Abstract
In crayfish claw closer muscle, the giant sarcomeres are 8.3 µm long at rest, four times longer than vertebrate striated muscle sarcomeres, and they are extensible up to 13 µm upon stretch. Invertebrate connectin (I-connectin) is an elastic protein which holds the A band at the center of the sarcomere. The entire sequence of crayfish I-connectin was predicted from cDNA sequences of 53 424 bp (17 352 residues; 1960 kDa). Crayfish I-connectin contains two novel 68- and 71-residue repeats, and also two PEVK domains and one kettin region. Kettin is a small isoform of I-connectin. Immunoblot tests using antibody to the 68-residue repeats revealed the presence of I-connectin also in long sarcomeres of insect leg muscle and barnacle ventral muscle. Immunofluorescence microscopy demonstrated that the two repeats, the long spacer and the two PEVK domains contribute to sarcomere extension. These regions rich in charged amino acids, occupying 63% of the crayfish I-connectin molecule, may allow a span of a 3.5 µm distance as a new class of composite spring.
Keywords: connectin/D-titin/elasticity/kettin/titin
Introduction
Titin/connectin is a giant spring-like protein that is responsible for passive tension generation and for positioning the thick filament at the center of vertebrate striated muscle sarcomeres (for reviews see Maruyama, 1994; Gautel et al., 1999; Gregorio et al., 1999; Linke, 2000). One titin/connectin molecule [3000 kDa (cardiac muscle) to 3700 kDa (soleus muscle)] spans a half sarcomere ∼1 µm long at rest length from the Z line to the M line in the A band. It consists of the N-terminal Z line-binding region (80 kDa), the I band region (800–1500 kDa) and a myosin-binding region (2100 kDa) (Labeit and Kolmerer, 1995; Freiburg et al., 2000). The extensible I band region is composed of a number of tandem immunoglobulin (Ig) domains and one PEVK domain. Within the physiological range of sarcomere lengths (<3.5 µm), entropic elasticity of the Ig domains (straightening of interdomains) and entropic and enthalpic elasticity of the PEVK region are responsible for reversible extension (cf. Linke, 2000).
In contrast, the giant sarcomere of crayfish claw muscle, the length of which is ∼8 µm at rest, is extensible up to 13 µm. How are the thick filaments kept in position around the center of the giant sarcomere regardless of stretch? Would a putative giant molecule responsible for this function also contain other sequence motifs found in vertebrate titin/connectin, such as long stretches of repetitive myosin-binding region and a protein kinase domain? These questions are important to understand the evolution of muscle architecture as well as that of biomechanically active elastic proteins. We have previously described a 3000 kDa protein found in crayfish claw muscle (Manabe et al., 1993). When the giant sarcomeres of crayfish claw opener muscle are stretched, passive tension is generated, but this tension is diminished when the fibers are treated mildly with trypsin to digest the 3000 kDa protein. The presence of very thin filaments connecting the Z line and the thick filament are observed with an electron microscope. The observations strongly suggest that the very thin filament formed by the 3000 kDa protein functions similarly to titin/connectin in vertebrate skeletal muscle. To understand whether and how the 3000 kDa protein was able to span as great a distance as 3.5 µm, we began to sequence the cDNA encoding this protein of the crayfish claw closer muscle, in which the myofibrils are composed solely of giant sarcomeres. The present work has demonstrated that the molecular mass (1960 kDa) calculated from the entire predicted sequence is much lower than that (3000 kDa) estimated by mobility on SDS–PAGE.
During the present study and when the work was near completion, several related papers were published. An autoimmune serum of a scleroderma patient stained chromosomes of human epithelial cells and also Drosophila muscle sarcomeres, and the partial cDNA sequence from a Drosophila cDNA library was similar to that of Ig repeats and the elastic PEVK domain in titin/connectin (Machado et al., 1998). The putative protein was called Drosophila titin (D-titin). Subsequent investigations on the sequence of Drosophila kettin revealed that the Ig domains found in D-titin are derived from kettin (Hakeda et al., 2000; Kolmerer et al., 2000). Recently, the entire sequence of D-titin has been predicted from the Drosophila genomic database: 16 215 residues giving a 1800 kDa protein (Zhang et al., 2000), and 17 903 residues with 1900–2000 kDa (Machado and Andrew, 2000). The entire cDNA sequence of the crayfish protein is similar to but distinctly different from the predicted sequence of D-titin. Both proteins lack regions specific to vertebrate titin/connectin, especially the kinase domain and long myosin-anchoring region. Since a similar protein was also found in barnacle ventral muscle, beetle leg muscle and fly larva muscle in this study, we propose to call these distinct proteins invertebrate connectin (I-connectin).
Results
cDNA cloning of crayfish I-connectin
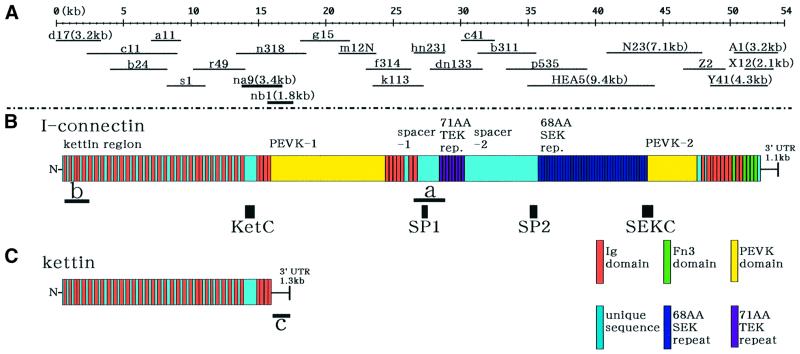
Using affinity-purified polyclonal antibody to the 3000 kDa protein (Manabe et al., 1993), immunoscreening was carried out with a crayfish closer muscle cDNA expression library. The longest cDNA in the positive clones, clone N23 (7.1 kb), was sequenced, and walking in the 3′ or 5′ direction was performed. The nucleotide sequence of the 68 amino acid repeat region was highly conserved (see below) and very unstable in the λZAP II phage clone. Because it was difficult to screen the cDNA clones from the cDNA library, RT–PCR was adopted using specific primers from both the portions just outside the repeat region, resulting in HEA5 clone isolation. As shown in Figure 1A, successive cDNAs were obtained including the 5′- and 3′-untranslated regions (UTRs). A stop codon (nucleotides 52 286–52 288, TAA) of I-connectin was found in the same position in Y41 and X12, and, in addition, there were polyadenylation and polyadenine sequences in A1. Hence the stop codon in Y41 or X12 was regarded as the 3′ end of the open reading frame of crayfish I-connectin cDNA. Thus, an ∼54 kb cDNA (53 424 bp) coding 17 352 amino acid residues was finally elucidated (DDBJ/EMBL/GenBank accession No. AB055861). This is the first complete transcript analysis of an invertebrate giant protein.
