Abstract
Proanthocyanidins (PAs), also called condensed tannins, can protect plants against herbivores and are important quality components of many fruits. Two enzymes, leucoanthocyanidin reductase (LAR) and anthocyanidin reductase (ANR), can produce the flavan-3-ol monomers required for formation of PA polymers. We isolated and functionally characterized genes encoding both enzymes from grapevine (Vitis vinifera L. cv Shiraz). ANR was encoded by a single gene, but we found two highly related genes encoding LAR. We measured PA content and expression of genes encoding ANR, LAR, and leucoanthocyanidin dioxygenase in grape berries during development and in grapevine leaves, which accumulated PA throughout leaf expansion. Grape flowers had high levels of PA, and accumulation continued in skin and seeds from fruit set until the onset of ripening. VvANR was expressed throughout early flower and berry development, with expression increasing after fertilization. It was expressed in berry skin and seeds until the onset of ripening, and in expanding leaves. The genes encoding LAR were expressed in developing fruit, particularly in seeds, but had low expression in leaves. The two LAR genes had different patterns of expression in skin and seeds. During grape ripening, PA levels decreased in both skin and seeds, and expression of genes encoding ANR and LAR were no longer detected. The results indicate that PA accumulation occurs early in grape development and is completed when ripening starts. Both ANR and LAR contribute to PA synthesis in fruit, and the tissue and temporal-specific regulation of the genes encoding ANR and LAR determines PA accumulation and composition during grape berry development.
The polyphenolic compounds known as condensed tannins, or proanthocyanidins (PAs), are plant secondary metabolites synthesized via the flavonoid biosynthetic pathway. They occur in a wide range of plants and play an important role in defense against herbivores (Harborne and Grayer, 1993; Peters and Constabel, 2002). PAs can protect ruminants against pasture bloat (McMahon et al., 2000) and act as antioxidants with beneficial effects for human health including protection against free radical-mediated injury and cardiovascular disease (Bagchi et al., 2000; Lin et al., 2002; Cos et al., 2004). PAs also contribute to the astringency and taste of many fruits and other plant products, such as fruit juices, tea (Camellia sinensis), and wine. In red wine, PAs play an important role in wine quality, contributing to mouthfeel and color stability (Glories, 1988). Thus, from both a nutraceutical and a food quality perspective, it is important to understand the mechanisms leading to the formation of PA polymers and how this is regulated by the plant.
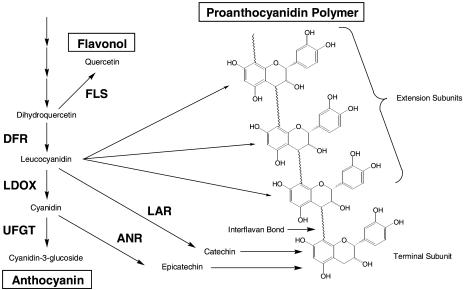
PA biosynthesis is part of the flavonoid pathway that also produces anthocyanins and flavonols. The genetics and biochemistry of the flavonoid pathway have been characterized in several plant species (Shirley et al., 1992; Holton and Cornish, 1995; Boss et al., 1996; Winkel-Shirley, 2001). The synthesis of PAs and anthocyanins share common steps in the flavonoid pathway (Fig. 1), leading to the synthesis of flavan-3,4-diols (such as leucocyanidin), which are precursors for anthocyanin synthesis but that also contribute the extension units of the PA polymer (Stafford, 1990). Synthesis of PA polymers is believed to occur by addition of an intermediate derived from a flavan-3,4-diol (such as leucocyanidin) to a flavan-3-ol terminal unit (such as catechin or epicatechin) with sequential addition of further extension subunits as the polymer lengthens (Fig. 1). In this model, production of larger PA polymers, as occurs in many plant tissues, requires a much higher flux to the flavan-3,4-diols, which produce the extension subunits, than to the flavan-3-ol terminal units produced further down the pathway (Haslam, 1998).
Figure 1.
Scheme of the flavonoid pathway leading to synthesis of anthocyanins and PA polymers. Leucocyanidin is a precursor to synthesis of anthocyanins such as cyanidin-3-glucoside but also contributes the extension subunits of the PA polymer. LAR catalyzes reduction of leucocyanidin to catechin and ANR (BANYULS) catalyzes reduction of cyanidin to epicatechin, both of which can form the terminal unit of the PA polymer. FLS, Flavonol synthase.
Flavan-3-ol biosynthesis had traditionally been considered to be the product of leucoanthocyanidin reductase (LAR), converting leucocyanidin to catechin with an epimerase then converting catechin to epicatechin (Stafford, 1990). LAR activity has been reported in several plants and its activity correlated with PA accumulation (Stafford, 1990; Joseph et al., 1998; Marles et al., 2003). Furthermore, Tanner et al. (2003) purified the LAR protein from leaves of Desmodium uncinatum and isolated the gene encoding this protein. The LAR gene was expressed in Desmodium leaves concomitant with PA accumulation, and the recombinant LAR protein was shown to catalyze the conversion of leucocyanidin, leucodelphinidin, or leucopelargonidin to the corresponding 2,3-trans-flavan-3-ol (Tanner et al., 2003). This demonstration that LAR catalyzes the conversion of leucocyanidin to catechin clearly established its role in PA biosynthesis.
In Arabidopsis (Arabidopsis thaliana), the transparent testa (tt) mutants that have an altered seed coat color define most reactions in flavonoid biosynthesis as well as several specifically involved in PA synthesis (Shirley et al., 1992; Abrahams et al., 2002). The tt genes necessary for PA accumulation include the transcription factors TT8, TT2, and TTG1 (Nesi et al., 2000, 2001; Baudry et al., 2004); three genes, TT19, TT12, and AHA10, involved in transport processes (Debeaujon et al., 2001; Kitamura et al., 2004; Baxter et al., 2005); and the BANYULS gene that catalyzes the conversion of anthocyanidins to the corresponding 2,3-cis-flavan-3-ols, (−)-epiafzelechin, (−)-epicatechin, or (−)-epigallocatechin (Xie et al., 2003). The observation that anthocyanidin reductase (ANR) utilizes cyanidin as a substrate rather than leucocyanidin is also consistent with the observation that leucoanthocyanidin dioxygenase (LDOX) is essential for PA synthesis in Arabidopsis (Abrahams et al., 2003). Thus, in the current model of PA biosynthesis (Fig. 1), LAR and ANR provide two separate pathways for the synthesis of the terminal units for PA polymers. It is not clear whether ANR and LAR provide alternate pathways to PA biosynthesis in different plant tissues or if both branches can be active in some tissues that accumulate high levels of PA.
To date, LAR has been characterized only in Desmodium leaves and ANR only in Arabidopsis and Medicago truncatula. In Arabidopsis, expression of the BANYULS gene was restricted to the endothelium of immature seeds (Devic et al., 1999), while expression in Medicago was observed in young seeds and in flowers and leaves (Xie et al., 2003). While Arabidopsis has no obvious LAR ortholog and seems to use exclusively epicatechin as a PA precursor, many other plants produce both epicatechin and catechin-based PAs in a range of different tissues (Tanner et al., 2003). One such plant is the grapevine Vitis vinifera, which produces PAs in the seeds and skin of the fruit and in the leaves (Souquet et al., 1996). Furthermore, the amount and composition of PAs varies in different parts of the fruit and at different developmental stages of the berry (Kennedy et al., 2000, 2001; Downey et al., 2003a).
In this report, we present the isolation and functional characterization of genes encoding ANR and two LAR isoforms from grapevine. We have measured the PA content of the different tissues and gene expression of VvLAR1, VvLAR2, and VvANR during berry development. The results suggest that both enzymes contribute to PA biosynthesis in grapes, and, to our knowledge, this is the first time that both genes have been isolated and characterized from the same plant species. The understanding of their temporal and tissue-specific regulation will be important for future attempts to alter PA biosynthesis in fruit.
