Abstract
Ambiguous germplasm identification; difficulty in tracing pedigree information; and lack of integration between genetic resources, characterization, breeding, evaluation, and utilization data are constraints in developing knowledge-intensive crop improvement programs. To address these constraints, the International Crop Information System (www.icis.cgiar.org), a database system for the management and integration of global information on genetic resources and crop improvement for any crop, was developed by genetic resource specialists, crop scientists, and information technicians associated with the Consultative Group for International Agricultural Research and collaborative partners. The International Rice Information System (www.iris.irri.org) is the rice (Oryza species) implementation of the International Crop Information System. New components are now being added to the International Rice Information System to handle the diversity of rice functional genomics data including genomic sequence data, molecular genetic data, expression data, and proteomic information. Users access information in the database through stand-alone programs and Web interfaces, which offer specialized applications and customized views to researchers with different interests.
International germplasm exchange was the engine of the Green Revolution. In the past, however, much of the important information generated from this exchange was accessible only locally, in field books or researchers' files. Although major international initiatives for germplasm collection and conservation followed the Green Revolution, much collected material is still not used because it is difficult to access. As a result, the potential impact upon agriculture has not yet been realized. Now the free exchange of information, through international crop information systems, should provide the foundation for a second revolution that adds value to germplasm by seamlessly uniting its conservation, evaluation, utilization, and exchange.
Furthermore, new technologies in molecular biology and genomics mean that traditional phenotypic information must be linked to large quantities of sequence and genetic information so that functional genomics and allele mining activities can speed up germplasm enhancement.
The International Maize and Wheat Improvement Center (CIMMYT) devised an information strategy and developed software on a mainframe computer during the 1980s to facilitate unambiguous identification of wheat (Triticum species and related species) germplasm, thereby establishing links between information from different sources. The read-only International Wheat Information System compact disk (Fox et al., 1996) duplicated data-querying capabilities and some of the genealogical diagnostics of the mainframe version. In 1995, CIMMYT and the International Rice Research Institute (IRRI) canvassed other Consultative Group for International Agricultural Research (CGIAR) centers to establish a project to develop an International Crop Information System (ICIS; Fox and Skovmand, 1996) applicable to a wide range of crops. Extensive communication among CGIAR centers highlighted the economies to be gained by collaborating on the development of an information system that could be used for many crops. IRRI has subsequently developed a database and software to support this generic design and deployed it in the form of the International Rice Information System (IRIS; Bruskiewich et al., 2003).
THE ICIS
Several CGIAR centers, national agricultural research systems, and advanced research institutes are collaborating to develop ICIS as a generic system that will accommodate all data sources for any crop and breeding system. The vision of ICIS is to integrate different data types in both private and public datasets into a single information system and provide specialized views and applications that operate on the single integrated data platform. After all phases of development are complete, ICIS will support a range of activities from germplasm conservation, evaluation, functional genomics, allele mining, breeding, testing, and release. Data will be accessible from CD-ROM or the World Wide Web, and users may either adopt the complete system or link only to its innovative genealogical features.
The driving force behind ICIS is accessing and sharing data rather than providing analytical and statistical tools. This is because the major bottleneck to intelligent data integration and utilization was not statistical software but rather the drudgery of finding, extracting, preparing, and managing the data. ICIS exports managed data in formats designed to make full use of external statistical software.
Functionality
The ICIS system is fast, user friendly, PC based, and distributable on CD-ROM or via the Internet. It contains (1) a genealogy management component to capture and process historical genealogies as well as to maintain evolving pedigrees, and to provide the basis for unique identification and internationally accepted nomenclature conventions for each crop; (2) a data management system (DMS) for genetic, phenotypic, and environmental data generated by evaluation and testing, as well as for providing links to genomic maps; (3) links to geographic information systems that can manipulate all data associated with latitude and longitude (e.g. international, regional, and national testing programs); (4) applications for maintaining, updating, and correcting genealogy records and tracking changes and updates; (5) applications for producing field books and managing sets of breeding material, and for diagnostics such as coefficients of parentage and genetic profiles for planning crosses; (6) tools to add new breeding methods, new data fields, and new traits; and (7) tools for submitting data to crop curators and for distributing data updates via CD-ROM and electronic networks.
