Abstract
We report the analysis and annotation of 146,075 expressed sequence tags from Vitis species. The majority of these sequences were derived from different cultivars of Vitis vinifera, comprising an estimated 25,746 unique contig and singleton sequences that survey transcription in various tissues and developmental stages and during biotic and abiotic stress. Putatively homologous proteins were identified for over 17,752 of the transcripts, with 1,962 transcripts further subdivided into one or more Gene Ontology categories. A simple structured vocabulary, with modules for plant genotype, plant development, and stress, was developed to describe the relationship between individual expressed sequence tags and cDNA libraries; the resulting vocabulary provides query terms to facilitate data mining within the context of a relational database. As a measure of the extent to which characterized metabolic pathways were encompassed by the data set, we searched for homologs of the enzymes leading from glycolysis, through the oxidative/nonoxidative pentose phosphate pathway, and into the general phenylpropanoid pathway. Homologs were identified for 65 of these 77 enzymes, with 86% of enzymatic steps represented by paralogous genes. Differentially expressed transcripts were identified by means of a stringent believability index cutoff of ≥98.4%. Correlation analysis and two-dimensional hierarchical clustering grouped these transcripts according to similarity of expression. In the broadest analysis, 665 differentially expressed transcripts were identified across 29 cDNA libraries, representing a range of developmental and stress conditions. The groupings revealed expected associations between plant developmental stages and tissue types, with the notable exception of abiotic stress treatments. A more focused analysis of flower and berry development identified 87 differentially expressed transcripts and provides the basis for a compendium that relates gene expression and annotation to previously characterized aspects of berry development and physiology. Comparison with published results for select genes, as well as correlation analysis between independent data sets, suggests that the inferred in silico patterns of expression are likely to be an accurate representation of transcript abundance for the conditions surveyed. Thus, the combined data set reveals the in silico expression patterns for hundreds of genes in V. vinifera, the majority of which have not been previously studied within this species.
On a worldwide basis, grapes (Vitis species) are both the most widely cultivated and economically important fruit crop, encompassing approximately 8 million hectares of arable land (Vivier and Pretorius, 2002). Although the grape berry is used for multiple purposes, including juice, fresh and dried fruit, and distilled liquor, wine produced from cultivars of Vitis vinifera has the highest economic value (Mullins et al., 1992). Worldwide wine production in the year 2000 was 271,212,000 hectoliters (http://www.wineinstitute.org). In addition to its economic impact, there is increasing interest in wine as a source of health-promoting secondary metabolites (e.g. Bagchi et al., 2003; Spiller et al., 2003), such as antioxidant polyphenols (Ferguson, 2001; Aldini et al., 2003). For example, the polyphenol resveratrol has gained particular attention due to its ability to increase longevity in yeast through the stimulation of anti-aging enzymes known as sirtuins (Howitz et al., 2003).
In contrast to many other crop species where genotypic variation is a tool for crop improvement, in wine grapes, constancy of the genotype or variety is often the desired goal. Varietal integrity is maintained through vegetative propagation. As a consequence, intensive crop management practices (i.e. viticulture) are more important to maintaining quality characteristics than are traditional breeding methodologies, which have been limited in their application in grapes relative to other major crop species. Genomics approaches are likely to have particular value for grape improvement because they have the potential to identify transcriptional, biochemical, and genetic pathways that contribute to agronomic properties. Examples include revealing transcriptional pathways that are correlated with berry quality (e.g. metabolism of sugars, organic acids, and flavonoids) and disease resistance (e.g. specific resistance genes and downstream transcriptional pathways) and determining how viticultural practices impact these molecular phenotypes. The application of such knowledge to grape improvement is likely to take the form of improved viticultural practices and precise molecular breeding. Approaches such as marker-assisted selection and transgenesis will facilitate transfer of genes for desirable traits into elite or classic cultivars of V. vinifera, with the goal of improving agronomic performance while preserving traditional quality traits (Bisson et al., 2002; Vivier and Pretorius, 2002).
The strategies outlined above will be aided by genomics efforts to characterize the gene content of grapes, such as those involving expressed sequence tag (EST) screens. ESTs currently represent the most abundant nucleotide commodity from plant genomes, providing a resource that can be exploited for gene discovery, genome annotation, and comparative genomics (Rudd, 2003). When EST sequencing is combined with a methodical cDNA library construction effort, in silico analysis of EST frequency can be used to identify genes whose expression is correlated with particular points in development or induced in response to abiotic or biotic factors. Moreover, the identification of coregulated genes provides the basis for analyzing presumptive gene networks and facilitates use of “guilt by association” logic to infer the function of unknown or imprecisely annotated genes (Journet et al., 2002).
Although EST collections are currently available for several crop and model plant species, until recently, most ESTs from Vitis species were not publicly available. Reports of nonpublic data include the analysis of 5,000 ESTs generated from V. vinifera cv Chardonnay leaf and berry tissue (Ablett et al., 2000), cv Shiraz berries at various stages of development (Terrier et al., 2001), and the analysis of over 4,000 ESTs from dormant buds of V. vinifera cv Purple Cornichon (Pacey-Miller et al., 2003). However, an international effort from several research groups worldwide has dramatically increased the availability of EST data from grapes. In 2001, there were just over 400 sequences deposited in GenBank. As of September 30, 2003, 146,075 sequences were deposited to the National Center for Biotechnology Information (NCBI) for several Vitis species. Here, we describe the analysis of this transcript data set, with emphasis on organization and annotation of the unigene set and analysis of differentially expressed genes.
RESULTS
The Vitis subsp. Data Sets
In total, 146,075 Vitis sequences were deposited into GenBank (NCBI) as of September 30, 2003. Eighty percent of these sequences were generated by the authors, with the majority of the remaining sequences deposited into public data repositories by five different research groups (Supplemental Table I). The most economically important species of Vitis, V. vinifera, was the source of the majority of the sequences, with 1,471 sequences corresponding to transcript and genomic DNA sequences and 135,541 ESTs from 58 cDNA libraries representing seven cultivars. The depth of sequencing of V. vinifera cDNA libraries ranged from eight to 24,400 ESTs (Supplemental Table I), and libraries represented numerous cultivars, organs, plant developmental stages, and stress treatments as shown in Table I. The remaining Vitis species (Supplemental Table I) were represented by 8,957 ESTs and 106 genomic or expressed transcripts.
Table I.
Distribution of V. vinifera EST data set
| Cultivars | Total ESTs (%) | Plant Organ | Total ESTs (%) | Developmental Stage | Total ESTs (%) | Stress Treatment | Total ESTs (%) |
|---|---|---|---|---|---|---|---|
| Cabernet | 63,799 | Berry | 48,806 | Preveraison | 34,751 | Abiotic | 51,492 |
| Sauvignon | (47) | (36) | (25.6) | (40) | |||
| Chardonnay | 56,037 | Leaf | 42,953 | Postveraison | 17,872 | Biotic | 3,650 |
| (41.3) | (31.7) | (13.2) | (3.0) | ||||
| Pinot noir | 5,681 | Flower | 11,326 | Veraison | 10,889 | ND | 74,399 |
| (4.2) | (8.4) | (8.0) | (57.0) | ||||
| Shiraz | 7,393 | Roots | 7,271 | Pre-anthesis | 6,117 | ||
| (5.5) | (5.4) | (4.5) | |||||
| Regent | 1,755 | Compound bud | 6,112 | Anthesis | 4,428 | ||
| (1.3) | (4.5) | (3.3) | |||||
| Ugni Blanc | 675 | Berry without seeds | 5,576 | Postmeiosis | 3,748 | ||
| (0.5) | (4.1) | (2.8) | |||||
| Chasselas | 261 | Stem | 4,700 | Bud swelling | 1,750 | ||
| (0.2) | (3.5) | (1.3) | |||||
| Petiole | 4,491 | NDa | 56,046 | ||||
| (3.3) | (41.3) | ||||||
| Berry pedicle | 2,722 | ||||||
| (2.0) | |||||||
| Shoot tip | 857 | ||||||
| (0.6) | |||||||
| Seeds | 478 | ||||||
| (0.4) | |||||||
| Berry skin | 309 | ||||||
| (0.2) |
ND, Not determined.
To facilitate analysis across the EST data set, we developed a controlled and structured vocabulary to classify cDNA libraries according to three main categories: namely, genotype, plant development, and stress. Each category was further subdivided according to accepted hierarchical relationships using vocabulary in common use for viticulture, as shown by example in Figure 1 (for complete detail, see Supplemental Table II). The vocabulary was used to generate an online query tool (http://cgf.ucdavis.edu/) to facilitate identification of genes that may be differentially or uniquely expressed under specified conditions. One can identify genes whose expression is enhanced in a particular Vitis species or cultivar, under biotic or abiotic stress, in a particular organ, or at a specific plant developmental stage. For example, several genes expressed only in grapes infected with the bacterial pathogen Xylella fastidiosa have been identified using this tool (F. Goes da Silva and D. Cook, unpublished data), demonstrating the utility of this structured vocabulary.
Figure 1.
A controlled vocabulary for description of the Vitis species cDNA libraries. Libraries are organized into three main categories, (A) genotype, (B) development, and (C) stress, which can be further subdivided as shown. Terms for the Cabernet Sauvignon Leaf (CA12LI) are shown by way of example, with detailed information for all libraries available in Supplemental Tables I and II.
Generation of Vitis Unigene Sets
For purposes of generating a unigene set, ESTs and expressed transcripts obtained en masse from NCBI were organized into contigs (also called tentative consensus sequences [TCs]) and singleton sequences by means of MegaBLAST and CAP3 (Liang et al., 2000). Clustering was performed separately for species represented by more than 250 sequences (i.e. V. vinifera, V. aestivalis, and V. arizonica × V. rupestris). Genomic DNA sequences as well as sequences from species represented by <250 sequences were used to produce a separate singleton data set. In total, 25,696 unigenes were predicted for V. vinifera, 1,977 for a Vitis rupestris × V. arizonica hybrid, and 1,314 for V. aestivalis (Table II).
Table II.
Summary of the Vitis spp. unigene sets
Clustering was performed individually for data sets from species represented by at least 250 sequences available at the NCBI database.
| Species | TCsa | Average TC Length | Singletons | Average Singleton Length | Total Unigenes |
|---|---|---|---|---|---|
| V. vinifera | 13,307 | 873 bp | 12,389 | 527 bp (range, 100–7,891 bp)b | 25,696 |
| V. rupestris × V. arizonica | 907 | 808 bp | 1,070 | 515 bp (range, 100–952 bp)a | 1,977 |
| V. aestivalis | 269 | 840 bp | 1,045 | 598 bp (range, 103–1,465 bp)a | 1,314 |
Includes only EST data.
Includes non-EST sequences.
The occurrence in a single TC of paired-end (i.e. 5′ and 3′ direction) reads from the same cDNA clone was taken as experimental evidence in support of computationally determined TC structure. Approximately 72% of V. vinifera TCs (9,599 TCs, with an average size of 1,020 bp) were composed of paired-end clones, of which 54% (5,146 TCs) of the paired-end cDNA sequences started within 40 bp of the 5′ most nucleotide. These results demonstrate that the majority of unigenes were captured within a single cDNA clone.
Annotation of the Unigene Sets
To identify Vitis unigenes that potentially encode homologs of known proteins, we conducted BLASTX (Altschul et al., 1997) against GenBank's nonredundant protein database. Sixty-nine percent (18,259) of the V. vinifera unigenes showed significant similarity to proteins in the database based on an E value cutoff of ≤ 1e−5, and 5,064 of the protein homologs were annotated as unknown protein, hypothetical protein, or expressed protein. This result is not surprising, considering that a great number of sequences in the NCBI database represent uncharacterized genes and proteins. BLASTX analysis for the remaining Vitis species revealed similar trends.
More detailed functional annotation was provided by mapping unigenes onto the Gene Ontology Consortium structure using The Institute for Genomic Research (TIGR) Vitis vinifera Gene Index (http://www.tigr.org/tdb/tgi/vvgi/) version 3.1 (Quackenbush et al., 2001). Gene Ontology provides a structured and controlled vocabulary to describe gene products according to three ontologies: molecular function, biological process, and cellular component (Gene Ontology Consortium, 2000). In total, 1,962 unigenes could be mapped to one or more ontology, with multiple assignments possible for a given protein within a single ontology. Thus, 2,362 assignments were made to the molecular function ontology, with 68% of these in the catalytic activity and binding categories (Fig. 2A) and annotations such as RNA helicases, kinases, ubiquitin, and nucleotide binding proteins. By way of example, the branch child terms for transporter activity and transcription regulator activity revealed several genes implicated in carbohydrate transport (e.g. CTG1030456 and CTG1030972) as well as predicted transcription factors with putative roles in berry ripening (e.g. myb-related regulatory genes CTG1028317 and CTG1028387 and MADS box genes CTG1028624 and CTG1030218). Under the biological process ontology, the vast majority of the 2,056 assignments were to the physiological process category, with frequent subclassification into the response to stress, response to external stimulus, and cell growth and maintenance categories (Fig. 2B). The abundance of stress- and growth-related annotations is not surprising, considering that a significant portion of the ESTs analyzed were derived from Vitis tissues responding to biotic or abiotic stress or from tissues in early stages of development. Finally, for cellular components, the vast majority of the assignments (96%) were to the cell category (Fig. 2C).
Figure 2.
Distribution of V. vinifera unigenes with putative functions assigned through annotation using gene ontology. Molecular function (A), biological process (B), and cellular component (C). Assignments are based on the data available at the TIGR Vitis vinifera Gene Index (http://www.tigr.org/tdb/tgi/vvgi/) version 3.1 (November, 2003).
Characterizing the Functional Gene Space in Vitis sp.
To determine the extent to which the functional gene space had been sampled, we searched the unigene set for homologs of structural enzymes involved in primary, intermediate, and secondary metabolic pathways. The targets of our analysis were biochemical pathways of known topology involved in the biosynthesis of flavonoids and their precursors, including the Embden-Meyerhoff-Parnas pathway (EMP pathway or glycolysis), the Krebs cycle (citric acid cycle, tricarboxylic acid cycle cycle), the oxidative/nonoxidative pentose phosphate pathway, the shikimic acid or aromatic amino acid pathway, and the general phenylpropanoid and flavonoid pathways. The selected metabolic network converts the simple six-carbon structure of hexoses into the more complex 6:3:6 basic carbon structures of flavonoids. Flavonoids are of particular interest in grape biology because they represent an important metabolic sink for carbon, particularly in organs and cells actively accumulating phenylpropanoids, such as cells in the exocarp and outer mesocarp of ripening grapevine berries (Coombe, 1987; Hardie et al., 1996).
Table III (and Supplemental Table III) shows TCs and singletons from V. vinifera and other non-vinifera Vitis subsp. predicted to code for structural enzymes of the selected pathways. Most pathways are fully represented by Vitis ESTs, and most enzymes are supported by a significant degree of redundancy, both in terms of the number of ESTs (expression) and the number of TCs and singletons (putative paralogous genes). This redundancy suggests numerous moderately sized gene families in the grapevine genome.
Table III.
