Abstract
Mcm 2–7 are essential replication proteins that bind to chromatin in mammalian nuclei during late telophase. Here, we have investigated the relationship between Mcm binding, licensing of chromatin for replication, and specification of the dihydrofolate reductase (DHFR) replication origin. Approximately 20% of total Mcm3 protein was bound to chromatin in Chinese hamster ovary (CHO) cells during telophase, while an additional 25% bound gradually and cumulatively throughout G1-phase. To investigate the functional significance of this binding, nuclei prepared from CHO cells synchronized at various times after metaphase were introduced into Xenopus egg extracts, which were either immunodepleted of Mcm proteins or supplemented with geminin, an inhibitor of the Mcm-loading protein Cdt1. Within 1 hour after metaphase, coincident with completion of nuclear envelope formation, CHO nuclei were fully competent to replicate in both of these licensing-defective extracts. However, sites of initiation of replication in each of these extracts were found to be dispersed throughout the DHFR locus within nuclei isolated between 1 to 5 hours after metaphase, but became focused to the DHFR origin within nuclei isolated after 5 hours post-metaphase. Importantly, introduction of permeabilized post-ODP, but not pre-ODP, CHO nuclei into licensing-deficient Xenopus egg extracts resulted in the preservation of a significant degree of DHFR origin specificity, implying that the previously documented lack of specific origin selection in permeabilized nuclei is at least partially due to the licensing of new initiation sites by proteins in the Xenopus egg extracts. We conclude that the functional association of Mcm proteins with chromatin (i.e. replication licensing) in CHO cells takes place during telophase, several hours prior to the specification of replication origins at the DHFR locus.
Keywords: Mammalian nuclei, Mcm proteins, Replication licensing, ODP, Cell cycle
Introduction
The minichromosome maintenance proteins (Mcm 2–7) have been shown to be key participants in the mechanism that limits eukaryotic replication to once-per-cell-cycle (Tye, 1999). In particular, the association of Mcm proteins with chromatin appears to be the final step in the formation of pre-replication complexes (pre-RCs) in a process referred to as replication licensing (Rowles and Blow, 1997). The molecular details of pre-RC assembly are beginning to emerge (Kelly and Brown, 2000; Leatherwood, 1998). The origin recognition complex (ORC) recruits Cdc6 which, along with Cdt1 (Maiorano et al., 2000b; Nishitani et al., 2000; Tada et al., 2001), is required for the subsequent loading of the Mcm 2–7 complex (Coleman et al., 1996; Romanowski et al., 1996; Rowles et al., 1996; Tanaka et al., 1997). The formation of pre-RCs takes place during late mitosis and early G1-phase and is prevented by the presence of cyclin-dependent kinase (CDK) activity (Coverley et al., 1996; Noton and Diffley, 2000) and geminin, a protein that binds to and inhibits Cdt1 (McGarry and Kirschner, 1998; Tada et al., 2001; Wohlschlegel et al., 2000). No activities have been identified that are required for the formation of pre-RCs after the binding of Mcm proteins. Hence, the binding of Mcm 2–7 to chromatin likely represents functional licensing and the culmination of pre-RC assembly. In late G1-phase, CDK levels rise sharply, resulting in the initiation of replication and the conversion of the pre-RC into an active replication complex. CDK activity and geminin levels rise throughout S-phase, until their destruction during the following metaphase-anaphase transition. Thus, once-per-cell-cycle replication is ensured by the sequential and mutually exclusive conditions necessary for the assembly of pre-RCs and their conversion to active replication complexes.
In cultured mammalian cells, replication initiates at specific sites (Aladjem et al., 1998; Giacca et al., 1994), however, no specific DNA sequences have been identified that direct the assembly of pre-RCs. We have shown that site-specific initiation of replication within the Chinese hamster ovary (CHO) dihydrofolate reductase (DHFR) locus can be achieved in Xenopus egg extracts using intact, late-G1-phase nuclei as a substrate (Gilbert et al., 1995). With late-G1-phase nuclei that have been permeabilized during preparation, or with intact, early-G1-phase nuclei, replication initiates at random sites (Dimitrova and Gilbert, 1998). At a distinct point during G1-phase, CHO nuclei experience a transition (origin decision point, ODP) that selects which of many potential chromosomal sites will function as an origin of replication in the upcoming S-phase (Wu and Gilbert, 1996). The ODP is unlikely to represent the association of hamster ORC with chromatin, since pre-ODP nuclei replicate efficiently and at random sites when introduced into Xenopus egg extracts that lack ORC proteins (Yu et al., 1998), indicating that non-specific origin selection is mediated by hamster ORC. Since the binding of Mcm proteins probably represents the culmination of pre-RC formation at replication origins, we were interested in determining whether the ODP represents the completed assembly of pre-RCs at specific sites. We have previously shown that the Chinese hamster homologue of Mcm2 is loaded gradually and cumulatively throughout G1-phase, beginning as soon as nuclei have formed during late telophase (Dimitrova et al., 1999). Thus, it remained possible that different origins could become licensed throughout G1-phase, as Mcm proteins bound to an increasing fraction of the chromatin. Alternatively, it was possible that the initial association of Mcm proteins with chromatin prior to the ODP could be non-functional, with functional Mcm complexes being formed only at the ODP. For example, partial Mcm complexes have been shown to associate with chromatin, but only the complete heterohexamer forms a functional pre-RC (Maiorano et al., 2000a; Prokhorova and Blow, 2000). This possibility seems to be supported by recent evidence that replication of G1-phase CHO nuclei in Xenopus egg extracts depends on the presence of Xenopus ORC and Mcm proteins if nuclei are isolated earlier than 2 hours post-metaphase (Natale et al., 2000).
Here, we have evaluated the capacity of licensing-deficient Xenopus egg extracts to replicate genomic DNA and to initiate replication specifically at the DHFR origin within CHO nuclei prepared at different times during G1-phase. We found that extracts that have been immunodepleted of Mcm proteins or supplemented with geminin are unable to replicate chromatin from CHO cells synchronized in metaphase, prior to Mcm binding. However, CHO nuclei prepared at all stages of G1-phase, including those from telophase cells, in which the first fraction of Mcm proteins had bound to chromatin, were replicated with similar efficiency. Furthermore, replication at the DHFR locus initiated at random sites with intact pre-ODP nuclei and at specific sites within post-ODP nuclei, regardless of the presence of Mcm proteins in the Xenopus egg extract. We conclude that Mcm proteins are incorporated into functional pre-RCs during late telophase and that a sufficient quantity of Mcm proteins have associated with chromatin at that time to support efficient genome replication. However, the licensing of mammalian chromatin is not sufficient to dictate which of many potential origins of replication will be selected for initiation in the upcoming S-phase.
Materials and Methods
Cell culture and synchronization
CHOC 400 is a CHO cell derivative, in which a 243 kb segment of DNA containing the DHFR gene has been amplified ~500-fold by stepwise selection in methotrexate (Milbrandt et al., 1981). Cells were cultured and synchronized as described (Dimitrova and Gilbert, 1998; Dimitrova et al., 1999).
