Abstract
Controversy exists in the literature over the involvement of the endoplasmic reticulum (ER) in the delivery of membrane proteins to peroxisomes. In this study, the involvement of the ER in the trafficking of two Arabidopsis (Arabidopsis thaliana) peroxisomal membrane proteins was investigated using confocal laser scanning microscopy of living cells expressing fusions between enhanced yellow fluorescent protein (eYFP) and AtPEX2 and AtPEX10. The fusion proteins were always detected in peroxisomes and cytosol irrespective of the location of the eYFP tag or the level of expression. The cytosolic fluorescence was not due to cleavage of the eYFP reporter from the C-terminal fusion proteins. Blocking known ER transport routes using the fungal metabolite Brefeldin A or expressing dominant negative mutants of Sar1 or RabD2a had no effect on the trafficking of AtPEX2 and AtPEX10 to peroxisomes. We conclude that AtPEX2 and AtPEX10 are inserted into peroxisome membranes directly from the cytosol.
Peroxisomes are eukaryotic organelles that are surrounded by a single membrane and contain no DNA, so acquire all their protein complement by import of cytosolically synthesized proteins. Proteins required for peroxisome biogenesis are termed peroxins and are encoded by genes with the nomenclature PEX (Distel et al., 1996). In yeasts the nomenclature is PEXN (gene), PexNp (protein), and pexn (mutant); in mammals and plants, it is PEXN (gene), PEXN (protein), and pexn (mutant). Over the past two decades enormous progress has been made in understanding the targeting and import of matrix proteins. There is a generally agreed model for the import of peroxisome matrix proteins, although there appear to be some species-specific variations in peroxisome assembly in different organisms (for review, see Purdue and Lazarow, 2001; Sparkes and Baker, 2002).
Much less is understood about the import of membrane proteins. Researchers have used a range of model systems, including various yeasts, mammals, and plants, and often reached contradictory conclusions. In particular, there are currently conflicting views concerning the role of the endoplasmic reticulum (ER) in the biogenesis of peroxisomes, which are summarized in recent reviews (Lazarow, 2003; Tabak et al., 2003). In the yeast Yarrowia lipolytica, a model has been proposed in which many peroxisomal membrane proteins (PMPs) are targeted initially to the ER. Vesicles containing these proteins are proposed to bud from the ER by a coatomer protein II (COPII)-dependent pathway, and then these vesicles fuse and grow by the subsequent posttranslational import of matrix proteins (for review, see Titorenko and Rachubinski, 2001a, 2001b). In Hansenula polymorpha it was reported that Brefeldin A (BFA), which inhibits the formation of COPI-coated vesicles, results in the accumulation of newly synthesized peroxisome membrane and matrix proteins in the ER (Salomons et al., 1997). Cottonseed (Gossypium hirsutum) peroxisomal ascorbate peroxidase (APX) was reported to be localized to peroxisomes and a circular/reticular compartment termed pER in tobacco (Nicotiana tabacum) Bright Yellow 2 (BY2) cells, and its sorting to peroxisomes was inhibited by BFA (Mullen et al., 1999). In mouse dendritic cells, the native PMPs PEX13 and PMP70 were detected by immuno electron microscopy in specialized domains of ER continuous with a peroxisome reticulum (Geuze et al., 2003).
In contrast, experiments using human fibroblast cell lines have shown that blocking COPI and COPII vesicle transport using BFA and dominant negative mutants of SAR1 (secretion-associated and ras superfamily-related gene 1) did not prevent the correct trafficking of PMPs to peroxisomes, including PEX3 (South et al., 2000) and PEX3, PEX16, and PEX2 (Voorn-Brouwer et al., 2001). In Saccharomyces cerevisiae, inactivation of Sec61p, the central component of the ER translocon or its homolog Ssh1p, did not have any effect on peroxisome biogenesis (South et al., 2001).
To resolve the issue of whether some PMPs are targeted to the ER and subsequently sorted to peroxisomes as part of their normal biogenetic pathway, it is important to be able to chase a protein from the ER to the peroxisome. This has been done for only one integral membrane protein, Pex2p from Y. lipolytica, which is also apparently glycosylated (Titorenko et al., 1997; Titorenko and Rachubinski, 1998). As the model of peroxisome biogenesis proposed for Y. lipolytica is very different from that derived from studies on mammalian cells and other species of yeast (for review, see Purdue and Lazarow, 2001), it is important to investigate this question in other systems.
To address this question, we investigated the trafficking pathway of Arabidopsis (Arabidopsis thaliana) PEX2 (AtPEX2) and PEX10 (AtPEX10) using time-resolved confocal microscopy on living cells. PEX2 and PEX10 are PMPs that contain a functionally important C3HC4 RING finger domain. In mammals and yeasts, PEX2 and PEX10 are located in peroxisome membranes and are required for import of peroxisome matrix proteins (Kalish et al., 1995; Okumoto et al., 1998), possibly for the reexport of PEX5, the receptor for the major peroxisomal matrix targeting signal PTS1 (Platta et al., 2004). In Arabidopsis, both have been shown to be localized to peroxisomes and are required for embryo development (Hu et al., 2002; Sparkes et al., 2003). Although Arabidopsis PEX10 was reported to be peroxisomal (Sparkes et al., 2003), Flynn et al. (2005) concluded that PEX10 resides in the ER and Schumann et al. (2003) reported abnormal lipid bodies and ER in the aborted embryos of an Arabidopsis pex10 null mutant, although it is unclear if these effects are a cause or a consequence of the loss of PEX10. Intriguingly, PEX10 contains a C-terminal motif (-YHSDF) that functions as an ER retrieval motif when fused to an artificial ER-targeted reporter protein (McCartney et al., 2004), which raises the following question: Does PEX10 cycle between the ER and peroxisomes?
In this study, we have established that Arabidopsis PEX10 and PEX2 can be detected only in cytosol and peroxisomes, and that inhibiting known ER transport routes does not result in accumulation in ER. Our data strongly suggest that Arabidopsis PEX2 and PEX10 are inserted directly from the cytosol into peroxisomes. This is discussed in relation to the possible role of the ER in peroxisome biogenesis.
RESULTS
AtPEX2 and AtPEX10 Fusions to Enhanced Yellow Fluorescent Protein Locate to the Peroxisome in Tobacco Cells
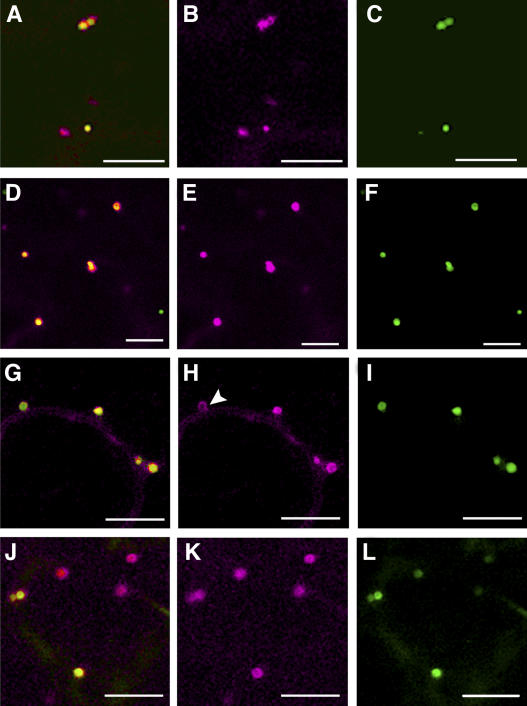
Both N- and C-terminal fusions of AtPEX2 and AtPEX10 to enhanced yellow fluorescent protein (eYFP) were expressed under the control of the 35S promoter. All four fusion proteins were located in punctate motile structures and to some extent in the cytosol when expressed transiently in tobacco epidermal cells (Fig. 1, B, E, H and K). A peroxisomal matrix marker, CFP fused to SKL (CFP-SKL; Fig. 1, C, F, I, and L), colocalized with both N- and C-terminal AtPEX2/10 fusions to eYFP (Fig. 1, A, D, G, and J). Additionally, AtPEX2 and AtPEX10 fusions to eYFP were frequently localized to the rim of the peroxisome with the matrix marker inside the peroxisome structure (Fig. 1H, arrowhead; Fig. 4, E and H; Fig. 6, D and G; Supplemental Fig. 1A).
Figure 1.
