Abstract
The sterol 14α-demethylase (CYP51) is the most widely distributed cytochrome P450 gene family being found in all biological kingdoms. It catalyzes the first step following cyclization in sterol biosynthesis, leading to the formation of precursors of steroid hormones, including brassinosteroids, in plants. Most enzymes involved in the plant sterol biosynthesis pathway have been characterized biochemically and the corresponding genes cloned. Genes coding for enzymes promoting substrate modifications before 24-methylenelophenol lead to embryonic and seed defects when mutated, while mutants downstream the 24-methylenelophenol intermediate show phenotypes characteristic of brassinosteroid mutants. By a differential display approach, we have isolated a fertilization-induced gene, encoding a sterol 14α-demethylase enzyme, named CYP51G1-Sc. Functional characterization of CYP51G1-Sc expressed in yeast (Saccharomyces cerevisiae) showed that it could demethylate obtusifoliol, as well as nontypical plant sterol biosynthetic intermediates (lanosterol), in contrast with the strong substrate specificity of the previously characterized obtusifoliol 14α-demethylases found in other plant species. CYP51G1-Sc transcripts are mostly expressed in meristems and in female reproductive tissues, where they are induced following pollination. Treatment of the plant itself with obtusifoliol induced the expression of the CYP51G1-Sc mRNA, suggesting a possible role of this transient biosynthetic intermediate as a bioactive signaling lipid molecule. Furthermore, treatments of leaves with 14C-labeled obtusifoliol demonstrated that this sterol could be transported in distal parts of the plant away from the sprayed leaves. Arabidopsis (Arabidopsis thaliana) CYP51 homozygous knockout mutants were also lethal, suggesting important roles for this enzymatic step and its substrate in plant development.
Sterols are ubiquitous components of the plasma membrane, where they play an important role in membrane fluidity and permeability (Hartmann and Benveniste, 1987; Hartmann, 1998) and as precursors of the brassinosteroids (BRs), a group of plant growth regulators known to affect a wide variety of physiological processes. BRs are involved in stem elongation, root growth inhibition, leaf bending and unrolling, pollen tube growth, photomorphogenesis, tracheary element differentiation, promotion of 1-aminocyclopropane-1-carboxylic acid production, and cell elongation mediated either through microtubule reorientation or through alteration of the mechanical properties of the cell wall (Clouse and Sasse, 1998; Clouse, 2002a, 2002b). Sitosterol, campesterol, and stigmasterol are the most abundant sterols in the plant membrane. More than 30 enzymatic steps catalyze the sequential reactions of sterol biosynthesis (Benveniste, 2004), resulting in the wide variety of sterols and biosynthetic intermediates found in plants. More than 60 sterols and derivatives have been identified in maize (Zea mays) seedlings (Guo et al., 1995), while animals and fungi contain a less complex sterol mixture, composed mainly of cholesterol and ergosterol, respectively (Edwards and Ericsson, 1999).
In vertebrates, cholesterol and steroid derivatives are involved in adequate embryonic development and in keeping homeostasis in adulthood (Farese and Herz, 1998). Intermediates in the sterol biosynthesis pathway can also act as signaling molecules (Farese and Herz, 1998; Ponting and Aravind, 1999), while lipid rafts composed of lipids and sterols create a microenvironment in the membrane that activates signal transduction by promoting clustering of specific membrane-associated proteins, such as channels, cytoskeletal-associated proteins, and receptor proteins (Simons and Toomre, 2000). In plants, structural platforms, such as lipid rafts, have recently been identified (Mongrand et al., 2004). In yeast (Saccharomyces cerevisiae), the ERGOSTEROL-DEFICIENT6 (ERG6) gene, the homolog of Arabidopsis (Arabidopsis thaliana) STEROL METHYLTRANSFERASE 1 (SMT1), affects protein transport and polar localization at the plasma membrane (Fischer et al., 2004). Furthermore, in the smt1 mutant, cell polarity and PIN protein positioning are affected (Willemsen et al., 2003). Some homeodomain transcription factors of the HD-ZIP family, such as ATHB-8, expressed in Arabidopsis embryo procambial cells (Baima et al., 1995; Sessa et al., 1998), as well as PHAVOLUTA, PHABULOSA, and REVOLUTA, involved in leaf adaxial/abaxial patterning (McConnell et al., 2001), contain a steroidogenic acute regulatory transfer domain (or START domain) in addition to their DNA binding domain. In animals, the START domain binds sterols that modulate their regulatory function (Ponting and Aravind, 1999). The functionality of START domains in plant HD-ZIP proteins remains to be proven.
Interestingly, in the plant sterol biosynthesis pathway, mutations that affect enzymes upstream of 24-methylenelophenol impair embryo patterning and seedling development, while mutations in enzymes located downstream do not (Clouse, 2000, 2002a, 2002b). Overexpression, down-regulation, and mutant analyses of enzymes involved in sterol biosynthesis before the step leading to sitosterol and campesterol synthesis (SMT1, C24-methyltransferase; CYP51, C14-demethylase; FACKEL, C14-reductase; HYDRA1, Δ8-Δ7 sterol isomerase) influence the amount of campesterol and the balance between sitosterol and BRs (Schaller et al., 1998; Jang et al., 2000; Schrick et al., 2000; Kushiro et al., 2001; Sitbon and Jonsson, 2001; Holmberg et al., 2002; Souter et al., 2002). The smt1 mutant also shows a defect in vascular patterning (Carland et al., 2002), and smt1, fk, and hyd1/2 cannot be rescued by exogenous application of BRs. While lack of sitosterol and campesterol in these mutants does not seem to explain the embryo defect, abnormal accumulation of biosynthetic intermediates (Jang et al., 2000) or lack of an intermediate sterol as a signal molecule (Schrick et al., 2000) could impair early plant development. The cvp1/smt2 mutant is slightly different because although it does not exhibit the altered embryogenesis of fk, hyd1, and smt1, it also cannot be rescued by BR treatment like the more downstream mutants (Schaeffer et al., 2001; Carland et al., 2002).
The sterol 14α-demethylase is a member of the cytochrome P450 gene family, which catalyzes the oxidative removal of the 14α-methyl group of lanosterol in mammals and yeast, 24-methylene-24,25-dihydrolanosterol in fungi, and obtusifoliol in plants and some bacteria. In these organisms, this enzymatic oxidative step is considered to be one of the key steps of sterol biosynthesis. Sterol 14α-demethylation involves three successive monooxygenation steps of the C14α-methyl group to be removed to produce a C14-double bond and requires NADPH (Werck-Reichhart and Feyereisen, 2000). Genomic analysis revealed >280 P450 enzymes in Arabidopsis, which is approximately 5 times more than in any nonplant genome sequenced yet (Schuler and Werck-Reichhart, 2003; Nelson et al., 2004). Out of these P450 enzymes, CYP51 is the only one that shows conservation between animal, fungi, plant, and bacteria phylum (Lepesheva and Waterman, 2004). The enzymatic pathway involving sterol 14α-demethylase (CYP51) leads to cholesterol formation in mammals, ergosterol in yeast and fungi, and sitosterol and BRs in plants. Yeast and mammal CYP51s act upstream of the biosynthetic pathway immediately after the production of lanosterol, the first cyclized intermediate, and can oxidize a variety of substrates. In contrast, plant CYP51s are operating several enzymatic steps later and, in relation with their location in the biosynthetic scheme, generally show a strict substrate specificity toward obtusifoliol (Taton and Rahier, 1991; Lamb et al., 1998). Specificity of plant 14α-demethylase has been shown in maize (Taton and Rahier, 1991), wheat (Triticum aestivum; Cabello-Hurtado et al., 1997), sorghum (Sorghum bicolor; Bak et al., 1997), and recently in Arabidopsis (Kushiro et al., 2001). The lack of activity of plant CYP51s toward fungi and mammal substrates has been attributed to a structural feature common to all these nonplant sterol substrates: the presence of a 4β-methyl group in lanosterol and eburicol, precursors of C14-desmethylsterols, could hinder the binding and/or demethylation of these sterols (Taton and Rahier, 1991).
Here, we report the cloning and characterization of a sterol 14α-demethylase from Solanum chacoense (CYP51G1-Sc) that can C14-demethylate obtusifoliol, lanosterol, and eburicol, thus being a plant CYP51 enzyme with a wider substrate specificity. Expression analyses, translocation of obtusifoliol away from the site of application, and Arabidopsis knockout insertion mutant phenotypes suggest that CYP51G1-Sc plays a role in embryogenesis as other members of the sterol pathway located above 24-methylenelophenol and that obtusifoliol could act as a lipid signaling molecule.
RESULTS
Gene Cloning and Sequence Analysis
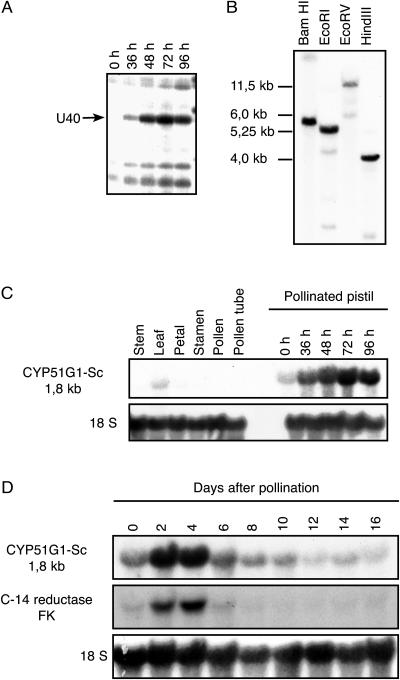
We used a differential display approach to isolate genes whose transcription is modulated by pollination and/or fertilization events in pistil tissues. The mRNA profile of pollinated pistils was compared with those of control pistils 24, 36, 48, 72, and 96 h after pollination. First-strand cDNA produced from those RNA were amplified with a combination of seven-nucleotide arbitrary primers (H-AP1–16) and a 12-nucleotide anchor primer (H-T11-N) that both included a HindIII sequence at their 5′ end. One candidate gene, amplified with H-AP-9 (AAGCTTCATTCCG) and H-T11-A (AAGCTTTTTTTTTTTA; Fig. 2A), showed up-regulation on a differential display gel after pollination, peeking at 72 h postpollination. This candidate, named U40 (for up-regulated 40), was excised from the polyacrylamide gel, reamplified with the H-AP-9 and H-T11-A primer pair, and cloned. Sequence information from this partial cDNA revealed that it belonged to the cytochrome P450 monoxygenase gene family. The initial sequence obtained by differential display RT-PCR covered the 3′ untranslated region (UTR) and a small part of the protein C-terminal region. In order to obtain a full-length clone, a cDNA pistil library made from 48 h postpollinated pistil tissues was probed with the U40 cDNA fragment, and a 1,747-bp clone was isolated (GenBank accession number AY552551). The size of this cDNA corresponded to the size estimated on RNA gel-blot analysis (1.8 kb), indicating that the cDNA is full length or near full length (Fig. 2C). Two in-frame stop codons are found upstream of the longest open reading frame in the 100-nucleotide 5′ UTR, indicating that the deduced protein sequence is complete. The gene codes for a 487-amino acid protein with a predicted Mr of 55.2 kD and a pI of 8.70. The cDNA shared strong sequence identities with sterol 14α-demethylase genes found in yeast, plants, and mammals and was renamed CYP51G1-Sc for S. chacoense cytochrome P450-sterol 14α-demethylase, following the new nomenclature for the CYP51 family (Nelson et al., 2004). At the amino acid level (similarity values are shown in parenthesis), CYP51G1-Sc shared 93% sequence identity (97%) with the CYP51 deduced protein from Nicotiana tabacum; 82% (91%) with the Arabidopsis CYP51A2, recently renamed CYP51G1; 77% (87%) with the CYP51G1 from Oryza sativa; 76% (87%) with the CYP51 from S. bicolor; 75% (83%) with the CYP51 from T. aestivum; 37% (55%) with the mouse CYP51; 35% (55%) with the human CYP51; 33% (47%) with the CYP51 from Mycobacterium tuberculosis; 32% (53%) with the CYP51 from Trypanosoma brucei; 30% (49%) with the CYP51 from the fission yeast (Schizosaccharomyces pombe); and 26% (42%) with the CYP51 from yeast (S. cerevisiae).