Cell fractionation and immunoblotting
Cell fractionation and immunoblotting were performed as described (Dimitrova et al., 1999). For the immunoblots displayed in Fig. 3, the cells were permeabilized with 80 μg/ml digitonin and incubated in Xenopus egg extracts at a concentration of 10,000 nuclei/μl for 1 hour at 21°C. The extracts were supplemented with 100 μg/ml aphidicolin (CalBiochem) to prevent DNA replication and release of Mcm proteins. The nuclei were then washed with cold cytoskeleton buffer (CSK), extracted for 5 minutes on ice with CSK containing 0.5% Triton X-100 (Sigma), washed twice with CSK buffer and processed for western blotting as described (Dimitrova et al., 1999). Mcm2 proteins (Xenopus and hamster) were detected with an affinity-purified rabbit polyclonal anti-human Mcm2 antibody (Todorov et al., 1995), Mcm3 proteins (Xenopus and hamster) – with a rabbit polyclonal serum raised against Xenopus Mcm3 (Prokhorova and Blow, 2000) and Xenopus ORC2 – with an affinity-purified rabbit polyclonal antibody (Yu et al., 1998).
Fig. 3.
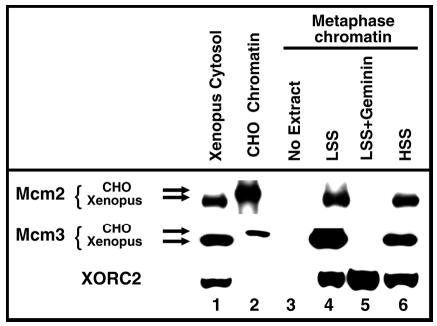
Geminin prevents binding of Xenopus Mcm proteins to hamster chromatin. Anti-HsMcm2 and anti-XMcm3 antibodies recognize both Xenopus (lane 1, Xenopus egg cytosol) and hamster (lane 2, Triton-extracted CHOC 400 G1-phase nuclei) Mcm proteins, which can be distinguished by their slightly different electrophoretic mobility. The anti-XORC2 antibody does not crossreact with hamster ORC2. (lanes 3–6) CHOC 400 metaphase cells were permeabilized with 80 μg/ml digitonin and incubated for 1 hour at 21°C in LSS (lanes 4,5) or HSS (lane 6) Xenopus egg extracts (supplemented with 100 μg/ml aphidicolin). Soluble or loosely bound proteins were removed by Triton extraction as described in Materials and Methods and the washed chromatin was subjected to immunoblotting analysis with antibodies specific for Mcm2, Mcm3 and ORC2. In lane 5, the LSS extract was supplemented with 2 μg/ml of purified Xenopus geminin. Aliquots of Xenopus egg extract (lane 1) or Triton-extracted CHOC 400 G1 nuclei (lane 2) were run in parallel and served as markers for the mobility of the respective proteins of hamster or frog origin.
Analysis of DNA replication in Xenopus egg extracts
Activated low speed (LSS) and high speed (HSS) Xenopus egg extracts were prepared as described (Chong et al., 1995; Goldberg et al., 1995). Extract depletions were carried out by a modification (T.A.P., unpublished) of the previously described procedure (Chong et al., 1997). Briefly, prior to the immunodepletion, the anti-Mcm3 antibody-coupled Protein A beads were pre-equilibrated in previously depleted extract. This decreased the extract dilution factor and helped to preserve better the nuclear assembly and DNA replication activity of the depleted extracts. Purified Xenopus geminin (McGarry and Kirschner, 1998) was used at final concentrations between 0.2–2 μg/ml. This destruction box-negative variant of geminin was stable during long incubation periods. Rates and extent of DNA replication with CHOC 400 nuclei were evaluated by measuring the amount of acid-precipitable [α-32P]dATP (Dimitrova and Gilbert, 1998; Gilbert et al., 1995). Specificity of initiation within the DHFR locus in CHO G1 nuclei was measured by the early-labeled fragment hybridization (ELFH) assay (Dimitrova and Gilbert, 1998; Gilbert et al., 1995). Each batch of G1 cells was tested for origin specificity before use in other experiments. Typically, the ODP transition occurred between 5–6 hours post-metaphase.
Results
Mcm3 binds to CHO chromatin gradually and cumulatively throughout G1-phase, beginning in telophase
We have previously shown that approximately 20% of the total Mcm2 proteins in CHO cells are loaded onto chromatin during telophase, and that an additional 25% is loaded gradually and cumulatively throughout the remainder of G1-phase (Dimitrova et al., 1999). To determine whether this was true for another Mcm subunit, Mcm3, CHO (CHOC 400) cells were synchronized in metaphase with nocodazole and released into G1-phase. Immunoblotting experiments were performed with either whole cell extracts, intact nuclei, permeabilized nuclei or detergent-extracted nuclei. Our anti-Mcm3 antibody detected a single band with an apparent molecular mass of ~100 kDa (Fig. 1A), consistent with the predicted size of mammalian Mcm3. The total amount of Mcm3 per cell did not vary significantly throughout the cell cycle and remained exclusively nuclear during interphase, however, the majority of Mcm3 was released from nuclei by Triton extraction (Fig. 1). Exit from mitosis is not synchronous for all cells and over a period of ~1 hour after removal of nocodazole there is a mixed population of cells at various stages of mitosis and early-G1. We used classical morphological criteria for their distinction: mitotic figures, degree of chromosome decondensation and dispersal, nuclear morphology, extent of nuclear lamina reassembly, completion of cytokinesis. Detergent-resistant association of Mcm3 proteins with chromatin was first observed 40 minutes after metaphase, coincident with the appearance of telophase cells in the population (Fig. 1B). Within 1 hour post-metaphase most of the cells have completed mitosis and are already in G1-phase. By this time, approximately 20% of total Mcm3 was loaded onto chromatin. Additional amounts of Mcm3 were loaded as cells proceeded through G1-phase (Fig. 1C). Later, as cells traversed S-phase, Mcm3 was released from chromatin. These results closely resemble previous results obtained for Mcm2 in CHO cells (Dimitrova et al., 1999). We conclude that Mcm2 and Mcm3 associate with chromatin with similar kinetics, consistent with their participation in a multi-subunit pre-RC complex.
Fig. 1.

Cell cycle-regulated association of hamster Mcm3 with chromatin. (A) Western blot of total CHOC 400 cellular protein extract probed with an Mcm3-specific antibody. Positions of molecular weight standards (indicated in kDa) are marked on the left. (B,C) Variations in the amount of chromatin-bound Mcm3 during different stages of the cell cycle in CHOC 400 cells. Synchronized cell populations were resuspended in cytoskeleton buffer (for Triton extractions) or transport buffer (for digitonin permeabilization) and incubated for 5 minutes on ice with or without addition of permeabilizing agents as described in Materials and Methods. The cellular or nuclear pellets (P) were separated from the soluble fractions (S) by centrifugation. The proteins from each fraction (only the pellets were analyzed in the case of digitonin extractions) were separated by electrophoresis in 10% SDS-polyacrylamide gels (amounts corresponding to 5×105 cells were loaded in each lane), transferred to nylon membranes and probed with an anti-Mcm3 antibody. In B, the percentages of cells in different stages of mitosis, determined microscopically after staining aliquots of the synchronized cells with 0.1 μg/ml 4′, 6-diamidino-2-phenylindole (DAPI), are indicated on the right for each time point. In C, the relative amounts of chromatin-bound Mcm3 proteins at each time point analyzed were estimated by comparing serial dilutions of the soluble fractions run in parallel to an aliquot of the Triton-resistant fraction and are indicated on the right.
