Abstract
The extensive germplasm resource collections that are now available for major crop plants and their wild relatives will increasingly provide valuable biological and bioinformatics resources for plant physiologists and geneticists to dissect the molecular basis of key traits and to develop highly adapted plant material to sustain future breeding programs. A key to the efficient deployment of these resources is the development of information systems that will enable the collection and storage of biological information for these plant lines to be integrated with the molecular information that is now becoming available through the use of high-throughput genomics and post-genomics technologies. The GERMINATE database has been designed to hold a diverse variety of data types, ranging from molecular to phenotypic, and to allow querying between such data for any plant species. Data are stored in GERMINATE in a technology-independent manner, such that new technologies can be accommodated in the database as they emerge, without modification of the underlying schema. Users can access data in GERMINATE databases either via a lightweight Perl-CGI Web interface or by the more complex Genomic Diversity and Phenotype Connection software. GERMINATE is released under the GNU General Public License and is available at http://germinate.scri.sari.ac.uk/germinate/.
Since the 1960s, successful worldwide initiatives have been developed to establish and maintain genetic resource collections of the world's major crop species and their close relatives (Marshall and Brown, 1975; Williams, 1984). These collections are the repository of millions of years of natural selection and contain the genetic diversity necessary for plant breeding efforts to cope with the recurring pressure of pathogen evolution and global changes in climate and soil. Such collections typically contain thousands of plant samples per species, usually termed accessions, and in some cases contain more than 100,000 distinct lines or accessions (Chang et al., 1989; Williams, 1989; Hoisington et al., 1999). The concept of an accession can vary between both communities and institutions. Within the genetic resources community, an accession is most commonly considered a distinct germplasm sample that is maintained in a collection (Sackville Hamilton et al., 2002). The composition of the germplasm sample can be narrow or wide and ranges from a distinct inbreeding line to a population. The unit of management in a collection depends partly on the species of the sample and the collection in which it is maintained.
Extensive documentation systems have been put in place to maintain and allow the use of these collections efficiently in plant breeding programs worldwide. These are currently evolving to incorporate developments in information management, such as the use of formal ontologies (http://www.plantontology.org/; Mouder and Stoner, 1989; Plant Ontology Consortium, 2002; Stein et al., 2004). The genetic resources community has developed a standard set of descriptive information to be recorded for any new line or accession, known as “passport” information (Williams, 1984). Passport information itself is evolving to make use of new technologies. For example, the use of global positioning satellite systems by plant collectors has made available precise geographic location information for new collections, which in turn means that climatic and edaphic information can be more precisely associated with genotypic and phenotypic information for a given plant line.
There is now a move toward increasing the quality of the information available for germplasm collections through a systematic approach to documentation. This includes the development of concepts and procedures for efficient GenBank management, such as reducing the number of duplicate accessions and establishing representative “core collections” (Engels and Visser, 2003). The goal of these efforts is the efficient management and utilization of the resources by plant breeding programs in both developed and developing countries. A key element in this strategy has been the development of high-throughput molecular technologies, which is enabling scientists to describe the genotypes of a significant proportion of accessions from genetic resource collections in addition to the conventional evaluation trials that describe phenotypes (Milbourne et al., 1998). An example of a large-scale application of this approach is the Generation Challenge Program, an international effort to use molecular biology and the rich stocks of genetic resources available for many major crop species to provide a new generation of crops to meet the needs of resource-poor people around the globe (http://www.generationcp.org).
The developments in molecular genotyping have passed though a number of phases as the genotyping technologies have become increasingly more sophisticated and higher throughput, contributing to the quantity and quality of data associated with plant genetic resources. Technology has ranged from protein polymorphism data based on isozymes in the 1960s to DNA markers such as restriction fragment length polymorphisms (Kochert, 1991; Brettschneider, 1998) and random amplified polymorphic DNA (Powell et al., 1995; Edwards, 1998) in the 1980s and 1990s and in the last decade, to the advent of amplified fragment length polymorphisms (Vos et al., 1995; Matthes et al., 1998), simple sequence repeats (SSRs; Ciofi et al., 1998; Li et al., 2000), and single nucleotide polymorphisms (SNPs; Wang et al., 1998). The rapid increase in the number of expressed sequence tag sequences available for many important crop plants has provided the information required to design and deploy functional SNP markers in known genes of many major crops, and many SNP development programs are currently under way. In parallel, a new generation of genotyping technologies based on arrays and other formats has evolved to score many thousands of data points in a single experiment, such as retrotransposon-based insertion polymorphisms (RBIPs; Flavell et al., 1998) and DarT (Jaccoud et al., 2001). These new technologies have been matched by high-throughput methods for DNA extraction (Aharoni and Vorst, 2002; Schnable et al., 2004), resulting in large-scale genotyping of plant lines being routinely possible.
A major challenge for plant genetics is how to both integrate and analyze this rapidly accumulating volume of information and concurrently cope with the rapid development of new technologies for genotyping and phenotyping. A key component of this process is the formal recording and storage of plant genotype information in a database or data warehouse context. Currently deployed database systems address the storage and management of germplasm and genetic resource collections (Knupffer, 1990; Bruskiewich et al., 2003; http://www.ars-grin.gov/), interspecies comparative genomics (Ware et al., 2002; Matthews et al., 2003; Sanchez-Villeda et al., 2003), and specific marker technologies (Lawrence et al., 2004; Warburton et al., 2004a, 2004b). While each of these is very useful in its own sphere, these databases have limitations that restrict their versatility for dealing with modern germplasm collection data. For example, they do not address the storing of genotype and phenotype data in a generic manner that can accommodate rapidly evolving technologies. Furthermore, existing databases are not flexible enough to deal with the wide variety of genetic systems exhibited by plants, such as breeding system (inbreeding, outbreeding) or ploidy levels. To address this issue more generically, a more comprehensive approach to the description of plant genotype information is needed that allows for a broad range of marker technologies in the very diverse genetic systems found in plants, including polyploidy and varying levels of inbreeding.
We present here the GERMINATE database, which aims to describe genetic and phenotypic information generically and can be used for any type of genetic system. Our aim is to fill a gap that currently exists in the community. To achieve this goal, we have based our database design around the elemental concept of a generic marker and its different forms or alleles. This can then be translated both to and from marker technologies and genetic systems by the use of structured metadata, allowing for the storage of genotype information without prior knowledge of either the technology or genetic system.
