Abstract
DNA replication is the process by which cells make one complete copy of their genetic information before cell division. In bacteria, readily identifiable DNA sequences constitute the start sites or origins of DNA replication. In eukaryotes, replication origins have been difficult to identify. In some systems, any DNA sequence can promote replication, but other systems require specific DNA sequences. Despite these disparities, the proteins that regulate replication are highly conserved from yeast to humans. The resolution may lie in a current model for once-per-cell-cycle regulation of eukaryotic replication that does not require defined origin sequences. This model implies that the specification of precise origins is a response to selective pressures that transcend those of once-per-cell-cycle replication, such as the coordination of replication with other chromosomal functions. Viewed in this context, the locations of origins may be an integral part of the functional organization of eukaryotic chromosomes.
Transmission of genetic information from one cell generation to the next requires the accurate and complete duplication of each DNA strand exactly once before each cell division. Typically, this process begins with the binding of an “initiator” protein to a specific DNA sequence or “replicator.” In response to the appropriate cellular signals, the initiator directs a local unwinding of the DNA double helix and recruits additional factors to initiate the process of DNA replication. This paradigm describes most of the currently tractable replication systems and, although derived from prokaryotic and viral systems, there is no compelling reason to doubt that it will apply to all eukaryotic organisms. In fact, the proteins that regulate replication are highly conserved from yeast to humans, including the origin recognition complex (ORC), which binds directly to replication origin sequences in budding yeast (1, 2). However, in several eukaryotic replication systems, it appears that any DNA sequence can function as a replicator. Those outside the field are often perplexed as to how investigators of different eukaryotic systems can work with assumptions that range from very specific to completely random origin sequence recognition, yet all agree on the basic mechanism regulating DNA replication. This review summarizes our current understanding of eukaryotic replication origins and then presents some simple guidelines to help demystify these seemingly disparate observations, providing a framework for understanding eukaryotic origins that includes all existing data.
Conserved Initiator, Divergent Replicator
The structure of replication origins that have been subjected to genetic analysis in various eukaryotic species is summarized in Fig. 1. In Saccharomyces cerevisiae, origins [autonomously replicating sequences, (ARS)] consist of an essential 11-base pair (bp) ARS consensus sequence (ACS) and several additional elements that contribute to initiation activity and are interchangeable between origins but are not conserved in sequence. Several single–base pair mutations in the ACS can abolish initiation activity (3, 4). Saccharomyces cerevisiae ORC binds in vivo to ARS elements throughout the cell cycle, and purified ORC binds specifically to the ACS in an ATP-dependent manner (2, 5). High-resolution mapping of ARS1 has delimited the initiation point to a single nucleotide adjacent to the ORC binding site (6). In Schizosaccharomyces pombe, origins are much larger and consist of multiple elements that contribute partially to origin activity (2, 7). Although these elements do not share a consensus sequence, they contain asymmetric AT stretches and can be replaced with artificial asymmetric AT stretches (e.g., A40) that reconstitute full origin activity (7). In vivo, S. pombe ORC associates with at least two separate fragments within the ARS 2004 (8).
Fig. 1.
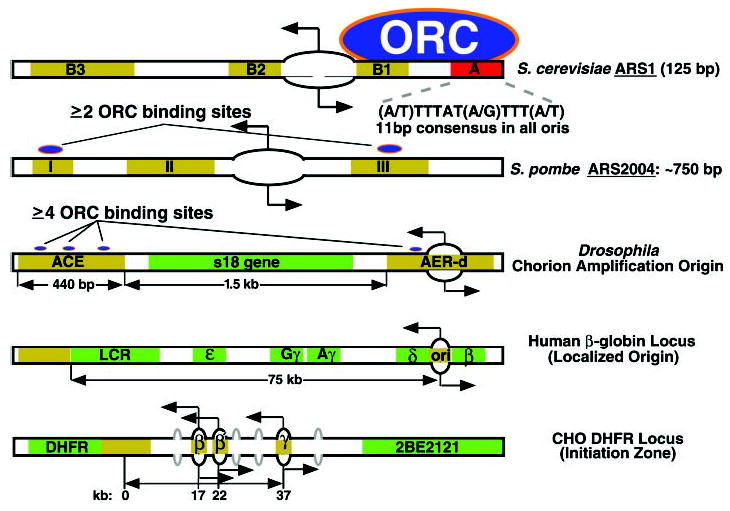
Structure of genetically dissected replication origins in eukaryotes. Consensus ORC binding sites are indicated in red. Additional sequences important for origin activity are shown in brown. Transcription units and regulatory sequences are shown in green. Sites of ORC binding, where known, are indicated. Sites of initiation of replication are indicated with a bidirectional arrow passing through a bubble.
