Abstract
Telomeres across the genus Drosophila are maintained, not by telomerase, but by two non-LTR retrotransposons, HeT-A and TART, that transpose specifically to chromosome ends. Successive transpositions result in long head-to-tail arrays of these elements. Thus Drosophila telomeres, like those produced by telomerase, consist of repeated sequences reverse transcribed from RNA templates. The Drosophila repeats, complete and 5′-truncated copies of HeT-A and TART, are more complex than telomerase repeats; nevertheless these evolutionary variants have functional similarities to the more common telomeres. Like other telomeres, the Drosophila arrays are dynamic, fluctuating around an average length that can be changed by changes in the genetic background. Several proteins that interact with telomeres in other species have been found to have homologues that interact with Drosophila telomeres. Although they have hallmarks of non-LTR retrotransposons, HeT-A and TART appear to have a special relationship to Drosophila. Their Gag proteins are efficiently transported into diploid nuclei where HeT-A Gag recruits TART Gag to chromosome ends. Gags of other non-LTR elements remain predominantly in the cytoplasm. These studies provide intriguing evolutionary links between telomeres and retrotransposable elements.
Keywords: Chromosome evolution, Retrotransposon evolution, Telomere maintenance, Nuclear localization, Gag protein
Introduction
Until recently telomeres were of interest primarily to cytologists and those concerned with how DNA polymerase replicates the extreme ends of linear DNA. Now a wealth of data ties these chromosome ends to cellular senescence, cell cycle checkpoints, organismal aging, and tumorogenesis (for review see Blackburn 2001, Greider 1996, Collins 2000). It is apparent that telomere biology is relevant to many areas of biology.
A new and unexpected mechanism of DNA replication was discovered when it was shown that an enzyme, telomerase, with an internal RNA template, copied part of this template repeatedly into DNA to extend chromosome ends (Greider & Blackburn 1987, Blackburn 1992). This extension circumvents the gradual shortening that should result from the inability of DNA polymerase to replicate the last few nucleotides of the chromosome.
It has been puzzling why cells devote so many resources to stopping chromosomal end erosion. Human germline cells, for example, have ~15 kbp of telomeric DNA on each end, considerably more than needed to support cell division over the life of the organism. Linear viruses solve the problem of replicating DNA ends in a variety of ways, all seemingly simpler than the chromosomal solution. Nevertheless telomerase extension is found in almost all animals, plants, and single-celled eukaryotes. In each case, telomeres are long and their length is regulated in species-specific and cell type-specific ways. Accumulating evidence shows that the large amount of DNA in telomeres does not simply buffer chromosome ends; this DNA also binds specific proteins and protein complexes with multiple functions, such as preventing telomeres from behaving like broken ends. Typically, broken ends trigger apoptosis or fuse to other ends, creating dysfunctional chromosomes. Studies on yeast and mammalian chromosomes show that length is only one factor in appropriate telomere function (Blackburn 2001, Blasco 2002, Karlseder et al. 2002). It appears that the formation of a telomere structure is necessary for having appropriate telomere function. What we now know may well be only the beginning of the story.
Drosophila telomeres, conserved evolutionary variants in telomere maintenance.
The molecular biology of telomeres is more complex –-and more interesting -- than it appeared in early studies. Understanding such complex biological problems is greatly assisted by insight from studies of evolutionary variants. Drosophila is such a variant (see Pardue & DeBaryshe 2003). Drosophila has no telomerase and it lacks the short telomeric repeats of other organisms. Instead, studies on D. melanogaster revealed two telomere-specific retrotransposable elements, HeT-A and TART, present in multiple copies on normal telomeres and able to “heal” broken chromosome ends (Fig. 1).
Figure 1.

Diagram of the two D. melanogaster telomere retrotransposons, drawn as their putative RNA transposition intermediates. Coding regions and 3′ UTRs are labeled. (A)n indicates the poly(A) tail on the RNAs. It is the source of the (dA/T)n that joins each DNA copy to the chromosome when the element transposes. HeT-A elements are ~ 6 kb. The 5′ end of TART has not yet been completely defined but subfamilies appear to be 10–12 kb.
HeT-A and TART are non-LTR retrotransposons that transpose specifically onto chromosome ends. Successive transpositions produce head-to-tail arrays of mixed complete and 5′-truncated elements on the ends of Drosophila chromosomes (fig. 2). These arrays appear to be analogous to the arrays of repeats produced by telomerase in other organisms but the Drosophila repeats are much longer and more complex than the other repeats. HeT-A is ~6 kb and TART is more than 10 kb; telomerase repeats tend to be very short (5–8 bp in most organisms).
Figure 2.

Model for extension of chromosome ends by telomeric retrotransposons. Retrotransposons yield sense-strand transcripts that serve as both mRNAs and transposition intermediates. This diagram shows our current model for the path of these RNAs from transcription until they are reverse transcribed to add another repeat onto the telomere array. Gray arrows represent HeT-A (dark) and TART (light) elements attached to the end of the chromosome. A poly(A) sense-strand RNA is transcribed from a member of the array (step 1). For the telomeric retrotransposons, there is evidence suggesting that this RNA must be translated (step 2) before serving as a template (step 3) for telomere additions. This suggestion is now supported by the finding that translation products (Gags) of these RNAs appear capable of delivering the transposition template specifically to its target at the telomere. Gray circles in diagram represent Gags of either HeT-A or TART. Analogy with retroviruses suggests that reverse transcriptase is also included in the Gag-RNA complex; however, there is no evidence on this point. Reproduced from The Journal of Cell Biology, 2002, 158:398 by copyright permission of the Rockefeller University Press.