RESULTS
Database Design
Overview
The primary goal of GERMINATE is to develop a robust database that may be used for storage and retrieval of a wide variety of data types for a broad range of plant species. GERMINATE focuses on genotype, phenotype, and passport data, but has been designed to potentially handle a much larger range of data. We have aimed to provide a versatile database structure that can be simple, require little maintenance, may be run on a desktop computer, and yet has the potential to be scaled to a large, well-curated database running on a server. The design of GERMINATE provides a generic database framework from which interfaces ranging from simple to complex may be used as a gateway to the data.
PostgreSQL (http://www.postgresql.org/) was selected for development of the GERMINATE database because it is relatively quick when used for small databases and lightweight interfaces, but is extremely scalable and can be used for large databases with complex interfaces. In addition, PostgreSQL is distributed under the BSD license (based on the Berkeley Software Distribution license; see http://www.postgresql.org/license.html), which allows GERMINATE to be released within an open-source project. This is particularly important for the worldwide genetic resource community, which includes many institutions with limited financial resources. Furthermore, releasing GERMINATE in an open-source project will enable others to develop tools and interfaces to the database, thus enhancing its versatility. The final reason for adopting PostgreSQL is the fact that it allows strict constraints, triggers, and foreign key relationships on objects in the database allowing data integrity checks on data loaded into the database, whereas many alternative open-source database languages do not fully implement these features.
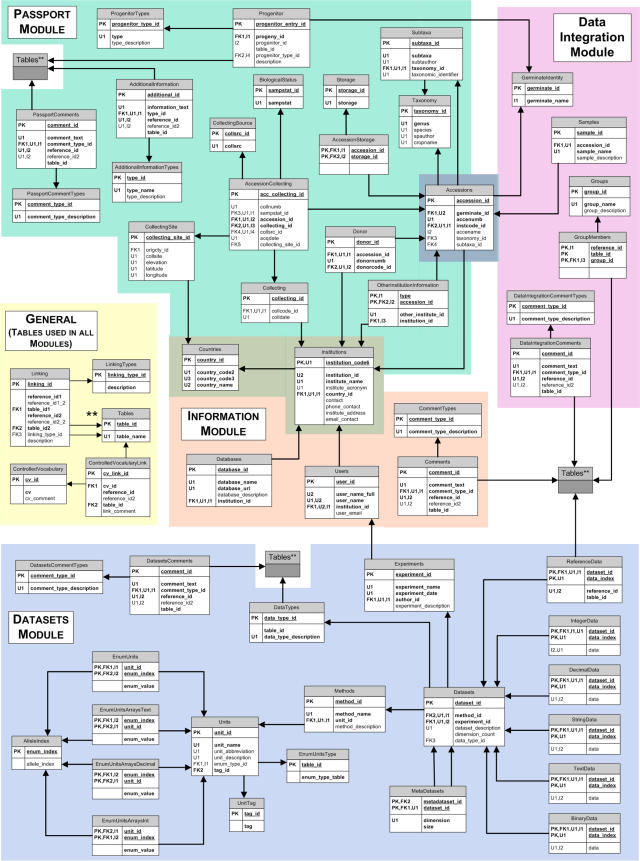
The GERMINATE database is currently divided into four modules, Data Integration, Information, Passport Data, and Datasets, and a set of general tables used by all modules (Fig. 1). Expansion, modification, and addition of modules may be implemented as necessary.
Figure 1.
Graphic of the GERMINATE schema. Codes: PK, Primary Key; U, Unique index; I, Index; FK, Foreign Key (the numbers following indicate if multiple columns are a part of the same index or key). The four modules and 54 tables are depicted.
Data Integration Module
This module is designed to store information on the nature of the plant samples used for data collection. Data collection in plant genetics varies dramatically with respect to the plant or plants sampled. In some cases, data are associated with a single plant or tissue, pooled plants, an accession (typically one or a limited number of plants grown up from a seed bag), or pooled accessions. Sometimes, the data collected will be at the population level, for example, when obligate outbreeders are being studied.
The Data Integration Module accommodates these various approaches to data collection used in the plant genetic resources community and allows a single generic method for handling the different data types (Fig. 1). This is achieved by making the association of the data with a database entry, which can be an accession, sample, GerminateIdentity, or a group of any of these. GerminateIdentity entries are used to link data to germplasm that have not yet been assigned an identifying number in a collection. Germplasm can be assigned a GerminateIdentity number without requiring information about the germplasm to be entered into any other tables in the database, such as the Accessions table. Accessions are a subtype of GerminateIdentity entries that are recognized in GERMINATE by an identifying number, together with an institution code. Samples can represent multiple entries derived from a single accession, such as individual plants or different parts of a particular plant (e.g. roots or leaves). Groups can be pools of Accessions, Samples, or GerminateIdentity entries or groups of progenitors for tracking ancestral information. Every object inserted into the database is assigned a database ID in the table used for that type of object (e.g. Accession or Sample). All data are then linked to the accession or other entity by the database ID and table ID. To ensure data integrity, a trigger in the database checks that the ID is present in the proper table when the data are entered. As all data in GERMINATE are linked in this manner, this design provides the framework for linking between varied data types and datasets in a single analysis. The key to analysis will then be the tools and interface used to access the data.
The concept of accessions, samples, and groups can vary greatly between institutions and individuals and can therefore be defined within the context of the database. Users implementing local copies of GERMINATE will be able to define these terms to suit their needs; however, community standards are expected to be defined for commonly used terms and incorporated into the database.
Passport Module
This module stores descriptive information about the origin, ancestry, and species of an accession, collectively known as passport data. An important component of passport data is the multi-crop passport descriptors (MCPDs), which are standards developed jointly by the Food and Agriculture Organization and the International Plant Genetic Resource Institute (Alercia et al., 2001; http://www.ipgri.cgiar.org/). The MCPDs are considered to be the lowest common denominator between plants and are a widely recognized standard for most crop species. They are also likely to be the most accessed descriptors for the accessions in the database. The Passport Module holds the MCPDs with extensions to these standards to improve their generality (Fig. 1). Since the MCPDs are not an exhaustive list of all possible descriptors, the GERMINATE database includes an AdditionalInformation table to accommodate any additional descriptors that users may want to hold in the database (Fig. 1). This table makes the module broadly applicable and capable of describing species and individuals within different contexts. We are currently working with plant breeders and geneticists to formalize how accessions are described and to ensure the data structure is equally applicable to the genetic resources, breeding, and genetics communities.
