Abstract
Ara6 of Arabidopsis thaliana is a novel member of the Rab/Ypt GTPase family with unique structural features. It resembles Rab5 GTPases best, but lacks a large part of the C-terminal hypervariable region and the cysteine motif, and instead harbors an extra stretch of amino acid residues containing myristoylation and palmitoylation sites at the N-terminus. Ara6 is tightly associated with membranes and is expressed constitutively. In contrast, the conventional Rab5 ortholog, Ara7, is highly expressed only in actively dividing cells. Examination of green fluorescent protein (GFP)-tagged proteins indicates that both Ara6 and Ara7 are distributed on a subpopulation of endosomes and suggests their roles in endosomal fusion. The endosomal localization of Ara6 requires N-terminal fatty acylation, nucleotide binding and the C-terminal amino acid sequence coordinately. Proteins similar to Ara6 are found only in higher plants and thus represent a novel class of Rab GTPases regulating endocytic function in a plant- specific manner.
Keywords: Ara6/endosome/GFP/plant-specific gene/Rab
Introduction
It is well established that the molecular mechanisms and machinery of vesicular transport are very well conserved from yeast to higher eukaryotes. However, considering that membrane trafficking is required for a variety of cellular functions, evolutionary diversion of molecular components should also be important. In higher plants, vesicular transport is required for specialized phenomena as well as common house-keeping events. For example, tip growth of pollen tubes and root hairs utilizes strictly regulated secretion mechanisms. Secretion also plays a central role in cell plate formation. Plants may well have developed plant-specific mechanisms of vesicular traffic.
The Rab/Ypt family of GTPases are known as key players of vesicular transport. They act as molecular switches regulating the fusion of vesicles with target membranes through the conformational change between GTP- and GDP-bound forms (Novick and Zerial, 1997; Martinez and Goud, 1998). The Rab/Ypt GTPases have four consensus amino acid sequences, which are necessary for nucleotide binding and GTP hydrolysis. The C-terminal region is basically hypervariable but almost always ends in conserved motifs containing cysteine residues. These cysteine residues are the sites of isoprenylation, which is essential for membrane binding and function (Araki et al., 1991; Musha et al., 1992; Soldati et al., 1993; Ueda et al., 2000). The hypervariable region is also known to play a critical role in determining the subcellular localization of each protein (Chavrier et al., 1991; Brennwald and Novick, 1993). To date many Rab GTPases have been identified from higher plants (Ma, 1994; Verma et al., 1994; Uchimiya et al., 1998). Ara is the name given to a group of Rab GTPases from Arabidopsis thaliana. ARA1 was the first RAB/YPT gene identified in plants (Matsui et al., 1989). ARA2 to ARA5 were isolated subsequently by cross-hybridization using ARA1 as a probe (Anai et al., 1991). Two new members, ARA6 and ARA7, are described in this study.
For correct operation of vesicle targeting and fusion, the GTPase cycles of Rab/Ypt proteins have to be strictly regulated. Many regulator molecules have been identified including GDP dissociation inhibitor (GDI), guanine nucleotide exchange factor (GEF) and GTPase-activating protein (GAP) (Pfeffer et al., 1995; Pan and Wessling-Resnick, 1998; Scheffzek et al., 1998). We have discovered Arabidopsis GDI, AtGDI1 and AtGDI2, as molecules interacting with Ara4 GTPase (Ueda et al., 1996b, 1998).
Here we report a novel, plant-unique type of Rab GTPase, Ara6. Ara6 shows some homology to Rab5 GTPases, which play a critical role in endosomal homotypic fusion, but has a unique structure. We isolated a conventional Arabidopsis ortholog of Rab5 and named it Ara7. Comparison of Ara6 and Ara7 reveals differences in distribution, expression and regulation. Characterization of Ara6 should enhance our understanding of the reason and mechanism of evolution of plants including those of a unique system for the regulation of vesicular traffic.
Results
Ara6 represents a plant-specific subtype of Rab/Ypt GTPases
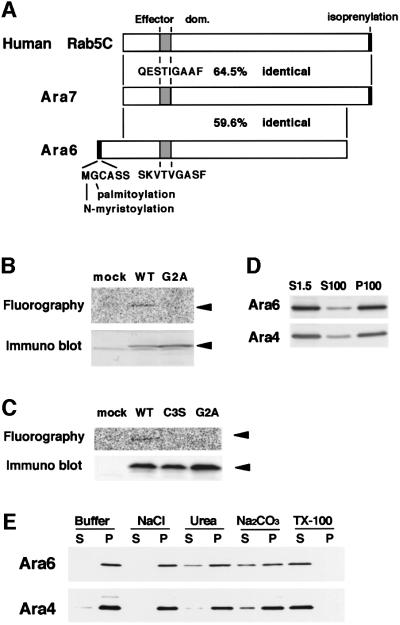
A novel member of the Arabidopsis Rab family was isolated fortuitously from a cDNA library of A.thaliana during screening for other genes. The nucleotide sequence predicted a protein of 202 amino acid residues that conserves the four consensus sequences characteristic of Rab/Ypt GTPases. We thus designated this cDNA ARA6 (DDBJ/EMBL/GenBank accession No. AB007766). The Ara6 protein is judged by homology analysis to be most similar to Rab5 proteins in the family (see Supplementary data, available at The EMBO Journal Online). However, Ara6 has some very unique features (Figure 1A). First of all, it lacks the C-terminal region containing part of the hypervariable region and the cysteine motif. The cysteine motif is conserved in almost all Rab/Ypt GTPases, and its isoprenylation is essential for attachment to membranes. The hypervariable region is important for their subcellular localization (Chavrier et al., 1991; Brennwald and Novick, 1993). Instead of this, Ara6 has an extra stretch of amino acids at the N-terminus, which contains putative N-myristoylation and palmitoylation sites. The amino acid sequence of the effector domain of Ara6, which is believed to specify the function of Rab/Ypt proteins (Brennwald and Novick, 1993; Dunn et al., 1993), does not match any other proteins of this family known to date. We examined whether Ara6 does indeed hydrolyze GTP using a fusion protein of Ara6 and GST. GST–Ara6 showed GTPase activity, and the rate of GTP hydrolysis was estimated to be ∼0.0065 mol GTP/min/mol (data not shown). Based on literature and database searches, homologs of Ara6 can be found in Lotus japonicus (Borg et al., 1997), Mesembryanthemum crystallinum (Bolte et al., 2000) and Oryza sativa (registered in a database), but this type of Rab/Ypt protein has never been reported in mammals or yeasts. The Rab subclass to which Ara6 belongs appears to be unique to plants.
Fig. 1. Ara6 is a peripheral membrane protein with a unique structure. (A) Schematic structures of human Rab5c, Ara7 and Ara6. (B) The N-terminus of Ara6 is N-myristoylated in vitro. In vitro transcription and translation of wild-type ARA6 (WT) and ARA6G2A (G2A) were performed in the presence of 14C-labeled myristic acid. The same reaction was carried out without the template (mock). (C) The N-terminus of Ara6 is palmitoylated in vitro. Ara6 (WT), Ara6C3S (C3S) and Ara6G2A (G2A) synthesized in vitro were immuno precipitated and incubated with Arabidopsis lysate in the presence of 14C-labeled palmitoyl-CoA. (D) Membrane binding of Ara6 and Ara4 was examined by western blotting. The total lysate prepared from Arabidopsis cultured cells (S1.5) was ultracentrifuged to separate the cytosolic fraction (S100) from the membrane fraction (P100), and analyzed by immunoblotting with anti-Ara6 polyclonal or anti-Ara4 monoclonal antibody. (E) The P100 fraction was incubated on ice in a buffer containing either salt, urea, alkali (Na2CO3) or Triton X-100. Samples were ultracentrifuged again, then the supernatant and the pellet were analyzed by immunoblotting with anti-Ara6 or anti-Ara4 antibody.
