Abstract
Previous UV cross-linking studies demonstrated that, upon integration of the U2 snRNP into the spliceosome, a 14 kDa protein (p14) interacts directly with the branch adenosine, the nucleophile for the first transesterification step of splicing. We have identified the cDNA encoding this protein by microsequencing a 14 kDa protein isolated from U2-type spliceosomes. This protein contains an RNA recognition motif and is highly conserved across species. Antibodies raised against this cDNA-encoded protein precipitated the 14 kDa protein cross-linked to the branch adenosine, confirming the identity of the p14 cDNA. A combination of immunoblotting, protein microsequencing and immunoprecipitation revealed that p14 is a component of both 17S U2 and 18S U11/U12 snRNPs, suggesting that it contributes to the interaction of these snRNPs with the branch sites of U2- and U12-type pre-mRNAs, respectively. p14 was also shown to be a subunit of the heteromeric splicing factor SF3b and to interact directly with SF3b155. Immuno precipitations indicated that p14 is present in U12-type spliceosomes, consistent with the idea that branch point selection is similar in the major and minor spliceosomes.
Keywords: branch site/pre-mRNA splicing/RRM/U2 snRNP/U11/U12 snRNP
Introduction
Recognition and selection of the pre-mRNA branch site are critical events in splicing. Pre-mRNAs are spliced in two sequential transesterification reactions by the spliceosome, a multicomponent ribonucleoprotein complex (Burge et al., 1999). In the first step, the 2′-OH of the branch site adenosine carries out a nucleophilic attack at the 5′ splice site phosphate, generating a free 5′ exon and the intron–3′ exon in the form of a lariat. In the subsequent step, the 3′-OH of the 5′ exon attacks the 3′ splice site leading to intron excision and ligation of the 5′ and 3′ exons. The branch site adenosine is thus involved chemically in the first step and also appears to contribute in a currently ill-defined way to the second step (Query et al., 1996, and references therein). As it serves as the nucleophile for the first transesterification, the branch site adenosine, together with any factors that intimately associate with it, are expected to form part of the spliceosome’s catalytic core.
Two different spliceosomes can be distinguished currently. The major or U2-dependent spliceosome catalyzes removal of U2-type introns and is formed by the ordered interaction of U1, U2, U5 and U4/U6 snRNPs, and numerous splicing factors with the pre-mRNA (Burge et al., 1999). U2-dependent spliceosome assembly involves the initial interaction of U1 snRNP with the 5′ splice site, followed by U2 snRNP association with the branch site to form the pre-spliceosome (complex A). Subsequently, U4/U6 and U5 interact as a pre-formed (U4/U6⋅U5) complex to form mature spliceosomes. The minor or U12-dependent spliceosome catalyzes splicing of relatively rare U12-type introns found in only a subset of metazoans (Burge et al., 1998). U12-dependent spliceosome assembly begins with the interaction of the 18S U11/U12 di-snRNP (Frilander and Steitz, 1999). U11 associates with the 5′ splice site and U12 with the branch site, and thus they are functional analogs of the U1 and U2 snRNPs, respectively (Hall and Padgett, 1996; Tarn and Steitz, 1996b; Kolossova and Padgett, 1997). Assembly is completed by the association of the U4atac/U6atac snRNP, a functional analog of U4/U6, and the U5 snRNP, which is present in both spliceosomes (Tarn and Steitz, 1996a,b).
The spliceosomal snRNPs are composed of snRNA complexed with seven so-called Sm proteins, which are present in each snRNP, and numerous snRNP-specific proteins (Will and Lührmann, 1997). The human 17S U2 snRNP contains, in addition to the Sm proteins, at least 11 U2-specific proteins (Behrens et al., 1993). Most, but not all, of the latter proteins have been characterized at the molecular level. These include U2-A′ and U2-B′′, which associate stably with U2 snRNA, and seven less stably associated proteins that constitute the essential splicing factors SF3a and SF3b. SF3a consists of subunits of 60, 66 and 120 kDa (also designated spliceosomal-associated proteins SAP61, SAP62 and SAP114, respectively; reviewed in Krämer, 1996), whereas SF3b contains known subunits of 49, 130, 145 and 155 kDa (Das et al., 1999; Krämer et al., 1999, and references therein). These four SF3b subunits are also present in the human U11/U12 snRNP and thus are conserved between the major and minor spliceosomes (Will et al., 1999). Orthologs of all characterized human 17S U2 proteins have also been identified in the yeast Saccharomyces cerevisiae, underscoring the evolutionary conservation of U2-associated proteins and the central role that many of them play in U2 function (Pauling et al., 2000, and references therein).
Branch site recognition is a key step in the early stages of spliceosome assembly. In S.cerevisiae, which contains only U2-type introns, the sequence UACUAAC (where A is the site of branch formation) is highly conserved. In mammals, the U2-type branch site (YNYURAC) is less well conserved, while that found in U12-type introns (UCCUUAAC) is similar to yeast in terms of its high degree of conservation (Burge et al., 1999). In both higher and lower eukaryotes, the U2-type branch site is recognized initially by the branch point-binding protein (BBP in yeast or mBBP/SF1 in mammals) (Abovich and Rosbash, 1997; Berglund et al., 1997). At the time of pre-spliceosome assembly, U2 snRNP interacts with the branch site, apparently displacing BBP. This interaction involves base pairing between U2 snRNA and the branch site, and the first step nucleophile is selected in part by being bulged from this duplex (Query et al., 1994, and references therein). A stable U2 snRNA–branch site interaction is dependent upon a number of proteins that recognize the pre-mRNA in this vicinity, including U2AF (Mud2p in yeast), which binds the polypyrimidine tract located 3′ of the branch site, and subunits of SF3a and SF3b (Brosi et al., 1993; Krämer et al., 1999). All SF3 subunits, except SF3b130, can be cross-linked to a 20 nucleotide region just 5′ of the branch site in pre-spliceosomes and spliceosomes (Gozani et al., 1996). In addition, SF3b155 cross-links both 5′ and 3′ of the branch site, indicating that it spans the branch site. SF3b155 also interacts directly with U2AF, which may also facilitate U2 snRNP recruitment to the branch site (Gozani et al., 1998)
Less is known about the mechanism of branch site recognition in U12-type introns. Analogously to U2 snRNA, U12 snRNA base-pairs with the U12-type branch site and a duplex is formed in which the adenosine that serves as the first step nucleophile is also bulged (Hall and Padgett, 1996; Tarn and Steitz, 1996b). Interactions between the U11 snRNP and the 5′ splice site are required to stabilize the U12–branch site interaction (Frilander and Steitz, 1999). As U12-type introns lack a polypyrimidine tract, U2AF does not appear to facilitate association of U12 with the branch site. Rather, SF3b subunits may also play this role in the minor spliceosome (Will et al., 1999).
To learn more about branch site recognition and about the composition of the spliceosome’s core, a number of cross-linking studies have been performed with U2-type pre-mRNAs. Using a highly reactive photoreagent placed specifically on the branch site base, we previously reported a strong cross-link to a 14 kDa protein (denoted p14) that appeared first in pre-spliceosomes (i.e. complex A) and persisted in spliceosomes; in addition, cross-links to several other proteins, which replaced each other during the assembly process, were also detected (MacMillan et al., 1994). UV cross-linking studies also revealed sequential contacts in the vicinity of the branch site (Gaur et al., 1995; Chiara et al., 1996), but it was not clear whether any of these proteins contacted the branch adenosine directly. However, in a study that focused specifically on UV cross-links to the latter nucleotide, solely a cross-link involving p14 was detected in both pre-spliceosomes and spliceosomes (Query et al., 1996). These results indicate that this protein contacts the branch site adenosine directly and that this interaction probably persists at least until the first catalytic step of splicing.