Putative genes in the Vitis spp. data sets coding for structural enzymes of key pathways of primary, intermediate, and secondary metabolism
| Pathways: Structural Enzymes | E.C. | Contigsa | Singletonsa | Total Nonredundant ESTsb |
|---|---|---|---|---|
| Glycolysis | ||||
| Hexokinase | 2.7.1.1 | 2 | 0 | 7 |
| Glucose-6-phosphoisomerase | 5.3.1.9 | 2 | 3 | 10 |
| 6-Phosphofructokinase | 2.7.1.11 | 4 | 1 | 22 |
| Fructose 1,6-biphosphate aldolase | 4.1.2.13 | 3 | 6 | 186 |
| Triose phosphate isomerase | 5.3.1.1 | 4 | 1 | 58 |
| Glyceraldehyde 3-P dehydrogenase | 1.2.1.12 | 2 | 5 | 26 |
| Phosphoglycerate kinase | 2.7.2.3 | 3 | 1 | 62 |
| Phosphoglycerate mutase | 5.4.2.1 | 1 | 1 | 31 |
| Enolase | 4.2.1.11 | 6 | 3 | 202 |
| Pyruvate kinase | 2.7.1.40 | 10 | 3 | 61 |
| Tricarboxylic acid cycle (Krebs cycle) | 24 | 13 | 218 | |
| Citrate synthase | 4.1.3.7 | 3 | 3 | 21 |
| Aconitase | 4.2.1.3 | 7 | 1 | 56 |
| Isocitrate dehydrogenase | 1.1.1.42 | 4 | 2 | 35 |
| α-Ketoglutarate dehydrogenase complex | No E.C. | No hit | No hit | No hit |
| Succinyl-CoA synthetase | 6.2.1.5 | No hit | No hit | No hit |
| Succinate dehydrogenase | 1.3.5.1 | 6 | 2 | 49 |
| Fumarase | 4.2.1.2 | 0 | 2 | 2 |
| Malate dehydrogenase | 1.1.99.16 | 4 | 3 | 55 |
| Oxidative/nonoxidative pentose phosphate pathway | 12 | 3 | 192 | |
| Glucose 6-P-1-dehydrogenase | 1.1.1.49 | 1 | 0 | 2 |
| 6-Phosphogluconolactonase | 3.1.1.31 | 3 | 0 | 22 |
| 6-Phosphogluconate dehydrogenase | 1.1.1.44 | 1 | 0 | 11 |
| Ribose-5-P isomerase | 5.3.1.6 | 1 | 1 | 8 |
| Ribose-5-P 3-epimerase | 5.1.3.1 | 1 | 0 | 48 |
| Transketolase | 2.2.1.1 | 2 | 2 | 87 |
| Transaldolase | 2.2.1.2 | 3 | 0 | 14 |
| Shikimic acid pathway: aromatic amino acid biosynthesis | 19 | 10 | 123 | |
| 3-Deoxy-d-arabino-heptulosonate 7-P synthase | 4.1.2.15 | 2 | 0 | 21 |
| 3-Dehydroquinate synthase | 4.2.3.4 | 1 | 2 | 11 |
| 3-Dehydroquinate dehydratase | 4.2.1.10 | 2 | 2 | 3 |
| Shikimate 5 dehydrogenase | 1.1.1.25 | 5 | ||
| Shikimate kinase | 2.7.1.71 | 4 | 0 | 21 |
| 5-Enolpyruvoylshikimate 3-P synthase | 2.5.1.19 | 2 | 0 | 8 |
| Chorismate synthase | 4.2.3.5 | 1 | 0 | 19 |
| Chorismate mutase | 5.4.99.5 | 1 | 1 | 3 |
| Prephenate dehydratase | 4.2.1.51 | 1 | 3 | 15 |
| Prephenate dehydrogenase | 1.3.1.12 | No hit | No hit | No hit |
| Aromatic amino acid transaminase | 2.6.1.57 | No hit | No hit | No hit |
| Anthranilate synthase | 4.1.3.27 | 1 | 0 | 1 |
| Anthranilate phosphoribosyl transferase | 2.4.2.18 | No hit | No hit | No hit |
| Phosphoribosylanthranilate synthase | 5.3.1.24 | No hit | No hit | No hit |
| Indol-3-glycerol phosphate synthase | 4.1.1.48 | 1 | 0 | 1 |
| Trp synthase | 4.2.1.20 | 3 | 2 | 15 |
| General phenylpropanoid pathway | 10 | 6 | 98 | |
| Phe ammonia-lyase | 4.3.1.5 | 5 | 3 | 43 |
| Cinnamate 4-hydroxilase (NADPH cytochrome P450 reductase) | 1.14.13.11 | 2 | 0 | 26 |
| 4-Coumarate coenzyme A ligase | 6.2.1.12 | 3 | 3 | 29 |
| General phenylpropanoid pathway: lignin biosynthesis | 38 | 16 | 228 | |
| Caffeate O-methyltransferase | No E.C. | 11 | 3 | 56 |
| Caffeoyl-CoA O-methyltransferase | 2.1.1.104 | 3 | 4 | 29 |
| Cinnamoyl-CoA reductase | No E.C. | 8 | 3 | 45 |
| Ferulate 5-hydroxylase | No E.C. | 1 | 1 | 6 |
| 5-Hydroxy feruloyl CoA O-methyltransferase | No E.C. | No hit | No hit | No hit |
| Cinnamyl alcohol dehydrogenase | 1.1.1.195 | 15 | 5 | 92 |
| 5-Hydroxy coniferyl alcohol O-methyltransferase | No E.C. | No hit | No hit | No hit |
| 5-Hydroxy coniferaldehyde O-methyltransferase | No E.C. | No hit | No hit | No hit |
| Flavonoid biosynthetic pathway: flavonol biosynthesis | 19 | 21 | 389 | |
| Naringenin chalcone synthase | 2.3.1.74 | 8 | 5 | 171 |
| Chalcone isomerase | 5.5.1.6 | 2 | 4 | 84 |
| Flavanone 3-hydroxylase | 1.14.11.9 | 3 | 3 | 89 |
| Flavonol 3-hydroxylase | 1.14.13.21 | No hit | No hit | No hit |
| Flavonol synthase | No E.C. | 3 | 4 | 25 |
| Flavonol 3-O-glucosyltransferase | No E.C. | 3 | 5 | 20 |
| Flavonoid biosynthetic pathway: anthocyanin biosynthesis | 10 | 13 | 107 | |
| Dihydroflavonol 4-reductase | 1.1.1.219 | 5 | 6 | 55 |
| Leucoanthocyanidin dioxygenase | No E.C. | 1 | 3 | 32 |
| UDP-flavonol 3-O-glucosyltransferase | 2.4.1.- | 2 | 4 | 13 |
| Anthocyanin 5-O-glucosyltransferase | No E.C. | 0 | 1 | 1 |
| Anthocyanin 5-aromatic acyltransferase | No E.C. | 1 | 0 | 2 |
| Anthocyanin permease | No E.C. | 1 | 0 | 4 |
| Flavonoid biosynthetic pathway: proanthocyanidin biosynthesis | 4 | 1 | 113 | |
| Anthocyanidin reductase | No E.C. | 1 | 0 | 99 |
| Leucoanthocyanindin reductase | No E.C. | 3 | 1 | 14 |
Putative genes were identified among contigs and singletons available at http://cgf.ucdavis.edu grape database based on BLASTX definition selecting only E values of E < 1.0e−5 (median of E = 1.26e−78).
Represents the total number of ESTs eliminating redundancy, i.e. clones with both 5′ and 3′ reads within a TC.
In Silico Characterization of Gene Expression in V. vinifera
For the purpose of inferring patterns of gene expression, we constructed a matrix containing nonredundant EST frequency across each cDNA library for all TCs. No TCs were observed with sequences present in all 58 cDNA libraries, which is not surprising given the diversity of tissue types sampled and the limited depth to which some cDNA libraries were sequenced. The most widely expressed TC encodes a putative Rubisco small subunit protein, with presence in 38 separate cDNA libraries. In total, 14 TCs were expressed in at least 30 cDNA libraries (Table IV). All of the broadly expressed TCs matched annotated proteins in the NCBI nonredundant database, with the exception of CTG1027353. Despite their presence in multiple cDNA libraries, certain of these broadly expressed TCs (e.g. CTG1026987 and CTG1027410, both annotated as a putative abscisic acid, stress, and ripening-induced protein implicated in sugar transport) were also predicted to be differentially expressed, as described below.
Table IV.
Annotation of ESTs with the widest distribution among V. vinifera cDNA libraries
| Contig No. | No. of Nonredundant ESTs in TCa | Tentative Annotation | E Value | Libraries Represented |
|---|---|---|---|---|
| CTG1026982 | 1,390 | Rubisco small subunit | 7.63E-70 | 38 |
| CTG1026987 | 532 | VvMSA, transcription factor that binds to the promoter of a hexose transporter | 8.21E-21 | 38 |
| CTG1027410 | 484 | VvMSA, transcription factor that binds to the promoter of a hexose transporter | 2.41E-19 | 38 |
| CTG1027454 | 290 | Elongation factor 1-α 1 | 0.00E+00 | 37 |
| CTG1027353 | 205 | No hit | NA | 37 |
| CTG1027439 | 766 | Early light-induced protein-like protein | 3.63E-48 | 36 |
| CTG1027458 | 320 | Unknown protein (Arabidopsis) | 9.38E-176 | 32 |
| CTG1027234 | 139 | Glyceraldehyde-3-P dehydrogenase | 8.74E-160 | 31 |
| CTG1027166 | 903 | Metallothionein-like protein | 7.80E-34 | 30 |
| CTG1027501 | 255 | Metallothionein-like protein | 1.63E-15 | 30 |
| CTG1026911 | 180 | Aquaporin PIP2-1 | 2.90E-161 | 30 |
| CTG1027477 | 101 | Hexameric polyubiquitin | 0.00E+00 | 30 |
| CTG1027494 | 78 | ADP-ribosylation factor | 2.40E-99 | 30 |
| CTG1027534 | 74 | S-adenosylmethionine synthetase 1 | 0 | 30 |
Represents the total number of ESTs eliminating redundancy, i.e. clones with both 5′ and 3′ reads within a TC.
To identify putative differentially expressed genes, we used the R statistic methodology described by Stekel et al. (2000). This analysis was restricted to libraries with over 1,000 ESTs (a total of 29 libraries), as libraries represented by relatively few ESTs are likely to confound the prediction of differentially expressed genes (Ewing et al., 1999). A total of 665 TCs with an R value ≥25 and false positive rate ≤1 (believability [B] ≥ 98.4%) were identified (Supplemental Table IV). Two-dimensional hierarchical clustering was performed using a correlation matrix constructed from EST frequencies for the 665 differentially expressed TCs. cDNA libraries clustered into two major clades: one relatively homogeneous clade representing all leaf libraries and a second more heterogeneous clade containing all other libraries (Fig. 3A). In general, libraries tended to cluster according to the stage of development and tissue sampled (Fig. 3A). An exception to this pattern is libraries constructed from preveraison berries, which exhibited very different positions within the cluster. It is noteworthy that several of these preveraison berry libraries were constructed from fruit that was carefully sampled to represent distinct stages of berry development, all prior to the onset of ripening (veraison). This differential clustering of preveraison libraries is suggestive of major shifts in gene expression during preveraison berry development (see below). The two root cDNA libraries included in the analysis also exhibited differential clustering, with abiotic stressed roots clustering separately from nonstressed roots. In total, the abiotic stressed Chardonnay root, leaf, and berry libraries contained 79, 26, and six library-specific TCs, respectively, with B > 98.0%. These putative stress-specific TCs were typically highly expressed (EST counts ranging from 8–215 ESTs) and in 72% of cases had either no similarity to sequences in the NCBI database nonredundant based on BLASTX or matched an unknown or hypothetical protein.
Figure 3.
Hierarchical cluster of grape cDNA libraries and TCs based on EST distribution. A, One-dimensional clustering of 29 grape cDNA libraries. B, Two-dimensional hierarchical clustering of 665 differentially expressed transcripts versus the 29 cDNA libraries shown in A. All 665 differentially expressed transcripts in B have a true positive rate of >98.4% (Stekel et al., 2000). Two-dimensional clustering is based on a Pearson's correlation coefficient matrix. Band intensity designates relative transcript abundance in a given library, as inferred from EST frequency within each TC. Black and white designate least and most abundant transcripts, respectively. ND, Not determined; NA, not available.
We further compared the patterns of expression predicted from the current in silico analysis with published results obtained by more traditional northern-blot analysis. Davies and Robinson (2000) reported 17 grape ripening-induced (GRIP) genes, all of which encode either putative cell wall or stress response proteins that are differentially expressed in ripening berries (Davies and Robinson, 2000). We identified the corresponding TCs for all 17 of these genes, 14 of which were included among the 665 differentially expressed TCs used for hierarchical clustering (Table V). As shown in Figure 4, in silico analysis confirms that expression of the GRIP genes was largely restricted to the veraison and postveraison libraries; moreover, this work extends the previous analysis of Davies and Robinson (2000) by demonstrating that expression of most GRIP genes was also absent from roots, buds, flowers, stems, petioles, and leaves, with the primary exception of GRIP 21 and VvTL2, which were expressed in 1-year-old root tissues of V. vinifera cv Pinot Noir. Examination of the hierarchical cluster populated by GRIPs 3, 13, 15, 21, and 61 identified four putatively coregulated genes (Table V). The annotations for several of these TCs suggest functional relatedness to the GRIPs, including a putative stress response thaumatin-like protein SCUTL2 from V. vinifera (CTG1028245) and a putative cell wall expansin (CTG1027667) with homology to a Glycine max gene. The fact that the patterns of expression predicted by in silico analysis are consistent with those obtained by alternative methods (Davies and Robinson, 2000) suggests that broader analysis of the 665 unigenes shown in Figure 3 is likely to be an accurate depiction of the expression for a significant fraction of the grape transcriptome.
Table V.
GRIP genes (Davies and Robinson, 2000) and coregulated TCs (with putative annotation) identified in the V. vinifera nonredundant data set (TCs and singletons)
TC numbers in bold represent those identified within the 665 differentially expressed transcripts based on R-statistics analysis (Stekel et al., 2000; Fig. 4). Contigs with a postscript letter were split from chimeric contigs by manual curation, as indicated in Supplemental Table IV. GB, GenBank accession number.
| Gene | Grip GB No. | TC Number | Total Nonredundant ESTsa |
|---|---|---|---|
| Cell wall protein | |||
| Grip 3 | AJ237981 | CTG1026936 | 1,137 |
| Grip 4 | AJ237982 | CTG1026928 | 440 |
| Grip 13 | AJ237983 | CTG1027228b | 85 |
| Grip 15 | AJ237984 | CTG1026914 | 220 |
| Grip 28 | AJ237985 | CTG1027503 | 199 |
| Grip 31 | AJ237986 | CTG1027500 | 204 |
| Grip 68 | AJ237987 | CTG1027627 | 75 |
| Stress induced | |||
| Grip 21 | AJ237988 | CTG1027987 | 42 |
| Grip 22 | AJ237989 | CTG1027566 | 139 |
| Grip 24 | AJ237990 | CTG1027357B | 160 |
| Grip 32 | AJ237991 | CTG1026897B | 65 |
| Grip 51 (VvTL1) | AJ237999 | CTG1027413 | 159 |
| VvTL2 | AJ237998 | CTG1027187 | 105 |
| Grip 55 | AJ237992 | CTG1033083 | 2 |
| Grip 58 | AJ237993 | CTG1028645 | 9 |
| Grip 61 | AJ237994 | CTG1027515 | 232 |
| gfh2 | AJ237995 | CTG1028904 | 10 |
| TCs coregulated with Grips 3, 13, 15, 21, and 61 | |||
| SCUTL2 (V. vinifera), thaumatin-like protein | AAF06347 | CTG1028245 | 26 |
| Expansin, G. max | AAO15998 | CTG1027667 | 79 |
| Isoflavone reductase-related protein, Pyrus communis | AAC24001 | CTG1026887 | 37 |
| Expressed protein, Arabidopsis | NP_177894 | CTG1027906 | 40 |
cDNA clones with both 5′ and 3′ reads within a TC were counted only once to produce a nonredundant EST count.
CTG1027228 is 96% identical to AJ237883, representing an allele or a close paralog.
Figure 4.
Expression of GRIP genes based on normalized EST frequency. A, Putative cell-wall-associated GRIP genes. B, Putative stress-response-associated GRIP genes. Gene expression is represented as percentage of distribution of ESTs in each of the corresponding libraries. ND, Developmental stage not determined. NCBI accession and contig numbers are given in Table V.
Towards a Compendium of Gene Expression during Flower-Berry Development
Grapevine berries are nonclimateric fruits (Giovannoni, 2001) with a characteristic double sigmoid growth curve (Fig. 5). The initial phase of exponential berry growth (stage I) is followed by a lag phase (stage II), with growth resumed after the onset of ripening or veraison (stage III). Berry development is characterized by changes in numerous biological processes, including cell division and enlargement, primary and secondary metabolism, and resistance/susceptibility to abiotic/biotic stresses. In particular, the transition to ripening, which distinguishes stage II from stage III, includes major shifts in the accumulation of anthocyanins, organic acids, and sugars and alterations in the relative susceptibility to pathogens, such as powdery mildew (Oidium tuckerii) and bunch rot (Botrytis cinerea), which affect berries primarily during stages I and III, respectively (Ollat et al., 2002). Transcriptional profiling should reveal the transcriptional correlates of this dynamic developmental system.
Figure 5.
