preS1 were incubated with DMSO-treated cells. The percentage of cells out of the total population that attached more than four beads per cell was calculated and is presented together with their respective standard deviation bars. (B) The experiment described in (A) was repeated with untreated HepG2 cells.
The 21–47 amino acid region of preS1 blocks HBV attachment to HepG2 cells. Antibody raised against this epitope has neutralizing activity (Figure 1C; Neurath et al., 1989). We subjected a corresponding synthetic peptide to BMA analysis. Here again, DMSO dramatically improved the attachment of these beads, but only ∼18% of the cells bound four or more beads (Figure 5). This number is comparable with that obtained with the recombinant preS1–beads. These data suggest that the attachment activity of the preS1 region is likely to be confined within the 21–47 amino acid sequence and that SVPs are twice as efficient in terms of cell attachment.
Fine mapping of the preS1 major determinant that mediates attachment
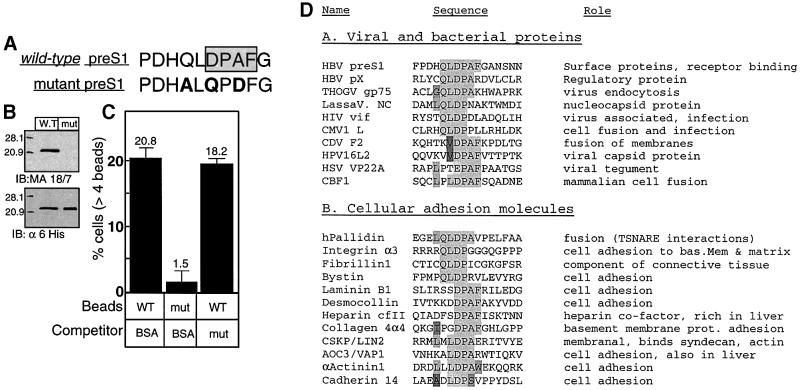
The major antigenic determinant of the HBV infection-neutralizing monoclonal antibody MA 18/7 is the preS1 DPAF sequence (Figure 2C; Sominskaya et al., 1992), suggesting that this sequence may be important for attachment (see below). We prepared a recombinant preS1 protein with a scrambled QLDPAF epitope to substantiate its role in cell attachment (Figure 6A). The wild-type and mutant are His-tagged preS1 proteins and, as expected, both are reactive with the His tag-specific antibody (Figure 6B). However, the MA 18/7 antibody did not recognize the mutant protein. These proteins were conjugated to beads and subjected to BMA assays. In contrast to the wild-type, the mutant shows poor attachment (Figure 6C). Moreover, a soluble fraction of this mutant is not capable of blocking the binding of wild-type preS1–beads to cells. Thus, the preS1 QLDPAF motif is the site that dominates HBV attachment.
Fig. 6. The QLDPAF motif in preS1 mediates preS1 attachment. (A) Sequence comparison of the wild-type QLDPAF preS1 and of the scrambled mutant protein. (B) Preferential detection of wild-type versus mutant preS1 by anti-preS1 (MA 18/7) (upper panel); there was equal detection by α6His. (C) Attachment efficiencies of beads conjugated to the indicated proteins in the presence or absence of pre-incubated mutant preS1 as a competitor. The attachment assay and the quantification of attached beads were performed as described in Figure 2. (D) Database analysis revealed that the QLDPA sequence is also found in pX, a second HBV protein. Also, a sequence similar to this motif was found in a number of viral bacterial and cellular proteins that are involved in cell adhesion, attachment and fusion.
Interestingly, we noticed that the X protein of HBV contains a sequence similar to the major preS1 attachment epitope (QLDPS/AR). Accordingly, we found that pX can be immunoprecipitated by the MA 18/7 anti-preS1 antibody (our unpublished data). This similarity raises the intriguing possibility that pX might also be involved in cell attachment. A search of the database revealed that this minimal epitope is shared by other viral, bacterial and cellular proteins that participate in cell adhesion, attachment and fusion (Figure 6D). It is tempting to suggest that the QLDPAF sequence, or part of it, is a general motif that plays a role in cell adherence and attachment.
Evidence for multivalent interaction
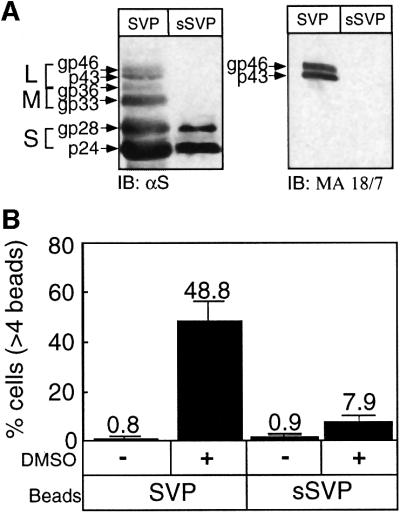
The fact that both recombinant preS1 and the synthetic 21–47 amino acid peptide show only 50% of the attachment activity of the SVP–beads strongly argues for the presence of a second region that plays a role in this process. To localize the second SVP attachment site, we prepared HBV SVPs composed of the small HBsAg and lacking the preS1 and preS2 regions (sSVP, Figure 7A). Interestingly, these beads display specific attachment in a DMSO-dependent manner (Figure 7B). However, the efficiency was ∼5 times lower than that obtained with SVP harboring the preS1 domain (7.9 versus 48.8). These results strongly argue that the small HBsAg has an additional and independent attachment site that interacts with a distinct cellular receptor.
