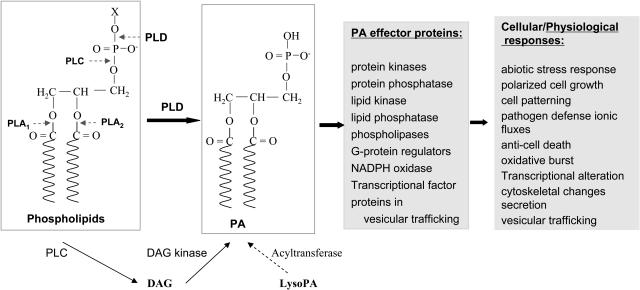
Phospholipase D (PLD) hydrolyzes membrane lipids to generate phosphatidic acid (PA) and a free-head group (Fig. 1), and this activity is widespread in plants. Recent results indicate that PLD plays multiple regulatory roles in diverse plant processes, including abscisic acid (ABA) signaling, programmed cell death, root hair patterning, root growth, freezing tolerance, and other stress responses (Fig. 1). In some cases, direct molecular targets of PLD and PA have been identified, providing insights into the mechanism by which the phospholipase and lipid messenger mediate plant functions.
Figure 1.
PLD-catalyzed hydrolysis of phospholipids, downstream targets, and cellular functions. The activity of PLD produces PA and free-head group (X). PA target proteins have been identified in animals, plants, and yeast (see also Table I). Multiple cellular effects of PLD and PA have been documented or implicated. Note that in addition to PLD, signaling PA can be generated from DAG kinase coupled to the activation of PLC and potentially from acylation reactions.
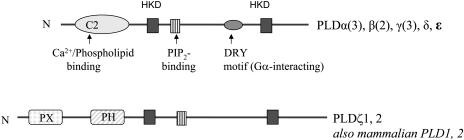
PLD is composed of a family of heterogeneous enzymes with distinguishable biochemical, regulatory, and structural properties. The PLD gene family in plants is more complex than that in other organisms: 12 PLD genes are in Arabidopsis (Arabidopsis thaliana), whereas two PLD genes are in mammals and one in baking yeast (Saccharomyces cerevisiae; Wang, 2002, 2004, and refs. therein). The 12 Arabidopsis PLDs can be classified into six types, PLDα(3), β(2), γ(3), δ, ɛ, and ζ(2) (Fig. 2). Based on the overall protein domain structures, PLDs can be divided into two subfamilies, C2-PLD and PX/PH-PLDs (Fig. 2). C2 is a Ca2+ and phospholipid-binding domain, and the PX and PH domains refer to two distinct phosphoinositide-interacting structural folds, phox homology and pleckstrin homology, respectively. Ten of the 12 Arabidopsis PLDs (α, β, γ, δ, and ɛ) contain the C2 domain. The PLDζs contain the PX and PH domains, and this domain structure is present in mammalian PLDs (Elias et al., 2002; Qin and Wang, 2002). The overall sequences of PLDζs are more similar to mammalian PLDs than to other Arabidopsis PLDs.
Figure 2.
Domain structures of Arabidopsis PLDs. Plant PLDs consist of two distinctive subfamilies: the C2-PLDs and the PX/PH-PLDs. Individual PLDs can differ in key amino acid residues in regulatory motifs, such as C2, PIP2 binding, and DRY. The duplicated HKD motifs are involved in catalysis. Note that the 12 Arabidopsis PLDs have now been classified into six types instead of five types; the only modification is that PLDα4 has been reclassified to PLDɛ because this PLD is quite distant from all the other PLDs.
In addition, new structural motifs have been identified in PLDs that interact with G proteins, Ca2+, and phosphoinositides (Fig. 2; Zheng et al., 2002; Pappan et al., 2004; Zhao and Wang, 2004). Individual PLDs differ in key amino acid residues in these regulatory motifs. These differences underlie a structural basis for the distinguishable biochemical and regulatory properties in different PLDs. The information on the molecular, structural, and biochemical heterogeneity of PLDs provides clues as to the regulation and diverse functions of PLDs in plants. This Update focuses on the recent developments toward understanding the function of specific PLDs and PA that they produce.
PLDα1: A CONNECTION BETWEEN G PROTEIN AND PROTEIN PHOSPHATASE 2C
The most abundant plant PLD requires high millimolar concentrations of Ca2+ for activity in vitro. PLDα1 possesses the common PLD activity (Wang, 2002). A gene knockout of PLDα1 abolishes the millimolar Ca2+-requiring PLD activity in Arabidopsis (Zhang et al., 2004), demonstrating that PLDα1 is the predominant one responsible for the common PLD activity in the plant. Arabidopsis deficient in PLDα1 displays alterations in various plant processes, such as reactive oxygen production, wound-induced accumulation of jasmonic acid, freezing tolerance, water loss, and abscisic acid signaling (for review, see Wang, 2002).