Figure 2.
Expression analysis and gene copy number of the S. chacoense CYP51 gene. A, Portion of the differential display gel showing the up-regulated number 40 band (U40). Reverse-transcribed total RNA was extracted from pollinated pistil tissues from 36 to 96 h after pollination, as well as from control unpollinated pistils (0 h), and amplified with the H-AP-9 and the H-T11-A primers. B, DNA gel-blot analysis of the CYP51G1-Sc gene. Genomic DNA (10 μg) isolated from S. chacoense leaves was digested with BamHI, EcoRI, EcoRV, or HindIII restriction enzymes, blotted on membrane, and probed with the complete 1.7-kb CYP51G1-Sc cDNA insert. Mr of the major detected fragments appears on the left. C, RNA gel-blot analysis of CYP51G1-Sc mRNA accumulation in mature tissues and in pistils at different times after pollination. Ten micrograms of total RNA from the tested tissues was probed with the CYP51G1-Sc cDNA insert. D, RNA gel-blot analysis of CYP51G1-Sc mRNA accumulation in isolated ovules (from 6–16 DPP) and ovaries without their pericarp (from anthesis to 4 DPP). A S. chacoense homolog of the C-14 reductase gene (FK) was also used to reprobe the same membrane after stripping.
Gene Copy Number of the CYP51G1-Sc 14α-Demethylase
A DNA gel blot of S. chacoense genomic DNA was probed with the complete CYP51G1-Sc cDNA insert (Fig. 2B). Only one strong hybridizing fragment could be detected for each of the four enzymes used. Since none of these enzymes has a cleavage site in the CYP51G1-Sc cDNA, this strongly suggests that CYP51G1-Sc is a single copy gene in S. chacoense. Although the hybridization and membrane washing conditions were stringent, faint bands could also be observed in some lanes. This could result from the presence of restriction cleavage sites in introns and/or the presence of a closely related gene in the S. chacoense genome. Indeed, a PCR analysis on genomic DNA with primers specific for 5′ and 3′ ends of the CYP51G1-Sc cDNA revealed that it had at least one intron in this region, spanning roughly 700 bp (data not shown). Furthermore, two genes in Arabidopsis and N. tabacum as well as 10 genes in the rice genome have been classified as CYP51 enzymes (Burger et al., 2003; Nelson et al., 2004). This suggests that there might also be related CYP51 isoforms in the S. chacoense genome.
CYP51G1-Sc Expression Profile
A thorough RNA expression profile was conducted in various vegetative tissues as well as in reproductive tissues before and after pollination. The CYP51G1-Sc transcript could be detected as a single 1.8-kb band on all tissues tested (Figs. 2, 3, and 5). CYP51G1-Sc transcripts were undetectable in stems, petals, stamens, pollen, and in-vitro-grown pollen tubes (Fig. 2C). The gene is weakly and similarly expressed in mature leaves and unpollinated pistils on the day of anthesis, as well as in roots (data not shown). Following fertilization, a 6-fold increase in mRNA levels is detected in pistils, peaking around 72 h postpollination (Fig. 2C). This increase is transient, as determined by a careful analysis of ovules (from 6–16 d postpollination [DPP]) and ovaries without their pericarp (from anthesis to 4 DPP). After peaking between 2 and 4 DPP, the CYP51G1-Sc mRNA levels steadily decrease thereafter (Fig. 2D). In the sterol biosynthesis pathway (Fig. 1), the next enzymatic step following the 14α-demethylation is the 14α-reduction, catalyzed by the sterol C14-reductase, encoded by the previously described FACKEL (FK) gene (Jang et al., 2000; Schrick et al., 2000). Using sequences selected from tomato (Lycopersicon esculentum) expressed sequence tags (AI484506, AI485167, and AW931153) similar to the Arabidopsis FK sequence, conserved primers derived from exons 9 and 13 according to the Arabidopsis sequence were synthesized and used to obtain a PCR fragment from reverse-transcribed ovule mRNAs corresponding to the S. chacoense FK gene. A 550-bp DNA fragment was obtained, and sequencing confirmed that the amplified fragment corresponded to the FK C14-reductase gene (data not shown). This fragment was used to reprobe the same membrane after complete stripping, and the result is shown in Figure 2D. An mRNA expression pattern identical to the one obtained with the CYP51G1-Sc probe could also be observed for the FK gene in isolated ovules, suggesting similar transcriptional regulation. The FK gene has been previously shown to be expressed in the embryo and is involved in cell division and cell expansion (Jang et al., 2000; Schrick et al., 2000).
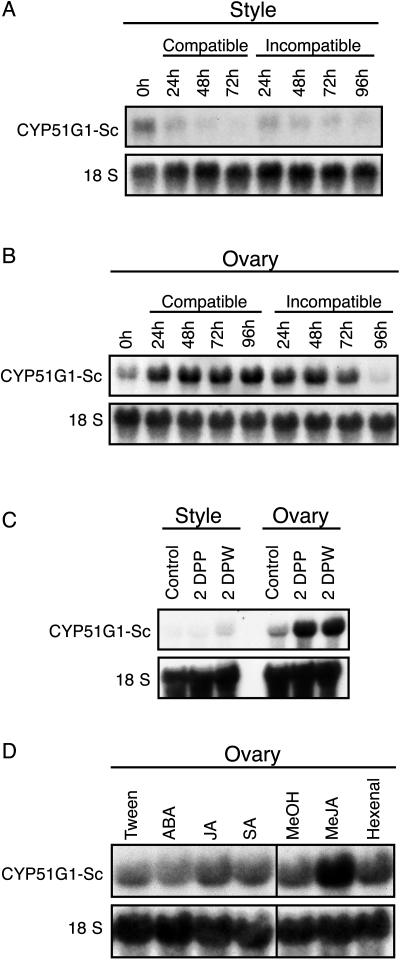
Figure 3.
RNA expression analysis of CYP51G1-Sc transcript levels in styles and ovaries following pollination and stress treatments. A and B, Effect of compatible and incompatible pollination on CYP51G1-Sc transcript levels. CYP51G1-Sc transcript levels were determined by RNA gel-blot analysis in unpollinated pistils on anthesis day (0 h) and in pistils collected from 24 to 96 h after a fully compatible (S11S13 × S12S14) pollination or a fully incompatible (S12S14 × S12S14) pollination. Styles and ovary mRNAs were extracted and blotted separately. C, CYP51G1-Sc transcript levels were determined by RNA gel-blot analysis of tissues 2 DPP or 2 d after wounding (DPW). Styles were either compatibly pollinated or slightly crushed with tweezers and collected 2 d later. Unpollinated and unwounded styles and ovaries served as controls. Ovaries from the same pistils were also collected either 2 DPP or 2 d after wounding. D, CYP51G1-Sc transcript levels were determined by RNA gel-blot analysis of unpollinated ovary tissues collected 48 h after various hormonal treatments. Spray treatments: abscisic acid (ABA) and jasmonic acid (JA) as 50 μm solutions in 0.05% Tween; salicylic acid (SA) as a 5 mm solution in 0.05% Tween. Flowers sprayed with a 0.05% Tween solution served as control. For treatment with volatiles, methyl jasmonate (MeJA) or trans-2-hexenal (Hexenal) were first diluted to 0.1 m in ice-cold methanol, and 100 μL of the compound (equivalent to 10 μL/L of air space) was added to a piece of Whatman paper suspended over the plant in an airtight container. Treatment with 100 μL of methanol (MeOH) served as control. For all RNA gel blots, 10 μg of total RNA from the various tissues was probed with the CYP51G1-Sc cDNA insert. To ascertain equal loading conditions, all RNA gel blots were stripped and reprobed with an 18S ribosomal cDNA probe from S. chacoense.
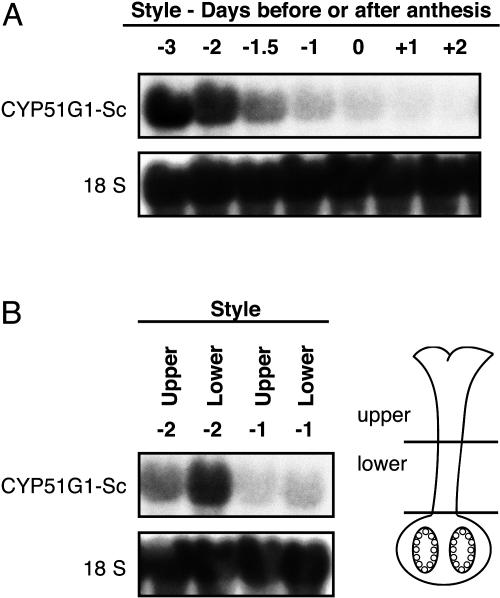
Figure 5.
Developmental expression pattern of CYP51G1-Sc mRNA levels in unpollinated pistil tissues. A, CYP51G1-Sc mRNA levels in styles of unpollinated pistil tissues were analyzed by RNA gel-blot analysis. Ten micrograms of total RNA from styles 3 d before anthesis to 2 d after anthesis were probed with the CYP51G1-Sc cDNA insert. B, RNA gel-blot analysis of CYP51G1-Sc mRNA levels in the upper and lower halves of styles from flowers 2 (−2) and 1 (−1) d before anthesis.
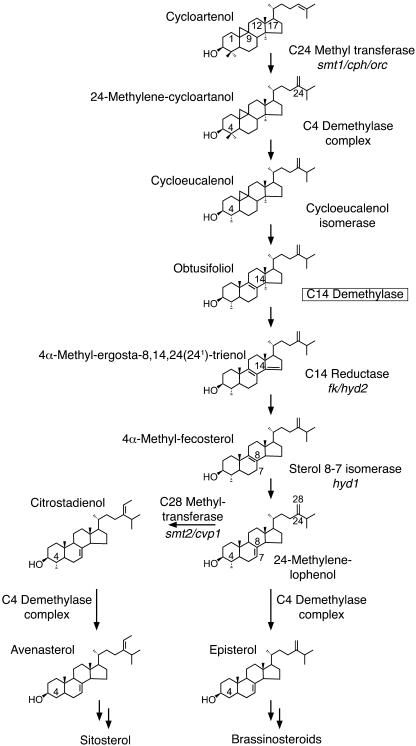
Figure 1.
Schematic representation of the sterol biosynthesis pathway in plants. Only the upper part of the pathway from the first cyclic precursor, cycloartenol, to the first branch point represented by 24-methylenelophenol is presented. Sterol molecules are labeled with their common names (see sterol nomenclature in “Materials and Methods”). Enzymes involved in the biosynthetic pathway are mentioned, and the corresponding genes identified by mutants, when characterized, are shown in italics underneath. The sterol C14-demethylation step is boxed.