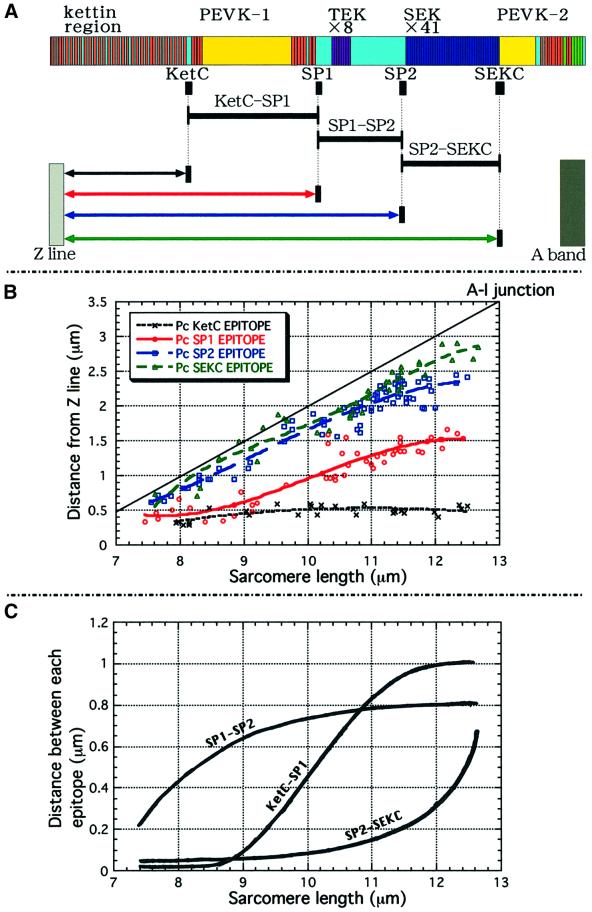
Fig. 1. The domain structure of crayfish I-connectin predicted from the cloned cDNAs. (A) cDNAs sequenced to reconstruct the cDNA contig encoding I-connectin. (B) The domain structure of I-connectin. Bars a and b, cDNAs used for northern blotting; KetC, SP1, SP2 and SEKC (black boxes), portions bacterially expressed and used for raising antibodies. (C) The domain structure of kettin cDNA. Bar c, 1.3 kb cDNA corresponding to the 3′-untranslated region (UTR) used for northern blotting.
Kettin is a small isoform of crayfish I-connectin
Figure 1B summarizes the entire sequence of crayfish I-connectin. The start codon (ATG; the first methionine) was found at position 230–232 of the clone d17. The arrangements of Ig domains and linkers in the N-terminal region (4822 residues) are remarkably similar to those of Drosophila kettin (4796 residues), a Z line-binding protein (Lakey et al., 1990, 1993; van Straaten et al., 1999; Hakeda et al., 2000; Kolmerer et al., 2000), although the identity of the amino acid sequence ranges from 46.5 to 77.4% for 35 Igs and from 11.1 to 63.4% for 34 linkers.
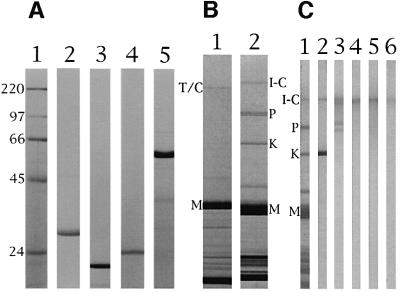
Kettin is expressed abundantly in crayfish claw muscle (Figure 4B, lane 2), and it was purified to homogeneity (Maki et al., 1995). Therefore, we tried northern blot tests in order to clarify whether kettin is an isoform or a proteolytic product of I-connectin using the I-connectin-specific domain (2170 nucleotides, 26442–28611, in Figure 1B, a) and the kettin region (1901 nucleotides, 262–2162, in Figure 1B, b) as probes. As seen in Figure 2, probe a reacted only with very large RNA (Figure 2A), whereas probe b reacted with very large and large RNAs (Figure 2B). The kettin mRNA is ∼15 kb in size and I-connectin mRNA is expected to be ∼54 kb in size. The stop codon of kettin was then sought downstream of the 3′ terminus of the kettin domain from the cDNA library. Two clones (na9 and nb1 in Figure 1A, bold underlines) were obtained containing a stop codon (TAA) followed by a polyadenylation signal and poly(A) sequences. Northern blotting using this kettin 3′-UTR sequence as a probe (1288 nucleotides in Figure 1C, c) showed that the probe reacted only with ∼15 kb RNA corresponding to kettin (Figure 2C). In addition, as shown below, polyclonal antibody to KetC (PcKetC) in crayfish I-connectin (Figure 1B) reacted with both I-connectin and kettin in the immunoblot tests (see Figure 4C, lane 2). Therefore, kettin is thought to be a small isoform of I-connectin (DDBJ/EMBL/GenBank accession No. AB055862). Zhang et al. (2000) and Machado and Andrew (2000) independently suggested that kettin is a splice variant or a proteolytic product of D-titin (I-connectin), and the present study shows that kettin is indeed a splice variant.
Fig. 4. Recombinant peptides used for raising antibodies and immunoblot tests of the reactions of the antibodies with crayfish claw closer muscle proteins. (A) SDS–PAGE patterns of purified recombinant peptides: lane 1, molecular weight markers (sizes shown in kDa); lanes 2–5, KetC, SP1, SP2 and SEKC recombinant peptides shown in Figure 1B. Electrophoresis was on 10% polyacrylamide gels. (B) SDS–PAGE patterns of rabbit cardiac muscle and crayfish claw closer muscle: lane 1, rabbit cardiac muscle; lane 2, crayfish claw closer muscle. T/C, titin/connectin; I-C, I-connectin; P, projectin; K, kettin; M, myosin heavy chain. Electrophoresis was on 2–6% polyacrylamide gels. (C) Immunoblot tests: lane 1, amido black stain of crayfish claw closer muscle; lanes 2–6, treated with antibodies PcKetC, PcSP1, PcSP2, PcSEKC and Pc3000K, respectively. Electrophoresis was on 2–6% polyacrylamide gels.
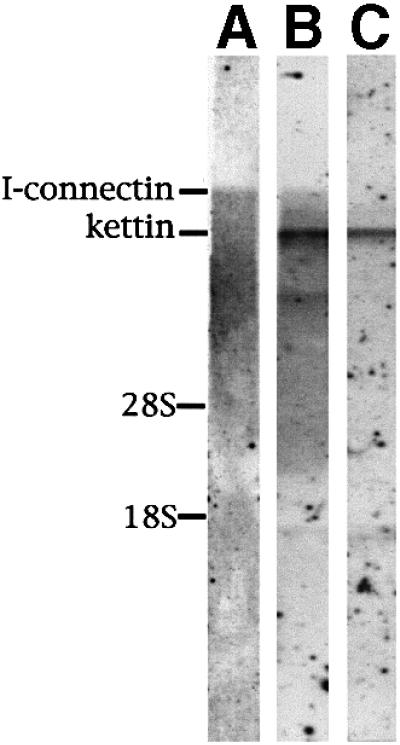
Fig. 2. Northern blot tests to detect the expression of kettin and I-connectin mRNAs in crayfish claw closer muscle. (A) Detectionof I-connectin mRNA by probe a in Figure 1B. (B) Detection of I-connectin and kettin mRNAs by probe b in Figure 1B. Note that kettin was expressed much more than I-connectin. (C) Detection of kettin mRNA by probe c in Figure 1C. 28S, vertebrate 28S rRNA; 18S, vertebrate 18S rRNA.