Telomeres have been studied in only a few insects; some have telomerase (Okazaki et al. 1993); however, a search for telomerase-type repeats across the phylum Arthropoda showed several branches of the phylogeny that apparently do not use this mechanism (Sahara et al. 1999)). How many of these groups use telomere-specific retrotransposons? It would seem that the rapid accumulation of genomic sequences would make it simple to answer this question. Unfortunately this is not the case. Chromosome ends present enormous challenges for sequence assembly because their sequences are highly repetitive. In addition to the terminal telomerase or retrotransposon repeats, the subtelomeric telomere associated sequences (TAS) are also repetitive. These regions are poorly represented in currently available sequence assemblies.
As is typical of retroelements, the sequences of HeT-A and TART evolve rapidly, making cross-species studies difficult. Both elements have long stretches of non-coding sequence (the 3′ UTR), which can differ significantly between copies of the element in the same Drosophila stock. Both elements also encode Gag proteins whose sequence can vary as much as the non-coding regions. Only the most conserved part of the D. melanogaster TART reverse transcriptase gene was able to cross-hybridize, at low stringency, with D. virilis DNA. This allowed us to isolate DNA fragments that provided entry into the D. virilis HeT-A/TART telomere arrays (Casacuberta & Pardue 2003a, 2003b).
Although D.virilis is separated from D. melanogaster by 60 million years, the HeT-A and TART elements in both species are present in telomere arrays and they are not found in euchromatic regions. As expected from the evolutionary distance between D.virilis and D. melanogaster (Beverly & Wilson 1984), the sequences of the D.virilis elements are significantly different from the D. melanogaster elements. However, they maintain so many of the unusual features that characterize the D. melanogaster elements that there is no doubt of their identity. The D. melanogaster features will be discussed below. They include long 3′UTRs, irregular A-rich repeats in the HeT-A 3′ UTR, the lack of reverse transcriptase coding sequences in HeT-A, and gag genes with motifs that are also found in retroviral gag genes. Although the high sequence divergence of the telomere elements makes it difficult to find these elements in new species, it also increases the probability that the conserved features are of biological importance.
The similarities between the telomeric retrotransposons in D. melanogaster and D. virilis, two of the most distantly related Drosophila species, argue that the telomeric roles of HeT-A and TART predate the separation of the genus Drosophila. Thus other insects may use this mechanism of telomere maintenance.
Drosophila telomeres, like those of other eukaryotic nuclear chromosomes, are dynamic structures.
It is notable that reverse transcription of RNA templates by telomerase provides an efficient way to cope with both gradual and sudden loss of chromosome end sequences. The Drosophila telomere is also a product of reverse transcription and has the same capabilities. Drosophila, like organisms with telomerase, can also use recombination or gene conversion to supplement this reverse transcription (Mikhailovsky et al. 1999, Kahn et al. 2000). Nevertheless, except in mutant individuals, RNA-templated (telomerase or retrotransposon) extension predominates, suggesting that this mechanism has advantages over recombination/gene conversion-based telomere extension.
One advantage of RNA templating may be that it gives a better control of telomere length and length control appears to be an important aspect of telomere metabolism. For example, telomere lengths in budding yeast fluctuate within narrow limits; however, those limits can be changed by genotype or by external conditions (see Blackburn 2001). A recent study identified >150 yeast genes that could cause the average telomere length to become either shorter or longer (Askree et al, 2004). In contrast, telomerase-null mutants of budding yeast have widely varying telomere lengths and poor growth (Lundblad & Blackburn 1993). Mammals regulate telomere length in specific cell types by regulating the expression of telomerase (Greider 1996). Some mammalian tumor cells that do not express telomerase use a recombination-based mechanism, Alternative Lengthening of Telomeres (ALT). Like telomerase-null yeast cells, these mammalian cells are marked by widely varying telomere lengths (see Henson et al. 2002).
Telomere lengths in D. melanogaster, like those in other organisms, are affected by genetic background. Mutations in three genes, Su(var)205 (Savitsky et al. 2002), E(tc) (Melnikova & Georgiev 2002) and Tel (Siriaco et al. 2002), have been shown to lead to increased telomere length. In addition, expression of HeT-A is strongly developmentally regulated (George & Pardue 2003, Walter & Biessmann 2004), although this regulation does not appear to be related to aging, as is the regulation in mammals. HeT-A RNA is found in diploid cells, which make up the tissues of the adult, rather than in the polyploid/polytene cells that form the larval body. Surprisingly, TART does not show this developmental regulation. TART RNA is abundant in larval cells as well as adult stages. Because studies of their Gag proteins (see below) suggest that HeT-A and TART act together for telomere targeting, it is not clear whether the expression of TART has consequences for larval cells where there is no HeT-A RNA.