Overview of berry developmental and stages sampled by EST sequencing in the UC Davis flower-berry developmental series. SI, stage I; SII, stage II; SIII, stage III. 1, Flower prebloom; 2, flower bloom; 3, berry stage I; 4, berry stage II green hard; 5, berry stage II green soft; 6, berry stage III, 19 brix. Brix is a measure of soluble solids and a standard measure of ripening.
To examine transcriptional responses during berry development, six cDNA libraries were prepared at UC Davis from a well-characterized developmental series that encompasses prebloom flowers through stage III ripe fruit (Fig. 5; Supplemental Table V). Each cDNA library was generated using an identical protocol and survey sequenced to an equivalent depth using a paired-end sequencing strategy that increases the utility of the resource for in silico analysis of gene expression. Based on R-statistics analysis, 87 differentially expressed genes were identified with a false positive rate ≤1 (B ≥ 98.8%; Supplemental Table V). As shown in Figure 6, two-dimensional hierarchical clustering reveals that each stage of flower and berry development is characterized by distinct patterns of gene expression, including numerous stage-specific transcripts (Supplemental Table V).
Figure 6.
Hierarchical cluster of differentially expressed berry transcripts. EST frequencies used in this analysis corresponds to those derived from sequencing of the UC Davis flower-berry developmental series (see Fig. 5). Contig (CTG) numbers on the vertical axis correspond to the 87 differentially expressed transcripts described in the compendium of berry gene expression. Color designates relative transcript abundance in a given library, with red and blue being the most and least abundant, respectively. Individual transcripts span the vertical axis. cDNA libraries are organized in a temporal series, horizontally from left to right, as follows: FpB, flower prebloom; FB, flower bloom; SI, stage 1 berries; SIIgh, stage II green hard berries; SIIgs, stage II green soft berries; SIII, stage III berries (postveraison).
Prior to detailed analysis of specific transcripts, we sought to validate the use of our EST frequency data as a measure of gene expression, specifically for this data set. Towards this end, canonical correlation analysis (CCA) was used to determine the extent of correlation between the UC Davis data set and a similar but independent data set compiled from data deposited at the NCBI EST database. Four developmental stages (flower, preveraison, veraison, and ripe berries) could be circumscribed from the nonredundant NCBI data, which served as a pseudoreplicate of the UC Davis developmental series. Only differentially expressed genes with B ≥ 98.8% were used for CCA. CCA variables were developmental stages, while EST frequencies in a given developmental stage served as the individual observations. The two data sets were related through the common unigene set described above.
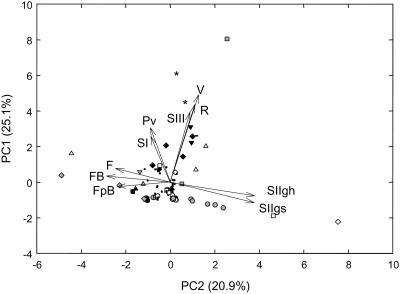
Taken together, the first and second canonical variates account for 89.52% of the variability in the EST data sets. Moreover, as shown in Table VI, CCA reveals a significant correlation between the UC Davis data set and the NCBI EST database pseudoreplicate, with correlations of 0.66 between the first canonical variates and 0.54 between the second canonical variates. The first canonical variates for both data sets are most strongly influenced by stage I (preveraison) and stage III (postveraison) libraries, while the second canonical variates in both data sets are most strongly influenced by stage III (veraison and ripe berry) libraries. Principal component analysis (PCA) of the combined data sets supports these conclusions, with a high level of congruence between libraries that sample similar stages of development (Fig. 7). In agreement with the results of two-dimensional hierarchical clustering, PCA also supports the distinct nature of gene expression during different stages of flower and berry development. Most striking among these differences is the opposite trend in EST frequencies observed between flowers and stage II berries, with a clear contrast along the second principal component. Unigenes having a major influence on PCA are indicated in the legend to Figure 7.
Table VI.
Correlations, standardized canonical coefficients, and canonical variates among the UC Davis College of Agricultural and Environmental Sciences libraries and other publicly available libraries, and cross-correlation analysis
The canonical correlations between the first and second canonical variables were 0.66 and 0.54, respectively. The first and second canonical variables accounted for 58.8% and 30.7% of the variability (cumulative = 89.5%). The approximate F test indicated that these two components were significant (P < 0.0001 and 0.001, respectively). CA&ES, UC Davis College of Agricultural and Environmental Sciences. Last two columns, cross-correlation analysis.
| Variable
|
First Variate
|
Second Variate
|
Canonical Variables
|
|||
|---|---|---|---|---|---|---|
| Correlation | Canonical Coefficient | Correlation | Canonical Coefficient | All1 | All2 | |
| CA&ES | ||||||
| Flower prebloom | 0.26 | 0.19 | −0.53 | −0.47 | 0.17 | −0.29 |
| Flower-bloom | 0.02 | 0.09 | −0.22 | 0.03 | 0.01 | −0.12 |
| Berry stage I | 0.92 | 0.85 | −0.18 | −0.32 | 0.61 | −0.10 |
| Berry stage II-gh | 0.07 | −0.19 | 0.07 | 0.02 | 0.05 | 0.04 |
| Berry stage II-gs | 0.04 | 0.33 | 0.02 | 0.01 | 0.02 | 0.01 |
| Berry stage III 19Brix | 0.49 | 0.35 | 0.81 | 0.86 | 0.32 | 0.43 |
| Other cDNA libraries | CA&ES1 | CA&ES2 | ||||
| Flower | 0.37 | 0.16 | −0.57 | −0.40 | 0.24 | −0.31 |
| Preveraison | 0.97 | 0.74 | −0.14 | −0.35 | 0.64 | −0.07 |
| Veraison | 0.45 | 0.73 | 0.81 | 1.20 | 0.30 | 0.44 |
| Ripen | 0.19 | −0.54 | 0.74 | −0.34 | 0.12 | 0.40 |
Figure 7.
PCA of 10 independent flower and berry EST data sets. F, Flower; FpB, flower prebloom; FB, flower bloom; SI, stage I berries; Pv, preveraison berries (approximately equivalent to stage I); SIIgh, stage II green hard berries; SIIgs, stage II green soft berries; SIII, stage III berries (postveraison); V, veraison; R, ripe (postveraison). Note that libraries that sampling similar developmental stages have highly similar principal components. Symbols are as follows: white diamond, chitinases; white square, nsLTPs; gray circle, allergens; white triangle, metallothioneins; *, putative transcription factors; white circle, Hsp; -, hidroxyproline-rich cell wall protein; gray diamond, photosynthesis-related; black inverted triangle, ripening induced proteins; gray square, plasma-membrane-associated proteins; black circle, ACC oxidase; black circle, caffeic acid OMT; black square, PPO; black triangle, chalcone isomerase; gray triangle, myo-inositol 1-P synthase; gray inverted triangle, aquaporin; and black diamond, no hit.
Taken together, the results of CCA and PCA indicate a strong statistical correlation between EST frequencies in the UC Davis and NCBI pseudoreplicate data sets, lending credence to the inference of gene expression patterns described below.
Functional Classification of Predicted Differentially Expressed Genes
For purposes of relating gene expression during grapevine flower and berry development to known changes in berry phenotypes, we assigned the 87 differentially expressed genes to eight functional categories based on the function of homologous proteins in Arabidopsis (Arabidopsis thaliana) or previously published work in grapes, as shown in Supplemental Table V. Categories included (1) pathogenesis-related (PR) proteins, (2) abiotic stress/cellular redox balance/detoxification-xenobiotic transport proteins, (3) primary metabolism, including CO2 assimilation, carbohydrate metabolism, lipid metabolism, and amino acid biosynthesis, (4) secondary metabolism, (5) berry growth, expansion, and water relations, (6) ethylene metabolism, (7) allergenic peptides and seed-specific proteins, and (8) other proteins of interest.
PR Proteins
Several PR proteins are predicted to be strongly up-regulated during stage II of berry development and during postveraison stages. Among these, chitinases and nonspecific lipid transfer and thaumatin-like proteins are the most abundant. From stage II of berry development through maturity, the protein content of the mesocarp cell walls increases >50% (Nunan et al., 1998). This increase in protein parallels the increase in soluble solid content of postveraison berries (Salzman et al., 1998). Approximately 90% of these proteins are PR proteins, especially thaumatin-like (osmotins) and chitinases (Salzman et al., 1998; Pocock et al., 2000). These proteins appear to be expressed under stress (Salzman et al., 1998; Pocock et al., 2000) and as part of normal berry development (Tattersall et al., 1997).
Chitinases constitute a large family of enzymes with hydrolytic activity against a linear polymer of β-1,4-N-acetylglucosamine, or chitin, which is a major component in the cell walls of most pathogenic fungi and in the exoskeleton of insect pests. In addition to their presumed importance in disease resistance, chitinases can be major allergens in grapes and wines (Pastorello et al., 2003), and they contribute to proteinaceous hazes that are detrimental to wine quality (Waters et al., 1998).
In silico analysis of EST frequencies (Fig. 6; Supplemental Table V) reveals the strong induction of an endochitinase (CTG1027444) during stage II pre- and post-berry softening and a class IV chitinase (CTG1027246) during postveraison development. These expression patterns are consistent with published measurements of enzyme activity and protein content in berries. Thus, chitinase activity has been observed to increase in proportion to the accumulation of soluble solids during ripening (Derckel et al., 1998), ultimately representing a substantial fraction of the total protein content of berries at industrial maturity (Derckel et al., 1998; Waters et al., 1998; Sarry and Gunata, 2004), although actual values depend on growth conditions and pathogen challenge during berry development (Monteiro et al., 2003).
Gene expression and enzyme activity of class IV chitinases have been observed in berry mesocarp at the onset of veraison, as well as in grapevine flowers (Robinson et al., 1997). Consistent with the former observation, these EST data reveal induction of transcript for a class IV chitinase in postveraison berries; however, homologous transcripts were not observed in EST libraries constructed from developing flowers (Supplemental Table V). Class I and class III chitinases have not been reported in developing berries, except upon pathogen challenge (Coviella et al., 2002), although they are well documented in cell culture and intact leaves following treatment with chemical elicitors of systemic acquired resistance (Busam et al., 1997; Aziz et al., 2003) and in response to fungal elicitors (Busam et al., 1997). The apparent absence of class I and class III chitinases during flower and berry development is also supported by EST frequency data analysis.
Nonspecific lipid transfer proteins (nsLTPs) are small apoplastic basic Cys-rich proteins that possess in vitro phospholipid transfer activity. nsLTPs belong to the PR-14 protein family (van Loon and van Strien, 1999), and they have been implicated in disease resistance as antimicrobial proteins (Ge et al., 2002) as well as in a proposed inducible defense mechanism (Kader, 1996; Broekaert et al., 1997; Jung et al., 2003). In addition to their induction in response to pathogen attack, nsLTPs accumulate in different plant organs during development (e.g. embryogenesis and in symbiotic root nodules of legumes) and in response to environmental cues (Thomas et al., 1994; Kader, 1996; Jung et al., 2003) and biotic/abiotic stresses (Jung et al., 2003). Specifically in grapes, nsLTPs were correlated with enhanced resistance to the causal agent of grape anthracnose Elsinoe ampelina (Jayasankar et al., 2003). In addition to their proposed function in disease resistance, nsLTPs may affect enological and edible properties of grape berries as they are implicated in detrimental proteinaceous haze formation in wines (Waters et al., 1998) and as plant-derived food allergens (Mills et al., 2002; Shewry and Halford, 2002). Together, the four contigs annotated as lipid transfer proteins in Table I were observed in all grape berry libraries but in neither flower library. Despite their apparent ubiquity during berry development, the level and tissue specificity of expression varied significantly between the different nsLTP homologs. Thus, CTG1027108 exhibited the second highest expression of any differentially expressed gene in stage II libraries but had low or undetectable expression in other tissues, similar to the expression pattern reported above for endochitinase (CTG1027444). Salzman et al. (1998) determined that a 9-kD nsLTP increases in grape berries at the onset of sugar accumulation, consistent with the high expression of CTG1027108 in stage II berry libraries. Interestingly, CTG1027108 is most closely related to a castor bean (Ricinus communis) nsLTP that has been observed in both peripheral tissues and seeds throughout castor bean development (Osafune et al., 1996). Lower levels and contrasting patterns of expression were observed for CTG1027465 and CTG1028016, for which transcripts were most abundant in stage I/preveraison and stage III berry libraries.
γ-Thionins or defensins are small basic Cys-rich proteins that are rapidly evolving and exhibit antimicrobial activity against a wide range of bacterial and fungal pathogens (Zhang and Lewis, 1997) and insect pests (Shade et al., 1994). Although defensin proteins accumulate in response to pathogens and their elicitors (Epple et al., 1997; Thomma et al., 2002), they can also be developmentally regulated and are frequently observed in plant reproductive tissues (Epple et al., 1997; Aluru et al., 1999) and are abundant in the nodules of Medicago truncatula (Graham et al., 2004). CTG1027862 (Supplemental Table V) encodes a putative γ-thionin that, in Cabernet Sauvignon, is expressed exclusively in ripening (stage III) berries. By contrast, CTG1027862 is expressed in preveraison, veraison, and ripening berries of cultivar Chardonnay (NCBI EST database), suggesting distinct patterns of gene expression between the two genotypes. Defensin proteins have not been previously reported from grapes.
A putative β-1,3-glucanase protein (CTG1029513) is predicted to be differentially expressed during stage I of berry development. This contrasts with the reports of Jacobs et al. (1999) and Robinson et al. (1997) in which β-1,3-glucanase activity was low or undetectable in preveraison berries. Although it is possible that transcript levels do not correlate with enzyme activity, this discrepancy could also be explained if developmental regulation of β-1,3-glucanase expression is genotype dependent, as we suggest above in the case of defensin transcripts. Alternatively, unrecognized biotic or abiotic factors may contribute to the difference between these two studies. Enhanced transcription of β-1,3-glucanases is well documented in grape berries in response to pathogens and their elicitors (Renault et al., 1996; Jacobs et al., 1999), and β-1,3-glucanase activity was strongly induced in preveraison berries by exogenous application of the ethylene precursor ethephon (Jacobs et al., 1999).
Lipoxygenases (LOXs; E.C.1.13.11.12) are a class of iron-containing dioxygenases that catalyze the hydroperoxidation of lipids containing a cis,cis-1,4-pentadiene structure. CTG1027316, which is predicted to encode a LOX protein, is relatively highly expressed in both prebloom and bloom flowers and at low or undetectable levels in all stages of berry development. Among Arabidopsis LOX proteins, the CTG1027316 protein product is most closely related to AtLOX2, which is implicated in the production of oxylipin volatiles, including the plant hormone jasmonic acid. In addition to its induction in response to herbivory (Van Poecke et al., 2001), AtLOX2 is highly expressed in anthers of Arabidopsis (Mandaokar et al., 2003), where jasmonic acid plays an essential role in development (McConn and Browse, 1996). Based on the flower-enhanced expression of CTG1027316 and its close homology to AtLOX2, we speculate that CTG1027316 functions in grape anther development and as a potential mediator of volatile signal production in response to wounding by insects.
Stress-Induced/Cellular Redox Balance/Detoxification or Xenobiotic Transport Proteins
The most abundant group of abiotic stress-induced proteins predicted to be differentially expressed in flowers and berries were heat shock proteins (Hsp). Eight out of the 10 Hsp reported in Supplemental Table V were observed almost exclusively at the onset of veraison, with 82 of 84 occurrences (98%) in stage II green soft berries. By contrast, CTG1027432 was strongly expressed in flowers at bloom, while CTG1029041 was most strongly expressed in stage II green hard and ripening stage III berries. Most Hsp function as molecular chaperones that aid adaptation to a range of internal and external stresses (Sabehat et al., 1998; Iba, 2002; Wang et al., 2003). Although classically defined based on their induction in response to temperature stress (Sabehat et al., 1998; Iba, 2002), many Hsp are subject to developmental regulation. In tomato (Lycopersicon esculentum) and strawberry (Fragaria spp.), a plastid-localized Hsp (pTOM111) increased severalfold in ripening fruit and in response to heat stress. pTOM111 has been implicated in the reorganization of thylakoid membranes during the transition from chloroplasts to carotenoid-accumulating chromoplasts, an event that coincides with the onset of ripening (Lawrence et al., 1997). The small tomato Hsp vis1 (AAM96946) has been implicated in cell wall pectin depolymerization at the onset of tomato ripening (Ramakrishna et al., 2003). In particular, VIS1 is hypothesized to stabilize pectinases that might otherwise be denatured by elevated daytime temperatures observed under field conditions (Ramakrishna et al., 2003). The V. vinifera homolog of vis1 (CTG1028022) is up-regulated at the onset of veraison (berry softening). The observation of the up-regulation of numerous Hsp specifically at the onset of ripening is consistent with the large redirection of development and metabolism that occurs at this stage and the likely need to stabilize preexisting and newly synthesized proteins in a changing physiochemical environment.
