Abstract
Two cytoplasmic mRNA-decay pathways have been characterized in yeast, and both are initiated by shortening of the 3′-poly(A) tail. In the major 5′-to-3′ decay pathway, the deadenylation triggers removal of the 5′-cap, exposing the transcript body for 5′-to-3′ degradation. An alternative 3′-to-5′ decay pathway also follows the deadenylation and requires two multi-complexes: the exosome containing various 3′-exonucleases and the Ski complex consisting of the RNA helicase Ski2p, Ski3p and Ski8p. In addition, Ski7p, which has an N-terminal domain and a C-terminal elongation factor 1α-like GTP-binding domain, is involved in the 3′-to-5′ decay. However, physical interaction between the exosome and the Ski complex, together with the function of Ski7p, has remained unknown. Here we report that the N domain of Ski7p is required and sufficient for the 3′-to-5′ decay. Furthermore, the exosome and the Ski complex interact with the different regions of Ski7p N domain, and both interactions are required for the 3′-to-5′ decay. Thus, Ski7p G protein appears to function as a signal-coupling factor between the two multi-complexes operating in the 3′-to-5′ mRNA-decay pathway.
Keywords: exosome/G protein/mRNA decay/SKI genes/yeast
Introduction
In addition to transcriptional regulation, mRNA decay plays an important role in the quality and quantity control of gene expression. mRNA degradation is carried out in the nucleus and the cytoplasm of budding yeast Saccharomyces cerevisiae (Bousquet-Antonelli et al., 2000). Two cytoplasmic mRNA-decay pathways have been characterized in yeast (for reviews see Caponigro and Parker, 1996; Mitchell and Tollervey, 2000a). In both, the initial step is shortening of the 3′-poly(A) tail of mRNAs to A10 or less. This step appears to involve the removal of a poly(A)-binding protein (PABP) from the poly(A) tail, to be degraded by a deadenylating enzyme (Coller et al., 1998). In the major pathway, the poly(A) shortening triggers the removal of the 5′-cap structure by a decapping enzyme, Dcp1p, exposing the transcript body to an exonuclease, Xrn1p, for the rapid 5′-to-3′ digestion (Muhlrad and Parker, 1992; Decker and Parker, 1993; Hsu and Stevens, 1993; Muhlrad et al., 1994, 1995; Beelman et al., 1996). An alternative 3′-to-5′ degradation pathway also follows the deadenylation and requires at least two multi-complex machineries: the exosome and a ternary complex consisting of Ski2p, Ski3p and Ski8p (we refer to the complex as the Ski complex) (Muhlrad et al., 1995; Jacobs Anderson and Parker, 1998; Brown et al., 2000).
The yeast exosome is a protein complex comprising at least 10 essential components. Among them, Rrp4p, Ski6p (Rrp41p) and Rrp44p (Dis3p) have been demonstrated to have 3′-to-5′ exoribonuclease activity in vitro, and the other members (Rrp40p, Rrp42p, Rrp43p, Rrp45p, Rrp46p and Mtr3p) except Csl4p (Ski4p) have high sequence homology to Escherichia coli 3′-to-5′ exoribonucleases, RNase PH and PNPase (for reviews see van Hoof and Parker, 1999; Mitchell and Tollervey, 2000b). The yeast exosome and its human counterpart, the PM-Scl complex, are present in both the cytoplasm and the nucleus (Mitchell et al., 1997; Allmang et al., 1999a). The cytoplasmic exosome degrades mRNAs, whereas the nuclear exosome processes small nuclear RNAs (snRNAs), small nucleolar RNAs (snoRNAs) and rRNAs, in addition to the degradation of pre-mRNA and pre-rRNA spacer (Mitchell et al., 1997; Jacobs Anderson and Parker, 1998; Allmang et al., 1999a, 2000; van Hoof et al., 2000a). The nuclear exosome has an additional subunit, Rrp6p, which also possesses 3′-to-5′ exoribonuclease activity (Briggs et al., 1998; Allmang et al., 1999b; Burkard and Butler, 2000). Consistent with its nuclear localization, RRP6 mutant strains do not exhibit defects in cytoplasmic mRNA turnover (van Hoof et al., 2000a).
On the other hand, the superkiller (SKI) genes encoded in yeast nuclear genome were initially identified from mutations that cause overexpression of a killer toxin encoded by the endogenous double-stranded RNA (Toh-e et al., 1978). Subsequent studies demonstrated that, in addition to the exosome, the products of SKI2, SKI3 and SKI8 genes are required for the 3′-mRNA degradation (Jacobs Anderson and Parker, 1998). Ski2p, Ski3p and Ski8p are a putative RNA helicase, a tetratricopeptide-repeat protein and a protein containing WD motif, respectively (Rhee et al., 1989; Matsumoto et al., 1993; Widner and Wickner, 1993). The three Ski proteins form a stable complex, and the complex is localized in the cytoplasm (Brown et al., 2000). Mutations in the three genes inhibit 3′-to-5′ mRNA decay, but do not affect the other function of the exosome (Jacobs Anderson and Parker, 1998; van Hoof et al., 2000a). Therefore, the Ski complex appears to be a cofactor for the exosome to degrade yeast mRNAs. However, direct interaction between the exosome and the Ski complex has not been observed (Brown et al., 2000).
More recently, another member of the Ski proteins, Ski7p, has been reported to be responsible for the 3′-to-5′ mRNA decay (van Hoof et al., 2000b). The SKI7 gene was initially identified as one of the SKI gene family (Benard et al., 1999). van Hoof et al. (2000b) reported that Ski7p is also required for 3′-to-5′ mRNA degradation and acts in the same pathway as Ski2p, Ski3p and Ski8p, because deletion of the SKI7 gene caused impaired 3′-mRNA decay similar to SKI2, SKI3 or SKI8 deletion. However, formation of the Ski complex, which had been impaired in ski2, ski3 or ski8 mutants, was still intact in the ski7 mutant, suggesting that Ski7p is not a member of the Ski complex and is not required for the formation of the Ski complex (Brown et al., 2000). Thus, the roles of Ski7p and the Ski complex members appear to be different in 3′-mRNA degradation.
Ski7p is a GTP-binding protein consisting of two separate domains; one is the C-terminal region homologous to the GTP-binding elongation factor 1α (EF1α), and the other is an extra N-terminal domain that was not present in EF1α. This domain structure is quite similar to that of the GTP-binding eukaryotic releasing factor 3 (eRF3). We previously indicated that eRF3 functions not only in translation termination, but also as an initiator of poly(A) shortening by interacting with PABP (Hoshino et al., 1998, 1999b). The N domain of eRF3 associated with PABP to unmask the poly(A) tail of mRNAs, presumably for their degradation (Hoshino et al., 1999b).
In order to understand the molecular details of the 3′-mRNA decay pathway, we investigated the role of Ski7p and its interacting molecules. Here we report that the N domain of Ski7p is required and sufficient, and its EF1α-like C domain is dispensable, for the 3′-to-5′ mRNA degradation. Furthermore, the Ski7p N domain was capable of interacting with the exosome and the Ski complex physically and functionally. Thus, the N domains of this G protein family appear to function in common as regulatory factors in mRNA-decay pathways.
Results
Ski7p is involved in the cytoplasmic 3′-to-5′ mRNA-decay pathway
A recent study has revealed that Ski7p is required for 3′-to-5′ degradation of yeast mRNAs (van Hoof et al., 2000b). We first confirmed the previous findings using an MFA2pG mRNA that contained a poly(G) tract inserted into its 3′-untranslated region. Since the poly(G) tract forms a stable structure that inhibits 5′-exoribonuclease activity (Decker and Parker, 1993), 5′-to-3′ degradation of the mRNA produces an intermediate that stretches from the poly(G) tract to the 3′ end of the mRNA. The resultant intermediate is normally degraded by the 3′-to-5′ mRNA-degradation pathway (Jacobs Anderson and Parker, 1998). Thus, mutants having a defect in 3′-to-5′ decay show a slow digestion of the 3′-end fragment, resulting in the accumulation of its fragments upon northern blot analysis. In accordance with the previous report (van Hoof et al., 2000b), deletion of the SKI7 gene (ski7Δ) caused accumulation of the 3′-end fragments of MFA2pG mRNA (data not shown, but see Figure 2).