Here, we have identified and characterized the 14 kDa branch site protein. It is highly conserved evolutionarily and associates with both U2 and U11/U12 snRNPs. We demonstrate a protein–protein interaction between p14 and an SF3b component, SF3b155, which probably mediates the association of p14 with U2 and U12 snRNPs. Further, we show that p14 is a stable component of SF3b. Finally, we provide evidence that p14 is also present in U12-dependent spliceosomes, consistent with the idea that p14 may contribute to branch site positioning in both the major and minor spliceosome.
Results
Cloning of the branch site p14 protein
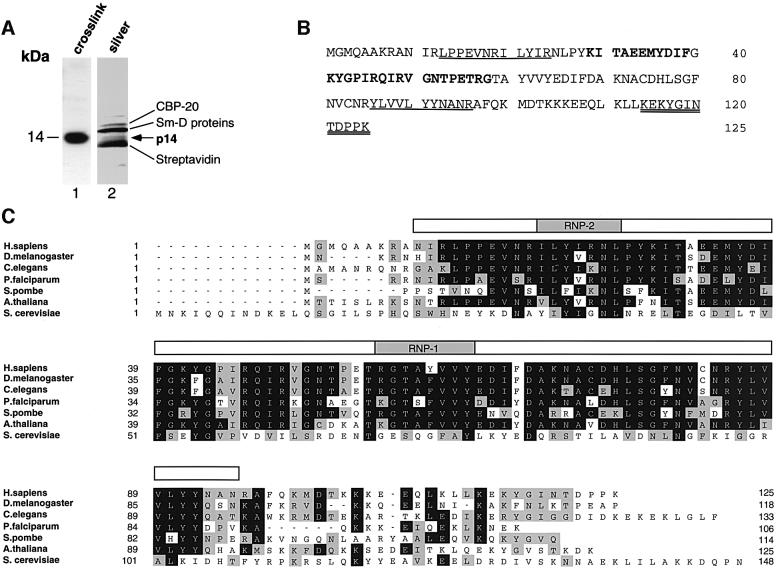
To isolate the branch site protein p14, spliceosomes were formed on a U2-type pre-mRNA containing biotin-tagged uridines and then purified by gel filtration followed by streptavidin–agarose selection. Proteins were eluted from the streptavidin–agarose under conditions that minimized release of streptavidin, which obscures proteins in the 14 kDa range, and separated by SDS–PAGE (Figure 1A). In the size range of the p14 cross-link, we could detect four bands (Figure 1A, lane 2), all of which were subjected to peptide sequencing. Three of these contained known proteins (the 20 kDa cap-binding protein, the SmD proteins and streptavidin). The fourth, which co-migrated with the cross-linked p14 band, yielded novel peptide sequences: KITAEEMYDIF and KYGPIRQIRVGNTPETRG (Figure 1B). These sequences matched the open reading frame of multiple cDNAs found in human expressed sequence tag (EST) libraries from a variety of tissues. The putative protein encoded by the longest EST is 125 amino acids long with a predicted mol. wt of 14.6 kDa. The identity of this cDNA was confirmed by several criteria. First, both peptide sequences obtained from the 14 kDa spliceosomal protein were present in the predicted amino acid sequence of the human cDNA (Figure 1B). Secondly, in vitro translation of the cDNA produced a protein that co-migrated with the original cross-linked species (not shown). Thirdly, antibodies raised against the cDNA-encoded protein specifically immunoprecipitated spliceosomes and snRNPs, and, more importantly, the original branch site-cross-linked protein (see below).
Fig. 1. p14 contains an RRM and is evolutionarily highly conserved. (A) The 14 kDa polypeptide region of purified U2-type spliceosomes. Proteins were separated on a 20% gel by SDS–PAGE and stained with silver (lane 2). For comparison, the branch site-cross-linked 14 kDa protein was separated in parallel and visualized by autoradiography (lane 1). (B) Amino acid sequence of human p14 (accession No. AF401310). Peptide sequences obtained by microsequencing of p14 from U2-dependent spliceosomes are indicated in bold, those from 17S U2 snRNPs are underlined, and those from 18S U11/U12 snRNPs are underlined twice. (C) Amino acid sequence alignment of human p14 and putative orthologs from D.melanogaster (#AC004767), C.elegans (#AF040642), P.falciparum (#AA550544/#AC004688), A.thaliana (#AB007727), S.pombe (#AL022299) and S.cerevisiae (#CAA86207.1). Residues identical in at least four sequences are boxed in black, and conserved residues (gray boxes) are grouped as follows: (D, E), (H, K, R), (A, F, I, L, M, P, V, W) and (C, G, N, Q, S, T, Y). The position of the RRM, including the highly conserved RNP-1 and RNP-2 motifs (shaded regions), is indicated above the alignment by an open bar. Sequence alignments were performed using the Clustal method and optimized by visual inspection.
p14 is highly conserved evolutionarily and contains an RRM
Database searches using the human p14 sequence identified likely orthologs in a wide variety of organisms, including Drosophila, nematodes (Caenorhabditis elegans), the malarial parasite Plasmodium falciparum, Arabidopsis thaliana, Schizosaccharomyces pombe and S.cerevisiae, which are aligned in Figure 1C. p14 is highly conserved evolutionarily, being 72 or 67% identical between humans and Drosophila or nematodes, respectively, and 49% identical between humans and S.pombe. The putative S.cerevisiae ortholog (denoted Snu17p) is less conserved, exhibiting 33% identity (45% similarity) with the human p14 protein. p14 contains one RNA recognition motif (RRM) (Figure 1C, indicated above the sequences by a bar), and a potential nuclear localization signal (residues 104–116 of the human protein). The majority of the sequence conservation lies within the RRM, with the C-terminus conserved only in general charged character. Comparing among these species, the overall domain structure is: a short N-terminal region that is not conserved; an RRM that is 67% identical between human and fission yeast; and a C-terminal charged region of variable length. Curiously, the RRM of the putative S.cerevisiae ortholog is more similar to the RRM of a predicted 37 kDa metazoan protein of unknown function (human protein AL050405); the RRM of Snu17p is 38% similar to that of human p14 and 74% similar to the RRM of AL050405. Nonetheless, a number of observations, including their similar lengths and that fact that both are associated with the U2 snRNP (see below; Gottschalk et al., 2001), support the idea that the yeast Snu17 protein is the ortholog of the human p14 protein (see Discussion).
The cDNA-encoded polypeptide is the p14 protein cross-linked to the branch site
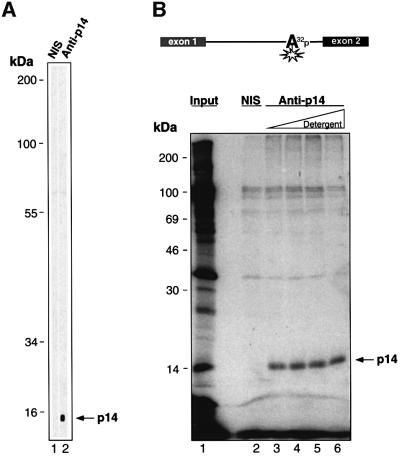
To verify that we had identified the branch site 14 kDa protein, we first raised antibodies against the cDNA-encoded protein. These anti-p14 antibodies, but not the pre-immune serum, reacted specifically with a 14 kDa protein in nuclear extract (Figure 2A). We next performed immunoprecipitations with proteins that had been cross-linked to the branch site adenosine of a U2-type pre-mRNA using the photoreactive agent benzophenone, and then analyzed the immunoprecipitated, cross-linked products by SDS–PAGE. The 14 kDa cross-linked protein was precipitated specifically by the anti-p14 antiserum (Figure 2B, lanes 3–6), but not by the pre-immune serum (lane 2). Immunoprecipitation of this protein was observed even in the presence of increasing amounts of detergents, added to ensure that all protein–protein interactions had been disrupted (lanes 3–6). Thus the cDNA identified indeed codes for the 14 kDa protein that contacts the branch site in the major spliceosome.