Telomere arrays in multicellular eukaryotes are typically much longer than the arrays in unicellular eukaryotes. D. melanogaster is no exception. The best estimate of Drosophila telomeres comes from the work of Abad, et al. 2004a, who have measured the length of the HeT-A/TART -containing BAC clones derived from the fly stock used by the Drosophila Genome Project. HeT-A/TART arrays from five different chromosome ends in that stock ranged between 147 kb and 10 kb, in the range reported for mammalian telomeres. We do not know whether these long arrays form a structure similar to the T-loops found at mammalian telomeres (Griffith et al. 1999); however, there are reasons to think that the HeT-A/TART arrays are in some special chromatin structure. For example, the many transposable elements that insert promiscuously in other parts of the genome have not been found in HeT-A/TART arrays except in small regions at the junctions between the HeT-A/TART arrays and subtelomeric regions. This spatial restriction suggests that the much of the array is in a structure that is inaccessible to insertion by other elements (J George, PG DeBaryshe & M-L Pardue, manuscript in preparation).
Sequence-specific binding proteins have been identified for yeast and human telomere repeats but none are known for HeT-A or TART. Two D. melanogaster proteins that bind telomeres, HP1 and HOAP, bind DNA ends, or something that binds DNA ends, rather than being specific for the retrotransposon sequences (Fanti et al. 1998, Siriaco et al. 2002, Cenci et al. 2003). Nevertheless, several proteins that affect telomere chromatin structure in other organisms also affect D. melanogaster telomeres. These include ATM, Mre11, and Rad 50, all of which are involved in preventing telomere fusion (see Purdy & Su 2004 for review). Ku, which affects telomeres in animals, plants, and yeast, has recently been shown to affect the frequency of HeT-A/TART addition to ends as well as terminal gene conversion (Melnikova et al. 2005). The evidence that Drosophila telomeres interact with several proteins that are homologous to telomere-related proteins in other species supports the idea that the Drosophila telomere DNA repeats are analogous to telomerase repeats.
HeT-A and TART are retrotransposons with an unusual relationship to Drosophila cells.
Sequence organization and replication of retrotransposable elements.
Three major groups of replication-competent retrotransposable elements are found in eukaryotic chromosomes: non-LTR retrotransposons, LTR retrotransposons, and retroviruses. Many elements in each of the three groups show a common theme of sequence organization. They also have very little sequence that does not code for polypeptides involved in their transposition and, in viruses, for extracellular survival and infectivity. HeT-A and TART are exceptions to this general pattern because they contain a significant portion of non-coding DNA (Fig 1). It seems likely that this non-coding DNA is related to their role at the telomere.
Coding regions of retroviruses have been studied intensively, originally by genetic analysis and more recently by use of recombinant DNA constructs to examine the function of viral products. Three major coding regions are common to all retroviruses: a 5′ gag gene encoding capsid proteins, a central pol gene encoding enzymes necessary for replication and integration, and a 3′ env gene encoding envelope proteins. The gag and pol genes of retrotransposons have been much less-well studied but the sequences of these two genes are very similar to the retroviral equivalents. This suggests that retroviral studies can inform studies of retrotransposons.
Both LTR retrotransposons and retroviruses carry long terminal repeats of a few hundred base pairs at each end of the element; these are involved in synthesis of double stranded DNA from the retroelement RNA and also in integration of this DNA into chromosomes. Because of these similarities in structure, replication, and integration mechanism, it is generally accepted that LTR retrotransposons and retroviruses are evolutionarily related by either gain or loss of ability to persist outside cells and to enter new cells. The direction of this evolution is debated; it is possible that both gains and losses have happened more than once.
Non-LTR elements differ notably from LTR-elements and retroviruses in their method of replication and integration. LTR elements and retroviruses typically are reverse transcribed in the cytoplasm and integrate into chromosomes as double-stranded DNA. Non-LTR elements are reverse transcribed directly onto the chromosome at the site of integration. Most non-LTR elements insert at nicks in DNA, using the exposed 3′ hydroxyl in the nick to prime reverse transcription of the retrotransposon RNA directly onto the chromosome (Luan et al. 1993). HeT-A and TART do not appear to require nicked DNA since they most probably are reverse transcribed directly onto the end of the chromosome. Thus the mechanism of extending the Drosophila telomere is directly analogous to that used by telomerase. In both cases DNA sequence is added by reverse transcription of an RNA template. The major difference is the size of the template. Telomerase templates tend to be 5–20 nt, depending on the species. The Drosophila templates are the much larger HeT-A and TART RNAs.
Targeting of HeT-A and TART to chromosome ends.
The D. melanogaster genome has some 60 families of known retrotransposable elements. Of these, only HeT-A and TART transpose to chromosome ends. Sequence analyses of 3′ junctions in HeT-A-TART arrays, as well as evidence that these elements can transpose to broken chromosome ends, provides evidence that targeting of these elements does not depend on the DNA sequence at the site of transposition. Instead the targeting seems to depend on the Gag proteins of the two elements, especially the HeT-A Gag protein (see Pardue & DeBaryshe 2003 for review).