Crayfish I-connectin is distinct from titin/connectin in domain structures
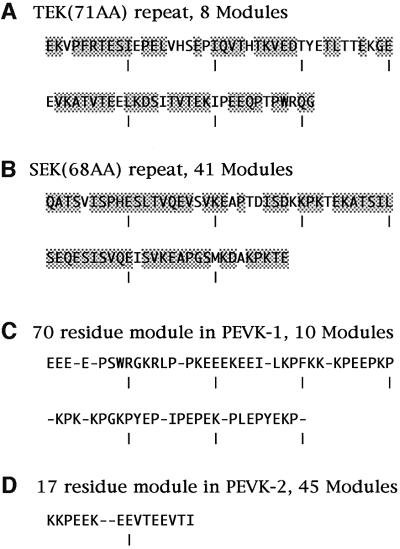
The kettin region is followed by a large PEVK-1 domain consisting of as many as 3364 residues [another shorter PEVK-2 domain (1310 residues) is positioned near the C-terminus] (Figure 1B). As shown in Table I, the content of proline (P), glutamic acid (E), valine (V) and lysine (K) is 63.1% for PEVK-1 and 68.4% for PEVK-2, which is somewhat lower than that (∼70%) of vertebrate titin/connectin (Labeit and Kolmerer, 1995). The V content is low for PEVK-1 and the P content is low for PEVK-2. In the two PEVK domains, there are a number of repetitive modules with consensus sequences. Ten 70-residue modules are present in PEVK-1 (Figure 3C) and forty-five 17-residue modules in PEVK-2 (Figure 3D). The former makes up 20% of PEVK-1 and the latter makes up 70% of PEVK-2. These consensus sequences are unique and distinct from the 25- to 30-residue modules in the vertebrate titin/connectin PEVK domain (Freiburg et al., 2000; Greaser, 2001; Gutierrez-Cruz et al., 2001). The proline content is 22% in the PEVK-1 module and 8% in the PEVK-2 module.
Table I. Contents of several amino acid residues of various regions of I-connectin.
| Region/residues (%) | P | E | V | K | Sum | D | R | Charged residues (E + K + D + R) |
|---|---|---|---|---|---|---|---|---|
| PEVK-1 (3364 amino acids) | 17.3 | 21.4 | 5.2 | 19.2 | 63.1 | 4.4 | 5.1 | 50.1 |
| Spacer-1 (467 amino acids) | 4.5 | 14.8 | 9.6 | 9.4 | 38.3 | 2.4 | 4.7 | 31.3 |
| TEK (71 × 8) (568 amino acids) | 7.9 | 18.1 | 9.9 | 9.2 | 45.1 | 4.1 | 2.5 | 33.9 |
| Spacer-2 (1666 amino acids) | 6.2 | 13.6 | 9.4 | 8.6 | 37.8 | 4.3 | 2.5 | 29.0 |
| SEK (68 × 41) (2788 amino acids) | 6.6 | 14.6 | 10.4 | 11.7 | 43.3 | 3.9 | 0.8 | 31.0 |
| PEVK-2 (1310 amino acids) | 8.1 | 27.9 | 13.2 | 19.2 | 68.4 | 2.1 | 2.8 | 52.0 |
Fig. 3. Amino acid sequences characteristic of crayfish I-connectin. (A) TEK repeats. Consensus sequences conserved in more than seven repeats out of eight are shaded. (B) SEK repeats. Consensus sequences conserved in >29 repeats out of 41 are shaded. (C) PEVK-1 module. Consensus sequences are >50% conserved. (D) PEVK-2 module. Consensus sequences are as in (C).
Following the PEVK-1 domain, seven Ig domains with unique sequence are contiguous to a moderately sized spacer-1 consisting of 467 residues. No similar sequence was listed in the EMBL database; this domain contains a large number of hydrophobic amino acids such as valine (9.6%), alanine (A; 7.9%) and leucine (L; 4.9%). As shown in Table I, charged amino acids account for 31.3%. The PEVK content is 38.3%, and therefore this spacer-1 is considered a PEVK-like domain.
Next, there are eight copies of a 71-residue repeat (Figure 1B). This novel repeat is rich in threonine (T; 14.8%), glutamic acid (E; 18.1%) and lysine (K; 9.2%), and is therefore called a TEK repeat. Seven repeats have 51 residues out of 71 in common (Figure 3A). The TEK repeat is contiguous to a very large spacer-2 comprising 1666 residues. A similar sequence was not found in the EMBL database. It contained 29% charged amino acids, and the PEVK content is calculated to be 37.8%. Again, this novel spacer-2, like spacer-1, is a PEVK-like domain.
The long spacer-2 is followed by 41 copies of 68-residue repeats (Figure 1B). Fifty-eight of the 68 amino acids are conserved in 29 repeats. This novel repeat contains 16.0% serine (S), 14.6% glutamic acid (E), 11.7% lysine (K) and 3.9% aspartic acid (D). Since S, E and K constitute 41.3%, it is called a SEK repeat (Figure 3B). The SEK repeat contains 43.3% PEVK and 28.1% hydrophobic residues.
After the SEK repeat there is the PEVK-2 domain consisting of 68.4% PEVK, followed by 10 Ig and five fibronectin type III (Fn3) domains. It is notable that there are only five Fn3 domains in I-connectin compared with 115 Fn3 domains in the thick filament-binding region of vertebrate titin/connectin. In addition, there is no kinase domain like that near the C-terminal region of titin/connectin.
Crayfish I-connectin spans from the Z line to the tip of the thick filament in giant sarcomeres
Four kinds of polyclonal antibody (PcKetC, etc.) were obtained using each fusion protein (KetC, SP1, SP2 and SEKC) as antigen. Figure 4A shows SDS–PAGE patterns of purified recombinant polypeptides corresponding to each part of crayfish I-connectin depicted in Figure 1B. The molecular masses (26, 18 and 24 kDa) of KetC, SP1 and SP2 recombinant peptides estimated from mobilities during SDS–PAGE were in agreement with those calculated from the sequences. However, the value based on the mobility of SEKC peptide (55 kDa) was much larger than that calculated (35 038 Da).
Immunoblot tests revealed that PcSP2 and PcSEKC reacted solely with I-connectin of crayfish closer muscle (Figure 4C, lanes 4 and 5). However, affinity-purified PcSP1 (Figure 4C, lane 3) reacted not only with I-connectin, but also with two bands larger and smaller than projectin. There may be unknown proteins that cross-react with PcSP1 in crayfish closer muscle. PcKetC reacted with both I-connectin and kettin (Figure 4C, lane 2).