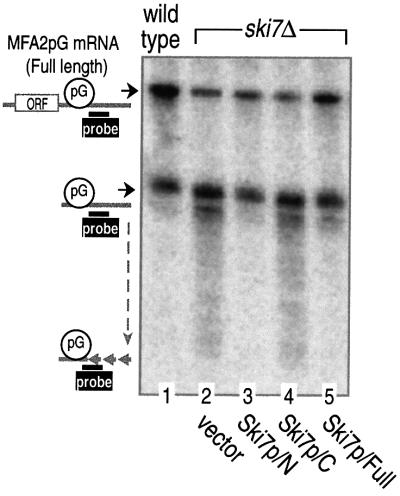
Fig. 2. The N domain of Ski7p is necessary and sufficient for the 3′-to-5′ degradation of mRNA. Yeast strains MC046 (vector), MC047 (Ski7p/N), MC048 (Ski7p/C) and MC049 (Ski7p/Full), carrying the GAL1:MFA2pG reporter, were grown in a galactose-containing medium and harvested at the mid-log phase (OD600 = 0.4–0.5). RNA extracted from the cells was subjected to polyacrylamide gel northern blot analysis as described in Materials and methods. Positions of the full-length and poly(G) (pG) to 3′-end fragments of MFA2pG mRNA are indicated on the left. ORF, open reading frame.
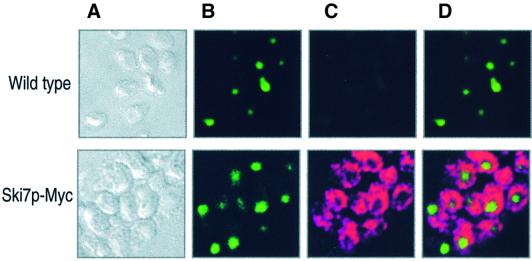
If Ski7p is directly involved in the cytoplasmic 3′-mRNA-decay pathway, at least a portion of this protein should be present in the cytoplasm. The chromosomal copy of SKI7 was tagged with nine Myc epitopes (Ski7p-Myc), and the Myc-tagged protein was produced under native transcriptional conditions. Figure 1 shows the cellular localization of Ski7p-Myc observed by indirect immunofluorescence, together with nuclear DNA stained with PicoGreen. Ski7p-Myc localized primarily in the cytoplasm and did not overlap with the nuclear signals. These observations suggested that Ski7p is directly involved in the 3′-to-5′ mRNA-decay pathway in yeast cytoplasm.
Fig. 1. Intracellular localization of Ski7p in yeast. A yeast strain expressing Myc-tagged SKI7 (MC028, top panels) and the wild type (W303a, bottom panels) were analyzed by confocal microscopy as described in Materials and methods. (A) Nomarski images of the cells. (B) DNA of the cells was stained with PicoGreen. (C) Ski7p-Myc was stained with anti-Myc (primary) and anti-mouse IgG coupled to Alexa-568 (secondary) antibodies. (D) Overlay of the two signals shown in (B) and (C).
Ski7p N domain is required and sufficient for the 3′-to-5′ mRNA degradation
Ski7p consists of two separated domains, the extra N-terminal domain (amino acid sequence 1–264) and the C-terminal EF1α-like domain (sequence 265–747). To identify which domain is responsible for the 3′-mRNA decay, each of the two domains was expressed in the ski7Δ strain. As shown in Figure 2, the N domain of Ski7p (Ski7p/N), together with the full-length form (Ski7p/Full), rescued the ski7Δ phenotype, which was characterized by the accumulation of 3′-end fragments of MFA2pG mRNA. However, such recovery was not observed with the EF1α-like C domain (Ski7p/C). This indicated that Ski7p/N is required and sufficient, and Ski7p/C is dispensable, for the 3′-mRNA decay.
Ski7p interacts physically with both the exosome and the Ski complex
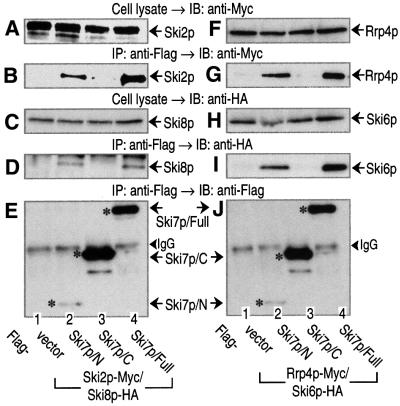
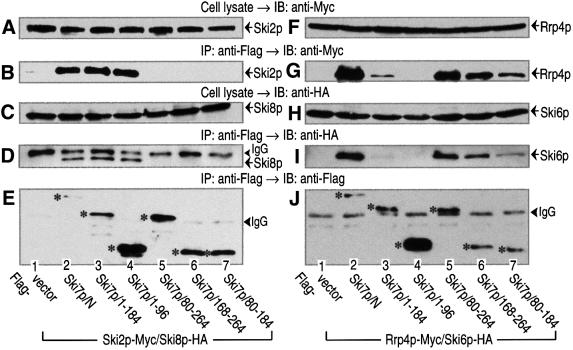
It has been reported that 3′-to-5′ mRNA degradation in yeast requires at least two multi-complexes: the exosome and the Ski complex consisting of Ski2p, Ski3p and Ski8p (Jacobs Anderson and Parker, 1998). Therefore, we investigated whether Ski7p was capable of interacting with these components (Figure 3). The chromosomal copies of SKI2 and SKI8 were tagged with Myc and hemagglutinin (HA) epitopes, respectively, and vectors expressing Ski7p-deletion variants under the control of the GAL1 promoter were introduced into the same yeast strain with a Flag epitope. Extracts were prepared from these strains and subjected to immunoprecipitation assay with an anti-Flag antibody. As shown in Figure 3B and D, both members of the Ski complex, Ski2p-Myc and Ski8p-HA, were co-precipitated with Ski7p/N and Ski7p/Full, but not with Ski7p/C. We also tagged the chromosomal copies of RRP4 and SKI6, the components of the exosome, with Myc and HA epitopes. As shown in Figure 3G and I, not only Rrp4p-Myc, but also Ski6p-HA, could be detected in the fractions precipitated with Ski7p/N and Ski7p/Full. These experiments were performed with the constructs overexpressing Ski7p variants. However, we found that the expression constructs rescued the phenotype of the SKI7-disruption strain (see Figure 2). Therefore, these interactions could be detected with a normal level of Ski7p. When yeast cell extract was fractionated on a gel-filtration column, Ski7p behaved as a complex (>300 kDa protein) larger than its predicted 85 kDa monomer, and it appeared to associate with the components of the Ski complex and the exosome (data not shown). Thus, the N domain of Ski7p is capable of interacting physically with the components of both the Ski complex and the exosome.
