Abstract
The cell expansion phase contributes in determining the major characteristics of a fleshy fruit and represents two-thirds of the total fruit development in tomato (Solanum lycopersicum). So far, it has received very little attention. To evaluate the interest of a genomic scale approach, we performed an initial sequencing of approximately 1,200 cell expansion stage-related sequence tags from tomato fruit at 8, 12, and 15 d post anthesis. Interestingly, up to approximately 35% of the expressed sequence tags showed no homology with available tomato expressed sequence tags and up to approximately 21% with any known gene. Microarrays spotted with expansion phase-related cDNAs and other fruit cDNAs involved in various developmental processes were used (1) to profile gene expression in developing fruit and other plant organs and (2) to compare two growing fruit tissues engaged mostly in cell division (exocarp) or in cell expansion (locular tissue surrounding the seeds). Reverse transcription-polymerase chain reaction analysis was further used to confirm microarray results and to specify expression profiles of selected genes (24) in various tissues from expanding fruit. The wide range of genes expressed in the exocarp is consistent with a protective function and with a high metabolic activity of this tissue. In addition, our data show that the expansion of locular cells is concomitant with the expression of genes controlling water flow, organic acid synthesis, sugar storage, and photosynthesis and suggest that hormones (auxin and gibberellin) regulate this process. The data presented provide a basis for tissue-specific analyses of gene function in growing tomato fruit.
After ovule fertilization, early tomato (Solanum lycopersicum) fruit development is characterized by a period of cell division followed by cell expansion, resulting in the formation of large vacuolated cells (Mohr and Stein, 1969). Cell expansion is the longest phase in fruit development and may contribute to 90% of the increase in fruit weight, depending on the cultivar. In addition, cell expansion contributes, along with ripening, to the major structural, biochemical, and physiological changes that characterize a fleshy fruit. The cell wall changes associated with cell enlargement and the continuous accumulation in the vacuole of water, sugars, organic acids, and other compounds are necessary to maintain the turgor pressure of the expanding cells. In addition, these physiological processes contribute significantly to the flavor, texture, and overall attractiveness of the ripe fruit. The succession of the different phases of tomato fruit development is not, however, as clear cut as described above. It varies greatly according to the fruit tissue considered (Joubès et al., 1999). Tomato fruit is a complex organ composed of two or more carpels separated by a radially orientated tissue called septum, which results from the fusion of two adjacent carpel walls (or pericarp). The pericarp encloses the locular cavity containing the seeds attached to a central parenchymatous axis or columella (for review, see Gillaspy et al., 1993). After successful fertilization of the ovules, the peripheral part of the columella, or placental tissue, develops into a tissue (called locular tissue or gel) that will eventually surround the seeds and fill the locular cavity. During the early stages of the cell expansion phase, the outer pericarp cell layers continue to divide and are thus composed of small cells, while the inner pericarp cells undergo a large expansion. Similar cell enlargement occurs in the locular tissue, which acquires a jelly-like appearance in the ripening fruit (Varga and Bruinsma, 1986). While absent from the fruits of some Solanaceae crop species bearing fleshy berries (e.g. eggplant [Solanum melongena] or pepper [Capsicum annuum]; see Knapp, 2002), tissues with a similar jelly-like appearance are also found in other prominent edible fleshy fruit species belonging to very diverse taxa, such as grape (Vitaceae). Interestingly, the amount of gel is comparatively much higher in the juicy, highly flavored, and highly seeded cherry tomato fruit, which is more closely related to the ancestral progenitor of tomato (Tanksley, 2004), than in the modern cultivated tomatoes. Domestication and ongoing tomato breeding efforts resulted in the strong development of the flesh, which is mostly composed of the carpel walls and columella in the modern tomato cultivars (Tanksley, 2004). Thus, fruit anatomy and tissue characteristics (proportion of large versus small cells and cell composition) will clearly have an influence on the overall characteristics of the ripe fruit. Since most of the studies to date have focused on the entire fruit or on the carpel wall, how the different fruit tissues contribute to fruit growth and affect fruit quality remains poorly understood. There are nevertheless some indications of early tissue specialization in the fruit, since structural and biochemical changes such as cell size (Mohr and Stein, 1969; Gillaspy et al., 1993), endoreduplication level (Joubès et al., 1999), photosynthesis (Laval-Martin et al., 1977; Meier et al., 1995), starch accumulation (Schaffer and Petreikov, 1997), cell wall polysaccharides (Cheng and Huber, 1996), and flavonoid composition (Muir et al., 2001) show great variation among the different fruit tissues. These data suggest coordinated expressions of genes with specific roles in the control of growth and regional differentiation in the various tissues of the developing fruit.