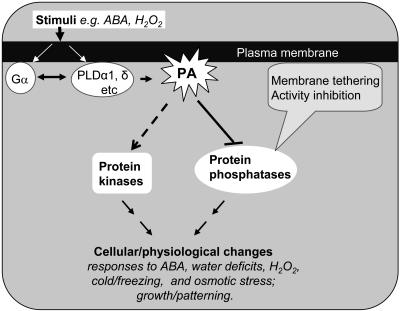
Recent results indicate that PLDα1 and its lipid product PA are intermediary links between important cellular regulators in plant cells (Fig. 3). PLDα1 interacts directly with Gα, the only canonical α-subunit of the heterotrimeric G protein in Arabidopsis (Zhao and Wang, 2004). PLDα1 binds to Gα through a sequence motif analogous to the DRY motif normally conserved in animal G-protein-coupled receptors. Mutation of amino acid residues in this motif abolishes the PLDα1-Gα binding and the Gα inhibition of PLDα1 activity. Meanwhile, the PLDα1-Gα interaction stimulates the intrinsic GTPase activity of Gα. These results indicate that the interaction reciprocally modulates the activities of PLDα1 and Gα.
Figure 3.
A working model depicting the activation of PLDs and the role of PA in regulating kinase and phosphatase activities in cell's response to ABA, H2O2, or other stimuli. Potential mediators in the PLD activation include changes in G protein function and membrane conformations and/or increases in cytosolic Ca2+, phopshoinositides, and oleic acid. PLD-produced PA may stimulate protein kinase cascades and also inhibit protein phosphatases. PA may do so by directly affecting the enzymatic activities and/or by tethering the signaling enzymes to specific membranes or regions of a membrane. These interactions modulate the cell's ability to respond to stresses and inhibit cell death.
Both PLDα1 and Gα are involved in ABA signaling. Gene suppression and overexpression results show that that PLDα1 plays a role in the ABA response (Sang et al., 2001). A recent study has unveiled a mechanism by which PLDα1 mediates such a response (Zhang et al., 2004). PLDα1-derived PA binds to ABI1 (ABA insensitive), a protein phosphatase 2C (PP2C) that is a negative regulator of ABA responses in Arabidopsis. The PA binding decreases PP2C activity and also appears to reduce its translocation to nuclei in response to ABA by tethering ABI1 to the plasma membrane. These results show that activation of PLDα1 inhibits the function of the negative regulator ABI1, thus promoting ABA signaling (Fig. 3).
PLDδ POSITIVELY REGULATES STRESS TOLERANCE
PLDδ exhibits several properties that distinguish it from other PLDs. It is activated by oleic acid and is tightly associated with the plasma membrane and microtubule cytoskeleton. The expression of PLDδ increases in severe dehydration, high salt, and during cold acclimation. Analyses of PLDδ-altered Arabidopsis suggest that PLDδ positively regulates plant tolerance to stresses such as freezing, oxidative assault, and ultra-violet irradiation (Zhang et al., 2003; Li et al., 2004). PLDδ-null Arabidopsis is less tolerant to freezing, whereas overexpression of PLDδ increases freezing tolerance (Li et al., 2004). Lipid profiling indicates that PLDδ activity produces selective PA species but does not result in substantial lipid hydrolysis during freezing stress.
By comparison, antisense suppression of PLDα1 increases freezing tolerance, and lipid profiling shows that PLDα1 activity results in a large decrease in phosphatidylcholine (PC) and an increase in PA (Welti et al., 2002). These results indicate that PLDα1 and δ are involved in the response to freezing via different mechanisms and also demonstrate that manipulation of different PLDs can have different phenotypic alterations. In addition, the PLDδ alterations do not result in changes in the expression of the cold-regulated genes COR47 or COR78, nor in any change in cold-induced increases in the levels of compatible osmolytes, Pro, or soluble sugars, which are known to play a role in plant freezing tolerance. These results suggest that PLDs and associated membrane lipid hydrolysis present a novel signaling pathway involved in mediating plant freezing tolerance.
PLD/PA IN REACTIVE OXYGEN RESPONSE, PRODUCTION, AND CELL DEATH
One mechanism by which PLDδ positively mediates plant stress tolerance is through its role in signaling resistance to damages inflicted by reactive oxygen species (ROS; Li et al., 2004). PLDδ is activated by hydrogen peroxide (H2O2), and the resulting PA functions to decrease H2O2-promoted programmed cell death (Zhang et al., 2003). Examining mitogen-activated protein (MAP) kinase activity suggests that PLDδ and PA play a role in H2O2-induced activation of MAP kinase cascades (Zhang et al., 2003). The ROS H2O2 is an important cellular mediator, and its concentration increases under various stress conditions, including freezing. Activation of MAP kinase cascades is involved in plant response to H2O2 and various stress responses. The involvement of PLDδ and its derived PA in these processes may present a mechanism by which PLDδ plays a positive role in stress response.