Fig. 7. Small SVPs (sSVPs) lacking the preS1 region contain an independent attachment region. (A) Recombinant sSVPs were produced by transfection of HEK 293 cells with the pMH8 plasmid. Western analysis with polyclonal anti-HBsAg (αS) and monoclonal anti-preS1 antibodies (MA 18/7) was performed to compare the composition of the various HBsAgs in sSVPs versus the recombinant SVPs. The various HBsAgs are indicated. (B) Both SVPs and the sSVPs were conjugated to beads and their ability to mediate bead attachment in the presence or absence of DMSO was determined. See Figure 2 for the structure of the different HBsAg proteins.
Evidence for synergistic cooperation between the distinct attachment sites
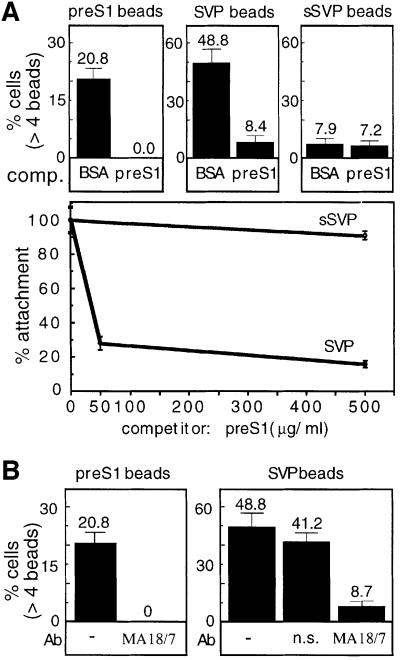
The preS1 region displays attachment efficiency of 50% as compared with SVP–beads, yet particles lacking the preS1 and preS2 regions exhibit only 8% attachment activity. This implies that either additional attachment epitopes are present that escaped our mapping or that the two regions identified by us act in a synergistic manner. To examine these possibilities, we determined the contribution of the preS1 domain in the context of SVPs. For this purpose, we performed ligand competition experiments using the soluble recombinant preS1 as a competitor. Cells were pre-incubated with increasing amounts of the preS1 protein prior to the addition of the conjugated beads. As expected, the preS1–bead attachment was completely blocked by the homologous soluble preS1 protein (Figure 8A). Interestingly, SVP–bead attachment was also competed by preS1, but ∼8% of the cells continued to show efficient SVP attachment even in the presence of a vast excess of the competitor (Figure 8A, lower panel). Under the same conditions, the attachment activity of the small SVPs remained refractory to the preS1 competitor, suggesting that this region binds a distinct receptor on the cell surface.
Fig. 8. Cooperative action of multivallent attachment sites in SVPs. (A) DMSO-treated HepG2 cells were pre-incubated for 2 h with 500 µg/ml of either BSA or recombinant preS1 as competitors prior to addition of preS1-conjugated beads. The soluble competitor proteins were pre-incubated at 100× the concentration of the conjugated ligand. The percentage of cells of the total population that attached more than four beads per cell is shown (upper panels). The experiment was carried out with an increasing amount of the competitor as indicated (lower panel). (B) The attachment of the preS1–beads was neutralized by pre-incubation of the beads for 2 h with either monoclonal anti-preS1 (MA 18/7) or the control monoclonal anti-HBcAg (ns, non-specific), as indicated.
Next we performed attachment-neutralizing experiments using the M/A 18/7 neutralizing monoclonal antibody. The SVP-conjugated beads were incubated for 2 h with the monoclonal MA 18/7 antibody prior to being subjected to the BMA assay. This antibody was effective in diminishing the attachment of preS1–beads (Figure 8B). Notably, here again the SVP–bead attachment was only partially blocked and ∼8% of the cells continued to display efficient attachment (Figure 8B, right panel). As a negative control, we used a specific anti-HBcAg monoclonal antibody, and no significant inhibition was observed. Thus, based on the competition assays and on the results obtained from the employment of the neutralizing antibody, we can conclude that the preS1 epitope is the major attachment epitope. As the small SVPs displays ∼8% attachment activity (Figure 7), which is the same level of activity that resisted competition, we suggest that in our assays, other HBsAg regions, such as preS2, play a minor role. Collectively, these results strongly argue that the distinct attachment regions of HBV surface antigen particles are likely to act in a synergistic manner.