Metallothioneins are small, Cys-rich proteins found in numerous organisms, including plants, fungi, and animals, where they are implicated in the detoxification of metal ions and reactive oxygen species as well as in the control of cellular redox potential. Conditions that promote oxidative stress are known to enhance metallothionein transcript accumulation (Navabpour et al., 2003), and the antioxidant properties of metallothioneins have been demonstrated recently (Akashi et al., 2004). Three putative metallothioneins were identified as differentially expressed based on EST frequency. The two most abundant contigs, CTG1027166 and CTG1027085, share 98% identity and may represent alleles of the same gene or close paralogs. Both genes are expressed throughout all stages of berry development, with transcripts most abundant in stage II berries and absent from the two flower libraries. Grip 24 (AJ237990), a probable allele of CTG1027166/CTG1027085 from cultivar Shiraz, encodes a ripening induced metallothionein that was shown by northern-blot analysis to be strongly up-regulated at the onset of ripening in grapes, although transcripts were also detected in earlier stages of berry development (Davies and Robinson, 2000; see also Table V). By contrast to CTG1027166/CTG1027085, the third metallothionein-encoding transcript (CTG1027501) is most highly expressed in flowers and stage I berries.
Thioredoxins constitute a group of small proteins involved in the regulation of the redox status of the cell (Gelhaye et al., 2004). They are involved in seed reserve breakdown during germination and cellular protection against oxidative stress. In addition, they act as electron donors for enzymes involved in protection against oxidative stress, such as peroxiredoxin and glutathione reductase. Thioredoxins possess a broad range of potential targets in plants, including many of the enzymes identified as differentially expressed during flower and berry development (e.g. endochitinases, nsLTPs, globulins, Hsp, aldolase, and enolase). CTG1027078 encodes a thioredoxin h homolog expressed throughout all stages of berry development and in post bloom flowers, with the highest levels of transcript observed in stage II green soft and stage III ripe berries. The fact that expression of CTG1027078 overlaps substantially with many of its potential target proteins is consistent with the possibility that CTG1027078 plays a central role in protein activation during berry development.
Plant peroxiredoxins are abundant and ubiquitous low-efficiency peroxidases (Dietz, 2003). As antioxidants, they protect against the reactive byproducts of photosynthesis and respiration, they modulate redox signaling during development, and they protect against oxidative stress (Dietz, 2003). A Vitis peroxiredoxin homolog (CTG1027597) was observed throughout all stages of berry development, with the highest occurrence in stage I berries. Peroxiredoxins are coupled to electron donors such as thioredoxin or glutaredoxin (Rouhier et al., 2004). The similarity of expression patterns between the Vitis homolog for thioredoxin h (CTG1027078) and peroxiredoxin (CTG1027597) supports the possibility that products from both transcripts are involved in similar processes. The closest homolog to the putative Vitis peroxiredoxin CTG1027597 was cloned from a poplar (Populus trichocarpa) xylem/phloem cDNA library (Rouhier, 2004). This thioredoxin- and glutaredoxin-dependent peroxidase is highly expressed in poplar plastids of the sieve tubes, where it is predicted to play a role in the regulation of the redox state of phloem cells (Rouhier, 2004).
CTG1027377 encodes a putative dehydrin. Dehydrin proteins have been reported in maturating seeds during desiccation (Nylander et al., 2001), in vegetative tissues treated with abscisic acid (Nylander et al., 2001; Parmentier-Line et al., 2002), and in plants exposed to environmental stress factors that result in cellular dehydration, such as water stress (Caruso et al., 2004) or cold stress (Caruso et al., 2004; Hara et al., 2004). In addition to seeds, dehydrins are also found among the major mesocarp proteins of ripe grapevine berries (Sarry and Gunata, 2004). The expression of CTG1027377, which is particularly elevated during stages II and III of berry development, may reflect coincident changes in water status, such as seed dehydration and osmotic stress in mesocarp tissue that results from hexose accumulation.
Polyphenol oxidases (PPO; E.C.1.14.18.1) catalyze the O2-dependent oxidation of monophenols and o-diphenols to o-quinones. These later compounds are highly reactive and mediate oxidative browning observed during plant organ senescence and in response to pathogen infection and wounding (Mayer and Harel, 1991). Preventing oxidative browning is a major concern in winemaking, particularly for white wine production (Boulton et al., 1998). CTG1026875 encodes a PPO that is predominantly expressed in prebloom flowers with low levels of transcript observed in other stages of flower and berry development. CTG1026875 is identical to Z27411 reported by Dry and Robinson (1994), a predicted chloroplast-localized PPO that is primarily expressed in immature tissues. In chloroplasts, PPO is associated with the chloroplast lumen, where it may participate in the Mehler reaction, photoreduction of oxygen by photosystem I, and metabolism of reactive oxygen species in the plastids (Thipyapong et al., 2004a, 2004b).
Transcripts Involved in Primary Metabolism
Several transcripts involved in primary metabolic pathways, such as those leading to CO2 assimilation, carbohydrate metabolism, and lipid and amino acid metabolism, are predicted to be differentially expressed. Taken together, the data described below highlight the complexity of metabolic changes taking place in distinct tissues and organs of flowers and berries at different developmental stages in grape.
Light and Dark Reaction of Photosynthesis—CO2 Assimilation
Rubisco was continually expressed throughout flower and berry development, although expression was significantly higher in floral tissues compared to berries. During early stages of development, Rubisco may be involved in light-mediated CO2 assimilation and refixation of CO2 released by respiration or other metabolic processes (Schwender et al., 2004), while after the onset of ripening, refixation of respiratory CO2 is likely to be the primary function (Blanke and Lenz, 1989; Aschan and Pfanz, 2003). Expression patterns derived from EST frequencies are consistent with the abundance of Rubisco in the berry mesocarp up to the onset of ripening, with trace levels after this point (Famiani et al., 2000). Other transcripts with predicted roles in photosynthesis are also highly expressed in flowers compared to berries. The expression patterns of CTG1027507, a putative photosystem II type I chlorophyll a/b-binding protein, is consistent with stages in flower-berry development where chlorophyll is abundant and flowers and berries fix CO2 (Blanke and Lenz, 1989). Similar patterns of expression were observed for an early light-induced-like protein (CTG1027439).
Carbohydrate Metabolism
Cytoplasmic Fru-bisphosphate aldolase (FPBA; E.C.4.1.2.13) is involved in the reversible glycolitic reaction leading to the formation of dihydroxyacetone and d-glyceraldehyde-3-P from Fru-1,6-biphosphate. d-Glyceraldehyde-3-P can be channeled to the remainder of the glycolitic pathway to generate, among other compounds, phosphoenolpyruvate, which can provide carbon skeletons to the phenylpropanoid-flavonoid pathway. The up-regulation of FBPA during stages I and III of berry development, when flavonoid biosynthesis is most active (Downey et al., 2003, 2004), is consistent with this relationship. The expression patterns of FBPA transcript deduced from EST frequency data agree well with that reported by Famiani et al. (2000), where FBPA protein was localized in the berry mesocarp using an immunohistochemical approach. In spite of the predicted high expression levels during ripening for CTG1037427, FBPA was not identified among the abundant proteins of the mesocarp of ripe berries (Sarry and Gunata, 2004) or in seeds after the onset of veraison (Famiani et al., 2000).
Enolase (E.C.4.2.1.11) participates in the conversion of the cytosolic pool of 3-phosphoglycerate to phosphoenolpryruvate (PEP). PEP can be channeled to aromatic amino acid biosynthesis via the shikimic acid pathway in the choloroplast stroma through a PEP:Pi transporter (Streatfield et al., 1999). Aromatic amino acids, in particular Phe, are the primary substrate for phenylpropanoid metabolism in the cytoplasm. The predicted up-regulation of an enolase encoding transcript during stages II and III of berry development may reflect an increased demand for PEP for different phenylpropanoids and flavonoids (e.g. proanthocyanidins and anthocyanins) that accumulate during these stages of berry development. Consistent with the expression pattern of CTG1027341, Sarry and Gunata (2004) used a proteomic approach to determine that several enolase isoforms are among the major proteins in the mesocarp of ripe grape berries.
The onset of ripening in grape berries is characterized by profound changes in the physiochemical properties of mesocarp cell walls. The most dramatic changes include the decrease in Gal/galactan content from pectins and the increased solubility of pectic polysaccharides (Nunan et al., 1998). These transformations are catalyzed by different cell wall modifying enzymes (Nunan et al., 2001; Ishimaru and Kobayashi, 2002). Among them, pectin methylesterase (PME) catalyzes the demethylesterification of homogalacturonan components of pectins in primary plant cell walls (Vorwerk et al., 2004). PME activity is higher during stages I and II of berry development and lower during ripening (Nunan et al., 2001). Despite contradictory reports (Ishimaru and Kobayashi, 2002), transcripts encoding a putative V. vinifera PME (VvPME1) were up-regulated soon after the onset of veraison (Nunan et al., 2001). In this study, VvPME1 transcript (CTG1028299) was detected in both stage II and stage III berry cDNA libraries but not classified as differentially expressed based on the R statistic (data not shown).
Inhibitor proteins often regulate the activity of enzymes involved in carbohydrate metabolism that are secreted out of the cytoplasmic compartment into the apoplast or to the vacuole (Juge et al., 2004). In addition, these proteinaceous inhibitors can also act against pathogen secreted cell-wall-degrading enzymes (Bellincampi et al., 2004). Examples of these proteinaceous inhibitors are invertase and PME inhibitors (Bellincampi et al., 2004). Plant PME and invertase inhibitors (PMEI) are members of a large family of proteins named PMEI-related proteins (PMEI-RPs; Hothorn et al., 2004). The molecular bases of target specificity and inhibitory mechanism are starting to be revealed (Hothorn et al., 2004). At the biochemical level, PMEI-RPs are likely to regulate the activity of cell wall invertases and PMEs; however, the precise biological role of PMEI-RPs during fruit ripening remains unknown. Moreover, PME activity can also be regulated by the expression of different isoforms and posttranslational mechanisms, such as changes in apoplastic pH. Consistent with a role for PMEI-RPs in PME regulation, in Actinidia chinensis fruit, the activity of PME opposes that of PMEI (Giovane et al., 1995). Interestingly, A. chinensis PMEI was only active against endogenous PME and not that secreted by fungal or bacterial pathogens. A putative invertase/PME inhibitor protein (CTG1029451) is highly represented in preveraison stage I berries but not detected during later stages of berry development. Inhibition of PME activity by PMEI may be essential to maintain high degrees of methylation in cell wall pectins, which are implicated in preformed resistance of the organ to fungal and bacterial pathogens (Le Cam et al., 1994). Inhibition of PME activity is also presumably important to prevent fruit softening during early development. Decreased PME and polygalacturonase activity induced by antisense repression was correlated with reduced pectin degradation but did not prevent fruit from softening (Tieman et al., 1992). However, fruit-specific expansins induced during ripening may also promote fruit softening independent of PME activity (Giovannoni, 2001). Interestingly, the predicted decrease in PMEI transcription contrasts with the up-regulation of a putative expansin (CTG1027667) during later stages of berry development.
Another potential target for PMEI is invertase, which functions in the degradation of apoplastic or vacuolar Suc into the monosaccharides Glc and Fru. Vacuolar invertase activity peaks around veraison, remaining at similar levels throughout stage III (Davies and Robinson, 1996; Dreier et al., 1998). The vacuolar invertase protein is found at all stages of development both in seeds and mesocarp cells (Famiani et al., 2000). In this study, two vacuolar invertase homologs (CTG1028100 and CTG1029156) were observed in developing berries but not scored as differentially expressed. Invertase leakage from vacuoles after the onset of ripening is believed to promote increased Suc flow and hexose accumulation in the berry mesocarp cells (Dreier et al., 1998). The activity of a specific invertase inhibitor (e.g. CTG1029451), operating in conjunction with cellular compartmentation, could explain the lack of invertase activity on Suc in early stages of berry development.
Two closely related contigs (CTG1027410 and CTG1026987) encode the putative transcription factor VvMSA, which is implicated in regulation of the hexose transporter VvHT1 (AF281656; Atanassova et al., 2003). CTG1027410 and CTG1026987 are highly represented in all berry libraries, in agreement with the report of Atanassova et al. (2003), with the most frequent occurrence in ripening (stage III) berries. By contrast, only low levels of expression were observed in flowers. Consistent with this pattern of expression, VvHT1 is one of the most abundant proteins in the mesocarp of ripe berries (Sarry and Gunata, 2004).
d-Myo-inositol-3-P synthase (MIPS; EC.5.5.1.4) catalyzes the first step in the synthesis of myo-inositol by converting d-Glc-6-P to d-myo-inositol-3-P (Loewus and Murthy, 2000). Different myo-inositol phosphates are involved in several aspects of cell and plant biology, acting as signal transduction molecules, osmoprotectants, cell wall constituents, mineral nutrient storage molecules, and in auxin metabolism (Loewus and Murthy, 2000; Hegeman et al., 2001). Contig CTG1027626 encodes a putative MIPS that is predicted to be differentially expressed during flower and stage I of berry development. Although the role of MIPS in flowers is not well understood, myo-inositol content is high in flowers of several species (Ichimura et al., 1999, 2000). Application of myo-inositol inhibited bloom in roses (Rosa hybrida; Ichimura et al., 1999, 2000), where it is proposed to function as an osmolite. In addition, both alkaline phytase and myo-inositol 1-P synthase occur in pollen grains (Loewus et al., 1984; Barrientos et al., 1994), and myo-inositol in particular is proposed as a nutritional substrate during pollen tube germination and growth (Kroh et al., 1970).
Lipid Metabolism
Grapevines accumulate considerable amounts of waxes after the onset of veraison (Rogiers et al., 2004). The accumulation of epicuticular waxes may enhance resistance to fungal pathogens such as B. cinerea (Comménil et al., 1997; Gabler et al., 2003) while simultaneously decreasing water loss (Rogiers et al., 2004) and mitigating the detrimental effects of solar UV radiation (Engeseth et al., 1996). Biosynthesis of waxes begins with the formation of fatty acids in the plastids and continues with fatty acid elongation to very long chains (C24-C34) that are ultimately processed into alkanes, secondary alcohols, ketones, primary alcohols, and wax esters (Engeseth et al., 1996). Very long chain fatty acid wax precursors and their derivatives are highly hydrophobic. Two protein families have been implicated in controlling the cytoplasmic pool of very long chain fatty acids: fatty acid binding proteins and acyl-CoA binding proteins. Cytosolic acyl-CoA binding proteins bind long-chain acyl-CoAs (Engeseth et al., 1996) and are believed to play a role in the intracellular transport and formation of cytoplasmic acyl-CoA pools (Knudsen et al., 1999). Contig CTG1027236, which encodes a putative acyl-CoA binding protein, is up-regulated at the onset of veraison (stage IIgs) and in ripening berries. Because this pattern of transcript accumulation correlates with stages of active wax accumulation during ripening, CTG1027236 is a candidate gene for this process.
Grape seed oil is rich in polyunsaturated fatty acids (PUFAs), accounting for up to 17% of the seed fresh weight. The major PUFA (72%–76%) is linoleic acid (ω6, C18:2; Cao and Ito, 2003). An additional 10% to 13% of PUFA content is the monounsaturated oleic acid (C18:1), with the remainder being saturated fatty acids, such as palmitic acid (C16:0, approximately 3.5% to 5%) and stearic acid (C18:0, approximately 1.5% to 2.3%). ω-6-Fatty acid desaturase catalyzes the addition of a double bond to position Δ12 of oleic acid, yielding linoleic acid (Jin et al., 2001). Contig CTG1028129, which encodes a putative ω-6-fatty acid desaturase, is first observed at the onset of veraison (stage IIgs), with the highest transcript levels attained in ripening berries (stage III). This expression pattern is in contrast to that of a grapevine seed-specific acyl-CoA carboxylase described by Walker et al. (1999).