PLDδ is activated by H2O2, but it is not involved in H2O2 production under the conditions tested (Zhang et al., 2003). By comparison, PLDα1 and PA are involved in the production of reactive oxygen species (Sang et al., 2001). These results indicate that specific PLDs function in different steps in plant oxidative stress pathways; whereas PLDα1 promotes the ROS production, PLDδ mediates plant responses to ROS. The role of PA in ROS production is supported by another study: PA treatment of whole leaves and single cells increased the levels of ROS and promoted leaf cell death (Park et al., 2004). This study also suggests that PA promotes the ROS production via activating the Rho-related small G protein GTPase-mediated pathway. The PA-induced cell death appears contradictory to the role of PLDδ-derived PA in mitigating H2O2-induced cell death. However, the location and timing of PA production and appropriate cellular concentrations and molecular species of PA are important determinants of PA function.
PLDζ1 AND PA IN ROOT HAIR GROWTH AND PATTERNING
Unlike C2-containing PLDs, the PX/PH-containing PLDζ1 is Ca2+ independent and PC specific (Qin and Wang, 2002). A study of the molecular target of the homeobox gene GLABRA2 (GL2) has provided insights into one physiological function of PLDζ1 (Ohashi et al., 2003). PLDζ1 is a direct target of GL2, a key component of a regulatory circuit composed of transcription factor genes that regulate the root hair pattern of Arabidopsis. GL2 is thought to be a negative regulator of root hair development. Inducible expression of PLDζ1 promoted ectopic root hair initiation, whereas inducible suppression inhibits root hair initiation (Ohashi et al., 2003). The results suggest that GL2 regulates root hair development through modulation of PLDζ1. However, loss of PLDζ1 does not cause obvious changes in root hair patterning (M. Li, C. Qin, and X. Wang, unpublished data). These results may indicate that more than one PLD could be involved in the root hair growth and development.
How PLD regulates the root hair patterning and initiation is not known. A recent study indicates that PA regulates a protein kinase pathway involved in root growth and initiation (Anthony et al., 2004). PA binds to the 3′-phosphoinositide-dependent kinase-1 (AtPDK1) and stimulates the activity of AGC2-1 in an AtPDK1-dependent manner. AGC refers to a kinase superfamily comprising the prototypes of the cAMP-dependent protein kinases A (PKA), cGMP-dependent protein kinases G (PKG), and protein kinases C (PKC). AGC2-1 is a target protein kinase of AtPDK1, and the AGC2-1 knockout plants exhibit a decrease in root hair length. It remains to be determined whether the AtPDK1-binding PA results from the activation of PLDs because signaling PA can be derived from diacylglycerol (DAG) kinase coupled to the activation of phospholipase C (PLC; den Hartog et al., 2003; De Jong et al., 2004) and potentially from acylation reactions (Fig. 1).
PLD INTERACTIONS WITH CYTOSKELETON
PLD and PA have been implicated in affecting both actin and tubulin cytoskeletons. Arabidopsis PLDβ1 binds to α-actin (Kusner et al., 2003). The effects of actin on the activities of the PLD are polymerization dependent; monomeric G-actin inhibits PLD activity, whereas polymerized F-actin augments PLD activity. Actin modulation of PLDβ1 has kinetic characteristics, efficacies, and potencies similar to those of human PLD1 (Kusner et al., 2003). This conservation of PLD-actin interaction between PLDs from plants and mammals indicates that other plant PLDs, such as PLDζs, may also bind to actin because of their close similarity to mammalian PLDs.
PA is reported to promote actin polymerization in soybean (Glycine max) cells (Lee et al., 2003). PA is found to bind a heterodimeric capping protein from Arabidopsis (AtCP; S. Huang and C.J. Staiger, personal communication). The PA-binding inhibits the activity of AtCP; in the presence of PA, AtCP is unable to cap the barbed or rapidly growing and shrinking end of actin filaments and fails to nucleate filament formation. The PA inhibition of capping-protein activity in plant cells results in stimulation of actin polymerization from a large pool of profilin-actin.
Following the finding that PLDδ binds to tubulin, two studies explored the effect of PLD on microtubule cytoskeleton organization. In one study, seeds and young seedlings of Arabidopsis were incubated with 1-butanol, and this treatment disrupted the organization of interphase cortical microtubules and also inhibited seed germination and seedling growth (Gardiner et al., 2003). In another study using tobacco (Nicotiana tabacum) Bright Yellow-2 cells expressing the microtubule reporter green fluorescent protein-MAP4, 1-butanol treatment promoted a partial depolymerization of cortical microtubules and the release of them from the plasma membrane (Dhonukshe et al., 2003). It has been postulated that PLD links microtubules via the PLD catalytic intermediate phosphatidyl moiety. Results from both of the studies suggest that activation of PLD is critical to triggering microtubule reorganization. It is likely that, besides PLDδ, other PLDs are involved in microtubule reorganization because the PLDδ-knockout plants grow and develop normally under regular growth conditions. However, PLDδ-null plants are less tolerant to stresses (Zhang et al., 2003; Li et al., 2004), and their role in cytoskeletal reorganization may contribute to the weakening of plant stress responses.