Functional significance of Mcm chromatin association
We next wished to distinguish between three possible explanations for the continued loading of Mcm proteins during G1. First, Mcm proteins loaded during telophase might not be functional: incompletely assembled and non-functional Mcm complexes have been observed to associate with chromatin (Maiorano et al., 2000a; Prokhorova and Blow, 2000). Second, continued Mcm loading could reflect the staggered assembly of functional pre-RCs onto an increasing number of origins during G1-phase. Third, the initial loading of Mcm proteins may be sufficient to license all origins for a new round of replication and the additional loading of Mcm-s during G1-phase may be superfluous (Mahbubani et al., 1997). To distinguish between these possibilities, we evaluated the efficiency of Mcm-depleted Xenopus egg extracts to initiate replication within nuclei prepared from cells synchronized at various times during early G1-phase. If early G1-phase hamster chromatin does not contain functional pre-RCs, then it should require the presence of Xenopus Mcm proteins in order to initiate replication. Likewise, an increase in the number of functional pre-RCs, assembled onto hamster chromatin during G1-phase, should result in a quantitative increase in the rate of replication in Mcm-deficient extracts. Results (Fig. 2) revealed that both intact nuclei and nuclei that had been permeabilized with digitonin to release the soluble pool of unbound Mcm proteins, replicated with similar efficiency when prepared from cells at all times during G1-phase, both in Mcm-depleted and mock-depleted extract. By contrast, Xenopus sperm chromatin (not shown) and CHO metaphase chromatin (Fig. 2), which are not associated with Mcm proteins, cannot be replicated by Mcm-depleted extracts. Our attempts to examine Triton-extracted nuclei were not successful (D.S.D., unpublished) as they are extremely poor substrates for DNA replication even in complete Xenopus egg extracts (Dimitrova and Gilbert, 1998).
Fig. 2.
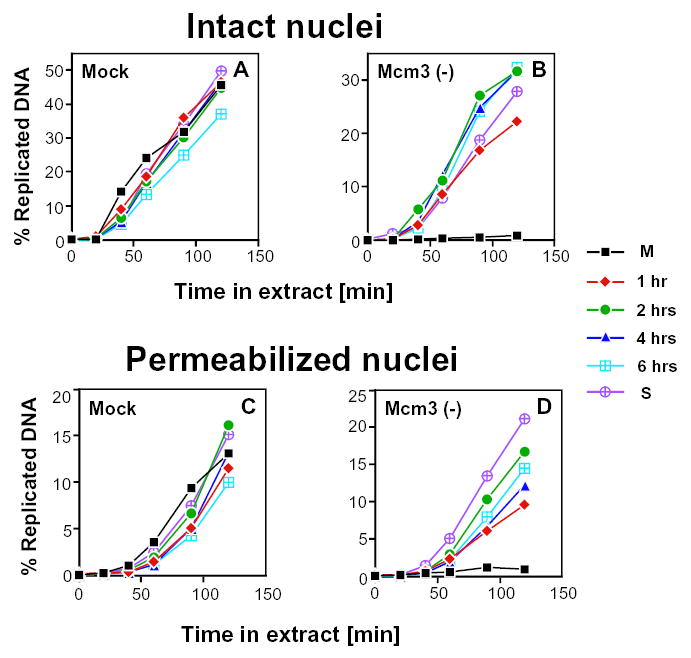
The replication capacity of CHOC 400 G1-phase nuclei is independent of the presence of XMcm3 in Xenopus egg extracts. Synchronized CHOC400 cells were permeabilized with 80 μg/ml (A,B) or 400 μg/ml (C,D) digitonin to prepare cells with intact or permeabilized nuclei, respectively. Permeabilized cells were introduced in mock-depleted (A,C) or XMcm3-depleted (B,D) Xenopus egg extracts supplemented with [α-32P]dATP. Aliquots were removed at the indicated times and the percentage of input DNA replicated was determined by acid precipitation as described (Dimitrova and Gilbert, 1998). Similar results were obtained in three independent experiments. Filled squares represent metaphase cells; diamonds, 1 hour; filled circles, 2 hours; triangles, 4 hours; hatched squares, 6-hour G1-phase nuclei; and hatched circles, G1/S-phase nuclei.
Recently, it has been shown that the protein geminin binds to Cdt1 and inhibits its ability to load Mcm proteins onto Xenopus sperm chromatin (McGarry and Kirschner, 1998; Tada et al., 2001; Wohlschlegel et al., 2000). We reasoned that, if geminin also prevented Xenopus Mcm proteins from being assembled onto hamster chromatin upon the introduction of permeabilized CHO cells into geminin-supplemented extracts, this would provide a simple means to study replication in the absence of licensing activity. In addition, the process of immunodepletion inevitably reduces the efficiency of replication in extracts due to the lengthy manipulations and the dilution of extract. To determine whether geminin can prevent the association of Xenopus Mcm proteins with hamster chromatin, CHO metaphase chromatin lacking hamster Mcm proteins (Fig. 3, lane 3) was introduced into a Xenopus egg extract in the presence or absence of geminin. After a 45 minute incubation, chromatin was washed with Triton X-100 and the association of XMcm2 and XMcm3 with chromatin was evaluated by immunoblotting. To exclude the possibility that Mcm proteins were loaded onto hamster chromatin and then released upon replication of that chromatin, the egg extracts were supplemented with aphidicolin, which blocks DNA synthesis and prevents the release of Mcm proteins (Dimitrova and Gilbert, 2000; Kubota et al., 1997). As a control, the binding of XORC2, which is not inhibited by geminin (Walter, 2000; Wohlschlegel et al., 2000), was also evaluated. Although plenty of XORC2, XMcm2 and XMcm3 were loaded onto metaphase chromatin in control Xenopus egg extract, only XORC2 associated with chromatin in geminin-supplemented extracts. In fact, consistent with results obtained previously with Xenopus sperm chromatin as a substrate (McGarry and Kirschner, 1998; Walter, 2000), the binding of XORC2 to metaphase chromatin was moderately enhanced by geminin (Fig. 3, lane 5). To verify that the concentrations of geminin employed in this study were sufficient to block DNA replication with unlicensed chromatin, either Xenopus sperm chromatin (Fig. 4A) or hamster metaphase chromatin (Fig. 4B) were introduced into a Xenopus egg extract supplemented with various concentrations of geminin. Whereas the replication of both these substrates was completely inhibited by 0.2 μg/ml of geminin, replication proceeded efficiently within control G1 nuclei (Fig. 4C) at concentrations up to 20 μg/ml of geminin. Again, CHO nuclei prepared at various times during G1-phase replicated with similar efficiency in geminin-supplemented Xenopus egg extracts (Fig. 4D-G). We conclude that functional pre-RCs are present in CHO nuclei from the very beginning of G1-phase.