CYP51G1-Sc Expression Is Regulated Both by Pollination and by Fertilization
Since fertilization in S. chacoense occurs from 36 h onward after pollination (O'Brien et al., 2002b), we next determined if the strong induction of the CYP51G1-Sc mRNA transcripts was triggered by pollination alone, or if fertilization was indeed needed to increase the steady-state levels of the CYP51G1-Sc mRNA. To address this issue, we carried out a closer time-course analysis and took advantage of the gametophytic self-incompatibility barrier expressed in S. chacoense stylar tissues. S. chacoense expresses stylar ribonucleases that can reject pollen having the same genetic constitution at the S locus, thus preventing inbreeding (Matton et al., 1997). In such an incompatible pollination, pollen tube growth is arrested in the first two-thirds of the style, and pollen tubes never reach the ovules. A pollination time course was conducted, and RNA gel-blot analyses were done on styles and ovaries separately (Fig. 3, A and B). Twenty-four hours after either a compatible or an incompatible pollination, CYP51G1-Sc mRNA levels were strongly induced in ovaries (Fig. 3B), indicating that the gene was pollination induced, at a distance, in the ovary. Furthermore, CYP51G1-Sc mRNA levels declined in compatibly pollinated styles (Fig. 3A), while they remained constant in styles following an incompatible pollination, albeit CYP51G1-Sc mRNA levels are only expressed at a very low level in both cases when compared to the CYP51G1-Sc mRNA levels found in the ovary. In a compatible pollination, the CYP51G1-Sc mRNA levels remained high in the ovary during the whole time-course period (until 4 d after pollination), while a drastic decrease was observed 3 d following an incompatible pollination, leading to nearly undetectable levels of CYP51G1-Sc mRNA 4 d after pollination (Fig. 3B), suggesting that the CYP51G1-Sc gene is regulated by both pollination and fertilization. Only during a compatible pollination leading to fertilization and seed set is a sustained accumulation observed (Figs. 2D and 3B), while an incompatible pollination only leads to the first and transient burst of accumulation, corresponding to the pollination-induced phase. This type of response has already been observed for some fertilization-responsive genes, and pollination-induced expression had been linked to a wounding effect (Lantin et al., 1999; O'Brien et al., 2002a). Indeed, in many species, pollination is known to induce deterioration and death of cells or tissues, including the secretory cells in the stigma and the transmitting tissue of the style (Cheung, 1996). To determine if cell death caused by wounding could also trigger CYP51G1-Sc mRNA accumulation at a distance in the ovary, the styles of young flowers were slightly crushed with tweezers, and the styles and ovaries were collected separately 48 h later. Figure 3C shows that wounding of the style also induced CYP51G1-Sc mRNA accumulation at a distance in the ovary (Fig. 3C, lane 2 DPW), to the same extent as a 48-h compatible pollination (Fig. 3C, lane 2 DPP). Wounding had only a modest effect in the style itself (Fig. 3C), as determined before for pollen tube growth in the style (Fig. 3A). Thus, cellular deterioration and death caused by pollination (either compatible or incompatible) or by wounding might be the initial trigger that induces CYP51G1-Sc mRNA accumulation in the ovary. In order to confirm this, various wound- or defense-related hormones were tested. None had a significant effect of CYP51G1-Sc mRNA accumulation in styles (data not shown), but in ovaries, methyl jasmonate treatment was able to mimic a wound- or pollination-induced response (Fig. 3D), suggesting that the first and transient CYP51G1-Sc mRNA accumulation phase was induced by transmitting tract cell death caused by pollen tube growth.
In Situ Analysis of CYP51G1-Sc mRNA Accumulation
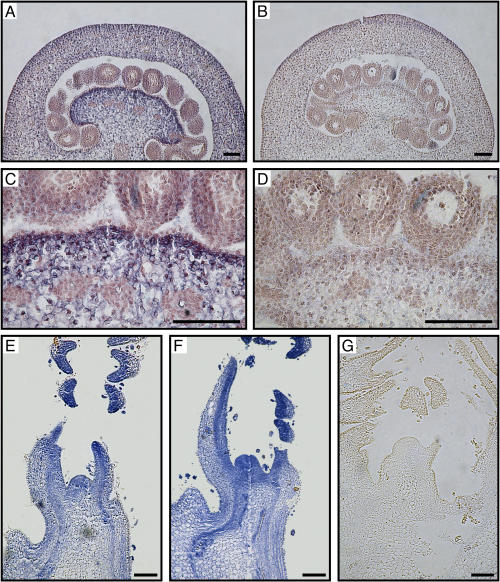
In order to determine more precisely the tissular expression pattern of the CYP51G1-Sc 14α-demethylase, in situ hybridizations were performed on 10-μm sections of fertilized ovaries 3 d after pollination, where peak accumulation occurs (Fig. 2, C and D). The CYP51G1-Sc antisense probe detected strong expression in the placenta, as well as in the developing pericarp, with much weaker staining in the ovule's integument (Fig. 4A). This expression pattern could explain the RNA gel-blot results shown in Figure 2D, where a steep decline was observed in CYP51G1-Sc mRNA levels in isolated ovules, from 6 d after pollination. A magnification of the major accumulation area detected in Figure 4A is shown in Figure 4C. Although CYP51G1-Sc mRNAs can be detected everywhere in the placenta and the pericarp, a darker staining band corresponding to thin-walled parenchymatous cells can be observed in the placenta layer immediately underneath the outermost cell layer that forms the placenta's epidermis. This layer, which does not express the CYP51G1-Sc mRNAs, is continuous with the ovule's integument epidermis, where no CYP51G1-Sc mRNAs could be detected, indicating a clear difference and specialization of this layer in both the placenta epidermis and the ovule's integument. As a control, similar sections were hybridized with the CYP51G1-Sc sense probe, and no significant staining could be observed (Fig. 4,B and D). Although CYP51G1-Sc mRNA levels were quite low in mature tissues, the fertilization-induced expression could suggest that CYP51G1-Sc mRNAs accumulate preferentially in rapidly growing tissues and meristematic cells, as observed for the FK gene (Jang et al., 2000; Schrick et al., 2000). In situ hybridizations were thus conducted on shoot apex containing shoot apical meristems. Figure 4, E and F, shows that CYP51G1-Sc mRNAs strongly accumulated in the shoot apical meristem, in young primordia, as well as in the young leaflets that surround the apex. Furthermore, a preferential accumulation can distinctly be observed on the adaxial side of the developing leaves. A control hybridization with a CYP51G1-Sc sense probe, showing the specificity of the staining pattern, is shown in Figure 4G. Although CYP51G1-Sc mRNA accumulation was shown to be very low in mature styles (Fig. 3, A and C), the strong accumulation observed in young developing tissues prompted us to determine if other actively growing tissues apart from the meristems and shoot apex would also accumulate substantial amounts of CYP51G1-Sc mRNAs. We had previously shown that a pistil-expressed aquaporin was strongly expressed in young developing styles and that peak accumulation coincided with the zones of fastest growth (O'Brien et al., 2002a). Other reports had also linked aquaporin expression with zones of cell division, cell elongation, and cell expansion in various tissues (Ludevid et al., 1992; Chaumont et al., 1998; Balk and de Boer, 1999). We thus used developing styles to determine the CYP51G1-Sc mRNA accumulation profile in a fast-growing tissue. Figure 5A shows that CYP51G1-Sc mRNA levels are highest in immature developing styles and that CYP51G1-Sc mRNAs are barely detectable in mature tissues after anthesis. Furthermore, an asymmetrical distribution can be observed with a preferential accumulation in the lower style region prior to anthesis (Fig. 5B).
Figure 4.
In situ localization of CYP51G1-Sc transcripts. A and B, Ovary section 72 h after pollination (approximately 36 h postfertilization). C and D, Magnification of the dark staining placenta layer shown in A and B. E to G, Shoot apex showing both shoot apical meristem and sections of young leaflets covering the shoot apex. A, C, E, and F, antisense probe. B, D, and G, control sense probe. Digoxigenin labeling is visible as a blueish staining. All hybridizations used 10-μm-thick sections and an equal amount of either CYP51G1-Sc sense or antisense probe. Bars = 100 μm.
erg11 Yeast Complementation
To further address the functionality of the CYP51G1-Sc enzyme, we assessed its potential to rescue the yeast erg11 mutant strain that is deficient in the CYP51 lanosterol demethylase enzyme. Under aerobic conditions, the homozygote erg11 mutant is deficient for lanosterol demethylation and is unable to grow without ergosterol supplementation (Kalb et al., 1987). Diploid ERG11/erg11 yeast strains transformed with the pYeDP60 vector only (control) and ERG11/erg11 yeast transformed with CYP51G1-Sc coding region in the pYeDP60 vector were grown on a sporulating liquid media to induce the production of haploid spores, hence revealing the mutant phenotype of the erg11 allele. Recombinant spores that harbored the pYeDP60 vector were selected through an adenine selection (Pompon et al., 1996). Spores were spread on a media supplemented with ergosterol as to obtain nonconfluent colony growth. Colonies were replicated and allowed to grow in a selective aerobic media without ergosterol. Percentage of viability was inferred by comparison of colony growth between the duplicated pair. The analysis was performed on five independently transformed series of yeast cells for which 36 randomly picked haploid colonies per series were analyzed. Mutant cells transformed with the empty vector only showed 48% viability (17.4 ± 2.3), which is near the expected value for sporulated heterozygous mutant cells (50%). A partial rescue of the mutant cells in the presence of the CYP51G1-Sc expressing plasmid was observed with a significant increase in cell viability to 81% (29.0 ± 1.2), indicating that the S. chacoense CYP51G1 enzyme could substitute for the yeast endogenous enzyme.
Demethylase Activity Profile of CYP51G1-Sc Expressed Protein
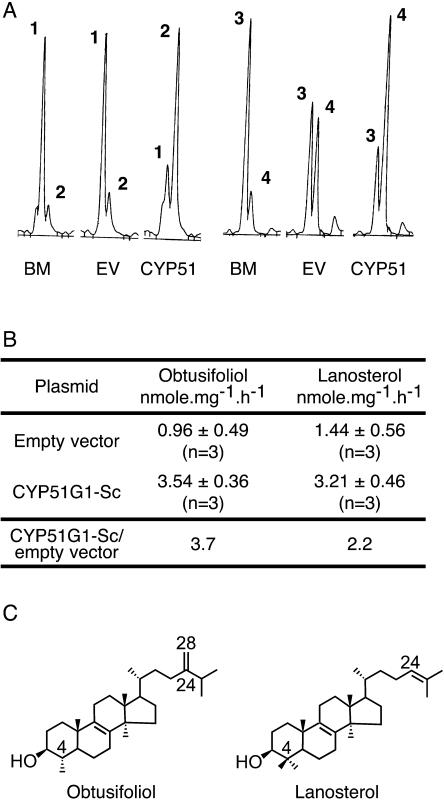
To confirm that the CYP51G1-Sc clone indeed coded for a C14-demethylase enzyme, the complete open reading frame was cloned in the pYeDP60 yeast expression vector, expressed in the WAT11 yeast strain, and tested for C14-demethylation activity on various substrates. Boiled membrane preparations (Fig. 6A, BM) were used as a control for the presence of endogenous demethylation products. Transformation of the WAT11 yeast strain with the empty pYeDP60 vector (EV) was used to monitor substrate transformation by endogenous CYP51 present in the yeast preparation. After transformation with this empty vector, in addition to the expected C14-demethylation of lanosterol (peak 3) into 4,4-dimethylzymosterol (peak 4; Fig. 6A, EV; 1.44 nmol mg−1 h−1) by the yeast endogenous ERG11p, a poorer but significant demethylation of obtusifoliol (peak 1) into 4α-methyl-ergosta-8,24(241)-dien-3βol (peak 2; Fig. 6A, EV) could be detected (0.96 nmol mg−1 h−1), in accordance with previous data showing that this 4α-methylsterol is a possible substrate of ERG11p (Aoyama and Yoshida, 1992). Expression of the CYP51G1-Sc construct in the WAT11 yeast strain led to a 4-fold increase of the demethylation rate of obtusifoliol into 4α-methyl-ergosta-8,24(241)-dien-3-β-ol (peak 2; Fig. 6A, CYP51; 3.54 nmol mg−1 h−1), indicating that CYP51G1-Sc indeed coded for an obtusifoliol 14α-methyl demethylase. Remarkably, when lanosterol was used as a substrate, its demethylation rate into 4,4-dimethyl-zymosterol (peak 4; Fig. 6A, CYP51) was also increased 2-fold following expression of the CYP51G1-Sc construct. This showed that the CYP51G1-Sc enzyme did not have a strict substrate specificity for the typical plant 4α-methylsterol physiological substrate (Taton and Rahier, 1991), being also able to C14-demethylate a 4,4-dimethylsterol (Fig. 6C). Similarly, we could also observe a significant increase for the C14-demethylation of another 4,4-dimethylsterol, eburicol, in the presence of the CYP51G1-Sc protein when compared to the WAT11 yeast strain transformed with the empty vector (data not shown). However, the observed higher relative increment of activity obtained with obtusifoliol strongly suggests a preference of CYP51G1-Sc for this substrate (Fig. 6B).