Fig. 2. p14 is the 14 kDa protein cross-linked to the branch adenosine. (A) Specificity of anti-p14 antibodies. Nitrocellulose strips containing proteins isolated from nuclear extract were immunostained with pre-immune serum (NIS, lane 1) or anti-p14 antibodies (lane 2). (B) Immunoprecipitation (IP) verifies that the p14 cDNA encodes the 14 kDa branch site-cross-linked species. Benzophenone was placed on the branch adenosine, and a radioactive phosphate at the adjacent nucleotide, of a U2-type pre-mRNA (depicted schematically at the top). After allowing for splicing complex formation, cross-linking and subsequent IPs were performed. Immunoprecipitated cross-linked proteins were fractionated by SDS–PAGE on a 16% gel and visualized by autoradiography. Lane 1, total cross-links from nuclear extract; lane 2, IP using pre-immune serum; lanes 3–6, IPs using anti-p14 serum and increasing amounts of detergent prior to IP (lane 3, 0.05% NP-40; lane 4, 0.5% NP-40; lane 5, 0.5% NP-40 and 0.5% deoxycholate; lane 6, 0.5% NP-40, 0.5% deoxycholate and 0.1% SDS).
p14 is associated with 17S U2 and 18S U11/U12 snRNPs
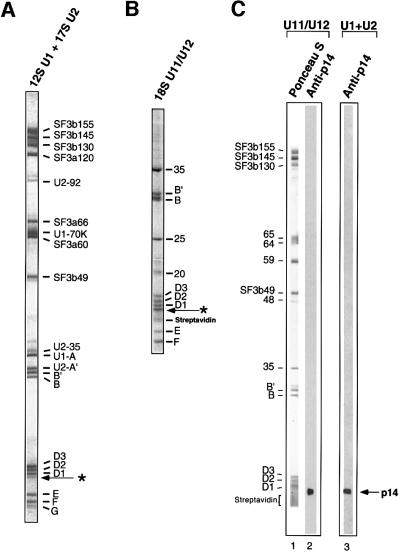
The fact that p14 is first cross-linked to the branch site upon integration of U2 snRNP into the pre-spliceosome suggested that it may be an integral U2 snRNP protein (MacMillan et al., 1994). Interestingly, snRNP preparations consisting of predominantly 17S U2 and 12S U1 snRNPs contained a polypeptide migrating in the 15 kDa range just below the SmD1 protein (Figure 3A). Significantly, this protein was recognized by a p14-specific antiserum (Figure 3C, lane 3). To determine its precise identity, peptide sequences were obtained by mass spectrometry. Three peptides (ILYIR, YLVVLYYNANR and LPPEVNR) were identified that are identical to peptide sequences found in p14 (Figure 1B). Taken together, these data indicate that p14 is associated with either 17S U2 or 12S U1 snRNPs.
Fig. 3. 17S U2-enriched snRNPs and 18S U11/U12 snRNPs contain a low molecular weight protein recognized by p14-specific antibodies. (A) The protein composition of 17S U2 and 12S U1 snRNPs, or (B) 18S U11/U12 snRNPs (only low molecular weight proteins are shown). The putative p14 band is indicated by an arrow. Proteins were separated by SDS–PAGE on 10/13% gels and stained with Coomassie Blue. (C) Proteins from 18S U11/U12 snRNPs (lanes 1 and 2) or a mixture of 17S U2 and 12S U1 snRNPs (lane 3) were stained with Ponceau S (lane 1) or anti-p14 antibodies (lanes 2 and 3). The identities of the 18S U11/U12 proteins are indicated on the left.
The recent finding that several proteins are shared by the U2 and U11/U12 snRNPs (Will et al., 1999) prompted us to investigate whether p14 was also an integral 18S U11/U12 protein. A protein migrating just below the SmD1 protein could also be detected in affinity-purified 18S U11/U12 snRNPs, when purification conditions minimizing contaminating streptavidin were used (Figure 3B). This protein was also recognized by antibodies directed against p14 (Figure 3C, lane 2). To confirm its identity, peptide sequences were obtained by microsequencing. One peptide (KYGINTDPPK) whose sequence matched that of p14 was identified (Figure 1B), demonstrating that p14 is also present in 18S U11/U12 snRNPs.
To distinguish whether p14 interacts with 17S U2 snRNP and/or 12S U1 snRNP, and to provide additional evidence that it is an integral 18S U11/U12 protein, immunoprecipitation experiments were performed with anti-p14 antibodies and nuclear extract. Immuno precipitated snRNPs were detected by 3′ end labeling the co-precipitated snRNAs with [32P]pCp (Figure 4). At low salt concentrations (150–250 mM), predominantly U2 snRNPs were precipitated by anti-p14 antibodies (Figure 4, lanes 6 and 7). In addition, under these conditions, U11 and U12 snRNPs were clearly precipitated by the anti-p14 serum, but not by the pre-immune serum (lanes 2 and 3, and 6 and 7); the low level of precipitation of these snRNPs relative to U2 is consistent with their low abundance in nuclear extract (∼1/100 the amount of U2). In contrast, the level of precipitation of U1, U4 and U5 snRNPs obtained with anti-p14 antibodies only minimally exceeded background levels observed with the pre-immune serum. At NaCl concentrations above 250 mM, the efficiency of U2, U11 and U12 precipitation was reduced (Figure 4, lanes 8 and 9), indicating that p14 associates in a salt-sensitive manner. Thus, p14 is a component of both U2 and U11/U12 snRNPs.
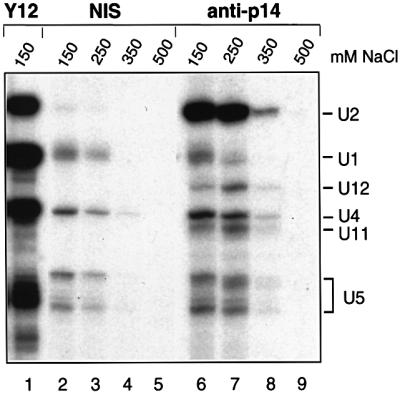
Fig. 4. Antibodies directed against p14 immunoprecipitate both U2 snRNPs and U11/U12 snRNPs. Immunoprecipitations were performed in the presence of 150–500 mM NaCl (as indicated above each lane) with HeLa nuclear extract and pre-immune serum (lanes 2–5), anti-p14 serum (lanes 6–9) or the anti-Sm protein, monoclonal Y12 (lane 1). Immunoprecipitated snRNAs were 3′ end labeled with [32P]pCp, fractionated on a 7 M urea–10% polyacrylamide gel and visualized by autoradiography. The identity of the snRNA species was confirmed by northern blotting.
p14 is a stable component of SF3b
We next asked whether p14 interacts with the snRNA and/or protein components of the U2 and U11/U12 snRNPs. Despite the presence of an RRM, we did not observe a specific direct interaction with the U2, U11 or U12 snRNAs, nor with pre-mRNA, when co-immunoprecipitation, gel mobility shift or GST pull-down experiments were performed with recombinant or in vitro translated p14 (not shown). This suggested that the association of p14 with U2 and U11/U12 snRNPs most probably was mediated by protein–protein interactions. Indeed 35S-labeled, in vitro translated p14 interacted predominantly with SF3b155 in far-western blots containing 18S U11/U12 snRNP proteins or 17S U2 snRNP proteins (not shown). In addition, when 17S U2 snRNPs were incubated under salt conditions where SF3b remains intact but dissociates from the U2 particle, and then immunoprecipitations were performed with anti-SF3b155 or anti-SF3b145 antibodies, both endogenous and exogenously added p14 were co-precipitated (not shown). These results indicated that p14 interacts directly with one or more subunits of SF3b under native conditions.
