Abstract
GrainGenes (http://wheat.pw.usda.gov) is an international database for genetic and genomic information about Triticeae species (wheat [Triticum aestivum], barley [Hordeum vulgare], rye [Secale cereale], and their wild relatives) and oat (Avena sativa) and its wild relatives. A major strength of the GrainGenes project is the interaction of the curators with database users in the research community, placing GrainGenes as both a data repository and information hub. The primary intensively curated data classes are genetic and physical maps, probes used for mapping, classical genes, quantitative trait loci, and contact information for Triticeae and oat scientists. Curation of these classes involves important contributions from the GrainGenes community, both as primary data sources and reviewers of published data. Other partially automated data classes include literature references, sequences, and links to other databases. Beyond the GrainGenes database per se, the Web site incorporates other more specific databases, informational topics, and downloadable files. For example, unique BLAST datasets of sequences applicable to Triticeae research include mapped wheat expressed sequence tags, expressed sequence tag-derived simple sequence repeats, and repetitive sequences. In 2004, the GrainGenes project migrated from the AceDB database and separate Web site to an integrated relational database and Internet resource, a major step forward in database delivery. The process of this migration and its impacts on database curation and maintenance are described, and a perspective on how a genomic database can expedite research and crop improvement is provided.
As crops improve year by year, so do crop databases. The GrainGenes database project was launched by the U.S. Department of Agriculture in 1992 to collect and distribute genetic data on the Triticeae (wheat [Triticum aestivum], barley [Hordeum vulgare], rye [Secale cereale], and triticale [x Triticosecale]) and oat (Avena sativa) crops to plant breeders, pathologists, geneticists, and molecular biologists (Matthews et al., 2003). Originally conceived as a stand-alone database, GrainGenes now encompasses an integrated database and Web site, working intensively with the small-grains research community to provide an Internet portal for numerous ancillary projects for Triticeae genetics and contributing to the development of informatics tools to support small-grains research. Like many other organism-focused databases, GrainGenes concentrates on genomic aspects; however, it is also an important repository for information concerning genetic resources, pathology, and colleagues.
The GrainGenes Database initially was operated using AceDB (http://www.acedb.org), a platform adopted by many early genome projects. AceDB served the GrainGenes project's needs very well with powerful schema, query languages, graphical displays, and the simplicity to be operated completely by a single biologist. The object-like data structures of AceDB worked well for handling the diverse underlying data, enabling straightforward connections between all data types and allowing the schema to be changed easily to accommodate new kinds of data. The AceDB software continued to develop and improve for several years and still has an active user community, providing an excellent resource for biological database development.
Eventually, the size of the GrainGenes database caused performance problems in the AceDB platform, due in part to the data from large-scale expressed sequence tag (EST) sequencing of wheat and barley (currently containing the most ESTs of any plant family, with more than a million public sequence records). In early 2003, a user committee (made up of 11 small-grains researchers and bioinformaticists) concurred with previous recommendations that GrainGenes should migrate to a relational database management system (RDBMS). The committee emphasized, however, the importance of maintaining the richness and depth of existing data types, while continuing to provide a high level of curation and community service.
Although it was clear that migration to a relational database would improve GrainGenes' ability to serve large datasets efficiently, the lack of graphical interfaces to display map data, a crucial part of the database, initially was a concern. However, map-viewing tools for relational databases have evolved significantly in the last few years. After reviewing several options for the graphical map-viewing software, CMap, developed by the Generic Model Organism Database group (http://www.gmod.org), was chosen to be the default map viewer for GrainGenes 2.0 and includes advanced features that were not available in the AceDB graphical displays.
This article provides an overview of the newly designed GrainGenes 2.0 database, a description of the hurdles encountered in the migration to a RDBMS format, and a discussion of the utility of the database from a user's perspective.
DATA CURATION IN GRAINGENES
Community service has always been the major focus of the GrainGenes staff. Curators are readily available to the small-grains research community, students, and the general public via feedback forms on the Web site or direct e-mail. Curators personally answer an average of 25 requests and queries a month from individual users on a wide variety of topics (for example, “Please add these references so they'll appear in my Author report,” and “How can I download a list of all mapped wheat ESTs and their map locations?”). It is also important for curators to avail themselves to the community to facilitate project development, whether temporal or with potential for permanent integration into the database. Although data is often mirrored in other databases, and vice versa, GrainGenes emphasizes information relevant to the small-grains community.