Figure 6.
CYP51G1-Sc enzymatic activity assay. A, The CYP51G1-Sc protein was expressed in the WAT 11 yeast strain. After incubation of substrate (50 μM) in the corresponding microsomal preparation and extraction procedure, a GC-MS analysis of the assay was performed. Substrates used are (1) obtusifoliol and (3) lanosterol, and products are (2) 4α-methyl-ergosta-8,24(241)-dien-3βol and (4) 4,4-dimethyl-zymosterol. BM, boiled microsome membrane; EV, pYeDP60 empty vector; CYP51, CYP51G1-Sc in pYeDP60 vector. B, The 14α-demethylation specific activity results from three independent experiments using distinct microsomal preparations. C, Chemical structures of obtusifoliol and lanosterol.
Obtusifoliol Can Act as a Signaling Molecule and Moves through the Phloem
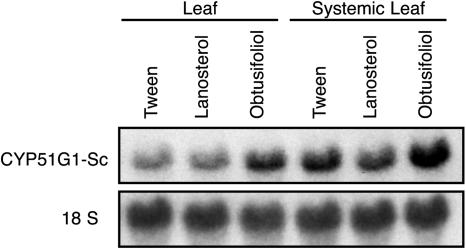
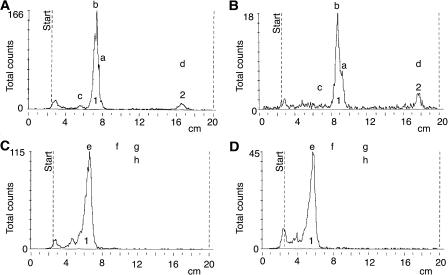
The results obtained with the fk mutant (sterol C14-reductase) prompted the authors to suggest that the synthesis of sterol signals in addition to BRs would be important in mediating cell growth and organization during embryo development (Jang et al., 2000; Schrick et al., 2000). This implies that sterols other than BRs would be active on their own as signaling molecules (Lindsey et al., 2003). In order to test if obtusifoliol could act as a signaling molecule, we tested if it could act locally and at a distance in systemic noninoculated leaves. Figure 7 shows an RNA gel-blot analysis probed with the CYP51G1-Sc cDNA insert. An isolated compound leaf was protected with a Whatman paper cone and gently sprayed with a solution of either lanosterol, obtusifoliol, or Tween as a control on different plants of the same age and kept in identical conditions in the greenhouse. Two days later, the sprayed leaf as well as an upper leaf were harvested and tested for CYP51G1-Sc mRNA levels. When compared to the control leaf, Tween and lanosterol had no effect, both locally and systemically. A significant and reproducible 2-fold increase could be observed with leaves treated with obtusifoliol, both locally and systemically, suggesting that the obtusifoliol sterol could itself act as a signaling molecule, or alternatively, that the effect could be mediated through another diffusible signal induced by the application of obtusifoliol. Consistent with previous results showing higher CYP51G1-Sc mRNA levels in actively growing tissues (Figs. 4 and 5), the basal level of CYP51G1-Sc mRNAs was higher in the younger systemic leaf, although an increase in steady-state level of CYP51G1-Sc mRNAs following obtusifoliol treatment was reproducibly observed both locally and at a distance in systemic leaf. To test if obtusifoliol could itself be a mobile signaling molecule, C14-radiolabeled obtusifoliol was produced and sprayed on a compound leaf again protected with a Whatman paper cone. After 2 d, detached leaves from the main stem were pressed and dried. Figure 8A shows the arrangement of the pressed leaves as they appear on the main stem. Compound leaf 5 was sprayed and exposed separately. Figure 8B shows an autoradiogram of the dried leaves. Leaves are numbered from the base, and the location of the spayed leaf is indicated with the duplication of its internode base (tagged and numbered 5; see also dotted line in Fig. 8A). Apart from residual C14-labeled obtusifoliol on the sprayed leaf, a high amount of labeling was also found in leaf 4, immediately below the sprayed leaf (leaf 5). By overlapping the two pictures, the labeling was detected in the stem as well as in portions of some leaflets in leaf 4. Lower but significant amount of labeling could also be observed in the stems of all leaves (from leaf 1–6), both positioned immediately above or below sprayed leaf 5. This suggested that obtusifoliol or a metabolite of this sterol was transported through the phloem and could act as a mobile signaling molecule. In order to determine the structure of the radiolabeled compound(s) found in leaves distal to the sprayed leaf, the total lipid fraction was isolated from both inoculated leaf 5 and distal leaf 4, where strong radiolabeling was observed. Thin-layer chromatography analysis of this fraction in the absence of addition of any carrier compounds indicated that the bulk of the radioactivity was associated in both cases with the 4α-methylsterol fraction (peak 1 in Fig. 9, A and B) and the steryl-ester fraction (peak 2 in Fig. 9, A and B). The 4α-methylsterol fraction was acetylated and further analyzed on AgNO3-impregnated SiO2-thin layer chromatography (TLC). This led in both cases to a single radioactive peak having a retardation factor corresponding to a sterol possessing two double bonds at C8(9) and C24(241) and comigrating with a standard of obtusifoliyl acetate (peak 1 in Fig. 9, C and D). Analysis of compound(s) present in peak 1 allowed a single compound to be directly and unequivocally identified as obtusifoliyl acetate by coupled capillary gas chromatography-mass spectrometry (GC-MS) analysis (i.e. by coincidental retention time and identical electron impact spectrum as authentic standard). After saponification, the radioactive steryl-ester fraction (peak 2 in Fig. 9, A and B) was further analyzed as above and allowed also a single compound, obtusifoliol, to be identified in the fraction.
Figure 7.
RNA expression analysis of CYP51G1-Sc transcript levels in treated and untreated systemic leaves. Lanosterol and obtusifoliol were emulsified with Tween 80 in water and sprayed (250 μL of a 0.5 mm solution) over a whole compound leaf. Tween application served as a control. After 48 h, the treated leaf and a younger (top) systemic leaf were harvested. Ten micrograms of total RNA from the tissues was probed with the CYP51G1-Sc cDNA insert. To ascertain equal loading conditions, all RNA gel blots were stripped and reprobed with an 18S ribosomal cDNA probe from S. chacoense.
Figure 8.
Treatment of S. chacoense intact leaves with [14C]-radiolabeled obtusifoliol. A, Compound leaf 5 was sprayed with a radiolabeled obtusifoliol solution (250 μL of a 2.7-mg/mL solution (5.66 mm), with a specific activity of 1,250,000 cpm mg−1) and left for 48 h on an intact plant in the greenhouse. Detached leaves were pressed and dried and arranged as they appear on the main stem. Location of the sprayed leaf is indicated with the duplication of its internode base (see dotted line). B, Autoradiogram of the radioactive pressed specimen in A after 1 week of exposure. The sprayed leaf (leaf 5) was exposed separately for 2 d. Bars = 2 cm.
Figure 9.
A and B, Radiochromatogram scans of TLC plates showing accumulation of radioactivity in the 4α-methylsterol (1) and steryl-ester (2) fractions from the unsaponifiable extract from both the initially inoculated compound leaf (A) and the distal compound leaf (B) corresponding to leaf 5 and leaf 4, respectively, in Figure 8. In the absence of added cold carrier obtusifoliol, the minor radioactivity peak on the starting line corresponds to degradation products of obtusifoliol during the feeding experiment and analytical procedure. C and D, Radiochromatogram scans of AgNO3-impregnated SiO2-TLC plates from the chromatography of acetylated 4α-methylsterol fractions showing accumulation of radioactivity in fraction 1 migrating as a standard of obtusifoliyl acetate in both the initially inoculated compound leaf (C) and the distal compound leaf (D) corresponding to leaf 5 and leaf 4, respectively, in Figure 8. Radioactivity on and near the starting line corresponds to degradation products due to the absence of addition of cold carrier obtusifoliol. GC-MS analysis of compounds present in peak 1 indicated the presence of a single compound, obtusifoliol acetate, in this fraction in both samples. Unlabeled standards in A and B: a, lanosterol; b, obtusifoliol; c, cholesterol; d, cholesteryl-stearate. Unlabeled standards in C and D: e, obtusifoliyl acetate; f, 4α, 14α-dimethyl-stigmasta-8,24-dienyl acetate; g, 4α,14α-dimethyl-cholest-8-enyl acetate; h, 4α, 14α-dimethyl-9β, 19-cyclo-ergostanyl acetate.
Physiological relevance of the observed effect of obtusifoliol was determined by the analysis of the recovered C14-labeled obtusifoliol fraction from the treated and distal leaves or stems. The C14-labeled obtusifoliol synthesized had a specific activity of 1.25 × 106 cpm mg−1. For sprayed leaf 5, 1,390 cpm of sterol per milligram of dry weight tissue was recovered, which corresponds to 1,112 μg of obtusifoliol per gram of dry weight tissue. For leaf 4, where most of the radioactivity was translocated, 233 cpm of sterol per milligram of dry weight tissue was recovered, which corresponds to 186 μg of obtusifoliol per gram of dry weight tissue. For the upper systemic leaf 6, which corresponds to the position of the leaf assayed in Figure 7, 3 cpm of sterol per milligram of dry weight tissue was recovered from the petiole, which corresponds to 2.4 μg of obtusifoliol per gram of dry weight tissue. Considering that, in plants, obtusifoliol represents from 0.1% to 1% of the total sterol fraction and that total sterols sum up to ≈2,500 to 4,000 μg per gram of dry weight tissue (Schmitt and Benveniste, 1979; Schaeffer et al., 2001; Holmberg et al., 2002; Schrick et al., 2004), this indicates that a normal physiological concentration of obtusifoliol would be ≈2.5 to 40 μg per gram of dry weight tissue, suggesting that the observed levels of C14-labeled obtusifoliol detected in leaves 4 and 6 would be in the physiological range of obtusifoliol concentrations.
The CYP51G1-At Homozygous Mutant Is Lethal
The Arabidopsis genome contains two CYP51-related genes, CYP51G1-At (formerly AtCYP51A2) and CYP51G2-At (formerly AtCYP51A1). The CYP51G1-Sc gene is most closely related to CYP51G1-At, sharing 82% sequence identity (91% similarity) with CYP51G1-At, compared to 67% sequence identity (79% similarity) with CYP51G2-At. In order to determine the role of the CYP51 14α-sterol demethylase in plants, a heterozygous T-DNA insertion line for CYP51G1-At was obtained from the SALK collection. This line (SALK_067630) has a T-DNA insertion in the CYP51G1-At open reading frame, interrupting the open reading frame around amino acid position 45. DNA primers derived from the CYP51G1-At gene confirmed that a T-DNA element was inserted at the reported position in this line (data not shown). After selfing, seeds from this mutant line were collected and germinated on agar plates. Out of 36 seeds, 5 did not germinate, 3 died at the cotyledon stage, and 28 showed a wild-type phenotype. PCR analysis carried out on genomic DNA extracted from cotyledons or leaves with primers bordering the insertion showed that the three seedlings that died at the cotyledon stage where homozygous for the T-DNA, while out of the 28 normal-looking plants, 16 where heterozygous for the T-DNA, and the remaining 12 were wild-type homozygous plants. As a control, 36 Arabidopsis wild-type plant seeds were also plated onto agar media. All germinated and gave normal-looking seedlings. The ratio obtained (8:16:12) in the selfed SALK_067630 line is quite close to the expected Mendelian ratio if the five seeds that did not germinate are counted as putative homozygous mutants. No PCR analysis was carried out on these nongerminated seeds because of the presence of tissues originating from the heterozygous parents. This indicates that the CYP51G1-At homozygous mutant is lethal. As for the fk mutant (Jang et al., 2000; Schrick et al., 2000), exogenous application of brassinolide at a concentration of 1 μm could not rescue the seedling-lethal phenotype observed in the CYP51G1-At homozygous mutant line (data not shown). Furthermore, this result also shows that CYP51G2-At is not redundant with the CYP51G1-At gene and must have evolved a new substrate specificity. Alternatively, it could have retained CYP51 activity, but its expression pattern might be completely different than that of CYP51G1-At, precluding complementation of the lethality phenotype observed. A SALK T-DNA insertion line for the CYP51G2-At (AtCYP51A1) gene was also available and was tested for its phenotype. This line (SALK_070949) has a T-DNA insertion in the 3′ UTR of the transcript. After selfing, 36 seeds were germinated on agar media. Thirty-five reached maturity, and only one did not germinate. Six plants were homozygous for the T-DNA, 21 were heterozygous, and eight were wild-type. No phenotype could be observed on plants homozygous for the T-DNA insertion, suggesting that, under the conditions used, the gene is dispensable, or alternatively, the T-DNA insertion has no effect on the transcript production or translation.