Location of the AtPEX2 and AtPEX10 fusions to eYFP. AtPEX2 and AtPEX10 fusions to eYFP were coexpressed with CFP-SKL in tobacco epidermal cells via transient expression. AtPEX2-eYFP (A–C) and eYFP-AtPEX2 (D–F) colocalize with the peroxisomal marker CFP-SKL. AtPEX10-eYFP (G–I) and eYFP-AtPEX10 (J–L) colocalize with CFP-SKL. AtPEX2 and AtPEX10 fusions localize to the rim of the peroxisomes (see arrowhead in H, for example). Images A, D, G, and J are merged images of the AtPEX2/10 fusions to eYFP (B, E, H, and K) with CFP-SKL (C, F, I, and L). eYFP fusions are in magenta; CFP-SKL is in green. Scale bar = 10 μm.
Figure 4.
AtPEX2 and AtPEX10 are not located in the ER. A cytosolic marker, free eYFP (A–C), AtPEX2-eYFP (D–F), eYFP-AtPEX2 (G–I), AtPEX10-eYFP (J–L), or eYFP-AtPEX10 (M–O) were transiently expressed in epidermal cells of stably transformed tobacco plants expressing an ER marker, GFP-HDEL (C, F, I, L, and O). Cytosolic eYFP is present throughout the cytosol. It flows around organelles in the cytoplasm, which are shown in negative contrast (arrowhead B), and surrounds the ER resulting in a similar pattern of fluorescence to the ER marker but is diffuse rather than extremely defined (compare region marked with arrow in B with C). AtPEX2 and AtPEX10 fusions to eYFP localized to peroxisomes and the cytosol. Cytosolic location is evident from organelles in negative contrast (arrowhead E and N) and the diffuse pattern of fluorescence. eYFP fusions are shown in magenta; GFPHDEL is shown in green. Scale bar = 10 μm.
Figure 6.
Sar1 H74L and RabD2a N121I have no affect on the localization of AtPEX2-eYFP, AtPEX10-eYFP, or eYFP-SKL. Tobacco epidermal cells transiently expressing ST-CFP either in stably transformed tobacco plants expressing AtPEX2-eYFP (A, D, G, J, and M), or wild-type plants transiently expressing AtPEX10-eYFP (B, E, H, K, and N) or eYFP-SKL (C, F, I, L, and O), were observed with and without coexpression of Sar1H74L (D–I) or RabD2a N121I (J–O). A to C, Localization of ST-CFP to Golgi bodies and AtPEX2-eYFP (A), AtPEX10-eYFP (B), and eYFP-SKL (C) to peroxisomes. D to F, Coexpression of Sar1 H74L affects ST-CFP localization which is now present in the ER rather than the Golgi, but does not affect the location of the peroxisomal fusions. G to I, The localization of the peroxisomal fusions shown in images D to F. All of the peroxisomal fusions are localized to the peroxisome and none are located in the ER. J to O, Coexpression of RabD2a N121I affects ST-CFP localization which is now present in the ER rather than the Golgi, but does not affect the location of the peroxisomal fusions. M to O, The localization of the peroxisomal fusions shown in images J to L. All of the peroxisomal fusions are localized to the peroxisome and none is located in the ER. Transient expression of Sar1H74L or RabD2a N121I results in a population of cells which do not express these constructs (see arrowheads in E and J). Comparison between these cells and cells in the same image which are expressing Sar1 H74L or RabD2a N121I clearly shows that the peroxisome markers are not affected by Sar1 H74L or RabD2a N121I expression. Where eYFP fusions are shown in the magenta channel and ST-CFP fusion is in the green channel. Scale bar = 10 μm.
Stable transgenic tobacco and Arabidopsis lines expressing either AtPEX2-eYFP or AtPEX10-eYFP were generated to determine whether a similar distribution of the fusion proteins was seen compared to transient expression in epidermal cells. Stable Arabidopsis plants expressing both AtPEX10-eYFP and GFP-SKL also show that AtPEX10 is present in the periphery of peroxisomes (Supplemental Fig. 1, A–C). AtPEX10-eYFP is also present in punctate motile structures in stable Arabidopsis (Supplemental Fig. 1D; Supplemental Movie 1) and tobacco (Supplemental Fig. 1E) lines.
Transgenic tobacco plants expressing AtPEX2-eYFP displayed lower levels of expression than AtPEX10-eYFP plants. However, more AtPEX2-eYFP was present in the periphery of punctate motile structures the size of peroxisomes than was seen under transient expression (Supplemental Fig. 1, J–L). Stable Arabidopsis plants expressing AtPEX2-eYFP show that AtPEX2 is present in the motile punctate structures of the expected size of peroxisomes (Supplemental Fig. 1, F–I, arrowhead). With all constructs expressed under both transient and stable transformation, diffuse nonperoxisomal fluorescence was seen to differing extents (Fig. 1; Supplemental Fig. 1).
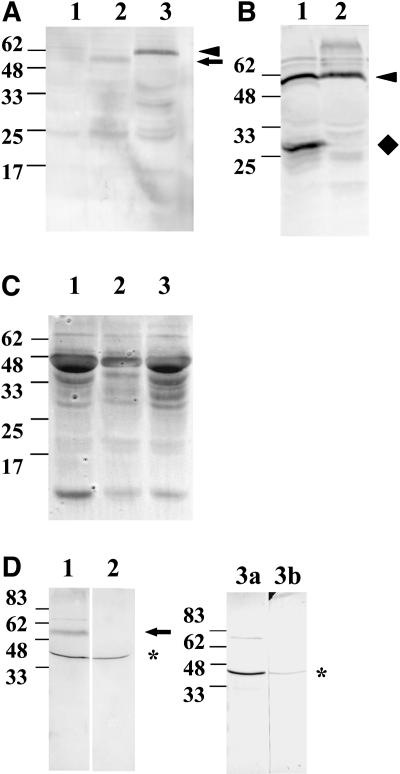
Nonperoxisomal fluorescence, as well as representing the full-length fusion protein, could be due to cleavage of the eYFP from the fusion proteins, leading to accumulation of fluorescent protein in the cytosol. An anti-green fluorescent protein (GFP) antibody, which cross reacts with eYFP, was used to probe leaf extracts derived from stable tobacco transformants (Supplemental Fig. 1) that express AtPEX10-eYFP (Fig. 2A, lane 3) or AtPEX2-eYFP (Fig. 2A, lane 2). Leaf extracts from untransformed tobacco were used as controls (Fig. 2A, lane 1). A band was detected in lane 3 (Fig. 2A), indicated by an arrowhead, which was slightly smaller than the expected size of AtPEX10-eYFP (69.6 kD). However, it is known that membrane proteins often migrate anomalously in SDS-PAGE gels. In lane 2 (Fig. 2A), a slightly smaller band was detected, indicated by an arrow, consistent with the smaller size of AtPEX2-eYFP (65.2 kD). Neither band was seen in the untransformed sample (Fig. 2A, lane 1). Figure 2C shows the Ponceau S-stained blot corresponding to Figure 2A. This confirms that the amount of protein from the wild-type plants was similar to or greater than the amount of protein from the transgenic samples. Thus, the absence of anti-GFP cross-reactive bands around 62 kD in the wild-type sample confirms that the bands of this size seen in the transformed samples are the YFP fusion proteins. Total protein extracts from stable tobacco plants expressing either eYFP-AtPEX10 (Fig. 2B, lane 1) or AtPEX10-eYFP (Fig. 2B, lane 2) were probed with the anti-GFP antibody. eYFP-AtPEX10 is not stable, and both the full-length (Fig. 2B, lane 1, arrowhead) and cleaved eYFP (Fig. 2B, lane 1, diamond) were detected. Crucially, there was no band corresponding to free eYFP at around 28 kD in total protein extracts from AtPEX2-eYFP (Fig. 2A, lane 2) or AtPEX10-eYFP (Fig. 2A, lane 3; Fig. 2B, lane 2). The band around 25 kD detected in all extracts does not represent free eYFP as it was also detected in protein extracts from wild-type plants (Fig. 2A, lane 1). Therefore, both AtPEX10-eYFP and AtPEX2-eYFP are stable fusion proteins and were used for subsequent experiments.
Figure 2.