It is worth noting that butanol treatment takes advantage of the unique, transphosphatidylation activity of PLD, which uses primary alcohols as substrates to form phosphatidylalcohol at the expense of PA. Because there is no specific, effective inhibitor for PLD, this approach has been useful in helping to determine the cellular functions of the PLD activity. However, primary alcohols stimulate rather than inhibit PLD activity. Also, the diversion of PA formation by alcohol is incomplete. Thus, an effect associated with an alcohol treatment needs to be interpreted with caution because it may result from other alcoholic effects, such as increases in lipid hydrolysis, changes in lipid composition, and/or release of lipid head groups that have regulatory functions (Chapman, 2004).
PLD IN POLLEN TUBE GROWTH AND PRO ACCUMULATION
Tobacco pollen germination and tube growth are stopped in the presence 0.5% 1-butanol, and this inhibition could be overcome by addition of exogenous PA, suggesting that PLD-derived PA plays a role in stimulating pollen germination and tube elongation (Potocky et al., 2003). In another study, the expansion of the pollen tube apical region is associated with a severalfold increase in PA, which is generated by PLD (Zonia and Munnik, 2004). Different phospholipid signals are differentially stimulated or attenuated in pollen tube volume changes in response to the osmotic perturbations. Hypo-osmotic stress induces rapid increases in PA, but hyperosmotic stress and cell shrinkage inhibit PLD activity and induce increases in the levels of phosphatidylinositides.
In other cells or tissues, however, PLD is activated under hyperosmotic stresses, such as high salinity and dehydration (Testerink and Munnik, 2005). Under these stresses, many plants accumulate Pro, which has been suggested to play a role in plant stress adaptation. A recent study probed the role of PLD activation in the regulation of Pro metabolism in Arabidopsis (Thiery et al., 2004). Application of 1-butanol stimulated Pro biosynthesis without hyperosmotic constraints and enhanced the Pro responsiveness of seedlings to mild hyperosmotic stress. Gene expression results suggest that the activation of PLD plays both positive and negative roles in hyperosmotic stress signaling in plants. These seemingly conflicting effects of PLDs are consistent with the hypothesis that different PLDs have unique functions during the stress responses (Fig. 4), as shown for the role of PLDα1 and PLDδ in the ROS stress and freezing tolerance (Sang et al., 2001; Welti et al., 2002; Zhang et al., 2003; Li et al., 2004).
Figure 4.
Unique roles of different PLDs, multifaceted functions of a PLD, and multiple modes of action of PA. Unique functions have been shown for PLDα1 and PLDδ. Box 1 lists some of the available findings that support different functions of PLDs. Box 2 indicates that a given PLD can be involved in different cellular processes, depending upon the nature of biotic and abiotic cues, such as the severity of stresses. Box 3 lists several effects that PA has on cell regulation. Documented examples are given in parentheses, and those in bold denote PA target proteins identified in plants. The symbol and function of the target proteins are explained in Table I and text. FFA, Free fatty acid; DAG-PPi, DAG-pyrophosphate.
NEW INSIGHTS ON CA2+ REGULATION OF PLD
All C2-PLDs require Ca2+ for activity, but how Ca2+ affects PLD activity is not well understood. It was previously reported that Ca2+ binds to the regulatory C2 domain that occurs in the n terminus of PLDs (Zheng et al., 2000). Using a C2-deleted PLDβ1 (PLDβcat) and other PLDβ1 domain constructs, a recent study has shown that Ca2+ also interacts with the catalytic regions of PLD (Pappan et al., 2004). The Ca2+-PLDβcat interaction increases the affinity of the protein for the activator, phosphatidylinositol 4,5-bisphosphate (PIP2), but not for the substrate, PC. This is in contrast to the effect of Ca2+ binding to the C2 domain, which stimulates PC binding but inhibits PIP2 binding of the domain. The PIP2-bound catalytic domain increases the enzyme's affinity for its substrate PC. These results demonstrate the contrasting and complementary effects of the Ca2+- and lipid-binding properties of the C2 and catalytic domains of plant PLD, and provide insight into the mechanism by which Ca2+ regulates PLD activity. A membrane-scooting model has been proposed to explain the regulation of PLD activity by changing cellular Ca2+ concentrations. These findings also account in part for why Ca2+ is required for the activity of the C2-PLDs but not the PX/PH-PLDs (Pappan et al., 2004).
UNIQUE AND MULTIFACETED FUNCTIONS OF DIFFERENT PLDS
The distinguishable phenotypes resulting from loss of different PLDs indicate that the loss of one PLD is not compensated for, at least completely, by the other 11 PLDs in Arabidopsis (Zhang et al., 2003, 2004; Li et al., 2004). Unique functions of different PLDs may occur via one or a combination of the following (Fig. 4): (1) Individual PLDs are regulated and activated differently in the cell; (2) they have different substrate preferences and have the potential to generate different PAs or other derivatives; (3) they are associated with different membranes; and (4) they have different temporal and spatial patterns of expression. The differences in regulation, activation, substrate preference, intracellular association, and expression patterns have been demonstrated for several PLDs (Wang 2002, 2004, and refs. therein). These differences ultimately regulate the location and timing of the PLD activities and PA produced by the enzymes. Spatial and temporal regulation is important to all signaling events, but it is particularly critical to intracellular lipid messengers because of their limited mobility.
