Abstract
PlantGDB (http://www.plantgdb.org/) is a database of plant molecular sequences. Expressed sequence tag (EST) sequences are assembled into contigs that represent tentative unique genes. EST contigs are functionally annotated with information derived from known protein sequences that are highly similar to the putative translation products. Tentative Gene Ontology terms are assigned to match those of the similar sequences identified. Genome survey sequences are assembled similarly. The resulting genome survey sequence contigs are matched to ESTs and conserved protein homologs to identify putative full-length open reading frame-containing genes, which are subsequently provisionally classified according to established gene family designations. For Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa), the exon-intron boundaries for gene structures are annotated by spliced alignment of ESTs and full-length cDNAs to their respective complete genome sequences. Unique genome browsers have been developed to present all available EST and cDNA evidence for current transcript models (for Arabidopsis, see the AtGDB site at http://www.plantgdb.org/AtGDB/; for rice, see the OsGDB site at http://www.plantgdb.org/OsGDB/). In addition, a number of bioinformatic tools have been integrated at PlantGDB that enable researchers to carry out sequence analyses on-site using both their own data and data residing within the database.
Plant genome sequence data have been accumulating from three major sources: whole-genome sequencing and assembly (Arabidopsis [Arabidopsis thaliana]: Lin et al., 1999; Mayer et al., 1999; Salanoubat et al., 2000; rice [Oryza sativa]: Goff et al., 2002; Yu et al., 2002; Medicago truncatula: http://medicago.org/), genome survey sequences (GSS; maize [Zea mays]: Palmer et al., 2003; Whitelaw et al., 2003; Fernandes et al., 2004; sorghum [Sorghum bicolor]: Bedell et al., 2005), and expressed sequence tags (ESTs; more than 50 species). This data flow is likely to continue, with a focus on complete sequencing of “reference species” (Arabidopsis, rice, maize, M. truncatula, and tomato [Lycopersicon esculentum]), draft sequencing of other selected species, and further EST and full-length cDNA sequencing. Considerable resources have been devoted to the development of public databases that provide access to plant genome data. However, finding ways to efficiently access and effectively analyze those sequence data remains a nontrivial challenge for many plant biologists.
PlantGDB (http://www.plantgdb.org/) is our ongoing effort to aid in the organization and interpretation of sequence data through the development and implementation of integrated databases and analytical tools. In this article, we discuss some of the unique sequence storage and analysis capabilities provided by PlantGDB and compare them to those made available through other online resources. All PlantGDB data and scripts described here are freely available from our download site (http://www.plantgdb.org/download/download.php) or by request.
DATA
PlantGDB is a plant sequence database. Its data consist of plant sequences and their associated annotations. There are mainly three types of plant sequences: complete genome sequences for Arabidopsis and rice, other kinds of sequences including EST and GSS extracted from public sequence repositories such as GenBank (Benson et al., 2005), and assembled EST and GSS contigs.
Data Sources and Updates
Plant sequences that are made available through public repositories compose the core PlantGDB sequence set. Currently, PlantGDB contains sequences from more than 24,000 plant species (belonging to more than 6,000 genera). Our sequence-processing scripts extract all plant nucleotide sequences from EST (http://www.ncbi.nlm.nih.gov/dbEST/), GSS (http://www.ncbi.nlm.nih.gov/dbGSS/), sequence tagged sites (http://www.ncbi.nlm.nih.gov/dbSTS/), high-throughput genomic (http://www.ncbi.nlm.nih.gov/HTGS/), and other genomic DNA sequence categories at GenBank and populate our relational database. All scripts are written in Perl and are available upon request. The extracted sequences are sorted by taxonomic classification to provide fast and easy access to subsets of sequences limited to an individual species or to a phylogenetically related group via the PlantGDB download site (http://www.plantgdb.org/download/download.php). To ensure that our data are synchronized with GenBank, this procedure is run daily. To maintain consistency with the version updates occurring at GenBank, our pipeline automatically tracks two GenBank data files, gbchg.txt and gbdel.txt (available from ftp://ftp.ncbi.nih.gov/genbank/), corresponding to sequence version changes and deletions, respectively. The entries in these two files are converted into corresponding “update/delete” SQL statements, which allow our database to stay synchronized with GenBank. Similarly, plant protein sequences are extracted from UniProt (Apweiler et al., 2004; http://www.pir.uniprot.org/). UniProt was chosen as the protein sequence source for PlantGDB because UniProt proteins are associated with Gene Ontology terms (Ashburner et al., 2000; http://www.geneontology.org/), which we use for provisional sequence annotation. Unlike GenBank, UniProt currently does not provide daily update data dumps. Instead, their records are dumped on a weekly basis. Therefore, PlantGDB downloads data from UniProt once a week and extracts all plant records, which are subsequently loaded into our database.