RESULTS
Cloning and Analysis of Grapevine ANR and LAR cDNAs
A cDNA fragment with homology to the BANYULS gene from Arabidopsis (At1g61720), which encodes ANR, was cloned by reverse transcription-PCR from RNA isolated from the skins of Shiraz berries sampled just after flowering. The 1,017-bp open reading frame (ORF), named VvANR, encodes a protein of 339 amino acids. VvANR has 66% identity to AtBANYULS at the nucleotide level and 83% amino acid similarity in the coding region. Southern-blot analysis indicated only a single ANR gene was detected in the Shiraz genome (see Supplemental Figure 2), similar to Arabidopsis (Devic et al., 1999) and Medicago (Xie et al., 2003).
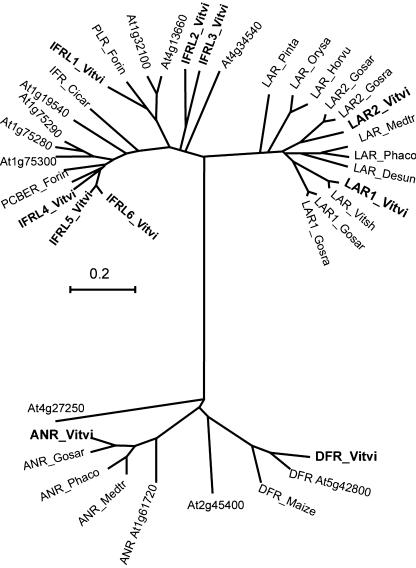
Possible LAR sequences were identified among plant expressed sequence tag (EST) sequences by querying the EST database with the Desmodium LAR (AJ550154; Tanner et al., 2003) protein sequence using the tBLASTn procedure. ESTs similar to Desmodium LAR were identified, assembled into contigs, and translated. When aligned (see Supplemental Figure 1) and displayed in a phylogenetic tree, those protein sequences that clustered together with Desmodium LAR and separately from all the isoflavone reductase (IFR)-like proteins in the Arabidopsis genome were regarded as putative LAR orthologs (Fig. 2). Complete putative LAR protein-coding cDNA sequences were obtained from the plants Hordeum vulgare, Phaseolus coccineus, Pinus taeda, Vitis shuttleworthii, and two each from Gossypium arboretum and Gossypium raimondii, while full gene sequences were obtained from M. truncatula and Oryza sativa. Incomplete LAR sequences were found among poplar (Populus spp.) genomic and sunflower (Helianthus annuus), blueberry (Vaccinium myrtillus), and spatterdock (Nuphar polysepalum) EST sequences (data not shown). Six full-length grape IFR-like proteins designated IFRL 1 to 6 were also identified in this process as the next best grape matches to the LAR sequence and were used in the phylogenetic analysis described later. Amino acid regions conserved in putative LAR sequences but absent from the related IFR proteins were identified. These motifs, indicated by the Desmodium sequence and position, were T(v/i)KRFLPSEFGHD (residues 113–125), ICCNSIA(g/a/s)WPY (residues 159–169), and THDIFI(n/k)GCQ (residues 275–284), and are highlighted in the alignment shown in Supplemental Figure 1. The motifs were termed RFLP, ICCN, and THD, respectively, after some of the conserved amino acids they contained. Degenerate oligonucleotides based on these motifs were designed to enable PCR amplification of LAR fragments from any LAR cDNA sequence. These degenerate primers were used to isolate a partial cDNA clone with homology to the Desmodium LAR gene from Shiraz grapevine cDNA. The 5′ and 3′ ends were identified by rapid amplification of 5′/3′ cDNA ends (5′/3′ RACE). The VvLAR1-1 (AJ865336) cDNA is 1,292-bp long and contains an ORF of 1,038 bp encoding a 346-amino acid protein. VvLAR1-1 and the Desmodium LAR share 66% identity at the nucleotide level and 57% identity and 77% similarity at the protein level. Comparative sequence analysis of VvLAR1-1 within The Institute for Genomic Research (TIGR) grape index (Quackenbush et al., 2000; http://www.tigr.org/tdb/tgi/) identified the tentative consensus (TC) sequence TC27478, which contains ESTs identical with the VvLAR1-1 sequence and of its putative allele VvLAR1-2 (AJ865335). In total, we found 15 nucleotide differences in the coding regions of VvLAR1-1 and VvLAR1-2, resulting in three conservative amino acid changes in the predicted proteins. We identified another sequence in the TIGR grape index, TC32909, which also contains the three LAR conserved motifs and has homology to the Desmodium LAR gene and VvLAR1. To isolate the Shiraz cDNA sequence of this putative LAR isoform, the 5′ end of a 5′ truncated EST clone from TC32909 was amplified by RACE. Primers were designed specific to the extreme 5′ and 3′ ends for the amplification of the 1,392-bp-long cDNA of VvLAR2 (AJ865334). The VvLAR2 cDNA contains a putative ORF of 1,086 bp encoding a 362-amino acid protein that shares 60% identity (77% similarity) with VvLAR1 and 55% identity (68% similarity) with Desmodium LAR. VvLAR2 contains the above-described conserved LAR motifs RFLP, ICCN, and THD with the exception of the first Thr of T(v/i)KRFLPSEFGHD.
Figure 2.
Phylogenetic tree showing LAR and IFR proteins as well as distantly related ANR and DFR proteins of the RED superfamily. The ClustalW multiple sequence alignment (shown in Supplemental Fig. 1) was formed using the default parameters of the MEGA package (Kumar et al., 2004). The tree was constructed from the ClustalW alignment using the unweighted pair group using arithmetic averages method by the MEGA program. The scale bar represents 0.2 substitutions per site. The tree includes all related proteins from the Arabidopsis genome (designated according to their Arabidopsis Genome Initiative number) to give an indication of how the proteins from other species fit into a complete genome. Also included are enzymes for which a catalytic function has been shown such as IFR, pinoresinol-lariciresinol reductase (PLR), and phenylcoumaran benzylic ether reductase (PCBER) in the IFR branch; Arabidopsis and Medicago ANR; and Arabidopsis and maize (Zea mays) DFR. The IFR branch includes six grape IFR-like (IFRL) proteins found in the grape EST collections (IFRL_Vitvi1-6, BN000706-11). The ANR/DFR branch shows only a small fraction of the proteins in this branch of the RED superfamily. The Arabidopsis proteins most closely related to ANR (At4g27250) and DFR (At2g45400) are also present to show that grape ANR and DFR are more closely related to their Arabidopsis orthologs than to the next most closely related Arabidopsis protein. The proteins are labeled according to their proposed catalytic activity followed by the species they come from, e.g. LAR_ Desun is LAR from D. uncinatum (Q84V83). Other species represented in the tree and database accession numbers are Cicer arietinum (IFR, Q00016), Forsythia x intermedia (PLR, AAC49608; PCBER, AAF64174), G. arboretum (ANR CAD91910; LAR1, BN000695; LAR2, BN000699), G. raimondii (LAR1, BN000700; LAR2, BN000701), H. vulgare (LAR, BN000696), M. truncatula (ANR, AAN77735; LAR, BN000703), O. sativa (LAR, BN000704), P. coccineus (ANR, CAD91909; LAR, BN000698), P. taeda (LAR, BN000697), V. shuttleworthii (LAR1, BN000702), grapevine (ANR, CAD91911; DFR, P51110; LAR1, AJ865336; LAR2, AJ865334), and Z. mays (DFR, CAA28734).
Phylogenetic Analysis
LAR and ANR are distantly related members of the reductase-epimerase-dehydrogenase (RED) protein superfamily. Phylogenetic analysis was performed on LAR, IFR (including catalytically verified proteins), ANR, and dihydroflavonol-4-reductase (DFR) protein sequences (Fig. 2; Supplemental Figure 1). The LAR/IFR region of the tree confirms the conclusions of Tanner et al. (2003) that, while LAR is most closely related to the IFR-like proteins, it forms a separate branch. The new tree confirms that the Arabidopsis genome does not encode a LAR because the new LAR proteins listed in Figure 2 are more closely related to each other than to any of the Arabidopsis IFR-like proteins. The distinction between LAR and IFR-like proteins is emphasized by the six grape IFR-like proteins included in the tree, which all fall within the IFR branch, and group with the Arabidopsis and catalytically verified IFR-like proteins. The lower half of the tree shows the relationship between ANR and DFR proteins. The grape ANR used in this work clusters with verified ANRs from Medicago and Arabidopsis and is distinct from the DFR cluster. Using proteins of the Arabidopsis genome as a framework, all the proteins within the ANR and DFR groups are more closely related to each other than to the next most closely related RED proteins in the Arabidopsis genome, namely At4g27250 and At2g45400, respectively, that are also included in the tree. The two grape LAR proteins, like the Desmodium LAR and the other putative LARs listed in Figure 2, consist of the core 315- to 320-amino acid region common to the IFR-like enzymes as well as a much less well conserved C-terminal region.