Membrane binding and expression of Ara6
To determine whether the N-terminus of Ara6 is in fact myristoylated, we performed in vitro transcription and translation of wild-type ARA6 and ARA6G2A, a mutant form in which the putative myristoylation site is destroyed, in the presence of 14C-labeled myristic acid. As the source of a transcription and translation system, we used wheat germ extract which contains N-myristoyltransferase (NMT) activity (Heuckeroth et al., 1988). As shown in Figure 1B, radioactivity was incorporated into the wild-type Ara6 but not the mutant Ara6, indicating that Gly2 is indeed the site of N-myristoylation. The palmitoylation of Ara6 was also observed when Ara6 protein synthesized in vitro was incubated with the Arabidopsis lysate and 14C-labeled palmitoyl-CoA. Incorporation of radioactivity into the Ara6C3S mutant protein was not observed, indicating that Ara6 is indeed palmitoylated at Cys3 (Figure 1C). The Ara6G2A mutant was not palmitoylated either, showing that myristoylation of Ara6 is a prerequisite for palmitoylation.
Next, we examined the membrane association of Ara6. Arabidopsis suspension cultured cells were harvested at a log phase, homogenized and centrifuged at 1500 g. The supernatant (S1.5) was centrifuged further at 100 000 g for 1 h to separate the crude membrane fraction (P100) from the cytosol (S100), which were subjected to immunoblotting analysis. As indicated in Figure 1D, most of the Ara6 protein was detected in the membrane fraction. A small amount was also found in the cytosol, as is the case for a conventional-type Ara protein, Ara4. Ara6 was not extracted from membranes by salt but was partially extracted by urea or alkali, and almost completely by Triton X-100 (Figure 1E). This property appears to be a little different from the case of Ara4, in which extrac tion by urea or alkali is less efficient. Such a weaker membrane association of Ara6 may reflect its unique lipid modification.
As an authentic Rab5 homolog in Arabidopsis, we cloned a novel cDNA by RT–PCR and named it ARA7 (DDBJ/EMBL/GenBank accession No. AB038491). Ara7 shares 64.5% identity at the amino acid level to human Rab5c, and is 90.5% identical to Rha1, another Rab5 homolog of Arabidopsis (Anuntalabhochai et al., 1991). In contrast to Ara6, the effector domain sequence of Ara7 and Rha1 is identical to that of Rab5c (Figure 1A), indicating that Ara7 and Rha1 are Rab5 orthologs. In the central part, Ara7 shares 59.6% identity with Ara6.
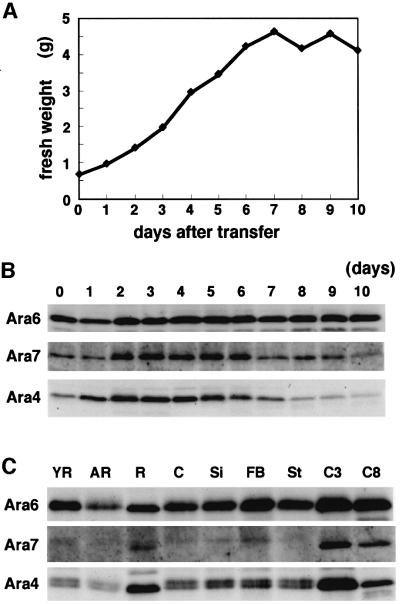
The expression of Ara6 and Ara7 in suspension cultured cells was examined by immunoblotting analysis. As shown in Figure 2A and B, Ara6 was expressed throughout the culture phase, and the level of protein was somewhat higher in the stationary phase. In contrast, Ara7 was highly expressed only when cells were actively dividing. A similar expression pattern was observed for Ara4, although the peak of expression was noted a little earlier than that of Ara7. Figure 2C shows the expression of these proteins in different organs of Arabidopsis. Ara6 was ubiquitously expressed in all organs examined, whereas Ara7 and Ara4 showed organ specificities. Ara7 and Ara4 were expressed abundantly in roots and suspension cultured cells. These results suggest that Ara6 has a more general role than Ara7 in plant cells.
Fig. 2. Expression of Ara6, Ara7 and Ara4 in Arabidopsis suspension cultured cells or plants. (A) The Arabidopsis suspension cultured cells were collected every day after transfer to the new medium for 10 days, and fresh weights were plotted. (B) The total protein samples prepared from a series of suspension cultured cells were subjected to immunoblotting analysis with anti-Ara6, anti-Ara7 or anti-Ara4 antibody. (C) The total protein samples were prepared from young rosettes (YR), adult rosettes (AR), roots (R), cotyledons (C), siliques (Si), flower-bud clusters (FB) and stems (St). These samples and total lysates from suspension cultured cells were collected over 3 (C3) or 8 days (C8) after transfer and were subjected to immunoblotting analysis with anti-Ara6, anti-Ara7 or anti-Ara4 antibody.
Cytosolic pool of Ara6 is independent of GDI
GDI is the best known regulator molecule for the Rab/Ypt GTPases. GDI controls both the nucleotide state and the subcellular localization of Rab/Ypt proteins (Pfeffer et al., 1995). The isoprenylation of Rab/Ypt proteins at their C-terminus is essential for their recognition by GDI (Araki et al., 1991; Musha et al., 1992; Soldati et al., 1993; Ueda et al., 2000). Since Ara6 lacks the C-terminal cysteine motif, its regulation should be independent of GDI. This was tested by using a yeast expression system. As we described previously, Arabidopsis GDI, AtGDI1, can replace the function of yeast GDI, Gdi1/Sec19 (Ueda et al., 1996b). Yeast temperature-sensitive sec19/gdi1 mutant cells transformed with AtGDI1 under the control of the galactose-inducible GAL1 promoter depend on the function of AtGDI for growth at high temperature. These cells were transformed further with a plasmid harboring ARA6 or ARA7 under the control of the constitutive TDH3 promoter. If GDI serves to recycle Ara proteins from membranes to the cytosol, depletion of GDI should lead to a decrease of Ara proteins in the soluble pool. The above transformant cells were cultured in galactose medium at 23°C for 6 h, shifted to 37°C and cultured for 4 h to inactivate yeast GDI, and then transferred to glucose medium to deplete AtGDI1. Cells were collected after 16 h culture at 37°C, lysed, separated into supernatants and pellets, and then analyzed for Ara6 or Ara7 by immunoblotting (Figure 3). When all GDI was depleted in glucose, the soluble pool of Ara7 was almost completely lost, indicating that AtGDI1 indeed serves to recycle Ara7. In contrast, almost the same amount of Ara6 was detected in the supernatant regardless of GDI depletion. Thus, GDI is not required for maintenance of the soluble pool of Ara6, suggesting that Ara6 has a unique system for the regulation of its GTPase cycle.
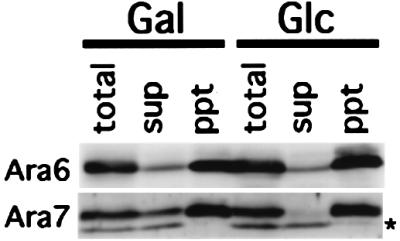
Fig. 3. Recycling of Ara6 is independent of GDI in yeast cells. ARA6 or ARA7 was expressed constitutively in yeast sec19/gdi1 mutant cells harboring AtGDI1 under control of the GAL1 promoter. Transformant cells were cultured in galactose medium at the permissive temperature, shifted up to the restrictive temperature and then transferred to new galactose medium (Gal) or glucose medium (Glc). The total cell lysates (total) were ultracentrifuged to obtain cytosol (sup) and membrane (ppt) fractions, and samples were subjected to immunoblotting analysis with anti-Ara6 or anti-Ara7 antibody. The asterisk indicates a cross-reacting band which is not related to Ara7.
