Abstract
ArrayExpress is a public microarray repository founded on the Minimum Information About a Microarray Experiment (MIAME) principles that stores MIAME-compliant gene expression data. Plant-based data sets represent approximately one-quarter of the experiments in ArrayExpress. The majority are based on Arabidopsis (Arabidopsis thaliana); however, there are other data sets based on Triticum aestivum, Hordeum vulgare, and Populus subsp. AtMIAMExpress is an open-source Web-based software application for the submission of Arabidopsis-based microarray data to ArrayExpress. AtMIAMExpress exports data in MAGE-ML format for upload to any MAGE-ML-compliant application, such as J-Express and ArrayExpress. It was designed as a tool for users with minimal bioinformatics expertise, has comprehensive help and user support, and represents a simple solution to meeting the MIAME guidelines for the Arabidopsis community. Plant data are queryable both in ArrayExpress and in the Data Warehouse databases, which support queries based on gene-centric and sample-centric annotation. The AtMIAMExpress submission tool is available at http://www.ebi.ac.uk/at-miamexpress/. The software is open source and is available from http://sourceforge.net/projects/miamexpress/. For information, contact miamexpress@ebi.ac.uk.
Following genome sequencing, DNA microarray technology has become a widespread tool for genome analysis. Microarray technology is currently producing very large quantities of gene expression data, which promise to provide insight into key biological processes such as gene function and interaction. However, unlike genome sequence data, which have standard formats for their presentation and widely available tools and databases for data sharing and comparison, microarray data have until recently lacked such data structuring and standards. It is important to note that gene expression data are inherently more complex than sequence data because the former are meaningful only in the context of the experimental conditions from which they were derived.
These considerations led to the development of the Minimum Information About a Microarray Experiment (MIAME) guidelines to describe a microarray experiment (Brazma et al., 2001). The MIAME guidelines were developed by the Microarray Gene Expression Data (MGED) Society (www.mged.org/Mission/index.html, on www.mged.org; June 1, 2005), a body primarily founded to promote standardization of gene expression experiments and latterly for other technologies. MIAME seeks to capture the minimum information that needs to be provided to interpret and reproduce the study. The Microarray Gene Expression-Object Model (MAGE-OM) and the related XML-format Microarray Gene Expression-Markup Language, or MAGE-ML (Spellman et al., 2002), was developed as an Object Management Group standard for representation of array-related experiments. MAGE-ML has become the standard platform independent microarray data exchange format used for file exchange between software applications and workers in this field.
Here, we describe some of the databases and software tools that have been developed to facilitate data exchange and comparison regarding microarray gene expression data at the European Bioinformatics Institute, and how they are useful for the Arabidopsis (Arabidopsis thaliana) research community. All these tools support the MIAME guidelines and support the standards and recommendations of the MGED Society. The complete suite of tools and databases consists of MIAMExpress, an online data submission tool; ArrayExpress, a public repository of microarray gene expression data; and the Data Warehouse, which contains public curated normalized data that supports gene-centric queries. Complementing these resources is Expression Profiler, a Web-based software tool for microarray gene expression and sequence data analysis. Here, for the purposes of this article, we will describe ArrayExpress and AtMIAMExpress.
AtMIAMExpress
Microarray experiments consist of an interrelated study made up of samples, array designs, and data files. MIAMExpress (M. Shojatalab, unpublished data) was designed as a tool for annotation of microarray experiments and was aimed at the bench biologist or those with minimal biocomputing support and presents a simplified solution to submitting MIAME-compliant data to ArrayExpress. The design of MIAMExpress was kept as generic as possible for users submitting data related to a range of organisms and domains. The AtMIAMExpress submission tool is a development of MIAMExpress enhanced to support the submission of Arabidopsis data with particular emphasis on the sample annotation of Arabidopsis. AtMIAMExpress has been implemented as a MySQL database with a Perl-CGI Web interface and exports data in MAGE-ML format for upload to any MAGE-ML-compliant application, such as ArrayExpress or Bioconductor.