In Drosophila, the elements required for amplification of the chorion genes have been analyzed extensively. Although the process of gene amplification involves re-replicating the same DNA segment many times within one cell cycle, initiation requires many of the same proteins as normal chromosomal replication does, including ORC (9). Amplification requires the 440-bp amplification control element (ACE), whereas the extent of amplification is stimulated by the presence of several amplification-enhancing elements (AERs), of which only the most origin-proximal (AER-d) is shown in Fig. 1. ORC associates with both ACE and AER-d in vivo, and purified Drosophila ORC exhibits preferential ATP-dependent binding to a fragment containing AER-d and three separate DNA fragments within the ACE (10). It is intriguing that replication initiates almost exclusively within the dispensable AER-d (11–13). Furthermore, when multiple tandem copies of the ACE are integrated at ectopic sites, they recruit additional ORCs to chromatin sites in the flanking DNA that are not otherwise occupied by ORC (10). The tandem ACEs promote amplification at these ectopic sites, but the primary initiation sites appear to reside in the flanking DNA (14, 15). Taken together, the ACE appears to contain several ORC binding sites that promote the interaction of ORC and the initiation of replication at specific adjacent sites.
Replication origins in multicellular organisms (metazoa) generally conform to one of two patterns. At some loci, initiation sites are localized to within a few kilobases. At other loci, multiple dispersed origins can be identified throughout “initiation zones” of 10 to 50 kb. Preliminary genetic dissection has been carried out at two of these loci, one representative of each class (Fig. 1). At the human β-globin locus, replication initiates within a few kilobases located between the adult δ- and β-globin genes. However, deletions of sequences greater than 50 kb from the origin, as well as deletions within the initiation site itself, abolish the activity of this origin (16, 17). When the β-globin origin is transferred to an ectopic site, it can direct site-specific initiation of replication, and this activity is dependent on specific segments of DNA within the 8-kb transferred fragment (18). Other loci where initiation sites appear to be confined to within a few kilobases have been identified [e.g., (19, 20)]. In fact, at the lamin B2 locus in human cells, a single nucleotide demarcates the major transition between leading and lagging DNA synthesis (20).
The Chinese hamster ovary (CHO) dihydrofolate reductase (DHFR) locus is representative of the second class of metazoan origins. The pattern of initiation sites at this locus has been the subject of much controversy. On the one hand, high-resolution mapping of the locations of small nascent DNA strands revealed only a single origin throughout a 6.1-kb fragment (21). On the other hand, two-dimensional gel electrophoresis (2D gel) analysis of replication intermediates identified DNA structures representing replication bubbles throughout the entire 55-kb intergenic zone between the DHFR gene and an adjacent gene (2BE2121) of unknown function (22). These results can be reconciled by considering the nature of the data obtained with each origin mapping technique. The 2D-gel method can search a larger area for initiation activity but cannot accurately discern the number or precise location of initiation sites within a fragment. By contrast, small nascent strand detection analyzes a focused area in detail. When the latter method was extended to cover an additional 6 kb of DNA, a second initiation site was revealed approximately 5 kb from the first (23). The 2D-gel method predicts that many more such sites will eventually be identified, constituting an initiation zone. Similar broad initiation zones have been identified at other metazoan loci (24–26).
Genetic analysis at the DHFR locus suggests the existence of specific elements that influence origin activity. When the most active origin is deleted (ori-β), adjacent replication origins retain or increase their activity (27). However, deletion of sequences near the 3′ end of the DHFR gene renders the entire locus inactive for early S phase–initiation activity. Like the β-globin origin, DHFR ori-β retains initiation activity when moved to ectopic sites, and deletions of specific sequences within ori-β can influence this activity (28). Thus, origins found in both broad and localized initiation regions contain specific sequences favorable for initiating DNA replication. However, the number and distribution of origins varies considerably at different loci.