Fig. 3. The N domain of Ski7p interacts with the components of the Ski complex and the exosome. Cell extracts were obtained from the indicated yeast strains expressing Ski2p-Myc and Ski8p-HA (A–E) or Prp4p-Myc and Ski6p-HA (F–J). The yeast strains also expressed the indicated forms of Flag-tagged Ski7p (A–E, lanes 1–4 for MC146–MC149; F–J, lanes 1–4 for MC151–MC154). The cell extracts were immunoprecipitated (IP) with an anti-Flag antibody. The precipitated proteins (B, D, E, G, I and J), together with the whole-cell extracts (A, C, F and H), were immunoblotted (IB) with anti-Myc (9E10), anti-HA (12CA5), and anti-Flag (M2) antibodies to detect Ski2p (or Rrp4p), Ski8p (or Ski6p) and Ski7p. The asterisks in (E) and (J) indicate Flag-tagged Ski7p mutants expressed in the various strains. The arrowheads indicate the positions of IgG bands.
Ski7p interacts with the exosome independently of its interaction with the Ski complex
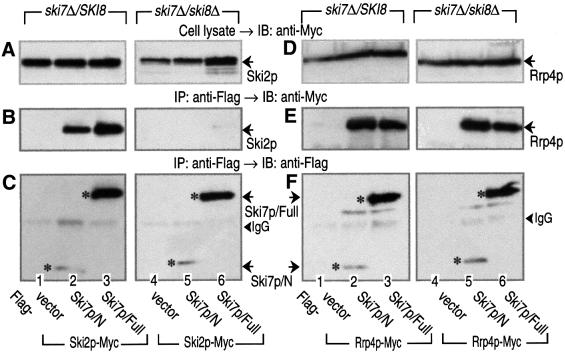
Disruption of SKI8 has been shown to abolish the association between Ski2p and Ski3p (Brown et al., 2000). Therefore, we tested whether the deletion of SKI8 could exert its influence on the Ski7p–Ski2p and/or Ski7p– Rrp4p interaction. As shown in Figure 4B, the association of Ski7p/N (or Ski7p/Full) with Ski2p observed in Ski8p-expressing strains was completely abolished in SKI8-disruption strains (ski8Δ). However, the interaction between Ski7p/N (or Ski7p/Full) and Rrp4p was still present in the ski8Δ strains (Figure 4E). These results indicated that an intact form of the Ski complex is required for the association between Ski7p and the Ski complex, and that the interaction between Ski7p and the exosome is unaffected by the presence or absence of the Ski complex.
Fig. 4. Interaction of the N domain of Ski7p with Rrp4p is maintained in the Ski complex-disrupted yeast strains. Cell extracts were prepared from wild-type SKI8 or ski8Δ strains carrying Ski2p-Myc (A–C, lanes 1–6 for MC052, MC053, MC055, MC130, MC131 and MC133) or Rrp4p-Myc (D–F, lanes 1–6 for MC126, MC127, MC129, MC134, MC135 and MC137). The yeast strains also expressed the indicated forms of Flag-tagged Ski7p. The cell extracts were immunoprecipitated (IP) with the anti-Flag antibody. The precipitated proteins (B, C, E and F), together with the whole-cell extracts (A and D), were immunoblotted (IB) with anti-Myc (9E10) and anti-Flag (M2) antibodies to detect Ski2p (or Rrp4p) and Ski7p, respectively. The asterisks in (C) and (F) indicate Flag-tagged Ski7p mutants expressed in the various strains. The arrowheads indicate the positions of IgG bands.
The Ski complex and the exosome interact with the different regions of the Ski7p N domain
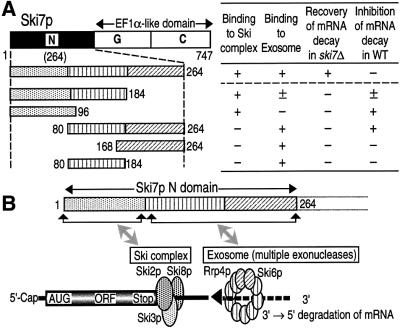
Since Ski7p appeared to interact with the Ski complex and the exosome independently through the N domain, we mapped binding regions for the two multi-complexes on the N domain. Yeast strains expressing five different Flag-tagged variants of Ski7p/N were prepared (see Figure 8), and the association with the Ski complex and the exosome was investigated by immunoprecipitation assay with the anti-Flag antibody. As shown in Figure 5B and D, the Ski complex-binding region, which had been measured by Ski2p-Myc and Ski8p-HA binding, was located in the amino acid sequence 1–96 of Ski7p/N. On the other hand, either of the two sequences, 80–184 or 168–264 of Ski7p, was sufficient for the association with the exosome measured by Rrp4p-Myc and Ski6p-HA binding (Figure 5G and I). It is unlikely that the exosome-binding site was present in only the overlapped region 168–184 between the above independent two sequences, since the more narrow sequences 80–176 and 177–264 were both capable of binding to the exosome (data not shown). Taken together, these results indicated that different regions on Ski7p/N are involved in the interaction with the exosome and the Ski complex.
Fig. 8. The N domain of Ski7p interacting with the Ski complex and the exosome. (A) Deletion mutants of Ski7p used in this study and the summary of the present results obtained in Figures 5, 6 and 7. (B) A proposed model for the Ski complex- and the exosome-interacting sites of the Ski7p N domain.
Fig. 5. The Ski complex and the exosome interact with the different regions of the Ski7p N domain. Cell extracts were obtained from the indicated yeast strains expressing Skip2-Myc and Ski6p-HA (A–E) or Prp4p-Myc and Ski2p-HA (F–J). The yeast strains (MC146, MC147, MC151, MC152, MC155–164) also expressed the various deletion mutants of Ski7p N domain with a Flag tag (lanes 1–7). The cell extracts were immunoprecipitated (IP) with the anti-Flag antibody. The precipitated proteins (B, D, E, G, I and J), together with the whole-cell extracts (A, C, F and H), were immuno blotted (IB) with anti-Myc (9E10), anti-HA (12CA5) and anti-Flag (M2) antibodies to detect Ski2p (or Rrp4p), Ski8p (or Ski6p) and Ski7p. The asterisks in (E) and (J) indicate Flag-tagged Ski7p mutants expressed in the various strains. The arrowheads indicate the positions of IgG bands.
Interaction of Ski7p/N with both the exosome and the Ski complex is required for 3′-to-5′ mRNA decay
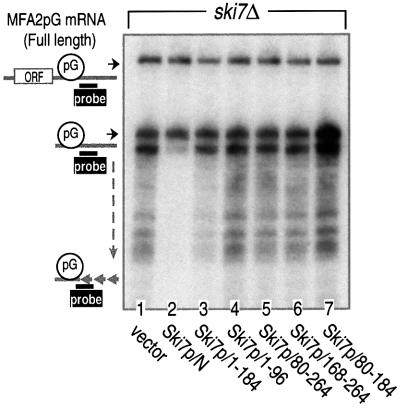
We investigated further which regions on Ski7p/N were responsible for the 3′-to-5′ mRNA decay. The five Ski7p/N-deletion variants used in Figure 5 were expressed in the ski7Δ strain, and their effects on the MFA2pG mRNA decay were monitored by northern blot analysis. As shown in Figure 6, all five deletion variants failed to rescue the ski7Δ phenotype; the accumulation of 3′-end fragments of MFA2pG mRNA was only abolished by the presence of the whole Ski7p/N sequence (lane 2). These results are consistent with the theory that Ski7p functions in 3′-to-5′ mRNA decay through its interaction with both the Ski complex and the exosome.
Fig. 6. Both the Ski complex- and the exosome-binding regions of Ski7p N domain are required for the 3′-to-5′ degradation of mRNA. The ski7Δ strain was transformed with plasmids carrying the indicated deletion mutants of Ski7p N domain (lanes 1–7 for MC046, MC047 and MC138–MC142). The transfected cells were grown in the galactose-containing medium and harvested at mid-log phase. RNA extracted from the cells was subjected to polyacrylamide gel northern blot analysis as described in Figure 2. Positions of the full-length and poly(G) to 3′-end fragments of MFA2pG mRNA are indicated on the left.