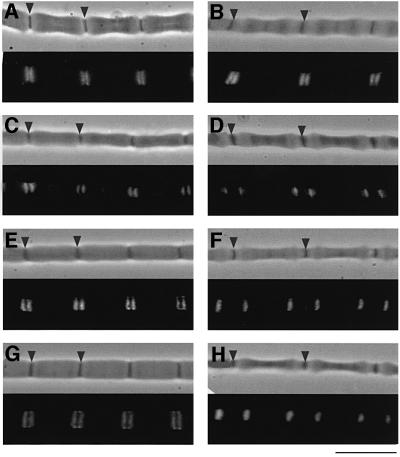
Immunofluorescence observations using the four kinds of antibodies revealed that KetC and SP1 are located near the Z line, and SP2 and SEKC are located near the AI junction at rest [sarcomere length (SL) ∼8.0 µm], as shown in Figure 5. Upon stretch to an SL of 12 µm, the distance between each epitope (SP1, SP2 and SEKC) and the Z line increased markedly. However, the epitope of KetC did not move appreciably, suggesting that the N-terminal 540 kDa kettin region is localized within and near the Z line, as shown in Drosophila flight muscle (Lakey et al., 1993; van Straaten et al., 1999). To locate the C-terminal portion of I-connectin, antibodies to the C-terminal Fn3 domains were raised, but these antibodies reacted not only with I-connectin but also with projectin. Therefore, although the antibodies bound to the edge of the myosin filament, we could not locate the site on I-connectin that binds to the thick filament. Since Ig and Fn3 domains of titin/connectin bind to the myosin filament (Labeit et al., 1992), the C-terminal region containing 10 Ig and five Fn3 domains is very likely to be responsible for the binding of I-connectin to the thick (myosin) filament in crayfish claw muscle. These observations demonstrate that the I-connectin molecule spans from the Z line to the tip of the thick filament.
Fig. 5. Immunofluorescence microscopic observations of giant sarcomeres of crayfish claw closer muscle using antisera against recombinant peptides of crayfish I-connectin. (A and B) Treated with PcKetC: sarcomere length (SL), (A) 9.2 µm; (B) 11.8 µm. (C and D) Treated with PcSP1: SL, (C) 8.7 µm; (D) 11.8 µm. (E and F) Treated with PcSP2: SL, (E) 8.9 µm; (F) 12.0 µm. (G and H) Treated with PcSEKC: SL, (G) 8.5 µm; (H) 12.0 µm. Upper, phase- contrast image; lower, fluorescence image. Arrowheads indicate the Z line. Bar, 10 µm.
Three-quarters of the I-connectin molecule is passively extensible
What are the mechanical properties of this unusually complex composite spring and, more specifically, of the novel sequence motifs we identified here? To determine this, we used biomechanical assays together with sequence-specific antibody labeling. A fluorescent stripe due to binding of each antibody to the corresponding epitope in the I-connectin molecule in situ was seen in a half sarcomere at rest (8.3 µm): KetC and SP1, near the Z line; and SP2 and SEKC, near the AI junction region (Figure 5). These observations provide important information on the physiological state and localization of several domains of I-connectin in the giant sarcomere at rest. First, it appears that the 540 kDa kettin region is localized within and near the Z line. Secondly, the long PEVK-1 domain is completely contracted judging from the location of SP1 near the Z line. Thirdly, the localization of both SP2 and SEKC at the AI junction suggests that both the SEK repeats and the PEVK-2 domain are also highly contracted.
To determine the extensibility of the I-connectin molecule, closer muscle fibers were stretched to various extents and the distance between each fluorescent stripe and the Z line was measured (Figure 6B). The distance between each epitope calculated from Figure 6B is shown in Figure 6C. The distance between SP1 and the Z line became greater (0.5–1.5 µm) when the SL was elongated from 8.3 to 12 µm. These changes were due primarily to extensibility of the PEVK-1 domain. The distance between SP2 or SEKC and the Z line became progressively greater during the stretch of sarcomeres: from 0.6 to 2.3 µm (SP2), and from 0.6 to 2.8 µm (SEKC). At the same time, the distance from SP2 to SEKC (SEK repeats) was increased from 0.02 to 0.6 µm when the SL was elongated. This region was the most resistant to extension, and the relationship between the length of the region and the SL was similar to that between passive tension generation and the SL (see Figure 7A). The distance between KetC and SP1 (PEVK-1) was only 0.1 µm at rest length; this extended remarkably to 0.8 µm at an SL of 11 µm and reached a plateau (1 µm) at an SL of ∼12 µm. The distance between SP1 and SP2 (from spacer-1 through TEK repeats to spacer-2) was distinct from the other regions: it was already extended to 0.5 µm at rest length, and elongated appreciably to 0.8 µm at an SL of 12 µm. Between SLs of 8 and 12 µm, the PEVK-1 domain (KetC-SP1) was most extensible (∼1 µm).
Fig. 6. Extensibility of various regions of the crayfish I-connectin molecule linking the Z line and the A band. (A) Positions of the four epitopes in the entire sequence of I-connectin. (B) Changes in the distance between each epitope and the Z line at increasing SL. Crosses, KetC; open circles, SP1; open squares, SP2; open triangles, SEKC. (C) Changes in the distance between each epitope at increasing SL.
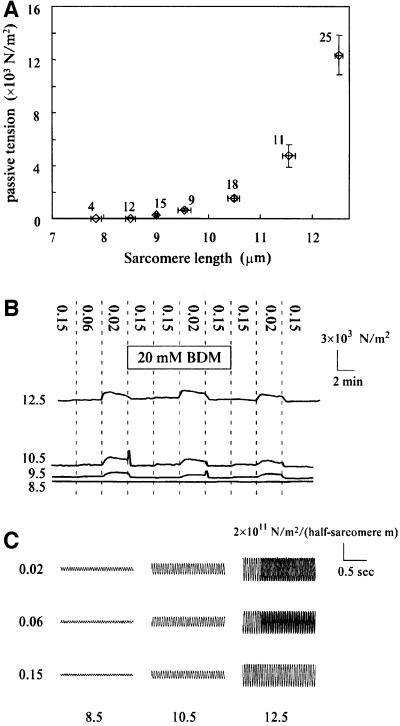
Fig. 7. (A) The relationship between passive tension and sarcomere length. Horizontal and vertical bars attached to each symbol indicate the standard error of means and the number of measurements. (B) Traces of passive tension showing a reversible effect of ionic strength. Sarcomere length (SL) is shown on the left in micrometers. The dotted lines indicate the changes in the incubating solution, the ionic strength of which is shown by the vertical numbers. BDM (20 mM) was added to the solution during the period shown by the rectangle. The cross-sectional area of the fiber was 40 000, 33 000, 76 000 and 56 000 µm2 for the traces at SL of 12.5, 10.5, 9.5 and 8.5 µm, respectively. (C) Representative traces of tension oscillation in response to 20 Hz sinusoidal length perturbation of 2% fiber length amplitude. Ionic strength is shown on the left and SL is shown below the traces in micrometers. For the traces at SL 12.5 µm, those obtained in the presence of 20 mM BDM were superimposed on the last three-quarters of the traces in the absence of BDM at ionic strength 0.02 and 0.06. All the traces in the figure were obtained from a specimen with 17 000 µm2 cross-sectional area.