Fig. 4.
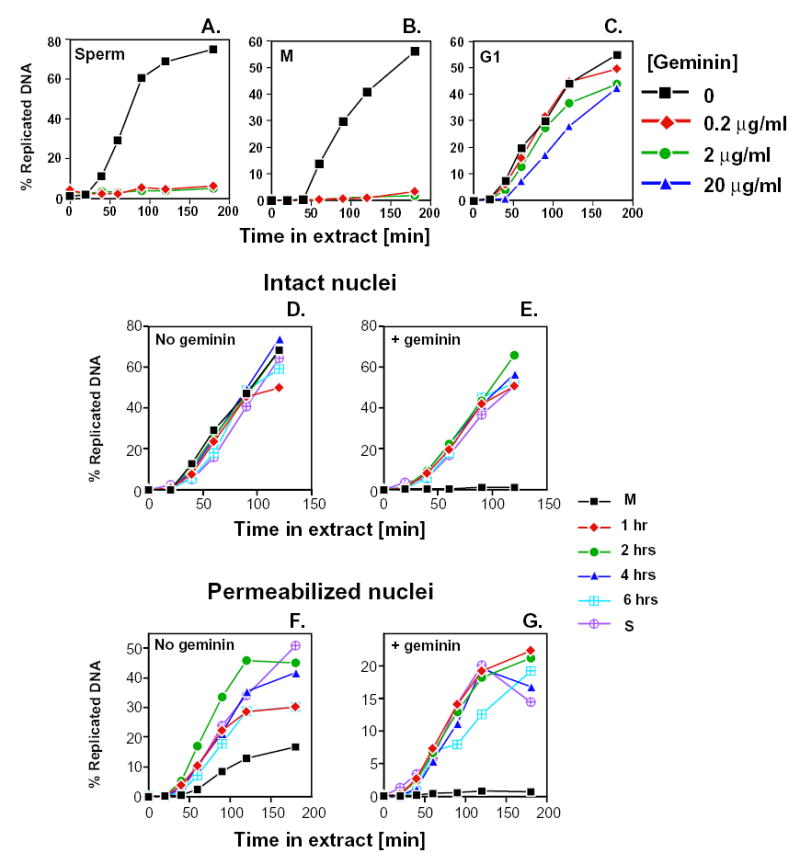
Hamster chromatin becomes licensed for replication at the end of mitosis. (A-C) Geminin prevents replication of Xenopus sperm (A; 1,000 nuclei/μl) and CHO metaphase chromatin (B; 10,000 nuclei/μl), but has no effect on hamster G1 nuclei (C; 6 hours in G1, 10,000 nuclei/μl). The indicated substrates were incubated in Xenopus egg extracts with (diamonds, 0.2 μg/ml; circles, 2 μg/ml; and triangles, 20 μg/ml) or without (squares) the addition of purified geminin. Aliquots were removed at the indicated times and the amount of replicated DNA was determined by acid precipitation as in Fig. 2. (D-G) Synchronized CHOC 400 cells were permeabilized as in Fig. 2 and incubated in control (D,F) or geminin-supplemented (E,G; 2 μg/ml) Xenopus egg extracts. The percentage of replicated DNA at various time points was determined as in Fig. 2. Similar results were obtained in four independent experiments.
Dispersed initiation within pre-ODP nuclei and site-specific intitiation within post-ODP nuclei are independent of Xenopus Mcm or Cdt1 proteins
When CHO nuclei are introduced into Xenopus egg extracts, replication initiates at the same specific sites used in cultured CHO cells, providing that nuclei are prepared with intact nuclear envelopes from cells that have passed a specific time point during G1-phase, termed the ODP. With nuclei isolated prior to the ODP, Xenopus egg extracts initiate replication at sites dispersed throughout the DHFR locus. It has been proposed that the ODP transition may represent the assembly of Mcm-containing pre-replication complexes on hamster chromatin (Natale et al., 2000). This proposal was based on data suggesting that pre-ODP nuclei cannot replicate in ORC-and Mcm-depleted extracts. However, our findings presented in Figs. 1–4 indicate that nuclei isolated from cells as early as 1 hour after metaphase are fully competent to replicate in licensing-defective extracts. This implies that replication licensing and the ODP are independent G1 events. Since this previous study did not directly measure the specificity of initiation in depleted extracts, we examined whether the pattern of initiation sites within the DHFR locus was influenced by the loading of Xenopus Mcm proteins. CHOC 400 cells were synchronized at 2 hours and 6 hours after metaphase, representative of the pre-ODP and post-ODP stages of G1-phase. Intact nuclei from these cells were introduced into either Mcm-depleted or geminin-supplemented extracts and the sites of in vitro initiation of replication in the DHFR locus were mapped. These results (Fig. 5) demonstrate clearly that efficient initiation of replication at dispersed sites throughout the DHFR locus within pre-ODP nuclei was not mediated by the loading of Xenopus Mcm proteins. Random initiation before the ODP is a property of the CHO nuclei and is mediated by hamster pre-RC proteins. Similarly, site-specific initiation within post-ODP nuclei was independent of Xenopus Mcm loading.
Fig. 5.
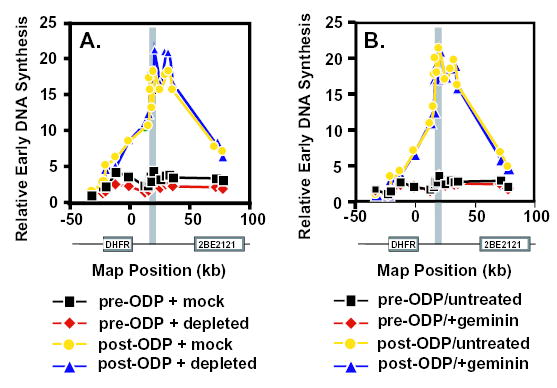
Replication origin selection in the DHFR locus in CHOC 400 G1 nuclei is independent of Xenopus Mcm proteins. Intact CHOC 400 pre-ODP (2 hours in G1; squares and diamonds) or post-ODP (6 hours in G1; circles and triangles) nuclei were introduced into mock-depleted (A; squares and circles), Mcm3-depleted (A; diamonds and triangles), control untreated (B; squares and circles) or geminin-supplemented (B; diamonds and triangles) Xenopus egg extracts. Specificity of initiation in the DHFR locus was determined by the ELFH assay as described in Materials and Methods.