Identification of Contaminants
Sequence sets downloaded from GenBank can contain “contaminants”. Contaminants typically only pose a problem when unrecognized (for an example of clever use of nonnative sequences derived in Drosophila sequencing projects that resulted in the serendipitous assembly of three Wolbachia bacterial genomes, see Salzberg et al., 2005). Sequences that represent contamination include cloning vectors, bacterial host DNA, DNA from plant-associated microbes, and even sequences from the human researchers who prepare plant DNA for sequencing. At PlantGDB, efforts are undertaken to identify and remove all contaminant DNA so that a “clean” dataset is made available to researchers. For example, to derive EST sets that are limited to species-specific nuclear transcripts, we use the Vmatch program (Abouelhoda et al., 2004; http://www.vmatch.de/; option settings: −l 50 −exdrop 1 −identity 90) to compare EST sequences (1) against UniVec (http://www.ncbi.nih.gov/VecScreen/UniVec.html) for vector contaminations, (2) against three available Escherichia coli genomes (obtained from ftp://ftp.ncbi.nih.gov/genbank/genomes/Bacteria/) for bacterial DNA, and (3) against plant mitochondrial and plastid genomes (obtained from http://www.ncbi.nlm.nih.gov/genomes/ORGANELLES/plants.html and http://www.ncbi.nlm.nih.gov/genomes/ORGANELLES/plastids.html, respectively) to identify plant organelle-encoded sequences.
Identification of Repetitive Sequences
The Institute for Genomic Research (TIGR) plant repeat database (http://www.tigr.org/tdb/e2k1/plant.repeats/; Ouyang and Buell, 2004) is used to identify and label repetitive sequences (using Vmatch with options: −l 100 −exdrop 2 −identity 80). All ESTs that match to known repetitive elements are excluded from the assembly. The reasons for this are both theoretical and practical. Theoretically, it is a problem for any assembly program to deal with repeats because it is virtually impossible to reliably reassemble the set of unique transcripts from which the set of repetitive ESTs was derived. In practice, large numbers of repetitive elements also can waste both time and space for the computational resources utilized for their assembly. All sequences labeled as contaminants or repeats are kept as individual records in the database and are listed on corresponding Web pages (http://www.plantgdb.org/prj/ESTCluster/contamination.php and http://www.plantgdb.org/prj/ESTCluster/repeat.php, respectively).
EST Contig Assembly and Annotation
EST sequences are valuable data for gene discovery, especially for plant species with large genomes that have not been fully sequenced, and they provide a convenient means of accessing the transcriptome of a given species. However, ESTs generally correspond to only partial cDNA sequences, and EST samples are typically highly redundant (especially if EST sets are not derived from normalized EST libraries). Therefore, the assembly of overlapping ESTs into putative unique transcript contigs on a frequent and regular basis constitutes the first step for all EST analyses performed at PlantGDB (for more details, see http://www.plantgdb.org/prj/ESTCluster/progress.php). A similar analysis is provided by the TIGR gene indices for selected species with sufficiently large numbers of ESTs (http://www.tigr.org/tdb/tgi/plant.shtml; Lee et al., 2005).
EST assembly remains a computational challenge given the large number of EST sequences currently available. For example, with more than 400,000 maize ESTs, CAP3 (one of the most popular assembly programs; Huang and Madan, 1999) would require roughly eight gigabytes of computer memory to generate an assembly. Such memory requirements suggest that most current computer systems will be unable to keep up with the explosive growth of new EST data. In this context, it can be appreciated that the aforementioned screening for vector contaminants and repetitive sequences is also necessary for assembly because such sequences would generate huge and irrelevant clusters that would severely tax computer resources during assembly. To further reduce computational requirements, PlantGDB uses the parallel EST clustering program PaCE (Kalyanaraman et al., 2003; http://bioinformatics.iastate.edu/bioinformatics2go/PaCE/) to preassemble EST sets before the final CAP3 assembly.
In addition to piecing together significantly overlapping fragments, EST assembly can be considered to be an initial step toward reducing the redundancy that exists in available EST datasets. Because no EST assembly can be guaranteed to be error free, we caution researchers to consider searches against the PlantGDB assemblies to be complementary and exploratory steps in gene discovery relative to more comprehensive analyses of promising targets. One obvious advantage of searching a database of EST contigs (rather than unassembled ESTs) is that the likelihood of finding a complete match against one's query should be increased (because EST contigs are longer, on average, than raw EST sequences).