These measurements clearly show that approximately three-quarters of the I-connectin molecule (from PEVK-1 to PEVK-2) is extensible when myofibrils are stretched.
Generation of passive tension by crayfish I-connectin
Passive tension generation by crayfish claw closer myofibrils was measured at various SLs (Figure 7A). Skinned fibers of crayfish closer muscle had sarcomeres with rest lengths ranging from 7.5 to 9.3 µm, with an average of 8.3 ± 0.13 µm for 22 fibers. Moderate extension of fibers up to 10 µm SL developed a small passive tension that rapidly decayed to a steady level. The greater tension generated instantly by further extension decayed rapidly and then more slowly. Therefore, a steady level was estimated from an exponential decay curve. The results shown in Figure 7A are in good agreement with those obtained for crayfish opener muscle (Manabe et al., 1993).
The effect of ionic strength on the passive tension of skinned fibers was observed to reveal possible contributions of electrostatic interactions in the I-connectin molecule. Tension development increased when ionic strength was lowered from 0.15 to 0.02, but the extent varied greatly depending upon the SL (Figure 7B). The effect was most marked at an SL of 9.5 µm (an ∼50% increase) and was less at longer SLs: 25% at 10.5 µm and 10% at 12.5 µm. These results suggest that electrostatic interactions contribute significantly to the elasticity of the PEVK-1 domain (KetC-SP1), but not of the SEK repeat domain (SP2-SEKC).
Next, the effect of ionic strength on fiber stiffness was investigated to demonstrate the electrostatic contribution to the viscous part of passive tension. Figure 7C shows fiber stiffness measured as tension in response to 20 Hz sinusoidal length perturbations. The stiffness decreased markedly (50%) around slack length (SL, 8.5 µm), when the ionic strength was increased from 0.02 to 0.15 (Figure 7C). In contrast, stiffness decreased only by 10% at an SL of 12.5 µm. The viscoelasticity that was insensitive to ionic strength became dominant in fibers extended beyond 10.5 µm.
Invertebrate connectin occurs in arthropod striated muscles
Is the giant protein and its specific modules we identified in crayfish a mere oddity, or rather an evolutionary achievement of broad distribution in invertebrate? The species distribution of I-connectin was shown by immunoblot tests with several types of invertebrate striated muscle, using antibody to the SEKC region of crayfish I-connectin. As shown in Figure 8, ∼3000 kDa bands in barnacle ventral muscle, beetle leg muscle and fly larva muscle clearly reacted with PcSEKC. The SLs of these muscles were 5–7 µm (Ohtani et al., 1996). I-connectin was also present in crab claw and leg muscles, and in other insect leg muscles (grasshopper and bumblebee) (data not shown). Therefore, I-connectin is widely distributed in arthropod muscles with long sarcomeres.
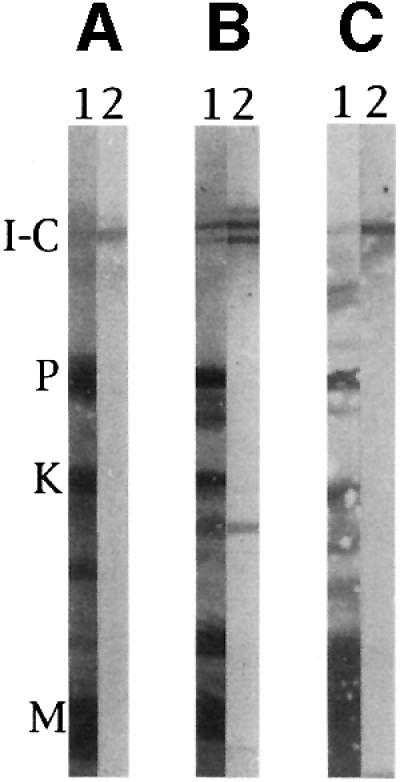
Fig. 8. Immunoblot detection of invertebrate connectin (I-connectin) in several arthropod striated muscles. SDS–PAGE was carried out using 2.3–4% polyacrylamide gels. (A) Barnacle (Dolichopus nitidus) ventral muscle; (B) beetle (Allomyrina dichotoma) leg muscle; (C) fly (Calliphora lata) larva muscle. Lane 1, amido black stain; lane 2, treated with Pc SEKC (cf. Figure 1B). I-C, I-connectin; P, projectin; K, kettin; M, myosin heavy chain.
Discussion
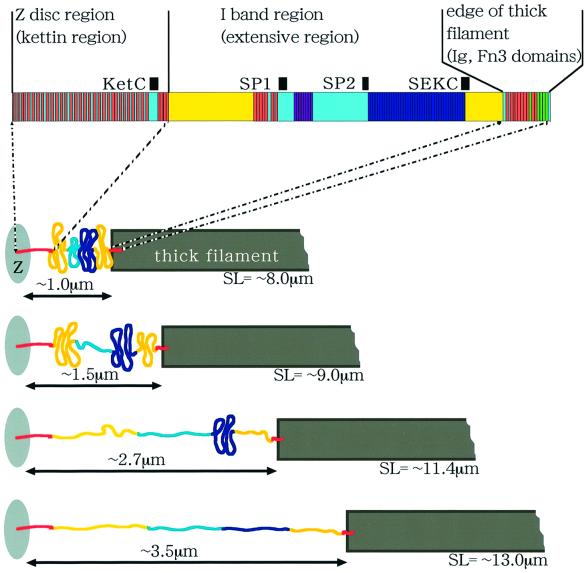
In the giant sarcomere of crayfish claw closer muscle in the resting state, the SL reaches 8.3 µm. The width of the I band is ∼1.2 µm and can be increased to 3.5 µm when the muscle fiber is stretched almost maximally to an SL of 13 µm (the A band width is a constant 6 µm). Therefore, if there is a titin/connectin homolog in the giant sarcomere, its extensible region in the I band must range from 1.2 to 3.5 µm. The present study has clearly demonstrated that this is the case. The localization and the differing extensibility of various regions of I-connectin in the giant sarcomere are summarized schematically in Figure 9.
Fig. 9. Scheme of structural changes of the I-connectin molecule in a giant sarcomere of crayfish claw muscle at varied sarcomere length. The N-terminal kettin domain and the C-terminal Ig and Fn3 domains are practically inextensible. At rest, all the extensible regions (PEVK-1 to PEVK-2) are largely retracted. At SL ∼9 µm, the region spacer-1–TEK repeats–spacer-2 (SP1–SP2) is greatly extended. At SL ∼11.4 µm, PEVK-1 is almost entirely extended. At SL ∼13 µm, SEK repeats (SP2–SEKC) are nearly completely elongated (cf. Figure 6C).