Geminin prevents the licensing of illegitimate replication origins that are activated when post-ODP nuclei are permeabilized
We have previously shown that, even under conditions that preserve overall replication efficiency, permeabilization of the nuclear envelope of post-ODP CHO nuclei results in a complete loss of origin specificity when these nuclei are introduced into Xenopus egg extracts (Dimitrova and Gilbert, 1998). This loss in specificity could be due to the loss from CHO nuclei of soluble proteins required for origin specification, the disruption of a multiprotein complex within nuclei, or to the inappropriate entry of a Xenopus factor(s) that can license illegitimate origins. To distinguish between these possibilities, intact and permeabilized nuclei were prepared from the same population of post-ODP cells and introduced into either complete extracts or the same extracts supplemented with geminin to prevent loading of Xenopus Mcm-s and unauthorized assembly of Xenopus pre-RCs on CHO chromatin. Consistent with the results presented in Fig. 5B, geminin had no effect on the site-specific pattern of initiation within intact nuclei (not shown). However, whereas initiation within permeabilized post-ODP nuclei was dispersed throughout the DHFR locus in control extracts, a significant degree of specificity was retained when these extracts were supplemented with geminin. Hence, an intact nuclear envelope is required to prevent the entry of a Xenopus activity that can license illegitimate origin sites. Importantly, the parallel examination of permeabilized nuclei prepared at 2 hours in G1-phase (pre-ODP) revealed that inhibition of the origin licensing potential of Xenopus egg extracts by geminin was not sufficient to create prematurely a specific initiation pattern within pre-ODP nuclei (Fig. 6, triangles and hatched circles). This result once again confirms that the licensing of CHO chromatin for replication is independent of the selection of specific origin sites. We expect that the introduction of permeabilized mammalian nuclei or chromatin preparations into geminin-supplemented Xenopus egg cytosol would provide a valuable in vitro system for dissection of the components of mammalian pre-RCs.
Fig. 6.
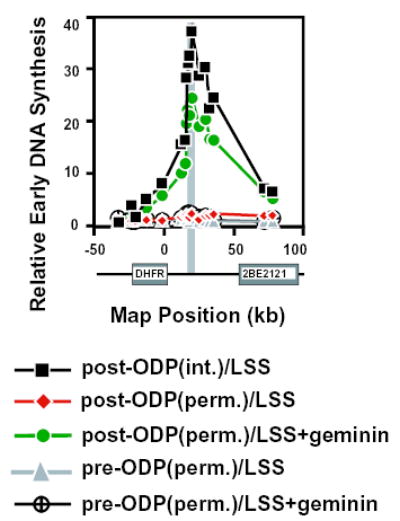
Replication initiates with a significant degree of specificity within permeabilized CHO post-ODP nuclei incubated in licensing-deficient Xenopus egg extracts. Digitonin-permeabilized pre-ODP (2 hours in G1; triangles and hatched circles) or post-ODP (6 hours in G1; diamonds and circles) nuclei were incubated in control untreated (diamonds and triangles) or geminin-supplemented (filled circles and hatched circles) Xenopus egg extracts and the specificity of initiation within the DHFR locus was measured by the ELFH assay. Squares represent intact post-ODP nuclei incubated in control untreated egg extracts. Similar results were obtained in three independent experiments. The relative specificity of initiation at ori-β within permeabilized post-ODP nuclei (measured as the average specificity of probes C, D, E and R, mapping to the region of peak initiation activity) varied in different experiments between 25 to 60–70% of that within intact post-ODP nuclei.
Discussion
We demonstrate for the first time that functional pre-RCs are assembled in CHO cells at the onset of G1-phase, several hours prior to the specification of the DHFR replication origin at the ODP. Mcm proteins bind to chromatin during late telophase and, coincident with this binding, nuclei become competent to replicate in licensing-defective Xenopus egg extracts. This indicates that the initial amount of Mcm-s loaded during telophase is sufficient for licensing of CHO nuclei. Similar to our previous observations with Mcm2, we found that additional amounts of hamster Mcm3 proteins are loaded throughout G1-phase. Interestingly, a similar gradual loading of XMcm2 and XMcm3 proteins onto chromatin was observed during incubation of demembranated Xenopus sperm nuclei in Xenopus egg extracts (Coue et al., 1998; Thommes et al., 1997). Increase in the amount of chromatin-bound Mcm proteins has been documented in S. cerevisiae (Zou and Stillman, 2000), S. pombe (Ogawa et al., 1999) and human nuclei (Mendez and Stillman, 2000). The additional Mcm-s loaded on chromatin during G1-phase may represent backup copies that can take over should replication forks become stalled. The Mcm hexamers may function as a replicative helicase (You et al., 1999) and the excess of chromatin-loaded proteins may serve to increase the replicative fidelity, not the replication rates. Consistent with this idea, although 10–20 copies of Mcm 2–7 were loaded onto each replication origin on sperm nuclei incubated in Xenopus extracts, maximal replication rates were still observed when this was reduced to only ~2 copies per origin (Mahbubani et al., 1997). Alternatively, these could be Mcm complexes assembled on origins that do not fire. It has been demonstrated that in S. cerevisiae pre-RC proteins are bound to both active and silent chromosomal replication origins (Santocanale and Diffley, 1996). It is possible that eukaryotic nuclei assemble more origins than necessary as a means to ensure that the whole genome would be replicated even if some origins failed to fire. A third possibility is that a fraction of the chromatin-bound Mcm proteins are involved in genomic processes other than DNA replication. For example, Mcm proteins have been shown to be associated with transcription factors (DaFonseca et al., 2001; Zhang et al., 1998) and with components of the general transcription machinery (Yankulov et al., 1999) or to bind the promoters of several cell cycle-regulated genes (B. Tye et al., unpublished). Regardless of other potential functions of the Mcm proteins, our study demonstrates that the initial amount of Mcm-s loaded during telophase is fully functional and sufficient to license the CHO chromatin for replication.
Replication licensing is clearly insufficient to specify the DHFR origin of replication, as active replication forks were efficiently assembled within CHO pre-ODP nuclei introduced into licensing-deficient extracts. Initiation under these conditions takes place at sites distributed throughout the DHFR locus and must be mediated by functional pre-RCs assembled by CHO nuclei prior to nuclear isolation in order to initiate replication within licensing-defective extracts. Interestingly, we observed increased loading of XORC2 onto CHO metaphase chromatin in Xenopus egg extracts supplemented with geminin (Fig. 3, compare lanes 4 and 5). Identical results have been reported when Xenopus sperm chromatin was incubated in geminin-supplemented extracts (McGarry and Kirschner, 1998; Walter, 2000). These observations raise the possibility that loading of increasing amounts of Mcm-s onto chromatin serves to prevent ORC binding to illegitimate sites or unwanted ‘sliding’ of ORC along chromatin. Alternatively, the increased loading of Mcm-s throughout G1-phase could weaken the interaction of ORC proteins with chromatin and cause the dissociation of ORC from sites with low affinity, thus focusing initiation to more specific chromosomal sites with high affinity for ORC. In support of this hypothesis, Rowles et al. found that licensing (i.e. loading of Mcm-s on chromatin) dramatically changed XORC1 affinity for Xenopus sperm chromatin so that it became sensitive to extraction with high salt (Rowles et al., 1999). The ODP could represent the culmination of this process. Studies of ORC and Mcm interactions along the entire length of defined replicons in mammalian nuclei during different stages of G1-phase will be needed to further explore this hypothesis.