New genes encoding proteins involved in fruit traits determined during early fruit development, such as morphology (ovate regulatory protein; Liu et al., 2002), size (FW2.2 repressor of cell division; Frary et al., 2000), or solid soluble content (LIN5 cell wall invertase; Fridman et al., 2000), have been identified through genetic analyses of natural tomato diversity (Tanksley, 2004). A wide range of tomato fruit mutants are available in the existing resources of induced tomato mutants (Menda et al., 2004). However, identification through a direct genetic approach of most of the genes underlying the fruit phenotypes will probably await completion of the tomato genome sequence. On the other hand, reverse genetic approaches for studying early fruit development await the identification of candidate genes. Based on studies on Arabidopsis (Arabidopsis thaliana) fruit, a dry dehiscent silique, likely candidates for fruit morphogenesis and development have been identified and studied in the tomato fruit context (Ferrandiz et al., 1999; Giovannoni, 2004). In addition, new candidate genes with functions more specific to fleshy fruit development and regulation should be identified. Gene expression profiling, widely used for evaluating genetic or environmental effects on plants, is a proven tool for gene discovery (Schnable et al., 2004). The transcriptome approach is particularly useful when applied to specific tissues or cells, where it allows discovery of new genes and comprehensive analysis of gene function (Leonhardt et al., 2004). The recent large-scale tomato expressed sequence tag (EST) sequencing projects have provided approximately 184,000 ESTs, among them approximately 37,000 fruit ESTs, enabling expression profiling of fruit development (Van der Hoeven et al., 2002; Alba et al., 2004) and identification of candidate genes for targeted functional analysis in planta. Previous studies in our laboratory (Joubès et al., 1999) and from others (Cong et al., 2002) have shown that the transition from cell division to the cell expansion period, which typically occurs at 7 to 14 DPA, is crucial for fruit development. However, in the tomato EST databases, only approximately 4,000 immature green fruit ESTs cover the 5 (cell division) to 35 DPA (approximately mature green stage) period, i.e. 60% to 70% of the total length of the fruit development period. In comparison, approximately 10,000 ESTs from ovary/carpel <5 DPA and approximately 23,000 ESTs from ripening fruit are present in the tomato EST databases and are available for studies of the cell division and ripening phases of tomato fruit.
We undertook the identification of genes expressed in early developing tomato fruit (transition from cell division to cell expansion, and cell expansion phase) and compared their expression in two tissues with contrasted developmental fates: (1) exocarp (mostly dividing cells) and (2) locular tissue (large cells accumulating water and storage compounds). Here, we demonstrate that new targeted EST analyses of selected stages of early fruit development (8, 12, and 15 DPA) can reveal up to 35% of new transcripts not previously identified in tomato. Using DNA microarrays and RT-PCR, we further show that different tissues from expanding fruit indeed have distinct and characteristic gene expression programs associated with tissue specialization. The analyses of these differences in gene expression patterns provide insights into the programs of cell differentiation in tomato fruit and their regulation.