Instead of deriving EST assembly parameters specific to each species, we use a common set of conserved assembly criteria for most assemblies: ESTs are initially clustered whenever they share a minimum overlap of 40 bases with at least 95% identity (these initial clusters may split into several contigs based on overall similarity; Huang and Madan, 1999). Therefore, when a biologist identifies a contig containing his or her gene of interest at PlantGDB, he or she should check the regions of overlap manually to ensure that the identified contig is reasonable. Because member EST sequence alignments are easily accessible through contig display pages, biologists can reassemble any contig's member ESTs using different criteria to determine whether the assembly of that contig is robust.
Groups working on a particular organism often carefully generate their own species-specific EST contigs, and some groups have asked to deposit their assemblies into PlantGDB to gain easy access to our annotations and integrated analysis tools. For example, the barley (Hordeum vulgare) EST contigs at PlantGDB are mirrored from the HarvEST Triticeae database (http://harvest.ucr.edu/). The PlantGDB display includes mapping of oligomer probes for microarray expression studies and links to expression data at BarleyBase (http://www.barleybase.org/). For species with EST sequence sets that are assembled at PlantGDB directly, feedback from researchers is used to determine the build release schedule: if a researcher has a need to gain access to a new build for a given species' EST assembly, that dataset can be given priority for a speedy build and release. After a new assembly has been created, the deprecated assemblies remain accessible online and can still be viewed on the Web (this serves as an historic record to enable long-term accessibility). However, deprecated assemblies cannot be accessed by the PlantGDB data analysis tools (e.g. BLAST@PlantGDB).
For each contig sequence, a putative function is assigned based upon sequence similarity to gene products that have been previously functionally annotated. This is accomplished by means of an automated BLAST search (Altschul et al., 1997) against the entire UniProt database. The annotated functions of the top three hits below an expectation value of E-20 are assigned as the putative function, along with all the Gene Ontology terms associated with the similar protein sequences identified. This procedure is not ideal due to the inherent danger of transitively propagating annotation errors (Gilks et al., 2000), but it is currently the only practical choice in view of the large and quickly changing datasets.
PlantGDB was designed to be a Web-based research workbench. Thus, all records are linked to tools that allow for immediate retrieval of raw data, and all data are made available alongside related data and applications that can be used to recalculate curated records using updated or proprietary data. For example, both the EST assembly and associated BLAST hit annotation can be updated by researchers using only data present at PlantGDB or data present at PlantGDB in conjunction with additional data from elsewhere (e.g. sequencing reads the researcher has not yet submitted to GenBank), or using parameters other than default via tools embedded within the sequence record display pages.
GSS Contig Assembly and Annotation
Roughly 2.6 million sequences have entered the current PlantGDB maize GSS assembly, which was generated using the PCAP program with default parameters (Huang et al., 2003). GSS sequences included in the assembly were generated using a variety of methods, including gene-enrichment approaches such as methylation filtration (Palmer et al., 2003) and high-CoT selection (Yuan et al., 2003), as well as random genome sequencing. In the absence of a fully sequenced maize genome, the maize GSSs provide the best dataset to study the gene space and genome organization of maize. Two complementary versions of maize GSS assembly are described elsewhere (Whitelaw et al., 2003; Emrich et al., 2004). The PlantGDB assembly implements a bottom-up annotation protocol, which seeks to identify contigs containing complete maize genes with accurate exon-intron gene structures annotated (Fig. 1). Here, “complete” refers to the encoded translation product, not necessarily including all the untranslated transcript regions and promoter and terminator regions. Rather than relying upon ab initio or BLAST-like similarity searches to assign gene structure and putative function to GSS contigs, the PlantGDB annotation pipeline is based upon accurate spliced alignment of contigs to homologous protein sequences using the GeneSeqer suite of programs (Usuka and Brendel, 2000; Brendel et al., 2004). So far, we have confidently derived 4,062 maize genes that contain a full-length or near full-length protein-coding region based on high-quality alignment with 5,116 annotated Arabidopsis and 8,016 annotated rice proteins. These identified maize genes belong to 32 superfamilies and 252 gene families and provide a significant addition to our current knowledge of the maize gene space. Note that gene families and superfamilies are not defined or redefined at PlantGDB. Similarity searches carried out at PlantGDB to assign such designations to maize genes rely upon the Arabidopsis gene family categorizations made available at The Arabidopsis Information Resource (http://www.arabidopsis.org/info/genefamily/genefamily.html). The research community can initiate both BLAST and keyword searches among identified maize full-length genes at http://www.plantgdb.org/prj/GSSAssembly/zeamays/. In addition, we have assembled roughly a half million sorghum GSSs (approximately 98% of which were generated by Orion Genomics using the methylation filtration approach and are also available at GenBank; Bedell et al., 2005). In the derived set of 79,343 contigs, we identified 1,561 genes that contain a full-length or near full-length protein-coding region (http://www.plantgdb.org/prj/GSSAssembly/sorghumbicolor/). A total of 903 Arabidopsis and 1,199 rice proteins matched both the maize and sorghum full-length gene sets.