Discussion
We report here that DMSO-treated hepatoblasoma HepG2 cells can be infected efficiently by HBV, as determined by expression of the viral proteins and by the formation of HBV RC, ss and cccDNA, the HBV replication markers. The latter provides strong evidence for active HBV replication. Naive HepG2 cells can support HBV transcription and replication of the transfected HBV DNA; therefore, lack of infection is very likely to be due to inefficient HBV attachment and entry. Our finding that DMSO sensitizes HepG2 to HBV infection raises the possibility that DMSO overcomes these restrictive steps. This hypothesis was tested here by developing a tool that quantitatively measures virus attachment. Although previous reports have indicated that DMSO treatment of the primary cultured human hepatocytes enhances HBV (Gripon et al., 1988) and DHBV infection (Pugh and Summers, 1989), the underlying mechanisms remained unexplored. DMSO allows sustained highly differentiated stages of the cultured cells (Guguen-Guillouzo et al., 1986, 1988; Isom et al., 1987; Pugh and Summers, 1989) and arrests primary hepatocyte cultures (Rumin et al., 1996) and HepG2 cells at the G1 phase (our unpublished observation). It is therefore possible that DMSO acts at multiple levels. Nevertheless, the possibility that it improves expression and presentation of differentiation-specific viral receptors, a process we generally refer to as receptor activation, remained particularly difficult to investigate. The major problems were our lack of information on the nature of the involved receptors and the difficulty in measuring virus attachment quantitatively.
In this study, we have utilized synthetic beads with conjugated viral proteins to evaluate virus attachment quantitatively. This method was used previously for the analysis of adhesion-mediated interactions (Levenberg et al., 1998, and references therein). The use of such beads has several advantages. (i) Beads are readily visible and their attachment is not followed by immediate entry into the cells and, therefore, is more reliably measurable. Image analysis systems can be developed for automated scoring of virus attachment. Thus, the viral epitopes involved and the cellular receptors interacting with it can be determined easily. (ii) Using this method, attachment is evaluated at single-cell resolution. (iii) Susceptible cells can be sorted out easily from the rest of the cells in a heterogeneous cell population. Furthermore, cells can be categorized and separated based on their attachment capacity. (iv) The high concentrations of the conjugated ligands on their surface may increase the binding avidity and permit detection of otherwise weak attachment. (v) Use of beads which can be visualized readily allows real time analysis. Thus the direct effect of viral attachment on cell behavior and morphology can be monitored. (vi) The physical properties of the beads allows the development of simple receptor purification protocols. (vii) Lastly, we show here that this method can be used to determine the effectiveness of neutralizing antibodies, sequestering peptides as well as inhibitory drugs.
Utilizing the BMA assay, specific but poor attachment of HBV SVPs to HepG2 cells was detected. Remarkably, DMSO treatment improved the attachment by >200-fold. DMSO increased both the number of attached beads per cell and the number of the cells engaged in attachment. About 5% of the cells do not attach SVPs at all, ruling out the involvement of ubiquitous factors in this process. The other 95% of cells show either low (1–4 beads per cell) or high (>4 beads per cell) attachment capacity. As HepG2 is a cell line, this rules out a genetic basis for this variation and therefore these subpopulations may represent cells with discrete concentrations of activated receptors. In that case, the level and presentation of these putative receptors must be highly regulated in order to account for the variations obtained. The mode of regulation of expression of the putative HBV receptors awaits clarification, but it might be regulated in part by the associated particles (Klingmuller and Schaller, 1993; Bruns et al., 1998). The particles may increase the concentration of anchored receptors by mediating receptor aggregation or by interfering with their proper recycling. The facts that the HBsAg SVPs regulate infection of Dane particles (Bruns et al., 1998) and that a relatively long incubation time, >10 h, is required for full productive infection lend further support to this model.
We show that activation of virus attachment correlates nicely with the rate of infection. We further show that DMSO significantly improved the cell attachment via this preS1 sequence, both as an isolated domain and as an integral component of the viral particle. Previously, it has been reported that preS1–cell interaction is essential for HBV attachment and infection (De Meyer et al., 1997; Le Seyec et al., 1999). We therefore suggest that the observed attachment of SVPs to DMSO-treated HepG2 cells is mediated via authentic HBV receptors. This conclusion is also supported by the fact that the 21–47 amino acid epitope of preS1 is involved in this process, in line with the reports of Neurath et al. (1986, 1989). So far, several cellular proteins have been isolated that bind the 21–47 amino acid epitope of preS1 (De Meyer et al., 1997). Unfortunately, no data are available as yet regarding the exact amino acid residues in preS1 that mediate these interactions and their relevance to virus–cell interactions in vivo. Studies on the interaction of the DHBV preS with carboxypeptidase D (gp180), the putative receptor, have revealed a relatively broad region in preS which mediates attachment (Urban et al., 2000). Although multiple epitopes may contribute sequentially to adhesion strengthening, our study shows that a rather small epitope plays the major role in attachment.
TEM and SEM studies reveal that improved attachment of SVPs to DMSO-treated cells is accompanied by bead internalization. Endocytosis was suggested previously as the mechanism for HBV entry into cells (Hagelstein et al., 1997). Although the preS1 attachment epitope is required for the observed internalization (data not shown), utilizing the same experimental approach it will be interesting to determine the importance of additional regions in this process.