The C-terminal 180 kDa region containing Ig and Fn3 domains, which is ∼60 nm (4 × 15 nm) long, is thought to be located on the tip of the thick filament. In the crayfish claw muscle sarcomere, projectin, a twitchin-, titin/connectin-related protein (cf. Benian et al., 1999), is localized on the thick filament, except for its central region (Hu et al., 1990; Manabe et al., 1993). It is likely that projectin binds to the C-terminal Ig and Fn3 domains of I-connectin since projectin is rich in Ig and Fn3 domains (Daley et al., 1998), resulting in stabilization of I-connectin binding to the thick filament. It is worthy of note that projectin retracted to the Z line together with I-connectin when crayfish claw myofibrils were treated with 0.6 M KI to dissolve myosin and actin filaments (Manabe et al., 1993).
The amount of I-connectin is only a tenth or less the amount of kettin and is likely to be less than one-third the amount of titin/connectin in rabbit skeletal muscle. The fine structure of the giant sarcomere of crayfish claw muscle (opener and closer) is more disorganized than that of vertebrate skeletal muscle, e.g. the AI junction is not always ordered in the same sarcomeres (cf. Govind, 1985). This rather irregular assembly of the giant sarcomere may be due to a small number of I-connectin filaments supporting the sarcomere structure, while a larger amount of kettin may maintain the Z line structure.
Sequence and extensibility of crayfish I-connectin
Manabe et al. (1993) estimated the molecular mass of crayfish I-connectin to be 3000 kDa based on mobility during SDS–PAGE, which is similar to that of vertebrate titin/connectin. However, the calculated value from the entire cDNA is 1960 kDa, in agreement with that of Drosophila titin [1800 kDa (Zhang et al., 2000) and 1900–2000 kDa (Machado and Andrew, 2000)] predicted from the genomic sequence. In view of the large discrepancy between the values of mobility-based molecular mass (55 kDa) and the calculated chain weight (35 038 Da) of recombinant SEK repeats (cf. Figure 4A, lane 5), the molecular mass of crayfish I-connectin based on the mobility in SDS–PAGE appears to be much larger than the value predicted from the sequence. This is consistent with the report that the predicted molecular weight (51 467 Da) of a recombinant PEVK peptide (469 residues) of human fetal skeletal muscle titin/connectin is much smaller than the mobility-based molecular mass (86 kDa) (Gutierrez-Cruz et al., 2001). I-connectin contains two long PEVK domains.
The size of I-connectin is two-thirds that of titin/connectin and yet it can span a distance as long as 3.5 µm. In addition, the N-terminal kettin region (540 kDa) and the C-terminal Ig and Fn3 domains (∼180 kDa) are thought to be most inextensible. Hence the other ∼1200 kDa portion is expected to cover a distance of ∼3.5 µm. This is possible if some 10 000 amino acid residues are stretched to a maximum extent (0.38 nm × 10 000 = 3.8 µm). Note that titin/connectin consists of ∼90% Ig and Fn3 domains, and ∼10% extensible regions (PEVK and N2-B domains).
PEVK-1 (KetC-SP1, ∼3364 residues) is progressively extensible up to 1 µm when sarcomeres are stretched from 8 to 12 µm of SL. This is within the calculated contour length of 1.3 µm. The two spacers and eight TEK repeats (SP1–SP2, ∼2350 residues, contour length 0.9 µm) are considerably extended at rest and are elongated to a small extent up to an SL of 12 µm. The distance SP1–SP2 at an SL of 12 µm is 0.8 µm, which is comparable with 0.9 µm contour length. The SEK repeats (41 × 68 residues; contour length 1.06 µm) are extended from 0.02 to >0.6 µm when the SL is stretched from 8.0 to 12.6 µm. PEVK-2 (1310 residues; contour length, 0.5 µm) is extensible from 0.2 to 0.4 µm when stretched from an SL of 8 to 12 µm.
The above observations strongly suggest that the extensible regions of crayfish I-connectin can be extended almost to the limit of their contour lengths. Examinations of the sequences of each extensible region indicated that there are clusters of charged amino acids, especially glutamic acid (E) and lysine (K), as in the PEVK domain of human titin/connectin (Labeit and Kolmerer, 1995). The contents of PEVK and charged amino acids, including aspartic acid (D) and arginine (R), are listed in Table I. The charged amino acids account for 50.1% in PEVK-1 and 52.0% in PEVK-2. The contents of charged amino acids ranged from 29.0 to 33.9% in spacer-1 and -2, and TEK and SEK repeats. The entire extensible region (10 935 residues; contour length ∼4.1 µm) is made up of 38.9% charged amino acids. These facts suggest that electrostatic interactions tend to let I-connectin retract at rest and that external forces counteract these to extend the polypeptide chain. Evidence for this assumption is that an increase in ionic strength resulted in a decrease in passive force of crayfish closer fibers generated by stretch. The decrease in tension generation by an increase in ionic strength was marked in the PEVK-1 domain, but less so in the SEK repeat domain (Figure 7B and C). Thus, the tension generation was not explained entirely by electrostatic interactions, suggesting that hydrophobic and hydrogen-bonding interactions are also involved. Entropic as well as enthalpic elasticity, as proposed for the vertebrate PEVK domain (cf. Linke, 2000), appear to be involved in the reversible extensibility and tension generation in I-connectin of the giant sarcomere of crayfish claw muscle. It is important to determine the three-dimensional structure of the extensible regions of crayfish connectin using bacterially expressed polypeptides.
Recently, the modular repeats consisting of 25–30 (average 28) residues in the human titin/connectin PEVK domain have been reported (Freiburg et al., 2000; Greaser, 2001; Gutierrez-Cruz et al., 2001). We analyzed two PEVK domain sequences of I-connectin and found distinct modules in both domains (Figure 3C and D). Ten 70-residue modules were distributed throughout the whole of PEVK-1 (20% of the domain). Forty-five 17-residue modules in tandem occupied the N-terminal region of PEVK-2 (70% of the domain). In Drosophila titin, there are seven 45- to 50-residue modules in tandem in the N-terminal region of PEVK-1, and there are four conserved 71-residue modules in tandem in the N-terminal region of PEVK-2 (Machado et al., 1998; this study). Surprisingly, there is no similarity in size or sequence between the titin/connectin PEVK and these PEVK modules. Gutierrez-Cruz et al. (2001) showed the presence of a polyproline II helix in the PEVK modules of titin/connectin. It is not certain whether this structure occurs in crayfish I-connectin or in Drosophila titin.
The 68-residue SEK repeat is mainly responsible for passive tension generation in crayfish closer muscle. The recombinant peptide is shown to consist of 40–50% β-sheet and 20% β-turn, suggesting a β-barrel structure from circular dichroic spectra (data not shown). This implies a rather compact structure for these repeats and suggests that unlike in titin/connectin, domain unfolding in I-connectin might be a major mechanism for tension generation of the protein.