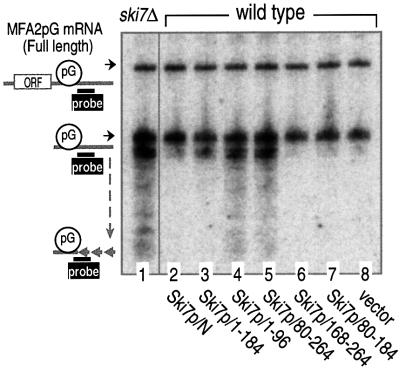
This theory predicted that interfering with either the interaction of Ski7p with the Ski complex or the exosome would stabilize 3′-to-5′ mRNA degradation. To test this prediction, we overexpressed the deletion variants of Ski7p/N in a wild-type strain containing MFA2pG mRNA. As shown in Figure 7, expression of the two mutants induced significant inhibition of 3′-to-5′ mRNA decay. One mutant was the sequence 1–96, identified as the Ski complex-binding region (see Figure 5B and D), and the other mutant was the sequence 80–264, which includes both exosome-binding regions (see Figure 5G and I). In addition, the sequence 1–184, which contains the Ski complex-binding region and one of the two exosome-binding regions, also inhibited the 3′-mRNA decay, but to a lesser extent. The lesser effect of the sequence 1–184 might be due to its low-level expression under the present experimental conditions (data not shown, but see Figure 5E and J). We thus concluded that the interaction between Ski7p/N and the two multi-complexes, the Ski complex and the exosome, is required for 3′-to-5′ mRNA degradation.
Fig. 7. Inhibition of 3′-to-5′ mRNA degradation by overexpressing either the Ski complex- or exosome-binding region of Ski7p N domain. The yeast strain MC035 (SKI7; wild type) was transformed with plasmids carrying the indicated deletion mutants of Ski7p N domain (lanes 1–8 for MC046, MC043, MC118–MC122 and MC042). The transfected cells were grown in the galactose-containing medium and harvested at mid-log phase. RNA extracted from the cells was subjected to polyacrylamide gel northern blot analysis as described in Figure 2. Positions of the full-length and poly(G) to 3′-end fragments of MFA2pG mRNA are indicated on the left.
Discussion
A recent study has revealed that Ski7p is a new member involved in the 3′-to-5′ mRNA-decay pathway in yeast cytoplasm, and that its role in mRNA decay may be different from those of the other members of the SKI gene family (van Hoof et al., 2000b). This idea was confirmed and extended further by our present findings. First, Ski7p did indeed exhibit cytoplasmic distribution in yeast (Figure 1). Secondly, the N-terminal domain of Ski7p is required and sufficient for 3′-to-5′ mRNA degradation. Its EF1α-like C domain was dispensable for the function of Ski7p (Figure 2). Thirdly, the exosome and the Ski com plex, which are known to be multi-complex machineries involved in 3′-mRNA decay, interacted physically with the different regions of Ski7p/N (Figures 3, 4 and 5). Fourthly, interaction of Ski7p/N with the two complexes appeared to be required for 3′-to-5′ mRNA decay (Figures 6 and 7). We thus concluded that the N domain of Ski7p mediates 3′-to-5′ mRNA degradation through its interaction with the exosome and the Ski complex (Figure 8).
Possible roles of Ski7p in the cytoplasmic mRNA decay of yeast
In the present study, we indicated that both the Ski complex and the exosome are capable of interacting with Ski7p/N, although direct binding of their constituent components to Ski7p/N has not been determined yet. The interacting sites of the two complexes were mapped on the different regions of Ski7p/N. Moreover, Ski7p/N was essential and sufficient for cytoplasmic 3′-to-5′ mRNA decay. These findings gave rise to an important question of how Ski7p/N regulates mRNA decay. There may be at least two independent mechanisms in Ski7p-induced mRNA degradation. One is that Ski7p/N binds to the Ski complex and the exosome in order to stimulate their intrinsic enzyme activities, such as RNA helicase of Ski2p and exoribonucleases in the exosome. Although the effects of Ski7p/N on the enzymatic activities have not been investigated in the present study, the above idea would be consistent with the following observations. Two RNA helicases, RhlB and eIF4A, associated with RNase E and eIF4B, respectively, and these interactions appeared to result in stimulation of their ATPase activities (Rozen et al., 1990; Vanzo et al., 1998).
Alternatively, Ski7p/N may function as a coupling factor between the Ski complex and the exosome for mRNA decay. In E.coli, the degradosome, a complex of RNase E, a polynucleotide phosphorylase and the RNA helicase RhlB, is required for 3′-to-5′ degradation of mRNA (Carpousis et al., 1994; Py et al., 1996). The C-terminal half of RNase E constitutes binding sites for each of the other two degradosomal components (Kaberdin et al., 1998; Vanzo et al., 1998). In vitro analysis of the E.coli degradosome indicates that RhlB is required for degradation of structured RNA substrates in a manner dependent on ATP (Py et al., 1996; Coburn et al., 1999). Similarly, Suv3p, a yeast mitochondrial RNA helicase, is a component of the mtEXO complex, which also contains the 3′-exoribonuclease Dss1p (Msu1p) and is required for intron degradation (Margossian et al., 1996). These findings may suggest that the RNA helicase (Ski2p of the Ski complex) and the exoribonucleases in the exosome might form a multi-complex via Ski7p/N for 3′-mRNA degradation. However, the Ski7p–Ski complex and Ski7p–exosome complex appeared to be present separately in yeast cytoplasm, since Ski7p was, but Ski2p (the component of the Ski complex) was not, co-immunoprecipitated with Rrp4p of the exosome (data not shown). These observations imply that a ternary complex consisting of Ski7p, the Ski complex and the exosome may not exist at the same time. This is consistent with a previous report showing that there is no direct interaction between the Ski complex and the exosome (Brown et al., 2000). Thus, it is very likely that the two complexes associating with Ski7p are formed separately, probably in sequential steps of the mRNA-decay pathway. Although further experiments are required to elucidate the molecular mechanism, our present report is the first indication that Ski7p may function as a signal-coupling factor between the RNA helicase and the exoribonucleases.
Comparison between cytoplasmic and nuclear mRNA degradation in yeast
The degradation of yeast mRNAs takes place in the nucleus as well as in the cytoplasm (Bousquet-Antonelli et al., 2000). There are many similarities between the nuclear pre-mRNA-degradation and cytoplasmic mRNA-decay pathways. Both pathways involve 3′-to-5′ degradation by their exosomes, while 5′-to-3′ degradation is carried out by two homologous exoribonucleases: Rat1p in the nucleus and Xrn1p in the cytoplasm (Hsu and Stevens, 1993; Muhlrad et al., 1994, 1995; Johnson, 1997; Jacobs Anderson and Parker, 1998; Bousquet-Antonelli et al., 2000). Therefore, we predict that the two 3′-to-5′ decay pathways may be carried out in a similar fashion. Both processing and degradative activities in their exosomes have been shown to require substrate-specific cofactor(s) (van Hoof and Parker, 1999). The Ski complex and Ski7p are thus cofactors for the cytoplasmic mRNA decay induced by the exosome, although factors for nuclear pre-mRNA degradation have not yet been determined. It is unlikely that the Ski complex and Ski7p also function in pre-mRNA degradation because of their cytoplasmic localization.