When the Gag proteins of HeT-A and TART are tagged with GFP (Green Fluorescent Protein) and transiently expressed in cultured D. melanogaster cells, both proteins are very efficiently moved into the nucleus and localized to well-defined sites, dots for HeT-A Gag (Het-dots) and clusters for TART Gag (Fig. 3). The efficient nuclear localization seen for HeT-A and TART Gags is very different from what happens to the Gags of other D. melanogaster retrotransposons. Doc and jockey are closely related to TART and HeT-A on the basis of coding sequence analysis. I Factor is more distantly related. When Gag proteins from these non-telomeric elements are tagged with GFP and expressed in the cultured cells, little if any, of the protein moves into the nucleus. It seems reasonable to suppose that the cytoplasmic retention of Gags from Doc, jockey, and I Factor is a reflection of the cell’s struggle to keep these “parasitic’ elements from increasing their copy number in the genome. Thus the nuclear localization of the Gags of HeT-A and TART reflects a relationship with D. melanogaster cells that is very different than the relationship of non-telomeric retrotransposons (Rashkova et al. 2002a, 2002b).
Figure 3.

Intracellular localization of GFP (Green Fluorescent Protein)-tagged Gags in transiently transfected cultured Drosophila cells of the SL2 line. Fluorescence micrographs show each cell in two panels. DNA in all cells is stained with DAPI (false-colored red). Left panels show merged GFP and DAPI superimposed on the DIC image. Right panels show GFP alone. Transfectants shown: a. HeT-A Gag – a cell with large Het-dots in the nucleus and Het-body in the cytoplasm; b. TART Gag--small clusters in the nucleus, more diffuse than Het-dots. c. Doc (a non-telomeric retrotransposon) Gag – irregular clusters in cytoplasm, more concentrated near the nucleus.
The nuclear localization of HeT-A and TART Gags depends not only on the Gag proteins but also on components of the host cell (Fig. 4). The host cell requirements became obvious when we tried to express HeT-A Gag in polytene cells so that we could use the giant polytene chromosomes to map the sites of where Gag protein associates. As mentioned earlier, HeT-A is not expressed in polytene cells and therefore we expressed Gag-GFP from a transgene that could be driven in these cells. The protein was expressed very well but none localized to the nucleus. Instead the protein accumulated in the cytoplasm, showing that these cells lacked the components needed for nuclear localization of this protein. The sites of cytoplasmic localization differed in different polyploid cell types, further emphasizing the difference in cell-type contributions to the localization of the proteins.
Figure 4.

Fluoresence micrographs of cells from transgenic Drosophila flies expressing HeT-A Gag-GFP protein under the control of Gal4 driven by the daughterless promoter. Gag protein does not enter the nuclei of polyploid cells (a, b). In polytene salivary gland cells (a), a large part of Gag-GFP is in an irregular aggregation (possibly the Golgi apparatus) adjacent to the nucleus. In polyploid tubule cells (b) the protein is entirely cytoplasmic and forms small clusters of approximately equal size. In diploid cells (c) the protein makes Het-dots in the nucleus with a small possible Het-body in the cytoplasm (but notice the magnification compared to a and b). Gag-GFP protein is green. DNA is stained with DAPI (red). These data stacks were deconvolved using Deltavision software.
In interphase diploid cells, HeT-A Gag localizes to well-defined Het-dots in the nucleus. The number and distribution of these dots is that expected for chromosome ends but, because the dots dissociate when chromosomes condense at metaphase, the possible association must be studied by colocalization with something that marks telomeres in interphase nuclei. The protein HOAP is such a marker. Currently HOAP is the only protein known to localize predominantly to D. melanogaster telomeres in both interphase and metaphase cells. When transfected cells are reacted with antibodies against HOAP, Het-dots colocalize with HOAP staining (Rashkova et al.2002b).
Even when cells are centrifuged onto slides to break nuclei and spread their contents, the colocalization of HeT-A Gag and HOAP survives the force. The increased resolution in the spread nuclei also shows that some spots, although overlapping, are not completely coinciding. This suggests that the two proteins are associated, not with each other, but with something in the same region of the chromosome. This suggestion is supported by comparison of the sizes of the spots. Metaphase preparations show that the amount of HOAP staining varies in a chromosome-specific way. In spread interphase nuclei, both Het-dots and HOAP spots vary in size; colocalized spots and dots are not always of the same relative sizes. If the localization were due to binding between the two proteins we would expect the amounts of the two proteins to vary together (Rashkova et al. 2002b).
There is one intriguing feature of the localization of HeT-A Gag; cells with nuclear Het-dots frequently show one well-defined cytoplasmic cluster of the protein that we have named the Het-body (Fig 3). These bodies form only after there has been significant accumulation of Gag protein in the nucleus, suggesting that the bodies reflect saturation of some aspect of the nuclear localization of the protein. We have not been able to associate Het-bodies with any known cytoplasmic organelle. Het-bodies are very specific to the full-length HeT-A Gag. We have expressed many proteins in these cells and only cells expressing full-length HeT-A Gag form Het-bodies.
Although TART Gag has the same striking nuclear sequestration as HeT-A Gag, its intranuclear localization is different. TART Gag forms many smaller clusters and they are less regularly distributed than those of HeT-A Gag. When cells expressing TART Gag are centrifuged onto slides and stained with HOAP antibody, TART Gag clusters show no association with the telomeres that are detected by HOAP staining. Instead the clusters dissociate and the protein spreads fairly evenly throughout the nucleus, Thus TART Gag differs from HeT-A Gag in both its nuclear distribution and its molecular associations. This difference is puzzling because the two elements form mixed arrays on telomeres and thus might be expected to have similar telomere associations.