RESULTS
Tomato ESTs from Early Developing Tomato Fruit
In order to increase the number of tomato fruit ESTs for studying early fruit growth, we constructed three cDNA libraries from whole cherry tomato fruit (cv WVa106) at selected stages of early fruit development: 8 DPA (tissue differentiation), 12 DPA (cell expansion), and 15 DPA (cell expansion and synthesis of storage compounds). The use of developing cherry tomato fruit, which contains a high proportion of locular tissue with large cells (Joubès et al., 1999), allowed the preferential isolation of cell expansion-related genes. A total of 1,248 cDNA clones was randomly selected for single-pass sequencing, and 1,089 individual 5′-ESTs (423 from 8 DPA, 340 for 12 DPA, and 326 for 15 DPA) were retained after elimination of poor quality sequence data (average sequence length of 658 bp). Of these, 265 ESTs were classified in 99 consensus and 824 ESTs were classified as singletons. Only seven consensus included more than five EST sequences, and the calculated EST redundancy (number of ESTs included in consensus/total number of ESTs) was 24.3%. This analysis allowed us to build a nonredundant set of ESTs for each library, containing 387 ESTs for 8 DPA, 311 ESTs for 12 DPA, and 296 ESTs for 15 DPA, and a global nonredundant set of 923 sequences. The EST sequences, available as supplemental data, have been deposited in the EMBL database and have been integrated into the Sol Genomics Network (SGN) and The Institute for Genomic Research (TIGR) tomato EST databases.
An analysis of sequence similarities was performed by searching available databases, including the tomato EST databases, Swiss-Prot, TrEMBL, EMBL, and dbEST (data not shown). A putative function could be assigned to <60% of the sequences. Fifteen to nineteen percent of the ESTs shared significant similarity to genes with unknown function. In addition, 11% to 16% of the ESTs matched with hypothetical proteins deduced essentially from Arabidopsis genomic sequences, thus validating their annotation as coding regions. The proportion of ESTs presenting no significant homology with known sequences was strikingly different between the cDNA libraries: 21.5% in the 8 DPA library, 10% in the 12 DPA library, and 4% in the 15 DPA library (data not shown). The comparison of our data with tomato EST resources emphasized the strong proportion of new tomato ESTs present in our cDNA libraries, which ranged from 34.6% in 8 DPA to 12.9% in 15 DPA fruit libraries.
Few Fruit-Specific Genes Are Expressed during the Early Stages of Fruit Development
How the fleshy fruit trait is acquired by the fruit is an intriguing question. One possibility is that it depends on the specific expression in the early developing fruit of a number of genes with functions specific to fleshy fruit development. Alternatively, it may depend on the coordinated regulation in the fruit of genes already expressed in other organs, where they function in related developmental programs, such as cell elongation. To address this question, we performed comparative expression profiling of developing fruit (8 and 15 DPA) and other plant tissues (developing seed, leaf, and root) using tomato cDNA microarrays that we designed for studying early tomato fruit development. The microarrays were generated from (1) the nonredundant ESTs (923) from the three fruit cDNA libraries, (2) cDNAs (165) preferentially expressed in various phases of early fruit development and previously isolated in our laboratory (Lemaire-Chamley et al., 2000), and (3) cDNAs (303) selected from tomato EST databases according to their putative role in early fruit development and in fruit quality. These mainly included genes involved in sugar and organic acid transport and metabolism (Causse et al., 2004), in hormonal regulation, and in other developmental processes.
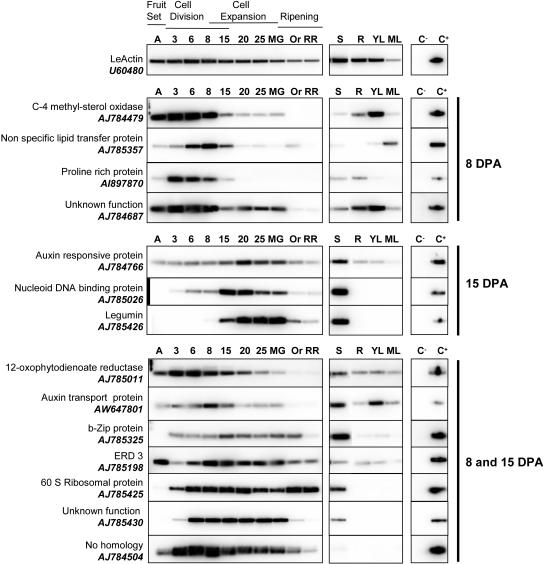
The comparison of 8-DPA fruit, seed, leaf, and root to 15-DPA fruit (see experimental design in Fig. 1) allowed the detection of 925 clones showing a significant (P value < 0.005) variation of expression (see supplemental data for the list of clones, raw data, and detailed analyses). Among them, 850 clones displayed a signal value (normalized intensity) higher than the cutoff value (see “Materials and Methods”). Among the 607 transcripts expressed in the fruit, 63 transcripts could be considered as fruit specific since they were not detected (normalized intensity < cutoff value) in other plant organs analyzed. Almost 50% of them encode proteins with unknown functions. However, close examination of the corresponding ESTs and expression profiles in the SGN and TIGR databases indicates that most of the transcripts considered as fruit specific according to our analyses are actually also expressed in other plant tissues not included in our study (e.g. flower bud and trichomes). As shown in Figure 2, RT-PCR analysis of 14 genes selected according to their microarray expression profiles and putative functions in fruit development (hormonal signaling and cell growth processes) confirmed the reliability of the microarray results. Most of the selected genes display a distinct temporal expression consistent with the succession of the different phases of fruit development (Gillaspy et al., 1993). Particularly evident is the transition between 8 and 15 DPA (see 8 and 15 DPA data in Fig. 2), which corresponds to the shift from cell division to cell expansion in most fruit tissues in the WVa106 cultivar (Joubès et al., 1999).