The amount of work involved in data curation for GrainGenes is dependent upon the data type. Some data classes, such as Sequence, acquire content largely via software scripts and require little additional curator input; other classes, such as the Quantitative Trait Loci (QTL) class, contain records created individually by the curator based on information gleaned from the source publications and contact with researchers generating the data. Below are some of the major GrainGenes data classes and a brief description of the mechanism involved for data curation.
Maps
Interactive genetic and physical maps (summarized at http://wheat.pw.usda.gov/ggpages/maps) are a well-used GrainGenes resource. The Map Data record provides links to all information available for a particular mapping study, including parent germplasm, remarks on map construction, and raw mapping data (when available). Currently, there are 136 Map Data records, comprising more than 1,200 linkage groups containing loci from DNA-based probes, QTL, and genes.
Map curation is a labor-intensive process. GrainGenes curators constantly monitor the literature for new maps; however, important maps are published faster than they can be added to the collection. To address this, tools are being developed to streamline data submission and involve the mapping laboratories to a greater degree. Once a mapping study is selected, the authors are contacted and requested to provide a spreadsheet with the marker names, map positions, and raw mapping data. While occasionally researchers request that their maps be placed in GrainGenes as the sole publisher without a journal publication, more often the GrainGenes staff is contacted by authors prior to publication with a request to add the new maps to the database. In these cases, a URL is created for inclusion in the publication, and curators assist in assuring the fidelity of assigned marker names.
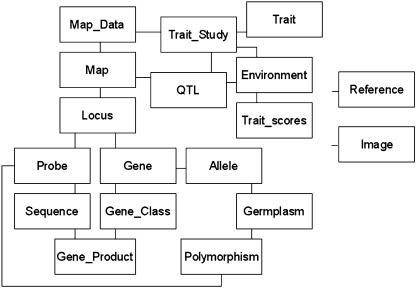
Curation of gene and probe names is the most labor-intensive part of map curation. For consistency with the literature, GrainGenes uses locus names exactly as shown in the publications. This is part of the rationale for separating Locus from Probe and Gene as separate classes in the data structure (Fig. 1), and curation is done at the level of the gene and probe names connected to each locus. GrainGenes users rely on the database for detailed information on probes for molecular markers, including PCR primer sequences, PCR conditions, and repeat sequences for simple sequence repeat (SSR) probes, and curators strive to add as much information to these records as possible.
Figure 1.
The core GrainGenes data classes. The conceptual structure of the GrainGenes database showing primary interconnections between classes. The Reference and Image classes connect to many others.
A controlled vocabulary is used for genes and a controlled set of naming rules for probes. Gene names are established by the authorities of the small-grains communities, e.g. Catalogue of Gene Symbols for Wheat (WGC; http://wheat.pw.usda.gov/ggpages/wgc/2003/). The nomenclature for probes, including all kinds of molecular markers (RFLP clones and primers for SSRs, sequence tagged sites, amplified fragment length polymorphisms, random-amplified polymorphic DNAs, etc.) is established essentially within GrainGenes by the curators (as guided by the users).
All maps in GrainGenes are now visible via the CMap graphical viewing tool. CMap provides an efficient Internet interface for comparing maps among different populations, homoeologous chromosomes within polyploid species, or homologous regions from other species. In addition, the curated maps are exported to other databases (Gramene [http://www.gramene.org] and the National Center for Biotechnology Information's [NCBI; http://www.ncbi.nih.gov] Plant Genomes Central [http://www.ncbi.nih.gov/genomes/PLANTS/PlantList.html]) for integration with their data using their own graphical displays. One of the GrainGenes curators also serves as wheat curator for Gramene, primarily for maps and QTL.
Sequence
Since the beginning of the International Triticeae EST Cooperative initiative in 1998 (http://wheat.pw.usda.gov/genome) when only six sequences from wheat were represented in the NCBI dbEST database (http://www.ncbi.nlm.nih.gov/dbEST/), the publicly available Triticeae EST collection has grown to more than one million sequences. The Triticeae and oat have some of the largest genomes of the grass species, e.g. wheat has an estimated 13,500 megabases, about 40 times larger than rice (Arumuganathan and Earle, 1991). Therefore, without a complete Triticeae genome sequence to anchor the EST sequences, it is a challenge to assure that all of the available data is represented.