AtMIAMExpress was developed primarily to provide a data submission tool for the Compendium of Arabidopsis Gene Expression (CAGE) project, but it also available to the wider Arabidopsis community. CAGE (www.psb.rug.ac.be/CAGE/index.htm, on www.psb.rug.ac.be; June 1, 2005) consortium members contributed to the specification of the software, and the tool was developed with their active consultation. The development of AtMIAMExpress was conducted with the aim of generating the Arabidopsis-specific interface as an add-on implementation to the existing infrastructure rather than generating a new tool. This will allow the future development of the current version of MIAMExpress to be readily implemented in AtMIAMExpress.
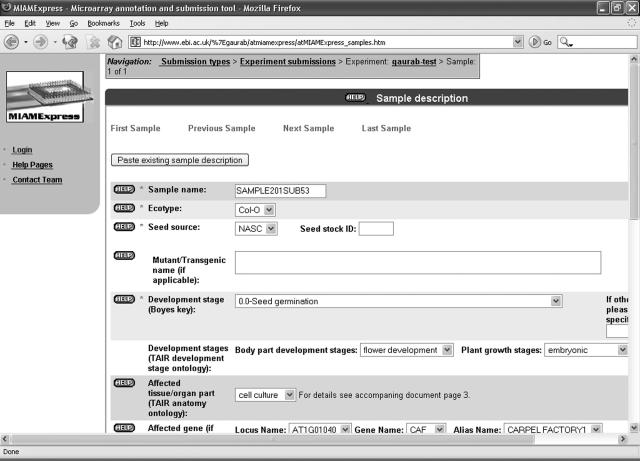
A particular feature of AtMIAMExpress has been the provision of a highly structured growth protocol to improve the annotation of protocol parameters such as temperature, photoperiod, humidity, and light intensity that was required by the Arabidopsis community. An Arabidopsis-specific sample Web form (Fig. 1) was developed to enhance the annotation of the sample. This includes fields to describe the origin of the sample, such as seed stock center name and accession. A key implementation was the integration of The Arabidopsis Information Resource (TAIR) anatomy ontology (Rhee et al., 2003) and Boyes key growth stages (Boyes et al., 2001) within the tool to aid submitters in the correct annotation of samples. Furthermore, Arabidopsis Gene Index locus identifiers and gene names have been provided as selectable terms from lists that the user can use to annotate their samples. AtMIAMExpress also utilizes JavaScript-based checking to ensure consistent annotation of submissions. AtMIAMExpress has extensive help pages to guide submitters through the submission process. The help pages have been written after feedback from users regarding the previous versions of help pages available in MIAMExpress.
Figure 1.
The sample page of AtMIAMExpress. Note the presence of pull-down lists and controlled vocabularies to reflect consistent annotation.
The Submission Process
AtMIAMExpress is a set of related Web forms that leads the user progressively through the submission process. Users provide MIAME-compliant data by annotating fields in the Web forms supported by extensive online and contextual help. The tool does not require knowledge of either MIAME or the MAGE-OM to be used. The user may also start the submission process before the experiment has been concluded and the results derived, which means that AtMIAMExpress may be used as an online laboratory notebook where daily results can be entered and then saved for access at a later date. A new group login feature has been implemented that will enable a particular user to retain editing privileges for the group's submission. This feature will also allow a particular group of submitters to have viewing but not editing privileges.
AtMIAMExpress allows three types of submissions: protocols, array designs, and experiments. If a commercial or previously submitted array design has been used, then the submitter needs only to state this in the submission so that this information can be reused rather than having to supply repetitive information. However, if the array design used has been developed in-house, then the user will need to supply the information as an Array Description File (ADF). This is spreadsheet format with predefined columns containing information on the locations of the spots on the array together with the associated sequence annotation. There is extensive online help regarding ADF creation together with examples covering the common spotter file formats, such as the widely used gal file format.
AtMIAMExpress encourages the use of controlled vocabularies to facilitate the submission process by providing drop-down menus for many fields. The terms available are obtained from the MGED Ontology (MO; mged.sourceforge.net/ontologies/MGEDontology.php, on mged.sourceforge.net; June 1, 2005), an ontology developed by the MGED Society to provide terms for describing the experimental conditions for a microarray gene expression experiment. The MO aims to be as generic as possible in its structure and content and allows the referencing of external ontologies whenever possible. An example is the class OrganismPartDatabase, which is defined as a database of organism part information. An instance of the class is the TAIR anatomy ontology. Therefore, a user may annotate a sample used in the experiment with a TAIR identifier to denote the anatomical part of the plant that was assayed in the course of the experiment. Users are also encouraged to suggest instances of new terms and classes to the MO to extend the ontology via a term tracker Web page available from the above MGED Web site. By limiting free text annotation in this way, AtMIAMExpress aids users in providing consistent annotation; therefore, querying and mining of the data is easier when it is exported to other applications. This approach will facilitate the design of query tools that can mine the data for ontology terms, such as TAIR terms, and identifiers to return particular gene expression data sets.