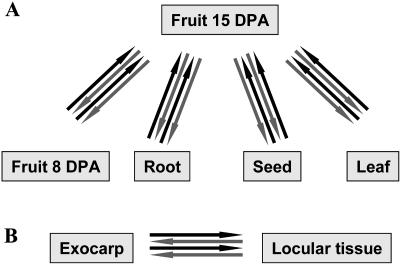
Figure 1.
Design of microarray experiment. A, Comparison of transcript abundance in developing tomato fruit, root, leaf, and seed from cherry tomato (cv WVa106). RNA was extracted from each organ sample (two RNA samples per organ), reverse transcribed, Cy labeled (Cy3 and Cy5 dye-swap for each RNA sample), and hybridized to tomato cDNA arrays for comparison with 15-DPA fruit sample, used as common reference (for each comparison, four slides, i.e. eight subarrays analyzed). B, Comparison of transcript abundance in exocarp and locular tissue from expanding tomato fruit (cv Ferum). Cytological observations and comparison of growth curves indicate that the developmental stage of 22-DPA Ferum fruit is equivalent to that of 15-DPA WVa106 fruit (data not shown). Exocarp and locular tissue samples were obtained by dissecting and pooling fruit tissues from 150 Ferum fruit harvested at 22 DPA. RNA was extracted from pooled tissue samples (two RNA samples per tissue), reverse transcribed, Cy labeled (Cy3 and Cy5 dye-swap for each RNA sample), and hybridized to tomato cDNA arrays (four slides, i.e. eight subarrays analyzed).
Figure 2.
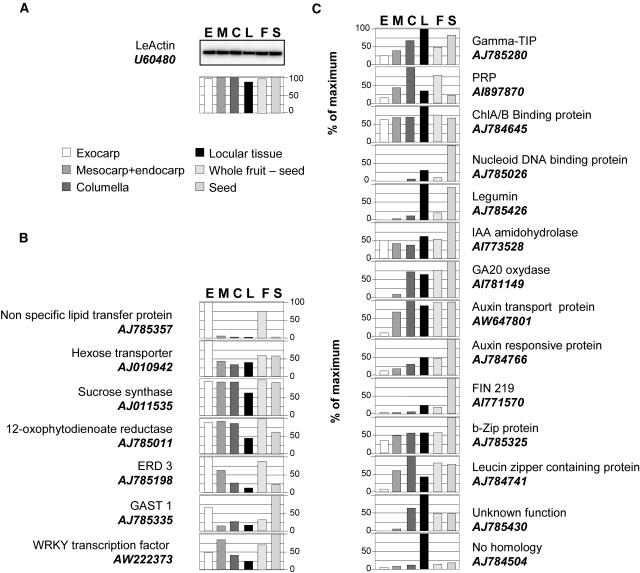
RT-PCR analyses of developing fruit confirm microarray data. The expression of 14 genes preferentially expressed in 8-DPA fruit, in 15-DPA fruit, or at both stages of fruit development, according to the microarray data, was assayed in developing fruit by semiquantitative RT-PCR using gene-specific primers. Total RNA was isolated from fruits harvested at different developmental stages (anthesis [A], 3, 6, 8, 15, 20, and 25 DPA; mature green [MG], orange [Or], and red ripe [RR]) and from seed (S), root (R), young leaf (YL), and mature leaf (ML). LeActin (U60480) was used as a control. C−, PCR-negative control without added DNA; C+, PCR-positive control using purified cDNA insert.