To test whether p14 is a bona fide component of SF3b, immunoaffinity chromatography was carried out with nuclear extract and affinity-purified, anti-peptide antibodies specific for SF3b155. To ensure that primarily SF3b and not 17S U2 snRNPs were affinity purified, selections were performed at 600 mM salt and the extract was first passed over an anti-m3G and then an anti-SF3a column to remove U2 snRNPs. Bound SF3b complexes were eluted with an excess of peptide and their protein composition was analyzed by SDS–PAGE. As shown in Figure 5 (lane 1), the eluate contained stoichiometric amounts of proteins that exhibited molecular weights identical to those of known SF3b subunits and also p14; the identity of these bands was confirmed by western blot analysis (not shown). A minor 120 kDa band of unknown identity, similar to that seen previously in other SF3b preparations, was also observed (Das et al., 1999; Krämer et al., 1999). Importantly, only trace amounts of U2 snRNA were detected in the eluate, indicating that primarily SF3b complexes had been affinity selected (not shown). When the eluate was subjected to glycerol gradient centrifugation, p14 sedimented together with the other SF3b subunits in an ∼10S complex (Figure 5, lanes 2–13). These results demonstrate that p14 is a stable component of the heteromeric splicing factor SF3b.
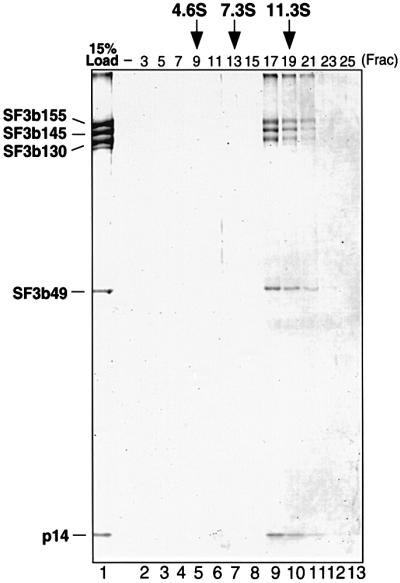
Fig. 5. p14 is a component of SF3b. SF3b was affinity purified with anti-SF3b155 antibodies and subjected to glycerol gradient centrifugation (see Materials and methods). Proteins were isolated from the affinity column eluate (lane 1, 15% of material loaded onto the gradient) or gradient fractions as indicated at the top of each lane (lanes 2–13) and analyzed as in Figure 3B. The identity of the bands, determined by western analysis, is indicated on the left. The S-values of human serum albumin (4.6), rabbit skeletal muscle aldolase (7.3) and bovine liver catalase (11.3), separated in parallel, are indicated at the top.
p14 interacts with SF3b155
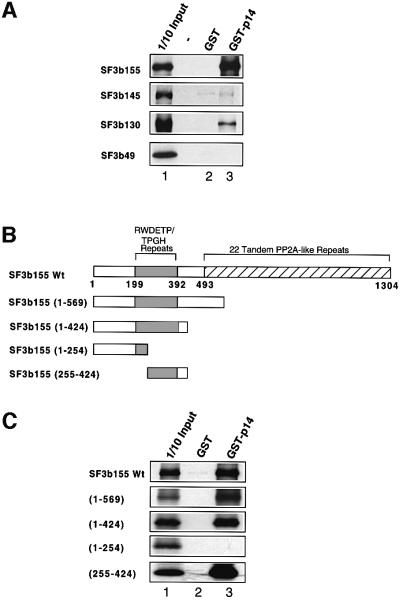
GST pull-downs were performed subsequently to identify p14’s interaction partner(s). GST or GST–p14 was incubated together with in vitro translated, 35S-labeled SF3b155, SF3b145, SF3b130 or SF3b49, precipitated with glutathione–Sepharose and co-precipitated proteins were analyzed by SDS–PAGE. Whereas no co-precipitation was observed with GST alone, significant amounts of SF3b155 and a low level of SF3b130 were co-precipitated with the GST–p14 fusion protein (Figure 6A, lanes 2 and 3). These interactions were also observed at salt concentrations up to 600 mM NaCl, demonstrating that the association of p14 with these proteins is stable (not shown). To map the region of SF3b155 that interacts with p14, several SF3b155 deletion mutants (Figure 6B) were translated in vitro and used in GST pull-down experiments (Figure 6C). Efficient co-precipitation was observed upon deletion of amino acids 425–1304, indicating that the C-terminal two-thirds of SF3b155 are dispensable for interaction with p14 [Figure 6C; mutants (1–569) and (1–424)]. However, a deletion mutant consisting of the N-terminal 234 amino acids was no longer co-precipitated, demonstrating that a region within residues 255–424 is required for p14/SF3b155 binding. To determine whether this region was not only necessary, but also sufficient for binding, we tested an SF3b155 deletion mutant consisting of residues 255–424. Significantly, this mutant was precipitated efficiently together with GST–p14 but not with GST alone (Figure 6C). Thus p14 interacts predominantly with SF3b155 and the p14 interaction region lies within amino acids 255–424.
Fig. 6. p14 interacts directly with SF3b155 in GST pull-down assays. (A) GST pull-downs were performed with 35S-labeled, in vitro translated SF3b155, 145, 130 or 49 (as indicated on the left) and GST (lane 2) or GST–p14 (lane 3). Co-precipitated proteins or 10% of input (lane 1) were analyzed by SDS–PAGE on 10/13% gels and visualized by fluorography. (B) Schematic of SF3b155 domain structure and mutants used in (C). Regions containing RWDETP and TPGH repeats (shaded box) or tandem PP2A-like repeats (striped box) (Wang et al., 1998) are indicated. (C) Delineation of the p14 interaction domain in SF3b155. Wild-type SF3b155 or various deletion mutants (indicated on the left) were incubated with GST (lane 2) or GST–p14 (lane 3), and glutathione–Sepharose-precipitated proteins or 10% of input were analyzed as in (A).
p14 is a component of both U2- and U12-dependent spliceosomes
The presence of p14 in U11/U12 snRNPs suggested that p14 is also a component of the U12-dependent spliceosome. To test this, splicing was performed in vitro with a pre-mRNA containing a U12-type intron (P120) (Figure 7B), or as a control with a U2-type intron (MINX) (Figure 7A), and immunoprecipitations were subsequently carried out at various time points with anti-p14 serum. Significant precipitation of the MINX pre-mRNA was observed with anti-p14 antibodies (Figure 7A, lanes 3 and 6), but not with the pre-immune serum (lanes 2 and 5), after 5 and 20 min of splicing. As pre-spliceosomes typically assemble on the MINX pre-mRNA within 5 min and mature spliceosomes after 10 min (not shown), these results are consistent with previous cross-linking studies demonstrating that p14 is present in both U2-dependent pre-spliceosomes and spliceosomes (MacMillan et al., 1994). Similarly, in the presence of an anti-U2 oligonucleotide that inhibits assembly of the major spliceosome, anti-p14 antibodies precipitated the P120 pre-mRNA to a greater extent than the pre-immune serum after both 30 and 240 min of incubation (Figure 7B, lanes 2 and 3, and 8 and 9). Significantly, when assembly of the minor spliceosome was blocked by addition of an oligonucleotide complementary to U12 (Tarn and Steitz, 1996b), P120 pre-mRNA precipitation by anti-p14 antibodies did not significantly exceed the background level observed with the pre-immune serum (Figure 7B, cf. lanes 5 and 6, and 11 and 12). Due to the inefficiency of splicing complex formation on the P120 pre-mRNA, pre-spliceosomes typically assemble after 20 min and spliceosomes after 60 min (Tarn and Steitz, 1996b; not shown). Although in the absence of precipitation of splicing intermediates or products we cannot conclude unequivocally that spliceosomes, as opposed to pre-spliceosomes, have been precipitated at the 240 min time point, these results nonetheless demonstrate that p14 associates with U12-type pre-mRNAs early during splicing complex formation, and further suggest that it remains associated throughout spliceosome assembly.