The BMA assay was also used for fine mapping of the sequence within preS1 that mediates cell attachment. A monoclonal antibody raised against the 21–47 peptide could elicit the neutralizing effect (Neurath et al., 1989) and block attachment (this study). Fine epitope mapping of this antibody has indicated that the DPAF sequence within this region is the major antigenic determinant (Sominskaya et al., 1992). We therefore expected this motif to play a role in HBV attachment. Accordingly, we found that a recombinant preS1 mutated at this region is inefficient in attachment. Analysis of the database revealed that this minimal epitope is shared by other viral, bacterial and cellular proteins that participate in cell adhesion, attachment and fusion. This suggests that the QLDPAF sequence or part of it may play a role in cell adherence and attachment. Variations in this epitope and the adjacent sequences may confer a higher order of specification such as tissue tropism and species specificity, as exemplified in HBV. To test this hypothesis, more detailed mutagenesis studies are required.
Interestingly, the X protein of HBV contains a similar sequence to the major preS1 attachment epitope (QLDPS/AR) at position 8–13. In agreement with this possibility, we found that pX can be immunoprecipitated by the MA 18/7 anti-preS1 antibody with low efficiency (our unpublished data). This region of pX is highly conserved, yet mutational studies so far have assigned no clear function to this region. The sequence similarity to the preS1 attachment domain raises the intriguing possibility that pX might be involved in cell attachment. We have identified two similar EGF repeat-containing proteins that bind the recombinant preS1. Furthermore, the expression of one of them is induced dramatically in DMSO-treated HepG2 cells (our unpublished data). Interestingly, one of these proteins was reported to bind pX (Sun et al., 1998), lending additional support to the hypothesis that pX and preS1 might share some functions.
We report here that the virus contains a secondary attachment region at the small HBsAg. This conclusion is based mainly on the fact that the HBsAg–beads lacking the preS1 and preS2 domains attached cells in a specific manner. This is in agreement with the reported cooperation in attachment between the preS1 and the non-preS1 components (Neurath et al., 1986; Gripon et al., 1988; Pugh and Summers, 1989; Petit et al., 1991; Page et al., 1992; Ishikawa et al., 1994; Qiao et al., 1994; Breiner et al., 1998; Urban et al., 1998). This cooperation may also have post-attachment roles affecting virus entry (Pugh et al., 1995). Furthermore, the fact that this attachment is not competed out by recombinant preS1 argues strongly that a distinct receptor on the cell surface specifically recognized the secondary HBsAg attachment site. The nature of such a receptor was not resolved but a few candidates were reported (reviewed in De Meyer et al., 1997). In any case, our data show that DMSO also activates this putative receptor. Thus, HBV displays multivalent binding to multiple receptors. Multivalent binding is expected to increase the binding avidity and, for many viruses, a secondary binding step has been shown to follow the initial binding (for a review see Haywood, 1994).
Quantitative evaluation of the attachment activity of the particles made of small HBsAg revealed that they retain only 15–20% activity as compared with particles containing the preS1 and preS2 regions. Yet beads conjugated to the recombinant preS1 protein display only 50% and not the remainding 80–85% activity. The difference might be the result of the involvement of an additional region, such as preS2. As preS2 has to be glycosylated (Heermann et al., 1984), we faced difficulties in directly evaluating its contribution to cell attachment. However, attachment competition and neutralization experiments both strongly argue that under the employed conditions, the preS2 contribution to cell attachment is minor if any. Thus, we are left with the possibility that the two attachment regions on the HBV surface proteins act synergistically rather than additively. Consequently, abolishing one of these affects the attachment rate beyond its intrinsic capacity. Based on these considerations, we propose a model whereby HBsAg brings the particles into close contact with the cell membrane to facilitate specific interaction of the preS1 domain with its receptor. The number of small HBsAgs on the HBV envelope is much higher than that of preS1 domains and they are therefore expected to form multiple contacts with the cell membrane and to increase the attachment rate. This binding, however, is probably unstable, but can be stabilized if followed by the interaction of preS1 epitope with its corresponding receptor.
Materials and methods
Cell culture
HepG2 and HEK 293 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Gibco) containing penicillin (100 U/ml) and streptomycin (100 µg/ml), supplemented with 8% fetal calf serum (Gibco). Transfections were carried out by the CaPi method as previously described (Haviv et al., 1995). Cells were seeded 8–12 h prior to transfection. About 60% confluent plates were transfected with the desired plasmids. Carrier DNA plasmid was added to reach the final amount of 20 µg of total DNA per 10 cm diameter plate. For DMSO treatment, cells were seeded at 60% confluence, on 18 mm coverslips in a 12-well plate (Costar), and after 8–12 h DMSO was added to reach 2% (v/v) final concentration.