Use Cases
ICIS is designed to allow biologists to manage local data and query and view their own data fully integrated with global public information.
The first use case is simply to provide an information management environment that facilitates unique identification of germplasm, the management of germplasm lists, and the production and processing of field and lab books. New germplasm is automatically linked to existing germplasm through dynamic pedigree management, and characterization and evaluation data are unambiguously linked to germplasm and are fully documented and annotated with controlled vocabularies identifying properties or traits, scales, and methods.
As data accumulate, integrative queries become possible. Pedigree-based measures of relationships between germplasm are easily computed and can be used to enhance the power of predictive models. Germplasm with particular phenotypic or molecular characteristics can be identified and these characteristics associated with performance in specified environments.
Ultimately, it will be possible to query different ICIS crop implementations to achieve the goals of comparative biology and transfer of knowledge between crops.
Remote Users
One of the innovative features of ICIS is that it permits independent users to integrate their own local data with public central data. ICIS does this by allowing read-only access to the central database for a particular crop and supporting a local copy of the ICIS data model where the local data is stored. Apart from providing user-friendly access to the data, so that crop scientists can make informed decisions, the system provides a local data management environment for the user and captures relevant data for the crop. Periodic updates to the central database by local users makes their data available to all other users as well as browsers of the central database. All data are fully credited to users, and quality control is further ensured by a central crop curator who must reconcile duplicate germplasm entries coming from different users and integrate new traits, scales, and methods into a community-curated controlled vocabulary that ensures compatibility across users.
The Data Model
The data model and database system of ICIS are designed for maximum flexibility to cater for as wide a range of crops as possible. The model must be independently adopted for a specific crop and data entered to create an independent system for that crop.
The Genealogy Management System
The core of ICIS is a common genealogical data model called the Genealogy Management System (GMS), which is generically designed to accommodate a wide range of crops. The functions of GMS are to assign and maintain unique germplasm identification, retain and manage information on genealogy, and manage nomenclature and chronology of germplasm development.
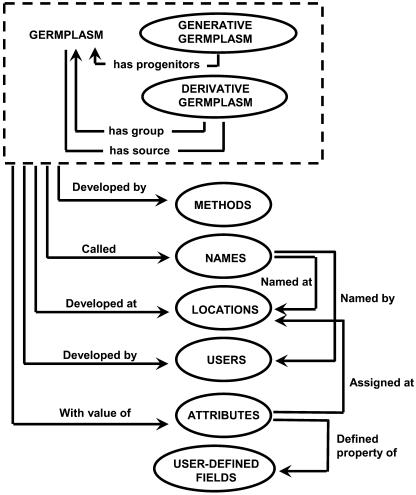
Each germplasm entity is identified by a controlled CropID (the domain), a controlled UserID (the authority), and a locally assigned GERMPLASM_ID (GID). The logical connection between a GID and a packet of seeds or other propagating material is that different packets that germplasm specialists would not mix get different GIDs. Information on method, location, date of genesis, and other attributes is managed through the data model shown in Figure 1. Each germplasm record is linked to its progenitors through their GIDs. Germplasm is divided into three categories: generative, derivative, and maintenance. Generative germplasm is produced by generative methods that tend to increase and combine genetic variation, such as crossing or mutation. Derivative germplasm is produced by derivative methods, such as selection, which tend to refine, target, and reduce genetic variation. Maintenance methods, such as seed increase or conservation, aim to maintain genetic status and produce maintained germplasm. Germplasm produced by generative methods may have any number of progenitors. Derivative and maintained germplasm are derived from a single germplasm source.