The Curation Process and Data Export
The curation team consists of trained biologists, and they check incoming submissions for MIAME compliance. Data sets are assessed regarding sample annotations, the protocols used and their content and accuracy, the extracts and labeled extracts together with the hybridizations, and associated data files. Data files are checked for their content as well as their format. The curator may contact the submitter at this point to clarify further points regarding the annotation. The data is exported from AtMIAMExpress as MAGE-ML and uploaded to ArrayExpress. Knowledge of the MAGE-OM and bioinformatics support is required to generate MAGE-ML from the data extracted from the MySQL database. In effect, the use of AtMIAMExpress allows submitters to circumvent the creation of MAGE-ML by themselves and allows the curation team to generate MAGE-ML and upload the submission to ArrayExpress.
In addition, curators may identify particular data sets that are of particular biological interest and provide extra annotation, thereby adding value. An example is the publicly available CATMA Arabidopsis thaliana 23K array version 2.3 array design, ArrayExpress accession number A-MEXP-120. This array design has been reannotated with Gene Ontology (GO) identifiers to facilitate data mining. Furthermore, curators may identify external data sets of particular interest and assist their submission to ArrayExpress. Examples of such Arabidopsis-based data sets are those produced by the AtGenExpress group hosted at TAIR (www.arabidopsis.org/info/expression/ATGenExpress.jsp on www.arabidopsis.org; June 1, 2005).
ArrayExpress
ArrayExpress is public repository of well-annotated gene expression data and contains submissions from a wide range of biological disciplines. There are two major methods of data submission to ArrayExpress, first via the Web-based MIAMExpress submission tool and second via the direct submission of MAGE-ML files from pipeline submitters. Pipeline submitters represent some of the larger microarray data centers with bioinformatics support. These include the Stanford Microarray Database, the Nottingham Arabidopsis Stock Centre, and The Institute for Genomic Research, as well as array manufacturers, such as Affymetrix and Agilent. As of May 2005, ArrayExpress contained 168 total plant experiments, including 122 Arabidopsis experiments (Fig. 2). The more commonly used array designs for this community are the two Affymetrix array designs, ATH1 and AG1, and also various versions of the Complete Arabidopsis Transcriptome MicroArray (CATMA) chip. Other plant-based chips are being added, including the recent Affymetrix wheat and rice array designs.
Figure 2.
Number of experiments according to organism present in ArrayExpress in May 2005.
There are three types of data submission in ArrayExpress: experiments, protocols, and array designs. Experiments contain information about the experiment study itself, its aim, the factors or variables under study, the nature and state of the samples, the RNA extracts obtained, how they were labeled together with the hybridizations, and associated raw and normalized data files. Protocols detail the actual methodology used in the study in a way that another worker is able to reproduce the methods used. These details include how the samples were grown and the treatments carried out, the RNA extraction procedure, the method of labeling the extracts, and the type of scanner used together with the software utilized for image acquisition and any further downstream analysis. Array designs contain data regarding the layout of the array or chip itself, the spots themselves, and the gene or sequence annotation. Where array designs are commercially available, the data is loaded into ArrayExpress with data supplied from the commercial vendor, such as Affymetrix or Agilent. Those submitters using self-spotted array designs are asked to supply the details of the array themselves. After the data set is loaded into ArrayExpress, a submitter is supplied with an accession number that identifies his or her particular submission in the repository. The data can be made publicly available in ArrayExpress at a particular date, for instance, to coincide with the simultaneous publication of the related journal article, or kept in a private area accessible only to those issued a password.