AtPEX2 and AtPEX10 fusions are stable. Total protein from wild-type tobacco plants (A, lane 1) or stable tobacco plants expressing AtPEX2-eYFP (A, lane 2), AtPEX10-eYFP (A, lane 3; B, lane 2), or eYFP-AtPEX10 (B, lane 1) were extracted. Equal volumes were separated by SDS-PAGE and blotted onto nitrocellulose. Blots in A and B were probed with anti-GFP polyclonal serum. Anti-GFP antibody cross-reacts with AtPEX2-eYFP (see arrow in A, lane 2), AtPEX10-eYFP (see arrowhead in A, lane 3; B, lane 2), eYFP-AtPEX10 (see arrowhead in B, lane 1), and a cleaved eYFP-AtPEX10 product (see diamond in B, lane 1). An image of the blot in A stained with ponceau prior to probing with the antibody is shown in C to indicate relative levels of protein. Total protein extracts from wild-type plants (D, lanes 2, 3a, and 3b) and wild-type tobacco plants transiently expressing AtPEX2-eYFP driven by the enhanced 35S promoter (D, lane 1) were separated by SDS-PAGE and transferred to nitrocellulose. Blots were probed with affinity-purified AtPEX2 peptide antibody (D, lanes 1, 2, and 3a) or affinity-purified peptide antibody preincubated with peptide (D, lane 3b). The affinity-purified AtPEX2 peptide antibody cross-reacts with native PEX2 (see asterisk in D) and AtPEX2-eYFP fusion protein (see arrow in D, lane 1). Preincubation of the affinity-purified antibody with peptide results in significantly reduced cross-reactivity (see asterisk and compare lanes 3a and 3b in D). The samples in lanes 3a and 3b are identical. Total protein was extracted from 200 mg of leaf material and equal volumes were loaded (20 μL). Expected sizes of proteins are AtPEX2-eYFP, 65.2 kD; AtPEX10-eYFP, 69.6 kD; eYFP-AtPEX10, 69.6 kD; and AtPEX2, 38.2 kD.
Polyclonal serum was raised against a 26-amino acid peptide corresponding to the amino terminus of AtPEX2. The resulting antibodies were affinity purified using the peptide. Due to the low levels of AtPEX2-eYFP expression in the stable tobacco line, the affinity-purified antibody was used to probe total protein extracts from tobacco plants transiently expressing AtPEX2-eYFP under the control of the enhanced 35S promoter (Fig. 2D, lane 1). Nontransformed tissue was used as a control (Fig. 2D, lane 2). The affinity-purified antibody cross-reacts with a product close to the predicted size for native AtPEX2, 34.6 kD, in both lanes 1 and 2 (Fig. 2D, asterisk) and a band corresponding to the fusion protein, AtPEX2-eYFP, in lane 1 only (Fig. 2D, arrow). To confirm antibody specificity, the affinity-purified antibody was preincubated with peptide to titrate out the antibody. The samples loaded in lanes 3a and 3b (Fig. 2D) are identical, where lane 3a was detected with affinity-purified antibody and lane 3b was detected with affinity-purified antiserum preincubated with peptide. As expected, the level of PEX2 detected in lane 3b (Fig. 2D, asterisk) is significantly less than in lane 3a (Fig. 2D, asterisk). An additional weaker cross-reactivity with a product around 70 kD was also detected.
Immunofluorescence on Arabidopsis cell culture using the AtPEX2 affinity-purified antibody results in punctate labeling (Fig. 3A) similar to that observed using antibodies against isocitrate lyase (ICL; Fig. 3B). Additionally, there was no evidence of cytosolic or reticular staining with the anti-PEX2 antibody. Dual labeling resulted in several punctate structures labeling with both anti-PEX2 and anti-ICL antibodies (see inset in Fig. 3C).
Figure 3.
Immunofluorescence of Arabidopsis cell culture. Arabidopsis cell culture was fixed, permeabilized, and subjected to single (A and B) and dual labeling (C–E) with primary antibodies against AtPEX2 (A and D) and ICL (B and E). Texas red-conjugated secondary antibody was used to detect PEX2 (magenta), and FITC conjugated secondary antibody was used to detect ICL (green).Merged image (C) shows colocalization of PEX2 and ICL to punctate structures (yellow). This is clearly seen in the enlarged insert in C. Scale bar = 5 μm.
AtPEX2 and AtPEX10 Fusions to eYFP Do Not Colocalize with an ER Marker
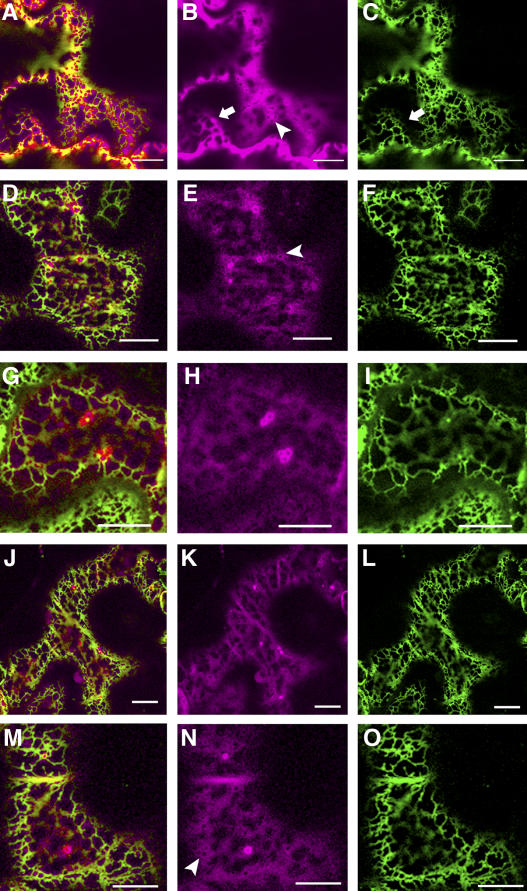
To ascertain whether AtPEX2 or AtPEX10 traffic via the ER, the first step was to determine whether the nonperoxisomal fluorescence colocalizes with an ER marker, GFP-HDEL. The cortical ER network in tobacco leaf epidermal cells forms an extensive network of polygonal tubules surrounded by the cytoplasm (Fig. 4, C, F, I, L, and O). These cells contain large vacuoles that confine the cytoplasm and organelles to the periphery of the cell (Fig. 4B). These spatial constrictions can restrict the cytosol to the region immediately surrounding the ER network (Fig. 4, B and C; see arrow) resulting in a similar yet diffuse pattern of fluorescence, or it can flow freely around other organelles in the cell, which appear in negative contrast (Fig. 4B; see arrowhead). Therefore, to be able to discern between ER and cytosolic location requires careful inspection and interpretation of the images.
N- or C-terminal fusions of AtPEX2 (Fig. 4, D–I) and AtPEX10 to eYFP (Fig. 4, J–O) were transiently expressed in stable tobacco plants expressing GFP-HDEL. The settings used on the microscope could distinguish between fluorescence from eYFP fusions and that from wild-type GFP fused to HDEL. GFP-HDEL labels the well-defined cortical ER network (Fig. 4, F, I, L, and O) and is seen in close association with the peroxisomes containing the fusion protein and the diffuse pool of peroxisomal fusion proteins (Fig. 4, D, G, J, and M). Careful analysis of the peroxisomal fusion proteins expressed in the GFP-HDEL plants showed that they did not colocalize with the ER but rather surrounded it in a diffuse pattern like that of free eYFP (compare Fig. 4, E, H, K, and N, to B). This cytosolic nature was further supported by observations of organelles in negative contrast (Fig. 4, E and N, arrowhead).
Similarly, epidermal cells in stable transgenic lines expressing AtPEX2-eYFP or AtPEX10-eYFP do not show ER localization. However, it is possible that the kinetics of trafficking may be such that the steady-state levels of these peroxisomal proteins in the ER are below detection, or that low levels in the ER may be masked by the levels in the cytosol. To address this, the export of proteins from the ER was blocked and the effect on trafficking of the AtPEX2 and AtPEX10 fusion proteins investigated.
Comparisons between transient and steady-state expression of AtPEX2-eYFP have shown that higher levels of the fusion protein are present in the peroxisome and less is detected in the cytosol in stably transformed lines. Therefore, subsequent studies relating to AtPEX2 trafficking were carried out using stable tobacco plants expressing AtPEX2-eYFP, since high cytosolic levels detected under transient expression could mask populations of fusion protein, which may locate to the ER upon treatment with BFA or expression of Sar1 H74L and RabD2a N121I. Studies involving AtPEX10-eYFP and a peroxisomal matrix marker, eYFP-SKL, are based on transient expression of these fusion proteins.