In addition, one PLD can mediate cell function via different modes of action, depending upon the nature of stimuli and severity of stresses (Fig. 4). These include (1) production of lipid mediators, such as PA, N-acyletanolamine, and other lipid derivatives; (2) direct interaction with other proteins, such as Gα, actin, and tubulin; (3) altering membrane lipid composition; and (4) membrane degradation. The role in changes of membrane lipid composition has been documented for PLDα1 in response to freezing (Welti et al., 2002). PLDα1 selectively hydrolyzes PC and alters the ratios of membrane lipids and contributes to membrane lipid degradation. Historically, PLD activity was associated with lipid degradation and membrane deterioration. This is perhaps best reflected by or attributed to a large increase of PA under conditions such as wounding, tissue handling, or uncontrolled lipid extraction. Such high PLD activity is likely to result from deregulation of PLD, such as disruption of cellular compartments. This deregulation may occur in plant tissues under severe stress conditions such as freezing, aging, or storage and thus is physiologically relevant. However, under normal plant growth, PLD activity is tightly regulated; for example, it is activated in response to specific stimuli, such as ABA and H2O2. These results indicate multifaceted functions of a PLD.
MOLECULAR TARGETS OF PLD-DERIVED PA
PLD is activated under various growth conditions, and PA is the direct lipid product of such activation (Welti et al., 2002; den Hartog et al., 2003; Potocky et al., 2003; Zhang et al., 2003, 2004; Li et al., 2004; Zonia and Munnik, 2004). Genetic and pharmacological manipulations of PLD-mediated PA formation suggest that PA mediates various signaling processes, as described here and reviewed earlier (Wang, 2002, 2004; Testerink and Munnik, 2005). It should be noted that signaling PA can be produced by another route (Fig. 1); the activation of the PLC/DAG kinase pathway can occur under certain stress conditions, such as hyperosmotic stress and pathogen elicitation (De Jong et al., 2004; Testerink and Munnik, 2005).
Major progress has been made recently in understanding how PA functions as a signaling messenger. In particular, direct molecular targets of PA have been identified in two specific plant processes. In ABA responses, PLDα1-derived PA has been shown to bind to ABI1 PP2C (Zhang et al., 2004). The amino acid residue required for PA-ABI1 interaction has been identified and resides in the n-terminal region of ABI1. The binding of PA to ABI1 decreases the PP2C activity and also tethers ABI1 to the plasma membrane. These changes reduce the translocation of ABI1 to the nucleus and thus inhibit the ABI1 function (Zhang et al., 2004). In mediating root growth and initiation, PA binds to AtPDK1 and stimulates the activity of a downstream kinase AGC2-1 (Anthony et al., 2004). In addition, a SnRK2 Ser/Thr protein kinase and the PP2A regulatory subunit RCN1 are potential PA targets, as they were identified through PA-affinity chromatography (Testerink et al., 2004; Table I).
Table I.
Examples of PA-binding proteins identified in plants, animals, and yeast
| PA-Binding Protein | Protein Function | Reference |
|---|---|---|
| From plants: | ||
| ABI1 | Protein phosphatase 2C like | Zhang et al. (2004) |
| AtPDK1 | 3′-Phopshoinositide-dependent kinase 1 | Anthony et al. (2004) |
| PEPC | Phosphoenolpyruvate carboxylase | Testerink et al. (2004)a |
| From yeast: | ||
| Opi1p | Soluble transcriptional repressor | Loewen et al. (2004) |
| From animals: | ||
| mTOR | Mammalian target of rapamycin | Fang et al. (2001) |
| Raf1 | MAP kinase kinase kinase | Ghosh et al. (2003) |
| Fgr | Tyr kinases expressed in neutrophils | Sergeant et al. (2001) |
| PKCz | Protein kinase Cζ | Cummings et al. (2002) |
| SHP-1 | SH2-containing protein-tyrosine phosphatase | Frank et al. (1999) |
| PP1 | Protein phosphatase-1 | Jones and Hannun (2002) |
| Type1 PI4P5K | Phosphatidylinositol 4P-5 kinase | Jenkins et al. (1994) |
| RGS4 | Regulators of G-protein signaling protein | Ouyang et al. (2003) |
| SPHK | Sphingosine kinase | Delon et al. (2004) |
| ARF | ADP-ribosylation factor | Manifava et al. (2001) |
| NSF | N-ethylmaleimide-sensitive factor | |
| Kinesin | Vesicular trafficking | |
| Phox 47 | A cytosolic component of NADPH oxidase | Karathanassis et al. (2002) |
| PDE4D3 | cAMP-specific phosphodiesterase | Grange et al. (2000) |
Several other proteins were also found to be associated with PA-affinity beads.