Figure 1.
A typical display of a maize GSS contig record at PlantGDB. The lower left diagram displays a schematic representation of a GSS contig and its gene structure. In this example, the GSS contig appears to encode a full-length maize ABC transporter (based upon spliced alignment with three similar Arabidopsis gene products). Predicted exons are shown as solid lines, and introns are represented by thin lines. Known repetitive elements are masked and appear along the contig as yellow Xs. This diagram can be manipulated using the “Image Control” box located in the upper left. Researchers can add, e.g. EST spliced alignments, to the display by checking the appropriate box within the “Image Control” diagram then clicking the button labeled “Redraw Image.” In the middle, the “Similar protein sequences” scrollable box displays descriptions of matched proteins. On the right, the “Utilities” box contains a set of analysis tools that can be applied to the contig. For example, researchers can choose to view the details of the spliced alignments, perform a BLAST search against chosen databases, see detailed descriptions of matched proteins, etc. Near the bottom, a “Help” message box explains the function of each analysis tool as the researcher moves the computer mouse across the tool links. Researchers can provide expert annotation for the gene by following the link to “Provide Annotation” near the bottom of the “Utilities” box.
Genome-Wide Gene Structure Annotation
The Arabidopsis EST and full-length cDNA collections were threaded onto the five established Arabidopsis chromosome sequences using the GeneSeqer program. Those data are stored in a specialized ancillary database called AtGDB (Arabidopsis thaliana Genome Database; http://www.plantgdb.org/AtGDB/). AtGDB incorporates 418,564 EST sequences, 64,840 full-length cDNA sequences, 31,971 predicted transcripts, and an ever-increasing number of user-contributed annotations (http://www.plantgdb.org/AtGDB/Annotation/UCAlist.php) into a workbench for genome informatics. Approximately 80% of the predicted protein-coding gene models are supported by EST or cDNA evidence, whereas 20% are based solely upon computational gene structure prediction. The degree of support for individual annotations in Arabidopsis is substantially better than that of rice, with 68% being supported to the extent that major changes are not likely (Tables I and II). However, 70% of the predicted Arabidopsis gene models exist in a genomic context such that corresponding EST and cDNA alignments reveal some form of incongruence, including incompletely annotated noncoding regions, alternative splicing, and erroneous gene predictions (Schlueter et al., 2005). Effects on individual annotations vary with the type of incongruence noted. Untranslated region annotation problems, for example, are quite different than the errant assignment of gene structures, though neither is unimportant. While correct gene structure assignment in the coding sequence is as much as most researchers require, it is important to provide accurate annotation of all gene features. This said, an astonishing 4,883 of the Arabidopsis gene models display incongruent gene structures, suggesting that various alterations to the coding sequence caused by alternative splicing, excluded exon regions, and an extended open reading frame have occurred (Table II; http://www.plantgdb.org/AtGDB/Annotation/gaeval/gaeval_lists.php).
Table I.
Evaluation of current gene annotation in Arabidopsis and rice
| Annotation Categorya
|
Annotation Support at AtGDB
|
Annotation Support at OsGDB
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Total
|
Coverageb
|
Total
|
Coverageb
|
|||||||
| =0% | <50% | <100% | =100% | =0% | <50% | <100% | =100% | |||
| Nonspliced | 5,331 | 1,659 | 266 | 658 | 2,748 | 12,633 | 8,144 | 512 | 1,089 | 2,888 |
| Spliced | ||||||||||
| All introns confirmed | 18,419 | – | 112 | 3,958 | 14,349 | 12,758 | – | 125 | 3,432 | 9,201 |
| ≥75% intron confirmation | 839 | – | 27 | 647 | 165 | 1,575 | – | 51 | 1,173 | 351 |
| ≥50% intron confirmation | 1,047 | – | 233 | 716 | 98 | 2,512 | – | 420 | 1,832 | 260 |
| >0% intron confirmation | 1,327 | – | 965 | 342 | 20 | 2,496 | – | 1,435 | 967 | 94 |
| No introns confirmed | 5,008 | 3,950 | 675 | 295 | 88 | 30,147 | 25,225 | 2,942 | 1,604 | 376 |
| Annotation totals | 31,971 | 5,609 | 2,278 | 6,616 | 17,468 | 62,121 | 33,369 | 5,485 | 10,097 | 13,170 |
Annotations with multi-exon structural definitions (spliced annotations) are categorized by the ratio of introns confirmed by EST and/or cDNA spliced alignment to the total number of predicted introns. For example, an annotation predicting a gene model with five exons would by definition have four introns. If three of these intron positions are confirmed, this annotation would be placed in the ≥75% category. If only two were confirmed, the annotation would fall into the ≥50% category.