An interesting model proposed previously was that the role of Mtr4p (Dob1p) could be analogous to that of Ski2p (de la Cruz et al., 1998). This prediction was based on the following observations. First, Mtr4p, a putative ATP-dependent RNA helicase, is highly homologous to Ski2p. Secondly, this molecule functions as an accessory factor for the nuclear form of the exosome, which is involved in 3′-to-5′ exonucleolytic processing of 5.8S rRNA, snoRNAs and U4 snRNA in the nucleus (de la Cruz et al., 1998; Allmang et al.,1999a; van Hoof et al., 2000a). Thirdly, mutations in MTR4 lead to the nucleolar accumulation of poly(A)+ RNAs (Liang et al., 1996). Finally, Mtr4p is localized throughout the nucleus (Liang et al., 1996). According to this model and our assumption, an additional cofactor equivalent to the cytoplasmic Ski7p G protein would be required for nuclear pre-mRNA degradation. Such a candidate may be Hbs1p, the only GTP-binding protein, structurally related to Ski7p and eRF3 in the genome of S.cerevisiae. HBS1 was identified as a suppressor in the slow growth of the ssb1 and ssb2 double mutant (Nelson et al., 1992). Hbs1p is highly conserved in mammals, as well as Mtr4p and the exosome, but is not required for the exosome-mediated RNA processing (van Hoof et al., 2000b). Further experiments should allow us to provide evidence for the above possibility.
Ski7p as a member of the eRF3 GTP-binding protein family
Ski7p appears to belong to the eRF3/GSPT family of GTP-binding proteins, since it has a two-domain structure with an extra N-terminal domain (∼260 amino acids) and the C-terminal (GC) domain homologous to the GTP-binding EF1α (Figure 8). We previously indicated that eRF3 functions not only in translation termination as an eRF1-carrying protein, but also in 3′-poly(A) shortening by interacting with PABP (Hoshino et al., 1998, 1999a,b). The two functions of eRF3 were mediated separately through the N and C domains: the N domain was capable of binding to PABP to inhibit its multimerization on the poly(A) tail, and only the EF1α-like C domain was required for the association with eRF1 (Hoshino et al., 1999b). This separated domain-function architecture would be quite comparable to the characteristics of Ski7p observed in the present study. The N domain of Ski7p was required and sufficient for 3′-to-5′ mRNA degradation. Moreover, Hbs1p, which was supposed to be involved in the nuclear pre-mRNA degradation (see above), may belong to this family. Thus, the eRF3 G protein family appear to function in common as regulators in mRNA-decay pathways through their N domains.
There was no marked change in the interaction between Ski7p/N and the exosome (or the Ski complex) when a non-hydrolyzable GTP analogue or GDP was added to the immunoprecipitation assay shown in Figure 3 (data not shown). Thus, the interaction appears not to be regulated by the GTP- or GDP-form of Ski7p G protein. This is consistent with our findings that the interaction between the N domain of eRF3 and PABP is insensitive to the nucleotide-bound forms of the G protein. Instead, the association between the C domain of eRF3 and eRF1 required GTP rather than GDP (T.Kobayashi, S.Hoshino and T.Katada, unpublished data). There are still many unresolved issues in the structure and function of Ski7p. For example, what is the role of Ski7p/C in mRNA decay? In this regard, it is very likely that Ski7p/C may interact with a component(s) responsible for poly(A) shortening, since the 3′-to-5′ mRNA decay follows deadenylation (Muhlrad et al., 1995). Furthermore, it will be very interesting to determine how GDP–GTP exchange and GTPase reactions are involved in the regulation of Ski7p function(s). These important issues are currently under investigation in our laboratory.
Materials and methods
Yeast strains and growth conditions
All yeast strains used in this study were derived from W303a, and are listed in Table I. The yeast cells were grown in standard culture media (Adams et al., 1997) and transformed with DNA by the lithium acetate method (Ito et al., 1983). Disruption of SKI7 and SKI8 genes was achieved using a PCR-based method with the genomic DNA and LEU2 marker gene of Candida glabrata, generous gifts from Y.Ohya (Baudin et al., 1993). Proofreading polymerases were used in all the PCR applications. Gene disruption was confirmed by the phenotypic analysis and/or PCR reactions with primers specific for the genes.
Table I. Yeast strains used in this study.
| Name | Relevant genotype |
|---|---|
| MC028 | SKI7-9Myc |
| MC035 | pGAL-MFA2pG |
| MC042 | pGAL-MFA2pG, pURAGAL1-His6-FLAG-FLAG |
| MC043 | pGAL-MFA2pG, pURAGAL1-His6-FLAG-SKI7/N |
| MC046 | ski7::cgHIS3, pGAL-MFA2pG, pURAGAL1-His6-FLAG |
| MC047 | ski7::cgHIS3, pGAL-MFA2pG, pURAGAL1-His6-FLAG-SKI7/N |
| MC048 | ski7::cgHIS3, pGAL-MFA2pG, pURAGAL1-His6-FLAG-SKI7/C |
| MC049 | ski7::cgHIS3, pGAL-MFA2pG,pURAGAL1-His6-FLAG-SKI7/Full |
| MC052 | ski7::cgHIS3, SKI2-9myc, pURAGAL1-His6-FLAG |
| MC053 | ski7::cgHIS3, SKI2-9myc, pURAGAL1-His6-FLAG-SKI7/N |
| MC055 | ski7::cgHIS3, SKI2-9myc, pURAGAL1-His6-FLAG-SKI7/Full |
| MC118 | pGAL-MFA2pG, pURAGAL1-His6-FLAG-SKI7/1-184 |
| MC119 | pGAL-MFA2pG, pURAGAL1-His6-FLAG-SKI7/1-96 |
| MC120 | pGAL-MFA2pG, pURAGAL1-His6-FLAG-SKI7/80-264 |
| MC121 | pGAL-MFA2pG, pURAGAL1-His6-FLAG-SKI7/168-264 |
| MC122 | pTRPGAL-MFA2pG, pURAGAL1-His6-FLAG-SKI7/80-184 |
| MC126 | ski7::cgHIS3, RRP4-9myc, pURAGAL1-His6-FLAG |
| MC127 | ski7::cgHIS3, RRP4-9myc,pURAGAL1-His6-FLAG-SKI7/N |
| MC129 | ski7::cgHIS3, RRP4-9myc, pURAGAL1-His6-FLAG-SKI7/Full |
| MC130 | ski7::cgHIS3, ski8::cgLEU2, SKI2-9Myc, pURAGAL1-His6-FLAG |
| MC131 | ski7::cgHIS3, ski8::cgLEU2, SKI2-9Myc, pURAGAL1-His6-FLAG-SKI7/N |
| MC133 | ski7::cgHIS3, ski8::cgLEU2, SKI2-9Myc, pURAGAL1-His6-FLAG-SKI7/Full |
| MC134 | ski7::cgHIS3, ski8::cgLEU2, RRP4-9Myc, pURAGAL1-His6-FLAG |
| MC135 | ski7::cgHIS3, ski8::cgLEU2, RRP4-9Myc, pURAGAL1-His6-FLAG-SKI7/N |