Amino Acid Biosynthesis
Cytoplasmic Asp aminotransferase (AAT; E.C. 2.6.1.1.), also known as Glu-oxaloacetate transaminase, catalyzes the reversible interconversion of Glu into Asp by transamination of either oxaloacetate or α-ketoglutarate. AAT protein content in the mesocarp and seeds of grape berries was shown to increase significantly during stages I and II of berry development, with protein remaining unchanged throughout the remaining stages of development (Famiani et al., 2000). CTG1027149 encodes a putative AAT that is highly expressed during stages I and II of berry development but absent from later-stage cDNA libraries. Thus, there is good agreement between the increase in AAT protein (Famiani et al., 2000) and CTG1027149 transcript, while the absence of CTG1027149 transcript later in development may indicate reduced turnover rates for ATT protein in ripening berries or the presence of a paralogous gene expressed late in berry development.
Transcripts Involved in Secondary Metabolism
Grapevine berries accumulate a wide range of secondary metabolites throughout development, often in a tissue-specific manner. Among the most abundant of these natural products are various phenylpropanoids, including hydroxycinnamic acids (e.g. caffeic acid), flavonoids, and stilbenoids. In the case of flavonoids, proanthocyanidins (also known as condensed tannins), anthocyanins, and flavonols are among the most abundant and biologically important groups. Different branches of the flavonoid pathway are temporally and spatially activated throughout berry development (Boss et al., 1996). For example, while proanthocyanidins in the berry skin are synthesized preveraison and postveraison (Downey et al., 2003, 2004), anthocyanin pigment biosynthesis and accumulation occur soon after the onset of fruit ripening. This EST frequency analysis identifies several transcripts encoding structural enzymes of the general phenylpropanoid and flavonoid pathways that are differentially expressed at various stages of flower and berry development.
Chalcone-flavanone isomerase (CHI; E.C.5.5.1.6) catalyzes the reversible isomerization of naringenin chalcone to naringenin flavanone, a key step in the biosynthesis of flavonoids (Dixon and Steele, 1999). CTG1026968 encodes a putative CHI that is expressed strongly in flowers and stage I berries as well as at the onset of veraison in stage II green soft berries. This bimodal pattern of expression agrees well with that reported by Boss et al. (1996) both for anthocyanin accumulation and gene expression.
Cytochrome b5 proteins are components of electron transport systems found in animals, plants, and yeast. The cytochrome b5 homolog, DIF-F, has been implicated as an alternative electron donor for the cytochrome P450 monoxygenase flavonoid 3′,5′hydroxylase (De Vetten et al., 1999), a key structural enzyme of the flavonoid pathway. The activity of flavonoid 3′,5′hydroxylase is implicated in the formation of dihydromyricetin, a trihydroxylated dihydroflavonol, and derived anthocyanic pigments such as delphinidin, petunidin, and malvidin. In petunia (Petunia hybrida) flowers, altered expression of the cytochrome b5 DIF-F resulted in novel flower pigmentation (De Vetten et al., 1999). The EST distribution of CTG1028576, a putative cytochrome b5 grape homolog, reveals a bimodal pattern of transcript accumulation, first in stage I and later in stage II soft berries. This pattern of expression is a subset of that observed for the chalcone isomerase homolog CTG1026968. To our knowledge, neither expression nor activity levels of cytochrome b5 grape homologs have been previously reported. However, the temporal coincidence of a putative cytochrome b5 transcript with that of a potentially upstream CHI gene suggests that the biosynthesis of 3-OH flavonoids is correlated with general flavonoid metabolism in berries.
A putative class II caffeic acid O-methyltransferase (COMTII; CTG1028406) is strongly up-regulated at bloom. Little is known about the function of O-methyltransferase in V. vinifera flowers, although in flowers of Rosa chinensis, COMTII homologs have been implicated in scent biosynthesis (Wu et al., 2003). By contrast, the COMTII tobacco (Nicotiana tabacum) homolog (AF484252) is strongly up-regulated upon infection with tobacco mosaic virus (Pellegrini et al., 1993) and thus behaves as a pathogenesis-related protein (Toquin et al., 2003).
In spite of the significant activity of the phenylpropanoid and flavonoid pathway in maturing berries, no other structural genes of the pathway were predicted to be differentially expressed based on EST frequency derived from the UC Davis College of Agricultural and Environmental Sciences flower-berry libraries. Nevertheless, low levels of transcript were observed for several key enzymes (e.g. Phe ammonia lyase and chalcone synthase), especially in flower and early-stage berry libraries. In support of these observations, none of the structural genes of the flavonoid pathway were identified among the major proteins of the berry mesocarp as revealed by two-dimensional gels (Sarry and Gunata, 2004). The combination of low protein and transcript levels with high metabolite biosynthesis is consistent with models that predict a high biosynthetic efficiency for this pathway (Burbulis and Winkel-Shirley, 1999).
Berry Growth, Expansion, and Water Relations
Permeation of water across biological membranes is facilitated and regulated by a set of ubiquitous membrane channel proteins known as aquaporins (Maurel, 1997). Transcripts of two putative V. vinifera aquaporins, VvPIP1a and VvPIP1b, were shown to accumulate in berries throughout berry development and particularly after the onset of veraison (Picaud et al., 2003). During berry ripening, aquaporins are implicated in the control of sugar/acid balance. CTG1026911 is a putative aquaporin transcript that was observed most frequently in flowers at bloom in the UC Davis data set and throughout berry development in the remaining public EST data.
Two transcripts encoding putative expansins are expressed at different stages of flower and berry development. Expansin proteins are essential components of acid-induced cell wall loosening in plants (Cosgrove, 1998). No fruit-related expansins have been previously reported in grapes. CTG1027667 encodes a putative expansin expressed at bloom and to lesser extent in berries during early stages after the onset of ripening. CTG1028151 is a homolog of the cytokinin-inducible soybean β-expansin Cim1 (Downes and Crowell, 1998) and was only detected in berries after the onset of veraison. Interestingly, exogenous applications of cytokinin analogs affect fruit set and final size (NeSmith, 2002; Stern and Flaishman, 2003) in fruit crops, including grapes (Reynolds et al., 1992). Based on expression patterns and the proposed cell-wall-associated function of their translation products, both expansins may be involved in cell wall loosening during stages of exponential berry growth.
Ethylene Metabolism
Grapevine berries are considered nonclimateric fruits (Giovannoni, 2001), where ethylene is not required to trigger the cascade of physiological processes that comprise the ripening phenotype. In contrast to this more traditional view, however, ethylene appears to mediate certain key aspects of grape berry development (Chervin et al., 2004). For example, application of 1-methylcyclopropene, a strong irreversible inhibitor of ethylene receptors, immediately prior to veraison was shown to reduce berry size and anthocyanin accumulation, while increasing total acidity (Chervin et al., 2004). Similarly, exogenous applications of ethephon, a commercially used ethylene precursor, are well documented to enhance coloration and ripening in grapes (Weaver and Montgomery, 1974; Shulman et al., 1985).
The first and last steps of ethylene biosynthesis are clearly up-regulated during flower and berry development. CTG1027535 codes for S-adenosylmethionine synthethase 2 (E.C. 2.5.1.6) and was observed in flowers at bloom, as well as stage I and stage III berries (Supplemental Table V). S-adenosylmethionine synthethase 2 represents an early step of ethylene biosynthesis, catalyzing conversion of S-Met to S-adenosylmethionine. However, S-adenosylmethionine also serves as an important methyl donor in a wide range of biosynthetic pathways mediated by O-methyltransferases (Chiang et al., 1996).
The oxidation of 1-aminocyclopropane-1-carboxylic acid (ACC) to ethylene is catalyzed by ACC oxidase (E.C. 1.14.17.4). Maximum ACC oxidase transcript accumulation has been observed immediately preceding veraison in coincidence with a peak in ACC accumulation and ethylene emission (Chervin et al., 2004). However, CTG1027790, which encodes an ACC oxidase homolog, appears to be strongly and specifically expressed at bloom. Although not reported previously, the abundance of ACC oxidase transcript at this stage of flower development suggests a corresponding peak in ethylene biosynthesis. In most commercial flowers, a peak in ethylene biosynthesis precedes petal senescence. For example, in carnation (Dianthus cariophyllus), pollen-pistil interactions during pollination trigger ethylene biosynthesis and promote petal senescence (Larsen et al., 1995). Moreover, several Arabidopsis ethylene response mutants exhibit delayed senescence and petal abscision (Patterson, 2001). High expression of transcripts involved in ethylene metabolism observed in flower libraries at bloom suggests a role for ethylene at this stage in development, perhaps in the stimulation of petal abscision during cap fall or in the abscision of nonpollinated flowers (Patterson, 2001).
During fruit development, rates of polyamine and ethylene biosynthesis are normally opposed. Early stages of fruit development are associated with higher levels of polyamines (Geny et al., 1997), while later in development, polyamine levels decrease as ethylene levels increase (Mehta et al., 2002). These patterns are in agreement with experimental evidence for the inhibitory effects of polyamines on ethylene biosynthesis and vice versa (Martin-Tanguy, 2001; Mehta et al., 2002). A key enzyme in polyamine biosynthesis is Arg decarboxylase (ADC; E.C. 4.1.1.19; Martin-Tanguy, 2001). A predicted ADC (CTG1028599) was highly and uniquely expressed in prebloom flowers. This enzyme, previously cloned from V. vinifera cell suspension cultures (Primikirios and Roubelakis-Angelakis, 1999), is a key regulatory enzyme of the putrescine biosynthetic pathway. High concentrations of polyamines can be found in Arabidopsis (Applewhite et al., 2000) and grapevine flowers before bloom (Geny et al., 1997; Aziz et al., 2001). Exogenous applications of spermidine prevented plants from bolting and flowering (Applewhite et al., 2000). Similarly, putrescine applications overcome short-day photoperiod inhibition, inducing Arabidopsis plants to flower (Applewhite et al., 2000). Polyamine concentrations decrease in advance of fruitlet physiological abscission during grape berry set (Aziz et al., 2001). Moreover, the exogenous applications of an ADC inhibitor lowered putrescine concentrations and stimulated abscission of fruitlets in grapes (Aziz et al., 2001). The role of putrescine and other polyamines as inducers of flowering and promoters of fruitlet abscission and the patterns of free polyamine accumulation in grape flowers (Aziz et al., 2001) are consistent with the observed accumulation of ADC transcripts in grape flowers before bloom.
CTG1029501 encodes a putative Leu-rich repeat protein kinase that is expressed specifically in prebloom flowers, during the period when pollination is believed to occur (Heazlewood and Wilson, 2004). CTG1029501 is most closely related (89% identity and 95% similarity) to TMK1 of Arabidopsis, a receptor-like kinase of unknown function. Among the extensive family of receptor-like kinases identified in Arabidopsis (Becraft, 2002), some are expressed specifically in floral organs (Takahashi et al., 1998) where they are implicated in pistil-pollen interactions, organ abscission, cell morphogenesis and differentiation, and gamethophyte development (Jinn et al., 2000; Becraft, 2002). By analogy, CTG1029501 may be involved in processes related to pollination in grapes, including petal abscission, commonly known as cap fall, which precedes bloom and thus has a unique chronology in grapes.
Allergenic Peptides and Seed-Specific Proteins
Ten transcripts encoding allergenic peptides and seed-specific proteins are differentially expressed during different stages of berry development. Among these, five transcripts, including three homologs of a globulin-like protein, an albumin precursor, and a conglutin γ-homolog, were strongly up-regulated during stage II of berry development. Transcripts encoding homologs of almond (Prunus amygdalus) globulins (storage proteins known as prunins) are abundant in cDNA libraries spanning stages of embryogenesis, consistent with the report of Garcia-Mas et al. (1995). More generally, seed proteins are known to increase approximately 10 d prior to veraison (stage II of berry development; Famiani et al., 2000).
The hazelnut (Coryllus avellana) 11S globulin-like protein homolog has been demonstrated to be a strong food allergen (Beyer et al., 2002), and humans with allergies to hazelnuts are frequently allergic to hazelnut pollen (Beyer et al., 2002). CTG1027989, a close homolog of the hazelnut 11S globulin, was detected specifically in prebloom flower cDNA libraries. Based on analogy to the hazelnut allergen and the expression of CTG1027989 in prebloom flowers, the CTG1027989 protein is a candidate allergen in grape pollen grains. CTG1027226 also encodes a globulin-like protein, with homology to the 7S globulin as well as γ-conglutin of lupin (Lupinus albus). The deduced proteins share similarity with a xyloglucan-specific endoglucanase inhibitor, a proteinaceous inhibitor present in tomato fruit cell walls (Qin et al., 2003). The strong up-regulation of CTG1027226 transcripts during stage II of berry development highlights a possible role in developmental- and/or pathogen-mediated cell wall metabolism.
Others Proteins of Interest
CTG1026914 is predicted to be one of the most highly up-regulated genes at and after the onset of ripening. This transcript encodes for a weak (E = 1e−12) homolog of a human multiple epidermal growth factor protein. EGF-like domains (Nakayama et al., 1998) participate in extracellular events, such as receptor-ligand interactions and cell adhesion. The deduced protein of CTG1026914 is identical to GRIP15, a putative Pro-rich cell wall protein reported by Davies and Robinson (2000).
A clathrin heavy chain-related (CTG1029148) transcript is predicted to be up-regulated during the prebloom stage in flowers. Clathrin-coated vesicles present in eukaryotic cells direct the selective translocation of receptor-ligand complexes, for example, during endocytic trafficking (Blackbourn and Jackson, 1996). In other plant systems, clathrin-coated vesicles localize to the plasma membrane at the apex of the growing pollen tube, where they are implicated in membrane recycling (Blackbourn and Jackson, 1996). Pollination in grape flowers takes place before dehiscence of the petals, referred to as cap fall (Heazlewood and Wilson, 2004). Based on homology and expression data, CTG029148 may encode a clathrin protein involved in pollination of grapes flowers.
DISCUSSION
Vitis species were represented by 146,075 sequences in the NCBI databases as of September 30, 2003. Eighty-eight percent of these sequences were derived from Cabernet Sauvignon and Chardonnay cultivars, the most globally important varieties of grape, while other sequencing efforts focused on varieties of regional importance, such as Pinot Noir (Burgundy). Much of the EST sequencing in grapes has focused on cDNAs derived from berries, irrespective of plant variety, consistent with the overriding interest of viticulturists in fruit quality. Nevertheless, a range of other tissues and developmental stages were sampled to increase gene discovery. Biotic and abiotic stresses were also emphasized in the sequencing efforts, representing over 45% of the EST sequences, including cDNA libraries constructed from stressed roots, berries, and leaves. In this study, we analyzed this complex data set of the Vitis transcriptome to gain insight into gene structure and content as well as to provide a preliminary assessment of gene expression in this important crop plant.
cDNA libraries were organized according to an accepted controlled vocabulary, allowing placement into distinct developmental windows and facilitating rational in silico analysis of gene expression. Of particular interest are the preveraison libraries, due to their heterogeneous distribution in the two-dimensional cluster analysis shown in Figure 3. In fact, the preveraison stage of grape berry development encompasses several distinct phases of development. Three of the preveraison cDNA libraries for cv Cabernet Sauvignon were selected to represent a chronological and developmental series including stage I, stage II green hard, and stage II green soft berries. Additional complexity for preveraison berries was provided by the contribution of preveraison berries without seed. The widely differing position of these cDNA libraries within the hierarchical cluster suggests that preveraison is characterized by large shifts in gene expression.
The unigene set for V. vinifera includes 13,307 contigs and 12,389 singletons. Significant support for the structure of a majority of the contigs was provided by analysis of paired-end sequences from single cDNA clones. Thus, 9,599 of the tentative consensus sequences can be validated as correct contig structures, including numerous predicted full-length or near-full-length clones, significantly increasing the value of the unigene data set. Although it is likely that additional sequencing would serve to condense some singletons into contigs, thus reducing the size of the unigene set, we suggest that this analysis identifies at least a portion of the majority of genes in the grape genome. Using Arabidopsis and rice (Oryza sativa) as benchmarks, where approximately 25,000 and approximately 38,000 genes are predicted from their high-quality draft genome sequences, respectively (Arabidopsis Genome Initiative, 2000; International Rice Genome Sequencing Project, 2005), this analysis is estimated to survey upwards of 67% of the grape transcriptome.