DISCUSSION
The recent identification of sterol mutants affecting pattern formation, cell polarity, embryogenesis, vascular patterning, and hormone signaling is shedding new light on the role of sterols in plant development (Clouse, 2002a, 2002b; Lindsey et al., 2003; Fischer et al., 2004). Apart from the well-described dwarf mutants resulting from reduced BR accumulation (Altmann, 1999), other mutants that cannot be rescued by BR addition show distinctive defects associated with the body organization of the embryo (Jang et al., 2000; Schrick et al., 2000, 2004). This suggests that sterols other than the BRs could also be active signaling molecules in plant development. Using a differential display approach to identify genes regulated by fertilization, we have isolated a cytochrome P450 gene of the CYP51 family, thereafter named CYP51G1-Sc. The sterol C14-demethylase (CYP51) is the most widely distributed cytochrome P450 gene family found in all biological kingdoms, and it is considered the most ancient member of the cytochrome P450 superfamily. In the sterol biosynthesis pathway, the obtusifoliol 14α-demethylase step is located immediately upstream of the C14-reductase, characterized through the fk/hyd2 mutants (Jang et al., 2000; Schrick et al., 2000), two steps upstream of the C-8,7 isomerase, characterized through the hyd1 mutant (Topping et al., 1997; Souter et al., 2002), and two steps downstream of the C24-methyltransferase characterized through the smt1/cph/orc mutants (Diener et al., 2000; Schrick et al., 2002). Loss-of-function mutations in all these genes lead to embryonic and seed defects that cannot be rescued by exogenous application of BR, unlike loss-of-function mutations downstream of the 24-methylenelophenol intermediate. Analysis of Arabidopsis insertional mutant plants for the ortholog of the S. chacoense CYP51G1-Sc, CYP51G1-At (82% amino acid sequence identity, 91% similarity), showed that homozygous insertional mutants were lethal and that seedlings, when they were obtained, died very early at the cotyledon stage (data not shown). Furthermore, the mutant could not be rescued by exogenous application of BR, as determined for the above-mentioned mutants. The analysis of this mutant line (SALK_067630) harboring an insertion at the n terminus of the coding region thus confirms previous results describing the embryo defective EMB 1738 mutant from an independent Syngenta line (Tzafrir et al., 2004). In this mutant line, the terminal phenotype was described as the cotyledon stage, while most embryos (64%) were arrested at the early transition to heart stage, and a high proportion of aberrant embryos were also detected (for details about this mutant, see http://www.seedgenes.org). This also clearly shows that the second gene classified as a CYP51 family member in the Arabidopsis genome cannot complement the mutant CYP51G1-At gene and that this second CYP51 enzyme has most probably evolved a distinct substrate specificity. A structural analysis of the various CYP51 isoforms also strengthens this view. Using various topology prediction tools for transmembrane spanning regions, two transmembrane helices for CYP51G1-Sc and CYP51G1-At were predicted. For both enzymes, a small stretch of amino acids resided in the cytoplasm between both transmembrane helices, which spanned 15 to 23 residues depending of the prediction tool used. The main core of these proteins remained in the endoplasmic reticulum environment where sterol metabolism occurs (Benveniste, 1986). As for Arabidopsis CYP51G2-At, all prediction tools revealed only one transmembrane-spanning region (data not shown). Furthermore, plants homozygous for a T-DNA insertion in the 3′ UTR of this second CYP51 gene showed no phenotype, suggesting that, under the conditions used, the gene is dispensable. Alternatively, the location of the T-DNA insertion might have no effect on transcript production or translation. Modulation of the C14-demethylase activity has also been attempted through antisense expression constructs in Arabidopsis (Kushiro et al., 2001) or a virus-induced gene silencing approach in Nicotiana benthamiana (Burger et al., 2003). In both cases, down-regulation of the obtusifoliol 14α-demethylase transcript level was incomplete, and the plants could survive, although the altered balance of sterols produced had an effect on the plant's growth and development. In both experiments, the plants had a severely reduced stature and a high obtusifoliol content. The antisense Arabidopsis plant also displayed a longer life span. Finally, studies performed in plants with a series of specific obtusifoliol-14DM inhibitors indicated a good correlation between their inhibition constants in vitro and their herbicidal activities in vivo (Rahier and Taton, 1997). All these results suggest that an altered sterol profile in plants affected at the C14-demethylation step is incompatible with normal growth requirements.
To our knowledge, until this study no expression data was available for plant C14-demethylases. Here, we have shown through RNA expression analyses that the CYP51G1-Sc gene is weakly expressed or undetectable in mature tissues, including vegetative and reproductive tissues (Fig. 2). RNA levels markedly increase in young developing tissues, like maturing styles (Fig. 5A), where an asymmetrical distribution could be observed with a preferential accumulation in the lower style region prior to anthesis (Fig. 5B). A kinematic analysis of gynoecial growth had previously showed that the basal style region was the predominant site of growth (Crone and Lord, 1991), confirming earlier results showing that cell divisions were only observed at the base of the style in early maturing petunia (Petunia hybrida) styles (Linskens, 1974). The CYP51G1-Sc gene is also expressed at significant levels in meristematic tissues as determined by in situ RNA analyses (Fig. 4E), where mRNAs strongly accumulate in the shoot apical meristems, in young primordia, as well as in the young leaflets that surround the apex. Although basal levels of CYP51G1-Sc mRNAs are generally quite low, a transient 6-fold increase in CYP51G1-Sc mRNA levels can be observed in ovaries immediately following fertilization (Fig. 2, C and D). A similar coordinate increase was also observed for the FK gene, corresponding to the enzymatic step immediately downstream of the CYP51G1-Sc enzyme (Fig. 2D). FK has been previously shown to be expressed in embryos and in actively growing cells (Jang et al., 2000; Schrick et al., 2000). In the ovary, this fertilization-induced mRNA accumulation can be seen by in situ hybridization to be diffusely localized in both the pericarp and placenta tissues (Fig. 4A) and more intensely in a single thin-walled parenchymatous cell layer immediately beneath the outermost cell layer that forms the placenta's epidermis. This layer, which does not express the CYP51G1-Sc mRNA, is continuous with the integument's epidermis, where no CYP51G1-Sc mRNAs could be detected, indicating a clear difference and specialization of this layer in both the placenta epidermis and in the ovule's integument. Since the obtusifoliol 14α-demethylase mutant is embryo defective, this suggests that sterols produced in specific cell layers might be translocated to the ovule and embryo during early fertilization steps. The increase in CYP51G1-Sc mRNA levels in the ovary can be best described as biphasic, since pollination alone also induced a transient increase in CYP51G1-Sc mRNA levels. This increase preceded the arrival of the pollen tube in the ovary and is both observed following compatible and incompatible pollinations, where pollen tubes are arrested in the first two-thirds of the style (Fig. 3B). This suggests that a long-distance signal, produced by the pollen tube itself or in the transmitting tract tissue of the style during pollen-pistil interaction, is perceived at a distance in the ovary. Among the treatments performed, only wounding of the style and methyl jasmonate application could fully mimic the effect of pollen tube growth on the accumulation of CYP51G1-Sc mRNAs at a distance in the ovary (Fig. 3, C and D). This suggests that cellular deterioration and death caused by pollination (either compatible or incompatible) or by wounding is the initial trigger that induces CYP51G1-Sc mRNA accumulation in the ovary and that this response is probably mediated by jasmonates, as for long-distance wound responses (Stratmann, 2003). Sustained levels of CYP51G1-Sc mRNAs for a longer period can then only be achieved if fertilization takes place (Figs. 2D and 3B).
A totally novel aspect of plant CYP51 substrate specificity was revealed by our biochemical characterization of the enzyme expressed in yeast (Fig. 6). All the previously characterized plant CYP51 enzymes from maize (Taton and Rahier, 1991), wheat (Cabello-Hurtado et al., 1997, 1999), sorghum (Kahn et al., 1996; Bak et al., 1997; Lamb et al., 1998), and Arabidopsis (Kushiro et al., 2001) were shown to have strict substrate specificity toward obtusifoliol. For the three monocot plants, this was determined enzymatically. In Arabidopsis, this was shown by the absence of complementation of the yeast erg11 mutant with the complete CYP51G1-At clone, while a chimera between the n terminus of the CYP51 yeast enzyme and the plant enzyme could rescue the erg11 mutant. Complementation of the yeast erg11 mutant with the complete CYP51G1-Sc clone leads to a significant increase in cell viability on media not supplemented with ergosterol, indicating that the S. chacoense CYP51G1 enzyme could substitute for the yeast endogenous enzyme and use lanosterol as a substrate. Biochemical characterization of the S. chacoense CYP51G1-Sc protein expressed in yeast confirmed that the enzyme could indeed use lanosterol as a substrate (as well as eburicol), albeit with lower efficiency than the plant endogenous substrate, obtusifoliol (Fig. 6). The lack of activity of plant CYP51 enzymes toward fungi and mammal substrates had been attributed to a structural feature common to all these nonplant sterol substrates: the presence of a 4β-methyl group in lanosterol and eburicol, precursors of C14-desmethylsterols (Taton and Rahier, 1991). Since only few plant CYP51 enzymes have been characterized at the biochemical level, sequence comparison and enzymatic characterization of other plant CYP51 combined with the expression of domain swap chimeras and site-directed mutagenesis should reveal the protein region(s) involved in substrate specificity (Lepesheva and Waterman, 2004).
Although a BR-independent role for sterols in plant development has only been suggested a few years ago, mounting evidence involving numerous mutants affected in early steps of sterol biosynthesis and showing severe defects in embryogenesis and cell patterning are now clearly highlighting new specific roles for sterols in plant development (Lindsey et al., 2003). Modification of the sterol balance as demonstrated for several mutants through chemical analysis of the sterol fraction could explain some of the observed phenotype. Alternatively, presence of biosynthetic intermediates that could be either toxic or that could act as signaling molecules is also considered. Exogenous treatments with natural plant sterols like sitosterol and stigmasterol or atypical sterols that accumulated in the fk mutant have been shown to be able to modulate the expression of genes involved in cell expansion or cell division (He et al., 2003). To address this issue, we have used purified obtusifoliol to determine its ability to modulate gene expression and to act as a signaling molecule. Since the CYP51G1-Sc gene was itself found to be highly active in developing tissues like maturing styles, shoot apices, and meristems (Figs. 4 and 5), we tested the ability of purified obtusifoliol to modulate the expression of its demethylating enzyme in a mature tissue where only a normally weak expression was detected. Application of an obtusifoliol emulsion on mature leaves lead to a local 2-fold increase of CYP51G1-Sc mRNA levels (Fig. 7). This response was specific and could not be mimicked by application of lanosterol, a structurally highly similar sterol. Thus, similarly to mammals, where genes involved in cholesterol homeostasis (including CYP51) are regulated by cholesterol or its metabolites (Stromstedt et al., 1996), the plant CYP51G1-Sc gene was also found to be regulated by sterols. The 2-fold increase observed following obtusifoliol application was not only highly reproducible but was also detected in nontreated systemic leaves, suggesting the long-distance transport or production of a signaling molecule triggering the expression of the CYP51G1-Sc mRNA. To further test if obtusifoliol could itself be a mobile signaling molecule, we used 14C-labeled obtusifoliol and monitored its distribution throughout the plant after a compound leaf was sprayed (Fig. 8). Not only was labeling detected in the petiole as well as in portions of some leaflets, both positioned immediately above or below the sprayed leaf, but a chemical analysis (Fig. 9) revealed that the radioactive compound detected at a distance from the sprayed leaf was almost exclusively composed of obtusifoliol without any other radioactive sterol. Furthermore, the concentrations of radiolabeled obtusifoliol found in the petiole of systemic leaf 6, where a 2-fold increase in CYP51G1-Sc mRNA levels was observed, was consistent with the physiological levels found in plants, strongly suggesting that obtusifoliol can act as a mobile signaling molecule involved in various aspects of plant growth and development, including pollination and fertilization.