The puzzle of TART Gag localization was resolved when the two Gags were tagged with different fluorescent proteins and co-expressed (Rashkova et al. 2002b). When both proteins are present in the same cell, HeT-A Gag dominates their localization and recruits TART Gag into both Het-dots and the Het-body. This cooperation may explain why we have never seen a Drosophila stock or species without both of these telomere-specific retrotransposons. The cooperation would allow HeT-A to provide the telomere targeting, which TART lacks, while TART provides the reverse transcriptase, which HeT-A lacks.
Although cooperation provides a convenient explanation for the two elements, it raises a second question: why has no recombinant element taken over the telomere? It would seem that an element with both HeT-A gag and TART pol should be a much more efficient solution, yet of all elements sequenced in several species only one complete and three partial copies of such a recombinant have been found (Abad et al. 2004b). These elements show that recombinants can form but their rarity argues that they are not able to compete with the two independent elements. The persistence of both HeT-A and TART strongly suggests that each contributes something beyond the telomere targeting and reverse transcription.
Evidence that HeT-A and TART Gag associate provides a possible answer to the question of where HeT-A obtains its reverse transcriptase. However there is still a question of how TART reverse transcriptase gets into the nucleus. Most retrotransposons have their Pol protein translationally linked to Gag either by a translational frame shift or by a leaky termination signal (Jacks, 1990). This translational linkage has two results. First, the pol gene is translated much less frequently than the gag gene, thus assuring lower levels of the catalytic pol products than of the structural gag products. Second, pol products are physically linked to the gag polypeptide; assuring that they are incorporated into the viral capsid formed by multimerization of gag products (Wills & Craven, 1992). TART has sequence between the gag and pol coding regions that seems to require reinitiation for translation of Pol. Thus Pol would not be physically linked to Gag, suggesting it must either associate with Gag post-translationally or be capable of making its own way to the telomere.
When TART Pol is transiently expressed in Drosophila cells from the vector used for the Gags, very little Pol is obtained and most, if not all, of this protein is cytoplasmic (Fig 5). It appears that Pol is unstable in the cells, possibly because it is not protected by association with Gag. Co-expression of TART Pol with both TART Gag and HeT-A Gag has not resulted in detectable telomere localization of Pol. Pol, like Gags of Doc and jockey, is detected in Het-bodies but not seen in Het-dots. It appears that these transfection experiments have not successfully replicated the Pol transport system.
Figure 5.
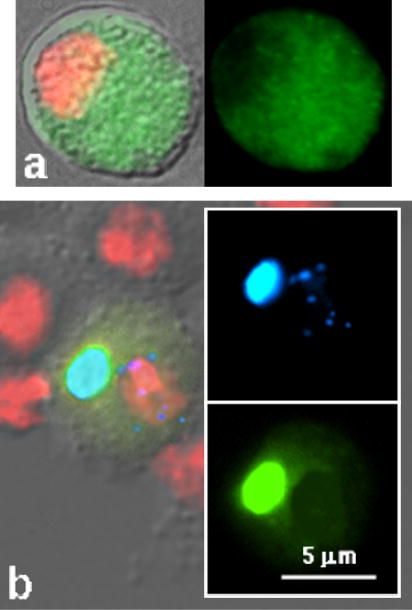
Intracellular localization of TART Pol-GFP in transiently transfected cultured Drosophila cells. (a) Left panel shows merged GFP and DAPI (red) superimposed on the DIC image. Right panel shows GFP alone. Pol-GFP is not detected in the nucleus. b. Left panel shows parts of four untransfected cells surrounding a cell co-transfected with TART Pol-YFP (Yellow Fluorescent Protein) and HeT-A Gag-CFP (Cyan Fluorescent Protein). Panels on right show YFP (shown as yellow-green) and CFP (shown as blue) only. HeT-A Gag forms nuclear Het-dots but does not recruit TART Pol into the nucleus; however the two proteins apparently have some interaction, as evidenced by the recruitment of TART Pol into the cytoplasmic Het-body.
Mapping domains of Gag involved in nuclear targeting.
Studies with deletion derivatives of the two Gags have been used to map parts of the protein involved in different steps in the telomere localization (Rashkova et al. 2003). The final step in telomere association of HeT-A Gag cannot be mapped to a single motif. Instead it requires information from several parts of the protein. In contrast, the association that colocalizes TART Gag maps to a single region (Fig. 6). This region contains two well-known amino acid motifs that characterize retroviral Gags (Wills & Craven 1992). These motifs are the Major Homology Region (MHR) and the zinc knuckles (or CCHC box). This region of retroviral Gags is also implicated in protein-protein associations.
Figure 6.