The PA targets in plants go beyond protein phophatases and kinases. Several additional proteins were associated with a PA-affinity column; these include phosphoenolpyruvate carboxylase, Hsp90, and 14-3-3 (Testerink et al., 2004; Table I). The binding to phosphoenolpyruvate carboxylase has been shown to be specific to PA. Several PA target proteins have been identified in other systems (Table I). In the budding yeast S. cerevisiae, PA on the endoplasmic reticulum directly bound to the soluble transcriptional repressor Opi1p to maintain it as inactive outside the nucleus (Loewen et al., 2004). A decrease in PA due to lipid biosynthesis releases Opi1p from the endoplasmic reticulum, allowing its nuclear translocation and repression of target genes. These results show that PA functions as both an essential metabolic intermediate and a signaling lipid. In animal cells, PLD-derived PA regulates mammalian target of rapamycin (mTOR), which belongs to the family of phosphatidylinositol kinase-like kinases and regulates translation initiation in mammals, and transcription, translation, and cytoskeletal rearrangement in yeast (Chen, 2004). PA directly interacts with the domain in the mTOR, and this binding is critical for mTOR's ability to activate downstream effectors (Fang et al., 2001).
It is important to note that PA may mediate cellular processes via different ways of action (Fig. 4). First, as a signaling messenger, PA can bind to its effector proteins, and this binding may activate or inhibit the function of its target proteins, as described above. Second, as a lipid in membranes, PA can tether its effector proteins to membranes, and the tethering may help to direct proteins to a specific type or region of a membrane. Third, PA may function via its structural effect on membranes to promote membrane fusion and trafficking. The space of PA's head group is smaller than that of its acyl chains, and this geometry renders PA a negative curvature and fusigenic lipid (Kooijman et al., 2005). PLD-derived PA is required for cell plate formation during meiosis and sporulation in yeast (Xie et al., 1998) and is a key facilitator of membrane vesicle trafficking events in different mammalian cells (Huang et al., 2005). In addition, PA serves as a substrate and/or an activator for enzymes that promote the formation of other lipid regulators, such as lysoPA, free fatty acids, DAG, DAG-pyrophosphate, and oxylipins (Wang 2002, 2004; Testerink and Munnik, 2005).
PERSPECTIVES
Substantial progress has been made recently toward understanding the regulation and function of PLDs. These advances present new opportunities to investigate the signaling mechanisms in plants. The finding of the novel signaling interaction between PLDα1 and Gα raises several intriguing questions. For instance, would PLDs function as GTPase-activating proteins or guanine nucleotide exchange factors? How would the PLD-Gα interaction affect the interactions of Gα with Gβγ subunits and with G-protein receptors? What would be the function and specificity of the PLD-G protein interaction in the cell? The observations that PA interacts with protein phosphatases and kinases warrant further investigation. PLD and PA may play an important role in the homeostasis of protein phosphorylation by concerted regulation of the two groups of signaling enzymes with opposite biochemical functions in a specific signaling response (Fig. 3). In the case of the PA-ABI1 interaction, it is conceivable that that PLD and PA may also affect the functions of other PP2Cs because Arabidopsis contains 69 PP2C-like genes.
A combination of the multiple routes of PA production, different PA species, diverse protein targets, and varied modes of action makes PA versatile mediators in cell regulation. Elucidation of the structural determinants that are required for PA-protein interaction will help to determine the specificity of the lipid-protein interaction and to understand how that interaction modulates structure, dynamics, and function in the ensuing lipid-protein complex. A better understanding of the regulation and function of PLDs and their derived messengers will advance not only the understanding of the major phospholipase family, but also the field of cell signaling and signaling networks in plant growth and stress responses.
This work was supported by grants from the National Science Foundation and the U.S. Department of Agriculture.
References
- Anthony RG, Henriques R, Helfer A, Meszaros T, Rios G, Testerink C, Munnik T, Deak M, Koncz C, Bogre L (2004) A protein kinase target of a PDK1 signalling pathway is involved in root hair growth in Arabidopsis. EMBO J 23: 572–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman KD (2004) Occurrence, metabolism, and prospective functions of N-acylethanolamines in plants. Prog Lipid Res 43: 302–327 [DOI] [PubMed] [Google Scholar]