Two significant differences exist between our data, presented here, and those presented previously by another lab (Natale et al., 2000). The first issue is whether the replication licensing and the ODP are separate events. In contrast to previous measurements of the ODP (Wu and Gilbert, 1996), Natale et al., concluded that the ODP occurs at 2 hours in G1-phase, a timepoint at which they first detected the assembly of functional pre-RCs. Our observations indicate that the ODP can, indeed, occasionally exhibit limited temporal shifts, depending on the cell cycle length and/or growth conditions (D.S.D., unpublished). Thus, the ODP must be determined individually for each origin in different cell lines, as well as for each batch of synchronized cells using the same cell line. We use aliquots of the same batch of synchronized cells to carry out all different types of experiments, which excludes variability due to batch-to-batch differences in the kinetics of cell cycle progression. However, we have never observed an ODP earlier than 4 hours in G1-phase. A close examination of the data published by Natale et al., provides an explanation for this apparent discrepancy. Whereas replication within the CHO nuclei becomes independent of Xenopus ORC and Mcm proteins at 2–3 hours post-metaphase [meaning that the licensing is completed within 2–3 hours, figures 1 and 8 in Natale et al. (Natale et al., 2000)], the selection of specific origin sites has just begun at this time, but is not completed until 4–5 hours post-metaphase [figure 2 in Natale et al. (Natale et al., 2000)], fairly close to the timing documented by us. This implies that in both studies the licensing of CHO nuclei occurred no less than 2 hours prior to the ODP transition. We conclude that the replication licensing of mammalian nuclei and the selection of specific origin sites are uncoupled. Taken together with our previous work on the establishment of a temporal program for replication of CHO chromosomal domains (Dimitrova and Gilbert, 1999) and the dynamics of replicon activation during S-phase (Dimitrova and Gilbert, 1999; Dimitrova et al., 1999), we have now demonstrated that the four major regulatory steps (replication licensing, replication timing, selection of preferred origin sites and firing of the origins) in the replication of the mammalian genome are temporally separated (Fig. 7).
Fig. 7.
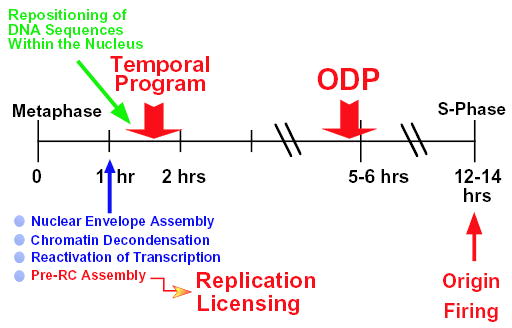
Establishment of a program for replication of mammalian nuclei. When CHOC 400 cells, blocked in metaphase with nocodazole, are released into G1-phase, nuclear assembly is completed within 1 hour [(Dimitrova and Gilbert, 1999; Dimitrova et al., 1999) and this study], coincident with the reactivation of general transcription (D.S.D., unpublished) and the licensing of chromatin for replication (this study). Within the next hour, chromosomal domains are directed to distinct and reproducible nuclear locations and by 2 hours into G1-phase (TDP, timing decision point) the temporal order for their replication in the upcoming S-phase is established (Dimitrova and Gilbert, 1999). At the ODP (5–6 hours in G1), specific chromosomal sites, where replication will initiate, are selected within the DHFR locus [(Wu and Gilbert, 1996) and this study]. Several hours later, at the G1/S transition (12–14 hours post-metaphase) the rise in S-phase promoting factor activity (S-phase cyclin-dependent kinases and dbf4/cdc7 kinase) leads to the assembly of replication factories and the firing of the earliest replication origins (Dimitrova et al., 1999). The four major events, which prepare the genome for replication, are highlighted in red.
The second important difference concerns the timing of assembly of functional pre-RCs. Natale et al. found that the competence to replicate in ORC- and Mcm-depleted extracts was not acquired until 2–3 hours after metaphase, whereas we observed efficient replication in Mcm-depleted or geminin-supplemented extracts as early as 1 hour after metaphase, when nuclear assembly has just been completed. One possibility is that this subtle difference could be due to differences in the handling of synchronized cells after mitotic shake-off. We have found that both the length of nocodazole treatment and the temperature at which mitotic cells are maintained between shake-off and re-plating are critical to the timely resumption of mitosis and normal progression into G1. Furthermore, we routinely monitor the exit of our mitotic cell preparations from mitosis and discard preparations that do not exhibit ≥90% G1-phase cells within 1 hour (Fig. 1). We have also confirmed that Mcm proteins are loaded on chromatin invariantly during late telophase in mammalian cells of different origin (human, hamster, rat and mouse) by indirect immunofluorescent labeling of Triton-extracted cells (D.S.D., unpublished). The characteristic mitotic figures allow for an extremely precise distinction of cells at different stages of mitosis even within asynchronous cell populations (Dimitrova et al., 1999), thus eliminating any possibility that the use of mitotic synchronization might have created artefacts in our experiments. As Natale et al. also observed tight binding of the Mcm proteins to chromatin within 1 hour post-metaphase, the only actual difference in results concerns the functional status of the pre-RCs assembled in late telophase. Despite our extensive efforts, we could not find conclusive evidence to account for this puzzling discrepancy. However, we believe that it may, at least in part, be a consequence of alterations introduced in the egg extract preparation protocol by these investigators (Li et al., 2000), resulting in unusual performance of their extracts. In particular, Li et al. found that upon introduction of hamster G1 nuclei in Xenopus egg extracts the radioactive label was incorporated into DNA strands 2–4 kb in length that do not grow as would be expected of active replication forks. By contrast, our extracts efficiently elongate nascent DNA (Dimitrova and Gilbert, 1998) (D.S.D. and D.M.G., unpublished), suggesting that the extracts used in the previous studies may have suffered a reduced replication competence. For unknown reasons, the earliest G1 nuclei might be more sensitive to the egg extract deficiencies and fail to replicate altogether. Even late G1 nuclei replicate at reduced levels under those conditions [only 10–20% of the genome within intact nuclei replicated Natale et al. (Natale et al., 2000) vs. 80–90% in our studies], once again pointing to suboptimal in vitro replication conditions.
Two observations suggest that our results are not due to deficiencies in our extracts: first, the Mcm-depleted extract can be rescued by purified Mcm complexes (T.A.P. and J.J.B., unpublished); second, we obtain similar results by simply adding geminin to the extract, which does not reduce extract activity or disrupt efficient nuclear envelope assembly (with Xenopus sperm or CHO metaphase chromatin). We conclude that licensing of mammalian chromatin takes place in late telophase, coincident with the stable association of Mcm proteins with chromatin, and is distinct from the specification of replication origin sites, which occurs several hours later, at the ODP.