Annotations are further categorized by coverage. Coverage percentages denote the fraction of the annotation-defined exon regions overlapped by a sequence alignment (the fraction represented by physical sequence in the form of EST and/or cDNA).
Table II.
Incongruent gene annotation in Arabidopsis and rice
| Annotation Category | Annotation Support at AtGDB | Annotation Support at OsGDB |
|---|---|---|
| Incongruent gene structurea | 4,883 | 26,291 |
| Incomplete untranslated region definitionb | 4,458 | 10,948 |
| Complex structural incongruencec | 286 | 3,499 |
Annotations with gene structure definitions, which are inconsistent with overlapping EST and/or cDNA.
Annotations with incomplete untranslated region definitions based on aligned EST and/or cDNA evidence (minimum 100 base variance).
Annotations that may require complex alterations, including annotation of polycistronic messages, documentation of alternative cleavage/polyA sites, or restructuring into multiple gene transcripts.
OsGDB (http://www.plantgdb.org/OsGDB/) is the rice equivalent to AtGDB. ESTs and full-length cDNAs stored at OsGDB are threaded onto the full set of rice bacterial artificial chromosome (BAC) sequences. Altogether, 298,857 EST sequences, 32,136 full-length cDNAs, 3,453 BAC sequences, 66,224 gene models (defined as GenBank file features), and 62,121 transcription unit (TU) models (defined by TIGR; http://www.tigr.org/tdb/e2k1/osa1/irgsp/eukan_routine_irgsp.shtml) are on display at OsGDB. Of the current TU gene model predictions, about one-half are supported by EST or cDNA evidence, whereas the remaining one-half are based on computational gene prediction alone (Table I). Similar to what was observed for Arabidopsis, a large percentage of rice TUs reveals some level of incongruence with local EST and cDNA alignments (Table II; http://www.plantgdb.org/OsGDB/Annotation/gaeval/gaeval_lists.php).
DATABASE DEVELOPMENT
PlantGDB is built upon a relational database. Underlying PlantGDB is the MySQL (http://www.mysql.com/) database management system running on the open-source RedHat Linux operating system (http://www.redhat.com/). The PlantGDB database schema can be accessed at http://www.plantgdb.org/document/public/schema.php. The schema was designed to efficiently store sequence data and associated annotations to facilitate meaningful biological queries. For example, for all the raw plant DNA sequences downloaded from GenBank, we extract and reorganize the information originally stored at GenBank as ASN.1 flat-files and store those records in tables. Such information extraction and reorganization enables researchers to construct meaningful biological queries that can be carried out in a precise and flexible environment, a process that is not always possible at GenBank. For example, using the National Center for Biotechnology Information (NCBI) Entrez tool, a researcher cannot construct queries like “show me all maize promoter sequences,” even though promoter region annotations are embedded in GenBank records. If a researcher attempted to use a query like “Zea mays[ORGN] AND Promoter,” he or she would only end up with a list of sequence records that contain the word “Promoter.” The word could appear anywhere in the sequence record (e.g. in the “comment” field). Furthermore, it is impossible to extract just the promoter regions from all the sequences and the output list provided at GenBank. However, at PlantGDB, the promoter (and all other GenBank feature) information is stored within a relational table, enabling researchers to accurately specify that only promoter regions be retrieved. This sort of flexibility is made possible by PlantGDB's TableMaker tool (http://www.plantgdb.org/search/query/TableMaker.php; Fig. 2). Furthermore, the PlantGDB TableMaker does not require that the researcher have any knowledge of SQL: TableMaker translates the researcher's specifications to SQL query statements and presents the results in a tabular format. For advanced users, direct SQL access to the PlantGDB backend database is also available online (http://www.plantgdb.org/search/query/websql.php).
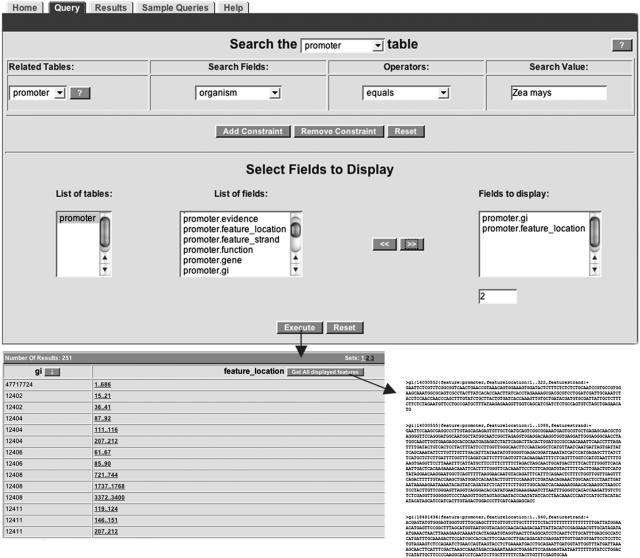
Figure 2.