The adaxial/abaxial polarity of leaves is established through the spatially restricted (adaxial) expression of the PHABULOSA (PHB)/PHAVOLUTA (PHV) genes in the leaf primordium (Bowman et al., 2002). PHB and PHV encode type III HD-ZIP transcription factors that contain a START domain, which in animals binds sterols and modulates the activity of the DNA binding domain. Proper adaxial/abaxial development of the primordium is also known to depend on signal(s) emanating from the meristem to the leaf. The presence of a START domain in the PHB and PHV proteins has prompted the authors to suggest that a lipid/sterol molecule would be involved in the acquisition of adaxial identity through interaction with these HD-ZIP transcription factors (McConnell et al., 2001). Interestingly, our in situ analysis has revealed that the CYP51G1-Sc gene is also preferentially expressed on the adaxial side of developing leaves (Fig. 4, E and F). This, combined with the ability of obtusifoliol to move away from its site of application (Fig. 8) and to induce the transcription of the C14-demethylase (Fig. 7), creating a localized positive feedback loop, suggest that this sterol intermediate could be involved in such processes.
Interestingly, mammalian sterols homologous to the sterols produced by the plant C14-demethylase and C14-reductase (FK) enzymes have been shown to play important roles in reproduction. In mammals, meiosis of follicule oocytes is maintained in the prophase of the first meiotic division, and oocytes do not resume meiosis spontaneously during oocyte growth and follicule development (Byskov et al., 2002; Tsafriri et al., 2005). In vitro, this can be overcome by the addition of meiosis activating sterols, and these were found to be 4,4-dimethyl-5α-cholesta-8,14,24-trien-3β-ol, also named FF-MAS (for follicular fluid meiosis activating sterol), and 4,4-dimethyl-5α-cholesta-8,24-dien-3β-ol, also named T-MAS (for testis meiosis activating sterol; Byskov et al., 1995). These sterols correspond to the transformation product of lanosterol by the CYP51 C14-demethylase enzyme (FF-MAS) and by the subsequent enzymatic step, the C14-reductase (T-MAS), suggesting that these sterol molecules might have been retained by plants and animals as signaling molecules. Furthermore, lanosterol has also been found to be a candidate endogenous ligand for the farnesoid X receptor β, a new mammalian nuclear receptor (Otte et al., 2003), strengthening the role of these sterols in signal transduction. Because lipids are hydrophobic, lipids involved in long-distance signaling have to be transported by soluble macromolecules like lipid-binding proteins or lipid-transfer proteins. Since these are quite numerous in the plant genome and have been shown to be involved in defense-response signaling and somatic embryogenesis (Blein et al., 2002), it is quite conceivable that sterol intermediates, like obtusifoliol, could also act as a long-distance signaling molecule in plants.
MATERIALS AND METHODS
Plant Material and Treatments
The diploid and self-incompatible wild potato Solanum chacoense Bitt. (2n = 2× = 24) was grown in the greenhouse with 14 to 16 h of light per day. The genotypes used were V22 (S allele S11 and S13) as pollen donor and G4 (S allele S12 and S14) as female progenitor. Phytohormone treatments were done as previously described (Lantin et al., 1999; O'Brien et al., 2002a), and wounding of the style was done by gentle tweezing of the stigma and upper style with forceps. Each hormone application was boosted 24 h after the initial treatment, and flower tissues were harvested 48 h after the initial treatment. For obtusifoliol treatment, an emulsion of the compound was performed in Tween 80. The desiccated obtusifoliol (see below for obtusifoliol production) was dissolved in chloroform (1–5 mg/mL final concentration) and mixed with a stock solution (50 mg/mL) of Tween 80 in absolute ethanol. After vigorous mixing, the mixture was desiccated in a speed vacuum apparatus. For leaf treatment, the mixture was dissolved in sterile water to a final concentration of 1 mg/mL (2.34 mm), and 500 μL was sprayed on one compound leaf. In order to prevent the mist from falling on other leaves, a large Whatman paper cone was placed over the treated leaf. The obtusifoliol in vivo mobile assay was performed in duplicate with C14-radiolabeled obtusifoliol (see below) and gave identical results. The GC-MS chemical analysis of the radiolabeled compound that was isolated from the sprayed leaf and from the distant leaves was performed on one of these duplicates that showed the best preservation of the pressed and dried leaves.
Isolation of the U40 Fragment by Differential Display and the CYP51G1-Sc cDNA
We followed the differential display procedure described in the RNAimage protocol (GenHunter), with some modifications as described previously (O'Brien et al., 2002a). The PCR fragment obtained was cloned in a homemade T-vector derived from a pBS II plasmid vector cut with EcoRV (Marchuk et al., 1991). To validate the differential display autoradiography, the radiolabeled PCR fragment was used to probe an RNA gel blot made with the same mRNAs used for the differential display RT-PCR procedure. After confirmation, the probe was used to screen a cDNA library derived from 48 h postpollinated pistil tissue mRNAs (Lantin et al., 1999) to obtain a full-length cDNA clone. For the S. chacoense C14-reductase homolog (FK), a PCR fragment was obtained either from reverse-transcribed ovule mRNAs or leaf genomic DNA with primer sequences selected from tomato (Lycopersicon esculentum) expressed sequence tags (AI484506, AI485167, and AW931153) similar to the Arabidopsis thaliana FK sequence. Forward primer was 5′-GTCTTTATACCGTTCACC-3′, and reverse primer was 5′-CTTCTCTGCACATCTAGC-3′.
Measure of C-14α-Demethylase Activity and GC-MS Analysis
For activity assays, the complete CYP51G1-Sc open reading frame plus vector flanking sequences from the bacterial pBluescript SK− vector was digested with the EcoRI and BamHI restriction enzymes and cloned downstream of the GAL10-CYC1 promoter into the yeast shuttle vector pYeDP60 (Pompon et al., 1996; Cabello-Hurtado et al., 1999). Expression of the CYP51G1-Sc protein was performed in the yeast strain WAT11 (Pompon et al., 1996) and tested for enzymatic activity (demethylation assay) on various substrates. Yeast microsomal preparations were prepared as described previously (Darnet and Rahier, 2003). Synthesis of obtusifoliol and measurements of sterol 14α-demethylase activity of obtusifoliol, lanosterol, and eburicol were performed as described previously (Taton and Rahier, 1991).
Yeast Complementation
Diploid Saccharomyces cerevisiae ERG 11/erg11 mutant cells, designation YHR007C BY4743 (MATa/MATα his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 lys2Δ0/+ met15Δ0/+ ura3Δ0/ura3Δ0 ΔERG11), were obtained from the Saccharomyces Genome Deletion Project through Research Genetics. The corresponding ATCC number for the strain is 4026604. For liquid cultures, the strain was maintained under selection with 200 mg/L of geneticin. Transformation followed the TRAFO procedure (Gietz and Woods, 2002). Yeast strain recombinants for the pYeDP60 vector were selected with the ADE2 selection on a media lacking adenine (Pompon et al., 1996). Sporulation of yeast cells was induced in sporulation media (http://www.umanitoba.ca/faculties/medicine/biochem/gietz/Trafo.html), and spores were grown on solid media lacking adenine and supplemented with 20 mg/mL ergosterol (Sigma-Adrich; E-6510). Colonies were replicated on solid media lacking adenine and scored for viability. Values and standard deviation were derived from five independent experiments, each including the viability assessment of 36 haploid yeast colonies grown on media lacking ergosterol.
DNA and RNA Gel-Blot Analysis
Total RNA was isolated as described previously (Jones et al., 1985) or with the Plant RNeasy RNA extraction kit from Qiagen. RNA concentration was determined by measuring its absorbance at 260 nm. Concentration and RNA quality were verified on agarose gel following ethidium bromide staining. For each tissue tested, 10 μg of RNA were separated on a formaldehyde/MOPS gel (Sambrook et al., 1989). RNA was then transferred onto Hybond N+ membranes (GE Healthcare) and cross-linked (120 mJ/cm2) with a Hoefer UVC 500 UV cross-linker. To confirm equal loading between RNA samples, a 1-kb fragment of S. chacoense 18S RNA was PCR amplified and used as a control probe. Prehybridization was performed at 45°C for 3 h in 50% formamide solution (50% deionized formamide, 6× SCC, 5× Denhardt solution, 0.5% SDS, and 200 μg of denatured salmon sperm DNA). Hybridization of the membranes was performed overnight at 45°C in 50% formamide solution. Genomic DNA isolation was performed by a modified cetyl-trimethyl-ammonium bromide extraction method (Reiter et al., 1992) or with the Plant DNeasy kit from Qiagen. Complete digestion of DNA (10 μg) was made overnight with 5 units/μg of either BamHI, EcoRI, EcoRV, or HindIII as recommended by the supplier (New England Biolabs). DNA gel-blot analysis was performed as previously described (Sambrook et al., 1989). DNA was capillary transferred to Hybond N+ membranes prior to cross-linking. Prehybridization (3 h) and hybridization (overnight) were performed as described for the RNA gel-blot analyses except that the temperature was set at 42°C. Probes for RNA and DNA gel-blot analyses were synthesized by random labeling using the Strip-EZ DNA labeling kit (Ambion) in the presence of α-32P-dATP (ICN Biochemicals). Following hybridization, membranes were washed 30 min at 25°C and 30 min at 35°C in 2× SSC/0.1% SDS, 30 min at 45°C and 30 min at 55°C in 1× SSC/0.1% SDS, and a final stringent wash for 10 min at 55°C in 0.1× SSC/0.1% SDS. Prior to 18S cRNA probe hybridization, RNA gel blots were stripped as recommended by the manufacturer (Ambion), and post-hybridization washes were done twice at 60°C for 30 min in 0.1× SSC/0.1% SDS. Membranes were exposed either at room temperature on a europium screen and scanned on a Typhoon 9200 phosphor imager (Amersham Biosciences) or exposed at −86°C on Kodak Biomax MR film (Interscience). For quantitative measurements, densitometric scans were performed and normalized against their individual 18S rRNA hybridization level using the ImageQuant software on a Typhoon 9200 phosphor imager (GE Healthcare).
In Situ Hybridization
Ovaries (cut in halves) were fixed in FAA (60% ethanol, 5% acetic acid, and 5% formaldehyde) at 4°C overnight. After dehydration with tert-butanol and embedding in paraffin, samples were cut into 10-μm sections and mounted on slides coated with AES (3-aminopropyltriethoxy-silane; Sigma-Aldrich). Tissue sections were treated and hybridized as described previously (Lantin et al., 1999).