BFA Does Not Affect the Trafficking of AtPEX2-eYFP, AtPEX10-eYFP, or a Peroxisomal Matrix Marker
Treatment of cells with BFA results in the redistribution of Golgi markers back to the ER (Ritzenthaler et al., 2002; Saint-Jore et al., 2002). This is thought to be due to the disruption of COPI-mediated trafficking at the Golgi and/or Golgi ER interface. Therefore, to assess what role COPI vesicular traffic may play in the trafficking of peroxisomal markers, AtPEX2-eYFP, AtPEX10-eYFP, and a peroxisomal matrix marker eYFP-SKL were coexpressed with a Golgi marker protein and treated with BFA.
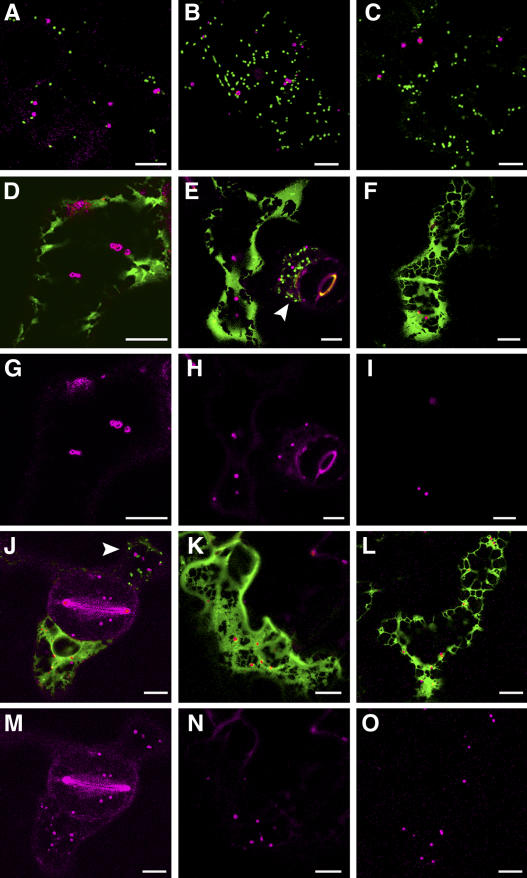
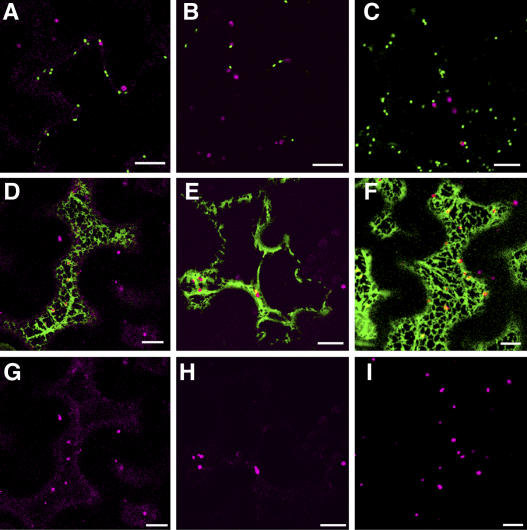
Tobacco epidermal cells transiently expressing a Golgi marker, sialyltransferase fused to CFP (ST-CFP), and a peroxisomal marker were treated with BFA (100 μg/mL) for 30 min. The Golgi marker acts as an internal control as BFA treatment should result in the redistribution of this marker to the ER. Images were taken prior to BFA treatment and then after 30 min incubation in BFA. Figure 5 shows that prior to treatment ST-CFP and the peroxisomal markers, AtPEX2-eYFP (Fig. 5A), AtPEX10-eYFP (Fig. 5B), and a peroxisomal matrix marker eYFP-SKL (Fig. 5C), label punctate structures. ST-CFP (green) and the peroxisomal markers (magenta) do not colocalize and are present in two separate populations of punctate structures, which represent the Golgi and peroxisomes. After 30 min treatment, the peroxisomal markers are still in motile punctate structures (Fig. 5, D–I), whereas the Golgi marker ST-CFP has been redistributed to the ER. The peroxisomal markers in merged images (Fig. 5, D–F) are shown for clarity in a single-channel image (Fig. 5, G–I). The affects of BFA are reversed after 5 h incubation in water (data not shown). The effects of BFA treatment on AtPEX10-eYFP and ST-CFP trafficking are shown in Supplemental Movie 2.
Figure 5.
BFA does not affect the localization of AtPEX2-eYFP, AtPEX10-eYFP, or eYFP-SKL. ST-CFP was transiently expressed in stably transformed tobacco plants expressing AtPEX2-eYFP (A, D, and G), or wild-type plants transiently expressing AtPEX10-eYFP (B, E, and H) or eYFP-SKL (C, F, and I). Leaf sections expressing these constructs were incubated in BFA (100 μg/mL) for 30 min. Images were taken prior to incubation (A–C) and after BFA treatment (D–I). BFA alters the localization of the Golgi marker ST-CFP to the ER (reticular structure) but has no affect on the localization of the peroxisomal markers (punctate structures). Images G, H, and I show the peroxisomal markers in the merged images D, E, and F, respectively. Scale bar = 10 μm.
Dominant Negative Inhibitors of ER-to-Golgi Trafficking Do Not Affect the Localization of AtPEX2-eYFP, AtPEX10-eYFP, or a Peroxisomal Matrix Marker
Sar1 and RabD2a (formally known as Rab1b) are small G-proteins involved in ER-to-Golgi transport. Transient expression of NtSar1H74L, a GTP-locked mutant (Andreeva et al., 2000), or Arabidopsis RabD2a N121I, a nucleotide-free mutant (Batoko et al., 2000), resulted in the accumulation of ST-GFP, a Golgi marker, and a secreted form of GFP, Sec-GFP, in the ER. This in vivo assay forms the basis for assessing the potential effects of blocking ER-to-Golgi trafficking on peroxisome biogenesis as the altered location of the ST fluorescent fusion acts as a marker for the expression of the dominant negative forms of the G proteins, Sar1 and RabD2a.
In cells where ST-CFP (green) was coexpressed with either AtPEX2-eYFP, AtPEX10-eYFP, or eYFP-SKL (magenta), the peroxisome markers were detected in distinct cellular structures (Fig. 6, A–C, respectively). Coexpression of the Sar1 H74L mutant had no effect on the localization of the peroxisomal markers but did affect ST-CFP as expected (Fig. 6, D–I). For clarity, images in Figure 6, G to I, show the peroxisomal markers in Figure 6, D to F. Peroxisomes containing the fluorescent fusion proteins in cells that coexpress Sar1 H74L are similar in size and display similar movement characteristics to cells not expressing the Sar1 H74L mutant (compare cell marked with arrow in Fig. 6E with the adjacent cell in the same section).
Cells coexpressing the RabD2a N121I mutant with the peroxisomal markers (magenta) and ST-CFP (green) are shown in Figure 6, J to O. The ST-CFP Golgi marker control is located in the ER due to the effects of RabD2a N121I, whereas the peroxisomal marker proteins are not affected. Images in Figure 6, M to O, display the peroxisomal marker fusions in the corresponding merged images with ST-CFP in Figure 6, J to L. Peroxisomes containing the fluorescent peroxisomal marker fusions are similar in size and display similar motility to cells not expressing the RabD2a N121I (compare cell marked with arrow in Fig. 6J with adjacent cell in the same section).
DISCUSSION
Previous work has raised questions concerning the subcellular localization of Arabidopsis PEX10 and the route by which PEX2 is trafficked to peroxisomes, with conflicting data reported for mammals and yeasts. Resolving these issues is important, both in terms of understanding the functions of these particular proteins and in addressing the following questions: Are peroxisomes made de novo, and, if so, how?
AtPEX2 and AtPEX10 Are Not Located to the ER
Transient expression of eYFP-AtPEX2, AtPEX2-eYFP, eYFP-AtPEX10, and AtPEX10-eYFP in tobacco epidermal cells resulted in colocalization with a peroxisomal matrix marker, CFP-SKL (Fig. 1). Transient expression does not allow for a uniform level of expression across the leaf epidermis, resulting in some cells displaying higher levels of expression than others. In these high-expressing cells, there are cytosolic pools of the fusion proteins. Such cytosolic accumulation of fluorescent proteins is also present in the early stages of expression. This is probably due to saturation of the machinery required for PMP import caused by overexpression of the fusion proteins, competition between these fusions and the native proteins in the cell, and the kinetics of import, which allows for the cytosolic accumulation. Transient expression of AtPEX2-eYFP and eYFP-AtPEX2 resulted in lower levels in the peroxisome and what appeared to be a larger cytosolic pool in comparison to AtPEX10 fusions to eYFP. When AtPEX10-eYFP or AtPEX2-eYFP was expressed in stable transgenic plants, they displayed a similar localization pattern to that seen in transient expression, peroxisomal with some cytosolic fluorescence. Western-blot analysis of total protein extracts from the stable transgenic lines expressing AtPEX10-eYFP and AtPEX2-eYFP confirmed that the cytosolic fluorescence was not due to cleavage of the fusion proteins resulting in free cytosolic eYFP (Fig. 2). To assess whether any of the nonperoxisomal fusion proteins could be located in the ER, the fusions were expressed in a stable tobacco plant expressing an ER marker, GFP-HDEL. Careful characterization of these expression studies showed that the fusion proteins were present in the diffuse pools reminiscent of cytosolic localization and not in well-defined ER strands (Fig. 4).