Ara6 functions in the endocytic pathway
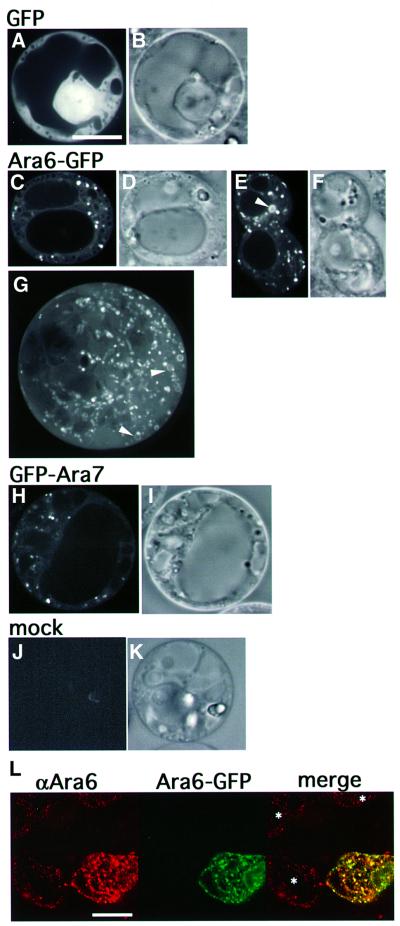
To examine the subcellular localization of Ara6, we transiently expressed a fusion protein of Ara6 and green fluorescent protein (GFP) in protoplasts prepared from Arabidopsis suspension cultured cells. The GFP fluorescence was observed with a high-speed confocal laser scanning microscope developed in our laboratory. Still images of Ara6–GFP expressed under the control of the constitutive 35S promoter are shown in Figure 4C, E and G. Fluorescent dots moved around in the cytoplasm very rapidly. They did not look like the Golgi apparatus, because GFP-tagged Golgi proteins give larger signals in Arabidopsis cells that move much more slowly (Takeuchi et al., 2000). Ring-like structures were also observed, often in association with a bright dot along the ring (Figure 4E and G). In addition, faint fluorescence was also recognized on the plasma membrane and the endoplasmic reticulum (ER) in some cells (Figure 4C). No fluorescence was observed in cells that were not treated with plasmids (Figure 4J). To verify that the punctate localization of Ara6–GFP reflects the localization of endogenous protein, immunofluorescence analysis was carried out. To obtain the dual-color image, the cells were viewed with a confocal laser microscope fitted with a spectrophotometer for emission band wavelength selection. The anti-Ara6 antibody and Ara6–GFP stained the identical punctate organelles in transformed cells, and the staining pattern by the anti-Ara6 antibody was very similar between transformed and untransformed cells (Figure 4L). These results led us to conclude that Ara6–GFP is localized correctly in Arabidopsis cells.
Fig. 4. Subcellular localization of Ara6–GFP and GFP–Ara7. ARA6 and ARA7 were tagged with GFP and expressed transiently in the protoplasts of Arabidopsis suspension cultured cells. (A and B) An Arabidopsis cell expressing GFP. (C–G) Ara6–GFP was localized on motile dots and ring-like organelles (arrowheads). (H and I) GFP–Ara7 was localized predominantly on the motile dotted organelles. (J and K) Protoplasts not treated with the plasmid showed no fluorescence. (A, C, E, H and J) Confocal images. (B, D, F, I and K) Nomarski images. (G) Projection of serial confocal images taken at 0.25 mm intervals. (B–K) Same scale as (A). Bar, 10 µm. (L) Distribution of Ara6 examined by immunofluorescence analysis. Protoplasts, some of which are expressing ARA6–GFP, were fixed and stained with the anti-Ara6 antibody. Asterisks indicate untransformed cells. Bar, 10 µm.
For comparison, we examined the localization of Ara7 tagged with GFP at the N-terminus (GFP–Ara7). If Ara7 is a true counterpart of Rab5, it is expected to reside on early endosomes. As shown in Figure 4H, GFP–Ara7 was localized predominantly on the rapidly moving punctate organelles. Ring-like structures were observed occasionally. Thus, Ara6–GFP and GFP–Ara7 yielded very similar fluorescent structures, which were most probably early endosomes.
The lipophilic styryl dye, FM4-64, is known to follow the endocytic pathway, from the plasma membrane via endosomes to the vacuole, when exogenously added to yeast cells (Vida and Emr, 1995). We tried to use this chemical to analyze endocytosis in plant cells. The protoplasts of the Arabidopsis suspension culture cells were incubated with 50 µM FM4-64 for 10 min, washed and resuspended in the culture medium containing 0.4 M mannitol. Just after labeling, FM4-64 stained the plasma membrane (Figure 5A) and invaginations of the plasma membrane in some populations of cells (Figure 5B). Half an hour later, this dye was internalized and localized to small punctate structures in almost all cells (Figure 5C and E), whose size and motility were almost indistinguishable from those visualized by Ara6–GFP and GFP–Ara7. Ring-like structures emerged following the punctate organelles (Figure 5C and E), which were also seen in the case of Ara6–GFP and GFP–Ara7. At 3 h after labeling, vacuolar membranes became fluorescent and, at 8 h, >85% of cells showed vacuolar membrane staining (Figure 5E). After 16 h of incubation, FM4-64 was completely transported to the vacuole (Figure 5D). These results strongly support the notion that Ara6 and Ara7 are localized to the organelles in the early step of the endocytic pathway.
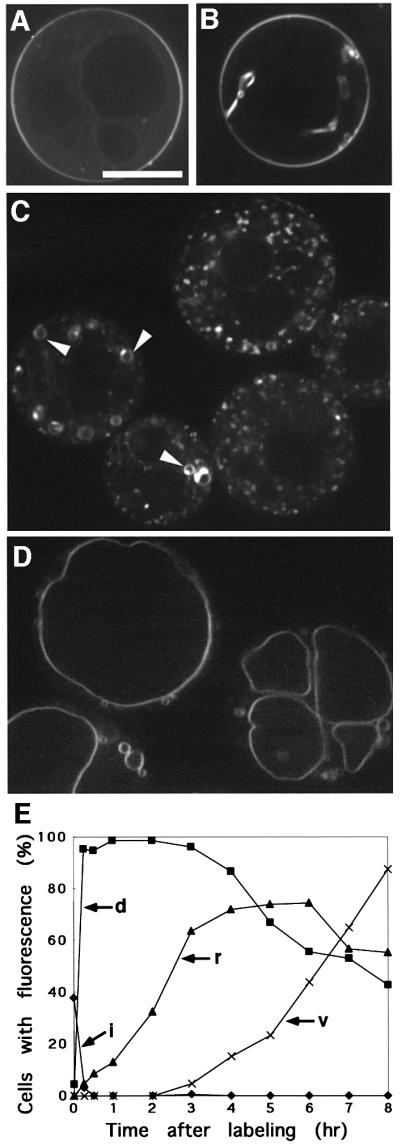
Fig. 5. Endocytic pathway of the Arabidopsis cell visualized with FM4-64. FM4-64 was internalized from the plasma membrane (A and B), via motile dotted organelles (C) and ring-like organelles (C, arrowheads), to the vacuole (D). Bar, 10 µm. (E) Time course of the internalization of FM4-64. Percentages of cells with stained plasma membrane invaginations (i), dotty endosomes (d), ring-like structures (r) and vacuole (v) are plotted.
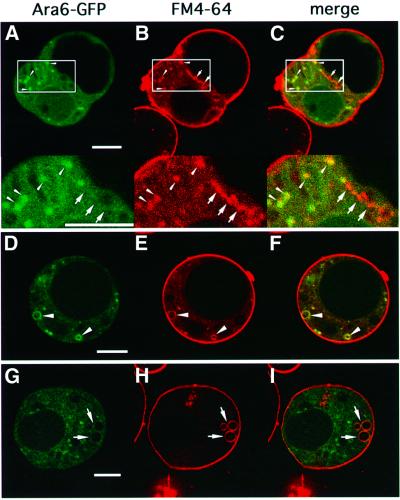
To confirm the endosomal localization of Ara6–GFP, we examined Ara6–GFP and FM4-64 simultaneously. Arabidopsis protoplasts transiently expressing Ara6–GFP were labeled with FM4-64, incubated for 30 min and viewed with a confocal microscope. At this time point, FM4-64 stained only the organelles in the early endocytic pathway (Figure 5E). As shown in Figure 6A–F, the small motile dots and the ring-like structures visualized by Ara6–GFP were both stained with FM4-64. This demonstrates that the organelles where Ara6–GFP resides are indeed early endosomes. Interestingly, Ara6–GFP was not localized on all early endosomes visualized by FM4-64 (Figure 6A–C). Ring-like structures, which were marked by FM4-64 but barely stained by Ara6–GFP, were also observed (Figure 6G–I). These findings suggest that Arabidopsis has at least two distinctive populations of early endosomes.