Fig. 7. p14 is present in both major and minor spliceosomes. (A) Anti-p14 antibodies precipitate U2-dependent spliceosomes. Splicing was performed for 5 (lanes 1–3) or 20 min (lanes 4–6) with MINX pre-mRNA and immunoprecipitations (IPs) carried out with pre-immune (NIS; lanes 2 and 5) or anti-p14 (lanes 3 and 6) serum. Twenty percent of the splicing reaction used for IP is also shown (lanes 1 and 4). (B) Anti-p14 antibodies precipitate U12-dependent spliceosomes. Splicing was performed for 30 (lanes 1–6) or 240 min (lanes 7–12) with P120 pre-mRNA in the presence of an oligonucleotide against U2 (lanes 1–3 and 7–9) or U12 (lanes 4–6 and 10–12). IPs were carried out with pre-immune (NIS; lanes 2, 5, 8 and 11) or anti-p14 (lanes 3, 6, 9 and 12) serum. Twenty percent of the splicing reaction used for IP is also shown (lanes 1, 4, 7 and 10). RNA was analyzed as described in Materials and methods and visualized by autoradiography.
Discussion
We have identified and characterized the 14 kDa protein that directly contacts the pre-mRNA branch site adenosine—the nucleophile for the first catalytic step of splicing. This protein, denoted p14, is highly conserved evolutionarily and is shared between the U2- and U12-dependent spliceosomes, by virtue of being a component of both U2 and U11/U12 snRNPs. p14 is a component of the essential splicing factor SF3b, interacting strongly with the U2 and U11/U12 protein SF3b155, and via this interaction may contribute to interactions of U2 and U12 with their respective branch sites.
p14 contains an RRM and is evolutionarily highly conserved
We identified the p14 cDNA by microsequencing a 14 kDa protein isolated from U2-type spliceosomes and confirmed its authenticity by several criteria, most importantly that anti-p14 antibodies immunoprecipitated spliceosomes, U2 or U11/U12 snRNPs, and, under denaturing conditions, the original branch site-cross-linked protein. Alignment of apparent orthologs in diverse organisms demonstrated that p14 is highly conserved evolutionarily, consistent with p14 playing a conserved role in splicing. p14 contains an RRM, and the highest degree of sequence conservation is found in this region, which accounts for nearly 70% of the human protein.
We also identified a probable ortholog in S.cerevisiae, YIR005W (also denoted Snu17). Several observations support this relationship. First, there are only four S.cerevisiae proteins smaller than 25 kDa that have RRMs (Proteome Database, www.proteome.com), and Snu17p is the only member of this group whose human ortholog has not yet been identified; thus it is the only likely candidate. Secondly, the most compelling evidence comes from recent studies demonstrating that Snu17p is a U2 snRNP protein (Gottschalk et al., 2001). Thirdly, there is no other abundant protein detected in human U2 snRNPs similar to Snu17p (C.Will and R.Lührmann, unpublished results). While a functional correspondence has not yet been demonstrated, these two proteins are homologous in sequence and are both U2 snRNP proteins, and we thus conclude that p14 and Snu17p are likely to be orthologs.
Unexpectedly, the RRM of Snu17p shares a greater degree of similarity with the RRM of a different human protein (AL050405), but several observations suggest that they are not orthologous in terms of splicing. First, similar length has a high predictive value for most orthologs, and Snu17p is much more similar in length to human p14 than to the AL050405 protein (148 amino acids versus 125 and 322, respectively). Secondly, the expression of AL050405 is consistent with a more rare function; RNA levels are ∼50- to 100-fold lower than for p14 or other U2-type spliceosomal components (not shown). Thirdly, and most importantly, we could not detect any AL050405 protein in HeLa snRNPs using antiserum directed against recombinant AL050405 (not shown), arguing that AL050405 is not the mammalian U2 snRNP ortholog of Snu17p. It is conceivable, however, that Snu17p is related to both proteins and carries out two functions, as is the case for the RRM-containing Snf protein in Drosophila (Polycarpou-Schwarz et al., 1996), but additional experiments are needed to clarify this relationship.
p14 is a U2 and U11/U12 snRNP component and interacts with SF3b155
Complementary approaches demonstrated that p14 is an integral component of the 17S U2 snRNP and 18S U11/U12 di-snRNP. The former finding is consistent with previous data that p14 first interacts with the branch site at the time of U2 addition to the pre-spliceosome and that this interaction is dependent on the presence of U2 snRNP (MacMillan et al., 1994). The initial characterization of 18S U11/U12 snRNPs did not detect p14 due to high levels of streptavidin, which obscured proteins in the 14 kDa range (Will et al., 1999). Similarily, p14 was not detected in the original 17S U2 snRNP preparations (Behrens et al., 1993), the likely explanation being that p14 was poorly resolved from the D1 protein. Nonetheless, our results conclusively demonstrate that p14 is associated with both U2 and U11/U12 snRNPs, and thus, together with the four other subunits of SF3b, represents the fifth example of a protein shared by these functionally analogous snRNPs. These results indicate that p14 enters the spliceosome and associates with the pre-mRNA branch site as part of the 17S U2 snRNP or, in the case of the minor spliceosome, as part of the 18S U11/U12 snRNP complex, and thus may facilitate the interaction of these snRNPs with their respective branch sites.
Despite the fact that it contains an RRM, p14 associates with U2 and U11/U12 snRNPs via protein–protein interactions. In vitro binding studies demonstrated that p14 interacts stably with SF3b155 and to a lesser extent with SF3b130. Consistent with this finding, both p14 and SF3b dissociate from U2 snRNP at moderate salt concentrations (i.e. typically >250 mM; Behrens et al., 1993). Indeed, p14 could be detected in an immunoaffinity-purified complex with SF3b proteins in the absence of U2 snRNA or U2 core proteins, demonstrating that it represents a novel SF3b component. Although the composition of the SF3b complex was previously reported to consist of four subunits with mol. wts of 49, 130, 145 and 155 kDa, information about lower molecular weight proteins was not presented in these studies (Das et al., 1999; Krämer et al., 1999). The presence of p14 in SF3b, coupled with its affinity for both SF3b130 and SF3b155, suggested that it may mediate the interaction of the latter SF3b subunits. However, binding studies with in vitro translated SF3b proteins failed to reveal a bridging role for p14 in the assembly of the SF3b complex (not shown).