Functional Analysis of Grapevine ANR and LAR Genes
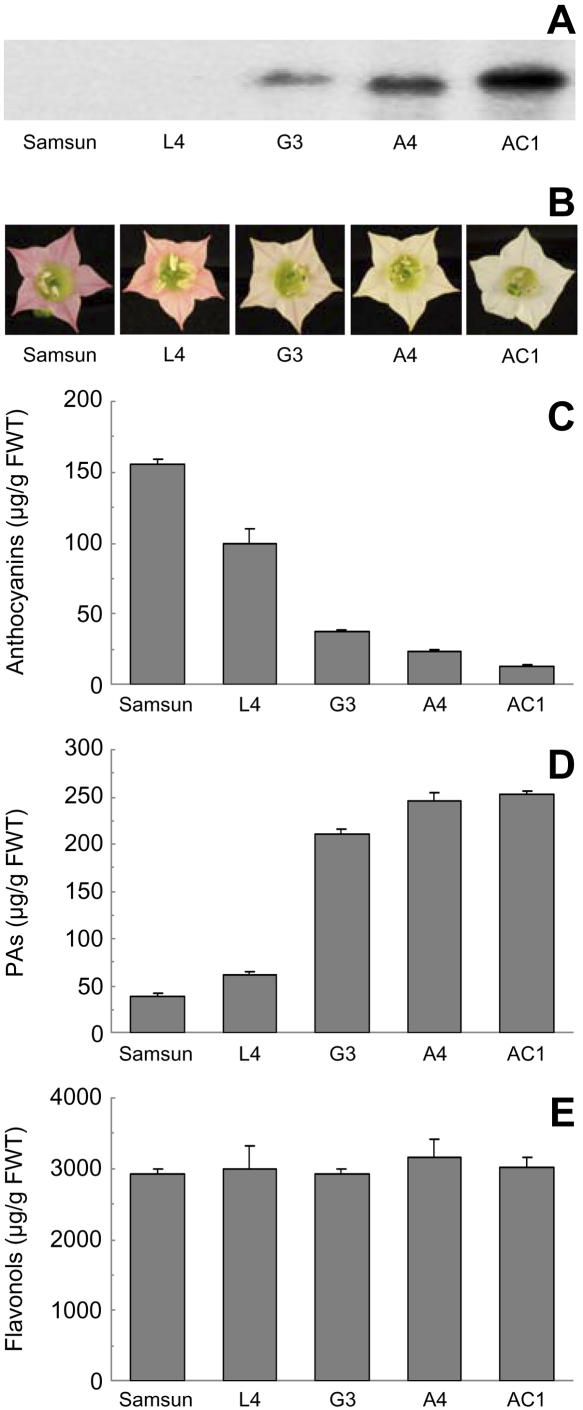
To test the function of VvANR, it was introduced into tobacco (Nicotiana tabacum) under the control of the 35S cauliflower mosaic virus promoter. Several transgenic lines were generated and the petals of the flowers showed visibly decreased color (Fig. 3B). Northern analysis showed that petal color was inversely related to the level of expression of the VvANR transgene, and the petals of lines having the highest expression, such as AC1, were virtually white (Fig. 3, A and B). HPLC analysis of the flavonoids in the petals confirmed the decreased anthocyanin levels, and in the AC1 line the level of anthocyanin was only 8% of that in the petals of untransformed Samsun plants (Fig. 3C). Dimethylaminocinnamaldehyde (DMACA) is a useful reagent for molecules of the PA pathway because it reacts with PA monomers such as flavan-3,4 diols and flavan-3-ols as well as their polymers to form a blue chromophore but does not react with anthocyanidin derivatives. The level of PAs in the petals of transgenic lines, quantitated with DMACA, was inversely proportional to the level of anthocyanins (Fig. 3D), and in the A4 and AC1 lines, PA was more than 6-fold higher than in the petals of the untransformed Samsun plants. There was a stoichiometric inverse relationship between the level of anthocyanins and PAs (Fig. 3, C and D), indicating diversion of anthocyanidin to PA (Fig. 1). Analysis of the PAs in extracts from the transgenic petals by solvent partitioning, HPLC, and thin-layer chromatography indicated the presence of PA monomers cochromatographing with epigallocatechin and gallocatechin and possibly dimers or modified, possibly glycosylated, monomers, but gave no evidence that larger PA polymers were present (data not shown). Flavonols, which were present at much higher levels than anthocyanins or PAs, were not significantly changed in the transgenic lines (Fig. 3E).
Figure 3.
Functional characterization of VvANR by its ectopic expression in tobacco. Flowers of untransformed tobacco (Samsun) and four transgenic lines were analyzed for expression of VvANR by northern analysis (A), flower color (B), anthocyanins by HPLC (C), PAs by DMACA (D), and flavonols by HPLC (E).
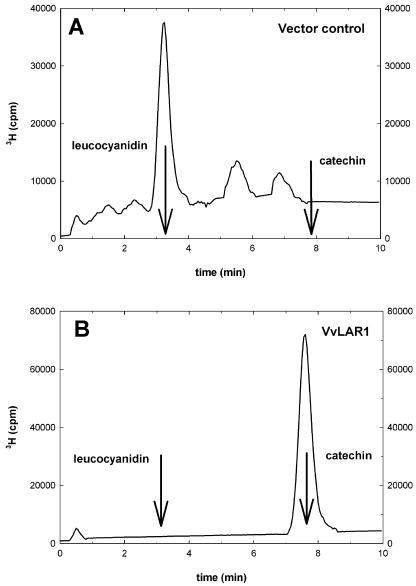
The function of the protein encoded by VvLAR1 was determined by expressing the gene in Escherichia coli and determining enzymic activity of the recombinant protein in the assay described by Tanner et al. (2003). When expressed in E. coli the VvLAR1 protein produced levels of LAR activity comparable to those produced by the recombinant D. uncinatum LAR protein. All the leucocyanidin substrate was converted to catechin by 1 μL of either enzyme preparation, but no detectable catechin was produced by 20 μL of a control E. coli extract (Fig. 4).
Figure 4.
Enzyme activity of recombinant grapevine LAR (VvLAR1). HPLC chromatography of LAR reactions of E. coli transformed with a control pET plasmid (A) or the pET LAR1_Vitvi plasmid (B). The chromatograms show the elution profiles of the 3H-labeled reaction mixture obtained and processed as described in “Material and Methods.” The retention times for the substrate (leucocyanidin) and the product (catechin) are indicated by arrows.
Expression of VvANR, VvLAR1, and VvLAR2, and PA Accumulation in Grapevine Tissues
The expression of VvLAR1, VvLAR2, and VvANR was investigated throughout grape berry development during the season 2000 to 2001 by real-time PCR. Expression of the gene encoding LDOX, which is required for synthesis of epicatechin and anthocyanins (Fig. 1), was also determined to monitor flavonoid pathway activity (Boss et al., 1996). VvUbiquitin1 (BN000705) is a putative ubiquitin extension protein with a ribosomal L40 C-terminal (Kato et al., 2001). It was chosen for normalization of gene expression because TIGR Gene Indices (Quackenbush et al., 2000) showed that its transcripts are less abundant than transcripts of the polyubiquitin genes, and its expression was found to be relatively constant throughout grape berry development (Downey et al., 2003b). This was verified by comparison of the normalized real-time PCR data with northern-blot analysis of VvANR expression throughout berry development (see Supplemental Figure 2). Additionally, the expression patterns for the anthocyanin biosynthetic genes UDP Glc:flavonoid-3-O-glucosyltransferase (UFGT) and LDOX were measured by real-time PCR during berry development, and we obtained very similar patterns of expression to those described by Boss et al. (1996) using northern analysis.