The majority of passport descriptors are divided into related groups that are reflected in the table structure of the Passport Module. Each descriptor is assigned to a field in the appropriate table in the Passport Module (Fig. 1). For ease of tracking, the descriptor names used in the MCPDs list are used as the field names in tables. For example, the Taxonomy table includes descriptors genus, species, spauthor (species authority), and cropname (Fig. 1). Certain passport descriptors are associated in this fashion as multiple accessions frequently share the same sets of descriptors, which will often be requested as a group in queries, and not all descriptors will be defined for any one accession. This associative approach simplifies and accelerates access to the passport descriptors, compared to the overcomplicated approach of accessing each descriptor individually by its association with an accession or the oversimplified approach of storing them all in a single table that would then contain many undefined fields. In addition, the database is streamlined by avoiding multiple database entries of descriptors duplicated between accessions (such as genus and species). Our approach follows common normalization guides for databases. The groups of descriptors in a table are then linked to accessions by associating an ID for the table entry with the accession ID.
The values of several passport descriptors are constrained by public standards. For example, the collecting or acquisition source (collsrc) uses a controlled vocabulary for the location where the accession was collected, such as wild habitat, farm, market, or institute. Each of these permitted values is assigned a number, defined in the MCPDs list, and this number is submitted as the value of the collsrc descriptor. GERMINATE adheres to these standards by limiting entries for these descriptor fields, using checks, triggers, or foreign keys to a look-up table that holds all current values. Look-up tables are used whenever possible, as they permit easy addition of new values as public and database standards evolve.
Passport data also includes geographical information about sites where accessions were collected. GERMINATE uses a data format for geographical data that is consistent with formats used by many Geographic Information System (GIS) programs. We have been working with the developers of DIVA-GIS (Hijmans et al., 2001; http://diva.riu.cip.cgiar.org/index.php) to ensure GERMINATE can be integrated with DIVA-GIS to display GIS information. Although other programs can display GIS data, DIVA is a key open-source GIS program in the genetic resources community and will be the default for displaying GIS data in GERMINATE.
Datasets Module
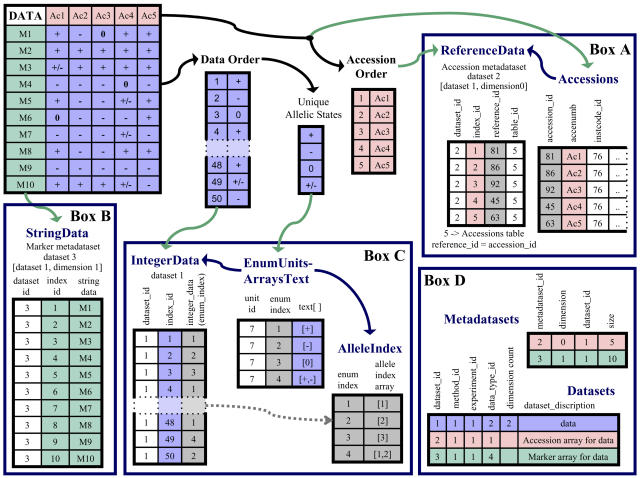
The Datasets Module stores genotype, phenotype, and trait data for the accessions in the database. An example of how molecular marker data are stored is shown in Figure 2. GERMINATE can accommodate integer, decimal, short and long text, and binary (large object) data in addition to array types for text, integer, and decimal types. Array types are a list of data stored in a single field in a table. Arrays permit easy access to any number of alleles per locus or marker for an accession. The arrays could also be used to store a haplotype for an accession across a set of loci. These arrays are assigned an integer ID, which is then stored in the IntegerData table because searching integers rather than text will speed up queries for large datasets. The array types are also able to accommodate datasets for plants of any ploidy level. In autopolyploids where multivalents still form and more than two alleles are inherited in an individual at a particular locus, there is no upper limit on the number of alleles for a loci that can be stored in the database. Alternatively, in allopolyploids or diploidized polyploids where homoeologous loci segregate independently, the distinct allele set at each homoeologous locus would be stored in the database, and these homoeologous loci can be linked together using the Linking table (described in the “Database Information” section). The chromosomal assignment of each homoeologous locus is pertinent for analysis on such loci, and this information is stored in a separate dataset that can then be linked back to the dataset that stores the alleles for the individual accessions at a particular locus. This is a particular advantage of the GERMINATE database structure. Furthermore, the ability to store multiple alleles for a specific marker/sample combination also means GERMINATE can store datasets derived from pooled individual plants from a population. Many databases deal with multiple alleles at a locus by storing all the allele values as a text string. This slows down access and queries on the allele values since it is necessary to decode the text string.
Figure 2.
Genetic dataset loading example. The Data table represents a sample of how molecular marker data are typically submitted: a set of markers analyzed in a set of accessions. The arrows in the figure show flow of information as it is inserted into the database. Black arrows indicate data are being held temporarily, green indicates the insertion to the database, and blue that data already inserted are being used to insert information into another table. In the latter case, IDs assigned by the database are used to trace back to the original data. The colors in the tables follow the dataset and metadatasets through the process of being inserted into the database. The peach color denotes the Accession metadataset, green denotes the Marker metadataset, and purple denotes the allele data. Box A represents the Accession data and metadata inserted into GERMINATE. On entry, each accession is assigned an accession_id that is unique in the database, and this ID is used to reference the appropriate accession in the accession metadataset. The order or number of accession_ids has no influence on the order of accessions in the metadataset. The ReferenceData table uses a data index to track the correct order of the accession_ids. Box B indicates where the marker information is inserted into the database, again retaining the order in the original dataset by the data index value. Box C demonstrates how the allelic state of the accession by marker is translated into an integer ID (enum_index). This ID is stored in appropriate order in the IntegerData table. The enum_index can then be used to translate back to the actual allele value or to an allele index if only the relative allele states between accessions are required in a query. The AlleleIndex table was created to speed up queries where technology is unimportant and the relative allele values will suffice to answer the question. Box D displays the metadata information recorded in the database required to recreate the dataset. This includes the number of dimensions for a dataset and relates the metadatasets to the dataset.
The Datasets Module is metadata driven; for a given dataset, any number of metadatasets can be associated with it. The metadatasets store the information to describe the data values. For example, in a genotyping dataset where the alleles are the data values, the metadatasets would describe the markers and accessions in which they were evaluated. The metadatasets are associated to their dataset by linking the dataset_id field to the metadataset_id fields in the Metadatasets table (Figs. 1 and 2). The data values for a dataset can then be associated with any other object in the database (such as Accession) using the ReferenceData table. These relationships are described in more detail below. The metadataset also has its own dataset entry with associated data and can have its own metadatasets if required, allowing the flexibility for storage of any level of complexity of data. Additional metadata information, such as details about the experiment, method, and markers used to generate the data, are associated to each dataset via an ID.