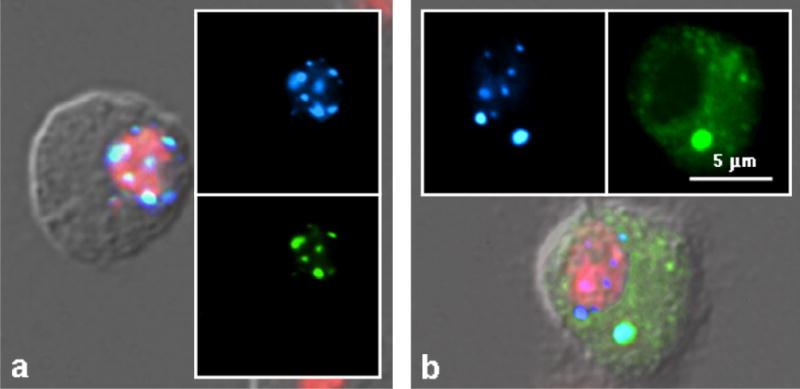
HeT-A Gag recruits TART Gag, but not Doc Gag, to nuclear Het-dots. (a) Cell co-transfected with HeT-A Gag-CFP and TART Gag-YFP. Left panel shows merged CFP, YFP and DAPI (red) superimposed on the DIC image. Right panels show CFP and YFP alone. HeT-A Gag recruits TART Gag to both nuclear Het-dots and cytoplasmic Het-body (although this cell has no Het-body). (b) Cell co-transfected with HeT-A Gag-CFP and Doc Gag-YFP. Bottom panel shows merged CFP, YFP and DAPI (red) superimposed on the DIC image. Top panels show CFP and YFP alone. HeT-A Gag does not carry Doc Gag into the nucleus but is associated with it in the cytoplasmic Het-body.
The MHR-zinc knuckle region is the most conserved part of HeT-A and TART Gags in different Drosophila species (Casacuberta & Pardue 2005). It is also fairly conserved in other Drosophila retrotransposons belonging to the clade with which TART and HeT-A group (Rashkova et al. 2003). Do these other retrotransposons exploit this similarity to foil cell defenses by hitching a ride on HeT-A or TART? To test this, tagged proteins from Doc, jockey, and I Factor were coexpressed with HeT-A Gag (Fig. 6). None of the proteins were transported into nuclei; however, the two most closely related Gags, from Doc and jockey, were incorporated into the cytoplasmic Het-body. Of the many proteins that have been co-expressed, these are two of only three (including TART Pol, mentioned above) that have been incorporated into Het-bodies without also being transported into nuclear Het-dots. It may be that the interaction with HeT-A Gag is not strong enough to withstand transport into the nucleus. Alternatively, something about the Doc/jockey protein might trigger more specific inhibition of nuclear entry.
Hypothetical evolution of the Drosophila telomere and its implications for chromosome evolution.
Because some insects have telomerase (Okazaki et al. 1993, Sahara et al, 1999) Drosophila must have shared ancestors with organisms that have the more common telomere. How did Drosophila come to have its telomeres maintained by transposable elements, a class of elements previously considered to be “parasitic” or “selfish” DNA? There are several possible hypotheses: Drosophila might have lost its telomerase and had the role taken over by a “parasitic” element, converting a wild element to domestic use. Alternatively, the telomere transposable elements might have evolved from telomerase. Conversely, transposable elements might be the retention of an ancestral mechanism. The first hypothesis, domestication of a pre-existing transposon, is less attractive because it requires the element to have acquired a tendency to associate with the chromosome ends and also to have completely lost the ability to transpose into euchromatic regions. (We do not find even small remnants of HeT-A or TART sequences in euchromatin, although Drosophila euchromatic regions contain many other transposable elements.)
Several observations suggest that the telomere transposable elements may be evolutionarily derived from telomerase (Pardue et al. 1996). The mechanism for extending the Drosophila telomere is basically the same as that used by telomerase: in each case, a reverse transcriptase extends the end of the chromosome by adding DNA copied from an RNA template. Telomerase is a ribonucleoprotein that repeatedly copies a segment of its RNA component to yield long tandem arrays. In Drosophila, the enzyme copies the entire RNA template. These same observations are compatible with the hypothesis that the Drosophila telomere is more like the ancestral telomere.
This evolutionary hypothesis is intriguing because it would relate retrotransposons – and the related retroviruses – to a cellular element, the telomerase complex. We have posited that gene rearrangement joined the 5′ end of a gene for the RNA component of telomerase to the 3′ end of a gene for a telomerase protein component that targeted the telomerase complex to the chromosome end (a “proto”-Gag protein). The transcript of such a chimeric gene would look like HeT-A. The 3′ end of this new RNA would already be a template for reverse transcription, because it serves that role in telomerase. The compound transcript might be translated into a protein (“proto-Gag”) that would escort the RNA to the telomere where it could do double duty by being reverse transcribed into DNA. This is still our working hypothesis for the origin of HeT-A. Our finding that TART belongs to a subgroup of non-LTR elements with PNTR [perfect non-terminal repeats (see Pardue & DeBaryshe 2003)] suggests that the two elements may be products of convergent evolution. TART may have evolved from a pre-existing retrotransposon and been co-opted to serve with HeT-A by acquiring a related gag gene.