Perhaps the most enigmatic aspect of the field is that, in many eukaryotic systems, replication seems to initiate within any DNA sequence. It appears that any cloned plasmid DNA will replicate autonomously in Caenorhabditis elegans (29) and Paramecium (30). In cultured animal cells, systematic searches for ARS elements analogous to those successfully carried out in yeast have generally failed to identify specific sequences that confer a significant replication advantage when reintroduced into cells (31). In one study with human cells, virtually every DNA fragment greater than 15 kb promoted autonomous and once-per-cell-cycle replication with equal efficiency, and initiation sites were distributed throughout the plasmid sequences (32). Similar results were obtained in cultured Drosophila cells (33). In Xenopus and Drosophila embryos, any DNA sequence will efficiently replicate once per cell cycle up to the blastula stage of development whether microinjected into embryos or introduced into egg extracts (34). Likewise, replication of embryonic chromosomes appears to initiate within any DNA sequence and does not become focused to specific sites until the midblastula transition, when transcription and differentiation commence (35, 36). Despite a random origin site selection, in both Xenopus and Drosophila extracts, initiation of replication requires the ATP-dependent DNA binding activity of ORC (2, 37). How can such a precisely regulated process be carried out without the requirement for specific start sites? The explanation is revealed in the mechanism by which DNA replication is coordinated with the various phases of the cell cycle.
Regulated Replication Is Independent of Sequence
In all eukaryotic systems that have been amenable to study, replication is regulated by the assembly of a prereplication complex (pre-RC) of highly conserved proteins at ORC-bound DNA sites shortly after metaphase. After cells pass through the R point or START, a sharp rise in the activities of S phase–promoting kinases (SPK: Cdc7/Dbf4 and B-type cyclin-Cdk) triggers the conversion of the pre-RC to an active replication complex. These high levels of Cdk activity (38), as well as a protein called geminin (1, 39, 40), persist from the onset of S phase through metaphase and prevent the assembly of new pre-RCs. Both of these activities are destroyed by proteolysis during anaphase, allowing pre-RCs to reassemble. Hence, mutually exclusive periods of the cell cycle that promote either pre-RC formation or initiation ensure that replication can only initiate once per cell cycle. This model (Fig. 2A) does not invoke any requirement for specific origin sequences to accomplish accurate duplication of the genome.
Fig. 2.

Once-per-cell-cycle genome duplication is independent of the positions or density of replication origins. (A) Duplication of DNA exactly once-per-cell division is achieved with two mutually exclusive periods of the cell cycle during which either pre-RCs can be assembled but replication cannot initiate or replication can initiate but pre-RCs cannot be assembled. Regardless of where pre-RCs assemble, DNA is completely replicated and cannot be re-replicated until after cell division. (B) The assembly of extraneous pre-RCs can ensure that the entire segment of DNA is replicated in a timely fashion, without the need for specific DNA sequences to optimize origin spacing. Any pre-RCs that do not initiate are destroyed by passage of the replication fork and so cannot re-initiate on the replicated strands.
The origin spacing problem.
Although initiation once per cell cycle does not require specific sequences, the positions of origins cannot be distributed randomly, as this would run the risk that some origins might be too far apart to complete replication of the intervening DNA within the length of a single S phase (41). One way to solve this problem is to direct ORC to specific DNA sequences spaced at appropriate intervals. However, the origin spacing problem can also be solved without the need for specific sequence recognition. Indeed, rapid replication in early Xenopus development is accomplished by initiating replication at sites that appear random with respect to sequence but are regularly spaced every 9 to 12 kb (41). The mechanism that establishes this regular origin spacing is unknown but, under conditions where chromatin is saturated with pre-RCs, any mechanism that prevents more than one pre-RC from assembling or firing per 10 kb would produce the observed spacing.
More pre-RCs than needed.
An alternative means to solve the origin spacing problem is to assemble more pre-RCs than are necessary (Figs. 2B and 3), which appears to be the case in many eukaryotic systems. In budding yeast, many ARS elements function efficiently to promote the autonomous replication of plasmid DNA but do not normally function as origins in the chromosome or are utilized significantly less than once per cell cycle (42–44). However, pre-RCs are assembled on both active and silent origins (45). In Xenopus egg extracts, as the concentration of sperm nuclei is increased, the number of active origins per nucleus decreases but the number of ORC- and Mcm-DNA complexes assembled per nucleus remains constant (46). In mammalian cells, pre-RCs are assembled during telophase but pre-RC assembly is not sufficient to specify which sites will function as replication origins (47). Together, these results suggest that the number of pre-RCs assembled in eukaryotic cells exceeds the number of origins activated in each cell cycle; additional factors must select which of these pre-RCs will initiate replication.