The PlantGDB TableMaker query tool. This display shows an example of constructing and executing a simple query to “Find all maize promoter sequences” (translated as “promoter organism equals Zea mays”). Once the fields that should be displayed in the output have been specified (middle panel of the same display), the researcher presses the button to execute the query. The output list (lower left) shows all sequences that match the query. Pressing the button to “Get all displayed features” downloads the features in FASTA format (bottom right). All query fields are based on GenBank annotation features. Detailed help and a tutorial resource are available from the TableMaker Web page.
For species that have a complete genome sequence available, specialized databases (i.e. AtGDB for Arabidopsis and OsGDB for rice) have been created to make available annotation of detailed exon-intron structures for protein-coding genes, based upon the threading of EST and full-length cDNAs onto chromosomes. Spliced alignments of ESTs and cDNAs as well as the recent annotation of the Arabidopsis and rice genomes are parsed and imported into database tables, and an elaborate Web interface was developed and is made available to provide a visual assessment of annotated gene structure (Zhu et al., 2003). Researchers can browse the rice or Arabidopsis genome and can query the database based upon description, identifier, or sequence-based similarity. The Web interface enables researchers to browse a genomic region within the context of any and all available annotation resources. A graphical representation displays multiple sources of alignment information relative to one another such that each is color coded based upon its specific annotation source. Sequence data, analysis tools, and related external links are stored for each EST/cDNA alignment and annotated transcript, and are made available on each data display page. To evaluate sequence incongruence, an additional context view has been developed that incorporates the aligned nucleotide sequences for the chromosome, BAC, EST, and cDNA sequences. Dynamic content fields provide the researcher with quality values and descriptions for individual sequence features, and the Web interface has an established framework enabling inductive analysis through visualization of alternative splicing, noncanonical introns, transcriptional expression, and gene family relationships.
Correct gene annotation must leverage the expertise that exists within the research community. To truly involve the research community, a simple feedback form is not sufficient. Instead, well-designed and easy-to-use annotation tools must be made available to researchers. To address this need, researchers using AtGDB can contribute updated annotations of their own to a shared community annotation collection through the use of Web-based annotation tools (Schlueter et al., 2005). These tools were developed to allow researchers to easily access ab initio predictions, native and homologous sequence alignments, open reading frame estimations, and other useful data analysis tools for gene structure determination. The results of these individual analyses are presented in the annotation tool in such a way that the users can easily select compatible exons to build their own complete transcript model. This “User-Contributed Annotation” is credited to the researcher who contributed the annotation, but (subsequent to a validation step) becomes owned by the research community at large. In this way, any member of the community can alter annotations in light of new evidence.
ANALYSIS TOOLS
PlantGDB makes available data analysis tools to help the research community gain access to the wealth of information that can be gleaned through the analysis of sequence data. Three online sequence analysis tools that are unique to PlantGDB are the BLAST@PlantGDB, GeneSeqer@PlantGDB, and PatternSearch@PlantGDB tools.
Although nearly all sequence databases provide an online BLAST server, most only allow researchers to search against one database at a time. For example, the current NCBI BLAST server (http://www.ncbi.nlm.nih.gov/BLAST/) requires that only one database be selected from a predefined set of database options (e.g. “nr,” “est,” “gss,” etc.). In addition, at NCBI researchers can further choose to search against either all organisms or a group of species that share a given taxonomic rank. This is not always convenient if a researcher wishes to search against multiple databases (e.g. EST and GSS) or to search against multiple species (e.g. maize and rice but not sorghum). BLAST@PlantGDB (http://www.plantgdb.org/PlantGDB-cgi/blast/PlantGDBblast/) serves as a much-needed supplement to NCBI's BLAST service, providing selection flexibility for both the database and species data source options (e.g. only rice or maize ESTs, or both; or all monocot GSSs; or rice ESTs and all cereal EST contigs, etc.). The BLAST@PlantGDB server implements the standard NCBI BLAST stand-alone search engine (Altschul et al., 1997). As an enhancement, the PlantGDB server also provides online batch search capabilities, enabling researchers to upload a maximum of 100 query sequences in one file for efficient simultaneous searches.