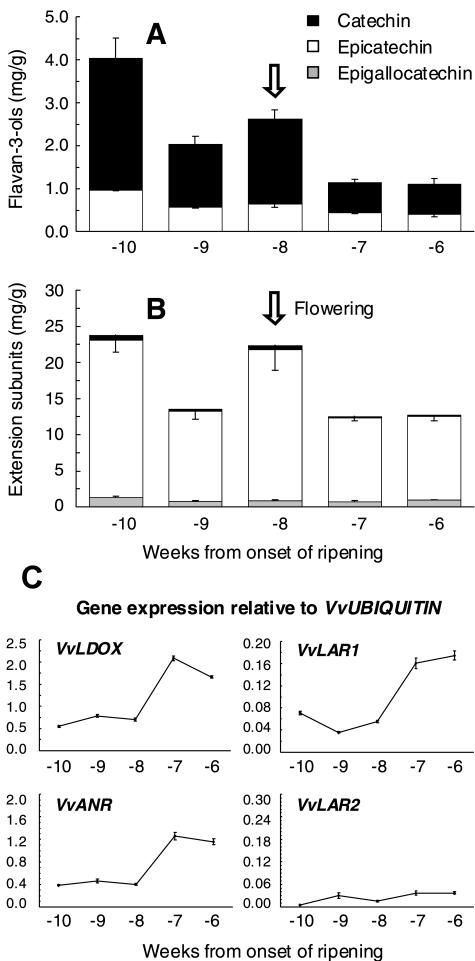
Figure 5 shows PA levels and expression patterns for VvLDOX, VvANR, VvLAR1, and VvLAR2 in grapes early in berry development, when the different tissues such as skin and seeds could not be readily separated. PAs were measured by acid-catalyzed cleavage of the polymers, and data are presented for total extension subunits and for flavan-3-ols, which includes terminal subunits and free monomers (Downey et al., 2003a). The concentration of PAs was maximal 2 weeks prior to flowering, which occurred 8 weeks before véraison, where véraison is defined as the onset of ripening in grapes. In developing flowers and berries, the PAs comprised polymers with a mean degree of polymerization (mDP) varying from 10 subunits in preflowering samples to 25 subunits 1 week after flowering. These polymers consisted of predominantly epicatechin extension subunits (Fig. 5B), while the flavan-3-ols were a mixture of catechin and epicatechin (Fig. 5A). The grapevine genes encoding LDOX and ANR were expressed before flowering, but there was a significant increase in their expression in the 2 weeks after flowering (Fig. 5). Expression of VvLAR1 showed a similar pattern to VvLDOX and VvANR, increasing after flowering, whereas expression of VvLAR2 was very low and showed less of an increase after flowering.
Figure 5.
Accumulation of PAs and gene expression of VvLDOX, VvANR, VvLAR1, and VvLAR2 in grapes during the early stages of berry development. Flowering (indicated by down arrow) occurred 8 weeks before véraison, which is the onset of ripening in grapes. A, Flavan-3-ol accumulation and composition. B, Accumulation and composition of PA extension subunits. C, Gene expression of VvLDOX, VvANR, VvLAR1, and VvLAR2. Expression was determined by real-time PCR and is shown relative to expression of VvUbiquitin1 in each sample. All data is presented as mean of three replicates.
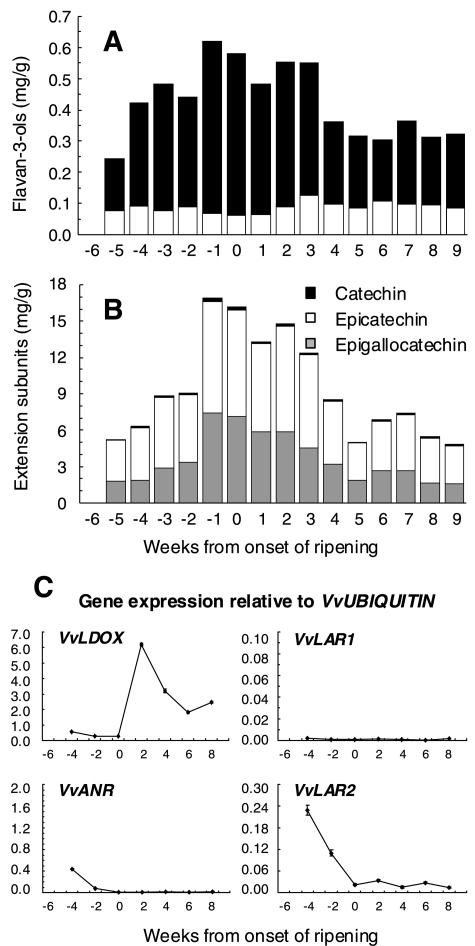
From 6 weeks before véraison, the grapes were large enough to allow the skins and seeds to be analyzed separately, although it was not possible to obtain samples of skin from berries 6 weeks before véraison. PA levels and expression patterns for VvLDOX, VvANR, VvLAR1, and VvLAR2 in grape skins are shown in Figure 6. In grape skin, the PAs were polymers of 25 to 40 subunits, consisting of epicatechin and epigallocatechin extension subunits in similar proportions (Fig. 6B). In contrast, catechin was the predominant flavan-3-ol in grape skins (Fig. 6A). The concentration of both extension subunits and flavan-3-ols increased from 5 weeks before véraison, reaching a maximum around the time ripening commenced, then declining during ripening. Expression of VvLDOX decreased from 6 weeks before véraison to very low levels in the weeks just before véraison, then increased significantly following véraison, concomitant with the beginning of anthocyanin synthesis during ripening (Boss et al., 1996). Expression of VvANR also decreased from 6 weeks before véraison and was not detected from véraison onwards. VvLAR1 was detected 4 weeks before véraison in skin but not at any later stages, whereas VvLAR2 expression increased to a maximum 4 weeks before véraison then declined to a low level at véraison and was maintained at this level throughout ripening.
Figure 6.
Accumulation of PAs and gene expression of VvLDOX, VvANR, VvLAR1, and VvLAR2 in grape skin during berry development. No skin sample was obtained for grapes 6 weeks before the onset of ripening. Time point −6 is included only for better comparison with Figure 5. A, Flavan-3-ol accumulation and composition. B, Accumulation and composition of PA extension subunits. C, Gene expression of VvLDOX, VvANR, VvLAR1, and VvLAR2. Expression was determined by real-time PCR as described for Figure 5.
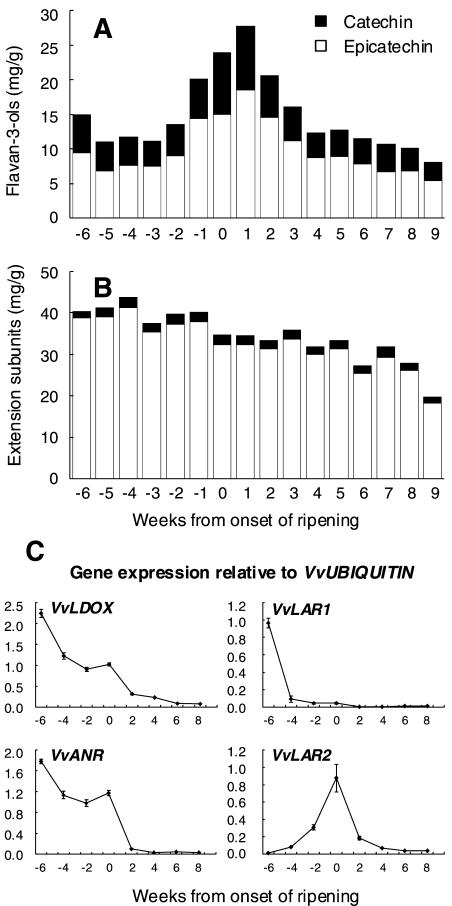
The pattern of PA accumulation and gene expression in seeds (Fig. 7) differed from that in skins. PAs in seed were much smaller polymers than in skin with a mDP of 4 to 6 subunits and comprised predominantly epicatechin, particularly in the extension subunits. The concentration of flavan-3-ols increased to a maximum just after véraison, whereas the level of extension subunits was relatively constant until véraison then declined during ripening (Fig. 7). In seeds, expression of VvLDOX and VvANR was maximal 6 weeks before véraison, with both genes expressed up until véraison, then expression declined to low levels during ripening. The two forms of LAR showed quite different expression patterns, with VvLAR1 expressed 6 weeks before véraison then declining to low levels, whereas VvLAR2 expression increased to a maximum at véraison then declined.