Fig. 3.

Exclusion of pre-RCs from specific regions could create the need for origin focusing mechanisms. In this model, passage of the transcription apparatus before replication depletes transcription units (black boxes) of pre-RCs (yellow stars). (A) Without transcription, there is no selective pressure to focus initiation to specific sites as the assembly of pre-RCs at many sites ensures timely genome replication. (B) When transcription units are sparse, the assembly of multiple pre-RCs at many sites within the intergenic regions is sufficient to accomplish genome replication. (C) When intergenic regions are few and far between, specific DNA sequence recognition elements are required to ensure that at least one pre-RC is assembled at intervals appropriate to accomplish the timely replication of the DNA segment.
Potentially, extraneous pre-RCs could effect the reduplication of portions of the genome if they were to persist on daughter DNA strands after replication. However, evidence suggests that pre-RCs are destroyed by the passage of replication forks from active origins (Fig. 2B). Silent or infrequently utilized budding yeast origins become activated when replication forks are prevented from passing through them (48, 49). In both budding and fission yeast, when two or more origins are found in close proximity only one appears to be utilized within any given S phase (50–54). The mechanism that determines the frequency with which a given pre-RC will be activated is not known, however, chromosomal context and epigenetic elements clearly play a role. When identical ARS elements were placed in close proximity, one of them was utilized more frequently than the other, with the determinants of origin preference localized to flanking DNA sequences (52, 54). Mutations that disrupt silent chromatin at telomeres in budding yeast activate a normally silent telomeric origin (55) and manipulations that alter the positioning of nucleosomes on origins can directly influence the efficiency with which origins fire (56). Hence, each pre-RC has a potential to initiate replication that is influenced by a combination of local chromatin structure and the probability that it will be activated before the passage of a replication fork from adjacent origins.
Origin Choice Is Developmentally Regulated
The gradual transition from random to specific origin site selection after the midblastula transition (MBT) during Xenopus (36) and Drosophila (35) development provides a dramatic example of origin choice during development. One possible explanation for this transition is that the higher concentration of ORC in preblastula embryos could result in a more relaxed binding of ORC to DNA, which would then become site-specific as the ORC:DNA ratio decreases after the MBT. Both purified (10) and recombinant (57) Drosophila ORC bind preferentially to specific DNA segments found near origins of gene amplification. In one case (57), the resolution was sufficient to conclude that replication initiates at the border of the ORC binding site, as in budding yeast (6). So far, the only sequence motifs shared by these ORC binding segments are short asymmetric AT stretches, reminiscent but not as prominent as those found in budding and fission yeast origins. Hence, it is presently difficult to predict the extent to which the affinity of ORC for specific DNA sequences, at any ORC:DNA ratio, can account for the specificity of initiation found in metazoan chromosomes. Changes in chromosome architecture that take place at the MBT, including chromatin condensation, the appearance of histone H1, cellular differentiation, and the gradual onset of transcription could also play a role in focusing initiation to specific sites (36, 58). Differences in origin specificity have also been observed between cell lines from different tissues. In murine non–B cells, the entire IgH locus is replicated from a single replication fork that proceeds gradually through the locus from an origin located downstream of the constant region genes (59). However, in pre–B cells, the entire locus is duplicated during the first hour of S phase, indicating that one or more additional origins are activated.