We suggest that the more typical transposable elements, the “parasites” that do not appear to be under rigorous cellular control, might actually be escapees that have managed to capture the mechanism for their own transposition and can now behave as independent agents (although most eventually fall under some sort of host regulation that keeps them from overwhelming their host.) This does not assume that all parasitic elements originated in Drosophila. If transposable elements can be derived from telomerase in Drosophila, they could also be derived from telomerase in organisms where they have not replaced telomerase. It is interesting that transposable elements were first observed by McClintock in experiments in which she was creating broken chromosome ends. She found genetic activity that she eventually traced to the movement of transposable elements. McClintock thought that the stress of the chromosome breaks effected mobilization of the elements (McClintock 1978). These elements were what we now hypothesize to be escapees. If the cell responds to chromosome breakage by hyperactivating its telomerase, any escapees in that cell might still be able to respond to those hyperactivating signals. Thus, studies in both corn and Drosophila suggest an evolutionary link between telomeres and retrotransposable elements.
Acknowledgments
We thank Helena Kashevsky for making the transgenic HeT-A Gag D. melanogaster stock and Dan Rines for assistance with the Deltavision microscope. The work of our laboratory was supported by National Institutes of Health Grant GM50315.
References
- Abad JP, De Pablos B, Osoegawa K, De Jong PJ, Martin-Gallardo A, Villasante A. Genomic analysis of Drosophila melanogaster telomeres: full-length copies of HeT-A and TART elements at telomeres. Mol Biol Evol. 2004a;21:1613–1619. doi: 10.1093/molbev/msh174. [DOI] [PubMed] [Google Scholar]
- Abad JP, De Pablos B, Osoegawa K, De Jong PJ, Martin-Gallardo A, Villasante A. TAHRE, a novel telomeric retrotransposon from Drosophila melanogaster, reveals the origin of Drosophila telomeres. Mol Biol Evol. 2004b;21:1620–1624. doi: 10.1093/molbev/msh180. [DOI] [PubMed] [Google Scholar]
- Askree SH, Yehuda T, Smolikov S, et al. A genome-wide screen for Saccharomyces cerevisiae deletion mutants that affect telomere length. Proc Natl Acad Sci USA. 2004;101:8658–8663. doi: 10.1073/pnas.0401263101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beverley SM, Wilson AC. Molecular evolution in Drosophila and the higher Diptera II. A time scale for fly evolution. J Mol Evol. 1984;21:1–13. doi: 10.1007/BF02100622. [DOI] [PubMed] [Google Scholar]
- Blackburn EH. Telomerases. Annu Rev Biochem. 1992;61:113–129. doi: 10.1146/annurev.bi.61.070192.000553. [DOI] [PubMed] [Google Scholar]
- Blackburn EH. Switching and Signaling at the telomere. Cell. 2001;106:661–673. doi: 10.1016/s0092-8674(01)00492-5. [DOI] [PubMed] [Google Scholar]
- Blasco MA. Telomerase beyond telomeres. Nat Rev Cancer. 2002;2:627–633. doi: 10.1038/nrc862. [DOI] [PubMed] [Google Scholar]
- Casacuberta E, Pardue M-L. Transposon telomeres are widely distributed in the Drosophila genus: TART elements in the virilis group. Proc Natl Acad Sci USA. 2003a;100:3363–3368. doi: 10.1073/pnas.0230353100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casacuberta E, Pardue M-L. HeT-A elements in D. virilis: Retrotransposon telomeres are conserved across the Drosophila genus. Proc Natl Acac Sci USA. 2003b;100:14091–14096. doi: 10.1073/pnas.1936193100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casacuberta E, Pardue M-L (2005) HeT-A and TART, two Drosophila retrotransposons with a bona fide role in chromosome structure for more than 60 million years. Cytogen. and Gen. Res. (special issue “Retransposable elements and genome evolution”) (in press). [DOI] [PMC free article] [PubMed]
- Cenci G, Siriaco G, Raffa GD, Kellum R, Gatti MM. The Drosophila HOAP protein is required for telomere capping. Nat Cell Biol. 2003;5:82–84. doi: 10.1038/ncb902. [DOI] [PubMed] [Google Scholar]
- Collins K. Mammalian telomeres and telomerase. Curr Opin Cell Biol. 2000;12:378–383. doi: 10.1016/s0955-0674(00)00103-4. [DOI] [PubMed] [Google Scholar]
- Fanti L, Giovinazzo G, Berloco M, Pimpinelli S. The heterochromatin protein 1 prevents telomere fusions in Drosophila. Mol Cell. 1998;2:527–538. doi: 10.1016/s1097-2765(00)80152-5. [DOI] [PubMed] [Google Scholar]
- George JA, Pardue M-L. The promoter of the heterochromatic Drosophila telomeric retrotransposon, HeT-A, is active when moved into euchromatic locations. Genetics. 2003;163:625–635. doi: 10.1093/genetics/163.2.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greider CW. Telomere length regulation. Annu Rev Biochem. 1996;65:337–365. doi: 10.1146/annurev.bi.65.070196.002005. [DOI] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH. The telomere terminal transferase of Tetrahymena is a ribonucleoprotein enzyme with two kinds of primer specificity. Cell. 1987;51:887–898. doi: 10.1016/0092-8674(87)90576-9. [DOI] [PubMed] [Google Scholar]
- Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- Henson JD, Neumann AA, Yeager TR, Reddel RR. Alternative lengthening of telomeres in mammalian cells. Oncogene. 2002;21:598–610. doi: 10.1038/sj.onc.1205058. [DOI] [PubMed] [Google Scholar]
- Jacks T. Translational suppression in gene expression in retroviruses and retrotransposons. Curr Top Microbiol Immunol. 1990;157:93–124. doi: 10.1007/978-3-642-75218-6_4. [DOI] [PubMed] [Google Scholar]
- Kahn T, Savitsky M, Georgiev P. Attachment of HeT-A sequences to chromosomal termini in Drosophila melanogaster may occur by different mechanisms. Mol Cell Biol. 2000;20:7634–7642. doi: 10.1128/mcb.20.20.7634-7642.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlseder J, Smogorzewska A, de Lange T. Senescence induced by altered telomere state, not telomere loss. Science. 2002;295:2446–2449. doi: 10.1126/science.1069523. [DOI] [PubMed] [Google Scholar]
- Luan DD, Korman MH, Jakubczak JL, Eickbush TH. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- Lundblad V, Blackburn EH. An alternative pathway for yeast telomere maintenance rescues est1− senescence. Cell. 1993;73:347–360. doi: 10.1016/0092-8674(93)90234-h. [DOI] [PubMed] [Google Scholar]
- McClintock B. Mechanisms that rapidly reorganize the genome. Stadler Genet Symp. 1978;10:25–47. [Google Scholar]
- Melnikova L, Georgiev P. Enhancer of terminal gene conversion, a new mutation in D. melanogaster that induces telomere elongation by gene conversion. Genetics. 2002;162:1301–1312. doi: 10.1093/genetics/162.3.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikova L, Biessmann H, Georgiev P. The ku protein complex is involved in length regulation of Drosophila telomeres. Genetics. 2005;169:034538. doi: 10.1534/genetics.104.034538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhailovsky S, Belenkaya T, Georgiev P. Broken chromosome ends can be elongated by conversion in Drosophila melanogaster. Chromosoma. 1999;108:114–120. doi: 10.1007/s004120050358. [DOI] [PubMed] [Google Scholar]
- Okazaki S, Tsuchida K, Maekawa H, Ishikawa H, Fijiwara H. Identification of a pentanucleotide telomeric sequence (TTAGGG)n in the silkworm, Bombyx mori, and other insects. Mol Cell Biol. 1993;13:1424–1432. doi: 10.1128/mcb.13.3.1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardue M-L, DeBaryshe PG. Retrotransposons provide an evolutionarily robust non-telomerase mechanism to maintain telomeres. Ann Rev Genetics. 2003;37:485–511. doi: 10.1146/annurev.genet.38.072902.093115. [DOI] [PubMed] [Google Scholar]
- Pardue M-L, Danilevskaya ON, Lowenhaupt K, Slot F, Traverse KL. Drosophila telomeres: new views on chromosome evolution. Trends Genet. 1996;12:48–52. doi: 10.1016/0168-9525(96)81399-0. [DOI] [PubMed] [Google Scholar]
- Purdy A, Su TT. Telomeres: not all breaks are equal. Cur Biol. 2004;14:R613–614. doi: 10.1016/j.cub.2004.07.042. [DOI] [PubMed] [Google Scholar]
- Rashkova S, Karam SE, Pardue M-L. Element-specific localization of Drosophila retrotransposon Gag proteins occurs in both nucleus and cytoplasm. Proc Natl Acad Sci USA. 2002a;99:3621–3626. doi: 10.1073/pnas.032071999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashkova S, Karam SE, Pardue M-L. Element-specific localization of Drosophila retrotransposon Gag proteins occurs in both nucleus and cytoplasm. Proc Natl Acad Sci USA. 2002b;99:3621–3626. doi: 10.1073/pnas.032071999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashkova S, Athanasiadis A, Pardue M-L. Intracellular targeting of Gag proteins of the Drosophila telomeric retrotransposons. J Virol. 2003;77:6376–6384. doi: 10.1128/JVI.77.11.6376-6384.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahara K, Marec F, Traut W. TTAAGG telomeric repeats in chromosomes of some insects and other arthropods. Chromosome Res. 1999;7:449–460. doi: 10.1023/a:1009297729547. [DOI] [PubMed] [Google Scholar]
- Savitsky M, Kravchuk O, Melnikova L, Georgiev P. Heterochromatin protein 1 is involved in control of telomere elongation in Drosophila melanogaster. Mol Cell Biol. 2002;22:3204–3218. doi: 10.1128/MCB.22.9.3204-3218.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siriaco GM, Cenci G, Haoudi A, et al. Telomere elongation (Tel), a new mutation in Drosophilla melanogaster that produces long telomeres. Genetics. 2002;160:235–245. doi: 10.1093/genetics/160.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter MF, Biessmann H. Expression of the telomeric retrotransposon HeT-A in D. melanogaster is correlated with cell proliferation. Dev Genes Evol. 2004;214:211–219. doi: 10.1007/s00427-004-0400-x. [DOI] [PubMed] [Google Scholar]
- Wills JW, Craven RC. Form, function, and use of retroviral Gag proteins. Aids. 1992;5:639–654. doi: 10.1097/00002030-199106000-00002. [DOI] [PubMed] [Google Scholar]