The p14 interaction site on SF3b155 was mapped to a region within amino acids 255–424. Interestingly, this region also contains those amino acids (267–369) required for the interaction of SF3b155 with U2AF35 and U2AF65 (Gozani et al., 1998). However, whether these interactions are mutually exclusive is not clear presently. Nonetheless, since the SF3b155–U2AF interaction has been proposed to assist in association of U2 snRNP with the branch site, it is possible that the SF3b155–p14 interaction may be replaced by the SF3b155–U2AF interaction during pre-spliceosome assembly. Intriguingly, the N-terminal 493 amino acids of SF3b155 can serve as a substrate for cyclin E–cyclin dependent kinase (cdk) 2 in vitro (Seghezzi et al., 1998) and SF3b155 is phosphorylated within spliceosomes prior to catalytic step II of splicing (Wang et al., 1998). SF3b155 amino acids 255–424 contain numerous potential cdk phosphorylation sites (S/T-P-X-basic) and thus its interaction with p14 might be modulated by phosphorylation/dephosphorylation cycles.
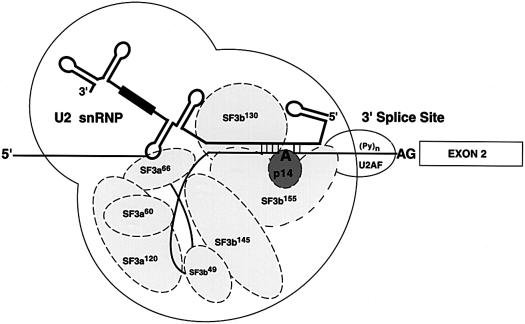
These data provide additional insight into the poorly defined higher order structure of the major spliceosome. The interaction between p14 and SF3b155 is compatible with previous cross-linking data from several groups. In pre-spliceosomes, p14 was cross-linked directly to the adenosine, whereas SF3b155 was cross-linked on both sides of the p14 cross-link site, at positions –6 and +5 from the adenosine (Query et al., 1996; Gozani et al., 1998). All of the remaining SF3b proteins, except SF3b130, and all subunits of SF3a have been shown to UV cross-link to a 20 nucleotide region immediately 5′ of the branch site (Gozani et al., 1996). Together, these data argue that in pre-spliceosomes SF3a and SF3b proteins are positioned near the branch site adenosine, with p14 centrally located and SF3b155 directly surrounding p14 (Figure 8). Although some of the SF3 proteins appear to be rearranged during the progression from pre-spliceosomes to spliceosomes (MacMillan et al., 1994; Staknis and Reed, 1994), p14 remains localized at the reactive adenosine in spliceosomes and SF3b155 still contacts both sides of the branch site during the catalytic steps of splicing (MacMillan et al., 1994; Wang et al., 1998). Thus, these two proteins most probably are core components of the spliceosome. Because p14 is located between SF3b155 and the branch site, this arrangement may facilitate proper positioning of the branch site–U2 duplex for interactions within the catalytic core. As both proteins are conserved between the major and minor spliceosomes, a similar three-dimensional structure could be envisaged to form in the U12-dependent spliceosome.
Fig. 8. Model of p14 interactions at the branch site in pre-spliceosomes. The U2 snRNP including U2 snRNA (thick solid line) and its Sm site (black box), and a subset of U2-specific proteins (ellipses) are shown schematically. Exon 2 is depicted by a box, intron sequences by a thin solid line, the polypyrimidine tract by (Py)n and the branch adenosine (A) and 3′ splice site AG dinucleotide are in bold.
p14 is conserved between U2- and U12-dependent spliceosomes
We demonstrated that pre-mRNAs that engage the U2- or the U12-dependent splicing machinery could be immunoprecipitated with antibodies directed against p14. The former result is consistent with previous cross-linking studies demonstrating contacts between p14 and the U2-type branch site in both pre-spliceosomes and spliceosomes (MacMillan et al., 1994), as well as our identification of p14 in purified U2-type spliceosomes. In our immunoprecipitation studies, we did not detect splicing intermediates or products, suggesting that p14 might dissociate from spliceosomes prior to the first catalytic step. However, previous studies demonstrated that p14 is present in the major spliceosomal complex C, which has undergone the first catalytic step of splicing (MacMillan et al., 1994). Thus, given that the spliceosome undergoes major structural arrangements prior to catalysis, a more likely explanation is that the p14 epitope is no longer accessible to the antibody in catalytically activated spliceosomes.
The finding that p14 is a component of both major and minor spliceosomes, coupled with its evolutionary conservation, is consistent with a conserved role in splicing. Indeed, in yeast, deletion of the gene encoding the apparent p14 ortholog, Snu17p, leads to a slow growth phenotype and inhibition of pre-mRNA splicing both in vivo and in vitro (Gottschalk et al., 2001). Additionally, analysis of splicing complex formation in a null mutant strain revealed a role for Snu17p in spliceosome formation (Gottschalk et al., 2001). To analyze p14’s role in metazoan splicing further, we performed in vitro depletion/complementation studies. However, while we could immunodeplete p14 quantitatively from HeLa splicing extracts using anti-p14 antibodies, and splicing in these p14-depleted extracts was significantly (but not completely) reduced, we were not able to restore splicing activity by the addition of recombinant p14 (not shown). This failure to complement splicing activity was probably due to the absence of other SF3b subunits, which were co-depleted with p14 even when depletions were performed at salt concentrations up to 1 M, consistent with our finding that p14 is a stable SF3b component.
In general, conservation between U2- and U12-dependent spliceosomes suggests similar molecular mechanisms. Many of the components that have been identified in both systems to date are those interacting in the major spliceosome at or near the sites where chemical events happen, for example the U5-specific 220 kDa protein (hPrp8) at the 5′ and 3′ splice sites (Collins and Guthrie, 1999; Luo et al., 1999; Siatecka et al., 1999), SF3b subunits near the branch site (Gozani et al., 1996; Will et al., 1999) and p14 at the branch site (this study). In particular, the conservation of p14 and surrounding SF3b subunits between both spliceosomes suggests that the mechanisms of branch site selection are similar in the major and minor spliceosomes. This interpretation is also compatible with the view that the structure of the active site responsible for the first transesterification reaction is conserved between both spliceosomes.
Materials and methods
Purification of p14 from spliceosomes and sequence analysis
Spliceosomes formed on biotinylated AdML pre-mRNA were isolated by Sephacryl S-500 gel filtration, followed by affinity selection with streptavidin–agarose (Bennett et al., 1992), using a high salt wash (250 mM NaCl, 20 mM Tris pH 7.8, 0.1% Triton X-100, 2.5 mM EDTA). RNase A (10 U; Boehringer Mannheim) was added to the resin-bound complex and this was incubated for 30 min at 30°C. Proteins were eluted from the beads by incubation in 1 ml of 20 mM Tris pH 7.8, 20 mM dithiothreitol (DTT) and 0.2% SDS at 65°C for 5 min, precipitated with acetone, fractionated by SDS–PAGE on a 15% gel and stained with Coomassie Blue. Proteins in the 14 kDa range were excised from the gel and peptide sequences obtained by microsequencing (MIT Biopolymers Lab). p14 peptides were used to search the NCBI EST database for full-length human cDNAs. Multiple ESTs were identified and sequenced on both strands using cycle sequencing.