Figure 7.
Accumulation of PAs and gene expression of VvLDOX, VvANR, VvLAR1, and VvLAR2 in grape seeds during berry development. A, Flavan-3-ol accumulation and composition. B, Accumulation and composition of PA extension subunits. C, Gene expression of VvLDOX, VvANR, VvLAR1, and VvLAR2. Expression was determined by real-time PCR as described for Figure 5.
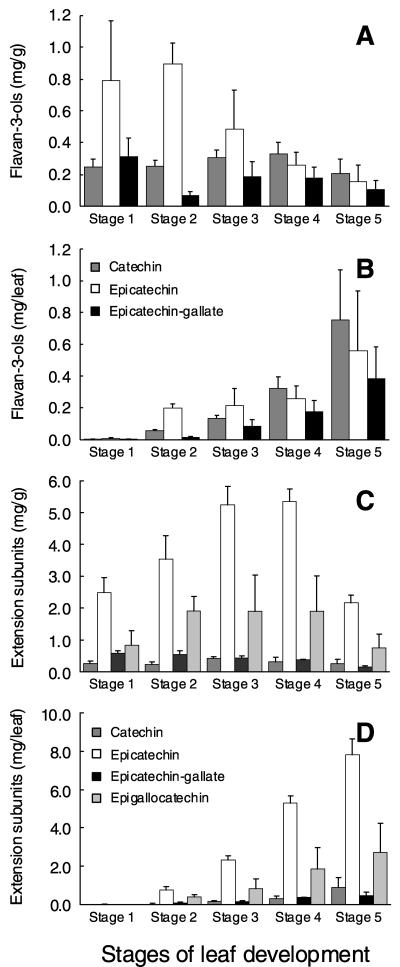
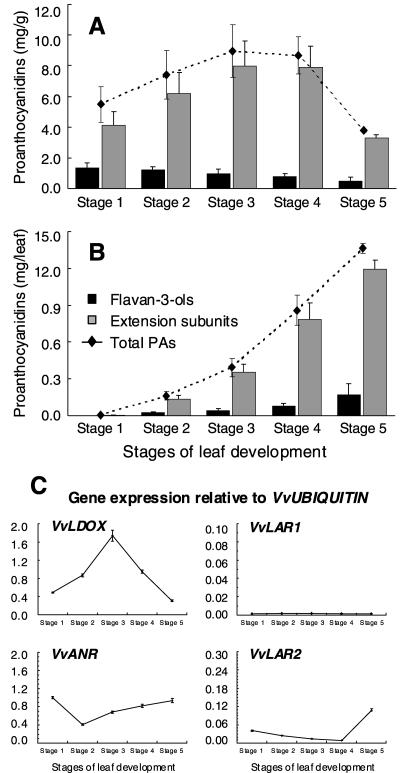
Grapevine leaves also contain significant levels of PAs, but the composition differed from that in fruit (Fig. 8). The leaf PA extension subunit composition was similar at all stages of leaf development, with epicatechin the major subunit followed by epigallocatechin, with small amounts of catechin and epicate chin-gallate (Fig. 8). Leaf PA comprised smaller polymers than berry skins, with the mDP ranging from 4 subunits in Stage 1 leaves up to 14 subunits in Stage 4 leaves (data not shown). The total amount of PAs per leaf increased throughout leaf expansion but the concentration of PAs was highest in Stage 3 and 4 leaves and was lower in fully expanded Stage 5 leaves, indicating that tannin synthesis declined in the later stages of leaf development (Fig. 9). VvLDOX was expressed at all stages of leaf development with maximal expression in Stage 3 leaves, whereas VvANR expression was highest in Stage 1 leaves, declined in Stage 2 leaves then increased in later stages (Fig. 9). By comparison with berries, expression of VvLAR1 was very low at all stages of leaf development, and expression of VvLAR2 was also very low in the first four stages of leaf development but increased in fully expanded Stage 5 leaves (Fig. 9).
Figure 8.
Composition of PAs in grape leaves during development. A, Flavan-3-ol content as mg/g. B, Flavan-3-ol content as mg per leaf. C, Extension subunits of PAs shown as mg/g. D, Extension subunits of PAs shown as mg per leaf. The stages of leaf development are defined in “Materials and Methods.” All data is presented as mean of three replicates.
Figure 9.
Accumulation of PAs and gene expression of VvLDOX, VvANR, VvLAR1, and VvLAR2 in grape leaves during development. A, PAs measured as mg/g. B, PAs measured as mg per leaf. C, Gene expression of VvLDOX, VvANR, VvLAR1, and VvLAR2. Expression was determined by real-time PCR as described for Figure 5.
DISCUSSION
PAs are important quality components of many fruits, contributing to their taste and health benefits. However, little is known about the genes contributing to the synthesis of PAs in fruit and their regulation. In this study, we show that the genes encoding both LAR and ANR are expressed during PA synthesis in grapes and are regulated in a temporal- and tissue-specific manner.
The BANYULS gene encodes ANR and has been shown to be involved in PA synthesis in the Arabidopsis seed coat (Devic et al., 1999; Xie et al., 2003), while LAR has been shown to be involved in PA biosynthesis in leaves of Desmodium (Tanner et al., 2003). Nevertheless, up to now, ANR and LAR have not been characterized from the same plant species. Grapevines accumulate significant levels of PAs in their fruit and leaves and offer an opportunity to examine the contributions of LAR and ANR to PA biosynthesis in different tissues of the same plant. To this end, we isolated the cDNAs coding for LAR (VvLAR1 and VvLAR2) and ANR (VvANR) from grapevine and determined their expression during berry and leaf development. The functionality of VvANR was demonstrated by ectopic expression in tobacco, resulting in flowers with decreased anthocyanins and an equivalent increase in PAs (Fig. 3) as observed by Xie et al. (2003) for Arabidopsis BANYULS. This is consistent with ANR competing with UFGT for cyanidin, diverting metabolism away from production of the anthocyanin and toward production of epicatechin (Fig. 1). VvLAR was shown to be functional by measuring enzyme activity of the recombinant protein (Fig. 4).
Development of the grape berry occurs in two successive growth phases (Coombe and McCarthy, 2000; Robinson and Davies, 2000). Following fertilization, the first phase of berry growth is initiated and berry expansion and seed formation occur in parallel. Following véraison, ripening occurs during the second growth phase and results in sugar accumulation, decreased acid levels, softening of the berry, and accumulation of anthocyanins and flavor compounds.
As PA biosynthesis is a branch of the flavonoid pathway (Fig. 1), regulation of this pathway must occur to coordinate synthesis of different flavonoids in different tissues of the grape berry during early development and ripening. The flavonoid pathway has been well studied in relation to anthocyanin synthesis in grapes but not in relation to PA synthesis. In Shiraz grapes, most genes of the flavonoid pathway are expressed at flowering and during early berry development, then expression declines 4 to 6 weeks after flowering before increasing again at véraison, when anthocyanin synthesis commences and UFGT expression is induced (Boss et al., 1996). No anthocyanins are made before véraison, and there is no detectable expression of the gene encoding UFGT. Therefore, it was assumed that expression of flavonoid pathway genes early in berry development is related to synthesis of other flavonoids, such as flavonols and PAs (Boss et al., 1996). The concentration of flavonols in grapes and the expression of the gene encoding flavonol synthase were both found to be highest 1 week before flowering and decreased after fertilization (Downey et al., 2003b). Grapevine flowers also contain significant levels of PAs (Fig. 5), indicating that PA synthesis also occurs in the developing flower prior to pollination. This is consistent with the expression of most of the flavonoid pathway genes at this stage (Boss et al., 1996), including LDOX (Fig. 5), as the pathway to epicatechin involving ANR would also require LDOX expression (Fig. 1). In the 2 weeks following flowering, expression of LDOX increased (Fig. 5), indicating increased flavonoid pathway activity. There was also an increase in the expression of VvANR and VvLAR1 (Fig. 5), indicating increased PA synthesis. In grapes, it appears that both flavonol and PA accumulation occur during formation of the flower. Following fertilization, fruit development starts and flavonoid pathway activity is directed primarily toward PA synthesis, with expression of genes encoding both ANR and LAR being elevated (Fig. 5) while expression of the gene encoding flavonol synthase is decreased (Downey et al., 2003b). At these early stages of fruit development, it was not possible to dissect the skin and seed tissues from the berry, so localization of the expression of these genes will require other techniques, such as in situ hybridization.