Figure 1.
Data model for the ICIS GMS.
Germplasm methods can be added by local users if they are not found in the central list, but these are checked by the central curator and integrated into the methods ontology when local data are uploaded to the central database.
Each instance of germplasm, whether generative, derivative, or maintained, falls into a single germplasm group identified by a group source. For germplasm produced by a generative process (such as an F1 from a cross) or for germplasm of unknown genesis (such as a land race), this group is defined by its own GID. Germplasm produced by derivative or maintenance methods retains the group ID of its source, although the group is often known when the source is not. For example, the cross may be known even when the line source is missing.
Method definitions are stored with complete documentation, including bibliographic references. Some methods depend on parameters, which may vary each time the method is used, such as the number and mixing proportions of parents in the generation of a population. These parameters can be defined by assigning an attribute to GID of the germplasm being produced. Attributes are flexible user-definable data fields.
Germplasm collects a multitude of labels during the development-and-release process. These are all tracked as NAMES in GMS. One name must be identified as the preferred name for display purposes. For a given genetic entity, different preferred names can be used in local and central applications. Names may contain imbedded information, and this can be made accessible to application programs for specific name types by specifying a format for the name.
Attributes are text variables used to store information about the genesis, genealogy, nomenclature, or chronology of germplasm. Attribute types are defined and described as USER_DEFINED_FIELDS. Like names, attributes may contain imbedded information in the form of subfields or variables within the attribute text. A scale identifier specifies the units in which the attribute are required and links attributes to the controlled vocabulary of properties, scales, and methods.
Location information is stored to record the origin or destination of germplasm or the location of sites where information or data on germplasm was collected. Locations may be as precise as fields or plots or large like countries or even regions. Locations are defined by name and can be associated with latitude and longitude points or polygons to allow spatial analysis.
The DMS
The functions of the DMS are to store and manage documented and structured data from genetic resource, variety evaluation, and crop improvement studies; link data to specialized data sources such as GMS, soil, and climate databases; and facilitate inquiries, searches, and data extraction across studies according to structured criteria for data selection.
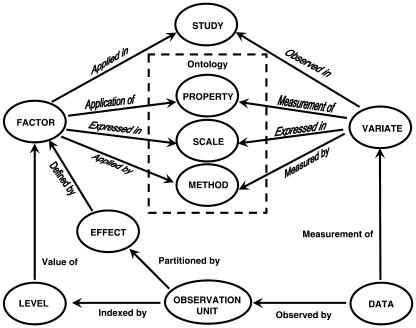
All types of data can be accommodated in DMS, including raw data, observed data, derived data, and summary statistics. Data may have continuous or discrete numeric values, or text or categorical character values. For example, observations on disease resistance or nutrient efficiency of a genotype can be numerical measurements, scored or calculated indices, or text data. More complex forms of data, such as pictures or documents, will also be considered. Figure 2 shows the basic data model of the DMS and shows the linkages between the entities described below.
Figure 2.
Data model for the ICIS DMS.
A study is the basic, reportable unit of research; it is synonymous with the notions of experiment, nursery, or survey. Since the DMS must deal with any of these, we use the term study. A study is characterized by a set of scientific objectives and testable hypotheses and results in the collection of one or more data sets. The division of data into sets is usually motivated by convenience; for example, data collected from different sampling scales are most conveniently treated in different data sets. Similarly, data collected at different times or from different locations are also often treated as different data sets, although it is feasible to treat these divisions in a single data set. The point is that the DMS is flexible enough to manage data in all the ways that researchers require.
Factors are classifying variables in a study, which take values from finite sets of discrete levels. These levels are usually labeled in some way to document the source and context of the data by expressing the conditions under which the data were collected or derived; for example, the names of treatments or design structures applying to the unit or units from which the data are recorded, or conditions such as the time and location of measurement. These labels are usually listed in columns in the data set. The study itself is treated in the data model as a factor with exactly one level, the study name. Hence, every study has at least one factor. A single factor is often represented by more than one set of labels.