Cross-linking of proteins to the branch site
Branch site-modified PIP85.b pre-mRNA was prepared, derivatized with benzophenone, incubated in HeLa nuclear extract for 30 min under in vitro splicing conditions, cross-linked and subsequently digested using RNase A as described previously (MacMillan et al., 1994). For immunoprecipitation, SDS was added to 0.15%, the reactions heated at 95°C for 2 min, then diluted 10-fold with IP100 buffer (100 mM NaCl, 50 mM Tris pH 7.5, 2 mM MgCl2, 0.5 mM DTT and 0.05% NP-40) or with IP100 buffer containing additional detergents (0.5% NP-40; 0.5% NP-40 and 0.5% deoxycholate; or 0.5% NP-40, 0.5% deoxycholate and 0.1% SDS) and added to 40 µl pf protein A–Trisacryl GF-2000 beads (Pierce) pre-bound with anti-p14 antibodies or pre-immune serum. After 2 h of mixing at 4°C, samples were washed three times with IP100 buffer containing the above detergents and then three times with IP150 buffer additionally containing 1 M urea, heated in SDS loading buffer and analyzed by SDS–PAGE on a 16% gel.
Purification of snRNPs and characterization of snRNP proteins
SnRNP preparations enriched in 17S U2 snRNPs were prepared essentially as described by Behrens et al. (1993). 18S HeLa U11/U12 snRNPs were affinity selected using biotinylated 2′-O-methyl oligonucleotides and streptavidin–agarose (Will et al., 1999). To minimize contaminating streptavidin, bound snRNPs were eluted with H buffer (20 mM HEPES pH 7.9, 700 mM KCl, 1.5 mM MgCl2, 5 mM EDTA) for 1 h at 25°C. SnRNP proteins were fractionated by SDS–PAGE on gels containing 10 (upper half) and 13% (lower half) polyacrylamide, and stained with Coomassie blue. Peptide sequences of the U11/U12- and U2-associated 14 kDa protein were obtained by Edman degradation (Toplab, Martinsried, Germany) and by mass spectrometry, respectively.
Immunoblotting and immunoprecipitation of snRNPs
To generate anti-p14 antibodies, rabbits were immunized with the p14 peptide KLLKEKYGINKDPPKC (positions 111–125 plus a cysteine) coupled to ovalbumin. For immunoblotting, proteins from nuclear extract or affinity-purified snRNPs were fractionated by SDS–PAGE, transferred to nitrocellulose and immunostained using an enhanced chemiluminescence (ECL) detection kit (Amersham). Immunoprecipitations were performed with HeLa nuclear extract as described by Teigelkamp et al. (1997).
Immunoaffinity purification of SF3b
To generate anti-SF3b155 and anti-SF3a120 antibodies for the preparative isolation of SF3b, rabbits were immunized with the SF3b155 peptide EQYDPFAEHRPPKIAC (positions 99–113 plus a cysteine) or the SF3a120 peptide CRWLEQRDRSIREKQS (positions 437–451 plus a cysteine), respectively, coupled to ovalbumin. The anti-pep.155 antibodies reacted specifically with SF3b155 on immunoblots (not shown). Anti-pep.155 and anti-pep.120 antibodies were affinity purified using a SulfoLink column (Pierce) containing immobilized SF3b155 or SF3b120 peptide, respectively, and then bound and covalently coupled to protein A–Sepharose beads with dimethylpimelimidate (Sigma). Nuclear extract was first subjected to anti-m3G immunoaffinity chromatography at a salt concentation (420 mM) where the majority of SF3b dissociates from U2. The salt concentration was then increased to 600 mM and the extract was passed over an anti-SF3a120 column (to remove any remaining 17S U2 snRNP), followed by an anti-SF3b155 column. The latter was washed with 50 vols of G600 buffer [20 mM HEPES pH 7.9, 600 mM KCl, 1.5 mM MgCl2, 5% glycerol, 0.5 mM DTT, 0.5 mM phenylmethylsulfonyl fluoride (PMSF)] and bound complexes were eluted with 0.1 M SF3b155 peptide in G600 buffer. Eluted SF3b complexes were fractionated on a 15–35% glycerol gradient containing G150 buffer, by centrifuging at 250 000 g for 16 h at 4°C.
In vitro splicing and immunoprecipitation of spliceosomes
U2- and U12-dependent splicing were performed with 32P-labeled MINX or P120 pre-mRNA as described previously (Will et al., 1996; Tarn and Steitz, 1996b). Nuclear extract was pre-incubated under splicing conditions (except in the absence of splicing substrate) for 10 min at 30°C followed by 5 min at 0°C with 5 µM of a 2′-O-methyl RNA oligonucleotide complementary to nucleotides 27–49 of U2 or 11–28 of U12, as indicated. Splicing complexes were immunoprecipitated as described by Luo et al. (1999). Immunoprecipitated RNA was recovered by proteinase K digestion followed by phenol extraction, and fractionated on a 14% (MINX) or 8% (P120) polyacrylamide–8 M urea gel.
In vitro translation
Coupled transcription and translation of the p14, SF3b155 and mutants thereof, SF3b145, SF3b130 or SF3b49 cDNAs were performed in rabbit reticulocyte lysate using the TNT system (Promega). The SF3b155 cDNA encodes a protein lacking amino acids 933–1014, as this region is toxic to Escherichia coli (Wang et al., 1998). For translation of SF3b155 deletion mutants (1–570), (1–423) and (1–254), the SF3b155 cDNA was linearized with NdeI, BstBI and BamHI, respectively. SF3b155 mutant (255–424), which contains an additional N-terminal methionine, was generated by PCR cloning.
GST pull-downs
The p14 cDNA was cloned into pGEX6P1 (Pharmacia Biotech) and GST–p14 or GST was expressed in E.coli (BL21) cells. Soluble protein was bound to glutathione–Sepharose beads, eluted with glutathione and dialyzed against G150 buffer containing 5% glycerol. One hundred picomoles of GST or GST–p14 in a total volume of 25 µl of G150 buffer were incubated with 8 µl of in vitro translate for 90 min at 0°C. The reactions were diluted with 200 µl of IPP150 buffer (20 mM Tris–HCl pH 8.0, 150 mM NaCl, 0.05% NP-40), added to 20 µl (packed volume) of glutathione–Sepharose, mixed by end-over-end rotation for 90 min at 4°C and the beads washed five times with IPP150 buffer. Bound proteins were eluted and analyzed by SDS–PAGE.
Acknowledgments
Acknowledgements
A.M.M. and C.C.Q. gratefully acknowledge Phillip Sharp at MIT and members of his laboratory where part of this work was initiated. We thank Ruth Santos, Gabi Heyne, Axel Badouin and Winfried Lorenz for excellent technical assistance, and Patrizia Fabrizio for helpful discussions and sharing data prior to publication. We are grateful to Robin Reed for kindly providing SF3b cDNA clones and SF3b antibodies and to Joan Steitz for plasmid encoding P120 pre-mRNA. This work was supported by the Gottfried Wilhelm Leibniz Program and a grant from the DFG (SFB 397/A6) to R.L. and RO1-GM57829 from the NIH to C.C.Q.
References
- Abovich N. and Rosbash,M. (1997) Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell, 89, 403–412. [DOI] [PubMed] [Google Scholar]
- Behrens S.E., Tyc,K., Kastner,B., Reichelt,J. and Lührmann,R. (1993) Small nuclear ribonucleoprotein (RNP) U2 contains numerous additional proteins and has a bipartite RNP structure under splicing conditions. Mol. Cell. Biol., 13, 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett M., Michaud,S., Kingston,J. and Reed,R. (1992) Protein components specifically associated with prespliceosome and spliceosome complexes. Genes Dev., 6, 1986–2000. [DOI] [PubMed] [Google Scholar]
- Berglund J.A., Chua,K., Abovich,N., Reed,R. and Rosbash,M. (1997) The splicing factor BBP interacts specifically with the pre-mRNA branchpoint sequence UACUAAC. Cell, 89, 781–787. [DOI] [PubMed] [Google Scholar]
- Brosi R., Hauri,H.P. and Kramer,A. (1993) Separation of splicing factor SF3 into two components and purification of SF3a activity. J. Biol. Chem., 268, 17640–17646. [PubMed] [Google Scholar]
- Burge C.B., Padgett,R.A. and Sharp,P.A. (1998) Evolutionary fates and origins of U12-type introns. Mol. Cell, 2, 773–785. [DOI] [PubMed] [Google Scholar]
- Burge C.B., Tuschl,T. and Sharp,P.A. (1999) Splicing of precursors to mRNAs by the spliceosomes. In Gesteland,R.F., Cech,T.R. and Atkins,J.F. (eds), The RNA World, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 525–560.