- Chen J (2004) Novel regulatory mechanisms of mTOR signaling. Curr Top Microbiol Immunol 279: 245–257 [DOI] [PubMed] [Google Scholar]
- Cummings R, Parinandi N, Wang L, Usatyuk P, Natarajan V (2002) Phospholipase D/phosphatidic acid signal transduction: role and physiological significance in lung. Mol Cell Biochem 234: 99–109 [PubMed] [Google Scholar]
- De Jong CF, Laxalt AM, Bargmann BO, De Wit PJ, Joosten MH, Munnik T (2004) Phosphatidic acid accumulation is an early response in the Cf-4/Avr4 interaction. Plant J 39: 1–12 [DOI] [PubMed] [Google Scholar]
- Delon C, Manifava M, Wood E, Thompson D, Krugmann S, Pyne S, Ktistakis NT (2004) Sphingosine kinase 1 is an intracellular effector of phosphatidic acid. J Biol Chem 279: 44763–44774 [DOI] [PubMed] [Google Scholar]
- den Hartog M, Verhoef N, Munnik T (2003) Nod factor and elicitors activate different phospholipid signaling pathways in suspension-cultured alfalfa cells. Plant Physiol 132: 311–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhonukshe P, Laxalt AM, Goedhart J, Gadella TW, Munnik T (2003) Phospholipase D activation correlates with microtubule reorganization in living plant cells. Plant Cell 15: 2666–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias M, Potocky M, Cvrckova F, Zarsky V (2002) Molecular diversity of phospholipase D in angiosperms. BMC Genomics 3: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Vilella-Bach M, Bachmann R, Flanigan A, Chen J (2001) Phosphatidic acid-mediated mitogenic activation of mTOR signaling. Science 294: 1942–1945 [DOI] [PubMed] [Google Scholar]
- Frank C, Keilhack H, Opitz F, Zschornig O, Boomer FD (1999) Binding of phosphatidic acid to the protein-tyrosine phosphatase SHP-1 as a basis for activity modulation. Biochemistry 38: 11993–12002 [DOI] [PubMed] [Google Scholar]
- Gardiner J, Collings DA, Harper JD, Marc J (2003) The effects of the phospholipase D-antagonist 1-butanol on seedling development and microtubule organisation in Arabidopsis. Plant Cell Physiol 44: 687–696 [DOI] [PubMed] [Google Scholar]
- Ghosh S, Moore S, Bell RM, Dush M (2003) Functional analysis of a phosphatidic acid binding domain in human Raf-1 kinase: mutations in the phosphatidate binding domain lead to tail and trunk abnormalities in developing zebrafish embryos. J Biol Chem 278: 45690–45696 [DOI] [PubMed] [Google Scholar]
- Grange M, Sette C, Cuomo M, Conti M, Lagarde M, Prigent AF, Nemoz G (2000) The cAMP-specific phosphodiesterase PDE4D3 is regulated by phosphatidic acid binding: consequences for cAMP signaling pathway and characterization of a phosphatidic acid binding site. J Biol Chem 275: 33379–33387 [DOI] [PubMed] [Google Scholar]
- Huang P, Altshuller YM, Hou JC, Pessin JE, Frohman MA (2005) Insulin-stimulated plasma membrane fusion of Glut4 glucose transporter-containing vesicles is regulated by phospholipase D1. Mol Biol Cell 16: 2614–2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins GH, Fisette PL, Anderson RA (1994) Type I phosphatidylinositol 4-phosphate 5-kinase isoforms are specifically stimulated by phosphatidic acid. J Biol Chem 269: 11547–11554 [PubMed] [Google Scholar]
- Jones JA, Hannun YA (2002) Tight binding inhibition of protein phosphatase-1 by phosphatidic acid: specificity of inhibition by the phospholipid. J Biol Chem 277: 15530–15538 [DOI] [PubMed] [Google Scholar]
- Karathanassis D, Stahelin RV, Bravo J, Perisic O, Pacold CM, Cho W, Williams RL (2002) Binding of the PX domain of p47phox to phosphatidylinositol 3,4-bisphosphate and phosphatidic acid is masked by an intramolecular interaction. EMBO J 21: 5057–5068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooijman EE, Chupin V, Fuller NL, Kozlov MM, de Kruijff B, Burger KN, Rand PR (2005) Spontaneous curvature of phosphatidic acid and lysophosphatidic acid. Biochemistry 44: 2097–2102 [DOI] [PubMed] [Google Scholar]
- Kusner DJ, Barton JA, Qin C, Wang X, Iyer SS (2003) Evolutionary conservation of physical and functional interactions between phospholipase D and actin. Arch Biochem Biophys 412: 231–241 [DOI] [PubMed] [Google Scholar]
- Lee S, Park J, Lee Y (2003) Phosphatidic acid induces actin polymerization by activating protein kinases in soybean cells. Mol Cells 15: 313–319 [PubMed] [Google Scholar]
- Li W, Li M, Zhang W, Welti R, Wang X (2004) The plasma membrane-bound phospholipase Ddelta enhances freezing tolerance in Arabidopsis thaliana. Nat Biotechnol 22: 427–433 [DOI] [PubMed] [Google Scholar]
- Loewen CJ, Gaspar ML, Jesch SA, Delon C, Ktistakis NT, Henry SA, Levine TP (2004) Phospholipid metabolism regulated by a transcription factor sensing phosphatidic acid. Science 304: 1644–1647 [DOI] [PubMed] [Google Scholar]