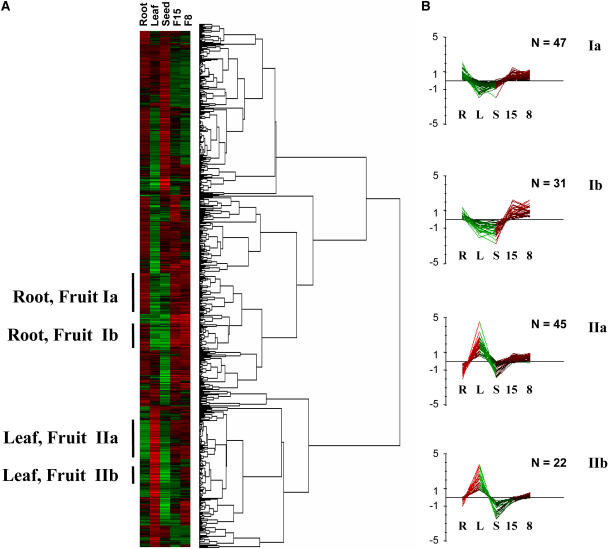
We then performed a hierarchical cluster analysis of all the transcripts expressed in the fruit (Fig. 3A). Figure 3B shows examples of groups of genes showing similar expression profiles in the different plant organs, e.g. Groups Ia and Ib that include genes up-regulated in fruit and roots and Groups IIa and IIb that include genes up-regulated in fruit and leaves. An appreciable proportion (30% to 40%) of the genes found in these groups encodes proteins with unknown functions. Genes with annotated functions fall into functional categories that are different according to the clustering group. For example, the 78 fruit- and root-preferentially expressed genes included in Groups Ia and Ib encode mostly proteins with functions related to growth processes, such as sugar transport and utilization, water transport, and cell wall synthesis and modification (data not shown). In contrast, the 67 genes found in Groups IIa and IIb encode proteins related to photosynthesis and metabolism that are mostly expressed in the leaf and, to a much lower extent, in the fruit. Close examination of the RT-PCR expression profiles of selected genes in fruit and different plant organs (Fig. 2) indicates that many transcripts analyzed are indeed detected in fruit but also in seed, root, young leaf, or mature leaf, confirming microarray results. Since gene-specific primers were used for RT-PCR, these experiments further indicate that cross-hybridizations between different members of multigene families, always possible with cDNA microarrays, cannot explain the large number of genes found to be expressed in both fruit and other plant organs. Thus, these results suggest that the acquisition of the fleshy fruit trait in tomato depends on the tight regulation during fruit development of the expression (timing, intensity, and spatial localization) of a set of genes that are also expressed in other plant organs.
Figure 3.
Hierarchical clustering of 607 tomato genes expressed in developing fruit. A, Each gene is represented by a single row of colored boxes. The five columns represent the different organs: fruit at 8 DPA (8), fruit at 15 DPA (15), leaf (L), root (R), and seed (S). Induction (or repression) ranges from pale to saturated red (or green). B, Four clusters showing distinctive expression profiles of genes preferentially expressed in fruit and root (Groups Ia and Ib) and in fruit and leaf (Group IIa and IIb) are presented. N indicates the number of transcripts in each group.