Sequence Analysis
Sequencing was performed on Applied Biosystems ABI Prism 310 or 3100 automated sequencer. Sequence alignments were performed with the ClustalW module in MacVector 7.2.3 (Accelrys). Possible signal peptide or membrane anchoring domain were predicted from the CBS prediction servers (http://www.cbs.dtu.dk/services/). Transmembrane spanning regions were predicted from various prediction servers as follows: http://sosui.proteome.bio.tuat.ac.jp/sosui_submit.html, http://www.enzim.hu/hmmtop, http://www.ch.embnet.org/software/TMPRED_form.html, and http://www.cbs.dtu.dk/services/.
Biosynthesis of [14C]-Obtusifoliol
The suspension cultures (0.8 L) of bramble cells (Rubus fruticosus) used have been described previously (Schmitt et al., 1982). At the beginning of the exponential phase of growth, the cells were incubated in the presence of a 14-demethylase inhibitor, CGA214372 [methyl 1-(2,2-dimethyllindan-1-yl)-imidazole-5 carboxylate], at a 5-mg/L final concentration in the medium and [2-14C]-acetate (600 μCi, 5.1 Ci/mol) for 7 d. The cells were harvested by filtration and lyophilized. The unsaponifiable matter was isolated and separated on TLC using CH2Cl2 (two runs) allowing the separation of 4α-methylsterols from other sterols. After acetylation, this fraction was analyzed on analytical AgNO3-impregnated SiO2-TLC, using CH2Cl2 as the developing solvent, allowing isolation of pure [14C]-obtusifoliyl acetate (1.6 mg): gas chromatography purity > 98% (DB5 column, J&W Scientific), mass spectrometry, mass-to-charge ratio (relative intensity) M+ = 468(35), 453(100), 425(8), 393(40), 341(7), 309(16), 227(22), and 215(18). Saponification yielded pure [14C]-obtusifoliol (1.4 mg): specific radioactivity (0.8 Ci/mol), gas chromatography purity > 98% (relative retention time = 1.128, DB1 column; relative retention time = 1.168, DB5 column), mass spectrometry, mass-to-charge ratio (relative intensity) M+ = 426(34), 411(100), 393(14), 342(3), 327(14), and 227(11).
Chemical Analysis of [14C]-Labeled Sterols from Potato Leaves
The dried leaves were extracted with a mixture of CH2Cl2/MeOH 2:1 for 2 h under reflux. After evaporation of the solvent, the residue was separated by TLC with CH2Cl2 as the solvent, allowing the radioactivity present in the total 4,4-dimethysterol, 4α-methylsterol, 4-desmethylsterol, and steryl-ester fractions to be measured. In order to be able to further identify by GLC-MS the radioactive components present in the 4α-methylsterol fraction, protecting cold carrier sterols were not added to this fraction. It was acetylated and further analyzed on analytical AgNO3-impregnated SiO2-TLC, using CH2Cl2 as the developing solvent, allowing the radioactivity present in fractions migrating as authentic standards of 4α-methylsterols to be determined. The single radioactive 4α-methylsterol fraction that showed a retardation factor identical to that of a standard of obtusifoliyl-acetate was eluted from silica, and the sterol(s) present in this fraction was unequivocally identified by coupled capillary GC-MS analysis (i.e. by coincidental retention time and identical electron impact spectrum as authentic standard; Rahier and Benveniste, 1989). The radioactive steryl-ester fractions were saponified under standard conditions, and the obtained free sterols were analyzed as described above.
Nomenclature
The official nomenclature of the sterols mentioned is as follows. Obtusifoliol, 4α,14α-dimethyl-ergosta-8,24(241)-dien-3β-ol; lanosterol, 4,4,14α-trimethyl-cholesta-8,24-dien-3β-ol; 4,4-dimethyl-zymosterol, 4,4-dimethyl-cholesta-8,24-dien-3β-ol; cycloartenol, 4,4,14α-trimethyl-9β,19-cyclo-cholest-24-en-3β-ol; 24-methylene-cycloartanol, 4,4,14α-trimethyl-9β,19-cyclo-ergost-24(241)-en-3β-ol; eburicol, 4,4,14α-trimethyl-ergosta-8,24(241)-dien-3β-ol.
Sequence data for the CYP51 sequence analyses can be found in the GenBank/EMBL data libraries under the following accession numbers: S. chacoense, AY552551; Nicotiana tabacum, AAL54888; Oryza sativa, AK060487; Sorghum bicolor, P93846; Triticum aestivum, CAA70475; Mus musculus, AAF74562; Homo sapiens, EAL24154; Mycobacterium tuberculosis, CAB02394; Trypanosoma brucei, AAK97386; Schizosaccharomyces pombe, Q09736; S. cerevisiae, AAA34546.
Acknowledgments
We would like to thank Danièle Werck-Reichhart for the pYeDP60 yeast shuttle vector. Special thanks to Annie Hoeft for technical assistance and to Gabriel Téodorescu for plant care and maintenance. M.O. and S.-C.C. are the recipients of PhD fellowships from the Natural Sciences and Engineering Research Council of Canada and from Le Fonds Québécois de la Recherche sur la Nature et les Technologies (Quebec). D.P.M. holds a Canada Research Chair in Functional Genomics and Plant Signal Transduction.
This work was supported by the Natural Sciences and Engineering Research Council of Canada and by the Canada Research Chair program.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.066639.
References
- Altmann T (1999) Molecular physiology of brassinosteroids revealed by the analysis of mutants. Planta 208: 1–11 [DOI] [PubMed] [Google Scholar]
- Aoyama Y, Yoshida Y (1992) The 4 beta-methyl group of substrate does not affect the activity of lanosterol 14 alpha-demethylase (P-450(14)DM) of yeast: difference between the substrate recognition by yeast and plant sterol 14 alpha-demethylases. Biochem Biophys Res Commun 183: 1266–1272 [DOI] [PubMed] [Google Scholar]
- Baima S, Nobili F, Sessa G, Lucchetti S, Ruberti I, Morelli G (1995) The expression of the Athb-8 homeobox gene is restricted to provascular cells in Arabidopsis thaliana. Development 121: 4171–4182 [DOI] [PubMed] [Google Scholar]
- Bak S, Kahn RA, Olsen CE, Halkier BA (1997) Cloning and expression in Escherichia coli of the obtusifoliol 14 alpha-demethylase of Sorghum bicolor (L.) Moench, a cytochrome P450 orthologous to the sterol 14 alpha-demethylases (CYP51) from fungi and mammals. Plant J 11: 191–201 [DOI] [PubMed] [Google Scholar]
- Balk PA, de Boer AD (1999) Rapid stalk elongation in tulip (Tulipa gesneriana L. cv. Apeldoorn) and the combined action of cold-induced invertase and the water-channel protein gammaTIP. Planta 209: 346–354 [DOI] [PubMed] [Google Scholar]
- Benveniste P (1986) Sterol biosynthesis. Annu Rev Plant Physiol 37: 275–308 [Google Scholar]
- Benveniste P (2004) Biosynthesis and accumulation of sterols. Annu Rev Plant Biol 55: 429–457 [DOI] [PubMed] [Google Scholar]
- Blein JP, Coutos-Thevenot P, Marion D, Ponchet M (2002) From elicitins to lipid-transfer proteins: a new insight in cell signalling involved in plant defence mechanisms. Trends Plant Sci 7: 293–296 [DOI] [PubMed] [Google Scholar]
- Bowman JL, Eshed Y, Baum SF (2002) Establishment of polarity in angiosperm lateral organs. Trends Genet 18: 134–141 [DOI] [PubMed] [Google Scholar]
- Burger C, Rondet S, Benveniste P, Schaller H (2003) Virus-induced silencing of sterol biosynthetic genes: identification of a Nicotiana tabacum L. obtusifoliol-14alpha-demethylase (CYP51) by genetic manipulation of the sterol biosynthetic pathway in Nicotiana benthamiana L. J Exp Bot 54: 1675–1683 [DOI] [PubMed] [Google Scholar]
- Byskov AG, Andersen CY, Leonardsen L (2002) Role of meiosis activating sterols, MAS, in induced oocyte maturation. Mol Cell Endocrinol 187: 189–196 [DOI] [PubMed] [Google Scholar]
- Byskov AG, Andersen CY, Nordholm L, Thogersen H, Xia G, Wassmann O, Andersen JV, Guddal E, Roed T (1995) Chemical structure of sterols that activate oocyte meiosis. Nature 374: 559–562 [DOI] [PubMed] [Google Scholar]
- Cabello-Hurtado F, Taton M, Forthoffer N, Kahn R, Bak S, Rahier A, Werck-Reichhart D (1999) Optimized expression and catalytic properties of a wheat obtusifoliol 14alpha-demethylase (CYP51) expressed in yeast. Complementation of erg11Delta yeast mutants by plant CYP51. Eur J Biochem 262: 435–446 [DOI] [PubMed] [Google Scholar]
- Cabello-Hurtado F, Zimmerlin A, Rahier A, Taton M, DeRose R, Nedelkina S, Batard Y, Durst F, Pallett KE, Werck-Reichhart D (1997) Cloning and functional expression in yeast of a cDNA coding for an obtusifoliol 14alpha-demethylase (CYP51) in wheat. Biochem Biophys Res Commun 230: 381–385 [DOI] [PubMed] [Google Scholar]
- Carland FM, Fujioka S, Takatsuto S, Yoshida S, Nelson T (2002) The identification of CVP1 reveals a role for sterols in vascular patterning. Plant Cell 14: 2045–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumont F, Barrieu F, Herman EM, Chrispeels MJ (1998) Characterization of a maize tonoplast aquaporin expressed in zones of cell division and elongation. Plant Physiol 117: 1143–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung AY (1996) The pollen tube growth pathway: its molecular and biochemical contributions and responses to pollination. Sex Plant Reprod 9: 330–336 [Google Scholar]
- Clouse SD (2000) Plant development: A role for sterols in embryogenesis. Curr Biol 10: R601–R604 [DOI] [PubMed] [Google Scholar]
- Clouse SD (2002. a) Arabidopsis mutants reveal multiple roles for sterols in plant development. Plant Cell 14: 1995–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse SD (2002. b) Brassinosteroids. In CR Somerville, EM Meyerowitz, eds, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, doi/10.1199/tab.0009, http://www.aspb.org/publications/arabidopsis/
- Clouse SD, Sasse JM (1998) BRASSINOSTEROIDS: essential regulators of plant growth and development. Annu Rev Plant Physiol Plant Mol Biol 49: 427–451 [DOI] [PubMed] [Google Scholar]
- Crone W, Lord EM (1991) A kinematic analysis of gynoecial growth in Lilium longiflorum: Surface growth patterns in all floral organs are triphasic. Dev Biol 143: 408–417 [DOI] [PubMed] [Google Scholar]
- Darnet S, Rahier A (2003) Enzymological properties of sterol-C4-methyl-oxidase of yeast sterol biosynthesis. Biochim Biophys Acta 1633: 106–117 [DOI] [PubMed] [Google Scholar]
- Diener AC, Li H, Zhou W, Whoriskey WJ, Nes WD, Fink GR (2000) Sterol methyltransferase 1 controls the level of cholesterol in plants. Plant Cell 12: 853–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards PA, Ericsson J (1999) Sterols and isoprenoids: signaling molecules derived from the cholesterol biosynthetic pathway. Annu Rev Biochem 68: 157–185 [DOI] [PubMed] [Google Scholar]
- Farese RV Jr, Herz J (1998) Cholesterol metabolism and embryogenesis. Trends Genet 14: 115–120 [DOI] [PubMed] [Google Scholar]
- Fischer U, Men S, Grebe M (2004) Lipid function in plant cell polarity. Curr Opin Plant Biol 7: 670–676 [DOI] [PubMed] [Google Scholar]
- Gietz RD, Woods RA (2002) Screening for protein-protein interactions in the yeast two-hybrid system. Methods Mol Biol 185: 471–486 [DOI] [PubMed] [Google Scholar]