Factors are named and described in each study. They have three main attributes: the property of the experimental material or survey units being manipulated or stratified, the method or procedure by which the levels are applied, and the scale or measurement units in which the levels are expressed. All levels of a particular factor are expressed in the same scale. The names of factors are consistent within studies and equivalent factors are linked across studies through common properties. Property is subject to a controlled vocabulary to facilitate this linkage.
Data sources such as field objects or sampling units are identified by combinations of levels of design or sampling factors. Data values such as treatment means are associated with level combinations of treatment factors, which do not correspond to field objects but that can be thought of as data sources. Both types of data sources, field objects and treatment combinations, are referred to as observation units. Observation units are conceptually equivalent to rows in a serially structured spreadsheet.
Each study involves the recording of data for one or more properties of some observation units. The data being recorded are described by variates and are often represented as data columns in spreadsheets. Each variate has the same three attributes as a factor: the property or trait being measured, the method or procedure by which the value is observed or derived, and the scale or measurement units in which the value is expressed. Variates are named and described within studies, and the name should be consistent throughout a study. The common vocabulary of properties links variates across studies in the same way that factors are linked across studies.
The central entity in the DMS data model is the datum, which links to exactly one observation unit and exactly one variate. The datum is conceptually equivalent to a cell in a variate column of a spreadsheet or field book. The most important attribute of a datum is its value, which is the recorded value of the associated variate for the associated observation unit.
Effects are sets of observation units in a study that are indexed by levels of subsets of the factors in the study. Effects form natural hierarchies according to the nesting of their index-factor subsets. Data associated with different effects may result from data collection at different sampling scales or on different field objects, but they often arise by statistical amalgamation of data values over related units in lower-level effects.
Correcting Data
Corrections and changes will inevitably be made in any database. Only authorized users can make these changes, and all changes are logged so that the sequence of changes can be traced and can be undone if required. Such log functions are increasingly necessary to comply with requirements associated with establishing intellectual property rights of germplasm.
Changes commonly occur when new information about an existing germplasm record is entered into a local database. If the existing record is in the local database then the local user can complete the changes. But if it is in the central database, changes cannot be completed until the central database is updated. Verifying and completing requested changes is part of the update process and sufficient information and justification needs to be recorded in the changes table to allow the process to be completed.
Verification and completion of changes to the central database may take some time but local users would like to see their changes reflected immediately. This is achieved by the software that always checks the local CHANGES table for central changes and applies them at run time for the specific installation where they are recorded.
User-Defined Data
The ICIS data model is extremely flexible and can accommodate new data types as required. Users are able to define new relationships between germplasm by specifying new breeding methods. They can specify new attributes of the germplasm to be stored, types of names, location descriptors and traits, scales, and methods for characterization and evaluation data. These new elements can be immediately applied at the local level, but are subject to curation when data is uploaded so that new elements are integrated into the controlled vocabularies for methods, properties, or traits and scales.
Stand-Alone Software Modules
Components of ICIS include a GMS, Set Generation Module including the External Pedigree Input Tool, Field Book Module, Trait Management System, DMS, a Work Book for data input and query, and a Data Retriever for cross-study data queries. The first four modules focus on germplasm and management of genealogy and nomenclature. The next three handle the management of evaluation and characterization data, and the Retriever provides access to both raw and processed data.
Web Access and Internet-Based Applications
The latest iteration of ICIS software is now being developed in the Java programming language and associated technologies. We have started this migration with emphasis on query tools since the Windows stand-alone tools are compatible with the new version and are still available. These will be ported as resources and demand dictate. The move of ICIS to Java was precipitated by a desire for greater operating system independence (the original system being Microsoft Windows dependent) and to achieve a greater degree of platform integration across stand-alone and Internet-based applications, tasks for which Java is well suited. Along the way, Java has also provided for greater internationalization of the system to accommodate the needs of developing country partners whose first language is not English.