The Sequence data class holds sequences from EST projects, genomic sequencing efforts, and contig assemblies. Basic information about all GenBank sequences for the Triticeae and oat is obtained from NCBI on a quarterly update cycle. Sequence records are also linked to corresponding records in external databases such as NCBI's GenBank and Unigenes, EMBL (http://www.embl.org), DNA Data Bank of Japan (http://www.ddbj.nig.ac.jp), The Institute for Genomic Research (http://www.tigr.org) Gene Indices, BarleyBase (http://www.barleybase.org), and Gramene where applicable.
Specific subsets of the Sequence data class are available for BLAST (Altschul et al., 1990) at http://wheat.pw.usda.gov/GG2/blast.shtml. These datasets include some that are regularly updated, such as “All GrainGenes sequences” and the “TREP Triticeae Repeats” sets, and some that are essentially frozen, e.g. the “Barley1 GeneChip exemplars.” Except for the “All GrainGenes sequences” set, these BLAST databases are also available for download as FASTA files at http://www.graingenes.org/blast_databases.html. The individual BLAST hits resulting from a search are Web-linked to appropriate databases within GrainGenes and elsewhere. For example, hits in the “Barley1 GeneChip exemplars” dataset link to the corresponding record in BarleyBase. Reciprocally, each barley record in BarleyBase has a link to “GrainGenes BLAST,” so it can be searched against any of GrainGenes datasets.
Sequences are being actively utilized by the research community to build comparative maps, compile unigene assemblies, develop genetic markers, and construct genome scaffolds. The GrainGenes curators work closely with the research community to assure these resources are available online in a timely manner. Results of bacterial artificial chromosome sequencing of gene-centric regions of the Triticeae will require new graphical tools to display the annotated data. The GBrowse viewer developed by Generic Model Organism Database will be implemented in GrainGenes to serve the wealth of bacterial artificial chromosome data that are expected in the coming years.
QTL
QTL data are gathered from a variety of sources such as the WGC (McIntosh et al., 2003), which publishes accepted QTL names and associated genetic markers, and the MASwheat project (http://maswheat.ucdavis.edu) for molecular markers used in marker-assisted selection in wheat. Barley QTL have been cataloged by the U.S. Barley Genome Project (Hayes et al., 2001) and BeerGenes (http://gnome.agrenv.mcgill.ca/bg/). Oat QTL are being compiled by Nick Tinker and Charlene Wight (personal communication), and rye QTL by Victor Korzun (personal communication). While the WGC serves as the authority for naming QTL in wheat and the rye community follows similar nomenclature when describing rye QTL, the GrainGenes curators are actively working with the community to assign acceptable names to the barley and oat QTL.
An effort is under way to represent all known QTL for the Triticeae and oats in GrainGenes and to relate these to similar rice (Oryza sativa) QTL cataloged by the Gramene project and maize (Zea mays) QTL managed within the MaizeGDB project (http://www.maizegdb.org). To correlate functionally similar QTL across species, it is essential to have a controlled vocabulary for describing traits. GrainGenes collaborates with Gramene, MaizeGDB, and other databases toward developing a Trait Ontology for this purpose, as well as ontologies for plant structure, growth stages, and environments required to describe experimental conditions in a structured, queryable way (http://www.gramene.org/plant_ontology/). Once these resources are developed, scientists will be able for the first time to use common phenotypes as a point of connection for QTL information from different cereal crop species.
Pathology
Much of the small-grains breeding effort worldwide is focused on improving resistance to diseases, pests, and abiotic stresses. The GrainGenes database contains records for 450 such pathology listings recognized as important in the Triticeae and oat. Many of these records are illustrated with photographs of symptoms. Originally compiled by a collaborating curator, Ken Kephart, from publications like the “Compendium of Wheat Diseases” (Wiese, 1987), these descriptions are maintained by monitoring new information from authoritative sources such as the American Phytopathological Society (http://www.apsnet.org). The GrainGenes Web site also includes a pathology page (http://wheat.pw.usda.gov/GG2/pathology) with links to online documents and other related sites.
References
The approximately 13,000 reference records in GrainGenes constitute a selective bibliography of Triticeae and oat genetics and genomics. Monthly updates of publication contents are provided by the U.S. Department of Agriculture National Agricultural Library using a set of keywords designed to enrich for the articles of interest. From this list, individual records are selected by a curator for inclusion in the GrainGenes reference collection and then processed through a data pipeline of Perl scripts and curator edits.