Interestingly, the stable Arabidopsis double transformant expressing AtPEX10-eYFP and GFP-SKL gave indications that the peroxisome population may be heterogeneous. Although most peroxisomes had both markers, some had detectable levels only of either AtPEX10-eYFP or GFP-SKL (Supplemental Fig. 1, A–C). Also, in the immunofluorescence studies, native PEX2 appears to be in many more punctuate structures than the matrix enzyme ICL (Fig. 3). These data could suggest heterogeneity of the peroxisome population and warrant further investigation.
Fluorescent Fusions to AtPEX2, AtPEX10, and −SKL Are Not Affected by the Inhibition of COPI- and COPII-Mediated Vesicle Trafficking
The trafficking of AtPEX2, AtPEX10, and the peroxisomal matrix marker were not effected by inhibiting COPI-mediated (BFA treatment) or COPII-mediated (Sar1 H74L mutant) vesicle trafficking. The trafficking was also not affected by coexpression of a mutant form of RabD2a. It is currently not known whether plant RabD2a is involved in the COPII-mediated trafficking pathway or an independent trafficking route. The effects of Sar1 H74L and RabD2a N121I were detected after 24 to 48 h expression, and the effects of BFA were seen after 30 min. In tobacco BY2 cells, γCOP (COPI component) is lost from Golgi cisternae within 5 min of BFA treatment, and a Golgi-ER hybrid compartment is formed with 15 to 20 min of treatment with only 10 μg/mL BFA (Ritzenthaler et al., 2002). When H. polymorpha cells were treated with 20 μg/mL BFA, mislocalization of peroxisomal matrix and membrane proteins to the ER was detectable after 1 h (Salomons et al., 1997). Since the rate of turnover of the peroxisomal proteins was not measured in our experiments, a caveat is that the effects of BFA may be faster than the rate of AtPEX2/10-YFP turnover, thus preventing the detection of any loss of fluorescence or localization to a compartment other than the peroxisome. However, since the BFA results are fully corroborated by the dominant negative inhibitors over a much longer time frame, protein turnover is unlikely to be an issue. In the report by Mullen et al. (1999), effects of BFA on the sorting of both a PMP (fusion between chloramphenicol acetyl transferase and peroxisomal APX) and matrix protein (chloramphenicol acetyl transferase-SKL) in BY2 cells were seen. Although the concentrations of BFA used were the same as in our experiments, these effects were seen at much later time points (8–10 h) and so may have been secondary effects.
In conclusion, transient and steady-state expression studies in tobacco epidermal cells show that AtPEX2 and AtPEX10 fusions to eYFP are localized to the peroxisome and to a certain extent in the cytosol. At no point have we detected AtPEX2 or AtPEX10 in the ER, even when ER export routes were blocked. Immunofluorescence studies of wild-type Arabidopsis cell culture confirmed the location of AtPEX2 in peroxisomes and did not highlight any additional pools in the cytosol or ER. The simplest interpretation of the data presented here is that AtPEX2 and AtPEX10 are imported into peroxisomes directly from the cytosol. Fusion of eYFP to either terminus does not affect trafficking; therefore the targeting signals are capable of functioning internally. A similar situation pertains in the 22-kD PMP (PMP22) of Arabidopsis, where a comprehensive mutagenesis study identified four distinct internal regions within PMP22 that functioned cooperatively to bring about direct insertion from the cytosol (Murphy et al., 2003).
Our results are in complete agreement with similar studies in mammalian cells where neither BFA nor inhibitors of COPI- and COPII-mediated transport affected the trafficking of PEX2, PEX3, and PEX16 (South et al., 2000; Voorn-Brouwer et al., 2001) and in contrast to what has been reported to date in plants (Mullen et al., 1999; Lisenbee et al., 2003a, 2003b; Flynn et al., 2005) and some yeasts (Salomons et al., 1997; Titorenko and Rachubinski, 1998). Flynn et al. (2005) used antibodies raised against Arabidopsis PEX10 to detect native PEX10 in Arabidopsis suspension culture cells and could detect PEX10 only “partially colocalized in BiP containing ER and in unidentified structures throughout the cytoplasm” (p. 648; Flynn et al., 2005). They could not detect any colocalization with catalase-containing peroxisomes, although it should be noted that IgG fraction rather than affinity-purified antiserum was used and that the antiserum cross reacted with an additional 63- to 66-kD polypeptide on western blots. Further, in their experiments, PEX10 that had been tagged with GFP, YFP, and various epitope tags was only detected in the cytosol in transiently transformed Arabidopsis suspension cells. This is in contrast to our previous results on PEX10 localization (Sparkes et al., 2003) and the more extensive data presented here, even though one of the constructs studied by Flynn et al. (2005) was identical to the AtPEX10-eYFP construct used in this study. Two possible explanations put forward by Flynn et al. (2005) for the observed differences can now be excluded. AtPEX10-eYFP and AtPEX2-eYFP behave identically in both tobacco and Arabidopsis cells, regardless of whether the constructs are transiently expressed or stable transformants are made. Neither the plant species nor the method of expression affected the results (compare Fig. 1 and Supplemental Fig. 1). Perhaps a more plausible explanation is that, in the transient expression experiments presented by Flynn et al. (2005), high levels of tagged PEX10 in the cytosol obscured any peroxisomal location, particularly as nonconfocal images were taken.
The last five amino acids of PEX10 (-YHSDF) can act as an ER retrieval motif when fused at the C terminus of an ER-targeted reporter protein (McCartney et al., 2004), but it was not tested in the context of the full-length native protein. Even if this motif could redirect PEX10 back to the ER, presumably this does not occur as the motif/protein may never encounter the ER retrieval machinery. We also fused the ER retrieval motif KKRY to the C terminus of eYFP-AtPEX2, but it had no effect on the targeting of this fusion protein to peroxisomes (data not shown). Similarly, the dilysine motif at the C terminus of PEX11 in trypanosomes and rat can bind coatomer in vitro (Passreiter et al., 1998). However, further studies highlighted this domain is not required for PEX11 function (Maier et al., 2000).
Is the ER Involved in Peroxisome Biogenesis?
Although the data presented here do not support a role for the ER in the sorting of AtPEX2 and AtPEX10, the argument for the involvement of the ER in peroxisome biogenesis remains open. In plants there is good evidence for non-COPII-mediated sorting of proteins from the ER to protein storage vacuoles (Hara-Nishimura et al., 1998) or to the cell surface (Tormakangas et al., 2001). It is conceivable that as yet undiscovered routes for protein traffic out of the ER exist and could play a role in peroxisome biogenesis.
In plants the best candidate for sorting to peroxisomes via ER is APX (APX3; Mullen et al., 1999; Nito et al., 2001; Lisenbee et al., 2003a, 2003b). However, direct evidence for movement of the ER-localized population to peroxisomes, as would be required if this was a biogenetic intermediate, has never been documented. Like Lisenbee et al. (2003b), we found overexpression of APX3 to result in mislocalization to many intracellular membranes, making the interpretation of these images problematic (data not shown). PEX3, one of the earliest peroxins to mark the identity of a forming peroxisome, has also been suggested to traffic via the ER and in a Hansenula pex3 mutant a PEX3-GFP fusion protein labels small vesicles that are derived from the nuclear membrane and can develop into functional peroxisomes when PEX3 is reintroduced (Faber et al., 2002). However, recent data indicate that the Arabidopsis PEX3 is also inserted into peroxisome membranes directly from the cytosol (Hunt and Trelease, 2004). SSE1, the Arabidopsis homolog of PEX16, another so-called early peroxin, is located to peroxisomes (Lin et al., 2004). The sse1 mutant lacks normal peroxisomes and has a severe defect in fatty acid biosynthesis and the formation of lipid bodies during seed development (Lin et al., 1999, 2004). While the development of seed oil bodies from ER is not in doubt, the argument that the sse1 and pex10 mutants affect this process directly (Schumann et al., 2003) is unproven. We are only beginning to understand the full range of metabolic capacities of peroxisomes, and it seems likely that derangement of peroxisome metabolism can have secondary effects elsewhere in the cell both in terms of compromised production of signaling molecules (Corpas et al., 2001; Theodoulou et al., 2005) or accumulation of potentially toxic metabolic products (Zolman et al., 2001). There are undoubtedly interactions between peroxisomes and ER, but whether this is at the level of protein traffic still remains an open question.