Fig. 6. Ara6–GFP resides on early endosomes. Protoplasts of Arabidopsis cultured cells expressing Ara6–GFP were labeled with FM4-64. (A–C) The motile dots with Ara6–GFP (arrowheads) were also stained by FM4-64. Note that some dots labeled by FM4-64 were not marked by Ara6–GFP (arrows). The lower panels are high magnification images of the boxed regions. (D–F) Ring-like organelles where Ara6–GFP resides (arrowheads) were also labeled with FM4-64. (G–I) Some structures (arrows) were marked by FM4-64 but barely by Ara6–GFP. Bar, 10 µm.
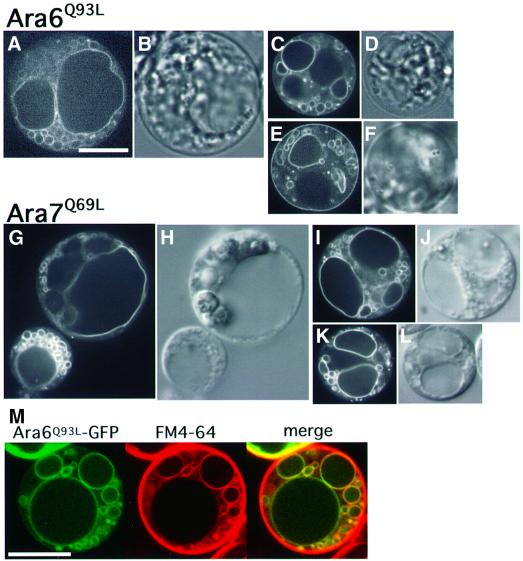
Next we investigated whether the mutations of Ara6 and Ara7 that alter their characteristics as GTPases affect their intracellular localization. We first constructed GFP fusions of Ara6Q93L and Ara7Q69L, which correspond to RasQ61L (Der et al., 1986) and are expected to be stabilized in the GTP-bound state. When these fusion proteins were expressed in Arabidopsis cells, predominant fluorescence was observed on aggregates of ring-like structures, the vacuole membrane and small dots (Figure 7). Fluores cence on the plasma membrane was also observed in some populations of cells expressing GFP–Ara6Q93L (Figure 7E). In contrast, Ara7Q69L–GFP never stained the plasma membrane. We again performed a double-labeling experiment, and confirmed that aggregating vesicles were positive for both markers before the large central vacuole became stained with FM4-64 (Figure 7M). These results suggest that the constitutive active mutants of Ara6 and Ara7 accelerate the fusion of early endosomes to form aggregates of larger membrane structures. These aggregating membranes may well represent late endosomes or the pre-vacuolar compartment, which are promoting fusions further and forming even larger vacuoles. Alternatively, this fluorescence may reflect faulty recycling of the mutant proteins. In the case of mammalian cells, the dominant-active mutant of Rab5 (Rab5Q79L), which is known as a key regulator of homotypic fusion of early endosomes, also promotes the formation of larger endosomes when overexpressed (Stenmark et al., 1994).
Fig. 7. GTP-bound mutant of Ara6 (Ara6Q93L) or Ara7 (Ara7Q69L) tagged by GFP resides on aggregates of ring-like structures, dots and the vacuole membrane. (A–F) Protoplasts of Arabidopsis suspension cultured cells expressing Ara6Q93L–GFP. (G–L) Protoplasts of Arabidopsis suspension cultured cells expressing Ara7Q69L–GFP. (A, C, E, G, I and K) Confocal images. (B, D, F, H, J and L) Nomarski images. (B–L) Same scale as (A). Bar, 10 µm. (M) The protoplast expressing Ara6Q93L was labeled with FM4-64 and viewed after 30 min. Bar, 10 µm.
Taken together, the above results led us to propose that the novel Rab/Ypt protein, Ara6, functions as a regulator of membrane fusion in the endocytic pathway of Arabidopsis cells. Ara7 should also be a regulator of endocytic membrane fusion. Currently we are examining whether Ara6 and Ara7 function on the same endosome. The endosomal localization of Ara7 seems more stringent than that of Ara6. We will discuss this point further below. The dominant effects of the GTP-bound mutants also suggest that GFP fusions of Ara6 and Ara7 retain their functions, as is the case for other Rab–GFP fusion proteins (Roberts et al., 1999; White et al., 1999; Bucci et al., 2000; Sönnichsen et al., 2000).
N-terminal modification is essential for correct localization of Ara6
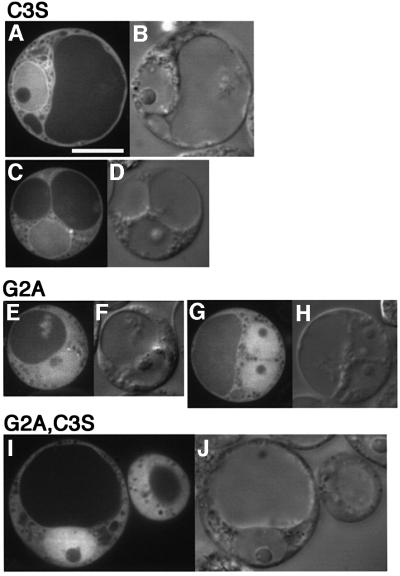
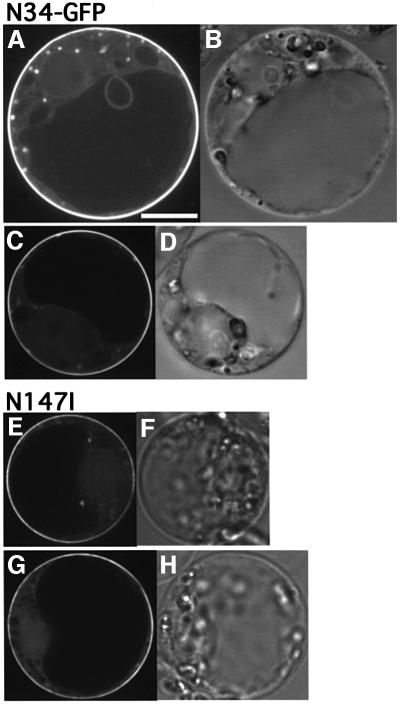
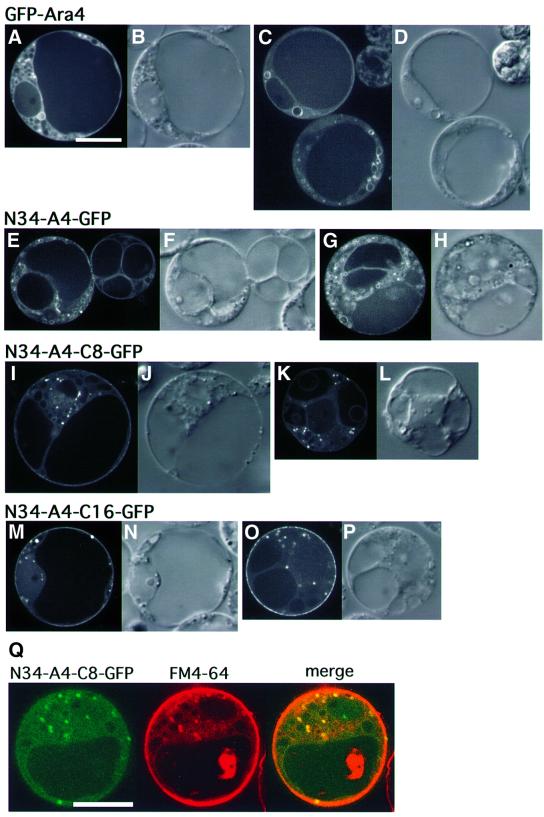
We also examined the role of N-terminal fatty acylation in the subcellular localization of Ara6. We constructed GFP-tagged mutant Ara6 which contained an amino acid substitution in Gly2 and/or Cys3, and expressed them transiently in Arabidopsis cells. Ara6C3S–GFP, which has Gly2 where N-myristoylation occurs but cannot be a substrate for palmitoylation, was localized mainly on the ER, but not on the endosomes (Figure 8A and C). This result indicates that N-myristoylation can direct Ara6 to the membrane, but palmitoylation is essential for the correct localization to the endosomes. Both Ara6G2A–GFP and Ara6G2A,C3S–GFP were localized in the cytosol and nucleus (Figure 8E–J), which is the same pattern as found for free GFP (Figure 4A), indicating that N-myristoylation is essential for membrane binding. Thus, Ara6 is attached to the membrane via N-terminal fatty acylation, and both N-myristoylation and palmitoylation are essential for localization of Ara6 on the endosomes. Then, is the N-terminal region sufficient to locate Ara6 on the endosomes? To answer this question, we analyzed whether the N-terminal 34 amino acids of Ara6 containing fatty acylation sites could direct the GFP fusion protein to the endosomes. The N-terminal 34 amino acids of Ara6 were added to the N-terminus of GFP (N34–GFP), which was then expressed in the suspension cultured cells. As shown in Figure 9A–D, it was localized mainly on the plasma membrane and occasionally on punctate structures. These results indicate that the N-terminal region of Ara6 is not sufficient for the predominant endosomal localization.