A variety of data storage methods are currently being implemented in the GERMINATE database. Two of the more common methods are described here (see Fig. 1 for details regarding the relationships between tables). Trait or phenotypic data are often recorded as observations of a trait in a defined context for a set of accessions. The trait data are stored in the appropriate data table, depending on the type of data submitted (e.g. integer type would be used for physical dimensions of a plant, or string type for flower color). The dataset ID indicates which dataset the information belongs to, and a data index keeps track of the order of data for the dataset. The accessions associated with the trait data are the only metadataset for the entry. The accession IDs are stored in the ReferenceData table in the same data index order as the trait values and associated with the accession dataset ID. The Metadatasets table then holds the information to link the metadataset (the accession dataset) to the dataset (the trait values) when the data are requested.
Genotyping data is another common data type submitted to GERMINATE. A typical dataset consists of a set of markers evaluated in a set of accessions (Fig. 2). For this type of data, a two-dimensional array is stored in the database, where the set of markers and the set of accessions are the two metadataset dimensions for the dataset. The primary dataset would be the allele values of each marker in each accession. Similarly to the trait data above, each dataset and metadataset is entered into the appropriate data table associated with its dataset ID, and a data index keeps track of the order of each set of data. The Metadatasets table tracks the dimensions associated with the primary dataset. To keep the database normalized, only a single instance of each marker and accession for a dataset is stored. Therefore, to recreate the dataset, a two-dimensional grid is set up with accessions in one dimension and markers in the other. The data are then iterated through to place them in their correct positions on the grid. If a user is only interested in a single data point, it can be easily accessed by a simple equation that relates the accession, marker, and data indices to each other. Alternatively, the user could employ a similar equation to access all allele values for a subset of markers or accessions. Additional metadatasets can be associated with the allele datasets or marker and accession metadatasets if required. For example, if the markers are SSR, a metadataset of quality score for the SSR can be defined that would have the same number of entries as the marker metadataset, or, if the allele values are sequences that require precisely defined start and end points, start and end sequence metadatasets can be added that would use the same index values as the allele dataset. The relationships of metadatasets to their primary datasets must then be defined within the database so that the dataset and with all of its metadatasets can be recreated.
Either of the two methods described above can also use one of the EnumUnits tables to store data (Fig. 1). These tables offer several advantages. For large datasets where the data are in text format but there are a limited number of possible options, the data could be translated to an integer index and the retrieval of data could make use of comparing integers rather than searching text fields, which could significantly speed up queries. These tables also make use of array types and are used if multiple alleles are possible for an accession. These tables translate the combination of alleles to a single integer index that is stored in the IntegerData table (Figs. 1 and 2). The individual allele values will still be easily searchable.
GERMINATE utilizes an AlleleIndex table (Figs. 1 and 2) to simplify and accelerate certain types of queries, such as those requiring only information for the relative allele values. The allele index indicates the relative alleles for a marker between accessions and whether the accessions are homozygous or heterozygous. When querying using the allele index, the user need not retrieve information about the units, methods, experiments, or actual allele values. This significantly speeds up some queries and does not generate any additional overhead for queries not using the allele index.
Information Module
The Information Module stores information on institution, country, and user, which does not need to be associated with an accession. This includes tables for institutions and countries, which are directly accessed via an ID from the Passport Module. A Users table is also contained within this module, to keep track of authors of data from the Datasets Module. Finally, a Databases table keeps links to any external databases that users may wish to access.
Database Information
This section includes information about features and tables contained in all the modules of the database. Each GERMINATE database module contains its own set of tables for comments and comment types. Comments on any aspect of any data item can be added, for example, remarks upon the performance of a particular microsatellite marker, suggested PCR conditions for an RBIP marker, clarification of a particular geographical location from which some accessions were collected, etc. A primary key ID and table ID combination indicates the database entry with which the remark is associated. A comment type (for example, if the remark is for a date or country) is also associated with the entry for ease of searching the tables. These tables in the Datasets Module can hold information such as that relevant to markers, for instance, sequences and PCR conditions. Having separate comment and comment type tables for each module allows their structures to be tailored to the needs of the specific module. The distribution of the comments tables to each module helps to keep the sizes of these tables under control and allows faster access. In addition, developers wishing to extract a single module from the GERMINATE database will encounter fewer problems if the modules are largely self-contained.
There are a few tables in the database shared by all modules. The first is the Tables table, which holds the names of all tables in the database associated with an ID, which is used as a reference for comments and reference tables. The GERMINATE database also holds a Linking table; this allows users to relate any entry in any table to any other entry in the same or different table and by any relationship. This may be used, for example, to associate a marker from one genetic map with a marker in another map, to indicate they are in fact the same marker, or to link accessions together if they have been discovered to be the same accession or to be related in some way.
Data Loading and Curation in GERMINATE
Loading and curation of information is crucial to proper functioning of any database. Concepts of referential integrity (ensuring invalid or inconsistent data is not entered or maintained in a database) and how to deal with duplicate entries or spelling errors must be methodically dealt with. GERMINATE is very flexible and efficient with regard to disparate data types, and relies upon breaking down the input data into its components and storing these in separate fields. Figure 2 illustrates a single example of how molecular marker data are loaded into GERMINATE, and further examples of data loading are provided and at our Web site (http://germinate.scri.sari.ac.uk/germinate/tutorial.html).
There are currently two options for loading data into the GERMINATE database. The first uses a combination of Perl and SQL scripts to extract data directly from Excel spreadsheets that follow the formats specified at the GERMINATE Web site (http://germinate.scri.sari.ac.uk/germinate/guidelines.html). There are different Perl scripts for use with different data types (e.g. genetic map data, phenotypic trait data, genotype data for multiple markers evaluated in multiple accessions). The output of each of these scripts is copied into a temporary table by a SQL script written in PostgreSQL. Similarly, there are separate SQL scripts for the various data types. Each of these SQL scripts has a set of variables to be set before the data can be loaded into the database. The SQL script then uses a series of pl/pgsql functions to check the data and load it into the appropriate tables in the database. The temporary table used to hold the data initially is then removed. Upon entry, the data are checked for referential integrity and accuracy where possible. Not all aspects of the data can be checked in an automated fashion, and manual curation will be required to check some of the details of the data.