| MC137 | ski7::cgHIS3, ski8::cgLEU2, RRP4-9Myc, pURAGAL1-His6-FLAG-SKI7/Full |
| MC138 | ski7::cgHIS3, pGAL-MFA2pG, pURAGAL1-His6-FLAG-SKI7/1-184 |
| MC139 | ski7::cgHIS3, pGAL-MFA2pG, pURAGAL1-His6-FLAG-SKI7/1-96 |
| MC140 | ski7::cgHIS3, pGAL-MFA2pG, pURAGAL1-His6-FLAG-SKI7/80-264 |
| MC141 | ski7::cgHIS3, pGAL-MFA2pG, pURAGAL1-His6-FLAG-SKI7/168-264 |
| MC142 | ski7::cgHIS3, pGAL-MFA2pG, pURAGAL1-His6-FLAG-SKI7/80-184 |
| MC146 | ski7::cgHIS3, SKI2-9Myc, SKI8-3HA, pURAGAL1-His6-FLAG |
| MC147 | ski7::cgHIS3, SKI2-9Myc, SKI8-3HA, pURAGAL1-His6-FLAG-SKI7/N |
| MC148 | ski7::cgHIS3, SKI2-9Myc, SKI8-3HA, pURAGAL1-His6-FLAG-SKI7/C |
| MC149 | ski7::cgHIS3, SKI2-9Myc, SKI8-3HA, pURAGAL1-His6-FLAG-SKI7/Full |
| MC151 | ski7::cgHIS3, RRP4-9Myc, SKI6-3HA, pURAGAL1-His6-FLAG |
| MC152 | ski7::cgHIS3, RRP4-9Myc, SKI6-3HA, pURAGAL1-His6-FLAG-SKI7/N |
| MC153 | ski7::cgHIS3, RRP4-9Myc, SKI6-3HA, pURAGAL1-His6-FLAG-SKI7/C |
| MC154 | ski7::cgHIS3, RRP4-9Myc, SKI6-3HA, pURAGAL1-His6-FLAG-SKI7/Full |
| MC155 | ski7::cgHIS3, SKI2-9Myc, SKI8-3HA, pURAGAL1-His6-FLAG-SKI7/1-184 |
| MC156 | ski7::cgHIS3, SKI2-9Myc, SKI8-3HA, pURAGAL1-His6-FLAG-SKI7/1-96 |
| MC157 | ski7::cgHIS3, SKI2-9Myc, SKI8-3HA, pURAGAL1-His6-FLAG-SKI7/80-264 |
| MC158 | ski7::cgHIS3, SKI2-9Myc, SKI8-3HA, pURAGAL1-His6-FLAG-SKI7/168-264 |
| MC159 | ski7::cgHIS3, SKI2-9Myc, SKI8-3HA, pURAGAL1-His6-FLAG-SKI7/80-184 |
| MC160 | ski7::cgHIS3, RRP4-9Myc, SKI6-3HA, pURAGAL1-His6-FLAG-SKI7/1-184 |
| MC161 | ski7::cgHIS3, RRP4-9Myc, SKI6-3HA, pURAGAL1-His6-FLAG-SKI7/1-96 |
| MC162 | ski7::cgHIS3, RRP4-9Myc, SKI6-3HA, pURAGAL1-His6-FLAG-SKI7/80-264 |
| MC163 | ski7::cgHIS3, RRP4-9Myc, SKI6-3HA, pURAGAL1-His6-FLAG-SKI7/168-264 |
| MC164 | ski7::cgHIS3, RRP4-9Myc, SKI6-3HA, pURAGAL1-His6-FLAG-SKI7/80-184 |
All strains are derivatives of W303a with the following full genotype: MATa, ade2-1, trp1-1, can1-100, leu2-3 112, his-11,15, ura3, GAL, psi+.
Epitope tagging of SKI2, SKI6, SKI7, SKI8 and RRP4 with nine Myc epitopes or three HA epitopes was performed by the one-step tagging method described by Knop et al. (1999). Transformants were checked by PCRs for correct integration. All epitope-tagged proteins expressed in this study were fully functional. The sequences of the oligonucleotides are as follows:
SKI2, TTGATTAAGAGAGATATTGTTTTCGCCGCAAGTTTGTATTTACGTACGCTGCAGGTCGAC and ACTTTTATAAACATGACTCACATTGAGAATAAATGAGCTCTATCGATGAATTCGAGCTCG;
SKI6, AGGAAACATGCTCAGAAAAGAGTCAGTAACGCCTCTGCTAGGCGTACGCTGCAGGTCGAC and ACTTTTATATAAACAGTGGCAATTAATGGCGTTTTTTATTATCGATGAATTCGAGCTCG;
SKI7, TAGATCATAAGGTTATCGCAGTTGGCAGAATTGCATGCCAGCGTACGCTGCAGGTCGAC and AATAAGTATGAATGCCTAGTATAATTTCTTAGTTGTAGGAATCGATGAATTCGAGCTCG;
SKI8, TTAGATAGAAGCATCAGGTGGTTTAGAGAAGCTGGCGGTAAACGTACGCTGCAGGTCGAC and GATGTAAGGTTACATGCAATATATCAAGATATTACTAGAGAATCGATGAATTCGAGCTCG;
RRP4, AGTGACATTCTTACCGCCGAAAAAATGAGAGGTAACGGCAACCGTACGCTGCAGGTCGAC and TGTAGTTTATAAATGTGTAAACAGCGGCTTTTGTATTCCTAATCGATGAATTCGAGCTCG.
Plasmid constructions
pURAGAL1 and pTRPGAL1 were derived from pRS316 and pRS314, respectively (Sikorski and Hieter, 1989). The NdeI site of URA3 was altered by site-directed mutagenesis as described previously (Kunkel et al., 1991) using the oligonucleotide GTATATATACGCATACGTAGTGTTGAAGAAA (the mutation position is underlined). These plasmids contained the promoter fragment of GAL1 gene and the transcription terminator of the CMK1 gene. To obtain pURAGAL1-His6-Flag and pTRPGAL1-His6-Flag, a DNA fragment was made by annealing eight oligonucleotides (AATTCACTATAATGA, GGTCATCACCATCACCATCACGGT, GACTACAAGGATGACGATGACAAGGGT, CATATGGCTCGAGTCGACTTGGATCCGTAGA, ATGGTGATGACCAGTCATTATAGT, CATCCTTGTAGTCACCGTGATGGTG, GACTCGAGCCATATGACCCTTGTCATCGT and AGCTTCTACGGATCCAAGTC). The fragment was then inserted into the region between the EcoRI and HindIII sites of pURAGAL1 and pTRPGAL1. pURAGAL1-His6-Flag and pTRPGAL1-His6-Flag can overexpress His6-Flag tagged proteins under the control of the GAL1 promoter.
Plasmids for expressing the full-length and truncated proteins of Ski7p were constructed as follows. These genes were amplified from the genomic DNA by PCR using the following oligonucleotides: full-length, CCGGATCCTTCATATGTCGTTATTAGAGCAATTAG and CCGTCGACTTACTGGCATGCAATTCTGCCAAC; N domain, CCGGATCCTTCATATGTCGTTATTAGAGCAATTAG and CCGTCGACT TAATGGGTGGCAATGAATGAATG; C domain, CGTCGACCGCATATGCCTCTGAATTTGACATGTTTG and CCGTCGACTTACTGGCATGCAATTCTGCCAAC; 1–184, CCGGATCCTTCATATGTCG TTATTAGAGCAATTAG and GGGTCGACTTAATTGTTATGCTTTTTTAAGC; 1–96, CCGGATCCTTCATATGTCGTTATTAGAGCAATTAG and GGGTCGACTTATCCTTGTTTCTCCAAATCAC; 80–264, CCGGATCCTTCATATGTTAAAGTTGTCTGCCTTAAAG and GGGTCGACTTAATGGGTGGCAATGAATGAATG; 168–264, CCGGATCCTTCATATGCCGCTATCGTCGCAGAATTC and GGGTCGACTTAATGGGTGGCAATGAATGAATG; 80–184, CCGGATCCTTCATATGTTAAAGTTGTCTGCCTTAAAG and GGGTCGACT TAATTGTTATGCTTTTTTAAGC. These fragments were then inserted into the region between the NdeI and SalI sites of pURAGAL1-His6-FLAG.
pGAL-MFA2pG was constructed essentially as described previously (Decker and Parker, 1993), with some modifications. To generate pGEM-MFA2, a DNA fragment containing MFA2 and its 3′ untranslated region (UTR) was amplified from the genomic DNA by PCR using two primers (CCGAATTCTCGAGCCATGCAACCGATCACCACTGC and GGGGTACCAAGCTTTTCCGTGACTGGTTG), and the fragment was subcloned into pGEM-T Easy vector (Promega). pGEM-MFA2/B178 was constructed by the one-day mutagenesis method (Imai et al., 1991) with two oligonucleotides (AATATCTACCCTTTCATTTATTACG and GATTAGATCTCTTGGTTGTCGTC) to create a new BglII site at B178 in the 3′-UTR of MFA2 (Muhlrad and Parker, 1992). To obtain pGEM-MFA2pG, two oligonucleotides were annealed and inserted into pGEM-MFA2/B178 previously digested with BglII (GATCTAGGAATTTGGGGGGGGGGGGGGGGGGAATTCCT and GATCAGGAATTCCCCCCCCCCCCCCCCCCAAATTCCTA). pGAL-MFA2pG was constructed by inserting MFA2pG into pTRPGAL1-His6-Flag. MFA2pG was isolated from pGEM-MFA2pG as a XhoI–KpnI fragment and inserted into pTRPGAL1-His6-Flag previously digested with XhoI and KpnI.