There are several means by which origin specification could be influenced or even regulated during development. Changes in chromatin structure that accompany key stages of development could influence both the sequences to which ORC will preferentially bind and the efficiency with which pre-RCs are activated. The onset of differentiation could also result in the expression of accessory factors that interact with ORC and target it to specific sites. For example, the ORC4 subunit in S. pombe contains an NH2-terminal extension with nine copies of the HMG-I(Y) related AT-hook motif, which is known to mediate binding to the minor groove of AT tracts (60). In metazoa, interaction with accessory factors such as HMG proteins could focus ORC to specific DNA sequences as, for example, HMG proteins increase the site-specific binding of steroid receptors to their cognate sites (61). In fact, human ORC subunits coimmunoprecipitate with many unidentified proteins (62) and are targeted to the Epstein-Barr virus (EBV) replication origin, apparently through an interaction with the viral origin-binding protein EBNA (63–65). Furthermore, extracts from differentiated Drosophila tissue culture cells contain activities that appear to increase the selectivity of ORC for specific DNA fragments (66). Assuming that there is a selective advantage to regulating the positions of origins during development, utilizing ancillary factors to regulate the specificity of ORC could have circumvented the need to evolve different initiators for different tissues.
Chromosomal Traffic Control
The fact that origins are localized in most species suggests that there is some selective pressure to initiate replication at particular sites. However, since once-per-cell-cycle replication per se does not require specific DNA sequences, the selective pressure must derive from considerations other than genome duplication. One potential source for this selective pressure is the need to coordinate transcription with replication (67). Indeed, transcription inhibits the autonomous replication of plasmids in human cells (68). There are at least two ways in which transcription and replication could be mutually antagonistic. First, head-on collisions of the replication and transcription machinery would create the need for both apparatuses to share the same DNA template temporarily. Bacterial genes are heavily biased toward an orientation that places the polarity of transcription and replication in the same direction (69, 70). In both budding yeast and humans, a physical barrier in the 3′ region of the ribosomal genes prevents replication forks from traveling in a direction that opposes RNA polymerase (25, 71, 72). Also, replication forks stall when they oppose the direction of yeast tRNA transcription but not when the tRNA genes are defective in transcription (73). However, there is some evidence in mammalian cells to suggest that transcription and replication do not take place simultaneously on the same DNA segments [reviewed in (74)], suggesting that polymerase collisions may not generally occur in mammalian cells.
A second antagonistic effect of replication and transcription could take place before replication, if pre-RCs are disrupted by passage of the transcription machinery. Although direct evidence for this mechanism is lacking, and exceptional cases exist (origins within transcription units and localized origins within transcriptionally silent regions), the explanatory power is intriguing (Fig. 3). In Xenopus embryos, there is no transcriptional activity before the MBT, so there is no selective pressure to place origins at particular locations. In contrast, the vast majority of the budding yeast genome is transcribed, and the locations of replication origins are almost exclusively restricted to intergenic regions (67). In this context, there would be a strong selective advantage for evolving specific DNA sequences that focus initiation to intergenic sites to ensure proper origin spacing (Fig. 3). This logic can also be applied to understand the different patterns of initiation at individual metazoan loci. Most solitary origin sites have been identified within loci containing multiple genes (18–20). By contrast, broad initiation zones consisting of multiple inefficient origins are observed at loci where there are large intergenic regions (22, 24, 25).
At present, it is difficult to predict the significance of transcription to origin localization. Clearly, we need to determine the sites where pre-RCs assemble during telophase and whether the onset of transcription after mitosis does, indeed, displace pre-RCs from transcription units. New applications for microarray analysis that reveal the genome-wide locations of DNA bound proteins have already been developed in budding yeast (75), and the answers to some of these questions in this simple model system should be forthcoming. In cases where replication does initiate within transcription units, it will be important to determine whether transcription is activated after replication. Finally, the coordination of transcription with replication provides a useful working model but other roles for origin placement should be considered. For example, ORC appears to play a central role in the assembly of heterochromatin (76) and chromosome condensation during mitosis (77, 78). The location of replication origins may be important to organize chromosomes for sister chromatid cohesion and/or chromosome condensation.
What is clear is that the need to duplicate the genome once per cell cycle does not itself impose selective pressure to initiate replication at specific sites. In fact, it could be argued that a more degenerate origin recognition system would favor rapid genome evolution, allowing more flexible rearrangement of sequences without risking large gaps of DNA without an origin, and allowing for the modulation of origin sites during development. With this revelation, the difficulties in identifying consensus origin sequences in metazoa should come as no surprise. The more pressing question becomes why origins are localized at all in most eukaryotic systems. The answer to this question may be different for different loci. Although this indeed complicates the analysis of eukaryotic origins, it should by no means discourage investigators from entertaining creative hypotheses and pursuing directions that will likely reveal new insights into the organization of complex genomes.