The second option available is a germinate loader, available through CropForge (http://cropforge.irri.org/projects/germinateloader/). This is a Java interface to the command line scripts, which allow users to set the variables without altering the SQL script. Many users will find this alternate more user friendly.
No matter which way users opt to load data into the database, the issues of curation and data integrity need to be considered. At the database level, we have made every attempt to use triggers, functions, and foreign keys to automatically screen for referential integrity while the data are being loaded. For example, triggers check that an accession has a genus and species entry before a subspecies may be added, and that tables contain the ID for reference tables. Foreign keys are used to look up tables, such as the collecting source table for passport information. An entry that is not currently in the table cannot be used. Additionally, functions are used to check if data, such as an accession entry, have already been entered into the database, thus preventing duplicate entries.
In many cases, however, the database alone cannot check for the accuracy of data. The data must also be curated by the user to check, for example, for spelling errors and decide if two similar accession names are in fact the same. A more user-friendly Web-based loading and curation interface is currently being designed that will give users without any bioinformatics experience the flexibility to load and curate data. Information on this will be posted at the GERMINATE Web site as it becomes available. We intend to deploy a curation tool similar to that used by MaizeGDB (Lawrence et al., 2004), which gives users variable levels of permission, limiting what they are allowed to do in the database. All users will be able to view data, but loading and curation will require special permissions to prevent unwarranted alterations.
Interfaces to GERMINATE
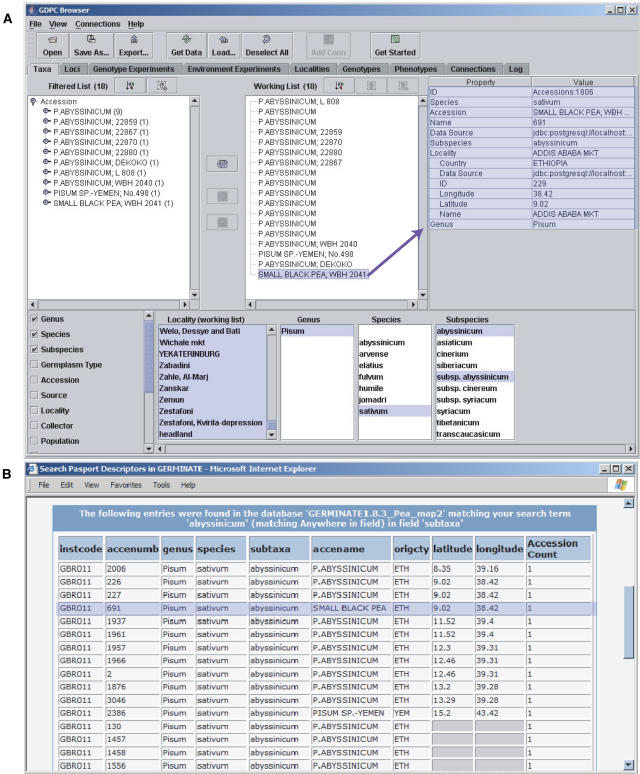
The main feature of user interaction with the GERMINATE database is the capability to link different types of data by association through accessions. This permits users to interact with multiple types of data in the same query and enables them to ask complex questions to the database. For example, a user could find all accessions that both share a particular allele for a given marker and that were collected from a specific geographical region. The complexity of queries allowed to the database will largely depend on the interface used to access the database. Users experienced in SQL could optionally interact with the database at the SQL level to construct any complexity of queries required for their work. Alternatively, other, more user-friendly interfaces are available. A light-weight Perl-CGI interface has been constructed for simple queries, and a more complex Java interface, the Genomic Diversity and Phenotype Connection (GDPC; Casstevens and Buckler, 2004; http://maizegenetics.net/gdpc/), has been connected to GERMINATE. Images of these interfaces are shown in Figure 3.
Figure 3.
Example of GDPC and the GERMINATE Perl-CGI interfaces returning information about Pisum sativum subspecies abyssinicum. A, The GDPC interface showing a taxa query that has retrieved accessions that are from P. sativum subspecies abyssinicum and that have a source geographical location. The properties shown are for accession number 691, Small Black Pea from Ethiopia, one of the accessions returned. B, The Perl-CGI interface showing a similar query. The passport descriptor subtaxa have been searched for abyssinicum; taxonomy information, some location information, accession name and number, and institution code have been returned. The same accession (691) highlighted in A is highlighted here for comparison.
The Perl-CGI interface is a Web-based user-friendly interface that allows users to perform tasks such as searching passport descriptors, accessions, markers, institutions, and species, retrieving all information available for a selected accession or retrieving datasets. This interface was designed primarily for retrieving passport data, but additional functionality was added as the need arose.
GDPC is a general-purpose Java-based interface that publishes data as Web services. Web services are well-defined objects that are published to the network using standard protocols that can be then used by other programs. One advantage of Web services over direct connections of tools to databases is that Web services are firewall friendly and can therefore be more widely used. In addition, multiple data sources can be configured to publish the same Web services, which can then be used by multiple programs. GDPC makes the data in a GERMINATE database available as a Web service by transferring XML-formatted data via standard Simple Object Access Protocol, which allows applications to exchange information in a platform- and language-independent manner over the Internet.
GDPC offers several useful features as an interface. In particular, it can connect to multiple databases simultaneously and permits cross-linking of the data therein. Using GDPC will expand the number of analysis and visualization tools that may be used with GERMINATE. Any programs that recognize GDPC Web services objects are able to access GERMINATE databases. This has the added advantage that tools made GDPC aware for use with other databases are also accessible to GERMINATE databases. A few GDPC-aware tools are already available and more will follow. One currently available tool, the GDPC browser, can be used to view data in the database, retrieve data based on property values, save sets of data as XML files that may be accessed later, and export data in certain formats (Casstevens and Buckler, 2004). TASSEL is also GDPC aware and can be used for analysis of trait associations, evolutionary patterns, and linkage disequilibrium (http://maizegenetics.net/bioinformatics/tasselindex.htm). Linkage disequilibrium analysis can be used to dissect complex traits, which is useful in the understanding of genomes (Flint-Garcia et al., 2003). Having TASSEL and other tools available for use with GERMINATE will make the database more useful to researchers.