Differential Gene Expression in Exocarp and Locular Tissues from Expanding Tomato Fruit
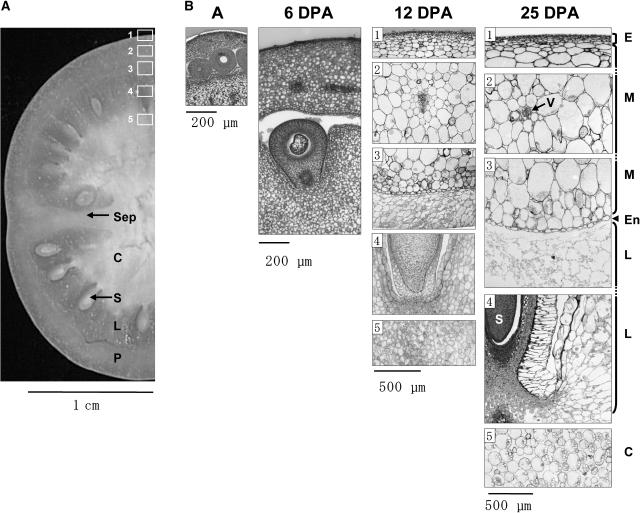
Since there are considerable variations in cell size, water, sugar, and organic acid accumulations within the fruit during the cell expansion phase (Baxter et al., 2005), most of the genes involved in the acquisition of the fleshy fruit trait should also exhibit a differential expression in the various fruit tissues. Therefore, we compared the gene expression profiles of two fruit tissues (Fig. 1): exocarp (mostly dividing cells) and locular tissue (large cells). For this study, we used Ferum, a tomato cultivar with medium sized fruits and thick pericarp that allows easy tissue dissection, rather than the cherry tomato fruit with its thin pericarp used above. As shown in Figure 4, the growing fruit is indeed constituted of distinct tissues. At 6 DPA, cell size is uniform in the various fruit tissues, since fruit growth is driven by cell division at this stage (Varga and Bruinsma, 1986; Gillaspy et al., 1993; Joubès et al., 1999). Later, during the cell expansion phase, i.e. from approximately 10 to 40 DPA in the Ferum cultivar shown, the cell fate differs depending on the fruit tissue. By 12 DPA and later, epidermal cells and outer pericarp cell layers close to the epidermis (exocarp) are still dividing, thus generating additional cell layers for fruit growth. Meanwhile, adjacent mesocarp cells enlarge considerably, to reach >500-μm length in 25-DPA fruit. As a consequence, from 6 to 25 DPA, the number of cell layers in the pericarp increases from 20 to 30, and the mean cell size increases from 14 to 250 μm in diameter (data not shown). In the inner parts of the fruit, tissue specialization is also apparent in the columella and in the locular tissue or gel, which originates from the placenta and fills the locule. The locular tissue, which differentiates early during fruit development, is composed of distinctly shaped, thin-walled, and highly vacuolated cells, which are very different from the small, dividing cells of the exocarp (Fig. 4).
Figure 4.
Structure of tomato fruit during early development. A, Fresh section from 22-DPA Ferum fruit. B, Tissue sections from ovary at anthesis (A) and from 6-, 12-, and 25-DPA fruit were cut from the equatorial region of the fruit (cv Ferum). Numbers 1 to 5 indicate fruit regions where samples from 12- and 25-DPA fruits were taken. P, Pericarp; Sep, septum; E, exocarp; M, mesocarp; En, endocarp; L, locular tissue; S, seed; C, columella; V, vascular bundles.
Among the 1,391 transcripts analyzed, 419 showed both significant variation of expression (P value < 0.005) in the exocarp and locular tissue (four independent slide hybridizations, including two dye-swaps and two replicates per slide; Fig. 1) and signal values > cutoff value. We selected 168 genes preferentially expressed in the exocarp and 129 in the locular tissue by using P < 0.005 and mean ratio > 1.6 as thresholds (Tables I and II). Many of these genes (up to 36% in the locular tissue) encode proteins with unknown functions or, alternatively, present no homology with known genes. Our findings correlate well for the few genes or enzymatic activities previously studied in the developing fruit (Laval-Martin et al., 1977; Schaffer and Petreikov, 1997; Rebers et al., 1999; Rodriguez-Conception and Gruissem, 1999; Joubès et al., 2000; Lemaire-Chamley et al., 2000; Muir et al., 2001; Guillet et al., 2002; Busi et al., 2003; Obiadalla-ali et al., 2004). Interestingly, the repartition of the known genes into the different functional categories is very different between the exocarp and the locular tissue. Some categories are overrepresented in one or the other tissue (e.g. cell-wall-related genes in the exocarp and photosynthesis-related genes in the locular tissue), reflecting tissue specialization in the expanding fruit. The putative roles in the developing fruit of the proteins encoded by the genes identified here will be addressed more thoroughly in the discussion.