Acknowledgments
I would like to thank K. Friedman, S. Fiering, M. Aladjem, J. Blow, M. Schmitt, M. Botchan, and S. Gerbi for helpful criticisms of this essay and H. Masukata, S. Gerbi, and M. Botchan for sharing unpublished information. Work in my laboratory is supported by NIH grant GM-57233-01, NSF grant MCB-0077507, and American Cancer Society grant RPG-97-098-04-CCG.
References
- 1.Diffley JF. Curr Biol. 2001;11:R367. doi: 10.1016/s0960-9822(01)00196-8. [DOI] [PubMed] [Google Scholar]
- 2.Kelly TJ, Brown GW. Annu Rev Biochem. 2000;69:829. doi: 10.1146/annurev.biochem.69.1.829. [DOI] [PubMed] [Google Scholar]
- 3.Marahrens Y, Stillman B. Science. 1992;255:817. doi: 10.1126/science.1536007. [DOI] [PubMed] [Google Scholar]
- 4.Van Houten J, Newlon CS. Mol Cell Biol. 1990;10:3917. doi: 10.1128/mcb.10.8.3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klemm RD, Bell SP. Proc Natl Acad Sci USA. 2001;98:8361. doi: 10.1073/pnas.131006898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bielinsky AK, Gerbi SA. Mol Cell. 1999;3:477. doi: 10.1016/s1097-2765(00)80475-x. [DOI] [PubMed] [Google Scholar]
- 7.Okuno Y, Satoh H, Sekiguchi M, Masukata H. Mol Cell Biol. 1999;19:6699. doi: 10.1128/mcb.19.10.6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ogawa Y, Takahashi T, Masukata H. Mol Cell Biol. 1999;19:7228. doi: 10.1128/mcb.19.10.7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spradling AC. Genes Dev. 1999;13:2619. doi: 10.1101/gad.13.20.2619. [DOI] [PubMed] [Google Scholar]
- 10.Austin RJ, Orr-Weaver TL, Bell SP. Genes Dev. 1999;13:2639. doi: 10.1101/gad.13.20.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delidakis C, Kafatos FC. EMBO J. 1989;8:891. doi: 10.1002/j.1460-2075.1989.tb03450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heck MM, Spradling AC. J Cell Biol. 1990;110:903. doi: 10.1083/jcb.110.4.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu L, Zhang H, Tower J. Genes Dev. 2001;15:134. doi: 10.1101/gad.822101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Cicco D, Spradling AC. Cell. 1984;38:45. doi: 10.1016/0092-8674(84)90525-7. [DOI] [PubMed] [Google Scholar]
- 15.Carminati JL, Johnston CG, Orr-Weaver TL. Mol Cell Biol. 1992;12:2444. doi: 10.1128/mcb.12.5.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cimbora DM, et al. Mol Cell Biol. 2000;20:5581. doi: 10.1128/mcb.20.15.5581-5591.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aladjem M, et al. Science. 1995;270:815. doi: 10.1126/science.270.5237.815. [DOI] [PubMed] [Google Scholar]
- 18.Aladjem MI, Rodewald LW, Kolman JL, Wahl GM. Science. 1998;281:1005. doi: 10.1126/science.281.5379.1005. [DOI] [PubMed] [Google Scholar]
- 19.Toledo F, et al. Nucleic Acids Res. 1998;26:2313. doi: 10.1093/nar/26.10.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abdurashidova G, et al. Science. 2000;287:2023. doi: 10.1126/science.287.5460.2023. [DOI] [PubMed] [Google Scholar]
- 21.Pelizon C, Diviacco S, Falaschi A, Giacca M. Mol Cell Biol. 1996;16:5358. doi: 10.1128/mcb.16.10.5358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dijkwel PA, Hamlin JL. Mol Cell Biol. 1995;15:3023. doi: 10.1128/mcb.15.6.3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kobayashi T, Rein T, DePamphilis M. Mol Cell Biol. 1998;18:3266. doi: 10.1128/mcb.18.6.3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ina S, Sasaki T, Yokota Y, Shinomiya T. Chromosoma. 2001;109:551. doi: 10.1007/s004120000112. [DOI] [PubMed] [Google Scholar]
- 25.Little RD, Platt TH, Schildkraut CL. Mol Cell Biol. 1993;13:6600. doi: 10.1128/mcb.13.10.6600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dijkwel PA, Mesner LD, Levenson VV, d’Anna J, Hamlin JL. Exp Cell Res. 2000;256:150. doi: 10.1006/excr.2000.4809. [DOI] [PubMed] [Google Scholar]
- 27.Kalejta RF, et al. Mol Cell. 1998;2:797. doi: 10.1016/s1097-2765(00)80294-4. [DOI] [PubMed] [Google Scholar]