Fig. 8. N-terminal fatty acylation is essential for the correct localization of Ara6. (A–D) Protoplasts of Arabidopsis cells expressing Ara6C3S–GFP, which retain Gly2 to be N-myristoylated but which cannot be the substrate for palmitoylation. (E–H) Protoplasts of Arabidopsis cells expressing Ara6G2A–GFP, which cannot be myristoylated. (I and J) Protoplasts of Arabidopsis cells expressing Ara6G2A,C3S–GFP, a double mutant of fatty acylation sites. (A, C, E, G and I) Confocal images. (B, D, F, H and J) Nomarski image. Bar, 10 µm.
Fig. 9. (A–D) The N-terminal region of Ara6 is not sufficient for endosomal localization. A fusion of the N-terminal 34 amino acids of Ara6 with GFP (N34–GFP) was expressed transiently in Arabidopsis suspension cultured cells. (E–H) Nucleotide binding is required for the endosomal localization of Ara6. The nucleotide-free mutant of Ara6 tagged by GFP, Ara6N147I–GFP, was expressed transiently in Arabidopsis suspension cultured cells. (A, C, E and G) Confocal images. (B, D, F and H) Nomarski images. Bar, 10 µm.
The C-terminal domain of Ara6 plays a crucial role in its localization
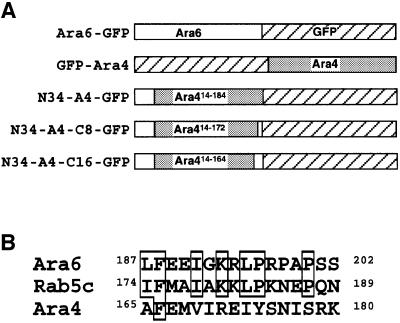
As hitherto demonstrated, the N-terminal sequence of Ara6 is important but not sufficient for localization on early endosomes. In the mutational analysis of Ara6, we found that the nucleotide-free mutant of Ara6, Ara6N147I, corresponding to the N116I mutant of RasH, did not localize to early endosomes. Interestingly, Ara6N147I–GFP resided predominantly on the plasma membrane like N34–GFP, suggesting that nucleotide binding is also important for the endosomal localization of Ara6 (Figure 9E–H). To determine the minimal regions of Ara6 that direct endosomal localization, we constructed a chimeric protein consisting of the N-terminal 34 amino acids of Ara6, the central part of Ara4 (from Lys14 to Ser184) and GFP (N34-A4–GFP; Figure 10A). This chimera was not localized to the endosomes but on unknown types of small structures (Figure 11E and G). GFP–Ara4 yielded patterns of ring-like structures and bright dots (Figure 11A and C). These dots most probably represent the Golgi apparatus as reported previously (Ueda et al., 1996a), and the rings may be identical to those visualized by Ara6WT–GFP or FM4-64. The structures on which N34-A4–GFP was localized also often looked like rings, but they were smaller in size than the ring-like FM4-64-stained organelles. The identities of these organelles have yet to be determined.
Fig. 10. (A) Schematic structures of chimeric proteins constructed to determine the minimal region required for endosome localization of Ara6. (B) Sequence comparison among the C-terminal 16 amino acids of Ara6, the corresponding region of human Rab5c and that of Ara4. Conserved amino acids are boxed.
Fig. 11. Subcellular localization of chimeric proteins expressed transiently in the protoplasts of Arabidopsis suspension cultured cells. (A–D) Arabidopsis cells expressing GFP–Ara4. (E–H) Arabidopsis cells expressing N34-A4–GFP. (I–L) Arabidopsis cells expressing N34-A4-C8–GFP. (M–P) Arabidopsis cells expressing N34-A4-C16–GFP. (A, C, E, G, I, K, M and O) Confocal images. (B, D, F, H, J, L, N and P) Nomarski images. (B–P) Same scale as (A). Bar, 10 µm. (Q) The protoplast expressing N34-A4-C8–GFP was labeled with FM4-64. Bar, 10 µm.
As a possible region of Ara6 that contains additional targeting information, we looked at its C-terminal part. The C-terminal hypervariable region is known to be the determinant of localization for conventional Rab/Ypt proteins (Chavrier et al., 1991; Brennwald and Novick, 1993; Dunn et al., 1993). To examine the role of this part, we replaced the C-terminal part of Ara4 in the N34-A4–GFP chimera by the corresponding region of Ara6 containing either eight or 16 amino acid residues (Figure 10B). We expressed these chimeric proteins in Arabidopsis cells, and found that the C-terminal eight amino acids of Ara6 can localize the chimeric protein (N34-A4-C8) to punctate organelles of high motility (Figure 11I and K). These motile organelles were labeled by FM4-64 after 30 min (Figure 11Q), demonstrating that N34-A4-C8 is localized on early endosomes. The chimera with the C-terminal 16 amino acids of Ara6 (N34-A4-C16) was also localized on the motile endosomes, although some populations occasionally marked the plasma membrane (Figure 11M and O). Thus, the C-terminal eight amino acids of Ara6 could locate the chimeric protein to the early endosomes. In this region, Ara6 shows some similarity to Rab5; three out of eight residues are conserved (Figure 10B). This conserved region may contain common information for early endosome localization.
Taking the above results together, Ara6 employs a unique mechanism for localization on early endosomes. N-terminal fatty acylations, nucleotide binding and the C-terminal region all appear to be required to determine the subcellular localization of Ara6 in a coordinated fashion.
Discussion
Membrane binding and subcellular localization of Ara6
The Rab/Ypt family of GTPases, key regulators of membrane fusion, have attracted the interest of many researchers, and much information continues to accumulate on their molecular functions. This family is extremely well conserved among divergent eukaryotes, including vertebrates, invertebrates, fungi and plants, indicating their essential roles in all eukaryotic kingdoms. The structures of Rab/Ypt GTPases are also highly conserved, which was thought to indicate that the rigidity of the structure was essential for their function. For example, C-terminal cysteine residues, which are conserved in virtually all previously known members of this family, and their isoprenylation were shown to be essential for membrane binding, interaction with GDI and, thus, for their functions (Araki et al., 1991; Musha et al., 1992; Soldati et al., 1993; Ueda et al., 2000). The hypervariable region, which is located just before the cysteine motif, was also known to have an important role in their subcellular localization (Chavrier et al., 1991; Brennwald and Novick, 1993). However, Ara6 has neither a cysteine residue at the C-terminus nor a complete hypervariable region; nevertheless, it is localized to the endosomes and probably regulates their fusion. This indicates that C-terminal isoprenylation as a membrane anchor is dispensable at least for this type of plant-specific Rab protein. In the case of Ara6, N-terminal myristoylation and palmitoylation appear to be employed as a substitute membrane anchoring system. Both of these modifications are essential for the correct localization of Ara6. This is different from the case of Arf GTPases, another class of small GTPases regulating vesicular traffic. In the case of Arfs, the myristoylation of the N-terminus is essential for their function, but the Cys3 residue to be palmitoylated is lacking. The non-myristoylated G2A mutant of human Arf6 is functionless, and C-terminal isoprenylation cannot remedy its defect (D’Sousa-Schorey and Stahl, 1995). Thus only N- myristoylation should be sufficient for the correct localization and functions of Arf GTPases. On the other hand, the substitution of isoprenylation by N-myristoylation also abolished the function of Rab5 (Li et al., 1995). The reason for such a difference in the dependence on fatty acylation remains unknown.