The GERMINATE public databases are accessible via the Web-based Perl-CGI interface at http://germinate.scri.sari.ac.uk/germinate/. GDPC can also be used with the public databases on the GERMINATE server; instructions on how to connect to the public GERMINATE databases via GDPC can be found on the GERMINATE Web site. Although GDPC and Java must be installed on the user's computer, the installation process is relatively simple, and the use of the software does not require informatics experience. Tutorials to get users started using either of these interfaces can be found at http://germinate.scri.sari.ac.uk/germinate/tutorial.html. The SQL scripts to build a local version of GERMINATE can also be downloaded at the GERMINATE Web site along with scripts to load various formats of data including passport, genetic data, trait data, and genetic maps. PostgreSQL is a prerequisite to creating a local GERMINATE database and is an open-source program that can be installed on any computer platform.
The GERMINATE database has to date been tested with pea, wheat, barley, and lettuce data at the partner institutions with various genetic marker data, genetic mapping data, and the ability to link between these data types. We have also tested the loading and retrieval of passport and phenotype data and querying via accessions between all the various data types. GERMINATE has now been distributed to interested collaborators for testing in a broader range of species with a wider variety of data. Collaborators include members of the Generation Challenge Program Consortium (http://www.generationcp.org/), who are testing GERMINATE for use with other species, including rice, potato, and maize.
Information Retrieval Example
The following example demonstrates the capabilities of the Perl-CGI for retrieving information from the GERMINATE database. An accompanying PowerPoint file is available as supplemental information online (http://germinate.scri.sari.ac.uk/germinate/tutorial.html). Another tutorial describing the use of the GDPC Browser for accessing data in GERMINATE (not described here) is also provided as supplemental information (http://germinate.scri.sari.ac.uk/germinate/tutorial.html). Figure 3 shows an example of each of these interfaces.
The public pea database can be accessed by navigating from the GERMINATE Web site (http://germinate.scri.sari.ac.uk/germinate/interface.html). The user can retrieve data from a genetic dataset and link to information about the accessions used in the experiment and information about the experiment, including original images used to score the data. The user can browse datasets with the “Browse Datasets in a Database” link. Selecting the pea public database (GERMINATE_Pea_Example_database) will return a list of all datasets in the database. Individual datasets describing separate experiments can then be retrieved. For this example, the dataset “281 × 44 for marker type RBIP” is followed in the tutorial, which includes data for a set of approximately 3,000 accessions evaluated by microarray for a single RBIP marker. This link retrieves information on the experiment and method used (marker method, primer sequences, marker locus sequence, and PCR annealing temperature). In addition, metadatasets are listed. For this example, a metadataset of the accessions used in the experiment is further linked to a metadataset of the spot numbers of the accessions on the microarray. The user may retrieve the entire dataset (including every marker score) or any one of the metadatasets via buttons. When a dataset containing accessions is retrieved, the accession number listed is provided with a link to information about this accession. Similarly, links are provided from the marker to associated information such as map location, sequences, passport data, and other information. Finally, the microarray image used to generate the dataset can be accessed by a button on the dataset information page.
The Perl-CGI interface can also be used for other queries such as retrieving all accessions from a specific geographical location or institute, searching accessions, markers, or species for word(s) or phrase(s), searching a passport descriptor and retrieving a distinct list of any combination of passport descriptors, or retrieving all accessions evaluated for a specific trait.
DISCUSSION
GERMINATE has been designed to be a versatile generic open-source relational database and interface for plant data. The GERMINATE database is intended for use with varying types and amounts of genetic and phenotypic data, but is also flexible enough to potentially store a much wider range of data and could be used outside the plant kingdom with minor modifications. The GERMINATE database is not designed to store large amounts of sequence data, such as complete genomes. Rather, it has been designed to handle multiple data types for many thousands of genetically distinct individuals and to cope with the high-throughput genotyping and phenotyping technologies that are common today. In addition, GERMINATE makes use of recent computer technology advances, such as Web services for the exchange of information between computer applications.
A particular goal of GERMINATE is to manage the higher standards and complexity of data collection that have arisen recently within the genetic resource community. It provides a framework for the implementation of emerging data standards, which includes quality assurance of data, and for the development of structured formats for describing experiments. These are primary requisites to enable the plant genetic resources, breeding, and genetics communities to fully realize the potential of new high-throughput technologies. GERMINATE also provides a platform for analytical and visualization methods through the integration of key datasets.
The method of storing datasets in GERMINATE is flexible and extensible. In particular, the storage of datasets is not designed around the technology used to create the dataset. This feature makes it easy to extend to new technology as it becomes available without altering the structure of the database. The GERMINATE database has also been designed to handle a wide variation in dataset complexity. The flexibility includes the option of having metadatasets for metadatasets, allowing storage of exceptionally complex information. Although the Datasets Module has been designed around the most common types of genotype and phenotype data, it has the flexibility to potentially hold a much broader range of data. This includes image data, which could be stored in the database as large objects or as links to an external image file.
Modularity and Interconnectibility
We expect most users will implement GERMINATE in its entirety. However, the division of the GERMINATE database into modules allows for the possibility that a subset of the database could be used within another database. This is a useful feature, as some user groups have databases specifically designed for certain tasks (such as holding passport information) that they would like to extend by importing modules from other databases, such as using the datasets module from GERMINATE to accommodate genotype data. GERMINATE currently includes four modules: Data Integration, Passport Data, Datasets, and Information. These modules will be extended and modified as needed.
We envisage that GERMINATE will be used in a variety of ways. Institutions with limited resources will be able to implement GERMINATE as a local, easily maintained database, and either the lightweight Perl-CGI interface or GDPC or both may used to interact with the local database. Such databases could be accessible via Web services and will not have to be operated in isolation. Web services may be used to merge data in a local copy of GERMINATE with those in public servers. This feature would be particularly useful to merge data within a local private copy, which contains data not yet public, with data in public databases, without publishing the local data as Web services. Alternatively, users experienced in SQL may wish to employ the powerful features of SQL to create complex queries across different datasets.
Computer Platforms
In principle, PostgreSQL allows GERMINATE to be set up on any computer platform. This was our intention from the start so that institutions with limited resources will not need to invest in new equipment to implement GERMINATE. We have compared a variety of platforms, including desktop computers running Windows or Linux, Macintosh desktops running OSX, and a Sun workstation. We ran a standard set of benchmarks on GERMINATE databases across a range of platforms. As a point of reference, the average query speed for selecting all accessions evaluated for a particular trait from the public pea database (comprising approximately 4,500 accessions and more than 300 datasets) using the Dell Optiplex machine is about 60 milliseconds. More complex queries, such as selecting a range of passport descriptors for a selection of accessions, take on average between 1 and 3 s. We find that performance of PostgreSQL and GERMINATE varies between platforms (Table I).