- 28.Altman AL, Fanning E. Mol Cell Biol. 2001;21:1098. doi: 10.1128/MCB.21.4.1098-1110.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mello CC, Kramer JM, Stinchcomb D, Ambros V. EMBO J. 1991;10:3959. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim CS, Preer JR, Polisky B. Dev Genet. 1992;13:97. doi: 10.1002/dvg.1020130202. [DOI] [PubMed] [Google Scholar]
- 31.Masukata H, Satoh H, Obuse C, Okazaki T. Mol Biol Cell. 1993;4:1121. doi: 10.1091/mbc.4.11.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krysan PJ, Smith JG, Calos MP. Mol Cell Biol. 1993;13:2688. doi: 10.1128/mcb.13.5.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith JG, Calos MP. Chromosoma. 1995;103:597. doi: 10.1007/BF00357686. [DOI] [PubMed] [Google Scholar]
- 34.Coverley D, Laskey RA. Annu Rev Biochem. 1994;63:745. doi: 10.1146/annurev.bi.63.070194.003525. [DOI] [PubMed] [Google Scholar]
- 35.Sasaki T, Sawado T, Yamaguchi M, Shinomiya T. Mol Cell Biol. 1999;19:547. doi: 10.1128/mcb.19.1.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hyrien O, Maric C, Mechali M. Science. 1995;270:994. doi: 10.1126/science.270.5238.994. [DOI] [PubMed] [Google Scholar]
- 37.I. Chesnokov, M. R. Botchan, Proc. Natl. Acad. Sci. U.S.A., in press.
- 38.Noton E, Diffley JF. Mol Cell. 2000;5:85. doi: 10.1016/s1097-2765(00)80405-0. [DOI] [PubMed] [Google Scholar]
- 39.Wohlschlegel JA, et al. Science. 2000;290:2309. doi: 10.1126/science.290.5500.2309. [DOI] [PubMed] [Google Scholar]
- 40.Tada S, Li A, Maiorano D, Mechali M, Blow JJ. Nature Cell Biol. 2001;3:107. doi: 10.1038/35055000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blow JJ, Gillespie PJ, Francis D, Jackson DA. J Cell Biol. 2001;152:15. doi: 10.1083/jcb.152.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamashita M, et al. Genes Cells. 1997;2:655. doi: 10.1046/j.1365-2443.1997.1530351.x. [DOI] [PubMed] [Google Scholar]
- 43.Friedman KL, Brewer BJ, Fangman WL. Genes Cells. 1997;2:667. doi: 10.1046/j.1365-2443.1997.1520350.x. [DOI] [PubMed] [Google Scholar]
- 44.A. Poloumienkol, A. Dershowitz, D. Jitakshi, C. S. Newlon, Mol. Biol. Cell, in press.
- 45.Santocanale C, Diffley J. EMBO J. 1996;15:6671. [PMC free article] [PubMed] [Google Scholar]
- 46.Walter J, Newport J. Science. 1997;275:993. doi: 10.1126/science.275.5302.993. [DOI] [PubMed] [Google Scholar]
- 47.Okuno Y, McNairn AJ, den Elzen N, Pines J, Gilbert DM. EMBO J. 2001;20:4263. doi: 10.1093/emboj/20.15.4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Y, Vujcic M, Kowalski D. Mol Cell Biol. 2001;21:4938. doi: 10.1128/MCB.21.15.4938-4948.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Santocanale C, Sharma K, Diffley JF. Genes Dev. 1999;13:2360. doi: 10.1101/gad.13.18.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wohlgemuth JG, Bulboaca GH, Moghadam M, Caddle MS, Calos MP. Mol Biol Cell. 1994;5:839. doi: 10.1091/mbc.5.8.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dubey D, Zhu J, Carlson D, Sharma K, Huberman J. EMBO J. 1994;13:3638. doi: 10.1002/j.1460-2075.1994.tb06671.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brewer BJ, Fangman WL. Proc Natl Acad Sci USA. 1994;91:3418. doi: 10.1073/pnas.91.8.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brewer BJ, Fangman WL. Science. 1993;262:1728. doi: 10.1126/science.8259517. [DOI] [PubMed] [Google Scholar]
- 54.Marahrens Y, Stillman B. EMBO J. 1994;13:3395. doi: 10.1002/j.1460-2075.1994.tb06642.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stevenson JB, Gottschling DE. Genes Dev. 1999;13:146. doi: 10.1101/gad.13.2.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lipford JR, Bell SP. Mol Cell. 2001;7:21. doi: 10.1016/s1097-2765(01)00151-4. [DOI] [PubMed] [Google Scholar]
- 57.A.-K. Bielinsky et al., Curr. Biol., in press.