Crayfish I-connectin is a Drosophila titin homologue
The D-titin (∼2000 kDa) sequence has been predicted from the genomic sequence of the third chromosome at 62C-2 (Machado and Andrew, 2000; Zhang et al., 2000). The N-terminal kettin region, two long PEVK domains and the C-terminal Ig and Fn3 domains are common to D-titin and crayfish I-connectin. Also, the protein kinase domain and A band spanning myosin-binding region are absent in both proteins. However, the SEK and TEK repeats are lacking in D-titin. Furthermore, the two kinds of PEVK modules in I-connectin are not like those in D-titin. Therefore, the two proteins are closely related but different. Mutants in the D-titin gene show defects in sarcomere assembly (Machado and Andrew, 2000; Zhang et al., 2000). D-titin was shown to be located in Drosophila flight muscle sarcomeres. In the regular size of sarcomeres (∼3.4 µm at rest), the D-titin must be largely contracted. Machado and Andrew (2000) emphasized the role of D-titin in compaction of chromosomes. The giant proteins, vertebrate titin/connectin, invertebrate connectin and D-titin, illustrate biodiversity in elastic structure and function.
Using the antibody to the characteristic SEK domain, it is shown that I-connectin is widely distributed in arthropod striated muscles: insect leg and larval muscles, crayfish and crab claw muscles, and barnacle ventral muscle. It is interesting that the SLs of these myofibrils at rest are fairly long (5–8 µm). This protein has a structure distinct from that of vertebrate titin/connectin, and connects the thick filament to the Z line in the long sarcomeres of some invertebrate striated muscles. Hence we propose to call this protein invertebrate connectin (I-connectin). Since the function of the giant protein we describe here is almost exclusively a mechanical one, connecting the thin and thick filaments in an elastic fashion, we feel that the term I-connectin is most appropriate, and sets it adequately apart from the functionally and structurally distinct vertebrate titin/connectin.
Materials and methods
DNA cloning and sequencing
Total RNA was prepared from crayfish claw closer muscle using TRIzol reagent (Gibco-BRL) and poly(A) RNA was purified from the total RNA using a Dynabeads Oligo (dT)25 mRNA Purification kit (Dynal) following the manufacturer’s instruction manual. cDNA was synthesized from poly(A) RNA primed with oligo(dT) or random hexameric primer using Superscript™ II RNase H– reverse transcriptase (Gibco-BRL). The cDNA fragments were ligated to λZAP II expression vector arms using a λZAP II Predigested Vector kit (Stratagene), and in vitro packaging into phage particles was carried out using Gigapack III Gold Packaging Extract (Stratagene). The cDNA library was screened with polyclonal antibody Pc3000K (Manabe et al., 1993). The positive clone N23 was used as a cDNA probe; 5′ and 3′ terminus-directed clones were isolated by standard cDNA hybridization screening of the cDNA library. The clone HEA5 was obtained by RT–PCR following a modification of the manufacturer’s instruction manual (TaKaRa LA Taq™ polymerase; TaKaRa) using a pair of primers, repA 5′: AAGGCTACAACTCATGTATCAA (DDBJ/EMBL/GenBank accession No. AB055861, 34 487–34 508) and PEAN 3′: TGATCATCACCTCTTCAGTGAC (DDBJ/EMBL/GenBank accession No. AB055861 in the opposite direction, 43 868–43 889). All the cDNAs shown in Figure 1A were sequenced in both strands with an ALFexpress DNA autosequencer (Amersham Pharmacia), using a Thermo Sequenase Fluorescent labeled primer cycle sequencing kit (7-deaza-dGTP and T3, T7 primer). Sequence analysis and homology search were performed mainly with a Genetyx Mac version 10.0 (SDC, Japan) and GenomeNet WWW server Sequence Similarity Search (BLAST).
Northern blotting
Crayfish closer muscle total RNA was prepared as described above. RNA was separated in a formaldehyde–0.8% agarose gel and transferred onto a Hybond-N+ membrane (Amersham Pharmacia) as described (Endo and Nadal-Ginard, 1987). The cDNAs shown in Figure 1B and C were used as a probe. Labeling, hybridization and detection were performed using the AlkPhos Direct Labeling and Detection System with CDP-Star (Amersham Pharmacia) following the manufacturer’s manual.
Recombinant protein expression and purification
Each cDNA construct was amplified by PCR from pBluescript SK–. The PCR products were cloned into pGEX6P-1 vector (Amersham Pharmacia). The soluble recombinant proteins tagged with glutathione S-transferase (GST) at their N-termini were expressed in Escherichia coli BL21 [DE3] pLysS and then the bacteria were harvested by centrifugation. Pellets were suspended with R buffer (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4 pH 7.4) and sonicated (TOMY UR-200P, output 1.5, 30 s, twice). After centrifugation, supernatants containing the soluble recombinant protein were incubated with glutathione–Sepharose 4B (Amersham Pharmacia) and the resin was washed twice in R buffer. The GST tag at the N-termini of the recombinant proteins was cleaved off by PreScission™ Protease (Amersham Pharmacia) in C buffer (50 mM Tris–HCl pH 7.0, 150 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol). Finally, the resin was centrifuged to obtain the recombinant protein in the supernatant. Purified proteins were subjected to SDS–PAGE.
Immunoblots
Samples in SDS were electrophoresed on polyacrylamide gels and transferred to a nitrocellulose sheet. The sheet was treated with antibodies to each recombinant protein and the bound antibodies were detected by treatment with horseradish peroxidase-conjugated anti-rabbit Ig (Dako).
Immunofluorescence microscopy
I-connectin recombinant peptides were injected into rabbits together with Freund’s incomplete adjuvant. Each antigen was coupled to Affi-Gel 10 or 15 (Bio-Rad) and specific antibodies were purified by affinity chromatography. Immunofluorescence observations were carried out as described (Manabe et al., 1993); Leitz Ortholax-2 and Zeiss Axioskop2 fluorescence microscopes were used. The phase-contrast and fluorescent images were recorded at various SLs. The antibody epitope spacing across the M line in a sarcomere was measured with image analysis software (Kodak Digital Science 1D) and the distance (SL – the epitope spacing) was divided by two to obtain the Z line–epitope distance in a half sarcomere. The distance between the epitope and the AI junction was calculated from the epitope spacing within a sarcomere using the A band width of 6 µm.
Tension and stiffness measurements
Skinned fibers were prepared mechanically in standard relaxing solution [26.11 mM CH3SO3K, 5.66 mM (CH3SO3)2Mg, 4.36 mM Na2ATP, 10 mM creatine phosphate, 10 mM EGTA, 1 mM phenylmethylsulfonyl fluoride (PMSF), 0.5 mM leupeptin and 20 mM PIPES (pH 7.0 at 5°C)]. Internal membranous structures were disrupted with 0.5% Triton X-100 for 15 min.
Tension and stiffness were measured as described by Yamaguchi and Takemori (2001) except that the stiffness was measured by 20 Hz and 2% fiber length sinusoidal oscillations. The effect of ionic strength was studied in relaxing solutions of 0.20, 0.15, 0.06 and 0.02 ionic strength: in addition to a common mixture of 2 mM EGTA, 1 mM PMSF, 0.5 mM leupeptin and 2 mM PIPES (pH 7.0 at 5°C), the solution of 0.20/0.02 ionic strength contained 179/0 mM CH3SO3K, 2.02/2.01 mM (CH3SO3)2Mg and 1.21/1.49 mM Na2ATP. The solutions of 0.20 and 0.02 ionic strength were mixed to attain 0.15 and 0.06 ionic strength. In some solutions, 20 mM 2,3-butanedione monoxime (BDM) was used to ensure that the tension was passive tension (Higuchi and Takemori, 1989).