GFP-tagged Ara6 and Ara7 are localized predominantly on early endosomes. GFP–Ara6 (both wild-type and mutant versions) resides on the plasma membrane and also on the ER, whereas Ara7 never shows such distribution. The broad range of localization sites exhibited by a series of Ara6–GFP variants would represent different stages of the authentic localization of Ara6. In subcellular localization experiments using sucrose density gradient fractionation, Ara6 was detected in multiple peaks, whereas Ara7 showed a single peak (T.Ueda and A.Nakano, unpublished results). This apparent multi-localization of Ara6 may reflect its targeting pathway. As demonstrated in Figure 9, the fusion protein comprising the N-terminal 34 amino acids of Ara6 and GFP is targeted predominantly to the plasma membrane. In mammalian cells, a myristoylated plus palmitoylated GFP variant is reported to be localized mainly to the plasma membrane (McCabe and Berthiaume, 1999). These results suggest that the plasma membrane is the default destination of such fatty acylated proteins in both plant and animal cells. Ara6C3S–GFP is localized mainly on the ER, suggesting that a myristoylated but not palmitoylated intermediate of Ara6 is formed on the ER. Little is known about the localization of palmitoyl transferase in plants, but it is possible that Ara6 may be targeted from the ER to the plasma membrane to be palmitoylated. Apparently, further information is necessary for Ara6 to be localized to the endosomes. Correct nucleotide binding is critical because the nucleotide-free mutant, Ara6N147I–GFP, is left behind in the plasma membrane. Whereas the GTP-bound mutant Ara6Q93L appears to cause aberrant fusion of endosomes, the GDP-bound mutant Ara6S47N again is located predominantly on the plasma membrane (T.Ueda and A.Nakano, unpublished), suggesting the cycling of Ara6 between the plasma membrane and the endosome during the GTPase cycle.
We have also demonstrated that the C-terminal eight amino acids of Ara6 are very important for its localization. This region shows some similarity between Ara6 and conventional Rab5 proteins. For Rab5, Chavrier et al. (1991) demonstrated that the C-terminal 35 amino acids containing highly variable C-terminal sequence followed by the cysteine motif specify its localization. Not all amino acid residues in this region are required for the determination of its localization, because eight or 13 C-terminal residues are exchangeable with the corresponding residues of Rab7 without affecting the localization on early endosomes. These pieces of evidence imply that Ara6 and Rab5 may share, at least in part, a common mechanism of localization on early endosomes.
The regulation system of Ara6 GTPase is also different from that of other Rab/Ypt proteins. The cytosolic pool of Ara6 is unaffected by depletion of GDI. GDI was reported to interact with all Rab/Ypt GTPases examined previously and to detach them from membranes and keep them soluble until the next GTPase cycle starts. Since GDI requires the C-terminal prenylation of Rab for interaction, it is reasonable that Ara6, which lacks this motif, should employ a different mechanism. Identification of regulatory components that act specifically on Ara6 would provide an exciting clue to understanding the role of such plant-specific Rab GTPases. A preliminary result of a gel filtration experiment indicated that Ara6 forms a large complex in the cytosol (T.Ueda and A.Nakano, unpublished). Characterization of this complex is currently underway. Other screening studies for Ara6-interacting molecules should also reveal how different mechanisms have evolved to regulate this type of Rab GTPases.
Do Ara6 and Ara7 function in the same way?
Both Ara6 and Ara7 appear to regulate fusion of early endosomes. However, several lines of evidence suggest that these molecules are involved in distinctive phenomena. First, temporal and spatial patterns of expression of these molecules are different. Ara6 is expressed constitutively all through the growth phases of suspension cultured cells, and ubiquitously in all organs examined. In contrast, Ara7 is expressed only when cells are actively dividing, and in an organ-specific manner in plant bodies. These expression patterns may imply that Ara6 plays a more general role than conventional Rab5 in vesicular traffic, which is required even in resting cells. The functional difference between Ara6 and Ara7 is also suggested by their amino acid sequences in the effector domain. The effector domain specifies the function of Rab/Ypt proteins (Brennwald and Novick, 1993; Dunn et al., 1993), and is regarded as an marker to classify the members of Rab/Ypt family into subfamilies. The effector domain of Ara7 is identical to that of human Rab5c. In contrast, the effector domain of Ara6 does not coincide with that of any subtype of Rab/Ypt GTPases identified previously, suggesting that Ara6 belongs to a bona fide novel subtype. The three plant homologs of Ara6 have exactly the same effector sequence. Based on this information, we speculate that the Ara6 subfamily is a novel class of Rab/Ypt GTPases which evolved in higher plants in parallel with the Rab5 group to regulate endosome fusion in a distinctive way.
The question remains as to whether Ara6 and Ara7 are on the same endosomes or on different populations. As demonstrated in the double-staining experiment, Arabidopsis cells have at least two distinct populations of early endosomes. One comprises the endosomes in which Ara6 resides, and the other the endosomes to which Ara6 is not targeted. It is an attractive idea that Ara6 and Ara7 may regulate the fusion of different endosomes, and we are planning more elaborate and careful experiments to address this hypothesis. The use of GFP variants with different colors and their simultaneous observation will give us much more conclusive information.
That higher plants have evolved a unique system of endocytic vesicular trafficking stimulates our imagination as to why it is required and how it is utilized. Attempts to answer these questions will be made in planta utilizing forward and reverse genetics.
Materials and methods
Antibody preparation
The full-length open reading frame (ORF) of ARA6 was amplified by PCR to generate BamHI sites at both ends and subcloned into pGEX-2T (Pharmacia Biotech, Piscataway, NJ). This recombinant plasmid, which directs synthesis of the Ara6 protein fused to the C-terminus of GST, was introduced into Escherichia coli. GST–Ara6 was induced by isopropyl-β-d-thiogalactopyranoside (IPTG), purified from the lysate by affinity chromatography on a glutathione–Sepharose 4B column (Pharmacia Biotech) and used to raise a polyclonal antibody in rabbits. The cross-reactivity of this antibody was tested for lysates from yeast cells expressing ARA6 and ARA7. Immunoblotting analysis detected Ara6 but not Ara7. For the anti-Ara7 antibody, a synthetic Leu185–Val194 peptide, in which Ara7 shows no homology to any other known Arabidopsis Rab proteins, was used to immunize rabbits. The antibody obtained was purified by protein G affinity column chromatography (Pharmacia Biotech) and was confirmed to have no cross-reactivity with Ara6.
In vitro modification and fractionation of Ara6
In vitro transcription and translation was carried out with the TNT Coupled Wheat Germ Extract System (Promega, Madison, WI) according to the manufacturer’s instructions. For myristoylation assay, linearized ARA6 and ARA6G2A cloned in the pBSIISK+ vector (Stratagene, La Jolla, CA) were used as templates, and 14C-labeled myristic acid was added to the reaction. After incubation, samples were separated by SDS–PAGE, dried and exposed to the imaging plate for 2 weeks. For palmitoylation assay, linearized ARA6, ARA6C3S and ARA6G2A in pBSIISK+ were used as templates. After the transcription–translation reaction was completed, the Ara6 protein was immunoprecipitated and incubated with the Arabidopsis lysate and 40 µM [14C]palmitoyl CoA (53 mCi/mmol) for 45 min. The immunocomplex was washed, separated by SDS–PAGE, dried and exposed to the imaging plate for 2 weeks. Autoradiograms were visualized with an imaging plate scanner BAS1000 (Fuji Film Co., Tokyo, Japan). As for fractionation, Arabidopsis suspension cultured cells (from Dr M.Umeda) were ground in GR buffer [50 mM HEPES–KOH pH 8.0, 330 mM sorbitol, 2 mM EDTA, 100 µM phenylmethylsulfonyl fluoride (PMSF), 10 µg/ml leupeptin and 10 µg/ml pepstatin A], filtered through four layers of cheesecloth and centrifuged at 1500 g to remove cell debris. Supernatant (S1.5) was centrifuged at 100 000 g for 1 h to separate the crude membrane fraction (P100) from the cytosol (S100). For solubilization of Ara6, the P100 fraction was resuspended in GR buffer plus 1 M NaCl, 2 M urea, 0.1 M Na2CO3 or 1% Triton X-100 and allowed to stand on ice for 1 h. Samples were then centrifuged at 100 000 g for 1 h to separate soluble from insoluble fractions, and subjected to immunoblotting analysis with an anti-Ara6 polyclonal or an anti-Ara4 monoclonal antibody (Anai et al., 1995).