GeneSeqer produces plant gene structure models based on spliced alignment to genomic sequences of both native and homologous EST, cDNA, and protein sequences. The GeneSeqer@PlantGDB (http://www.plantgdb.org/PlantGDB-cgi/GeneSeqer/PlantGDBgs.cgi) Web service allows researchers to “thread” EST/cDNA sequences onto genomic DNA across all plant species. Integration of the stand-alone GeneSeqer program with backend database operations allows researchers to conveniently align ESTs of specific quality or origin to their genomic counterparts. For a detailed description of this server, see Schlueter et al. (2003) and the online tutorial available at http://www.plantgdb.org/tutorial/.
The PatternSearch@PlantGDB tool (http://www.plantgdb.org/PlantGDB-cgi/vmatch/patternsearch.pl) allows researchers to conduct pattern searches, i.e. searches for relatively short matches possibly interspersed with mismatches and/or insertions/deletions (“indels”), against PlantGDB sequences (e.g. against the Arabidopsis and rice genome, or cDNAs, etc.). For example, if a researcher were to design primers (e.g. for reverse transcription-PCR or genomic PCR) to amplify a gene of interest, the primers might also hybridize with nontarget sequences. This researcher could use the PlantGDB PatternSearch tool to find out whether the primer sequences chosen are unique to the gene of interest. The underlying search engine for the PlantGDB PatternSearch is the Vmatch program, which is based on enhanced suffix arrays (Abouelhoda et al., 2004). In contrast to heuristic BLAST-like pattern search tools, our server provides complete and accurate matching results without requiring parameter fine-tuning. NCBI also provides a “Search for short, nearly exact matches” BLAST function (http://www.ncbi.nih.gov/BLAST/), but, with its default parameters, that tool is not guaranteed to find all the occurrences of the specified pattern, and researchers cannot specify the maximum number of mismatches or indels allowed. For PatternSearch@PlantGDB, users simply specify the number or percentage of allowed mismatches and indels.
EDUCATION AND OUTREACH
An educational site ancillary to PlantGDB provides a centralized repository for various Plant Genome Research “Outreach” Program (PGROP) materials and related activities (http://www.plantgdb.org/PGROP/pgrop.php; Baran et al., 2004). PGROP's intended audience includes (but is not limited to) high-school students and teachers, undergraduates (including special target groups like minorities), journalists, and the general public. It is the goal of the PGROP project to broaden participation of these various groups in plant genome research by helping them find materials, information, and answers to questions that are related to plant genomic research.
CONCLUSIONS AND FUTURE DIRECTIONS
Compared to the large number of finished prokaryotic and animal genomes, plant genomics is still in its infancy. However, the increasing importance of plant biotechnology is spearheading a dramatic increase in the resources available for plant genomics research. Thus, we can anticipate with certitude the complete genome sequencing of several crop species in the next few years. As is the case for the prokaryotic and animal genomics fields, comparative genomics will be a most valuable tool for turning the plant sequence information into knowledge of genome repertoire, organization, and function. Databases like PlantGDB that synthesize current knowledge will provide a requisite foundation for making rapid progress by leveraging existing knowledge to annotate and evaluate new genomes.
There are a number of services that we are working to improve at PlantGDB in the coming months. For instance, the creation of formally defined “versions” of both the data and database will help researchers to cite the database using links and version designators that will persist long after a manuscript's publication. We also are working to provide various case studies in tutorial form as examples of work flow using PlantGDB data and tools. We also plan to provide access to data and analysis tools via Web services to allow for better integration with other online resources.
Acknowledgments
We thank Ann Fu and Mike Brekke for their technical contributions to this work.
This work was supported by the National Science Foundation Plant Genome Research Projects (grant no. DBI–0321600 to V.B. and C.L.).