PAs appear to be synthesized during the first phase of grape berry development, and there is a decline in extractable PA after véraison and throughout ripening, which is thought to be the result of complexation of the PA polymers with other cellular components (Kennedy et al., 2000, 2001; Downey et al., 2003a). The level of PA detected in grapes thus represents a balance between accumulation of PA through synthesis and decreased extractability, and may not be a true reflection of biosynthesis in the fruit. The expression patterns of genes encoding ANR and LAR indicate that PA biosynthesis occurs before flowering and then increases following fertilization, in parallel with formation and maturation of the seeds in the developing fruit. PA synthesis appears to continue in the seed up until 2 to 4 weeks after véraison (Fig. 7), at which time the seed is generally mature and the seed coat has turned brown (Coombe and McCarthy 2000; Kennedy et al., 2000). The peak of expression of VvLAR2 in seeds at véraison occurred just before the peak in the level of flavan-3-ols (Fig. 7) and suggests that PA monomers were still being synthesized quite late in seed development. In contrast, PA accumulation in grape skins appeared to be complete by véraison, when expression of genes encoding ANR, LAR, and LDOX had declined to low levels (Fig. 6). The increase in LDOX following véraison was related to the onset of anthocyanin synthesis, where the expression of most genes in the pathway is increased, including the gene encoding UFGT (Boss et al., 1996). This indicates that in the skin of grape berries, synthesis of anthocyanin and PAs is temporally separated and anthocyanin synthesis occurs after PA accumulation is complete. A similar pattern of flavonoid biosynthesis was reported in bilberry (Vaccinium myrtillus), with two separate phases of expression of flavonoid pathway genes (Jaakola et al., 2002). The first phase, around flowering, coincided with flavonol and PA synthesis, and the second phase, later in fruit development, coincided with anthocyanin biosynthesis.
Grapevine leaves also contained significant levels of PAs, although the composition differed from that in grape berry skins and seeds (Fig. 8; Downey et al., 2003a). VvANR was expressed at high levels throughout leaf development, whereas expression of VvLAR1 was extremely low in leaves, and significant expression of VvLAR2 was only detected in mature leaves (Fig. 9). This expression, combined with the greater proportion of flavan-3-ols and extension subunits with 2,3-cis- stereochemistry (Fig. 8; Xie et al., 2003), indicates that ANR may have a more significant role in PA synthesis in grapevine leaves than LAR.
In Arabidopsis, expression of BANYULS (encoding ANR) was reported only in the endothelial layer of the developing seed coat (Devic et al., 1999), whereas in Medicago, expression was also detected in flower buds and at low levels in leaves (Xie et al., 2003). The grapevine homolog, VvANR, was expressed at similar levels in leaves and flowers as well as in the skin and seeds of developing berries. All of these tissues accumulate significant levels of PAs containing epicatechin-based subunits, although the PA polymer length and composition differs between the tissues. The results indicate that in grapevine, ANR has a role in PA synthesis in a number of different plant tissues. In Arabidopsis, expression of BANYULS in the developing seed coat is regulated synergistically by at least three separate regulatory genes, TT2, TT8, and TTG1 (Baudry et al., 2004). It will be of interest to determine if homologous regulators control expression of VvANR in grapevine, where the gene is expressed in a much wider range of tissues and temporal control of PA synthesis needs to be coordinated with production of other flavonoids, such as anthocyanins and flavonols.
The gene encoding LAR in Desmodium was expressed in leaves (Tanner et al., 2003), and while there are LAR homologs in many plants, it is not known whether these are expressed in other tissues such as the fruit. The two genes encoding LAR in grapevines showed different, and tissue-specific, patterns of expression. VvLAR1 appeared to be seed specific, with the highest expression occurring 2 weeks after flowering, while only very low levels of VvLAR1 expression were detected in berry skin and in leaves. Expression of VvLAR2 was also highest in seeds but maximum expression occurred at véraison. VvLAR2 was also detected in flowers, skin, and leaves, albeit at a lower level. The results indicate that in grapevine, LAR contributes to PA synthesis in the skin and seed of the fruit, as well as in leaves. The two grapevine LAR genes clearly have different patterns of expression, and it will be of interest to determine whether this represents two genes of LAR with differing properties.
In grapevine, it appears that both ANR and LAR contribute to PA synthesis in leaves, flowers, and in the skin and seeds of the developing fruit. These tissues contain PAs of different polymer length and subunit composition, which raises the question of how PA composition is influenced by the relative contributions of ANR and LAR to flavan-3-ol synthesis in these tissues. As there are two distinct pathways to the formation of catechin and epicatechin, manipulation of the relative activities of ANR and LAR may have the potential to modify both the content and composition of PAs in plant tissues (Schijlen et al., 2004).
MATERIALS AND METHODS
Plant Material
Grapevine tissues of Vitis vinifera L. cv Shiraz were collected from a commercial vineyard during the 2000 to 2001 season. Approximately 10 leaves and 100 berries from at least 20 bunches were collected at weekly intervals throughout berry development from floral initiation until harvest. Bunches were randomly collected from 120 vines on a modified Scott-Henry trellis, as described in Downey et al. (2003a). Whole berries were sampled and analyzed from 6 weeks before véraison, when seeds could be readily separated from the remainder of the berry. Four weeks before véraison, it was also possible to readily separate the skin from the rest of the berry. Leaves from Shiraz grapevines were collected at five stages of development. The first stage was a newly emerged leaf less than 2 cm2 and 0.09 g. The second stage was 5 to 6 cm2 and weighing 0.22 g. Shiraz leaf Stage 3 was approximately 15 cm2 and 0.44 g. Stage 4 was an expanded leaf around 40 cm2 and 0.99 g. The final leaf stage, Stage 5, was a fully expanded mature leaf approximately 160 cm2 in area, weighing 3.62 g. All samples were frozen in liquid nitrogen upon collection in the field and stored at −80°C until analyzed.
HPLC Analysis
Grape PAs were extracted into 70% acetone with 0.1% ascorbate and analyzed by HPLC following acid-catalyzed cleavage in the presence of excess phloroglucinol, as described by Downey et al. (2003a).
Anthocyanins and flavonols in transgenic tobacco (Nicotiana tabacum) petal tissues were extracted into methanol/HCl as described in Downey et al. (2004). Anthocyanin and flavonol content and composition were determined by reverse-phase HPLC using an HP1100 system (Agilent) with a Wakosil analytical column (150 mm×4.6 mm; 3μm packing; SGE International). The HPLC separation utilized a binary solvent gradient where Solvent A was 10% formic acid (v/v with water) and Solvent B was methanol. The gradient conditions were: zero minutes, 17% Solvent B; 15 min, 35% Solvent B; 40 min, 37% Solvent B; 42 min, 100% Solvent B; 44 min, 100% Solvent B; 45 min, 17% Solvent B; and 46 min, 17% Solvent B. The column was maintained at 40°C and the flow rate was 1.0 mL/min. Anthocyanins and flavonols were expressed as malvidin-3-glucoside and quercetin-3-glucoside equivalents, respectively, based on commercial standards (Extrasynthese).
Preparation of cDNAs
Total RNA was isolated from the various plant tissues as described in Downey et al. (2003b). The quality of RNA was verified by demonstration of intact ribosomal bands following agarose gel electrophoresis in addition to the absorbance ratios (A260/280) of 1.8 to 2.0. For cDNA synthesis, four micrograms of total RNA were reverse transcribed using oligo d(T)18 and SuperScript III reverse transcriptase (Invitrogen Life Technologies) following the protocol of the supplier.