Expression pattern of Ara6 protein
Total cell lysates (S1.5) were prepared as described above from the Arabidopsis suspension cultures collected daily after medium change. Samples were also prepared from young rosettes, adult rosettes, roots, cotyledons, siliques, flower-bud clusters and stems. Proteins (30 µg for Ara6 and Ara4, and 50 µg for Ara7) were separated by SDS–PAGE and analyzed by immunoblotting. The immune complexes were visualized with the ECL+Plus system (Amersham Pharmacia Biotech, Amersham, UK).
Expression of ARA6 and ARA7 in yeast and the effect of GDI depletion
Media for yeast cultures were described previously (Ueda et al., 2000). For the expression of ARA6 and ARA7 in yeast, their ORFs were first placed under control of the constitutive TDH3 promoter in pTU1 (Ueda et al., 2000). The fragment containing the TDH3 promoter, ARA6 or ARA7, and the CMK1 terminator was cut out by BamHI and SalI, and subcloned into pRS315, which is a single-copy plasmid containing the LEU2 marker. Cells of ANS19-4A (MATa sec19-1 ura3 leu2 his) (Nakano and Muramatsu, 1989) were transformed with this plasmid, and further transformed with a plasmid containing AtGDI1 under control of the GAL1 promoter and the URA3 marker (Ueda et al., 1998). Transformants were cultured in minimal galactose medium (MVGS minus uracil and leucine) at 23°C for 6 h, shifted to 37°C for 4 h and then transferred to minimal glucose medium (MVD minus uracil and leucine) or again to minimal galactose medium. Cells were collected after 16 h culture at 37°C, resuspended in Tris-buffered saline (TBS) containing 1% protease inhibitor cocktail (Sigma, St Louis, MO) and vortexed vigorously with glass beads. After centrifugation at 800 g for 5 min to eliminate debris, samples were adjusted to 5 mg protein/ml and centrifuged at 100 000 g for 30 min to obtain supernatant and pellet fractions. Each sample was analyzed by SDS–PAGE and immunoblotting.
Transient transformation of Arabidopsis suspension culture and immunofluorescent labeling
Wild-type and mutant versions of ARA6 and ARA7, which were constructed by PCR-directed mutagenesis, were fused to the ORF of GFP (from Dr Y.Niwa) in the directions 5′-ARA6-GFP-3′ and 5′-GFP-ARA7-3′. The fusion genes were subcloned into pHTS13 (from Dr H.Tsukaya), which is a derivative of pBSIIKS+ containing the cauliflower mosaic virus (CaMV) 35S promoter and the Nos terminator. Transient expression of GFP-fused Ara proteins in Arabidopsis suspension cultured cells was performed by the method described previously (Forreiter et al., 1997), with minor modifications. Cultured cells were incubated in enzyme solution [0.4 M mannitol, 5 mM EGTA, 1% (w/v) cellulase Y-C and 0.05% (w/v) pectolyase Y-23] for 1 h at 30°C under gentle agitation and then passed through a nylon mesh (125 µm pore). Protoplasts were collected by centrifugation at 500 g for 10 min, washed twice with solution A (0.4 M mannitol, 70 mM CaCl2 and 5 mM MES pH 5.7) and resuspended in MaMg solution (0.4 M mannitol, 15 mM MgCl2 and 5 mM MES pH 5.7). After the addition of plasmid and carrier DNA to 100 µl of protoplast solution, 0.4 ml of DNA uptake solution containing 40% (w/v) polyethylene glycol (PEG) 6000, 0.4 M mannitol and 0.1 M Ca(NO3)2 was added. The mixture was placed on ice for 20 min and then diluted with 10 ml of 0.4 M mannitol, 125 mM CaCl2, 5 mM KCl, 5 mM glucose and 1.5 mM MES pH 5.7. Protoplasts were collected by centrifugation at 500 g for 10 min, resuspended in Murashige and Skoog (MS) cell culture medium (Murashige and Skoog, 1962) supplemented with 0.4 M mannitol, and incubated under gentle agitation at 23°C for 16 h in the dark. Transformed cells were observed with a confocal laser microscope system (Olympus BX60 fluorescence microscope equipped with a confocal scanner Model CSU10 of Yokogawa Electric, Tokyo). For immunofluorescent labeling, Arabidopsis protoplasts transiently expressing Ara6–GFP were fixed with 3.7% formaldehyde and permeabilized by 1% NP-40. The cells were incubated in phosphate-buffered saline containing 0.1 M glycine and 1% bovine serum albumin and subsequently with anti-Ara6 antibody. Immunocomplexes were detected by Alexa 568 anti-rabbit IgG (Molecular Probes, Eugene, OR) and observed with a Leica (Heerbrugg, Switzerland) DMIRB/E microscope equipped with a TCS4D confocal scanner, TCS NT software and an Ar/Kr laser.
FM4-64 staining
Protoplasts of Arabidopsis suspension cultured cells were incubated with 50 µM FM4-64 (Molecular Probes) in MS cell culture medium plus 0.4 M mannitol for 10 min on ice, washed with MS medium plus 0.4 M mannitol, and resuspended in the same medium. Cells were incubated for appropriate periods and observed by confocal laser microscopy.
Double staining with GFP and FM4-64
ARA6–GFP or its mutant versions was introduced into the protoplasts of Arabidopsis suspension cultured cells as described above. After 16 h incubation in MS cell culture medium containing 0.4 M mannitol, protoplasts were collected by centrifugation, resuspended in MS cell culture medium containing 0.4 M mannitol and 50 µM FM4-64, and placed on ice for 10 min. After labeling, cells were washed twice, resuspended in the same medium minus FM4-64 and incubated for 30 min at 23°C. Samples were viewed by confocal laser microscopy.
Supplementary data
Supplementary data for this paper are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We are grateful to Masaaki Umeda (University of Tokyo) for the Arabidopsis suspension culture cell line, to Yasuo Niwa (University of Shizuoka) and Hirokazu Tsukaya (National Institute for Basic Biology, Okazaki) for GFP and expression plasmids, respectively, and to Chieko Saito of our laboratory for technical help in microscopy. This work was supported by a Grant-in-Aid from the Ministry of Education, Science, Sports and Culture of Japan, by a Special Coordination Fund for Promotion of Science and Technology from the Science and Technology Agency of Japan, by a grant from the Bioarchitect Project of RIKEN, and by a President’s Special Research Grant from RIKEN.