References
- Abouelhoda MI, Kurtz S, Ohlebusch E (2004) Replacing suffix trees with enhanced suffix arrays. J Discrete Algorithms 2: 53–86 [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apweiler R, Bairoch A, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, et al (2004) UniProt: the Universal Protein knowledgebase. Nucleic Acids Res 32: D115–D119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al (2000) Gene Ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baran S, Lawrence CJ, Brendel V (2004) Plant Genome Research Outreach Portal. A gateway to plant genome research “outreach” programs and activities. Plant Physiol 134: 889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedell JA, Budiman MA, Nunberg A, Citek RW, Robbins D, Jones J, Flick E, Rholfing T, Fries J, Bradford K, et al (2005) Sorghum genome sequencing by methylation filtration. PLoS Biol 3: e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL (2005) GenBank. Nucleic Acids Res 33: D34–D38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brendel V, Xing L, Zhu W (2004) Gene structure prediction from consensus spliced alignment of multiple ESTs matching the same genomic locus. Bioinformatics 20: 1157–1169 [DOI] [PubMed] [Google Scholar]
- Emrich SJ, Aluru S, Fu Y, Wen TJ, Narayanan M, Guo L, Ashlock DA, Schnable PS (2004) A strategy for assembling the maize (Zea mays L.) genome. Bioinformatics 20: 140–147 [DOI] [PubMed] [Google Scholar]
- Fernandes J, Dong Q, Schneider B, Morrow DJ, Nan GL, Brendel V, Walbot V (2004) Genome-wide mutagenesis of Zea mays L. using RescueMu transposons. Genome Biol 5: R82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilks WR, Audit B, De Angelis D, Tsoka S, Ouzounis CA (2000) Modeling the percolation of annotation errors in a database of protein sequences. Bioinformatics 18: 1641–1649 [DOI] [PubMed] [Google Scholar]
- Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296: 92–100 [DOI] [PubMed] [Google Scholar]
- Huang X, Madan A (1999) CAP3: a DNA sequence assembly program. Genome Res 9: 868–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Wang J, Aluru S, Yang SP, Hillier L (2003) PCAP: a whole-genome assembly program. Genome Res 13: 2164–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyanaraman A, Aluru S, Kothari S, Brendel V (2003) Efficient clustering of large EST data sets on parallel computers. Nucleic Acids Res 31: 2963–2974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Tsai J, Sunkara S, Karamycheva S, Pertea G, Sultana R, Antonescu V, Chan A, Cheung F, Quackenbush J (2005) The TIGR Gene Indices: clustering and assembling EST and known genes and integration with eukaryotic genomes. Nucleic Acids Res 33: D71–D74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Kaul S, Rounsley S, Shea TP, Benito MI, Town CD, Fujii CY, Mason T, Bowman CL, Barnstead M, et al (1999) Sequence and analysis of chromosome 2 of the plant Arabidopsis thaliana. Nature 402: 761–768 [DOI] [PubMed] [Google Scholar]
- Mayer K, Schuller C, Wambutt R, Murphy G, Volckaert G, Pohl T, Dusterhoft A, Stiekema W, Entian KD, Terryn N, et al (1999) Sequence and analysis of chromosome 4 of the plant Arabidopsis thaliana. Nature 402: 769–777 [DOI] [PubMed] [Google Scholar]
- Ouyang S, Buell CR (2004) The TIGR Plant Repeat Databases: a collective resource for the identification of repetitive sequences in plants. Nucleic Acids Res 32: D360–D363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer LE, Rabinowicz PD, O'Shaughnessy AL, Balija VS, Nascimento LU, Dike S, de la Bastide M, Martienssen RA, McCombie WR (2003) Maize genome sequencing by methylation filtration. Science 302: 2115–2117 [DOI] [PubMed] [Google Scholar]
- Salanoubat M, Lemcke K, Rieger M, Ansorge W, Unseld M, Fartmann B, Valle G, Blocker H, Perez-Alonso M, Obermaier B (2000) Sequence and analysis of chromosome 3 of the plant Arabidopsis thaliana. Nature 408: 820–822 [DOI] [PubMed] [Google Scholar]
- Salzberg SL, Dunning Hotopp JC, Delcher AL, Pop M, Smith DR, Eisen MB, Nelson WC (2005) Serendipitous discovery of Wolbachia genomes in multiple Drosophila species. Genome Biol 6: 402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlueter SD, Dong Q, Brendel V (2003) GeneSeqer@PlantGDB: gene structure prediction in plant genomes. Nucleic Acids Res 31: 3597–3600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlueter SD, Wilkerson MD, Huala E, Rhee SY, Brendel V (2005) Community-based gene structure annotation for the Arabidopsis thaliana genome. Trends Plant Sci 10: 9–14 [DOI] [PubMed] [Google Scholar]
- Usuka J, Brendel V (2000) Gene structure prediction by spliced alignment of genomic DNA with protein sequences: increased accuracy by differential splice site scoring. J Mol Biol 297: 1075–1085 [DOI] [PubMed] [Google Scholar]
- Whitelaw CA, Barbazuk WB, Pertea G, Chan AP, Cheung F, Lee Y, Zheng L, van Heeringen S, Karamycheva S, Bennetzen JL, et al (2003) Enrichment of gene-coding sequences in maize by genome filtration. Science 302: 2118–2120 [DOI] [PubMed] [Google Scholar]
- Yu J, Hu S, Wang J, Wong GK, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X, et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296: 79–92 [DOI] [PubMed] [Google Scholar]
- Yuan Y, SanMiguel PJ, Bennetzen JL (2003) High-Cot sequence analysis of the maize genome. Plant J 34: 249–255 [DOI] [PubMed] [Google Scholar]
- Zhu W, Schlueter SD, Brendel V (2003) Refined annotation of the Arabidopsis genome by complete EST mapping. Plant Physiol 132: 469–484 [DOI] [PMC free article] [PubMed] [Google Scholar]