ArrayExpress provides a query interface to query within three broad areas: experiments, protocols, and array designs. Protocols and array designs can be further queried with the type of protocol, i.e. growth, treatment, or scanning, while array designs may be searched for array design name and provider. Experiment data sets are highly annotated, and attributes such as organism and experiment design types, among others, may also be queried on the interface. For a full description of ArrayExpress, refer to Parkinson et al. (2005).
DATA ANALYSIS AND DATA WAREHOUSE
There are a number of applications and services that can import and export data in MAGE-ML format. Of interest to the Arabidopsis community are software analysis tools such as J-Express (Dysvik and Jonassen, 2001) and Expression Profiler (Kapushesky et al., 2004). The latter is the ArrayExpress Web-based integrated online analysis platform and offers a suite of data analysis tools. Bioconductor (Gentleman et al., 2004) is an open-source suite that can be used for the analysis of both Affymetrix and two color arrays. It includes a dedicated package RMAGEML (Durinck et al., 2004) that enables import of MAGE-ML data. This tool allows the analysis of microarray data and allows the export of the analysis to be appended to the original MAGE-ML data. Subsequently, this data set can be exported to a MAGE-ML-based database such as ArrayExpress. Currently, this tool is being used to analyze the AtGenExpress data sets as they become available in ArrayExpress. Commercial array manufacturers such as Affymetrix and Agilent can also supply their array designs in MAGE-ML format.
The Data Warehouse (www.ebi.ac.uk/aedw/ArrayExpress_main.html on www.ebi.ac.uk; June 1, 2005) supports gene-centric queries such as gene names, gene function (GO annotation), and additional annotations, including the gene family, domains and, motifs (Interpro terms) that the particular gene may belong to. At the first instance, the AtGenExpress data sets will be made available in the Data Warehouse as the first plant-based data set.
ONGOING DEVELOPMENT
The generic software tool MIAMExpress was primarily designed to capture data and annotation from experiments with typically 50 hybridizations associated with them. A number of in-house software tools are available to facilitate the submission of larger data sets. These include a tool termed Tab2Mage that allows data to be captured from spreadsheets, with columns providing the annotation required and rows separating the samples studied in the experiment. A batch uploader tool is also being developed to allow the submission of very large data sets comprising a few hundred hybridizations. The use of such tools will expedite the annotation of large data sets compared with the MIAMExpress submission tool. A further development will be the design and implementation of an Ontology Manager tool that will allow the full integration of external ontologies and controlled vocabularies and also allow their terms to be automatically updated.
Acknowledgments
The authors of this article have been funded from contributions from various sources, including CAGE, the European Commission (TEMBLOR grant), EMBL, and the International Life Sciences Institute.
References
- Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Görlach J (2001) Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13: 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, et al (2001) Minimum information about a microarray experiment (MIAME)—toward standards for microarray data. Nat Genet 29: 365–371 [DOI] [PubMed] [Google Scholar]
- Durinck S, Allemeersch J, Carey VJ, Moreau Y, De Moor B (2004) Importing MAGE-ML format microarray data into BioConductor. Bioinformatics 20: 3641–3642 [DOI] [PubMed] [Google Scholar]
- Dysvik B, Jonassen I (2001) J-Express: exploring gene expression data using Java. Bioinformatics 17: 369–370 [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapushesky M, Kemmeren P, Culhane AC, Durinck S, Ihmels J, Korner C, Kull M, Torrente A, Sarkans U, Vilo J, et al (2004) Expression Profiler: next generation—an online platform for analysis of microarray data. Nucleic Acids Res 32: W465–W470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson H, Sarkans U, Shojatalab M, Abeygunawardena N, Contrino S, Coulson R, Farne A, Lara GG, Holloway E, Kapushesky M, et al (2005) ArrayExpress—a public repository for microarray gene expression data at the EBI. Nucleic Acids Res (Database issue) 33: D553–D555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee SY, Beavis W, Berardini TZ, Chen G, Dixon D, Doyle A, Garcia-Hernandez M, Huala E, Lander G, Montoya M, et al (2003) The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res 31: 224–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman PT, Miller M, Stewart J, Troup C, Sarkans U, Chervitz S, Bernhart D, Sherlock G, Ball C, Lepage M, et al (2002) Design and implementation of microarray gene expression markup language (MAGE-ML). Genome Biol 3: RESEARCH0046 [DOI] [PMC free article] [PubMed]