Immunofluorescence microscopy
Immunofluorescence analysis of Ski7p-Myc localization was carried out essentially as described previously (Heyer et al., 1995), with some modifications. The wild-type yeast strain (W303a) and SKI7-9Myc (MC028) were grown in a selective medium to mid-exponential phase. Yeast cells were fixed with 4% formaldehyde at room temperature for 40 min. The Myc epitope was detected by a mouse monoclonal antibody, 9E10 (diluted 1:300). The secondary antibody was a goat anti-mouse IgG coupled to Alexa-568 (diluted 1:1000). DNA was stained with PicoGreen (Molecular Probes). Immunofluorescence images were obtained with a Zeiss Laser Scanning Microscope 510.
Immunoprecipitation of epitope-tagged proteins
Immunoprecipitation of Flag-tagged proteins and subsequent detection of co-precipitated proteins were performed as follows. Logarithmically growing cells (5 × 108) in the selective medium containing 2% galactose were resuspended in 250 µl of a lysis buffer consisting of 50 mM Tris–HCl pH 7.4, 150 mM NaCl, 5 mM EDTA, 1 mM dithiothreitol, 0.1% NP-40 and protease inhibitors. The cells were mixed with glass beads (1 g) and disrupted by 12 cycles of vortexing for 30 s followed by incubation on ice for 1 min. The cell extracts were obtained by two consecutive centrifugations (14 000 g for 5 min). After addition of anti-Flag antibody-conjugated beads (M2-AGAROSE-AFFINITY; Sigma), the extracts were incubated on a rotator at 4°C for 2 h. Beads were pelleted and washed extensively with the lysis buffer without protease inhibitors. Proteins binding to the beads were eluted with the SDS–PAGE sample buffer by boiling for 5 min. Equivalent amounts of the cell extract and the eluted proteins were separated by SDS–PAGE and immunoblotted with anti-Myc (9E10), anti-HA (12CA5) and anti-Flag (M2) monoclonal antibodies.
RNA analysis
The pGAL-MFA2pG plasmid was transformed in the ski7Δ strain as well as in the isogenic wild-type strain. The degradation of this reporter was assayed as described (Boeck et al., 1998). The ski7Δ strain as well as the wild-type strain were grown at 30°C to mid-log phase (OD600 = 0.4–0.5) in the galactose-containing selective medium. RNA that had been extracted from the frozen cell pellets by the hot phenol procedure (Boeck et al., 1998) was separated by PAGE and transferred to a Hybond N (Amersham Pharmacia) by electroblotting. The reporter was detected by northern blotting using the oligonucleotide oPR140, whose sequence is: MFA2pG, oPR140 (ATATTGATTAGATCAGGAATTCC) (Caponigro and Parker, 1995).
All experiments were performed at least three times with different samples of the yeast strains, and the results were fully reproducible. Hence, most of the data shown are representative of several independent experiments.
Acknowledgments
Acknowledgements
We are grateful to Yoshikazu Ohya, Yoshiko Kikuchi and Elmar Schiebel for generous gifts of various plasmids and yeast strains. This work was supported in part by research grants from the ‘Research for the Future’ Program of the Japan Society for the Promotion of Science (JSPS-RFTF 96L00505) and the Scientific Research Funds of the Ministry of Education, Culture, Sports, Science and Technology of the Japanese Government.
References
- Adams A., Gottschling,D.E., Kaiser,C.A. and Stearns,T. (1997) Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Allmang C., Kufel,J., Chanfreau,G., Mitchell,P., Petfalski,E. and Tollervey,D. (1999a) Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J., 18, 5399–5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allmang C., Petfalski,E., Podtelejnikov,A., Mann,M., Tollervey,D. and Mitchell,P. (1999b) The yeast exosome and human PM-Scl are related complexes of 3′→5′ exonucleases. Genes Dev., 13, 2148–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allmang C., Mitchell,P., Petfalski,E. and Tollervey,D. (2000) Degradation of ribosomal RNA precursors by the exosome. Nucleic Acids Res., 28, 1684–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin A., Ozier-Kalogeropoulos,O., Denouel,A., Lacroute,F. and Cullin,C. (1993) A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res., 21, 3329–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beelman C.A., Stevens,A., Caponigro,G., LaGrandeur,T.E., Hatfield,L., Fortner,D.M. and Parker,R. (1996) An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature, 382, 642–646. [DOI] [PubMed] [Google Scholar]
- Benard L., Carroll,K., Valle,R.C., Masison,D.C. and Wickner,R.B. (1999) The ski7 antiviral protein is an EF1-α homolog that blocks expression of non-poly(A) mRNA in Saccharomyces cerevisiae. J. Virol., 73, 2893–2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeck R., Lapeyre,B., Brown,C.E. and Sachs,A.B. (1998) Capped mRNA degradation intermediates accumulate in the yeast spb8-2 mutant. Mol. Cell. Biol., 18, 5062–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousquet-Antonelli C., Presutti,C. and Tollervey,D. (2000) Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell, 102, 765–775. [DOI] [PubMed] [Google Scholar]
- Briggs M.W., Burkard,K.T. and Butler,J.S. (1998) Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8 S rRNA 3′ end formation. J. Biol. Chem., 273, 13255–13263. [DOI] [PubMed] [Google Scholar]
- Brown J.T., Bai,X. and Johnson,A.W. (2000) The yeast antiviral proteins Ski2p, Ski3p and Ski8p exist as a complex in vivo. RNA, 6, 449–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkard K.T. and Butler,J.S. (2000) A nuclear 3′-5′ exonuclease involved in mRNA degradation interacts with poly(A) polymerase and the hnRNA protein Npl3p. Mol. Cell. Biol., 20, 604–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caponigro G. and Parker,R. (1995) Multiple functions for the poly(A)-binding protein in mRNA decapping and deadenylation in yeast. Genes Dev., 9, 2421–2432. [DOI] [PubMed] [Google Scholar]
- Caponigro G. and Parker,R. (1996) Mechanisms and control of mRNA turnover in Saccharomyces cerevisiae. Microbiol. Rev., 60, 233–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpousis A.J., Van Houwe,G., Ehretsmann,C. and Krisch,H.M. (1994) Copurification of E.coli RNAase E and PNPase: evidence for a specific association between two enzymes important in RNA processing and degradation. Cell, 76, 889–900. [DOI] [PubMed] [Google Scholar]
- Coburn G.A., Miao,X., Briant,D.J. and Mackie,G.A. (1999) Reconstitution of a minimal RNA degradosome demonstrates functional coordination between a 3′ exonuclease and a DEAD-box RNA helicase. Genes Dev., 13, 2594–2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller J.M., Gray,N.K. and Wickens,M.P. (1998) mRNA stabilization by poly(A) binding protein is independent of poly(A) and requires translation. Genes Dev., 12, 3226–3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker C.J. and Parker,R. (1993) A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev., 7, 1632–1643. [DOI] [PubMed] [Google Scholar]