We have previously shown that the permeabilization of CHO post-ODP nuclei results in loss of origin specificity even under conditions that preserve high replication rates (Dimitrova and Gilbert, 1998). There are at least three possible explanations for this observation. First, permeabilization may result in loss of soluble nuclear components, essential for specific origin activation. Second, the permeabilizing reagents affect the stability of endogenous hamster pre-RC complexes, which leads to their disassembly. Third, CHO pre-RCs remain stable after permeabilization, but factors in the Xenopus egg extracts gain access to the exposed CHO chromatin and re-arrange the existing pre-RCs or license illegitimate sites. The availability of licensing-defective extracts allowed us to address which of these possibilities were true. We found that a significant degree of DHFR origin specificity was preserved when digitonin-permeabilized post-ODP nuclei were allowed to initiate replication in geminin-supplemented extracts. The efficient site-specific initiation was supported by endogenous pre-RC proteins stably associated with CHO chromatin. This result supports primarily the third possibility, although a minor damage to endogenous hamster pre-RCs could not be ruled out, given the reduced degree of specificity, as compared to intact post-ODP nuclei. Our findings are consistent with a host of previous studies that have demonstrated the essential role of the intact nuclear envelope for properly regulated DNA replication. Importantly, the fact that initiation takes place at dispersed sites within pre-ODP nuclei, with or without the addition of geminin to the Xenopus egg extract, once again demonstrates that events taking place within CHO nuclei at a discrete point in G1-phase are responsible for focusing of initiation to specific origin sequences. Finally, our findings open up new possibilities for studies of the assembly of mammalian pre-RCs at specific chromosomal sites via the development of manipulatable systems for replication of mammalian chromatin. The role of individual pre-RC proteins can be evaluated by introducing chromatin preparations, from which specific pre-RC components have been removed, into geminin-supplemented Xenopus cytosol, which can support efficient replication without the unwanted licensing of additional initiation sites.
Footnotes
This work was supported by a Cancer Research Campaign (CRC) grant SP2385/0101 to J.J.B. and by NIH grant GM57233-01 to D.M.G.
References
- Aladjem MI, Rodewald LW, Kolman JL, Wahl GM. Genetic dissection of a mammalian replicator in the human β-globin locus. Science. 1998;281:1005–1009. doi: 10.1126/science.281.5379.1005. [DOI] [PubMed] [Google Scholar]
- Chong JPJ, Mahbubani HM, Khoo CY, Blow JJ. Purification of an MCM-containing complex as a component of the DNA replication licensing system. Nature. 1995;375:418–421. doi: 10.1038/375418a0. [DOI] [PubMed] [Google Scholar]
- Chong JP, Thommes P, Rowles A, Mahbubani HM, Blow JJ. Characterization of the Xenopus replication licensing system. Methods Enzymol. 1997;283:549–564. doi: 10.1016/s0076-6879(97)83043-1. [DOI] [PubMed] [Google Scholar]
- Coleman TR, Carpenter PB, Dunphy WG. The Xenopus Cdc6 protein is essential for the initiation of a single round of DNA replication in cell-free extracts. Cell. 1996;87:53–63. doi: 10.1016/s0092-8674(00)81322-7. [DOI] [PubMed] [Google Scholar]
- Coue M, Amariglio F, Maiorano D, Bocquet S, Mechali M. Evidence for different MCM subcomplexes with differential binding to chromatin in Xenopus. Exp Cell Res. 1998;245:282–289. doi: 10.1006/excr.1998.4271. [DOI] [PubMed] [Google Scholar]
- Coverley D, Wilkinson HR, Downes CS. A protein kinase-dependent block to reinitiation of DNA replication in G2 phase in mammalian cells. Exp Cell Res. 1996;225:294–300. doi: 10.1006/excr.1996.0179. [DOI] [PubMed] [Google Scholar]
- DaFonseca CJ, Shu F, Zhang JJ. Identification of two residues in MCM5 critical for the assembly of MCM complexes and Stat1-mediated transcription activation in response to IFN-gamma. Proc Natl Acad Sci USA. 2001;98:3034–3039. doi: 10.1073/pnas.061487598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova DS, Gilbert DM. Regulation of mammalian replication origin usage in Xenopus egg extract. J Cell Sci. 1998;111:2989–2998. doi: 10.1242/jcs.111.19.2989. [DOI] [PubMed] [Google Scholar]
- Dimitrova DS, Gilbert DM. The spatial position and replication timing of chromosomal domains are both established in early G1 phase. Mol Cell. 1999;4:983–993. doi: 10.1016/s1097-2765(00)80227-0. [DOI] [PubMed] [Google Scholar]
- Dimitrova DS, Gilbert DM. Temporally coordinated assembly and disassembly of replication factories in the absence of DNA synthesis. Nat Cell Biol. 2000;2:686–694. doi: 10.1038/35036309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova DS, Todorov IT, Melendy T, Gilbert DM. Mcm2, but not RPA, is a component of the mammalian early G1-phase prereplication complex. J Cell Biol. 1999;146:709–722. doi: 10.1083/jcb.146.4.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacca M, Zentilin L, Norio P, Diviacco S, Dimitrova D, Contreas G, Biamonti G, Perini G, Weighardt F, Riva S, et al. Fine mapping of a replication origin of human DNA. Proc Natl Acad Sci USA. 1994;91:7119–7123. doi: 10.1073/pnas.91.15.7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert DM, Miyazawa H, DePamphilis ML. Site-specific initiation of DNA replication in Xenopus egg extract requires nuclear structure. Mol Cell Biol. 1995;15:2942–2954. doi: 10.1128/mcb.15.6.2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg M, Jenkins H, Allen T, Whitfield WG, Hutchison CJ. Xenopus lamin B3 has a direct role in the assembly of a replication competent nucleus: evidence from cell-free egg extracts. J Cell Sci. 1995;108:3451–3461. doi: 10.1242/jcs.108.11.3451. [DOI] [PubMed] [Google Scholar]
- Kelly TJ, Brown GW. Regulation of chromosome replication. Annu Rev Biochem. 2000;69:829–880. doi: 10.1146/annurev.biochem.69.1.829. [DOI] [PubMed] [Google Scholar]
- Kubota Y, Mimura S, Nishimoto S, Masuda T, Nojima H, Takisawa H. Licensing of DNA replication by a multi-protein complex of MCM/P1 proteins in Xenopus eggs. EMBO J. 1997;16:3320–3331. doi: 10.1093/emboj/16.11.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leatherwood J. Emerging mechanisms of eukaryotic DNA replication initiation. Curr Opin Cell Biol. 1998;10:742–748. doi: 10.1016/s0955-0674(98)80117-8. [DOI] [PubMed] [Google Scholar]
- Li CJ, Bogan JA, Natale DA, DePamphilis ML. Selective activation of pre-replication complexes in vitro at specific sites in mammalian nuclei. J Cell Sci. 2000;113:887–898. doi: 10.1242/jcs.113.5.887. [DOI] [PubMed] [Google Scholar]