The overall architecture of the system has been redesigned in parallel with the move to Java. This has resulted in greater decoupling between applications and data sources, and also opening the system up for more efficient distributed computing. In this new system, the data access layers of the Java middleware provide for both local database access (using Java Database Connectivity) and remote access (using Internet Web services). Implicit in the new architecture is a move away from the local/central dichotomy of previous versions of ICIS toward global identification of data using Life Sciences Identifiers (see http://lsr.omg.org/), which will allow networks rather than pairs of databases to interoperate.
At the application layer, new stand-alone and Web applications are being developed (the latter using Java servlet, Java Server Pages, and associated technologies). In addition, Web service provider facilities are being added to complement data source level Web service client facilities. For maximum flexibility, current Web service facilities are not dogmatic about protocol; XML schema-based ICIS services and MOBY (www.biomoby.org) services coexist, and provisions are being made to accommodate Web service protocols from the Global Biodiversity Facility (www.gbif.org).
THE IRIS
IRIS is the rice (Oryza species) implementation of ICIS. The GMS of IRIS stores information on more than one and a half million varieties, breeding lines, and accessions of rice. This allows pedigree analysis to trace germplasm flows and relationships between lines that can be used to improve evaluation estimates or plan improvement programs. The IRIS DMS contains five million data values from more than 500 studies from breeding, screening, and international testing trials. This allows integrative analysis over different environments.
Curation of Rice Data into the IRIS
The vision of ICIS and its implementation in IRIS is to facilitate distributed curation of crop information by experts in the course of routine research and development activities. Gene banks can manage and document collections with ICIS, and the International Rice Germplasm Collection is in the final stages of transferring data management to an ICIS-based system. International testing networks, like the International Network for Genetic Evaluation of Rice, are able to mange nurseries and store and publish evaluation data through IRIS. Any crop improvement project can plan crossing, manage pedigree nurseries, and manage release of improved rice germplasm through IRIS as is done for all breeding projects at IRRI. In this way, genealogy, nomenclature, and evaluation data are automatically collected and can be easily shared with the international community by uploading local ICIS databases into the central database or opening access to local databases via the networking facilities of the ICIS Web interface. Distributed and shuttle breeding projects benefit particularly from the features of ICIS that allow remote activities. The South and Southeast Asian Rainfed Lowland Rice Shuttle Breeding Projects are able to share germplasm and information in a synchronous manner.
Specialized data sources such as that for the IR64 rice mutant collection and the Isozyme Characterization and Classification data set are being curated at IRRI as part of IRIS using the ICIS DMS structure. Web-based applications for searching and querying such specialized datasets are being implemented.
Central curation issues arise when users wish to publish data in the central IRIS database. These are issues of identity and quality. Although ICIS is designed to allow unique identification of germplasm, there is nothing intrinsic that prevents specification of the same germplasm in more than one local database. If these are then uploaded, the central curator must identify the duplicates and either make replacements, if they really are the same germplasm (i.e. users would be happy to mix the seed), or make pedigree connections if they are not (usually specifying a management method such as release or import).
As discussed earlier, local users may specify changes and corrections to central records. If these are presented to be uploaded, the central curator must verify the changes and accept or reject them. This is less onerous than it may appear, because it must be remembered that the local data managers are often the experts in their local area and can generally be trusted with changes in their own domain.
Quality of characterization and evaluation data is a difficult task, and the current approach of IRIS is to make sure all data is clearly attributed. This is not only essential to recognize intellectual contribution but also encourages quality. So far, no automatic quality control procedures are in place to check for outlying or spurious data, but these are envisaged and can be deployed as developed.