SPECIALIZED DATASETS AND COMMUNITY SERVICES
It must be noted that GrainGenes is not simply a database but seeks to be an information hub and portal for user communication. Through its history, GrainGenes has evolved into a resource that serves as a forum to present community standards, such as nomenclature (e.g. WGC [McIntosh et al., 2003]), periodicals (e.g. Barley Genetic Newsletter), workshops (e.g. International Triticeae Mapping Initiative), project initiatives (e.g. International Genome Research of Wheat), and a place for other news updates, such as meeting and employment announcements. The new GrainGenes homepage at http://wheat.pw.usda.gov serves as an integrated tool to access all resources.
The GrainGenes project also provides a portal for developing datasets and ancillary databases (Table I). Once matured, some of these datasets are then integrated into the GrainGenes database proper, molding its schema as necessary, while others are maintained as separate entities serving the specific needs of those producing and using the data.
Table I.
Ancillary GrainGenes projects
| Project | Organizer(s) | Content |
|---|---|---|
| Triticeae Repeat Sequence Database (http://wheat.pw.usda.gov/ITMI/Repeats) | Thomas Wicker | Comprehensive database of annotated Triticeae repetitive elements |
| SNP Discovery (http://wheat.pw.usda.gov/ITMI/wheatSNP) | Peter Isaac | SNPs in wheat-breeding germplasm to minimize duplication among laboratories |
| Triticeae EST-SSR Coordination (http://wheat.pw.usda.gov/ITMI/EST-SSR) | Rejeev Varshney, Nils Stein | EST-derived microsatellite (SSR) markers in the Triticeae |
| Genoplante/Institut National de la Recherche Agronomique Wheat SSR Club (http://wheat.pw.usda.gov/ggpages/SSRclub) | Pierre Sourdille | Distribution site for wheat SSR-containing sequences for primer development |
| BLAST server on GrainGenes (http://wheat.pw.usda.gov/GG2/) | GrainGenes Curators | BLASTable collections of Triticeae sequences (e.g. mapped wheat ESTs, Barley1 GeneChip exemplars) |
| Wheat EST Bin Mapping (http://wheat.pw.usda.gov/wEST) | Gerard Lazo | Results from deletion mapping on 146 aneuploid wheat stocks using 7,600 wheat EST probes for 16,000 genetic loci |
| Physical Mapping of the Wheat D Genome (http://wheat.pw.usda.gov/PhysicalMapping) | Jan Dvorak, Frank You | Development site to physically map the D genome (Aegilops tauschii), including bacterial artificial chromosome clone contigs and anchors |
| Transposon-Mediated Functional Genomics in Barley (http://wheat.pw.usda.gov/BarleyTNP) | Peggy Lemaux | Results from targeted mutagenesis in barley using Ac/Ds, and an interactive map with links to information on transposed lines and naked-eye phenotypes |
| U.S. Wheat and Barley Scab Initiative (http://www.scabusa.org) | Rick Ward | Clearinghouse for information about Fusarium head blight |
GrainGenes is an active partner in several current and planned research projects in the United States, including physical mapping of the wheat D genome and development of genome-specific single nucleotide polymorphisms (SNPs) for wheat. A particularly large effort now beginning is the creation of a database to manage large amounts of genotype (alleles of molecular markers, especially SSRs and SNPs) and phenotype data about cultivars of wheat and barley for use in marker-assisted breeding as well as association genetics for discovery of new genes and QTL. For this project, GrainGenes is collaborating with some of the producers of the data such as the U.S. Department of Agriculture genotyping laboratories (http://wheat.pw.usda.gov/GenotypingLabs/). GrainGenes is also actively working with Gramene to develop a database schema and new user interfaces for storing and presenting such diversity data, which will be increasingly abundant and important for all cereal crops, indeed for all agricultural species (http://www.maizegenetics.net/gdpdm/). Challenges in this work will include presenting maps with large numbers (thousands) of markers, visualizing SNP genotypes in context with the gene sequence features, and haplotype representation.
MIGRATION TO AN RDBMS
MySQL (http://www.mysql.com) was chosen as the RDBMS software platform for GrainGenes 2.0 after consideration of a number of factors, including cost, simplicity, speed, documentation, and size of the user base. The migration process involved four major activities: (1) translating the AceDB data model to MySQL schema, (2) exporting AceDB data to MySQL tables, (3) developing the Internet interface, and (4) developing tools for curation in MySQL.
Relational Schema
The overall conceptual structure of the existing GrainGenes database (Fig. 1) was considered acceptable, as it had been refined over approximately 10 years in the AceDB environment, where changes to data models are relatively simple. Therefore, it was decided to retain the existing data structure and implement it using relational tables, while satisfying the user group's charge to maintain the richness of data connections.