- 58.Gilbert DM, Miyazawa H, DePamphilis ML. Mol Cell Biol. 1995;15:2942. doi: 10.1128/mcb.15.6.2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ermakova OV, et al. Mol Cell. 1999;3:321. doi: 10.1016/s1097-2765(00)80459-1. [DOI] [PubMed] [Google Scholar]
- 60.Chuang RY, Kelly TJ. Proc Natl Acad Sci USA. 1999;96:2656. doi: 10.1073/pnas.96.6.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Boonyaratanakornkit V, et al. Mol Cell Biol. 1998;18:4471. doi: 10.1128/mcb.18.8.4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dhar SK, Dutta A. J Biol Chem. 2000;275:34983. doi: 10.1074/jbc.M006069200. [DOI] [PubMed] [Google Scholar]
- 63.Chaudhuri B, Xu H, Todorov I, Dutta A, Yates JL. Proc Natl Acad Sci USA. 2001;98:10085. doi: 10.1073/pnas.181347998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dhar SK, et al. Cell. 2001;106:287. doi: 10.1016/s0092-8674(01)00458-5. [DOI] [PubMed] [Google Scholar]
- 65.Schepers A, et al. EMBO J. 2001;20:4588. doi: 10.1093/emboj/20.16.4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.E. L. Beall, M. R. Botchan, personal communication.
- 67.Brewer BJ. Curr Opin Genet Dev. 1994;4:196. doi: 10.1016/s0959-437x(05)80045-0. [DOI] [PubMed] [Google Scholar]
- 68.Haase SB, Heinzel SS, Calos MP. Mol Cell Biol. 1994;14:2516. doi: 10.1128/mcb.14.4.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brewer BJ. Cell. 1988;53:679. doi: 10.1016/0092-8674(88)90086-4. [DOI] [PubMed] [Google Scholar]
- 70.Tillier ER, Collins RA. Nature Genet. 2000;26:195. doi: 10.1038/79918. [DOI] [PubMed] [Google Scholar]
- 71.Brewer BJ, Lockshon D, Fangman WL. Cell. 1992;71:267. doi: 10.1016/0092-8674(92)90355-g. [DOI] [PubMed] [Google Scholar]
- 72.Gerber JK, et al. Cell. 1997;90:559. doi: 10.1016/s0092-8674(00)80515-2. [DOI] [PubMed] [Google Scholar]
- 73.Deshpande AM, Newlon CS. Science. 1996;272:1030. doi: 10.1126/science.272.5264.1030. [DOI] [PubMed] [Google Scholar]
- 74.Cook P. Science. 1998;281:1466. doi: 10.1126/science.281.5382.1466. [DOI] [PubMed] [Google Scholar]
- 75.Ren B, et al. Science. 2000;290:2306. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 76.Pak D, et al. Cell. 1997;91:311. doi: 10.1016/s0092-8674(00)80415-8. [DOI] [PubMed] [Google Scholar]
- 77.Loupart M, Krause S, Heck MS. Curr Biol. 2000;10:1547. doi: 10.1016/s0960-9822(00)00844-7. [DOI] [PubMed] [Google Scholar]
- 78.Pflumm MF, Botchan MR. Development. 2001;128:1697. doi: 10.1242/dev.128.9.1697. [DOI] [PubMed] [Google Scholar]