Access via the World Wide Web
A Web interface for ICIS has been developed and deployed for IRIS (http://www.iris.irri.org). This interface is extending ICIS functionality into a broader range of biological datasets including mutant and expressed sequence tag clone data. In 2005 and beyond, it is anticipated that these data sets will also include rice gene expression and allele mining data. Each class of data is accessed through a specialized view that queries and displays the target data type in its biological context and cross-references it to related data, including traditional germplasm and evaluation data.
Special effort is being made to incorporate controlled vocabularies and ontologies from the Gene Ontology Consortium (www.geneontology.org) and Plant Ontology Consortium (www.plantontology.org, 2002), in particular, to capture traits and phenotyping. One particular context exploiting such ontology is the MutantView, with which researchers can specify mutant phenotypes as a means of identifying specific mutant stocks.
Links to Global Crop Informatics Resources
Other databases of agricultural information have close links with IRIS (or ICIS in general). The System-wide Information Network for Genetic Resources (SINGER; http://www.singer.cgiar.org) aims to provide global access to genetic resources data across all CGIAR-mandated crops and commodities. Germplasm records in ICIS that relate to accessions in the CGIAR collections are linked with SINGER, so the ICIS can provide information on the utilization and deployment of those genetic resources. Each individual ICIS database manages data for a particular crop and generally does not share data with other crops. The integrating role for genetic resources information across crops is played by SINGER. IRIS has also enjoyed collaboration with the U.S. Department of Agriculture-sponsored GrainGenes and Gramene database initiatives, with the International Rice Genome Sequencing Project (Rice Genome Research Program, The Institute for Genomic Research, and other partners), with Oryzabase (Japanese National Institute of Genetics), and with the Beijing Genome Institute. Linking these implementations to ICIS will provide access to sequence information and molecular maps, which will facilitate functional genomics and allele mining and integrate information across crops at the genomic end of the spectrum.
CONCLUSION
The technical challenge for plant scientists and software developers is to implement the type of system outlined here. The challenge for administrators is perhaps more difficult: to facilitate the continued free exchange of information and to nurture a scientific culture in which users take full advantage of the data of others and in turn contribute to shared databases.
ICIS Software Availability
The ICIS system is being developed under an open-source software model and is freely available. Stand-alone executable files and public data for several crops together with an installation script are available from the download section of the ICIS project Web site at www.icis.cgiar.org. Where possible, ICIS Web interface components are being developed by the adoption of public open-source database schemata and software components such as those from the Gene Ontology consortium (e.g. the Gene Ontology database schemata and the Amigo browser), Generic Model Organism Database (chado, gbrowse, and cmap at http://www.gmod.org), and the Open Bioinformatics Foundation (www.open-bio.org).
To promote the open, collaborative spirit of the project, two collaborative environments have been established. CropForge.irri.org hosts source code and community development of both Windows and Web components. CropWiki.irri.org hosts technical documentation, user support discussions, and collaborative development of documents and FAQs. There is also a developers e-mail list hosted at www.bioinformatics.org. Community participation is also promoted by open annual ICIS workshops.
References
- Bruskiewich R, Cosico A, Eusebio W, Portugal A, Ramos LR, Reyes T, Sallan MAB, Ulat VJM, Wang X, McNally KL, et al (2003) Linking genotype to phenotype: the International Rice Information System (IRIS). Bioinformatics (Suppl) 19: i63–i65 [DOI] [PubMed] [Google Scholar]
- Fox PN, Lopez C, Skovmand B, Sanchez H, Herrera R, White JW, Duveiller E, van Ginkel M (1996) International Wheat Information System (IWIS), Version 1 (CD-ROM). CIMMYT, El Batan, Mexico
- Fox PN, Skovmand B (1996) The International Crop Information System (ICIS) connects Genebank to breeder to farmer's field. In M Cooper, GL Hammer, eds, Plant Adaptation and Crop Improvement. CAB International, Wallingford, UK, pp 317–326