Plasmid constructions
For bacterial expression of preS1, the DNA containing the preS1 domain (amino acids 6–113) was cloned into the pRSET B vector (Invitrogen) at the XhoI–HindIII sites. Site-directed mutagenesis was performed by PCR using PWO Taq DNA polymerase (Boehringer Mannheim) and the following primers: 5′-CCCGATCATGCATTGCAGCCTGACTTCGG AGCC-3′ (sense) and 5′-GGCTCCGAAGTCAGGCTGCAATGCATG ATCGGG-3′ (antisense) (adw strain), and two vector primers 5′-CGC GGATCCTTGGCCGATTCATTAATGCAG-3′ and 5′-TCCCCGCGG GCTAACCAGATCCGGATATAGTTCCT-3′. The constructed mutants were analyzed by DNA sequencing.
Production and purification of HBV SVP surface proteins
For production of SVPs, CHO cells were stably transfected with the AL26 plasmid, and high HBsAg-producing cells were isolated. The culture medium was collected, and centrifuged (Rotor SS34 17 000 r.p.m., 30 min at 4°C) to remove cell debris, and SVPs were fractionated on CsCl gradients. The HBsAg level in the resulting fractions was determined by radioimmunoassay using 125I-labeled polyclonal αHBsAg antibody. Fractions enriched for HBsAgs were collected and SVPs were enriched by centrifugation through a 30% (w/v) sucrose cushion in phosphate-buffered saline (PBS) (16 h, 27 000 r.p.m., 4°C, SW28 rotor). Mass production and purification of SVPs was performed by Bio-Technology General (BTG; Israel). SVPs containing only the small HBsAg (sSVP) were produced by transfection of HEK 293 cells with pMH8 plasmid. Six days post-transfection, the culture medium was collected and centrifuged (17 000 r.p.m., 30 min at 4°C), to remove cell debris. The supernatant was then layered on top of a 30% (w/v) sucrose cushion (in PBS) and was subjected to ultracentrifugation (16 h, 27 000 r.p.m., 4°C, SW28 rotor). The pellet was resuspended in PBS, and analyzed for sSVP by western immunobloting using polyclonal anti-SVP antibodies.
Recombinant preS1 (wild-type or mutant) was produced using the pRSETB preS1 vector in BL-21::pLysS bacteria as described previously (Haviv et al., 1996). Protein expression was induced at OD600 0.8 by 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 30 min at 37°C. Cells were collected, washed and sonicated in 30 ml per 500 ml of original culture volume of lysis buffer [25mM HEPES–KOH pH 7.9, 5 mM MgCl2, 200 mM NaCl, 10 mM β-mercaptoethanol, 10% glycerol, 200 mM phenylmethylsulfonyl fluoride (PMSF), 100 mM benzamide and 10 mM benzamidine]. Soluble proteins were separated from the debris and inclusion bodies by centrifugation with SS-34 (16 000 r.p.m., 30 min), pre-incubated with 10 mM imidazole and loaded on a 1 ml Ni-NTA–agarose column (Qiagen) pre-equilibrated with the equivalent buffer containing 10 mM imidazole. Non-specific proteins were washed out by 150 column volumes of lysis buffer containing 10 mM imidazole. The preS1 proteins were step-eluted in lysis buffer containing 150 mM imidazole, and dialyzed against PBS. The identity and homogeneity of the purified proteins were determined by Coomassie staining and western immunoblotting.
Preparation of conjugated beads
Polybead amino microspheres (mean diameter 6 µm, Polysciences) were conjugated to the desired proteins according to the manufacturer’s protocols. Briefly, 108 beads were washed three times with PBS at pH 7.4, incubated with 8% glutaraldehyde in PBS for 16 h at room temperature with gentle mixing, washed with PBS and incubated with the desired protein for 5 h at room temperature. The beads were then incubated with 0.5 M ethanolamine in PBS for an additional 30 min, followed by incubation with 10 mg/ml BSA in PBS for 30 min. The beads were then resuspended in a storage buffer containing 10 mg/ml BSA, 0.1% NaN3 and 5% glycerol in PBS, pH 7.4, and stored at 4°C until used.
Antibodies and peptides
Polyclonal goat antibodies against SVPs (αS) were obtained from BTG (Israel), and were diluted 1:3000 (in PBS) for both antibody-mediated blocking assays and western analysis (PBS, 0.1% Tween 20 + 4% dried low fat milk). The monoclonal mouse anti-preS1 antibody (MA 18/7) (IgG 0.5 mg/ml) was diluted 1:5000 in PBS for antibody-mediated blocking assays. The monoclonal mouse anti-His6 (Sigma) antibody was diluted 1:3000 for western analysis. Monoclonal mouse anti-HBcAg antibodies were generated by injection of bacterially expressed FPLC-purified HBV core particles and selection of hybridoma-secreting αHBcAg antibodies. Polyclonal rabbit antibodies against HBV core protein were generated by repeated injection of bacterially expressed FPLC-purified HBV core particles. For antibody-mediated blocking assays, the ascites fluid was diluted 1:5000 in PBS. For western analysis, horseradish peroxidase-conjugated donkey anti-goat and goat anti-mouse antibodies (Jackson Immunoresearch Laboratories) were used.
A synthetic peptide encompassing the 21–47 amino acids of preS1 (PLGFFPDHQLDPAFGANSNNPDWDFNPGK) was synthesized, purified on a C18 column by HPLC, lyophilized and freshly resuspended in PBS before use. The peptide composition was confirmed by amino acid analysis.