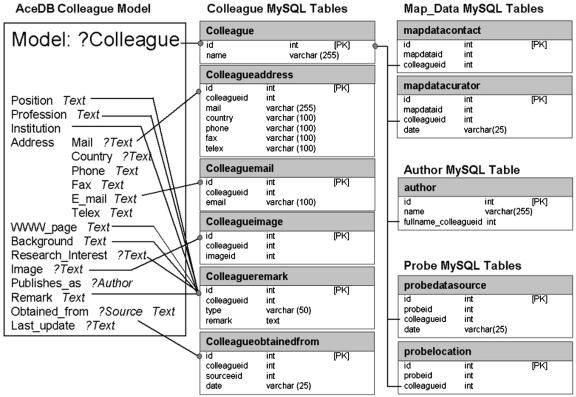
In AceDB, each data record is an independent object containing all the information specified by the model for its data class. For example, a colleague record contains the data indicated in the model for class ?Colleague (Fig. 2, left). Several features of AceDB models presented special issues that were important in the conversion to a relational database schema: (1) By default any data field can be multivalued; and (2) some fields have two or more subfields. For example, the Obtained_from field of ?Colleague comprises a reference to an object of class ?Source, followed by a text string (usually a date).
Figure 2.
Colleague schema in AceDB versus MySQL. Like most data classes in GrainGenes, the simple AceDB data model requires several MySQL tables. ID numbers for a data record, rather than object names, are then referenced by other tables (e.g. the colleagueid row in the tables on the right).
Many of the links between objects of different classes are many-to-many relations; e.g. a colleague can have more than one image, and an image record can link to multiple colleagues.
Implementing these features in a relational schema required the data from a single AceDB object to be broken into multiple tables (Fig. 2, right). Multivalued fields became tables like “colleagueemail,” which could contain multiple rows for each “colleagueid.” Fields with multiple subfields were also handled as separate tables, e.g. table “colleagueobtainedfrom.” Many-to-many relations were implemented with relation tables such as table “colleagueimage,” in which each row contains only the identification (ID) number of a row in the colleague table and the ID number of a row in the image table.
To reduce the number of tables required, some fields judged to be of low priority for querying purposes were combined into a single table with an extra column indicating which field each row was derived from. An example in Figure 2 is the “colleagueremark” table, where the values in the type column can be Position, Profession, Institution, WWW_Page, Background, Research_Interest, or Remark. Also to reduce the number of tables required, the database was analyzed to determine which fields actually contained multiple values, as well as fields that were unpopulated. Fields that were rarely used or only rarely multivalued were reevaluated, and the corresponding MySQL schema were simplified accordingly. The result was a total of 292 MySQL tables for the 38 principle AceDB data classes.
Exporting AceDB Data to MySQL
The AceDB query language (AQL; http://www.acedb.org/Software/whelp/AQL/) was used to extract tab-delimited text files of the data corresponding to each MySQL table. AceDB does not use numeric IDs; the primary key for each data object is its name. To assign the MySQL IDs, the list of object names in each AceDB class was loaded into the corresponding entity table for that class (e.g. table “colleague” in Fig. 2), using the auto_increment feature in MySQL to assign numbers in the ID column. These ID numbers were then substituted for the object names where necessary in the tab-delimited text files extracted from AceDB, for example, column “colleagueaddress.colleagueid.” The resulting files were then loaded into MySQL.
Currently, the direct curation of the database is still being done in AceDB, using its excellent tools for data editing and validation. To update the MySQL database, the set of AQL queries, ID substitutions, and other file manipulation steps involved in exporting and loading the data have been compiled into a single shell script. This script is executed once a week to recreate the MySQL database de novo from the AceDB database. Development of tools for curating and validating new data entry directly in MySQL (activity 4 above) is still under way.