- Mahbubani HM, Chong JP, Chevalier S, Thommes P, Blow JJ. Cell cycle regulation of the replication licensing system: involvement of a Cdk-dependent inhibitor. J Cell Biol. 1997;136:125–135. doi: 10.1083/jcb.136.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiorano D, Lemaitre JM, Mechali M. Stepwise regulated chromatin assembly of MCM2-7 proteins. J Biol Chem. 2000a;275:8426–8431. doi: 10.1074/jbc.275.12.8426. [DOI] [PubMed] [Google Scholar]
- Maiorano D, Moreau J, Mechali M. XCDT1 is required for the assembly of pre-replicative complexes in Xenopus laevis. Nature. 2000b;404:622–625. doi: 10.1038/35007104. [DOI] [PubMed] [Google Scholar]
- McGarry TJ, Kirschner MW. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell. 1998;93:1043–1053. doi: 10.1016/s0092-8674(00)81209-x. [DOI] [PubMed] [Google Scholar]
- Mendez J, Stillman B. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol Cell Biol. 2000;20:8602–8612. doi: 10.1128/mcb.20.22.8602-8612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milbrandt JD, Heintz NH, White WC, Rothman SM, Hamlin JL. Methotrexate-resistant Chinese hamster ovary cells have amplified a 135-kilobase-pair region that includes the dihydrofolate reductase gene. Proc Natl Acad Sci USA. 1981;78:6043–6047. doi: 10.1073/pnas.78.10.6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natale DA, Li CJ, Sun WH, DePamphilis ML. Selective instability of Orc1 protein accounts for the absence of functional origin recognition complexes during the M-G1 transition in mammals. EMBO J. 2000;19:2728–2738. doi: 10.1093/emboj/19.11.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitani H, Lygerou Z, Nishimoto T, Nurse P. The Cdt1 protein is required to license DNA for replication in fission yeast. Nature. 2000;404:625–628. doi: 10.1038/35007110. [DOI] [PubMed] [Google Scholar]
- Noton E, Diffley JF. CDK inactivation is the only essential function of the APC/C and the mitotic exit network proteins for origin resetting during mitosis. Mol Cell. 2000;5:85–95. doi: 10.1016/s1097-2765(00)80405-0. [DOI] [PubMed] [Google Scholar]
- Ogawa Y, Takahashi T, Masukata H. Association of fission yeast Orp1 and Mcm6 proteins with chromosomal replication origins. Mol Cell Biol. 1999;19:7228–7236. doi: 10.1128/mcb.19.10.7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokhorova TA, Blow JJ. Sequential MCM/P1 subcomplex assembly is required to form a heterohexamer with replication licensing activity. J Biol Chem. 2000;275:2491–2498. doi: 10.1074/jbc.275.4.2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanowski P, Madine MA, Rowles A, Blow JJ, Laskey RA. The Xenopus origin recognition complex is essential for DNA replication and MCM binding to chromatin. Curr Biol. 1996;6:1416–1425. doi: 10.1016/s0960-9822(96)00746-4. [DOI] [PubMed] [Google Scholar]
- Rowles A, Blow JJ. Chromatin proteins involved in the initiation of DNA replication. Curr Opin Genet Dev. 1997;7:152–157. doi: 10.1016/s0959-437x(97)80123-2. [DOI] [PubMed] [Google Scholar]
- Rowles A, Chong JP, Brown L, Howell M, Evan GI, Blow JJ. Interaction between the origin recognition complex and the replication licensing system in Xenopus. Cell. 1996;87:287–296. doi: 10.1016/s0092-8674(00)81346-x. [DOI] [PubMed] [Google Scholar]
- Rowles A, Tada S, Blow JJ. Changes in association of the Xenopus origin recognition complex with chromatin on licensing of replication origins. J Cell Sci. 1999;112:2011–2018. doi: 10.1242/jcs.112.12.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santocanale C, Diffley JF. ORC- and Cdc6-dependent complexes at active and inactive chromosomal replication origins in Saccharomyces cerevisiae. EMBO J. 1996;15:6671–6679. [PMC free article] [PubMed] [Google Scholar]
- Tada S, Li A, Maiorano D, Mechali M, Blow JJ. Repression of origin assembly in metaphase depends on inhibition of RLF-B/Cdt1 by geminin. Nat Cell Biol. 2001;3:107–113. doi: 10.1038/35055000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Knapp D, Nasmyth K. Loading of an Mcm protein onto DNA replication origins is regulated by Cdc6p and CDKs. Cell. 1997;90:649–660. doi: 10.1016/s0092-8674(00)80526-7. [DOI] [PubMed] [Google Scholar]
- Thommes P, Kubota Y, Takisawa H, Blow JJ. The RLF-M component of the replication licensing system forms complexes containing all six MCM/P1 polypeptides. EMBO J. 1997;16:3312–3319. doi: 10.1093/emboj/16.11.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todorov IT, Attaran A, Kearsey SE. BM28, a human member of the MCM2-3-5 family, is displaced from chromatin during DNA replication. J Cell Biol. 1995;129:1433–1445. doi: 10.1083/jcb.129.6.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tye BK. MCM proteins in DNA replication. Annu Rev Biochem. 1999;68:649–686. doi: 10.1146/annurev.biochem.68.1.649. [DOI] [PubMed] [Google Scholar]
- Walter JC. Evidence for sequential action of cdc7 and cdk2 protein kinases during initiation of DNA replication in Xenopus egg extracts. J Biol Chem. 2000;275:39773–39778. doi: 10.1074/jbc.M008107200. [DOI] [PubMed] [Google Scholar]
- Wohlschlegel JA, Dwyer BT, Dhar SK, Cvetic C, Walter JC, Dutta A. Inhibition of eukaryotic DNA replication by geminin binding to Cdt1. Science. 2000;290:2309–2312. doi: 10.1126/science.290.5500.2309. [DOI] [PubMed] [Google Scholar]
- Wu JR, Gilbert DM. A distinct G1 step required to specify the Chinese hamster DHFR replication origin. Science. 1996;271:1270–1272. doi: 10.1126/science.271.5253.1270. [DOI] [PubMed] [Google Scholar]
- Yankulov K, Todorov I, Romanowski P, Licatalosi D, Cilli K, McCracken S, Laskey R, Bentley DL. MCM proteins are associated with RNA polymerase II holoenzyme. Mol Cell Biol. 1999;19:6154–6163. doi: 10.1128/mcb.19.9.6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Z, Komamura Y, Ishimi Y. Biochemical analysis of the intrinsic Mcm4-Mcm6-Mcm7 DNA helicase activity. Mol Cell Biol. 1999;19:8003–8015. doi: 10.1128/mcb.19.12.8003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Wu JR, Gilbert DM. Analysis of mammalian origin specification in ORC-depleted Xenopus egg extracts. Genes Cells. 1998;3:709–720. doi: 10.1046/j.1365-2443.1998.00224.x. [DOI] [PubMed] [Google Scholar]
- Zhang JJ, Zhao Y, Chait BT, Lathem WW, Ritzi M, Knippers R, Darnell JE., Jr Ser727-dependent recruitment of MCM5 by Stat1alpha in IFN-gamma-induced transcriptional activation. EMBO J. 1998;17:6963–6971. doi: 10.1093/emboj/17.23.6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Stillman B. Assembly of a complex containing Cdc45p, replication protein A, and Mcm2p at replication origins controlled by S-phase cyclin-dependent kinases and Cdc7p-Dbf4p kinase. Mol Cell Biol. 2000;20:3086–3096. doi: 10.1128/mcb.20.9.3086-3096.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]