Cloning of VvANR and VvLAR
Degenerate PCR primers designed to the aligned sequences of the Arabidopsis (Arabidopsis thaliana) BANYULS cDNA and an EST sequence from cotton (Gossypium hirsutum) allowed the amplification and cloning of a fragment of the grapevine BANYULS homolog. Standard RACE-PCR reactions were used to identify the 5′ and 3′ ends of the coding sequence and a full-length clone was subsequently cloned into pGem-T-Easy (Promega) using standard protocols and primers designed to, and including, the initiation and termination codon of the cDNA. A 514-bp PCR-fragment of VvLAR1-1 was amplified from Shiraz skin cDNA (2 weeks after flowering) with the degenerate primers RFLP, 5′-GCIGTIAARMGITTYYTICCIWSIGARTTYGGICAYGAY-3′, and DHT (i.e. THD rev), 5′-GRCAICCITTDATRAADATRTCRTGIGT-3′. The 5′ and 3′ ends of VvLAR1-1 and the 5′ end of VvLAR2 were obtained from Shiraz skin cDNA using RACE-PCR (GeneRacer, Invitrogen) according to the supplier's protocol. The 3′ end of VvLAR1-1 was amplified with the gene-specific primer LAR1F2 (5′-GATGTCCGAACACTGAACAA-3′) in conjunction with the supplier's 3′-nested primer. The 5′-RACE of LAR1-1 was conducted using the gene-specific primer LAR1R1 (5′-TTGAGACAATTGCAAGATGG-3′) and the supplier's 5′-nested primer. The sequence of a 5′-truncated EST clone from TC32909 was used to design the gene-specific primer LAR2racen (5′-ATGTCGTGCCCAAATTCTGATGGAAGG-3′) for 5′-RACE of VvLAR2. The final full-length sequences of VvLAR1 and VvLAR2 were then amplified from Shiraz skin cDNA using PfuTurbo polymerase (Stratagene) and gene-specific primers directed to the putative 5′- and 3′-untranslated regions of the respective gene. All described PCR products were tailed using A-Addition kit (Qiagen), cloned into pDrive (Qiagen), and subjected to DNA sequencing.
Real-Time PCR for Expression Analysis of LAR and ANR
Expression analysis of the genes VvLAR1, VvLAR2, and VvANR was done by real-time PCR, using SYBER green method on a Rotor-Gene 2000 (version 4.2) real-time cycler (Corbett Research). Each PCR reaction (15 μL) contained 26 6-nm primer (each), cDNA (diluted 1:60), and 1× ABsolute QPCR SYBR Green ROX mix (ABgene House). The thermal cycling conditions were 95°C for 15 min followed by 95°C for 30 s, 58°C for 25 s, and 72°C for 25 s for 40 cycles, followed by a melt cycle from 50°C to 96°C. The primers LAR1F (5′-CAGGAGGCTATGGAGAAGATAC-3′) and LAR1R (5′-ACGCTTCTCTCTGTACATGTTG-3′) were designed to amplify across an intron of VvLAR1, giving products of 210 bp from cDNA and 297 bp from genomic DNA. This primer pair was used to check all used cDNAs for genomic contaminations. The EST clone TC32909 (TIGR database) was used to design the primers TC32F (5′-TCTCGACATAAATGATGATGTG-3′) and TC32R (5′-TGCAGTTTCTTTGATTGAGTTC-3′) amplifying a 166-bp DNA fragment from the 3′-untranslated region of VvLAR2. The primers LDOXF1 (5′-ACCTTCATCCTCCACAACAT-3′) and LDOXR2 (5′-AGTAGAGCCTCCTGGGTCTT-3′) were used for real-time PCR of VvLDOX (accession no. X75966) amplifying a 340-bp DNA fragment. The primers BANF (5′-CAATACCAGTGTTCCTGAGC-3′) and BANR (5′-AAACTGAACCCCTCTTTCAC-3′) were used for real-time PCR of VvANR amplifying a fragment of 141 bp. With all cDNAs used, the primer sets gave single PCR products, which were verified by determining the melt curves for the products at the end of each run, by analysis of the products using gel electrophoresis, and by comparing the DNA sequence of the PCR products with the gene sequence. The efficiency of the primers was tested in preliminary experiments with dilutions of the purified PCR product and maintained an r2 value ≥ 0.98. The expression of genes was normalized to VvUbiquitin1 (TC32075, TIGR database), and transcripts were detected by amplifying a 182-bp product with the primers VvUbiquitin Forward (5′-GTGGTATTATTGAGCCATCCTT-3′) and VvUbiquitin Reverse (5′-AACCTCCAATCCAGTCATCTAC-3′). All samples were measured in triplicate, every run included the VvUbiquitin1 control for each sample, and experiments were repeated. The difference between the cycle threshold (Ct) of the target gene and the Ct of Ubiquitin, ΔCt = CtTarget − CtUbiquitin, was used to obtain the normalized expression of target genes, which corresponds to 2−ΔCt. The Rotor Gene 2000 software (Corbett Research) and the Q-Gene software (Muller et al., 2002) were used to calculate the mean normalized expression of the genes.
Transformation of Tobacco with VvANR
The full-length VvANR cDNA was cloned into the vector pART7 and transformed into tobacco via the pART27 vector under the control of the CaMV 35S promoter. Expression of VvANR in transformed plants was determined by northern-blot analysis, as described by Boss et al. (1996). PAs in transgenic tobacco petals were determined by extracting the petal tissue into 70% acetone with 0.1% ascorbate and quantitating PAs with the DMACA reagent described by Nagel and Glories (1991) using catechin (Sigma) as a standard.
Expression of VvLAR1 in Escherichia coli
The coding region of VvLAR1-2 was amplified with Platinum Pfx Polymerase (Invitrogen Life Technologies) from Shiraz cDNA and introduced into the NdeI and BamHI sites of the expression vector pET14b (Novagen) using the primers LAROEF (5′-GCACATATGACTGTTTCTCCGGTTCCT-3′) and LAROER (5′-ATAGGATCCTCAAGCGCAGGTTGCAGTGACTT-3′). The sequence of the clone was verified by DNA sequencing. This plasmid was then introduced into the Rosetta (DE3) (pLysS) strain of E. coli. For expression, bacteria were grown in Luria-Bertani medium at 37°C for 2 h, isopropylthio-β-galactoside added to 1 mm, and the bacteria grown for another four hours. The cells were collected, disrupted by sonication in 5 volumes of Grinding buffer and insoluble material removed by centrifugation. LAR activity of 1-μL portions was determined by monitoring the conversion of [4-3H]-3,4-cis-leucocyanidin to [3H] catechin by HPLC as described by Tanner et al. (2003).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AJ865336 (VvLAR1-1), AJ865335 (VvLAR1-2), AJ865334 (VvLAR2), and CAD91911 (VvANR).
Supplementary Material
Acknowledgments
We wish to thank John and Di Harvey of Slate Creek vineyard, Willunga, for their exceeding generosity in providing access to grapevines; Nicole Cordon for HPLC analysis of the tobacco petals; and Karin Sefton for excellent technical assistance.
This research was supported by an Australian Postgraduate Award and by the Grape and Wine Research and Development Corporation.
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.064238.