Bead-mediated attachment (BMA) assays
Cells were seeded and, 2 h before addition of the conjugated beads, competitor antigens were added as described in the Figure legends. Subsequently 2 × 106 beads were added to the cells and incubated for an additional 14 h. The unbound beads were removed and cells were fixed in 4% paraformaldehyde solution and mounted in Aqua-polymount (Polysciences). Attached beads were visualized, documented and quantified by DIC light microscopy (Zeiss Axiophot). The average number of beads per cell was evaluated by monitoring ∼300 cells from 7–10 representative fields. Attachment efficiency was quantified for each representative microscopic field and the average attachment efficiency of a certain number of beads per cell was determined. The results were grouped into the following categories: cells that remained unoccupied (0 beads per cell), cells that attached 1–4 beads per cell and cells that attached more than four beads per cell. For each group, the average is presented together with the standard deviation.
HBV infection and DNA analysis
HBV-positive sera were used to infect cells as previously described (Tuttleman et al., 1986; Gripon et al., 1988; Pugh and Summers, 1989). Briefly, HepG2 cells were cultured on glass coverslips (18 mm in diameter, No.1) in a 12-well plate (Costar), and treated with 2% DMSO for 6–10 days or left untreated. Cells were incubated with HBV-positive sera (109 particles/ml) for 14 h at 37°C, washed extensively with PBS and fresh medium was added with or without DMSO. For PCR analysis, at 4 days post-infection cells were washed extensively in PBS, collected using a rubber policeman and lysed in a proteinase K lysis buffer for 6 h at 37°C, extracted with phenol–chloroform and precipitated with ethanol. Detection of cccDNA and RC DNA was performed as described (Ilan et al., 1999). The amount of DNA was determined by conducting semi-quantitative PCR analysis of a cellular gene (AML-2). Equal amounts of DNA were used for HBV RC and cccDNA analysis. A similar analysis was performed with the secreted virions.
For analysis of HBV replicative DNA forms, infected cells were harvested at various time intervals as indicated, and both total and extrachromosomal DNA were extracted and analyzed by agarose gel electrophoresis and Southern blotting (Tuttleman et al., 1986). HBV sequences were detected by employing an intact HBV DNA genome that was used as a template for production of a 32P-labeled DNA probe by random priming.
For detection of viral proteins, 4 days after infection cells were washed four times in cold PBS, fixed as described above and incubated with polyclonal αS and αCore antibodies. For immunostaining, fluorescein isothiocyanate (FITC)-conjugated anti-goat antibodies and rhodamine red X (RRX)-conjugated anti-rabbit antibodies (Jackson Immunoresearch Laboratories) were used. Slides were mounted in Aqua-polymount (Polysciences). Microscopic images were obtained using a Bio-Rad MRC-1024 confocal system, utilizing an argon–krypton mixed gas laser, and mounted on a Zeiss Axiovert microscope.
Scanning electron microscopy
Cells were cultured on glass coverslips (13 mm in diameter, No.1, Marienfeld) in a 12-well plate (Costar) for at least 4 days with or without DMSO before incubation with beads for 16 h. Cells were washed extensively with DMEM pre-warmed to 37°C. Cells were fixed for 1 h with Karnovsky’s fixative (3% paraformaldehyde, 2% glutaraldehyde, 5 mM CaCl2 in 0.1 M cacodylate buffer, pH 7.4 containing 0.1 M sucrose). Cells were rinsed and post-fixed for 1 h with 1% osmium tetroxide in 0.1 M cacodylate buffer followed by incubation with 1% tannic acid in water and then with 1% uranyl acetate in water. All the buffers and DMEM were freshly prepared and were filtered using a 0.2 µm filter before use. The slides were rinsed, dehydrated with a graded ethanol series, critically point dried (Pelco CPD2) and sputter coated with gold (S150 Edwards, USA). The specimens were examined at an accelerating voltage of 20–25 kV using a JEOL GMC 6400 scanning electron microscope.
Transmission electron microscopy
For TEM, cells were seeded in 35 mm Falcon dishes, for 4 days with or without DMSO, before adding the beads for an additional 16 h. Cells were fixed in Karnovsky’s fixative and post-fixed with 1% osmium tetroxide, 0.5% potassium dichromate and 0.5% potassium hexacyanoferrate in 0.1 M cacodylate buffer. Cells were stained en bloc with 2% aqueous uranyl acetate, followed by ethanol dehydration. The dishes were embedded in Epon 812 (Tuosimis, MD). Sections were cut using a diamond knife (Diatome, Biel) and examined using a Philips CM-12 transmission electron microscope at an accelerating voltage of 100 kV. Negative staining of SVP was performed as described (Laub et al., 1983).