- Guo DA, Venkatramesh M, Nes WD (1995) Developmental regulation of sterol biosynthesis in Zea mays. Lipids 30: 203–219 [DOI] [PubMed] [Google Scholar]
- Hartmann M-A (1998) Plant sterols and the membrane environment. Trends Plant Sci 3: 170–175 [Google Scholar]
- Hartmann M-A, Benveniste P (1987) Plant membrane sterols: isolation, identification and biosynthesis. Methods Enzymol 148: 632–650 [Google Scholar]
- He JX, Fujioka S, Li TC, Kang SG, Seto H, Takatsuto S, Yoshida S, Jang JC (2003) Sterols regulate development and gene expression in Arabidopsis. Plant Physiol 131: 1258–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmberg N, Harker M, Gibbard CL, Wallace AD, Clayton JC, Rawlins S, Hellyer A, Safford R (2002) Sterol C-24 methyltransferase type 1 controls the flux of carbon into sterol biosynthesis in tobacco seed. Plant Physiol 130: 303–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang JC, Fujioka S, Tasaka M, Seto H, Takatsuto S, Ishii A, Aida M, Yoshida S, Sheen J (2000) A critical role of sterols in embryonic patterning and meristem programming revealed by the fackel mutants of Arabidopsis thaliana. Genes Dev 14: 1485–1497 [PMC free article] [PubMed] [Google Scholar]
- Jones JDG, Dunsmuir P, Bedbrook J (1985) High level expression of introduced chimeric genes in regenerated transformed plants. EMBO J 4: 2411–2418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn RA, Bak S, Olsen CE, Svendsen I, Moller BL (1996) Isolation and reconstitution of the heme-thiolate protein obtusifoliol 14alpha-demethylase from Sorghum bicolor (L.) Moench. J Biol Chem 271: 32944–32950 [DOI] [PubMed] [Google Scholar]
- Kalb VF, Woods CW, Turi TG, Dey CR, Sutter TR, Loper JC (1987) Primary structure of the P450 lanosterol demethylase gene from Saccharomyces cerevisiae. DNA 6: 529–537 [DOI] [PubMed] [Google Scholar]
- Kushiro M, Nakano T, Sato K, Yamagishi K, Asami T, Nakano A, Takatsuto S, Fujioka S, Ebizuka Y, Yoshida S (2001) Obtusifoliol 14alpha-demethylase (CYP51) antisense Arabidopsis shows slow growth and long life. Biochem Biophys Res Commun 285: 98–104 [DOI] [PubMed] [Google Scholar]
- Lamb DC, Kelly DE, Kelly SL (1998) Molecular diversity of sterol 14alpha-demethylase substrates in plants, fungi and humans. FEBS Lett 425: 263–265 [DOI] [PubMed] [Google Scholar]
- Lantin S, O'Brien M, Matton DP (1999) Pollination, wounding and jasmonate treatments induce the expression of a developmentally regulated pistil dioxygenase at a distance, in the ovary, in the wild potato Solanum chacoense Bitt. Plant Mol Biol 41: 371–386 [DOI] [PubMed] [Google Scholar]
- Lepesheva GI, Waterman MR (2004) CYP51–the omnipotent P450. Mol Cell Endocrinol 215: 165–170 [DOI] [PubMed] [Google Scholar]
- Lindsey K, Pullen ML, Topping JF (2003) Importance of plant sterols in pattern formation and hormone signalling. Trends Plant Sci 8: 521–525 [DOI] [PubMed] [Google Scholar]
- Linskens HF (1974) Some observations on the growth of the style. Incompatibility Newsletter 4: 4–15 [Google Scholar]
- Ludevid D, Höfte H, Himelblau E, Chrispeels MJ (1992) The expression pattern of the tonoplast intrinsic protein γ-TIP in Arabidopsis thaliana is correlated with cell enlargement. Plant Physiol 100: 1633–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchuk D, Drumm M, Saulino A, Collins FS (1991) Construction of T-vectors, a rapid and general system for direct cloning of unmodified PCR products. Nucleic Acids Res 19: 1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matton DP, Maes O, Laublin G, Xike Q, Bertrand C, Morse D, Cappadocia M (1997) Hypervariable domains of self-incompatibility RNases mediate allele-specific pollen recognition. Plant Cell 9: 1757–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell JR, Emery J, Eshed Y, Bao N, Bowman J, Barton MK (2001) Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411: 709–713 [DOI] [PubMed] [Google Scholar]
- Mongrand S, Morel J, Laroche J, Claverol S, Carde JP, Hartmann MA, Bonneu M, Simon-Plas F, Lessire R, Bessoule JJ (2004) Lipid rafts in higher plant cells: purification and characterization of Triton X-100-insoluble microdomains from tobacco plasma membrane. J Biol Chem 279: 36277–36286 [DOI] [PubMed] [Google Scholar]
- Nelson DR, Schuler MA, Paquette SM, Werck-Reichhart D, Bak S (2004) Comparative genomics of rice and Arabidopsis. Analysis of 727 cytochrome P450 genes and pseudogenes from a monocot and a dicot. Plant Physiol 135: 756–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien M, Bertrand C, Matton DP (2002. a) Characterization of a fertilization-induced and developmentally regulated plasma-membrane aquaporin expressed in reproductive tissues, in the wild potato Solanum chacoense Bitt. Planta 215: 485–493 [DOI] [PubMed] [Google Scholar]
- O'Brien M, Kapfer C, Major G, Laurin M, Bertrand C, Kondo K, Kowyama Y, Matton DP (2002. b) Molecular analysis of the stylar-expressed Solanum chacoense small asparagine-rich protein family related to the HT modifier of gametophytic self-incompatibility in Nicotiana. Plant J 32: 985–996 [DOI] [PubMed] [Google Scholar]
- Otte K, Kranz H, Kober I, Thompson P, Hoefer M, Haubold B, Remmel B, Voss H, Kaiser C, Albers M, et al (2003) Identification of farnesoid X receptor beta as a novel mammalian nuclear receptor sensing lanosterol. Mol Cell Biol 23: 864–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pompon D, Louerat B, Bronine A, Urban P (1996) Yeast expression of animal and plant P450s in optimized redox environments. Methods Enzymol 272: 51–64 [DOI] [PubMed] [Google Scholar]
- Ponting CP, Aravind L (1999) START: a lipid-binding domain in StAR, HD-ZIP and signalling proteins. Trends Biochem Sci 24: 130–132 [DOI] [PubMed] [Google Scholar]
- Rahier A, Benveniste P (1989) Mass spectral identification of phytosterols. In WD Nes, E Parish, eds, Analysis of Sterols and Other Biologically Significant Steroids. Academic Press, San Diego, pp 223–250
- Rahier A, Taton M (1997) Fungicides as tools in studying postsqualene sterol synthesis in plants. Pestic Biochem Physiol 57: 1–27 [Google Scholar]
- Reiter RS, Young RM, Scolnik PA (1992) Genetic linkage of the Arabidopsis genome: methods for mapping with recombinant inbreds and Random Amplified Polymorphic DNAs (RAPDs). In C Koncz, N-H Chua, J Schell, eds, Methods in Arabidopsis Research. World Scientific Publishing, Singapore, pp 170–190
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Schaeffer A, Bronner R, Benveniste P, Schaller H (2001) The ratio of campesterol to sitosterol that modulates growth in Arabidopsis is controlled by STEROL METHYLTRANSFERASE 2;1. Plant J 25: 605–615 [DOI] [PubMed] [Google Scholar]
- Schaller H, Bouvier-Nave P, Benveniste P (1998) Overexpression of an Arabidopsis cDNA encoding a sterol-C24(1)-methyltransferase in tobacco modifies the ratio of 24-methyl cholesterol to sitosterol and is associated with growth reduction. Plant Physiol 118: 461–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt P, Benveniste P (1979) Effect of fenarimol on sterol biosynthesis in suspension cultures of bramble cells. Phytochemistry 18: 1659–1665 [Google Scholar]
- Schmitt P, Rahier A, Benveniste P (1982) Inhibition of sterol biosynthesis in suspension-cultures of bramble cells. Physiol Veg 20: 559–571 [Google Scholar]
- Schrick K, Fujioka S, Takatsuto S, Stierhof YD, Stransky H, Yoshida S, Jurgens G (2004) A link between sterol biosynthesis, the cell wall, and cellulose in Arabidopsis. Plant J 38: 227–243 [DOI] [PubMed] [Google Scholar]
- Schrick K, Mayer U, Horrichs A, Kuhnt C, Bellini C, Dangl J, Schmidt J, Jurgens G (2000) FACKEL is a sterol C-14 reductase required for organized cell division and expansion in Arabidopsis embryogenesis. Genes Dev 14: 1471–1484 [PMC free article] [PubMed] [Google Scholar]
- Schrick K, Mayer U, Martin G, Bellini C, Kuhnt C, Schmidt J, Jurgens G (2002) Interactions between sterol biosynthesis genes in embryonic development of Arabidopsis. Plant J 31: 61–73 [DOI] [PubMed] [Google Scholar]
- Schuler MA, Werck-Reichhart D (2003) Functional genomics of P450s. Annu Rev Plant Biol 54: 629–667 [DOI] [PubMed] [Google Scholar]
- Sessa G, Steindler C, Morelli G, Ruberti I (1998) The Arabidopsis Athb-8, -9 and -14 genes are members of a small gene family coding for highly related HD-ZIP proteins. Plant Mol Biol 38: 609–622 [DOI] [PubMed] [Google Scholar]
- Simons K, Toomre D (2000) Lipid rafts and signal transduction. Nat Rev Mol Cell Biol 1: 31–39 [DOI] [PubMed] [Google Scholar]
- Sitbon F, Jonsson L (2001) Sterol composition and growth of transgenic tobacco plants expressing type-1 and type-2 sterol methyltransferases. Planta 212: 568–572 [DOI] [PubMed] [Google Scholar]
- Souter M, Topping J, Pullen M, Friml J, Palme K, Hackett R, Grierson D, Lindsey K (2002) hydra mutants of Arabidopsis are defective in sterol profiles and auxin and ethylene signaling. Plant Cell 14: 1017–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratmann JW (2003) Long distance run in the wound response–Jasmonic acid is pulling ahead. Trends Plant Sci 8: 247–250 [DOI] [PubMed] [Google Scholar]
- Stromstedt M, Rozman D, Waterman MR (1996) The ubiquitously expressed human CYP51 encodes lanosterol 14 alpha-demethylase, a cytochrome P450 whose expression is regulated by oxysterols. Arch Biochem Biophys 329: 73–81 [DOI] [PubMed] [Google Scholar]
- Taton M, Rahier A (1991) Properties and structural requirements for substrate specificity of cytochrome P-450-dependent obtusifoliol 14 alpha-demethylase from maize (Zea mays) seedlings. Biochem J 277: 483–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topping JF, May VJ, Muskett PR, Lindsey K (1997) Mutations in the HYDRA1 gene of Arabidopsis perturb cell shape and disrupt embryonic and seedling morphogenesis. Development 124: 4415–4424 [DOI] [PubMed] [Google Scholar]
- Tsafriri A, Cao X, Ashkenazi H, Motola S, Popliker M, Pomerantz SH (2005) Resumption of oocyte meiosis in mammals: on models, meiosis activating sterols, steroids and EGF-like factors. Mol Cell Endocrinol 234: 37–45 [DOI] [PubMed] [Google Scholar]
- Tzafrir I, Pena-Muralla R, Dickerman A, Berg M, Rogers R, Hutchens S, Sweeney TC, McElver J, Aux G, Patton D, Meinke D (2004) Identification of genes required for embryo development in Arabidopsis. Plant Physiol 135: 1206–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werck-Reichhart D, Feyereisen R (2000) Cytochromes P450: a success story. Genome Biol 1: REVIEWS3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemsen V, Friml J, Grebe M, van den Toorn A, Palme K, Scheres B (2003) Cell polarity and PIN protein positioning in Arabidopsis require STEROL METHYLTRANSFERASE1 function. Plant Cell 15: 612–625 [DOI] [PMC free article] [PubMed] [Google Scholar]