References
- Anai T., Hasegawa,K., Watanabe,Y., Uchimiya,H., Ishizaki,R. and Matsui,M. (1991) Isolation and analysis of cDNAs encoding small GTP-binding proteins of Arabidopsis thaliana. Gene, 108, 259–264. [DOI] [PubMed] [Google Scholar]
- Anai T., Aspuria,E.T., Fujii,N., Ueda,T., Matsui,M., Hasegawa,K. and Uchimiya,H. (1995) Immunological analysis of a small GTP-binding protein in higher plant cells. J. Plant Physiol., 147, 48–52. [Google Scholar]
- Anuntalabhochai S., Terryn,N., Van Montagu,M. and Inze,D. (1991) Molecular characterization of an Arabidopsis thaliana cDNA encoding a small GTP-binding protein, Rha1. Plant J., 1, 167–174. [PubMed] [Google Scholar]
- Araki S., Kaibuchi,K., Sasaki,T., Hata,Y. and Takai,Y. (1991) Role of the C-terminal region of smg p25A in its interaction with membranes and the GDP/GTP exchange protein. Mol. Cell. Biol., 11, 1438–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte S., Schiene,K. and Dietz,K.J. (2000) Characterization of a small GTP-binding protein of the rab5 family in Mesembryanthemum crystallinum with increased level of expression during early salt stress. Plant Mol. Biol., 42, 923–936. [DOI] [PubMed] [Google Scholar]
- Borg S., Brandstrup,B., Jensen,T.J. and Poulsen,C. (1997) Identification of new protein species among 33 different small GTP-binding proteins encoded by cDNAs from Lotus japonicus and expression of corresponding mRNAs in developing root nodules. Plant J., 11, 237–250. [DOI] [PubMed] [Google Scholar]
- Brennwald P. and Novick,P. (1993) Interactions of three domains distinguishing the Ras-related GTP-binding proteins Ypt1 and Sec4. Nature, 362, 560–563. [DOI] [PubMed] [Google Scholar]
- Bucci C., Thomsen,P., Nicoziani,P., McCarthy,J. and van Deurs,B. (2000) Rab7: a key to lysosome biogenesis. Mol. Biol. Cell, 11, 467–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavrier P., Grovel,J.P., Stelzer,E., Simons,K., Gruenberg,J. and Zerial,M. (1991) Hypervariable C-terminal domain of rab proteins acts as a targeting signal. Nature, 353, 769–772. [DOI] [PubMed] [Google Scholar]
- Der C.J., Finkel,T. and Cooper,G.M. (1986) Biological and biochemical properties of human rasH genes mutated at codon 61. Cell, 44, 167–176. [DOI] [PubMed] [Google Scholar]
- Dunn B., Stearns,T. and Botstein,D. (1993) Specificity domains distinguish the Ras-related GTPases Ypt1 and Sec4. Nature, 362, 563–565. [DOI] [PubMed] [Google Scholar]
- D’Sousa-Schorey C. and Stahl,P.D. (1995) Myristoylation is required for the intracellular localization and endocytic function of ARF6. Exp. Cell Res., 221, 153–159. [DOI] [PubMed] [Google Scholar]
- Forreiter C., Kirschner,X. and Nover,L. (1997) Stable transformation of an Arabidopsis cell suspension culture with firefly luciferase providing a cellular system for analysis of chaperone activity in vivo. Plant Cell, 9, 2171–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuckeroth R.O., Towler,D.A., Adams,S.P., Glaser,L. and Gordon,J.I. (1988) 11-(Ethylthio)undecanoic acid. A myristic acid analogue of altered hydrophobicity which is functional for peptide N-myristoylation with wheat germ and yeast acyltransferase. J. Biol. Chem., 263, 2127–2133. [PubMed] [Google Scholar]
- Li G., Barbieri,M.A. and Stahl,P.D. (1995) Myristoylation cannot functionally replace the isoprenylation of Rab5. Arch. Biochem. Biophys., 316, 529–534. [DOI] [PubMed] [Google Scholar]
- Ma H. (1994) GTP-binding proteins in plants: new members of an old family. Plant Mol. Biol., 26, 1611–1636. [DOI] [PubMed] [Google Scholar]
- McCabe J.B. and Berthiaume,L.G. (1999) Functional roles for fatty acylated amino-terminal domains in subcellular localization. Mol. Biol. Cell, 10, 3771–3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez O. and Goud,B. (1998) Rab proteins. Biochim. Biophys. Acta, 1404, 101–112. [DOI] [PubMed] [Google Scholar]
- Matsui M., Sakamoto,S., Kunieda,T., Nomura,N. and Ishizaki,R. (1989) Cloning of ara, a putative Arabidopsis thaliana gene homologous to the ras-related gene family. Gene, 76, 313–319. [DOI] [PubMed] [Google Scholar]
- Murashige T. and Skoog,F. (1962) A revised medium for rapid growth and bioassay with tobacco tissue cultures. Physiol. Plant., 15, 473–497. [Google Scholar]
- Musha T., Kawata,M. and Takai,Y. (1992) The geranylgeranyl moiety but not the methyl moiety of the smg25A/rab3A protein is essential for the interactions with membrane and its inhibitory GDP/GTP exchange protein. J. Biol. Chem., 267, 9821–9825. [PubMed] [Google Scholar]
- Nakano A. and Muramatsu,M. (1989) A novel GTP-binding protein, Sar1p, in involved in transport from the endoplasmic reticulum to the Golgi apparatus. J. Cell Biol., 109, 2677–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P. and Zerial,M. (1997) The diversity of Rab proteins in vesicle transport. Curr. Opin. Cell Biol., 9, 496–504. [DOI] [PubMed] [Google Scholar]
- Pan J.Y. and Wessling-Resnick,M. (1998) GEF-mediated GDP/GTP exchange by monomeric GTPases: a regulatory role for Mg2+? Bioessays, 20, 516–521. [DOI] [PubMed] [Google Scholar]
- Pfeffer S.R., Dirac-Svejstrup,B. and Soldati,T. (1995) Rab GDP dissociation inhibitor: putting Rab GTPases in the right place. J. Biol. Chem., 270, 17057–17059. [DOI] [PubMed] [Google Scholar]
- Roberts R.L., Barbieri,M.A., Pryse,K.M., Chua,M., Morisaki,J.H. and Stahl,P.D. (1999) Endosome fusion in living cells overexpressing GFP–rab5. J. Cell Sci., 112, 3667–3675. [DOI] [PubMed] [Google Scholar]
- Scheffzek K., Ahmadian,M.R. and Wittinghofer,A. (1998) GTPase-activating proteins: helping hands to complement an active site. Trends Biochem. Sci., 23, 257–262. [DOI] [PubMed] [Google Scholar]
- Soldati T., Riederer,M.A. and Pfeffer,S.R. (1993) Rab GDI: a solubilizing and recycling factor for rab9 protein. Mol. Biol. Cell, 4, 425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sönnichsen B., De Renzis,S., Nielsen,E., Rietdorf,J. and Zerial,M. (2000) Distinct membrane domains on endosomes in the recycling pathway visualized by multicolor imaging of Rab4, Rab5 and Rab11. J. Cell Biol., 149, 901–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark H., Parton,R.G., Steele-Mortimer,O., Lütcke,A., Gruenberg,J. and Zerial,M. (1994) Inhibition of rab5 GTPase activity stimulates membrane fusion in endocytosis. EMBO J., 13, 1287–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi M., Ueda,T., Sato,K., Abe,H., Nagata,T. and Nakano,A. (2000) A dominant mutant of Sar1 GTPase inhibits protein transport from the endoplasmic reticulum to the Golgi apparatus in tobacco and Arabidopsis cultured cells. Plant J., 23, 517–525. [DOI] [PubMed] [Google Scholar]
- Uchimiya H., Anai,T., Aspuria,E.T., Matsui,M., Nakano,A. and Ueda,T. (1998) The biological roles of small GTPases and interacting proteins in plants. J. Plant Res., 111, 257–260. [Google Scholar]
- Ueda T., Anai,T., Tsukaya,H., Hirata,A. and Uchimiya,H. (1996a) Characterization and subcellular localization of a small GTP-binding protein (Ara4) from Arabidopsis: conditional expression under control of the promoter of the gene for heat-shock protein HSP81-1. Mol. Gen. Genet., 250, 533–539. [DOI] [PubMed] [Google Scholar]
- Ueda T., Matsuda,N., Anai,T., Tsukaya,H., Uchimiya,H. and Nakano,A. (1996b) An Arabidopsis gene isolated by a novel method for detecting genetic interaction in yeast encodes the GDP dissociation inhibitor of Ara4 GTPase. Plant Cell, 8, 2079–2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda T., Yoshizumi,T., Anai,T., Matsui,M., Uchimiya,H. and Nakano,A. (1998) AtGDI2, a novel Arabidopsis gene encoding a Rab GDP dissociation inhibitor. Gene, 206, 137–143. [DOI] [PubMed] [Google Scholar]
- Ueda T., Matsuda,N., Uchimiya,H. and Nakano,A. (2000) Modes of interaction between the Arabidopsis Rab protein, Ara4 and its putative regulator molecules revealed by a yeast expression system. Plant J., 21, 341–349. [DOI] [PubMed] [Google Scholar]
- Verma D.P.S., Cheon,C.I. and Hong,Z. (1994) Small GTP-binding proteins and membrane biogenesis in plants. Plant Physiol., 106, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vida T.A. and Emr,S.D. (1995) A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol., 128, 779–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J. et al. (1999) Rab6 coordinates a novel Golgi to ER retrograde transport pathway in live cells. J. Cell Biol., 147, 743–759. [DOI] [PMC free article] [PubMed] [Google Scholar]