Table I.
Comparison tests of computers running PostgreSQL and GERMINATE
*, Average Relative Query Speed is relative the Dell Optiplex machine and was determined by taking the average speed of a set of standard queries. See text.
| Machine | Operating System | Average Relative Query Speed* |
|---|---|---|
| SUN SunFire V880, dual Sparc processors, 4 GB RAM | Solaris 9 | 20 |
| Dell PowerEdge 1750, single 3.0 GHz Intel Xeon processor, 512 MB RAM | Fedora Core 2 | 1 |
| Dell Optiplex GX260, 2.8 GHz, 1 GB RAM | Windows XP | 1 |
| Dell Latitude C640, 2.0 GHz with 512 MB RAM | Windows XP | 1 |
| Apple PowerPC G4, 1.25 GHz with 1.25 GB RAM | Mac OSX 10.3.8 | 2 |
Datasets Tested
We have so far tested the database with a variety of data types and dataset sizes. Actual datasets loaded into the database have ranged in size from around 50 accessions evaluated with around 20 markers to around 3,000 accessions evaluated with 15 markers. In addition, we have tested the database with 26 randomly generated datasets of 3,000 accessions evaluated with 100 markers each.
We have also compared loading of small datasets into empty databases against loading into databases already containing large datasets, and find little change in the loading and retrieval times. A wide variety of data types have been tested in GERMINATE, including single locus codominant marker assays on 3,000 samples and hundreds of multiplex SSR and amplified fragment length polymorphism markers evaluated in multiple accessions. We have also tested the ability of the database to hold genetic mapping data, including multiple genetic maps associated with a single set of data. Dr. King's group at Rothamsted Research is also testing GERMINATE for use with genetic maps in Brassica. Furthermore, phenotypic trait datasets and large passport datasets (approximately 10,000 accessions) have been loaded into GERMINATE for testing. Some of the passport datasets have included descriptors not in the MCPDs, to test implementation of the additional passport information table. In addition, we have tested more complex genotype datasets by adding additional dimensions such as data quality, which can be associated with any data point(s). We have also linked markers from a genotype dataset to their genetic map positions in a genetic map dataset to test our implementation of the Linking table. In all these cases, GERMINATE has performed at a level equal to or greater than that required.
Future Goals
The main goals for the future of GERMINATE are the further development of flexible, user-friendly interfaces to the database, together with the identification or construction of a broad set of analysis and visualization tools to maximize the accessibility and usefulness of the stored data. A Graphical Genotype Tool is currently under development for use with GERMINATE data. This tool will display graphically the distribution of alleles at loci across taxa viewed by genetic linkage map position. As user needs are further defined, additional tools will be developed and connected to the database along with the connection of existing tools that would be useful to researchers. Programs such as DIVA for displaying geographical information and statistical analysis modules written in programming languages such as R are currently being considered for use with GERMINATE. In addition, a user-friendly loading interface will be constructed such that users who are not experienced in programming and SQL will be able to insert datasets into the GERMINATE database easily.
Availability
GERMINATE is freely available under the terms of the GNU general public license (http://germinate.scri.sari.ac.uk/germinate/distribution/). We envisage that this will form an ideal platform to enable us and other groups to develop a range of standard interfaces and analytical tools to interact with the underlying database. In addition, we hope that the future development of GERMINATE will see community and database standards come to realization, and that these will be accepted and expanded upon by those involved with development and utilization of GERMINATE.
Supplementary Material
Acknowledgments
We thank Terry Casstevens and Ed Buckler for the connection to GDPC, Robbie Waugh and Luke Ramsey at SCRI for valuable input, and colleagues from the Consultative Group on International Agricultural Research centers International Potato Center, International Rice Research Institute, International Center for the Improvement of Maize and Wheat, International Network for the Improvement of Banana and Plantain, and International Center for Tropical Agriculture and other members of the Generation Challenge Program for their valuable input. We are also grateful to our colleagues in the EC Framework 5 TEGERM project, particularly Alan Schulman, Albert Grit, She-May Tam, and Marie-Angèle Grandbastien, for providing datasets to test GERMINATE. Lastly, we thank Julie Hofer for comments on the manuscript and the members of the Computational Biology Group at SCRI, particularly Paul Shaw and Linda Milne.
This work was supported by the Biotechnology and Biological Sciences Research Council (grant no. 94/BEP17084) and the Scottish Executive Environment and Rural Affairs Department (grant no. FF00589), as part of the Bioinformatics and E-science program.
The online version of this article contains Web-only data.