Acknowledgments
Acknowledgements
We are indebted to Drs T.Endo and N.Yonezawa for their helpful advice. We thank Dr H.Yajima for his contribution during the earlier phase of this work. The technical assistance of Ms S.Maki and Ms Y.Ohtani is gratefully acknowledged. This work was supported by Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan to S.K.
References
- Benian G.M., Ayme-Southgate,A. and Tinley,T.L. (1999) The genetics and molecular biology of the titin/connectin-like proteins of invertebrates. Rev. Physiol. Biochem. Pharmacol., 138, 235–268. [DOI] [PubMed] [Google Scholar]
- Daley J., Southgate,R. and Ayme-Southgate,A. (1998) Structure of the Drosophila projectin protein: isoforms and implication for projectin filament assembly. J. Mol. Biol., 279, 201–210. [DOI] [PubMed] [Google Scholar]
- Endo T. and Nadal-Ginard,B. (1987) Three types of muscle-specific gene expression in fusion-blocked rat skeletal muscle cells: translational control in EGTA-treated cells. Cell, 49, 515–526. [DOI] [PubMed] [Google Scholar]
- Freiburg A. et al. (2000) Series of exon-skipping events in the elastic spring region of titin as the structural basis for myofibrillar elastic diversity. Circ. Res., 86, 1114–1121. [DOI] [PubMed] [Google Scholar]
- Gautel M., Mues,A. and Young,P. (1999) Control of sarcomeric assembly: the flow of information on titin. Rev. Physiol. Biochem. Pharmacol., 138, 97–137. [DOI] [PubMed] [Google Scholar]
- Govind C.K. (1985) Neural control of bilateral asymmetry in crustacean cheliped muscles. In Bush,B.M. and Clarac,F. (eds), Coordination of Motor Behavior. Cambridge University Press, Cambridge, UK, pp. 20–22.
- Greaser M. (2001) Identification of new repeating motifs in titin. Proteins, 43, 145–149 [DOI] [PubMed] [Google Scholar]
- Gregorio C.C., Granzier,H., Sorimachi,H. and Labeit,S. (1999) Muscle assembly: a titanic achievement? Curr. Opin. Cell Biol., 11, 18–25. [DOI] [PubMed] [Google Scholar]
- Gutierrez-Cruz G., Van Heerden,A., and Wang,K. (2001) Modular motif, structural folds and affinity profiles of PEVK segment of human fetal skeletal muscle titin. J. Biol. Chem., 276, 7442–7449. [DOI] [PubMed] [Google Scholar]
- Hakeda S., Endo,S. and Saigo,K. (2000) Requirements of kettin, a giant muscle protein highly conserved in overall structure in evolution, for normal muscle function, viability, and flight activity of Drosophila. J. Cell Biol., 148, 101–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi H. and Takemori,S. (1989) Butanedione monoxime suppresses contraction and ATPase activity of rabbit skeletal muscle. J. Biochem., 105, 638–643. [DOI] [PubMed] [Google Scholar]
- Hu D.H., Matsuno,A., Terakado,K., Matsuura,T., Kimura,S. and Maruyama,K. (1990) Projectin is an invertebrate connectin (titin): isolation from crayfish claw muscle and localization in crayfish claw muscle and insect flight muscle. J. Muscle Res. Cell Motil., 11, 497–511. [DOI] [PubMed] [Google Scholar]
- Kolmerer B. et al. (2000) Sequence and expression of the kettin gene in Drosophila melanogaster and Caenorhabditis elegans. J. Mol. Biol., 296, 435–448. [DOI] [PubMed] [Google Scholar]
- Labeit S. and Kolmerer,B. (1995) Titins: giant proteins in charge of muscle ultrastructure and elasticity. Science, 270, 293–296. [DOI] [PubMed] [Google Scholar]
- Labeit S., Gautel,M., Lakey,A. and Trinick,J. (1992) Towards a molecular understanding of titin. EMBO J., 11, 1711–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakey A., Ferguson,C., Labeit,S., Reedy,M., Larkins,A., Butcher,G., Leonard,K. and Bullard,B. (1990) Identification and localization of high molecular weight proteins in insect flight and leg muscle. EMBO J., 9, 3459–3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakey A., Labeit,S., Gautel,M., Ferguson,C., Barlow,D.P., Leonard,K. and Bullard,B. (1993) Kettin, a large modular protein in the Z-disc of insect muscles. EMBO J., 12, 2863–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linke W.A. (2000) Stretching molecular springs: elasticity of titin filaments in vertebrate striated muscle. Histol. Histopathol., 15, 799–811. [DOI] [PubMed] [Google Scholar]
- Machado C. and Andrew,D.J. (2000) D-Titin. A giant protein with dual roles in chromosomes and muscles. J. Cell Biol., 151, 639–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado C., Sunkel,C.E. and Andrew,D.J. (1998) Human autoantibodies reveal titin as a chromosomal protein. J. Cell Biol., 141, 321–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki S., Ohtani,Y., Kimura,S. and Maruyama,K. (1995) Isolation and characterization of a kettin-like protein from crayfish claw muscle. J. Muscle Res. Cell Motil., 16, 579–585. [DOI] [PubMed] [Google Scholar]
- Manabe T., Kawamura,Y., Higuchi,H., Kimura,S. and Maruyama,K. (1993) Connectin, giant elastic protein, in giant sarcomeres of crayfish claw muscle. J. Muscle Res. Cell Motil., 14, 654–665. [DOI] [PubMed] [Google Scholar]
- Maruyama K. (1994) Connectin, an elastic protein of striated muscle. Biophys. Chem., 50, 73–85. [DOI] [PubMed] [Google Scholar]
- Ohtani Y., Maki,S., Kimura,S. and Maruyama,K. (1996) Localization of connectin-like proteins in leg and flight muscles of insects. Tissue Cell, 28, 1–8. [DOI] [PubMed] [Google Scholar]
- van Straaten M., Goulding,D., Kolmerer,B., Labeit,S., Clayton,J., Leonard,K. and Bullard,B. (1999) Association of kettin with actin in the Z-disc of insect flight muscle. J. Mol. Biol., 285, 1549–1562. [DOI] [PubMed] [Google Scholar]
- Yamaguchi M. and Takemori,S. (2001) Activating efficiency of Ca2+ and cross-bridges as measured by phosphate analog release. Biophys. J., 80, 371–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Featherstone,D., Davis,W., Rushton,E. and Broadie,K. (2000) Drosophila D-titin is required for myoblast fusion and skeletal muscle striation. J. Cell Sci., 113, 3103–3115. [DOI] [PubMed] [Google Scholar]