- de la Cruz J., Kressler,D., Tollervey,D. and Linder,P. (1998) Dob1p (Mtr4p) is a putative ATP-dependent RNA helicase required for the 3′ end formation of 5.8S rRNA in Saccharomyces cerevisiae. EMBO J., 17, 1128–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer W.D., Johnson,A.W., Reinhart,U. and Kolodner,R.D. (1995) Regulation and intracellular localization of Saccharomyces cerevisiae strand exchange protein 1 (Sep1/Xrn1/Kem1), a multifunctional exonuclease. Mol. Cell. Biol., 15, 2728–2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino S., Imai,M., Mizutani,M., Kikuchi,Y., Hanaoka,F., Ui,M. and Katada,T. (1998) Molecular cloning of a novel member of the eukaryotic polypeptide chain-releasing factors (eRF). Its identification as eRF3 interacting with eRF1. J. Biol. Chem., 273, 22254–22259. [DOI] [PubMed] [Google Scholar]
- Hoshino S., Hosoda,N., Araki,Y., Kobayashi,T., Uchida,N., Funakoshi,Y. and Katada,T. (1999a) Novel function of the eukaryotic polypeptide-chain releasing factor 3 (eRF3/GSPT) in the mRNA degradation pathway. Biochemistry (Mosc), 64, 1367–1372. [PubMed] [Google Scholar]
- Hoshino S., Imai,M., Kobayashi,T., Uchida,N. and Katada,T. (1999b) The eukaryotic polypeptide chain releasing factor (eRF3/GSPT) carrying the translation termination signal to the 3′-poly(A) tail of mRNA. Direct association of eRF3/GSPT with polyadenylate-binding protein. J. Biol. Chem., 274, 16677–16680. [DOI] [PubMed] [Google Scholar]
- Hsu C.L. and Stevens,A. (1993) Yeast cells lacking 5′→3′ exoribo nuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol. Cell. Biol., 13, 4826–4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y., Matsushima,Y., Sugimura,T. and Terada,M. (1991) A simple and rapid method for generating a deletion by PCR. Nucleic Acids Res., 19, 2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda,Y., Murata,K. and Kimura,A. (1983) Transformation of intact yeast cells treated with alkali cations. J. Bacteriol., 153, 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs Anderson A.R. and Parker,R.P. (1998) The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J., 17, 1497–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A.W. (1997) Rat1p and Xrn1p are functionally interchangeable exoribonucleases that are restricted to and required in the nucleus and cytoplasm, respectively. Mol. Cell. Biol., 17, 6122–6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaberdin V.R., Miczak,A., Jakobsen,J.S., Lin-Chao,S., McDowall,K.J. and von Gabain,A. (1998) The endoribonucleolytic N-terminal half of Escherichia coli RNase E is evolutionarily conserved in Synechocystis sp. and other bacteria but not the C-terminal half, which is sufficient for degradosome assembly. Proc. Natl Acad. Sci. USA, 95, 11637–11642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M., Siegers,K., Pereira,G., Zachariae,W., Winsor,B., Nasmyth,K. and Schiebel,E. (1999) Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast, 15, 963–972. [DOI] [PubMed] [Google Scholar]
- Kunkel T.A., Bebenek,K. and McClary,J. (1991) Efficient site-directed mutagenesis using uracil-containing DNA. Methods Enzymol., 204, 125–139. [DOI] [PubMed] [Google Scholar]
- Liang S., Hitomi,M., Hu,Y.H., Liu,Y. and Tartakoff,A.M. (1996) A DEAD-box-family protein is required for nucleocytoplasmic transport of yeast mRNA. Mol. Cell. Biol., 16, 5139–5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margossian S.P., Li,H., Zassenhaus,H.P. and Butow,R.A. (1996) The DExH box protein Suv3p is a component of a yeast mitochondrial 3′-to-5′ exoribonuclease that suppresses group I intron toxicity. Cell, 84, 199–209. [DOI] [PubMed] [Google Scholar]
- Matsumoto Y., Sarkar,G., Sommer,S.S. and Wickner,R.B. (1993) A yeast antiviral protein, SKI8, shares a repeated amino acid sequence pattern with β-subunits of G proteins and several other proteins. Yeast, 9, 43–51. [DOI] [PubMed] [Google Scholar]
- Mitchell P. and Tollervey,D. (2000a) mRNA stability in eukaryotes. Curr. Opin. Genet. Dev., 10, 193–198. [DOI] [PubMed] [Google Scholar]
- Mitchell P. and Tollervey,D. (2000b) Musing on the structural organization of the exosome complex. Nature Struct. Biol., 7, 843–846. [DOI] [PubMed] [Google Scholar]
- Mitchell P., Petfalski,E., Shevchenko,A., Mann,M. and Tollervey,D. (1997) The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell, 91, 457–466. [DOI] [PubMed] [Google Scholar]
- Muhlrad D. and Parker,R. (1992) Mutations affecting stability and deadenylation of the yeast MFA2 transcript. Genes Dev., 6, 2100–2111. [DOI] [PubMed] [Google Scholar]
- Muhlrad D., Decker,C.J. and Parker,R. (1994) Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′→3′ digestion of the transcript. Genes Dev., 8, 855–866. [DOI] [PubMed] [Google Scholar]
- Muhlrad D., Decker,C.J. and Parker,R. (1995) Turnover mechanisms of the stable yeast PGK1 mRNA. Mol. Cell. Biol., 15, 2145–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson R.J., Ziegelhoffer,T., Nicolet,C., Werner-Washburne,M. and Craig,E.A. (1992) The translation machinery and 70 kd heat shock protein cooperate in protein synthesis. Cell, 71, 97–105. [DOI] [PubMed] [Google Scholar]
- Py B., Higgins,C.F., Krisch,H.M. and Carpousis,A.J. (1996) A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature, 381, 169–172. [DOI] [PubMed] [Google Scholar]
- Rhee S.K., Icho,T. and Wickner,R.B. (1989) Structure and nuclear localization signal of the SKI3 antiviral protein of Saccharomyces cerevisiae. Yeast, 5, 149–158. [DOI] [PubMed] [Google Scholar]
- Rozen F., Edery,I., Meerovitch,K., Dever,T.E., Merrick,W.C. and Sonenberg,N. (1990) Bidirectional RNA helicase activity of eucaryotic translation initiation factors 4A and 4F. Mol. Cell. Biol., 10, 1134–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh-e A., Guerry,P. and Wickner,R.B. (1978) Chromosomal superkiller mutants of Saccharomyces cerevisiae. J. Bacteriol., 136, 1002–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof A. and Parker,R. (1999) The exosome: a proteasome for RNA? Cell, 99, 347–350. [DOI] [PubMed] [Google Scholar]
- van Hoof A., Lennertz,P. and Parker,R. (2000a) Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol. Cell. Biol, 20, 441–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof A., Staples,R.R., Baker,R.E. and Parker,R. (2000b) Function of the ski4p (Csl4p) and ski7p proteins in 3′-to-5′ degradation of mRNA. Mol. Cell. Biol., 20, 8230–8243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanzo N.F., Li,Y.S., Py,B., Blum,E., Higgins,C.F., Raynal,L.C., Krisch,H.M. and Carpousis,A.J. (1998) Ribonuclease E organizes the protein interactions in the Escherichia coli RNA degradosome. Genes Dev., 12, 2770–2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widner W.R. and Wickner,R.B. (1993) Evidence that the SKI antiviral system of Saccharomyces cerevisiae acts by blocking expression of viral mRNA. Mol. Cell. Biol., 13, 4331–4341. [DOI] [PMC free article] [PubMed] [Google Scholar]