World Wide Web Interface to the Database
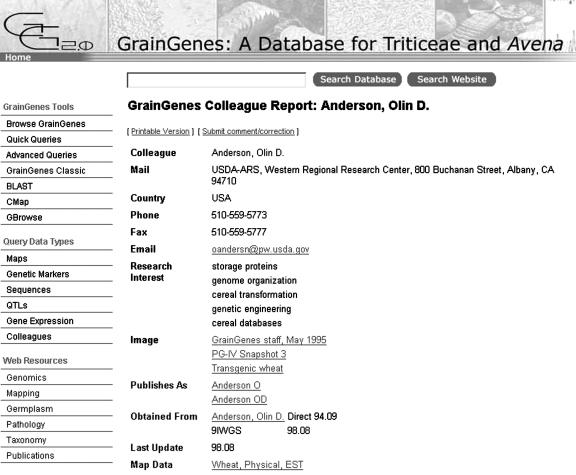
The design team decided that the new GrainGenes homepage, which would integrate the former Web site with a portal into the database, should feel familiar to its long-time user community, be intuitive for new users, and provide a great deal of information without appearing too obtrusive. The existing user interface to the AceDB database, AceBrowser (http://www.graingenes.org), was used as a starting point for design of a Perl CGI interface to the MySQL database. A report script recreated the presentation of each original AceDB object, assembling it from the individual SQL queries for each field of an object of that class (Fig. 3). To search for the user's desired data, four basic kinds of queries were implemented: the Class Browser, direct SQL, Quick Queries, and a full-text search engine.
Figure 3.
Report of a colleague record in GrainGenes 2.0. Colleague records contain contact information as well as links to data contributed by the researcher. The left menu bar appears on all GrainGenes 2.0 pages providing a common portal to the database query tools and information available via the Web site.
The Class Browser (http://wheat.pw.usda.gov/cgi-bin/graingenes/browse.cgi) allows searching for records by name, optionally restricted to a particular data class or in all data classes. The initial page is a query for anything (*) in all classes, providing users with the list of data classes and number of records in each class.
The direct SQL interface (http://wheat.pw.usda.gov/cgi-bin/graingenes/sql.cgi) allows advanced users to compose SQL “select…from…where…” queries to extract tables of any desired data. Some assistance is provided, including links to the schema and editable sample queries. A modified “batch” SQL interface further allows the user to paste in a list of words as arguments to the “where” clause of the query.
The Quick Queries page (http://wheat.pw.usda.gov/GG2/quickquery.shtml) is easy to use but powerful. Specific, sometimes complex, queries that have been requested by users are prewritten and can be executed from a simple Web form with text boxes for entering query terms. Operationally these quick queries are just special cases of the SQL interface; thus, the output is a table of data instead of only a list of hits, and the SQL code for each query is provided in an editable box for advanced users to customize. A popular example is the “Nearby Loci” query, which searches for all loci within a specified distance (e.g. 10 cM) of a specified locus on any map in the database.
The simplest, most inclusive query type is the “Search Database” box at the top of every GrainGenes 2.0 page (Fig. 3), which performs a search for the query string wherever it may occur in the contents of a database report. Currently, the search engine used for this purpose is ht://dig (http://www.htdig.org).
Results of the Migration
The RDBMS version of GrainGenes, although somewhat complex in schema, and the software required to deliver it to the Internet are overall quite satisfactory, with users reporting a substantial improvement. In part this is due to the custom script for generating the data record reports, which not only includes features that existed also in AceBrowser (e.g. links to corresponding records in external databases such as GenBank, TIGR, and Gramene) but also new ones like a shortcut to execute the “Nearby Loci” query immediately from the current Locus record. New features continue to be added and further enhancements are feasible within the system, e.g. possible import of data from other databases instantly via Web services.
In terms of performance, the AceBrowser system is also quite fast for 99% of queries. But for the largest data class (Sequence, approximately one million records), some useful queries take more than 5 min, resulting in Internet browsers timing out. The same queries execute in approximately 10 s in GrainGenes 2.0. The whole GrainGenes 2.0 system is sufficiently portable and is being mirrored daily to another public host site in France (kindly provided by the Institut National de la Recherche Agronomique; http://grain.jouy.inra.fr/GG2/).
A USER'S PERSPECTIVE OF GRAINGENES
As GrainGenes strives to keep relevant for its user base, the curator team is constantly asking the research community for input and perspective. The following is a perspective from one long-time GrainGenes user.
GrainGenes provides plant scientists insight into the current limitations of modern cultivars and genetic routes that might lead to crop improvement breakthroughs. The world's water resource is already strained. As populations increase, especially in the marginal and difficult environments of Africa and Asia, we will increasingly depend upon crop varieties that are productive in environments with increasingly limited water availability and quality. Improving drought tolerance consumes much of the international cereal improvement effort (for insight into the efforts of the Consultative Group for International Agricultural Research in this area, see http://www.generationcp.org) and GrainGenes serves as the primary information hub linking a worldwide user community to useful genes, germplasm, and analytical resources.