- Manifava M, Thuring JW, Lim ZY, Packman L, Holmes AB, Ktistakis NT (2001) Differential binding of traffic-related proteins to phosphatidic acid- or phosphatidylinositol (4,5)-bisphosphate-coupled affinity reagents. J Biol Chem 276: 8987–8994 [DOI] [PubMed] [Google Scholar]
- Ohashi Y, Oka A, Rodrigues-Pousada R, Possenti M, Ruberti I, Morelli G, Aoyama T (2003) Modulation of phospholipid signaling by GLABRA2 in root-hair pattern formation. Science 300: 1427–1430 [DOI] [PubMed] [Google Scholar]
- Ouyang YS, Tu Y, Barker SA, Yang F (2003) RGS4: insertion into model membranes and inhibition of activity by phosphatidic acid. J Biol Chem 278: 11115–11122 [DOI] [PubMed] [Google Scholar]
- Pappan K, Zheng L, Krishnamoorthi R, Wang X (2004) Evidence for and characterization of Ca2+ binding to the catalytic region of Arabidopsis thaliana phospholipase Dβ. J Biol Chem 279: 47833–47839 [DOI] [PubMed] [Google Scholar]
- Park J, Gu Y, Lee Y, Yang Z, Lee Y (2004) Phosphatidic acid induces leaf cell death in Arabidopsis by activating the Rho-related small G protein GTPase-mediated pathway of reactive oxygen species generation. Plant Physiol 134: 129–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potocky M, Elias M, Profotova B, Novotna Z, Valentova O, Zarsky V (2003) Phosphatidic acid produced by phospholipase D is required for tobacco pollen tube growth. Planta 217: 122–130 [DOI] [PubMed] [Google Scholar]
- Qin C, Wang X (2002) The Arabidopsis phospholipase D family: characterization of a Ca2+-independent and phosphatidylcholine-selective PLDζ1 with distinct regulatory domains. Plant Physiol 128: 1057–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang Y, Cui D, Wang X (2001) Phospholipase D- and phosphatidic acid-mediated generation of superoxide in Arabidopsis. Plant Physiol 126: 1449–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergeant S, Waite KA, Heravi J, McPhail LC (2001) Phosphatidic acid regulates tyrosine phosphorylating activity in human neutrophils: enhancement of Fgr activity. J Biol Chem 276: 4737–4746 [DOI] [PubMed] [Google Scholar]
- Testerink C, Dekker HL, Lim ZY, Johns MK, Holmes AB, Koster CG, Ktistakis NT, Munnik T (2004) Isolation and identification of phosphatidic acid targets from plants. Plant J 39: 527–536 [DOI] [PubMed] [Google Scholar]
- Testerink C, Munnik T (2005) Phosphatidic acid: a multifunctional stress signaling lipid in plants. Trends Plant Sci 10: 368–375 [DOI] [PubMed] [Google Scholar]
- Thiery L, Leprince AS, Lefebvre D, Ghars MA, Debarbieux E, Savoure A (2004) Phospholipase D is a negative regulator of proline biosynthesis in Arabidopsis thaliana. J Biol Chem 279: 14812–14818 [DOI] [PubMed] [Google Scholar]
- Wang X (2002) Phospholipase D in hormonal and stress signaling. Curr Opin Plant Biol 5: 408–414 [DOI] [PubMed] [Google Scholar]
- Wang X (2004) Lipid signaling. Curr Opin Plant Biol 7: 329–336 [DOI] [PubMed] [Google Scholar]
- Welti R, Li W, Li M, Sang Y, Biesiada H, Zhou HE, Rajashekar CB, Williams TD, Wang X (2002) Profiling membrane lipids in plant stress responses: role of phospholipase D alpha in freezing-induced lipid changes in Arabidopsis. J Biol Chem 277: 31994–32002 [DOI] [PubMed] [Google Scholar]
- Xie Z, Fang M, Rivas MP, Faulkner AJ, Sternweis PC, Engebrecht JA, Bankaitis VA (1998) Phospholipase D activity is required for suppression of yeast phosphatidylinositol transfer protein defects. Proc Natl Acad Sci USA 95: 12346–12351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Qin C, Zhao J, Wang X (2004) Phospholipase Dalpha1-derived phosphatidic acid interacts with ABI1 phosphatase 2C and regulates abscisic acid signaling. Proc Natl Acad Sci USA 101: 9508–9513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Wang C, Qin C, Wood T, Olafsdottir G, Welti R, Wang X (2003) The oleate-stimulated phospholipase D, PLDδ, and phosphatidic acid decrease H2O2-induced cell death in Arabidopsis. Plant Cell 15: 2285–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Wang X (2004) Arabidopsis phospholipase Dα1 interacts with the heterotrimeric G-protein a subunit through a motif analogous to the DRY motif in G-protein-coupled receptors. J Biol Chem 279: 1794–1800 [DOI] [PubMed] [Google Scholar]
- Zheng L, Krishnamoorthi R, Zolkiewski M, Wang X (2000) Distinct calcium binding properties of novel C2 domains of plant phospholipase Dα and β. J Biol Chem 275: 19700–19706 [DOI] [PubMed] [Google Scholar]
- Zheng L, Shan J, Krishnamoorthi R, Wang X (2002) Activation of plant phospholipase Dβ by phosphatidylinositol 4,5-bisphosphate: characterization of binding site and mode of action. Biochemistry 41: 4546–4553 [DOI] [PubMed] [Google Scholar]
- Zonia L, Munnik T (2004) Osmotically induced cell swelling versus cell shrinking elicits specific changes in phospholipid signals in tobacco pollen tubes. Plant Physiol 134: 813–823 [DOI] [PMC free article] [PubMed] [Google Scholar]