Acknowledgments
Acknowledgements
We gratefully acknowledge I.Sabanay and O.Yeger for their invaluable assistance in the electron microscopy work, S.Budilovsky for excellent assistance, and A.Cooper for the bacterially expressed HBV core particles and polyclonal anti-core antibodies. We thank Drs W.H.Gerlich for the anti-preS1 antibodies, N.Moav for the HBsAg SVPs, H.Giladi for pMH8 plasmid, and R.Zemel for pRSET B preS1 and for synthesis and purification of the preS1 peptide. N.P. is supported by the Dr Morton and Toby Mower graduate student research fund. B.G. holds the E.Neter chair in Cell and Tumor Biology. Y.S. holds the Oscar and Emma Getz Professorial chair.
Note added in proof
We found that combined treatments of the HepG2 cells with 2% DMSO + 100 mM 5-aza-2′-deoxycytidine further improved HBV attachment and infection.
References
- Breiner K.M., Urban,S. and Schaller,H. (1998) Carboxypeptidase D (gp180), a Golgi-resident protein, functions in the attachment and entry of avian hepatitis B viruses. J. Virol., 72, 8098–8104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruns M., Miska,S., Chassot,S. and Will,H. (1998) Enhancement of hepatitis B virus infection by noninfectious subviral particles. J. Virol., 72, 1462–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Meyer S., Gong,Z.J., Suwandhi,W., van Pelt,J., Soumillion,A. and Yap,S.H. (1997) Organ and species specificity of hepatitis B virus (HBV) infection: a review of literature with a special reference to preferential attachment of HBV to human hepatocytes. J. Viral Hepat., 4, 145–153. [DOI] [PubMed] [Google Scholar]
- Faktor O., De-Medina,T. and Shaul,Y. (1988) Regulation of hepatitis B virus S gene promoter in transfected cell lines. Virology, 162, 362–368. [DOI] [PubMed] [Google Scholar]
- Gripon P., Diot,C., Corlu,A. and Guguen-Guillouzo,C. (1989) Regulation by dimethylsulfoxide, insulin and corticosteroids of hepatitis B virus replication in a transfected human hepatoma cell line. J. Med. Virol., 28, 193–199. [DOI] [PubMed] [Google Scholar]
- Guguen-Guillouzo C., Bourel,M. and Guillouzo,A. (1986) Human hepatocyte cultures. Prog. Liver Dis., 8, 33–50. [PubMed] [Google Scholar]
- Guguen-Guillouzo C., Gripon,P., Vandenberghe,Y., Lamballe,F., Ratanasavanh,D. and Guillouzo,A. (1988) Hepatotoxicity and molecular aspects of hepatocyte function in primary culture. Xenobiotica, 18, 773–783. [DOI] [PubMed] [Google Scholar]
- Guo J.T. and Pugh,J.C. (1997) Monoclonal antibodies to a 55-kilodalton protein present in duck liver inhibit infection of primary duck hepatocytes with duck hepatitis B virus. J. Virol., 71, 4829–4831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagelstein J., Fathinejad,F., Stremmel,W. and Galle,P.R. (1997) pH-independent uptake of hepatitis B virus in primary human hepatocytes. Virology, 229, 292–294. [DOI] [PubMed] [Google Scholar]
- Haviv I., Vaizel,D. and Shaul,Y. (1995) The X protein of hepatitis B virus coactivates potent activation domains. Mol. Cell. Biol., 15, 1079–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haviv I., Vaizel,D. and Shaul,Y. (1996) pX, the HBV-encoded coactivator, interacts with components of the transcription machinery and stimulates transcription in a TAF-independent manner. EMBO J., 15, 3413–3420. [PMC free article] [PubMed] [Google Scholar]
- Haywood A.M. (1994) Virus receptors: binding, adhesion strengthening and changes in viral structure. J. Virol., 68, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heermann K.H., Goldmann,U., Schwartz,W., Seyffarth,T., Baumgarten, H. and Gerlich,W.H. (1984) Large surface proteins of hepatitis B virus containing the pre-s sequence. J. Virol., 52, 396–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilan E. et al. (1999) The hepatitis B virus-trimera mouse: a model for human HBV infection and evaluation of anti-HBV therapeutic agents. Hepatology, 29, 553–562. [DOI] [PubMed] [Google Scholar]
- Ishikawa T., Kuroki,K., Lenhoff,R., Summers,J. and Ganem,D. (1994) Analysis of the binding of a host cell surface glycoprotein to the preS protein of duck hepatitis B virus. Virology, 202, 1061–1064. [DOI] [PubMed] [Google Scholar]
- Isom I., Georgoff,I., Salditt-Georgieff,M. and Darnell,J.E.,Jr (1987) Persistence of liver-specific messenger RNA in cultured hepatocytes: different regulatory events for different genes. J. Cell Biol., 105, 2877–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingmuller U. and Schaller,H. (1993) Hepadnavirus infection requires interaction between the viral pre-S domain and a specific hepatocellular receptor. J. Virol., 67, 7414–7422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroki K., Eng,F., Ishikawa,T., Turck,C., Harada,F. and Ganem,D. (1995) gp180, a host cell glycoprotein that binds duck hepatitis B virus particles, is encoded by a member of the carboxypeptidase gene family. J. Biol. Chem., 270, 15022–15028. [DOI] [PubMed] [Google Scholar]