The working plant breeder knows that it's not the genes themselves that provide genetic gain, but rather key allelic variants that confer improved specific or general adaptation. When observed in a well-designed and implemented field experiment, these genes and their more useful alleles are indicated by QTL analysis. Improving drought tolerance typically demands greater reliance on local adaptation than does development of lines for irrigated environments (Larson et al., 1996; Yadav et al., 2004). Our collective future depends upon better utilizing our water resource base, and no endeavor better matches Swift's proposition (that he who makes two blades of grass grow where one did before deserves the gratitude of a hungry world) than making our rain-fed environments more sustainably productive.
Key traits that need to be improved in our cereal germplasm bases include rapid canopy closure (Lanning et al., 1997), the “stay green” character (Silva et al. 2000; Haussmann et al., 2002), improved carbon fixation under drought stress (Elouafi and Nachit, 2004; Forster et al., 2004), and a wide array of additional stress management strategies. These improvements cannot be made at the expense of grain quality, disease resistance, and insect resistance. Assembling useful alleles at many loci in appropriate genetic backgrounds is relatively straightforward if we utilize the tools and data available through GrainGenes.
Improving grain yield is another aim of plant breeders. Experts suggest that the world's human population will grow from the current 6.3 billion people to between 7.5 and 8.5 billion in 2025 (see http://www.un.org/esa/population/publications/wpp2002/WPP2002-HIGHLIGHTSrev1.PDF for details). This suggests that the world's grain producers will need to produce around two to four trillion additional calories per day to feed an additional one to two billion individuals. Since a kilogram of wheat flour contains about 2,800 calories, we may need almost an additional million tons of grain per day to support increased population, in total around 350 million tons per year. The grain-importing countries of the world currently import a little more than 100 million tons of grain per year (http://www.openi.co.uk/h050404.htm, data from 2003). While much of this increase in cereal and grain legume production will take place in the current major grain-exporting countries, local production requires less infrastructure development and demands less transportation investment. As a global grains research community, we need to do all we can to make small grains more productive in a wider array of environments in the next 10 to 20 years. GrainGenes, by providing practically useful information to cereals breeders around the world, fills a critical niche.
GrainGenes provides the only site that consolidates comparative linkage maps, sequence polymorphism, and QTL information in a way that plant breeders can readily access. Through its unique foci on QTL and comparative mapping, GrainGenes provides the basic information needed by breeders to deduce the locations of genes with specific alleles that might be beneficial within specific crop production contexts and the markers that will help track them during varietal development. As this resource expands, we will find interesting and uniquely useful alleles of many genes that, when assembled into appropriate genetic backgrounds, will result in breakthrough varieties of barley, wheat, oats, and rye. This has already been accomplished in barley (Hayes et al., 2000; Blake et al., 2002; Castro et al., 2003) and is rapidly being implemented by the world's wheat and oat improvement communities.
Knowing the chromosomal locations of genes that have alleles with significantly different effects on plant performance helps breeders develop “ideotypes,” postulated ideal genotypes for specific environmental or quality needs (Donald, 1979). Understanding the physiological mechanisms that mediate these important adaptations is central if we are to continue improving crop productivity and value. GrainGenes provides an information resource that should, and ideally does, link crop genetics with crop physiology (See et al., 2002; Mickelson et al., 2003). GrainGenes is not merely a data repository, but at its best is a center of hypothesis generation, a site where productive research collaborations are initiated and a conduit for direct interaction between GrainGenes curatorial staff and small-grains researchers.
FUTURE DIRECTIONS
GrainGenes currently serves the small-grains community by providing a common portal to access rapidly expanding data resources. Overlap among mapping experiments has led to the development of consensus linkage maps, and these will grow more dense and more interesting as researchers around the world increase their use of common molecular markers. Plant breeders need to know which genotypes carry useful alleles and how many functional alleles occur for each gene of interest. Comparative QTL analysis will be the starting point from which QTL allele tests will be performed. The GrainGenes team is actively working with the small-grains communities to assure that all described QTL are represented in the database, and this resource should be up to date by the end of 2005. A new QTL query interface will be designed, and enhanced QTL report pages will expand to include traits, trait_studies, environment descriptions, etc. Spotted microarrays and Affymetrix chips are beginning to provide enormous volumes of data concerning the regulation of gene expression and will hopefully lead us to a better understanding of development and how genetic variation may be utilized to enable crops to better fit the environment. GrainGenes will provide the portal small-grains researchers use to interface maps, QTL, and gene expression datasets, enabling development of meaningful and testable hypotheses that in turn will lead to development of better-adapted, more reliable, higher-quality small-grains varieties.