A feature of fully sequenced genomes of several eukaryotic species is the frequent occurrence of genes in gene families. Thus, 37% and 24% of the Arabidopsis and Caenorhabditis elegans genes, respectively, are members of gene families with at least five paralogs (Arabidopsis Genome Initiative, 2000). This level of duplication is likely to reflect complexity in genome function beyond that contributed by single copy genes, for example, due to subfunctionalization for gene expression (Adams et al., 2003) or protein function (Wagner, 2002) or by providing redundant function (e.g. Kirik et al., 2004; Ohgishi et al., 2004). In this analysis, we surveyed several well-characterized metabolic pathways to determine the extent to which such pathways were represented within the grape unigene set. In addition to the finding that most enzymatic steps are represented by grape homologs, we were also able to provide a preliminary estimate of gene duplication based on the occurrence of paralogous sequences. Excluding enzymatic steps where no homologs were identified, 31% of the reference enzymes are represented by a single V. vinifera homolog, whereas 21% are members of gene families with at least five paralogs. This is a conservative estimate of gene duplication in V. vinifera because singletons were not considered in this analysis and because deeper sequencing would likely discover additional paralogs. Defining such relationships and understanding functional diversification of paralogs will be an important task for grape biotechnologists interested in genomics-assisted crop improvement.
In total, homologous proteins were identified for approximately 70% of the Vitis unigene set, although the large fraction of unigenes with homology to imprecisely annotated proteins (e.g. expressed or hypothetical protein) reflects an extremely limited extent of functional characterization. Annotations derived from the definition lines of NCBI files are not standardized; moreover, they are frequently incorrect, and their use can propagate transitive errors. To partially mitigate this problem, we used the Gene Ontology assignments available at the TIGR Vitis vinifera Gene Index (http://www.tigr.org/tdg/tgi/vvgi) to develop more detailed annotations for approximately 8% of the unigenes. As examples of the resulting annotation, several unigenes annotated as MADS box genes (e.g. CTG1028624 and CTG1030218) were identified within the transcription regulator activity branch of biological process. Certain MADS box genes are regulators of flower development in Arabidopsis (e.g. Honma and Goto, 2001) and fruit development in tomato (Vrebalov et al., 2002). In silico expression analysis indicates that CTG1028624 is highly specific to berry development, whereas CTG1030218 is highly specific to flower development. These unigenes are paralogous to the berry-ripening-related MADS box genes reported by Boss et al. (2002). Thus, CTG1028624 and CTG1030218 would appear to represent previously undiscovered regulators of grape berry and flower development.
One of the main advantages of deep sequencing of non-normalized cDNA libraries is that it provides the basis for in silico analysis of gene expression based on the frequency of ESTs across libraries (for review, see Ohlrogge and Benning, 2000). Here, we used a combination of methodologies to identify and validate differentially expressed genes in grape. In particular, we used the R statistic (Stekel et al., 2000) and hierarchical clustering to identify and organize contigs composed of ESTs with a nonrandom distribution, similar to the approaches used in potato (Ronning et al., 2003) and M. truncatula (Journet et al., 2002). In total, 665 differentially expressed genes (TCs) revealed gene expression patterns reflective of plant developmental stage and environmental stimuli (Fig. 3). Not surprisingly, given the major physical and biochemical changes that define the ripening process in grape berries (Davies and Robinson, 2000), veraison and postveraison berry libraries formed a uniform group, while preveraison libraries (stage I and stage II) occupied heterogenous positions within the hierarchical cluster. Interestingly, berries at stage I of development (green tissue) grouped with other non-leaf green tissue libraries, such as stems, petioles, and compound buds (Fig. 3A). We were able to validate our clustering data based on comparison to patterns of gene expression described in the primary literature. Thus, genes for several GRIPs characterized by Davies and Robinson (2000) were also identified in this in silico analysis. Moreover, based on similar positions within the hierarchical cluster, we identified coregulated, but previously uncharacterized, genes; the fact that these genes had annotations similar to those of GRIP genes (i.e. stress- and cell-wall-related proteins) suggests a system of coregulated genes with related functions.
Despite the utility of the in silico approach for identifying candidate genes, the power of tag-based approaches, which include ESTs, Serial Analysis of Gene Expression, and Massively Parallel Signature Sequencing, is limited by the cost of conducting biological replicates to statistically validate results (Meyers et al., 2004). To circumvent this difficulty, we compared EST frequencies derived from a UC Davis flower-berry EST data set with a similar but independent data set compiled from the NCBI EST database. Multivariate statistical analyses suggest that EST frequencies in the two data sets are highly correlated, further validating the in-silico-derived expression patterns. By combining gene expression with knowledge of enzyme activities and biological processes taking place during berry development, we developed a compendium of gene expression during grape berry development.
In conclusion, the sequencing efforts of several research groups worldwide have placed grapes (particularly V. vinifera) among the best-characterized plant species with respect to ESTs. We demonstrated that this data can be utilized not only for gene discovery but also for comparative analysis of gene expression, both within grape tissues and to homologous genes in other plant species. These sequences have enabled the production of a first-generation public Affymetrix microarray for grapes, composed of approximately 14,000 distinct probe sets for V. vinifera and an additional approximately 1,700 probe sets for other Vitis species and hybrids, capable of surveying an estimated 30% to 50% of the grape transcriptome. Microarray mRNA expression profiling in grape should further contribute in important and novel ways to understanding fundamental aspects of grape biology, such as transcriptional responses correlated with biotic and abiotic stress, gene expression during fruit development, and transcriptional responses to particular viticultural practices. The outcomes of such studies will likely impact grape improvement in several ways: for example, the results may yield marker genes (e.g. genes correlated with particular physiological stresses) that can be used to establish diagnostic assays in the field, or they may identify candidate genes that contribute to the agronomic properties of grape, including disease resistance and fruit quality.
MATERIALS AND METHODS
cDNA Library Preparation
Because most of the grape EST sequencing efforts (>80% of available ESTs) described here was performed by the University of California and University of Nevada research groups, in this section, we only describe protocols for the libraries generated by those groups. Additional information for remaining libraries can be obtained at http://www.ncbi.nlm.nih.gov/UniGene/UGOrg.cgi?TAXID=29760.
University of Nevada cDNA Library Construction and Sequencing
Total RNA was extracted from abiotically stressed Vitis vinifera cv Chardonnay leaf, berry, and root tissue using a modified Tris-LiCl protocol (Tattersall et al., 2005). Homogenization buffer contained 200 mm Tris-HCl, pH 8.5, 1.5% (w/v) lithium dodecylsulfate, 300 mm LiCl, 10 mm sodium EDTA, 1% (w/v) sodium deoxycholate, and 1% (v/v) Nonidet P-40. Following autoclaving, 2 mm aurintricarboxylic acid, 20 mm dithiothreitol, 10 mm thiourea, and 2% (w/v) polyvinylpolypyrrolidone were added immediately before use. Following precipitation with sodium acetate and isopropanol precipitation, samples were extracted 1× with 25:24:1 phenol:chloroform:isoamyl and 1× with 24:1 chloroform:isoamyl prior to performing LiCl precipitations to remove DNA contamination. Poly(A)+ RNA was purified from 500 mg of total RNA using the Micro-FastTrak 2.0 mRNA isolation kit (Invitrogen) according to the manufacturer's instructions. cDNA was synthesized from 1 to 5 μg of poly(A)+ RNA using a Lambda Uni-Zap-XR cDNA synthesis kit according to the manufacturer's recommended protocol (Stratagene). The directionally cloned (EcoRI/XhoI) cDNA libraries generated were then mass-excised in vivo and the resulting plasmids (pBluescript II) propagated in the Escherichia coli SOLR host strain. Individual cDNA clones containing inserts were amplified using the TempliPhi amplification kit (Amersham Biosciences) and sequenced using the dideoxy chain-termination method on an Applied Biosystems 3700 automated DNA sequencing system using the Prism Ready Reaction Dyedeoxy Terminator Cycle Sequencing kit (Perkin-Elmer Applied Biosystems). The T3 primer (5′- GGGAAATCACTCCCAATTAA-3′) and the T7 primer (5′-GTAATACGACTCACTATAGGGC-3′) were used for 5′ reads and 3′ reads of cDNA clones, respectively. Oligo(dT) primer (T22M) was used for 3′ sequencing reads of cDNA clones containing poly(A) tails.
University of California cDNA Library Construction and Sequencing
Total RNA was extracted from tissues (compound bud, flower, leaf, stem, petiole, and berry) of V. vinifera cv Cabernet Sauvignon and leaves of Vitis sp. as described by Iandolino et al. (2004). Total RNA (1.5–3.5 μg) was used as starting material for cDNA library construction. cDNA was prepared using the cDNA synthesis kit from CLONTECH (Creator Smart cDNA library construction kit) according to the manufacturer's protocol. Only libraries having average insert sizes above 0.9 kb were further selected for sequencing at the UC Davis College of Agricultural and Environmental Sciences Genomics Facility (http://cgf.ucdavis.edu). Templates for DNA sequencing were prepared by rolling circle amplification using bacteriophage Phi29 DNA polymerase (TempliPhi amplification kits; Amersham Biosciences). Amplified DNA was used directly for cycle sequencing without purification. Sequencing reactions were performed with 2 μL of ABI PRISM BigDye Terminators v3.0 (Applied Biosystems) and 5 pmol of primer in 10 μL reaction volume, followed by sequencing reaction clean up to remove residual dye and enzyme. Sequencing primers were designed to flank the cDNA insertion site by 28 and 13 bp for the 5′ and 3′ ends, respectively. The primer for 5′ end sequencing was 5′-GTTATCAGTCGACGGTACC-3′ and the primer for 3′ end sequencing was 5′-GCCAAACGAATGGTCTAG-3′. Sequencing products were separated and analyzed with a 96-capillary sequencer (ABI 3730xl DNA analyzer; Applied Biosystems).
Data Sets Used, Clustering Analysis, and Annotation
All available Vitis sequences (including ESTs, expressed transcripts, and other DNA sequences in the NCBI database) were extracted from GenBank with Batch Entrez at NCBI (http://www.ncbi.nlm.nih.gov/). The sequences were cleaned by removing vectors, E. coli, mitochondria, chloroplast, and low quality sequences. The remaining sequences were clustered into contigs with the TIGR Gene Indices Clustering Tools (http://www.tigr.org/tdb/tgi/software/). Only sequences from species with over 250 entries at the NCBI EST and nonredundant databases were subject to clustering, i.e. V. vinifera, a V. rupestris × V. arizonica hybrid, and V. aestivalis. Sequences not clustering into contigs, as well as sequences from species with <250 entries at NCBI, were treated as singletons. For preliminary annotation of the unigene sets, sequences were compared to the NCBI nonredundant protein database using BLASTX (Altschul et al., 1997); the annotation of the top BLASTX hit was adopted and used to create a keyword list. The resulting information is available online at http://cgf.ucdavis.edu/. For functional annotation using Gene Ontology and Kyoto Encyclopedia of Genes and Genomes, we used the assignments described in the TIGR grape gene index (http://www.tigr.org/tdb/tgi/vvgi/) version 3.1 (November, 2003).
Digital Analyses of Gene Expression in V. vinifera
To characterize the functional gene space in Vitis, we searched the data sets to identify TCs and singletons homologous to genes coding for structural enzymes involved in primary, intermediate, and secondary metabolic pathways based on a BLASTX hit of at least E = 1e−5 (median of E = 1.26e−78). For purposes of in silico analysis of gene expression, cDNAs analyzed from both 5′ and 3′ ends were counted only once, and ESTs from normalized cDNA libraries were not considered. The remaining 13,311 TCs were organized into a matrix representing the frequency of ESTs across 58 cDNA libraries. Hierarchical clustering of differentially expressed TCs was performed using the Unweighted Pair Group Method with Arithmetic Mean algorithm based on Euclidean distances calculated from correlation coefficients as described by Ewing et al. (1999). R statistic and hierarchical clustering analyses were implemented in Matlab 6.5 (The MathWorks).
Sequence data from this article can be obtained from the NCBI GenBank EST database as all records for Vitis species submitted prior to September 30, 2003. All contigs (e.g. CTG1027266) mentioned in the text or referred to in tables and figures are referenced to representative EST sequence accession numbers in Supplemental Table IV.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge the contributions of John Quackenbush, Sirisha Sunkara, and the rest of the TIGR Gene Index bioinformatics team (Rockville, MD).
This work was supported in part by grants from the U.S. Department of Agriculture Agricultural Research Service (SCA 58–5302–2–788) and the California Department of Food and Agriculture (contract 02–0150; to D.R.C.) and from the National Science Foundation Plant Genome Program (DBI–58–5302–2–788), the American Vineyard Foundation, and the Nevada Agricultural Experiment Station (to G.R.C. and J.C.C.). This work is published as publication number 03055535 of the University of Nevada Agricultural Experiment Station.
The online version of this article contains Web-only data.