- Chiara M.D., Gozani,O., Bennett,M., Champion-Arnaud,P., Palandjian,L. and Reed,R. (1996) Identification of proteins that interact with exon sequences, splice sites, and the branchpoint sequence during each stage of spliceosome assembly. Mol. Cell. Biol., 16, 3317–3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins C.A. and Guthrie,C. (1999) Allele-specific genetic interactions between Prp8 and RNA active site residues suggest a function for Prp8 at the catalytic core of the spliceosome. Genes Dev., 13, 1970–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das B.K., Xia,L., Palandjian,L., Gozani,O., Chyung,Y. and Reed,R. (1999) Characterization of a protein complex containing spliceosomal proteins SAPs 49, 130, 145, and 155. Mol. Cell. Biol., 19, 6796–6802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frilander M.J. and Steitz,J.A. (1999) Initial recognition of U12-dependent introns requires both U11/5′ splice-site and U12/branchpoint interactions. Genes Dev., 13, 851–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur R.K., Valcarcel,J. and Green,M.R. (1995) Sequential recognition of the pre-mRNA branch point by U2AF65 and a novel spliceosome-associated 28-kDa protein. RNA, 1, 407–417. [PMC free article] [PubMed] [Google Scholar]
- Gottschalk A., Bartels,C., Neubauer,G., Lührmann,R. and Fabrizio,P. (2001) A novel yeast U2 snRNP protein, Snu17p, is required for the first catalytic step of splicing and for progression of spliceosome assembly. Mol. Cell. Biol., 21, 3037–3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozani O., Feld,R. and Reed,R. (1996) Evidence that sequence-independent binding of highly conserved U2 snRNP proteins upstream of the branch site is required for assembly of spliceosomal complex A. Genes Dev., 10, 233–243. [DOI] [PubMed] [Google Scholar]
- Gozani O., Potashkin,J. and Reed,R. (1998) A potential role for U2AF–SAP 155 interactions in recruiting U2 snRNP to the branch site. Mol. Cell. Biol., 18, 4752–4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall S.L. and Padgett,R.A. (1996) Requirement of U12 snRNA for in vivo splicing of a minor class of eukaryotic nuclear pre-mRNA introns. Science, 271, 1716–1718. [DOI] [PubMed] [Google Scholar]
- Kolossova I. and Padgett,R.A. (1997) U11 snRNA interacts in vivo with the 5′ splice site of U12-dependent (AU–AC) pre-mRNA introns. RNA, 3, 227–233. [PMC free article] [PubMed] [Google Scholar]
- Krämer A. (1996) The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu. Rev. Biochem., 65, 367–409. [DOI] [PubMed] [Google Scholar]
- Krämer A., Gruter,P., Groning,K. and Kastner,B. (1999) Combined biochemical and electron microscopic analyses reveal the architecture of the mammalian U2 snRNP. J. Cell Biol., 145, 1355–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H.R., Moreau,G.A., Levin,N. and Moore,M.J. (1999) The human Prp8 protein is a component of both U2- and U12-dependent spliceosomes. RNA, 5, 893–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMillan A.M., Query,C.C., Allerson,C.R., Chen,S., Verdine,G.L. and Sharp,P.A. (1994) Dynamic association of proteins with the pre-mRNA branch region. Genes Dev., 8, 3008–3020. [DOI] [PubMed] [Google Scholar]
- Pauling M.H., McPheeters,D.S. and Ares,M.,Jr (2000) Functional Cus1p is found with Hsh155p in a multiprotein splicing factor associated with U2 snRNA. Mol. Cell. Biol., 20, 2176–2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polycarpou-Schwarz M., Gunderson,S.I., Kandels-Lewis,S., Seraphin,B. and Mattaj,I.W. (1996) Drosophila SNF/D25 combines the functions of the two snRNP proteins U1A and U2B′ that are encoded separately in human, potato, and yeast. RNA, 2, 11–23. [PMC free article] [PubMed] [Google Scholar]
- Query C.C., Moore,M.J. and Sharp,P.A. (1994) Branch nucleophile selection in pre-mRNA splicing: evidence for the bulged duplex model. Genes Dev., 8, 587–597. [DOI] [PubMed] [Google Scholar]
- Query C.C., Strobel,S.A. and Sharp,P.A. (1996) Three recognition events at the branch-site adenine. EMBO J., 15, 1392–1402. [PMC free article] [PubMed] [Google Scholar]
- Seghezzi W., Chua,K., Shanahan,F., Gozani,O., Reed,R. and Lees,E. (1998) Cyclin E associates with components of the pre-mRNA splicing machinery in mammalian cells. Mol. Cell. Biol., 18, 4526–4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siatecka M., Reyes,J.L. and Konarska,M.M. (1999) Functional interactions of Prp8 with both splice sites at the spliceosomal catalytic center. Genes Dev., 13, 1983–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staknis D. and Reed,R. (1994) Direct interactions between pre-mRNA and six U2 small nuclear ribonucleoproteins during spliceosome assembly. Mol. Cell. Biol., 14, 2994–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarn W.Y. and Steitz,J.A. (1996a) Highly diverged U4 and U6 small nuclear RNAs required for splicing rare AT–AC introns. Science, 273, 1824–1832. [DOI] [PubMed] [Google Scholar]
- Tarn W.Y. and Steitz,J.A. (1996b) A novel spliceosome containing U11, U12, and U5 snRNPs excises a minor class (AT–AC) intron in vitro. Cell, 84, 801–811. [DOI] [PubMed] [Google Scholar]
- Teigelkamp S., Mundt,C., Achsel,T., Will,C.L. and Lührmann,R. (1997) The human U5 snRNP-specific 100-kD protein is an RS domain-containing, putative RNA helicase with significant homology to the yeast splicing factor Prp28p. RNA, 3, 1313–1326. [PMC free article] [PubMed] [Google Scholar]
- Wang C., Chua,K., Seghezzi,W., Lees,E., Gozani,O. and Reed,R. (1998) Phosphorylation of spliceosomal protein SAP 155 coupled with splicing catalysis. Genes Dev., 12, 1409–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will C.L. and Lührmann,R. (1997) snRNP structure and function. In Krainer,A.R. (ed.), Eukaryotic mRNA Processing. IRL Press at Oxford University Press, Oxford, UK, pp. 130–173.
- Will C.L., Rumpler,S., Klein Gunnewiek,J., van Venrooij,W.J. and Lührmann,R. (1996) In vitro reconstitution of mammalian U1 snRNPs active in splicing: the U1-C protein enhances the formation of early (E) spliceosomal complexes. Nucleic Acids Res., 24, 4614–4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will C.L., Schneider,C., Reed,R. and Luhrmann,R. (1999) Identification of both shared and distinct proteins in the major and minor spliceosomes. Science, 284, 2003–2005. [DOI] [PubMed] [Google Scholar]