References
- Abrahams S, Lee E, Walker AR, Tanner GJ, Larkin PJ, Ashton AR (2003) The Arabidopsis TDS4 gene encodes leucoanthocyanidin dioxygenase (LDOX) and is essential for proanthocyanidin synthesis and vacuole development. Plant J 35: 624–636 [DOI] [PubMed] [Google Scholar]
- Abrahams S, Tanner GJ, Larkin PJ, Ashton AR (2002) Identification and biochemical characterization of mutants in the proanthocyanidin pathway in Arabidopsis. Plant Physiol 130: 561–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagchi D, Bagchi M, Stohs SJ, Das DK, Ray SD, Kuszynski CA, Joshi SS, Pruess HG (2000) Free radicals and grape seed proanthocyanidin extract: importance in human health and disease prevention. Toxicology 148: 187–197 [DOI] [PubMed] [Google Scholar]
- Baudry A, Heim MA, Dubreucq B, Caboche M, Weisshaar B, Lepiniec L (2004) TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J 39: 366–380 [DOI] [PubMed] [Google Scholar]
- Baxter IR, Young JC, Armstrong G, Foster N, Bogenschutz N, Cordova T, Peer WA, Hazen SP, Murphy AS, Harper JF (2005) A plasma membrane H+-ATPase is required for the formation of proanthocyanidins in the seed coat endothelium of Arabidopsis thaliana. Proc Natl Acad Sci USA 102: 2649–2654; erratum Baxter IR, Young JC, Armstrong G, Foster N, Bogenschutz N, Cordova T, Peer WA, Hazen SP, Murphy AS, Harper JF (2005) Proc Natl Acad Sci USA 102: 5635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boss PK, Davies C, Robinson SP (1996) Analysis of the expression of anthocyanin pathway genes in developing Vitis vinifera L. cv Shiraz grape berries and the implications for pathway regulation. Plant Physiol 111: 1059–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombe BG, McCarthy MG (2000) Dynamics of grape berry growth and physiology of ripening. Aust J Grape Wine Res 6: 131–135 [Google Scholar]
- Cos P, De Bruyne T, Hermans N, Apers S, Berghe DV, Vlietinck AJ (2004) Proanthocyanidins in health care: current and new trends. Curr Med Chem 11: 1345–1359 [DOI] [PubMed] [Google Scholar]
- Debeaujon I, Peeters AJM, Léon-Kloosterziel KM, Koornneef M (2001) The TRANSPARENT TESTA12 gene of Arabidopsis encodes a multidrug secondary transporter-like protein required for flavonoid sequestration in vacuoles of the seed coat endothelium. Plant Cell 13: 853–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devic M, Guilleminot J, Debeaujon I, Bechtold N, Bensuade E, Koorneef M, Pelletier G, Delseny M (1999) The BANYULS gene encodes a DFR-like protein and is a marker of early seed coat development. Plant J 19: 387–398 [DOI] [PubMed] [Google Scholar]
- Downey MO, Harvey JS, Robinson SP (2003. a) Analysis of tannins in seeds and skins of Shiraz grapes throughout berry development. Aust J Grape Wine Res 9: 15–27 [Google Scholar]
- Downey MO, Harvey JS, Robinson SP (2003. b) Synthesis of flavonols and expression of flavonol synthase genes in developing grape berries of Shiraz and Chardonnay (Vitis vinifera L.). Aust J Grape Wine Res 9: 110–121 [Google Scholar]
- Downey MO, Harvey JS, Robinson SP (2004) The effect of bunch shading on berry development and flavonoid accumulation in Shiraz grapes. Aust J Grape Wine Res 10: 55–73 [Google Scholar]
- Glories Y (1988) Anthocyanins and tannins from wine: organoleptic properties. Prog Clin Biol Res 280: 123–134 [PubMed] [Google Scholar]
- Harborne JB, Grayer RJ (1993) Flavonoids and insects. In JB Harborne, ed, The Flavonoids: Advances in Research Since 1986. Chapman & Hall, London, pp 589–618
- Haslam E (1998) Polyphenols: structure and biosynthesis. In Practical Polyphenolics: from Structure to Molecular Recognition and Physiological Action. Cambridge University Press, Cambridge, pp 35–42
- Holton TA, Cornish EC (1995) Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 7: 1071–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaakola L, Maatta K, Pirttila AM, Torronen R, Karenlampi S, Hohtola A (2002) Expression of genes involved in anthocyanin biosynthesis in relation to anthocyanin, proanthocyanidin, and flavonol levels during bilberry fruit development. Plant Physiol 130: 729–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph R, Tanner G, Larkin P (1998) Proanthocyanidin synthesis in the forage legume Onobrychis vicifolia: a study of chalcone synthase, dihydroflavonol 4-reductase and leucoanthocyanidin 4-reductase in developing leaves. Aust J Plant Physiol 25: 271–278 [Google Scholar]
- Kato A, Nishi R, Ozaki M (2001) Isolation and characterization of two genes encoding ubiquitin fused to a ribosomal protein of 53 amino acids in rice. DNA Seq 12: 53–58 [DOI] [PubMed] [Google Scholar]
- Kennedy JA, Hayasaka Y, Vidal S, Waters EJ, Jones GP (2001) Composition of grape skin proanthocyanidins at different stages of berry development. J Agric Food Chem 49: 5348–5355 [DOI] [PubMed] [Google Scholar]
- Kennedy JA, Matthews MA, Waterhouse AL (2000) Changes in grape seed polyphenols during ripening. Phytochemistry 55: 77–85 [DOI] [PubMed] [Google Scholar]
- Kitamura S, Shikazono N, Tanaka A (2004) TRANSPARENT TESTA 19 is involved in the accumulation of both anthocyanins and proanthocyanidins in Arabidopsis. Plant J 37: 104–114 [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5: 150–163 [DOI] [PubMed] [Google Scholar]
- Lin LC, Kuo YC, Chou CJ (2002) Immunomodulatory proanthocyanidins from Ecdysanthera utilis. J Nat Prod 65: 505–508 [DOI] [PubMed] [Google Scholar]
- Marles MAS, Ray H, Gruber MY (2003) New perspectives on proanthocyanidin biochemistry and molecular regulation. Phytochemistry 64: 367–383 [DOI] [PubMed] [Google Scholar]
- McMahon LR, McAllister TA, Berg BP, Majak W, Acharya SN, Popp JD, Coulman BE, Wang Y, Cheng KJ (2000) A review of the effects of forage condensed tannins on ruminal fermentation and bloat in grazing cattle. Can J Plant Sci 80: 469–485 [Google Scholar]
- Muller PY, Janovjak H, Miserez AR, Dobbie Z (2002) Processing of gene expression data generated by quantitative real-time RT-PCR. Biotechniques 32: 1372–1379 [PubMed] [Google Scholar]
- Nagel CW, Glories Y (1991) Use of a modified dimethylaminocinnamaldehyde reagent for analysis of flavonols. Am J Enol Vitic 42: 364–366 [Google Scholar]
- Nesi N, Debeaujon I, Jond C, Pelletier G, Caboche M, Lepiniec L (2000) The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell 12: 1863–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesi N, Jond C, Debeaujon I, Caboche M, Lepiniec L (2001) The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell 13: 2099–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters DJ, Constabel CP (2002) Molecular analysis of herbivore-induced condensed tannin synthesis: cloning and expression of dihydroflavonol reductase from trembling aspen (Populus tremuloides). Plant J 32: 701–712 [DOI] [PubMed] [Google Scholar]
- Quackenbush J, Liang F, Holt I, Pertea G, Upton J (2000) The TIGR Gene Indices: reconstruction and representation of expressed gene sequences. Nucleic Acids Res 28: 141–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson SP, Davies C (2000) Molecular biology of grape berry ripening. Aust J Grape Wine Res 6: 175–188 [Google Scholar]
- Schijlen EGWM, De Vos CH, Van Tunen AJ, Bovy AG (2004) Modification of flavonoid biosynthesis in crop plants. Phytochemistry 65: 2631–2648 [DOI] [PubMed] [Google Scholar]
- Shirley BW, Hanley S, Goodman HM (1992) Effects of ionizing radiation on a plant genome: analysis of two Arabidopsis transparent testa mutations. Plant Cell 4: 333–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souquet J-M, Cheynier V, Brossaud F, Moutounet M (1996) Polymeric proanthocyanidins from grape skins. Phytochemistry 43: 509–512 [Google Scholar]
- Stafford HA (1990) Pathway to proanthocyanidins (condensed tannins), flavan-3-ols, and unsubstituted flavans. In Flavonoid Metabolism. CRC Press, Boca Raton, FL, pp 63–100
- Tanner GJ, Francki KT, Abrahams S, Watson JM, Larkin PJ, Ashton AR (2003) Proanthocyanidin biosynthesis in plants: purification of legume leucoanthocyanidin reductase and molecular cloning of its cDNA. J Biol Chem 278: 31647–31656 [DOI] [PubMed] [Google Scholar]
- Winkel-Shirley B (2001) Flavonoid biosynthesis: a colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol 126: 485–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie DY, Sharma SB, Paiva NL, Ferreira D, Dixon RA (2003) Role of anthocyanidin reductase, encoded by BANYULS in plant flavonoid biosynthesis. Science 299: 396–399 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.