References
- Ablett E, Seaton G, Scott K, Shelton D, Graham MW, Baverstock P, Lee LS, Henry R (2000) Analysis of grape ESTs: global gene expression patterns in leaf and berry. Plant Sci 159: 87–95 [DOI] [PubMed] [Google Scholar]
- Adams KL, Cronn R, Percifield R, Wendel JF (2003) Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proc Natl Acad Sci USA 100: 4649–4654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi K, Nishimura N, Ishida Y, Yokota A (2004) Potent hydroxy radical-scavenging activity of drought-induced type-2 metallothionein in wild watermelon. Biochem Biophys Res Commun 323: 72–78 [DOI] [PubMed] [Google Scholar]
- Aldini G, Carini M, Piccoli A, Rossoni G, Facino RM (2003) Procyanidins from grape seeds protect endothelial cells from peroxynitrite damage and enhance endothelium-dependent relaxation in human artery: new evidences for cardio-protection. Life Sci 73: 2883–2898 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aluru M, Curry J, O'Connell MA (1999) Nucleotide sequence of a defensin or gamma-thionin-like gene from habanero Chile. Plant Physiol 120: 63310447467 [Google Scholar]
- Applewhite PB, Kaur-Sawhney R, Galston AW (2000) A role for spermidine in the bolting and flowering of Arabidopsis. Physiol Plant 108: 314–320 [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 48: 796–815 [DOI] [PubMed] [Google Scholar]
- Aschan G, Pfanz H (2003) Non-foliar photosynthesis—a strategy of additional carbon acquisition. Flora 198: 81–97 [Google Scholar]
- Atanassova R, Leterrier M, Gaillard C, Agasse A, Sagot E, Coutos-Thévenot P, Delrot S (2003) Sugar-regulated expression of a putative hexose transport gene in grape. Plant Physiol 131: 326–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz A, Brun O, Audran J-C (2001) Involvement of polyamines in the control of fruitlet physiological abscission in grapevine (Vitis vinifera). Physiol Plant 113: 50–58 [Google Scholar]
- Aziz A, Poinssot B, Daire X, Adrian M, Bezier A, Lambert B, Joubert JM, Pugin A (2003) Laminarin elicits defense responses in grapevine and induces protection against Botrytis cinerea and Plasmopara viticola. Mol Plant Microbe Interact 16: 1118–1128 [DOI] [PubMed] [Google Scholar]
- Bagchi D, Sen CK, Ray SD, Das DK, Bagchi M, Preuss HG, Vinson JA (2003) Molecular mechanisms of cardioprotection by a novel grape seed proanthocyanidin extract. Mutat Res 523-524: 87–97 [DOI] [PubMed] [Google Scholar]
- Barrientos L, Scott JJ, Murphy PPN (1994) Specificity of hydrolysis of phytic acid by alkaline phytase from lily pollen plant. Plant Physiol 106: 1489–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becraft PW (2002) Receptor kinase signaling in plany development. Annu Rev Cell Dev Biol 18: 163–192 [DOI] [PubMed] [Google Scholar]
- Bellincampi D, Camardella L, Delcour JA, Desseaux V, D'Ovidio R, Durand A, Elliot G, Gebruers K, Giovane A, Juge N, et al. (2004) Potential physiological role of plant glyosidase inhibitors. Biochim Biophys Acta 1696: 265–274 [DOI] [PubMed] [Google Scholar]
- Beyer K, Grishina G, Bardina L, Grishin A, Sampson HA (2002) Identification of an 11S globulin as a major hazelnut food allergen in hazelnut-induced systemic reactions. J Allergy Clin Immunol 110: 517–523 [DOI] [PubMed] [Google Scholar]
- Bisson LF, Waterhouse AL, Ebeler SE, Walker MA, Lapsley JT (2002) The present and future of the international wine industry. Nature 418: 696–699 [DOI] [PubMed] [Google Scholar]
- Blackbourn HD, Jackson AP (1996) Plant clathrin heavy chain: sequence analysis and restricted localisation in growing pollen tubes. J Cell Sci 109: 777–787 [DOI] [PubMed] [Google Scholar]
- Blanke MM, Lenz F (1989) Fruit photosynthesis. Plant Cell Environ 12: 31–46 [Google Scholar]
- Boss PK, Davies C, Robinson SP (1996) Analysis of the expression of anthocyanin pathway genes in developing Vitis vinifera L. cv Shiraz grape berries and the implications for pathway regulation. Plant Physiol 111: 1059–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boss PK, Sensi E, Hua C, Davies C, Thomas MR (2002) Cloning and characterization of grapevine (Vitis vinifera L.) MADS-box genes expressed during inflorescence and berry development. Plant Sci 162: 887–895 [Google Scholar]
- Boulton RB, Singleton VL, Bisson LF, Kunkee RE (1998) Principles and Practices of Winemaking. Aspen Publishers, New York
- Broekaert WF, Cammue BPA, DeBolle MFC, Thevissen K, DeSamblanx GW, Osborn RW (1997) Antimicrobial peptides from plants. CRC Crit Rev Plant Sci 16: 297–323 [Google Scholar]
- Burbulis IE, Winkel-Shirley B (1999) Interactions among enzymes of the Arabidopsis flavonoid biosynthetic pathway. Proc Natl Acad Sci USA 96: 12929–12934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busam G, Kassemeyer HH, Matern U (1997) Differential expression of chitinases in Vitis vinifera L. responding to systemic acquired resistance activators or fungal challenge. Plant Physiol 115: 1029–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao XL, Ito YC (2003) Supercritical fluid extraction of grape seed oil and subsequent preparation of free fatty acids by high-speed counter-current chromatography. J Chromatogr A 1021: 117–124 [DOI] [PubMed] [Google Scholar]
- Caruso A, Morabito D, Delmotte F, Kahlem G, Carpin S (2004) Dehydrin induction during drought and osmotic stress in Populus. Plant Physiol Biochem 40: 1033–1042 [Google Scholar]
- Chervin C, El-Kereamy A, Roustan J-P, Latché A, Lamon J, Bouzayen M (2004) Ethylene seems required for the berry development and ripening in grape, a non-climateric fruit. Plant Sci 167: 1301–1305 [Google Scholar]
- Chiang PK, Gordon RK, Tal J, Zeng GC, Doctor BP, Pardhasaradhi K, McCann PP (1996) S-adenosylmethionine and methylation. FASEB J 10: 471–480 [PubMed] [Google Scholar]
- Comménil P, Brunet L, Audran J-C (1997) The development of the grape berry culticle in relation to susceptibility to bunch rot disease. J Exp Bot 48: 1599–1607 [Google Scholar]
- Coombe BG (1987) Distribution of solutes within the developing grape berry in relation to its morphology. Am J Enol Vitic 38: 120–127 [Google Scholar]
- Cosgrove DJ (1998) Cell wall loosening by expansins. Plant Physiol 118: 333–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coviella CE, Stipanovic RD, Trumble JT (2002) Plant allocation to defensive compounds: interactions between elevated CO2 and nitrogen in transgenic cotton plants. J Exp Bot 53: 323–331 [DOI] [PubMed] [Google Scholar]
- Davies C, Robinson SP (1996) Sugar accumulation in grape berries. Cloning two putative vacuolar invertase cDNAs and their expression in grapevine tissues. Plant Physiol 111: 275–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies C, Robinson SP (2000) Differential screening indicates dramatic change in mRNA profiles during grape berry ripening. Cloning and characterization of cDNAs encoding putative cell wall and stress response proteins. Plant Physiol 122: 803–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derckel J-P, Audran J-C, Haye B, Lambert B, Legendre L (1998) Characterization, induction by wounding and salicylic acid, and activity against Botrytis cinerea of chitinases and β-1,3-glucanases of ripening grape berries. Physiol Plant 104: 56–64 [Google Scholar]
- De Vetten N, Ter Host J, Van Schaik H-P, De Boer A, Mol J, Koes R (1999) A cytochrome b5 is required for full activity of flavonoid 3′,5′-hydroxylase, a cytochrome P450 involved in the formation of blue flower colors. Proc Natl Acad Sci USA 96: 778–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz K-J (2003) Plant peroxiredoxins. Annu Rev Plant Biol 54: 94–107 [DOI] [PubMed] [Google Scholar]
- Dixon RA, Steele CL (1999) Flavonoids and isoflavonoids: a goldmine for metabolic engineering. Trends Plant Sci 4: 394–400 [DOI] [PubMed] [Google Scholar]
- Downes BP, Crowell DN (1998) Cytokinin regulates the expression of a soybean β-expansin gene by a post-transcriptional mechanism. Plant Mol Biol 37: 437–444 [DOI] [PubMed] [Google Scholar]
- Downey MO, Harvey JS, Robinson SP (2003) Analysis of tannins in seeds and skins of Shiraz grapes throughout berry development. Aust J Grape Wine Res 9: 15–27 [Google Scholar]
- Downey MO, Harvey JS, Robinson SP (2004) The effect of bunch shading on berry development and flavonoid accumulation in Shiraz grapes. Aust J Grape Wine Res 10: 55–73 [Google Scholar]
- Dreier LP, Hunter JJ, Ruffner HP (1998) Invertase activity, grape berry development and cell compartmentation. Plant Physiol Biochem 36: 865–872 [Google Scholar]
- Dry IB, Robinson SP (1994) Molecular cloning and characterisation of grape berry polyphenol oxidase. Plant Mol Biol 26: 495–502 [DOI] [PubMed] [Google Scholar]
- Engeseth NJ, Pacovsky RS, Newman T, Ohlrogge JB (1996) Characterization of an acyl-CoA-binding protein from Arabidopsis thaliana. Arch Biochem Biophys 331: 55–62 [DOI] [PubMed] [Google Scholar]
- Epple P, Apel K, Bohlmann H (1997) ESTs reveal a multigene family for plant defensins in Arabidopsis thaliana. FEBS Lett 400: 168–172 [DOI] [PubMed] [Google Scholar]
- Ewing RM, Ben Kahla A, Poirot O, Lopes F, Audic S, Claverie JM (1999) Large-scale statistical analysis of rice ESTs reveal correlated patterns of gene expression. Genome Res 9: 950–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Famiani F, Walker RP, Tecsi L, Chen Z-H, Proietti P, Leegood RC (2000) An immunohistochemical study of the compartmentation of metabolism during the development of grape (Vitis vinifera L.) berries. J Exp Bot 51: 675–683 [PubMed] [Google Scholar]
- Ferguson LR (2001) Role of plant polyphenols in genomic stability. Mutat Res 475: 89–111 [DOI] [PubMed] [Google Scholar]
- Gabler FM, Smilanick JL, Mansour M, Ramming DW, Mackey BE (2003) Correlations of morphological, anatomical, and chemical features of grape berries with resistance to Botrytis cinerea. Phytopathology 93: 1263–1273 [DOI] [PubMed] [Google Scholar]
- Garcia-Mas J, Messeguer R, Arus P, Puigdomenech P (1995) Molecular characterization of cDNAs corresponding to genes expressed during almond (Prunus amygdalus Batsch) seed development. Plant Mol Biol 27: 205–210 [DOI] [PubMed] [Google Scholar]
- Ge XC, Chen JC, Lin Y, Sun CR, Cao KM (2002) Expression, purification and function of rice nonspecific lipid transfer protein. Acta Biochim Biophys Sin 34: 83–87 [PubMed] [Google Scholar]
- Gelhaye E, Rouhier N, Jacquot JP (2004) The thioredoxin h system in higher plants. Plant Physiol Biochem 42: 265–271 [DOI] [PubMed] [Google Scholar]
- Gene Ontology Consortium (2000) Gene Ontology: tool for the unification of biology. Nat Genet 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geny L, Broquedis M, MartinTanguy J, Bouard J (1997) Free, conjugated, and wall-bound polyamines in various organs of fruiting cuttings of Vitis vinifera L cv Cabernet Sauvignon . Am J Enol Vitic 48: 80–84 [Google Scholar]
- Giovane A, Balestrieri C, Quagliuolo L, Castaldo D, Servillo L (1995) A glycoprotein inhibitor of pectin methylesterase in kiwi fruti. Purification by affinity chromatography and evidence of a ripening-related precursor. Eur J Biochem 233: 926–929 [DOI] [PubMed] [Google Scholar]
- Giovannoni J (2001) Molecular biology of fruit maturation and ripening. Annu Rev Plant Physiol Plant Mol Biol 52: 725–749 [DOI] [PubMed] [Google Scholar]
- Graham MA, Silverstein KAT, Cannon SB, VandenBosch KA (2004) Computational identification and characterization of novel genes from legumes. Plant Physiol 135: 1179–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara M, Fujinaga M, Kuboi T (2004) Radical scavenging activity and oxidative modification of citrus dehydrin. Plant Physiol Biochem 42: 657–662 [DOI] [PubMed] [Google Scholar]
- Hardie WJ, O'Brien TP, Jaudzems VG (1996) Morphology, anatomy and development of the pericarp after anthesis in grape, Vitis vinifera L. Aust J Grape Wine Res 2: 97–142 [Google Scholar]
- Heazlewood JE, Wilson S (2004) Anthesis, pollination and fruitset in Pinot Noir. Vitis 43: 65–68 [Google Scholar]
- Hegeman CE, Good LL, Grabau EA (2001) Expression of D-myo-inositol-3-phosphate synthase in soybean. Implications for phytic acid biosynthesis. Plant Physiol 125: 1941–1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honma T, Goto K (2001) Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409: 525–529 [DOI] [PubMed] [Google Scholar]
- Hothorn M, Wolf S, Aloy P, Greiner S, Scheffzek K (2004) Structural insights into the target specificity of plant invertase and pectin methylesterase inhibitory proteins. Plant Cell 16: 3437–3447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, Zipkin RE, Chung P, Kisielewski A, Zhang LL, Scherer B, Sinclair DA (2003) Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature 425: 191–196 [DOI] [PubMed] [Google Scholar]
- Iandolino AB, da Silva FG, Lim H, Choi H, Williams LE, Cook DR (2004) High-quality RNA, cDNA, and derived EST libraries from grapevine (Vitis vinifera L.). Plant Mol Biol Rep 22: 269–278 [Google Scholar]
- Iba K (2002) Acclimative response to temperature stress in higher plants: approaches of gene engineering for temperature tolerance. Annu Rev Plant Biol 53: 225–245 [DOI] [PubMed] [Google Scholar]
- Ichimura K, Kohata K, Goto R (2000) Soluble carbohydrates in delphinium and their influence on sepal abscission in cut flowers. Physiol Plant 108: 307–313 [Google Scholar]
- Ichimura K, Mukasa Y, Fujiwara T, Katsunori K, Goto R, Suto K (1999) Possible roles of methyl glucoside and myo-inositol in the opening of cut rose flowers. Ann Bot (Lond) 83: 551–557 [Google Scholar]
- International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436: 793–800 [DOI] [PubMed] [Google Scholar]
- Ishimaru M, Kobayashi S (2002) Expression of a xyloglucan endo-transglycosylase gene is closely related to grape berry softening. Plant Sci 162: 621–628 [Google Scholar]
- Jacobs AK, Dry IB, Robinson SP (1999) Induction of different pathogenesis-related cDNAs in grapevine infected with powdery mildew and treated with ethephon. Plant Pathol 48: 325–336 [Google Scholar]
- Jayasankar S, Li ZJ, Gray DJ (2003) Constitutive expression of Vitis vinifera thaumatin-like protein after in vitro selection and its role in anthracnose resistance. Funct Plant Biol 30: 1105–1115 [DOI] [PubMed] [Google Scholar]
- Jin UH, Lee JW, Chung YS, Lee JH, Yi YB, Kim YK, Hyung NI, Pyee JH, Chung C-H (2001) Characterization and temporal expression of a ω-6 fatty acid desaturase cDNA from sesame (Sesamum indicum L.) seeds. Plant Sci 161: 935–941 [Google Scholar]
- Jinn T, Stone J, Walker J (2000) HAESA, an Arabidopsis leucine-rich repeat receptor kinase, controls floral organ abscission. Genes Dev 11: 108–117 [PMC free article] [PubMed] [Google Scholar]
- Journet EP, van Tuine D, Gouzy J, Crespeau H, Carreau V, Farmer MJ, Niebel A, Schiex T, Jaillon O, Chatagnier O, et al (2002) Exploring root symbiotic programs in the model legume Medicago truncatula. Nucleic Acids Res 30: 5579–5592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juge N, Svensson B, Henrissat B, Williamson G (2004) Plant proteinaceous inhibitors of carbohydrate-active enzymes. Biochim Biophys Acta 1696: 141 [Google Scholar]
- Jung HW, Kim W, Hwang BK (2003) Three pathogen-inducible genes encoding lipid transfer protein from pepper are differentially activated by pathogens, abiotic, and environmental stresses. Plant Cell Environ 26: 915–928 [DOI] [PubMed] [Google Scholar]
- Kader JC (1996) Lipid-transfer proteins in plants. Annu Rev Plant Physiol Plant Mol Biol 47: 627–654 [DOI] [PubMed] [Google Scholar]
- Kirik V, Simon M, Huelskamp M, Schiefelbein J (2004) The ENHANCER OF TRY AND CPC1 gene acts redundantly with TRIPTYCHON and CAPRICE in trichome and root hair cell patterning in Arabidopsis. Dev Biol 268: 506–513 [DOI] [PubMed] [Google Scholar]
- Knudsen J, Jensen MV, Hansen JK, Faergeman NJ, Neergaard TB, Gaigg B (1999) Role of acylCoA binding protein in acylCoA transport, metabolism and cell signaling. Mol Cell Biochem 192: 95–103 [DOI] [PubMed] [Google Scholar]