References
- Aharoni A, Vorst O (2002) DNA microarrays for functional genomics. Plant Mol Biol 48: 99–118 [DOI] [PubMed] [Google Scholar]
- Alercia A, Diulgheroff S, Metz T (2001) FAO/IPGRI Multi-Crop Passport Descriptors. http://www.ipgri.cgiar.org/publications (June 26, 2003)
- Brettschneider R (1998) RFLP analysis. In A Karp, PG Isaac, DS Ingram, eds, Molecular Tools for Screening Biodiversity. Chapman and Hall, London, pp 83–95
- Bruskiewich RM, Cosico AB, Eusebio W, Portugal AM, Ramos LM, Reyes MT, Sallan MA, Ulat VJ, Wang X, McNally KL, et al (2003) Linking genotype to phenotype: the International Rice Information System (IRIS). Bioinformatics (Suppl 1) 19: i63–i65 [DOI] [PubMed] [Google Scholar]
- Casstevens TM, Buckler ES (2004) GDPC: connecting researchers with multiple integrated data sources. Bioinformatics 20: 2839–2840 [DOI] [PubMed] [Google Scholar]
- Chang T-T, Dietz SM, Westwood MN (1989) Management and use of plant germplasm collections. In L Knutson, AK Stoner, eds, Biotic Diversity and Germplasm Preservation, Global Imperatives. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 127–159
- Ciofi C, Funk SM, Coote T, Cheesman DJ, Hammond RL, Saccheri IJ, Bruford MW (1998) Genotyping and microsatellite markers. In A Karp, PG Isaac, DS Ingram, eds, Molecular Tools for Screening Biodiversity. Chapman and Hall, London, pp 195–201
- Edwards KJ (1998) Randomly amplified polymorphic DNA (RAPDs). In A Karp, PG Isaac, DS Ingram, eds, Molecular Tools for Screening Biodiversity. Chapman and Hall, London, pp 171–175
- Engels JMM, Visser L, editors (2003) A Guide to Effective Management of Germplasm Collections. IPGRI Handbook for Genebanks Number 6. International Plant Genetic Resources Institute, Rome
- Flavell AJ, Knox MR, Pearce SR, Ellis TH (1998) Retrotransposon-based insertion polymorphisms (RBIP) for high throughput marker analysis. Plant J 16: 643–650 [DOI] [PubMed] [Google Scholar]
- Flint-Garcia S, Thornsberry JM, Buckler ES (2003) Structure of linkage disequilibrium in plants. Annu Rev Plant Biol 54: 357–374 [DOI] [PubMed] [Google Scholar]
- Hijmans RJ, Guarino L, Cruz M, Rojas E (2001) Computer tools for spatial analysis of plant genetic resources data: 1. DIVA-GIS. Plant Genet Resour Newsl 127: 15–19 [Google Scholar]
- Hoisington D, Khairallah M, Reeves T, Ribaut J-M, Skovmand B, Taba S, Warburton M (1999) Plant genetic resources: What can they contribute toward increased crop productivity? Proc Natl Acad Sci USA 96: 5937–5943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaccoud D, Peng K, Feinstein D, Kilian A (2001) Diversity arrays: a solid state technology for sequence information independent genotyping. Nucleic Acids Res 29: e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochert G (1991) Restriction fragment length polymorphism in plants and its implications. Subcell Biochem 17: 167–190 [DOI] [PubMed] [Google Scholar]
- Knupffer H (1990) The European barley database of the ECP/GR. Barley Genet Newsl 19: 37–38 [Google Scholar]
- Lawrence CJ, Dong Q, Polacco ML, Seigfried TE, Brendel V (2004) MaizeGDB, the community database for maize genetics and genomics. Nucleic Acids Res 32: D393–D397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YC, Fahima T, Korol AB, Peng J, Roder MS, Kirzhner V, Beiles A, Nevo E (2000) Microsatellite diversity correlated with ecological-edaphic and genetic factors in three microsites of wild emmer wheat in North Israel. Mol Biol Evol 17: 851–862 [DOI] [PubMed] [Google Scholar]
- Marshall DR, Brown AHD (1975) Optimum sampling strategies in genetic conservation. In OH Frankel, JG Hawkes, eds, Crop Genetic Resources for Today and Tomorrow. Cambridge University Press, Cambridge, pp 53–80
- Matthes MC, Daly A, Edwards KJ (1998) Amplified fragment length polymorphism (AFLP). In A Karp, PG Isaac, DS Ingram, eds, Molecular Tools for Screening Biodiversity. Chapman and Hall, London, pp 183–190
- Matthews DE, Carollo VL, Lazo GR, Anderson OD (2003) GrainGenes, the genome database for small-grain crops. Nucleic Acids Res 31: 183–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milbourne D, Russell J, Waugh R (1998) Comparison of molecular marker systems in inbreeding (barley) and outbreeding (potato) species. In A Karp, PG Isaac, DS Ingram, eds, Molecular Tools for Screening Biodiversity. Chapman and Hall, London, pp 371–381
- Mouder JD, Stoner AK (1989) Plant germplasm information systems. In L Knutson, AK Stoner, eds, Biotic Diversity and Germplasm Preservation, Global Imperatives. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 419–426
- Plant Ontology Consortium (2002) The Plant Ontology Consortium and plant ontologies. Comp Funct Genomics 3: 137–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell W, Orozco-Castillo C, Chalmers KJ, Provan J, Waugh R (1995) Polymerase chain reaction-based assays for the characterisation of plant genetic resources. Electrophoresis 16: 1726–1730 [DOI] [PubMed] [Google Scholar]
- Sackville Hamilton NR, Engels JMM, van Hintum TJL, Koo B, Smale M, editors (2002) Accession Management. Combining or Splitting Accessions as a Tool to Improve Germplasm Management Efficiency. IPGRI Technical Bulletin Number 5. International Plant Genetic Resources Institute, Rome
- Sanchez-Villeda H, Schroeder S, Polacco M, McMullen M, Havermann S, Davis G, Vroh-Bi I, Cone K, Sharopova N, Yim Y, et al (2003) Development of an integrated laboratory information management system for the maize mapping project. Bioinformatics 19: 2022–2030 [DOI] [PubMed] [Google Scholar]
- Schnable PS, Hochholdinger F, Nakazono M (2004) Global expression profiling applied to plant development. Curr Opin Plant Biol 7: 50–56 [DOI] [PubMed] [Google Scholar]
- Stein L, McCouch SR, Kellogg E, Rhee SY, Jaiswal P, Stevens P, Ware D, Vincent L, Polacco M, Reiser L, et al (2004) The Plant Ontology Consortium. Plant and Animal Genomes XII Conference. http://www.intl-pag.org/12/abstracts/ (September 12, 2005)
- Vos P, Hogers R, Bleeker M, van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23: 4407–4414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang DG, Fan JB, Siao CJ, Berno A, Young P, Sapolsky R, Ghandour G, Perkins N, Winchester E, Spencer J, et al (1998) Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science 280: 1077–1082 [DOI] [PubMed] [Google Scholar]
- Warburton M, Norgaard J, Lopez C, Alarcon JC (2004. a) CIMMYT Molecular Characterization Data for Wheat (CD-ROM). CIMMYT, Texcoco, Mexico
- Warburton M, Norgaard J, Lopez C, Alarcon JC, George ML, Regalado E (2004. b) CIMMYT Molecular Characterization Data for Maize (CD-ROM). CIMMYT, Texcoco, Mexico
- Ware D, Jaiswal P, Ni J, Pan X, Chang K, Clark K, Teytelman L, Schmidt SC, Zhao W, Cartinhour S, et al (2002) Gramene: a resource for comparative grass genomics. Nucleic Acids Res 30: 103–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JT (1984) A decade of crop genetic resources research. In JHW Holden, JT Williams, eds, Crop Genetic Resources: Conservation and Evaluation. George Allen and Unwin, London, pp 1–17
- Williams JT (1989) Plant germplasm preservation: a global perspective. In L Knutson, AK Stoner, eds, Biotic Diversity and Germplasm Preservation, Global Imperatives. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 81–96
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.