MATERIALS AND METHODS
Growth and Transformation of Arabidopsis and Tobacco Plants
Arabidopsis (Arabidopsis thaliana ecotype Columbia) plants were grown (Sparkes et al., 2003) and then transformed by floral dipping (Clough and Bent, 1998). Growth and transient transformation of tobacco (Nicotiana tabacum) were as previously described (Kotzer et al., 2004, and refs. therein). Agrobacteria were transformed by freeze-thawing (Hofgen and Willmitzer, 1988). Agrobacteria containing various constructs were infiltrated into tobacco leaf epidermal cells at the following OD600nm: CFP-SKL and eYFP-SKL, 0.04; AtPEX10 and AtPEX2 fusions to eYFP, 0.1; Sar1 H74L, 0.01 to 0.03; RabD2a N121I, 0.02 to 0.05; ST-CFP, 0.03 and 0.05 to 0.1. RabD2a N121I and Sar1 H74L were infiltrated either at the same time as the ST-CFP and peroxisomal eYFP fusions, or 24 h after initial infiltration with these constructs. Leaf segments (approximately 0.5 cm2) were excised from regions of the leaf expressing ST-CFP and a peroxisomal marker 3 to 4 d postinfiltration. Segments were immersed in BFA 100 μg/mL (Sigma-Aldrich; 10 mg/mL stock solution in dimethyl sulfoxide) for 30 min, mounted in water, and analyzed on the confocal microscope. After analysis samples were immersed in water to wash out BFA. Control experiments with samples immersed in water for the duration of the treatment were carried out in parallel.
Stable tobacco plants were generated as follows: Three days after infiltration, tobacco leaves were removed, placed in sterilization solution (1:1 hyperchlorite solution:water, 0.01% [v/v] Tween 20) for 5 min, washed three times in sterile distilled water, cut into small pieces using sterile forceps, and placed on shooting media (2.15 g/L Murashige and Skoog salts, 0.8% agar, 3% [w/v] Suc, 0.1 mg/L indole butyric acid [Sigma Aldrich, 1 mg /mL stock], 0.8 mg/L 6-benzylaminopurine [Sigma Aldrich, 1 mg /mL stock], 0.1 mg/L carbenicillin [Melford], 0.2 mg/L Ticarcillin/Clavulanic acid [Ducheva]) and selection for binary vector (hygromycin, 30 μg/mL). The leaf discs were left for 3 to 4 weeks for shooting to occur, and shoots were removed using sterile technique and placed on rooting media (same as shooting media without 6-benzylaminopurine and selection for binary vector, and 0.5 mg/L indole butyric acid) for approximately 10 d. After this time, plantlets were transferred to larger growth containers for screening.
Construction of Plant Expression Vectors
Standard molecular cloning procedures were used (Sambrook and Russell, 2001) for the construction of CFP-SKL. CFP-SKL was amplified using Pfx with primers containing XbaI (forward primer 5′ CGATCTAGAGCAGATCGATGGTGAGCAAGGGCGAGGAGCTG 3′) and SacI (reverse primer 5′ GCTGAGCTCGGCTAAAGTTTTGACTCCTTGTACAGCTCGTCCATGCC 3′) sites at the 5′ and 3′ ends, respectively. The resulting PCR product substituted ST-CFP in the plant binary vector pVKH18-EN6∷ST-CFP (Brandizzi et al., 2002b).
Gateway homologous recombination technology (Invitrogen) was used to clone the remaining constructs described herein.
AtPEX2 was amplified from expressed sequence tag N96573 using Pfx polymerase. Two AtPEX2 clones were amplified, one with a stop codon (forward primer 5′ GGGGACAAGTTTGTACAAAAAAGCAGGCTGGGGACAAGTTTGTACAAAAAAGCAGGCTTCCCGCCAATGACGCCGTCTACGCCTGCAGAC 3′, reverse primer 5′ GGGGACCACTTTGTACAAGAAAGCTGGGTGGGGACCACTTTGTACAAGAAAGCTGGGTTCATTTGCCACTTGAAACACCTTCCC 3′) and the other without (forward primer same as above, reverse primer 5′ GGGGACCACTTTGTACAAGAAAGCTGGGTGGGGACCACTTTGTACAAGAAAGCTGGGTCTTTGCCACTTGAAACACCTTCCCGTTG 3′), for the subsequent generation of C- and N-terminal fusions to eYFP. These clones were recombined into the entry vector pDONOR 207, and then recombined into the plant binary destination vectors 35S-Cassette B-eYFP-Nos∷pCAMBIA 1300 (Sparkes et al., 2003), 35S-eYFP-Cassette A-Nos∷pCAMBIA 1300, and En35S-Cassette B-eYFP-Nos∷pCAMBIA 1300 to generate 35S-AtPEX2-eYFP∷pCAMBIA 1300, 35S-eYFP-AtPEX2∷pCAMBIA 1300, and En35S-AtPEX2-eYFP-Nos∷pCAMBIA 1300, respectively. Enhanced 35S promoter was excised from pVKH18-En6 as a HindIII XbaI fragment and replaced the 35S promoter to generate En35S-Cassette B-eYFP-Nos∷pCAMBIA 1300. The 35S promoter-eYFP-gateway Cassette A-Nos terminator was constructed as follows: The cauliflower mosaic virus 35S promoter (800 bp) and nopaline synthase terminator (250 bp) DNA fragments in this construct originate from pBI121 (Jefferson et al., 1987). The GUS reporter gene located downstream of the 35S promoter in pBI121 was replaced by PCR-amplified eYFP (accession no. AF242870) using BamHI and SacI restriction sites. The eYFP fragment was amplified with the following oligonucleotides: forward strand, 5′-ATCGGATCCATATAAAACAATGGTGAGCAAGGGCGAGGA, and reverse strand, 5′-GTACCCGGGCTTGTACAGCTCGTCCAT. The amplified fragment was cloned into pGEM-Teasy (Promega) and then excised using a BamHI restriction site introduced in the forward-strand oligonucleotide and the SacI site present in the vector MCS. The Cassette A DNA fragment (Invitrogen) was then placed downstream of eYFP into a SmaI restriction site present in the reverse-strand oligonucleotide. The entire 5′-cauliflower mosaic virus 35S promoter-eYFP-Cassette A-nopaline synthase terminator construct was then transferred into the HindIII and EcoRI sites of pCAMBIA 1300 (8,958 bp).
The point mutation in AtPEX10, resulting in an E357K substitution (Sparkes et al., 2003), was corrected by amplification with Pfx proofreading DNA polymerase (Invitrogen) and the product ligated into pGEMT-easy (Promega). The point mutation was corrected by overlapping PCR using forward (P11 5′ ATGAGGCTTAATGGGGATTCG 3′, P3 5′ GATGAAGCTACTTGGACAG 3′) and reverse (P28 5′ GCATTCTTGCTTCTCGTTG 3′, P4 5′ CCCATTGTG CCTAAAAATCAG 3′) primers in PCR A (P11 P28) and PCR B (P3 P4), respectively, and primers P11 and P4 for the final PCR amplification. Subsequent amplification from this template with forward (5′ GGGGACAAGTTTGTACAAAAAAGCAGGCTTCACCATG AGGCTTAATGGGGATTCG 3′) and reverse (5′ GGGGACCACTTTGTACAAGAAAGCTGGGTTCTAAAAATCAGAATGATACAA 3′) primers using PfuTurbo hotstart DNA polymerase (Stratagene) was recombined into pDONOR 201 resulting in AtPEX10∷pDONOR 201, which was recombined with 35S-eYFP-Cassette A-Nos∷pCAMBIA 1300 to give 35S-eYFP-AtPEX10-Nos∷pCAMBIA 1300. All constructs were fully sequenced to establish authenticity.