- Kroh M, Miki-Hirosige H, Rosen W, Loewus F (1970) Incorporation of label into pollen tube walls from myoinositol-labeled Lilium longiflorum pistils. Plant Physiol 45: 92–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen PB, Ashworth EN, Jones ML, Woodson WR (1995) Pollination-induced ethylene in carnation (role of pollen tube growth and sexual compatibility). Plant Physiol 108: 1405–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence SD, Cline K, Moore GA (1997) Chromoplast development in ripening tomato fruit: identification of cDNAs for chromoplast-targeted proteins and characterization of a cDNA encoding a plastid-localized low-molecular-weight heat shock protein. Plant Mol Biol 33: 483–492 [DOI] [PubMed] [Google Scholar]
- Le Cam B, Massiot P, Rouxel F (1994) Cell wall polysaccharide-degrading enzymes produced by isolates of Mycocentrospora acerina differing in aggressiveness on carrot. Physiol Mol Plant Pathol 44: 187–198 [Google Scholar]
- Liang F, Holt I, Pertea G, Karamycheva S, Salzberg AL, Quackenbush J (2000) An optimized protocol for analysis of EST sequences. Nucleic Acids Res 28: 3657–3665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewus FA, Murthy PPN (2000) Myo-inositol metabolism in plants. Plant Sci 150: 1–19 [Google Scholar]
- Loewus MW, Bedgar DL, Loewus FA (1984) 1L-Myo-inositol 1-phosphate synthase from pollen of Lillium longiflorum—an ordered sequential mechanism. J Biol Chem 259: 7644–7647 [PubMed] [Google Scholar]
- Mandaokar A, Kumar VD, Amway M, Browse J (2003) Microarray and differential display identify genes involved in jasmonate-dependent anther development. Plant Mol Biol 52: 775–786 [DOI] [PubMed] [Google Scholar]
- Martin-Tanguy J (2001) Metabolism and function of polyamines in plants: recent development (new approaches). Plant Growth Regul 34: 135–148 [Google Scholar]
- Maurel C (1997) Aquaporins and water permeability of plant membranes. Annu Rev Plant Physiol Plant Mol Biol 48: 399–429 [DOI] [PubMed] [Google Scholar]
- Mayer AM, Harel E (1991) Phenoloxidases and their significance in fruit and vegetables. In PF Fox, ed, Food Enzymology. Elsevier, London, pp 373–398
- McConn M, Browse J (1996) The critical requirement for linolenic acid is pollen development, not photosynthesis, in an Arabidopsis mutant. Plant Cell 8: 403–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta RA, Cassol T, Li N, Ali N, Handa AK, Mattoo AK (2002) Engineered polyamine accumulation in tomato enhances phytonutrient content, juice quality, and vine life. Nat Biotechnol 20: 613–618 [DOI] [PubMed] [Google Scholar]
- Meyers BC, Vu TH, Shivakundan ST, Ghazal H, Matvienko M, Agrawal V, Ning J, Haudenschild CD (2004) Analysis of the transcriptional complexity of Arabidopsis thaliana by massively parallel signature sequencing. Nat Biotechnol 22: 1006–1011 [DOI] [PubMed] [Google Scholar]
- Mills ENC, Madsen C, Shewry PR, Wicher HJ (2002) Food allergens of plant origin—their molecular and evolutionary relationships. Trends Food Sci Technol 14: 145–156 [Google Scholar]
- Monteiro S, Picarra-Pereira MA, Teixeira AR, Loureiro VB, Ferreira RB (2003) Environmental conditions during vegetative growth determine the major proteins that accumulate in mature grapes. J Agric Food Chem 51: 4046–4053 [DOI] [PubMed] [Google Scholar]
- Mullins MG, Bouquet A, Williams LE (1992) Biology of the Grapevine. Cambridge University Press, Cambridge, UK
- Nakayama M, Nakajima D, Nagase T, Nomura N, Seki N, Ohara O (1998) Identification of high-molecular-weight proteins with multiple EGF-like motifs by motif-trap screening. Genomics 51: 27–34 [DOI] [PubMed] [Google Scholar]
- Navabpour S, Morris K, Allen R, Harrison E, Mackerness SA-H, Buchanan-Wollaston V (2003) Expression of senescence-enhanced genes in response to oxidative stress. J Exp Bot 54: 2285–2292 [DOI] [PubMed] [Google Scholar]
- NeSmith DS (2002) Response of rabbiteye blueberry (Vaccibium ashei Reade) to the growth regulators CPPU and gibberellic acid. HortScience 37: 666–668 [Google Scholar]
- Nunan KJ, Davies C, Robinson SP, Fincher GB (2001) Expression patterns of cell wall-modifying enzymes during grape berry development. Planta 214: 257–264 [DOI] [PubMed] [Google Scholar]
- Nunan KJ, Sims IM, Cacic A, Robinson SP, Fincher GB (1998) Changes in cell wall composition during ripening of grape berries. Plant Physiol 118: 783–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nylander M, Svensson J, Palva ET, Welin BV (2001) Stress induced accumulation and tissue-specific localization of dehydrins in Arabidopsis thaliana. Plant Mol Biol 45: 263–279 [DOI] [PubMed] [Google Scholar]
- Ohgishi M, Saji K, Okada K, Sakai T (2004) Functional analysis of each blue light receptor, cry1, cry2, phot1, and phot2, by using combinatorial multiple mutants in Arabidopsis. Proc Natl Acad Sci USA 101: 2223–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlrogge J, Benning C (2000) Unraveling plant metabolism by EST analysis. Curr Opin Plant Biol 3: 224–228 [PubMed] [Google Scholar]
- Ollat N, Diakou-Verdin P, Carde JP, Barrieu F, Gaudillere JP, Moing A (2002) Grape berry development: a review. J Int Sci Vigne Vin 36: 109–131 [Google Scholar]
- Osafune T, Tsuboi S, Ehara T, Satoh Y, Yamada M (1996) The occurrence of non-specific lipid transfer proteins in developing castor bean fruits. Plant Sci 113: 125–130 [Google Scholar]
- Pacey-Miller T, Scott K, Ablett E, Tingey S, Ching A, Henry R (2003) Genes associated with the end of dormancy in grapes. Funct Integr Genomics 3: 144–152 [DOI] [PubMed] [Google Scholar]
- Parmentier-Line CM, Panta GR, Rowland LJ (2002) Changes in dehydrin expression associated with cold, ABA and PEG treatments in blueberry cell cultures. Plant Sci 162: 273–282 [Google Scholar]
- Pastorello EA, Farioli L, Pravettoni V, Ortolani C, Fortunato D, Giuffrida MG, Garoffo LP, Calamari AM, Brenna O, Conti A (2003) Identification of grape and wine allergens as an endochitinase 4, a lipid-transfer protein, and a thaumatin. J Allergy Clin Immunol 111: 350–359 [DOI] [PubMed] [Google Scholar]
- Patterson SE (2001) Cutting loose. Abscission and dehiscence in Arabidopsis. Plant Physiol 126: 494–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini L, Rohfritsch O, Fritig B, Legrand M (1993) Molecular cloning and expression of a new class of ortho-diphenol-O-methyltransferases induced in tobacco (Nicotiana tabacum L.) leaves by infection or elicitor treatment. Plant Physiol 103: 509–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picaud S, Becq F, Dédaldéchamp F, Ageorges A, Delrot S (2003) Cloning and expression of two plasma membrane aquaporins expressed during the ripening of grape berry. Funct Plant Biol 30: 621–630 [DOI] [PubMed] [Google Scholar]
- Pocock KF, Hayasaka Y, McCarthy M, Waters EJ (2000) Thaumatin-like proteins and chitinases, the haze-forming proteins of wine, accumulate during ripening of grape (Vitis vinifera) berries and drought stress does not affect the final levels per berry at maturity. J Agric Food Chem 48: 1637–1643 [DOI] [PubMed] [Google Scholar]
- Primikirios NI, Roubelakis-Angelakis KA (1999) Cloning and expression of arginine decarboxylase in Vitis vinifera L. Planta 208: 574–582 [DOI] [PubMed] [Google Scholar]
- Qin Q, Bergmann CW, Rose JKC, Saladie M, Kolli VSK, Albersheim P, Darvill AG, York WS (2003) Characterization of a tomato protein that inhibits a xyloglucan-specific endoglucanse. Plant J 34: 327–338 [DOI] [PubMed] [Google Scholar]
- Quackenbush J, Cho J, Lee D, Liang F, Holt I, Karamycheva S, Parvizi B, Pertea G, Sultana R, White J (2001) The TIGR Gene Indices: analysis of gene transcript sequences in highly sampled eukaryotic species. Nucleic Acids Res 29: 159–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishna W, Deng Z, Ding CK, Handa AK, Ozminkowski RH Jr (2003) A novel small heat shock protein gene, vis1, contributes to pectin depolymerization and juice viscosity in tomato fruit. Plant Physiol 131: 725–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault A, Deloire A, Bierne J (1996) Pathogenesis-related proteins in the grapevine induced by salicylic acid and Botritys cinerea. Vitis 35: 49–52 [Google Scholar]
- Reynolds AG, Wardle DA, Zurowski C, Looney NE (1992) Phenylureas CPPU and thidiazuron affect yield components, fruit composition, and storage potential of four seedless grape selections. J Am Soc Hortic Sci 117: 85–89 [Google Scholar]
- Robinson SP, Jacobs AK, Dry IB (1997) A class IV chitinase is highly expressed in grape berries during ripening. Plant Physiol 114: 771–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogiers SY, Hatfield JM, Jaudzems VG, White RG, Keller M (2004) Grape berry cv. Shiraz epicuticular wax and transpiration during ripening and pre-harvest weight loss. Am J Enol Vitic 55: 121–127 [Google Scholar]
- Ronning CM, Stegalkina SS, Ascenzi RA, Bougri O, Hart AL, Utterbach TR, Vanaken SE, Riedmuller SB, White JA, Cho J, et al (2003) Comparative analyses of potato expressed sequence tag libraries. Plant Physiol 131: 419–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhier N, Gelhaye E, Gualberto JM, Jordy M-N, De Fay E, Hirasawa M, Duplessis S, Lemaire SD, Frey P, Martin F, et al (2004) Poplar peroxiredoxin Q: a thioredoxin-linked chloroplast antioxidant functional in pathogen defense. Plant Physiol 134: 1027–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhier N, Gelhaye E, Sautiere PE, Brun A, Laurent P, Tagu D, Gerard J, de Fay E, Meyer Y, Jacquot JP (2001) Isolation and characterization of a new peroxiredoxin from poplar sieve tubes that uses either glutaredoxin or thioredoxin as a proton donor. Plant Physiol 127: 1299–1309 [PMC free article] [PubMed] [Google Scholar]
- Rudd S (2003) Expressed sequence tags: alternative or complement to whole genome sequences? Trends Plant Sci 8: 321–329 [DOI] [PubMed] [Google Scholar]
- Sabehat A, Weiss D, Lurie S (1998) Heat-shock proteins and cross-tolerance in plants. Physiol Plant 103: 437–441 [Google Scholar]
- Salzman RA, Tikhonova I, Bordelon BP, Hasegawa PM, Bressan RA (1998) Coordinate accumulation of antifungal proteins and hexoses constitutes a developmentally controlled defense response during fruit ripening in grape. Plant Physiol 117: 465–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarry JE, Gunata Z (2004) Plant and microbial glycoside hydrolases: volatile release from glycosidic aroma precursors. Food Chem 87: 509–521 [Google Scholar]
- Schwender J, Goffman F, Ohlrogge JB, Shachar-Hill Y (2004) Rubisco without the Calvin cycle improves the carbon efficiency of developing green seeds. Nature 432: 779–782 [DOI] [PubMed] [Google Scholar]
- Shade RE, Schroeder HE, Pueyo JJ, Table LM, Murdock LL, Higgins TJV, Chrispeels MJ (1994) Transgenic pea seeds expressing the alpha-amylase inhibitor of the common bean are resistant to burchid beetles. Biotechnology 12: 793–796 [Google Scholar]
- Shewry PR, Halford NG (2002) Cereal seed storage proteins: structures, properties and role in grain utilization. J Exp Bot 53: 947–958 [DOI] [PubMed] [Google Scholar]
- Shulman Y, Cohen SC, Loinger S (1985) Improved maturation and wine quality of Carignane grapes by ethephon treatment. Am J Enol Vitic 36: 264–267 [Google Scholar]
- Spiller GA, Story JA, Furumoto EJ, Chezem JC, Spiller M (2003) Effect of tartaric acid and dietary fibre from sun-dried raisins on colonic function and on bile acid and volatile fatty acid excretion in healthy adults. Br J Nutr 90: 803–807 [DOI] [PubMed] [Google Scholar]
- Stekel DV, Git Y, Falciani F (2000) The comparison of gene expression from multiple cDNA libraries. Genome Res 10: 2044–2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern RA, Flaishman MA (2003) Benzyladenine effects on fruit size, fruit thinning and return yield of ‘Spadona’ and ‘Coscia’ pear. Sci Hortic (Amst) 98: 499–504 [Google Scholar]
- Streatfield SJ, Weber A, Kinsman EA, Hausler RE, Li J, Post-Beittenmiller D, Kaiser WM, Pyke KA, Flugge UI (1999) The phosphoenolpyruvate/phosphate translocator is required for phenolic metabolism, palisade cell development and plastid dependent nuclear gene expression. Plant Cell 11: 1609–1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, Mu JH, Gasch A, Chua NH (1998) Identification by PCR of receptor-like protein kinases from Arabidopsis flowers. Plant Mol Biol 37: 587–596 [DOI] [PubMed] [Google Scholar]
- Tattersall DB, Van Heeswijck R, Hoj PB (1997) Identification and characterization of a fruit-specific, thaumatin-like protein that accumulates at very high levels in conjunction with the onset of sugar accumulation and berry softening in grapes. Plant Physiol 114: 759–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tattersall EAR, Ergul A, AlKayal F, DeLuc L, Cushman JC, Cramer GR (2005) A comparison of methods for isolating high-quality RNA from leaves of grapevine. Am J Enol Vitic 56: (in press)
- Terrier N, Ageorges A, Abbal P, Romieu C (2001) Generation of ESTs from grape berry at various developmental stages. J Plant Physiol 158: 1575–1583 [Google Scholar]
- Thipyapong P, Hunt MD, Steffens JC (2004. b) Antisense downregulation of polyphenol oxidase results in enhanced disease susceptibility. Planta 220: 105–117 [DOI] [PubMed] [Google Scholar]
- Thipyapong P, Melkonian J, Wolfe D, Steffens JC (2004. a) Suppression of polyphenol oxidases increases stress tolerance in tomato. Plant Sci 167: 693–703 [Google Scholar]
- Thomas MR, Cain P, Scott NS (1994) DNA typing of grapevines: a universal methodology and database for describing cultivars and evaluating genetic relatedness. Plant Mol Biol 25: 939–949 [DOI] [PubMed] [Google Scholar]
- Thomma BPHJ, Cammue BPA, Thevissen K (2002) Plant defensins. Planta 216: 193–202 [DOI] [PubMed] [Google Scholar]
- Tieman DM, Harriman RW, Ramamohan G, Handa AK (1992) An antisense pectin methylesterase gene alters pectin chemistry and soluble solids in tomato. Plant Cell 4: 667–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toquin V, Grausem B, Geoffroy P, Legrand M (2003) Structure of the tobacco caffeic acid O-methyltransferase (COMT II) gene: identification of promoter sequences involved in gene inducibility by various stimuli. Plant Mol Biol 52: 495–509 [DOI] [PubMed] [Google Scholar]
- van Loon LC, van Strien EA (1999) The families of pathogenesis-related proteins, their activities and comparative analysis of PR-I type proteins. Physiol Mol Plant Pathol 55: 85–97 [Google Scholar]
- Van Poecke RM, Posthumus MA, Dicke M (2001) Herbivore-induced volatile production by Arabidopsis thaliana leads to attraction of the parasitoid Cotesia rubecula: chemical, behavioral, and gene-expression analysis. J Chem Ecol 27: 1911–1928 [DOI] [PubMed] [Google Scholar]
- Vivier MA, Pretorius IS (2002) Genetically tailored grapevines for the wine industry. Trends Biotechnol 20: 472–478 [DOI] [PubMed] [Google Scholar]
- Vorwerk S, Somerville S, Somerville C (2004) The role of plant cell wall polysaccharides composition in disease resistance. Trends Plant Sci 9: 203–209 [DOI] [PubMed] [Google Scholar]
- Vrebalov J, Ruezinsky D, Padmanabhan V, White R, Medrano D, Drake R, Schuch W, Giovannoni J (2002) A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (Rin) locus. Science 296: 343–346 [DOI] [PubMed] [Google Scholar]
- Wagner A (2002) Selection and gene duplication: a view from the genome. Genome Biol 3: reviews1012.1–1012.3 [DOI] [PMC free article] [PubMed]
- Walker RP, Chen ZH, Tecsi LI, Famiani F, Lea PJ, Leegood RC (1999) Phosphoenolpyruvate carboxykinase plays a role in interactions of carbon and nitrogen metabolism during grape seed development. Planta 210: 9–18 [DOI] [PubMed] [Google Scholar]
- Wang ZP, Deloire A, Carbonneau A, Federspiel B, Lopez F (2003) An in vivo experimental system to study sugar phloem unloading in ripening grape berries during water deficiency stress. Ann Bot (Lond) 92: 523–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters EJ, Hayasaka Y, Adams K, Tattersall DB, Williams PJ (1998) Sequence analysis of grape (Vitis vinifera) berry chitinases that cause haze formation in wines. J Agric Food Chem 46: 4950–4957 [Google Scholar]
- Weaver RJ, Montgomery R (1974) Effect of ethephon on coloration and maturation of wine grapes. Am J Enol Vitic 25: 39–41 [Google Scholar]
- Wu SQ, Watanabe N, Mita S, Ueda Y, Shibuya M, Ebizuka Y (2003) Two O-methyltransferases isolated from flower petals of Rosa chinensis var. spontanea involved in scent biosynthesis. J Biosci Bioeng 96: 119–128 [PubMed] [Google Scholar]
- Zhang Y, Lewis K (1997) Fabatins: new antimicrobial plant peptides. FEMS Microbiol Lett 149: 59–64 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.