- Laub O., Rall,L.B., Truett,M., Shaul,Y., Standring,D.N., Valenzuela,P. and Rutter,W.J. (1983) Synthesis of hepatitis B surface antigen in mammalian cells: expression of the entire gene and the coding region. J. Virol., 48, 271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Seyec J., Chouteau,P., Cannie,I., Guguen-Guillouzo,C. and Gripon,P. (1999) Infection process of the hepatitis B virus depends on the presence of a defined sequence in the pre-S1 domain. J. Virol., 73, 2052–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenberg S., Katz,B.Z., Yamada,K.M. and Geiger,B. (1998) Long-range and selective autoregulation of cell–cell or cell–matrix adhesions by cadherin or integrin ligands. J. Cell Sci., 111, 347–357. [DOI] [PubMed] [Google Scholar]
- Li J.S., Tong,S.P. and Wands,J.R. (1996) Characterization of a 120-kilodalton pre-S-binding protein as a candidate duck hepatitis B virus receptor. J. Virol., 70, 6029–6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neurath A.R., Kent,S.B., Strick,N. and Parker,K. (1986) Identification and chemical synthesis of a host cell receptor binding site on hepatitis B virus. Cell, 46, 429–436. [DOI] [PubMed] [Google Scholar]
- Neurath A.R., Seto,B. and Strick,N. (1989) Antibodies to synthetic peptides from the preS1 region of the hepatitis B virus (HBV) envelope (env) protein are virus-neutralizing and protective. Vaccine, 7, 234–236. [DOI] [PubMed] [Google Scholar]
- Page K.A., Stearns,S.M. and Littman,D.R. (1992) Analysis of mutations in the V3 domain of gp160 that affect fusion and infectivity. J. Virol., 66, 524–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit M.A., Strick,N., Dubanchet,S., Capel,F. and Neurath,A.R. (1991) Inhibitory activity of monoclonal antibody F35.25 on the interaction between hepatocytes (HepG2 cells) and preS1-specific ligands. Mol. Immunol., 28, 517–521. [DOI] [PubMed] [Google Scholar]
- Pontisso P., Ruvoletto,M.G., Gerlich,W.H., Heermann,K.H., Bardini,R. and Alberti,A. (1989) Identification of an attachment site for human liver plasma membranes on hepatitis B virus particles. Virology, 173, 522–530. [DOI] [PubMed] [Google Scholar]
- Pugh J.C. and Summers,J.W. (1989) Infection and uptake of duck hepatitis B virus by duck hepatocytes maintained in the presence of dimethyl sulfoxide. Virology, 172, 564–572. [DOI] [PubMed] [Google Scholar]
- Pugh J.C., Di,Q., Mason,W.S. and Simmons,H. (1995) Susceptibility to duck hepatitis B virus infection is associated with the presence of cell surface receptor sites that efficiently bind viral particles. J. Virol., 69, 4814–4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao M., Macnaughton,T.B. and Gowans,E.J. (1994) Adsorption and penetration of hepatitis B virus in a nonpermissive cell line. Virology, 201, 356–363. [DOI] [PubMed] [Google Scholar]
- Rumin S., Gripon,P., Le Seyec,J., Corral-Debrinski,M. and Guguen-Guillouzo,C. (1996) Long-term productive episomal hepatitis B virus replication in primary cultures of adult human hepatocytes infected in vitro. J. Viral Hepat., 3, 227–238. [DOI] [PubMed] [Google Scholar]
- Ryu C.J., Cho,D.Y., Gripon,P., Kim,H.S., Guguen-Guillouzo,C. and Hong,H.J. (2000) An 80-kilodalton protein that binds to the pre-S1 domain of hepatitis B virus. J. Virol., 74, 110–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sominskaya I., Pushko,P., Dreilina,D., Kozlovskaya,T. and Pumpen,P. (1992) Determination of the minimal length of preS1 epitope recognized by a monoclonal antibody which inhibits attachment of hepatitis B virus to hepatocytes. Med. Microbiol. Immunol., 181, 215–226. [DOI] [PubMed] [Google Scholar]
- Sun B.S., Zhu,X., Clayton,M.M., Pan,J. and Feitelson,M.A. (1998) Identification of a protein isolated from senescent human cells that binds to hepatitis B virus X antigen. Hepatology, 27, 228–239. [DOI] [PubMed] [Google Scholar]
- Tuttleman J.S., Pugh,J.C. and Summers,J.W. (1986) In vitro experimental infection of primary duck hepatocyte cultures with duck hepatitis B virus. J. Virol., 58, 17–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban S., Breiner,K.M., Fehler,F., Klingmuller,U. and Schaller,H. (1998) Avian hepatitis B virus infection is initiated by the interaction of a distinct pre-S subdomain with the cellular receptor gp180. J. Virol., 72, 8089–8097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban S., Schwarz,C., Marx,U.C., Zentgraf,H., Schaller,H. and Multhaup,G. (2000) Receptor recognition by a hepatitis B virus reveals a novel mode of high affinity virus–receptor interaction. EMBO J., 19, 1217–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]