Preparation of Protein Extracts
Fresh leaf tissue (0.2 g) was excised, placed in a 1.5 mL Eppendorf tube, frozen, and ground in liquid nitrogen using an Eppendorf grinder. The sample was placed on ice, 1 mL of extraction buffer (0.2 m NaOH, 2% β-mercaptoethanol), 10 μL protease inhibitor cocktail for plant cell extracts (Sigma-Aldrich) added, and mixed using the Eppendorf grinder. The homogenate was centrifuged for 10 min at 18,000g, and the soluble proteins were precipitated by adding an equal volume of 80% TCA, 20% acetone, left on ice for a minimum of 30 min, and spun at 18,000g for 30 min. The resulting pellet was washed twice with 100% ice-cold acetone, resuspended in 200 μL SDS-sample buffer (Sambrook and Russell, 2001), and heated for 5 min at 95°C prior to loading on the gel. Samples were separated on 12% acrylamide gels using the miniprotean III system (Bio-Rad) and subsequently blotted onto nitrocellulose (Sambrook and Russell, 2001).
Affinity Purification of AtPEX2 Peptide Antibody and Immunoblotting Conditions
A polyclonal antibody was raised in rabbits to a 26-amino acid peptide (MTPSTPADDAWIRSYQRLLPESQSLC) corresponding to the amino terminus of AtPEX2 (Genosphere). The peptide was covalently coupled to Sulfolink coupling gel (Pierce Biotechnology) and used to affinity purify the antibody according to manufacturer's instructions. Sample buffer was 50 mm Tris-HCl, pH 7.5, 150 mm NaCl, and elution buffer was 1 mm EGTA 0.3 m Glycine-HCl, pH 2.7.
Western blots were incubated in Ponceau S stain for 5 min. Blots were subsequently washed with phosphate-buffered saline containing 0.1% Tween (v/v; PBST), blocked in PBST 5% (w/v) milk for 1 h, and incubated either in affinity-purified AtPEX2 antibody (1:500 dilution) or a polyclonal raised against GFP (1:2,000 dilution, Molecular Probes) in 2% milk PBST for 16 h. Blots were subsequently washed five times for 5 min in PBST, incubated for 1 h in alkaline phosphatase-conjugated secondary antibody (1:1,000 dilution, DAKO), and washed again. Detection was performed using the 5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium color development substrate according to manufacturer's instructions (Promega).
AtPEX2 26-amino acid peptide was resuspended in distilled water (1 mg/mL) and 0.4 mg was incubated at 37°C with the affinity-purified AtPEX2 peptide antibody (1:5 dilution) in 2% milk PBST for 30 min. After this time the antibody solution was diluted to a final 1:500 antibody dilution and used as previously described.
Immunofluorescence Techniques
Arabidopsis cell culture was harvested 4 d after subculture, fixed, permeabilized, and stained according to Saint-Jore et al. (2002). For dual labeling, cells were labeled with affinity-purified anti-PEX2, detected with Texas red-conjugated secondary antibody, blocked with whole rabbit serum for 20 min (as both primary antibodies were raised in rabbits), washed in buffer, and incubated with anti-ICL detected with fluorescein isothiocyanate (FITC)-conjugated secondary. Cells were incubated either in affinity-purified anti-PEX2 (1:5) or anti-ICL (1:1,000) for 18 h, washed, and incubated in FITC-conjugated goat anti-rabbit (Sigma Aldrich, 1:40) or Texas red-conjugated goat anti-rabbit IgG (H+L; Molecular Probes; 1:100) for 2 h.
Sampling and Imaging
Confocal imaging was performed using a Zeiss inverted LSM510 laser scanning microscope with an argon laser, a helium neon laser, and 100×, 63×, and 40× oil immersion objective.
For imaging coexpression of wild-type GFP and eYFP, excitation lines 458 nm for GFP and 514 nm for eYFP were used alternately with line switching in multitracking mode of the microscope (Brandizzi et al., 2002a). Fluorescence was detected using a 458-nm/514-nm dichroic beam splitter with 515-nm dichroic filter and 475- to 525-nm band pass filter for GFP and 535- to 590-nm band pass filter for eYFP. Dual settings for imaging CFP and eYFP are as detailed above. For imaging FITC and Texas red excitation lines, 488 nm and 543 nm were used with frame switching in multitracking mode of the microscope. Fluorescence was detected using 488-nm/543-nm dichroic beam splitter and long pass 560-nm filter for Texas red and 505- to 530-nm band pass filter for FITC. Controls were carried out to prevent cross talk and bleed through of fluorescence. The pinhole was usually set to give a 1 to 1.5 μm optical slice.
Postacquisition image processing was done using the LSM 5 browser software (Zeiss) and Adobe Photoshop elements or creative suite premium.
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third party owners of all or parts of the material. Obtaining permission will be the responsibility of the requester.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AJ276134 (PEX10), AAG52254 (PEX2), AF210431 (Sar1), D01027, and U89959 (RabD2a).
Supplementary Material
Acknowledgments
We thank Barbara Johnson, Anne Kearns, Jan Evins, and Fiona Moulton for production and maintenance of plant lines, Dr. Steve Slocombe for constructing 35S-eYFP-Cassette A-Nos∷pCAMBIA 1300, Laura-Anne Brown for the pDONOR 201∷AtPEX10 construct, Dr. Ian Moore for the RabD2a N121I construct, Dr. Jaideep Mathur for eYFP-SKL∷pCAMBIA 1300, and Dr. John Runions for help with image preparation.
This work was supported by the Biotechnology and Biology Research Council (grant nos. C19029 and C19030 to A.B. and C.H.).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.065094.
References
- Andreeva AV, Zheng H, Saint-Jore CM, Kutuzov MA, Evans DE, Hawes CR (2000) Organization of transport from endoplasmic reticulum to Golgi in higher plants. Biochem Soc Trans 28: 505–512 [PubMed] [Google Scholar]
- Batoko H, Zheng H-Q, Hawes C, Moore I (2000) A Rab1 GTPase is required for transport between the endoplasmic reticulum and Golgi apparatus and for normal Golgi movement in plants. Plant Cell 12: 2201–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandizzi F, Fricker M, Hawes C (2002. a) A greener world: the revolution in plant bioimaging. Nat Rev Mol Cell Biol 3: 520–530 [DOI] [PubMed] [Google Scholar]
- Brandizzi F, Snapp EL, Roberts AG, Lippincott-Schwartz J, Hawes C (2002. b) Membrane protein transport between the endoplasmic reticulum and the Golgi in tobacco leaves is energy dependent but cytoskeleton independent: evidence from selective photobleaching. Plant Cell 14: 1293–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Corpas FJ, Barroso JB, del Rio LA (2001) Peroxisomes as a source of reactive oxygen species and nitric oxide signal molecules in plant cells. Trends Plant Sci 6: 145–150 [DOI] [PubMed] [Google Scholar]
- Distel B, Erdmann R, Gould SJ, Blobel G, Crane DI, Cregg JM, Dodt G, Fujiki Y, Goodman JM, Just WW, et al (1996) Unified nomenclature for peroxisome biogenesis factors. J Cell Biol 135: 1–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber KN, Haan GJ, Baerends RJS, Kram AM, Veenhuis M (2002) Normal peroxisome development from vesicles induced by truncated Hansenula polymorpha Pex3p. J Biol Chem 277: 11026–11033 [DOI] [PubMed] [Google Scholar]
- Flynn CR, Heinze M, Schumann U, Gietl C, Trelease RN (2005) Compartmentalization of the plant peroxin, AtPex10p, within subdomain(s) of ER. Plant Sci 168: 635–652 [Google Scholar]
- Geuze HJ, Murk JL, Stroobants AK, Griffith JM, Kleijmeer MJ, Koster AJ, Verkleij AJ, Distel B, Tabak HF (2003) Involvement of the endoplasmic reticulum in peroxisome formation. Mol Biol Cell 14: 2900–2907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara-Nishimura I, Shimada T, Hatano K, Takeuchi Y, Nishimura M (1998) Transport of storage proteins to protein storage vacuoles is mediated by large precursor-accumulating vesicles. Plant Cell 10: 825–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofgen R, Willmitzer L (1988) Storage of competent cells for Agrobacterium transformation. Nucleic Acids Res 16: 9877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu JP, Aguirre M, Peto C, Alonso J, Ecker J, Chory J (2002) A role for peroxisomes in photomorphogenesis and development of Arabidopsis. Science 297: 405–409 [DOI] [PubMed] [Google Scholar]