Acknowledgments
The GrainGenes staff would like to thank Mark Sorrells for his ongoing support of the GrainGenes project. We would also like to thank the members of the Liaison Committee for their guidance and suggestions throughout the migration process, and all those in the small-grains community who have contributed data.
This work was supported by the U.S. Department of Agriculture, Agricultural Research Service (project no. 5325–21000–010–01).
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Arumuganathan K, Earle ED (1991) Nuclear DNA content of some important plant species. Plant Mol Biol Rep 9: 208–219 [Google Scholar]
- Blake T, Bowman J, Hensleigh P, Boss D, Carlson G, Kushnak G, Eckhoff J (2002) Release of ‘Valier’ Barley. Crop Sci 42: 1748–1749 [Google Scholar]
- Castro AJ, Chen X, Hayes PM, Johnston M (2003) Pyramiding QTL alleles determining resistance to barley stripe rust: effects on resistance at the seedling stage. Crop Sci 43: 651–659 [Google Scholar]
- Donald CM (1979) A barley breeding programme based on an ideotype. J Agric Sci 93: 261–269 [Google Scholar]
- Elouafi I, Nachit MM (2004) A genetic linkage map of the Durum x T. dicoccoides backcross population based on SSRs and AFLP markers, and QTL analysis for milling traits. Theor Appl Genet 108: 401–413 [DOI] [PubMed] [Google Scholar]
- Forster BP, Ellis RP, Moir J, Talame V, Sanguineti MC, Tuberosa R, This D, Teulat-Merah B, Mariy S, Bahri H, et al (2004) Genotype and phenotype associations with drought tolerance in barley tested in North Africa. Ann Appl Biol 144: 157–168 [Google Scholar]
- Haussmann BIG, Mahalakshmi V, Reddy BVS, Seetharama N, Hash CT, Geiger HH (2002) QTL mapping of stay-green in two sorghum recombinant inbred populations. Theor Appl Genet 106: 133–142 [DOI] [PubMed] [Google Scholar]
- Hayes PM, Castro A, Marquez-Cedillo L, Corey A, Henson C, Jones B, Kling J, Mather D, Matus I, Rossi C, Sato K (2001) A summary of published barley QTL reports. BarleyWorld. http://www.barleyworld.org/northamericanbarley/qtlsummary.php (March 28, 2005)
- Hayes PM, Corey AE, Covel R, Karow R, Mundt C, Rhinart K, Vivar H (2000) Registration of ‘Orca’ Barley. Crop Sci 40: 849–851 [Google Scholar]
- Lanning SP, Talbert LE, Martin JM, Blake TK, Bruckner PL (1997) Genotype of wheat and barley affects light penetration and wild oat growth. Agron J 89: 100–103 [Google Scholar]
- Larson S, McDonald C, Blake TK (1996) Evaluation of barley chromosome 3 yield QTL in a backcross F2 population using PCR-STS markers. Theor Appl Genet 93: 618–625 [DOI] [PubMed] [Google Scholar]
- Matthews DE, Carollo VL, Lazo GR, Anderson OD (2003) GrainGenes, the genome database for small-grain crops. Nucleic Acids Res 31: 183–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh RA, Hart GE, Devos KM, Gale MD, Rogers WJ, Dubcovsky J, Morris CF (2003) Catalogue of gene symbols for wheat. GrainGenes. http://wheat.pw.usda.gov/wgc/2003 (March 28, 2005)
- Mickelson S, Fischer AM, Meyer FD, Garner JP, Blake TK (2003) Mapping of QTLs associated with nitrogen storage and remobilization in barley leaves. J Exp Bot 54: 801–812 [DOI] [PubMed] [Google Scholar]
- See D, Kanazin V, Kephart K, Blake T (2002) Mapping the genes controlling variation in barley grain protein content. Crop Sci 42: 680–685 [Google Scholar]
- Silva SA, Coimbra JLM, Vasconcellos NJS, Lorencetti C, Carvalho FIF, Caetano VR, Oliveira AC (2000) The genetic basis for stay-green in bread wheat. J New Seeds 2: 55–68 [Google Scholar]
- Wiese MV (1987) Compendium of Wheat Diseases. APS Press, St. Paul
- Yadav RS, Hash CT, Bidinger FR, Devos KM, Howarth CJ (2004) Genomic regions associated with grain yield and aspects of post-flowering drought tolerance in pearl millet across stress environments and tester background. Euphytica 136: 265–277 [Google Scholar]