- Hunt JE, Trelease RN (2004) Sorting pathway and molecular targeting signals for the Arabidopsis peroxin 3. Biochem Biophys Res Commun 314: 586–596 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) Gus fusions: beta-glucoronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalish JE, Theda C, Morrell J, Berg JM, Gould SJ (1995) Formation of peroxisome lumen is abolished by loss of Pichia pastoris Pas7p, a zinc-binding integral membrane protein of the peroxisome. Mol Cell Biol 15: 6406–6419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotzer AM, Brandizzi F, Neumann U, Paris N, Moore I, Hawes C (2004) AtRabF2b (Ara7) acts on the vacuolar trafficking pathway in tobacco leaf epidermal cells. J Cell Sci 117: 6377–6389 [DOI] [PubMed] [Google Scholar]
- Lazarow PB (2003) Peroxisome biogenesis: advances and conundrums. Curr Opin Cell Biol 15: 489–497 [DOI] [PubMed] [Google Scholar]
- Lin Y, Cluette-Brown JE, Goodman HM (2004) The peroxisome deficient Arabidopsis mutant sse1 exhibits impaired fatty acid synthesis. Plant Physiol 135: 814–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Sun L, Nguyen LV, Rachubinski RA, Goodman HM (1999) The pex16p homolog SSE1 and storage organelle formation in Arabidopsis seeds. Science 284: 328–330 [DOI] [PubMed] [Google Scholar]
- Lisenbee CS, Heinze M, Trelease RN (2003. a) Peroxisomal ascorbate peroxidase resides within a subdomain of rough endoplasmic reticulum in wild-type Arabidopsis cells. Plant Physiol 132: 870–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisenbee CS, Karnik SK, Trelease RN (2003. b) Overexpression and mislocalization of a tail-anchored GFP redefines the identity of peroxisomal ER. Traffic 4: 491–501 [DOI] [PubMed] [Google Scholar]
- Maier AG, Schulreich S, Bremser M, Clayton C (2000) Binding of coatomer by the PEX11 C-terminus is not required for function. FEBS Lett 484: 82–86 [DOI] [PubMed] [Google Scholar]
- McCartney AW, Dyer JM, Dhanoa PK, Kim PK, Andrews DW, McNew JA, Mullen RT (2004) Membrane-bound fatty acid desaturases are inserted co-translationally into the ER and contain different ER retrieval motifs at their carboxy termini. Plant J 37: 156–173 [DOI] [PubMed] [Google Scholar]
- Mullen RT, Lisenbee CS, Miernyk JA, Trelease RN (1999) Peroxisomal membrane ascorbate peroxidase is sorted to a membranous network that resembles a subdomain of the endoplasmic reticulum. Plant Cell 11: 2167–2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy MA, Phillipson BA, Baker A, Mullen RT (2003) Characterization of the targeting signal of the Arabidopsis 22-kD integral peroxisomal membrane protein. Plant Physiol 133: 813–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nito K, Yamaguchi K, Kondo M, Hayashi M, Nishimura M (2001) Pumpkin peroxisomal ascorbate peroxidase is localized on peroxisomal membranes and unknown membranous structures. Plant Cell Physiol 42: 20–27 [DOI] [PubMed] [Google Scholar]
- Okumoto K, Itoh R, Shimozawa N, Suzuki Y, Tamura S, Kondo N, Fujiki Y (1998) Mutations in PEX10 is the cause of Zellweger peroxisome deficiency syndrome of complementation group B. Hum Mol Genet 7: 1399–1405 [DOI] [PubMed] [Google Scholar]
- Passreiter M, Anton M, Lay D, Frank R, Harter C, Wieland FT, Gorgas K, Just WW (1998) Peroxisome biogenesis: involvement of ARF and coatomer. J Cell Biol 141: 373–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platta H, Girzalsky W, Erdmann R (2004) Ubiquitination of the peroxisomal import receptor Pex5p. Biochem J 384: 37–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purdue PE, Lazarow PB (2001) Peroxisome biogenesis. Annu Rev Cell Dev Biol 17: 701–752 [DOI] [PubMed] [Google Scholar]
- Ritzenthaler C, Nebenfuhr A, Movafeghi A, Stussi-Garaud C, Behnia L, Pimpl P, Staehelin LA, Robinson DG (2002) Reevaluation of the effects of Brefeldin A on plant cells using tobacco Bright Yellow 2 cells expressing Golgi-targeted green fluorescent protein and COPI antisera. Plant Cell 14: 237–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Jore CM, Evins J, Batoko H, Brandizzi F, Moore I, Hawes C (2002) Redistribution of membrane proteins between the Golgi apparatus and endoplasmic reticulum in plants is reversible and not dependent on cytoskeletal networks. Plant J 29: 661–678 [DOI] [PubMed] [Google Scholar]
- Salomons FA, van der Klei I, Kram AM, Harder W, Veenhuis M (1997) Brefeldin A interferes with peroxisomal protein sorting in the yeast Hansenula polymorpha. FEBS Lett 411: 133–139 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW (2001) Molecular Cloning: A Laboratory Manual, Ed 3. Cold Spring Harbor Laboratory Press, New York
- Schumann U, Wanner G, Veenhuis M, Schmid M, Gietl C (2003) AthPEX10, a nuclear gene essential for peroxisome and storage organelle formation during Arabidopsis embryogenesis. Proc Natl Acad Sci USA 100: 9626–9631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- South ST, Baumgart E, Gould SJ (2001) Inactivation of the endoplasmic reticulum protein translocation factor, Sec61p, or its homolog, Ssh1p, does not affect peroxisome biogenesis. Proc Natl Acad Sci USA 98: 12027–12031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- South ST, Sacksteder KA, Li XL, Liu YF, Gould SJ (2000) Inhibitors of COPI and COPII do not block PEX3-mediated peroxisome synthesis. J Cell Biol 149: 1345–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes IA, Baker A (2002) Peroxisome biogenesis and protein import in plants, animals and yeasts: enigma and variations? (review). Mol Membr Biol 19: 171–185 [DOI] [PubMed] [Google Scholar]
- Sparkes IA, Brandizzi F, Slocombe SP, El-Shami M, Hawes C, Baker A (2003) An Arabidopsis pex10 null mutant is embryo lethal, implicating peroxisomes in an essential role during plant embryogenesis. Plant Physiol 133: 1809–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabak HF, Murk JL, Braakman I, Geuze HJ (2003) Peroxisomes start their life in the endoplasmic reticulum. Traffic 4: 512–518 [DOI] [PubMed] [Google Scholar]
- Theodoulou FL, Job K, Slocombe SP, Footitt S, Holdsworth M, Baker A, Larson TR, Graham IA (2005) Jasmonic acid levels are reduced in COMATOSE ATP-binding cassette transporter mutants: implications for transport of jasmonate precursors into peroxisomes. Plant Physiol 137: 835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titorenko VI, Ogrydziak DM, Rachubinski RA (1997) Four distinct secretory pathways serve protein secretion, cell surface growth, and peroxisome biogenesis in the yeast Yarrowia lipolytica. Mol Cell Biol 17: 5210–5226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titorenko VI, Rachubinski RA (1998) Mutants of the yeast Yarrowia lipolytica defective in protein exit from the endoplasmic reticulum are also defective in peroxisome biogenesis. Mol Cell Biol 18: 2789–2803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titorenko VI, Rachubinski RA (2001. a) Dynamics of peroxisome assembly and function. Trends Cell Biol 11: 22–29 [DOI] [PubMed] [Google Scholar]
- Titorenko VI, Rachubinski RA (2001. b) The life cycle of the peroxisome. Nat Rev Mol Cell Biol 2: 357–368 [DOI] [PubMed] [Google Scholar]
- Tormakangas K, Hadlington JL, Pimpl P, Hilmer S, Brandizzi F, Teeri TH, Denecke J (2001) A vacuolar sorting domain may also influence the way in which proteins leave the endoplamic reticulum. Plant Cell 13: 2021–2032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorn-Brouwer T, Kragt A, Tabak HF, Distel B (2001) Peroxisomal membrane proteins are properly targeted to peroxisomes in the absence of COPI- and COPII-mediated vesicular transport. J Cell Sci 114: 2199–2204 [DOI] [PubMed] [Google Scholar]
- Zolman BK, Monroe-Augustus M, Thompson B, Hawes JW, Krukenberg KA, Matsuda SPT, Bartel B (2001) chy1, an Arabidopsis mutant with impaired beta-oxidation, is defective in a peroxisomal beta-hydroxyisobutyryl-CoA hydrolase. J Biol Chem 276: 31037–31046 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.