Table I.
Genes preferentially expressed in the exocarp
| Clone IDb
|
Accession No.
|
SGNa
|
Annotationc
|
Fold Change E/Ld
|
Adjusted P Valuee
|
|
|---|---|---|---|---|---|---|
| Identifier | E Value | |||||
| Cell cycle and DNA processing: DNA processing | ||||||
| LE12CB04 | AJ785151 | No hit | – | Helicase SKI2W | 1.96 | 9.57E-04 |
| Cell fate: growth regulators/regulation of cell size | ||||||
| LE12CD10 | AJ785177 | U212786 | 0.0 | 1-Aminocyclopropane-1-carboxylic acid (ACC) oxidase | 3.44 | 2.68E-04 |
| LE08DC03 | AJ784658 | U212955 | 0.0 | S-adenosyl-l-Met (SAM) synthetase | 3.42 | 2.67E-04 |
| LE08CF02 | AJ784613 | U212956 | 0.0 | SAM synthetase | 2.73 | 4.64E-04 |
| LE12AA11 | AJ785011 | U214814 | 0.0 | 12-Oxophytodienoate reductase 3 | 3.55 | 2.25E-05 |
| Cell rescue defense and virulence: detoxification | ||||||
| Cat1 | M93719 | U212687 | 0.0 | Catalase1 | 2.94 | 1.72E-04 |
| LE08DF09 | AJ784688 | U232447 | 0.0 | Metallothionein | 4.95 | 5.39E-07 |
| Cell rescue defense and virulence: disease, virulence, and defense | ||||||
| LE08CE03 | AJ784607 | U215854 | 0.0 | Glucan endo-1-3-β-d-glucosidase | 4.10 | 1.53E-04 |
| LE12CG12 | AJ785203 | U213405 | 0.0 | Chitinase-like protein 1 | 2.38 | 4.32E-04 |
| LE12DD01 | AJ785237 | U212849 | 7.0E-59 | Disease resistant gene (Mi-copy2) | 2.38 | 2.61E-04 |
| Cell rescue defense and virulence: stress response | ||||||
| LEEGp6-1 | AJ832090 | U213745 | 2.0E-100 | Dehydrin | 4.61 | 2.08E-03 |
| LE08EE11 | AJ784748 | U228119 | 0.0 | Snakin | 9.87 | 1.09E-06 |
| LE12CE01 | AJ785180 | U214534 | 0.0 | Late embryogenesis-like protein | 5.08 | 4.73E-04 |
| LE08ED02 | AJ784732 | U220274 | 0.0 | DnaJ domain-containing unknown protein | 2.04 | 1.94E-03 |
| Cellular communication/signal transduction mechanism: intracellular signaling | ||||||
| LE08BE10 | AJ784545 | U213537 | 1.0E-132 | Zinc-finger protein | 1.73 | 1.15E-03 |
| LE15CD10 | AJ785473 | U216752 | 0.0 | Zinc finger protein LSD1 | 2.19 | 6.84E-04 |
| cLEN7J4 | AW222373 | U220143 | 0.0 | WRKY-type DNA binding protein | 4.52 | 1.66E-06 |
| cLED26N22 | AI897173 | U215853 | 0.0 | WRKY transcription factor Nt-SubD48 | 1.75 | 3.99E-04 |
| LE15CC02 | AJ785459 | U217026 | 0.0 | DNA-binding protein 3 | 6.17 | 8.36E-05 |
| LE15CD02 | AJ785467 | U213644 | 0.0 | Transcription factor JERF1 | 2.42 | 1.32E-03 |
| LE08AA01 | AJ784436 | U216050 | 0.0 | EREBP-3 | 4.52 | 5.65E-05 |
| LE15BB06 | AJ785382 | U224784 | 0.0 | DNA-binding protein remorin 2 | 1.64 | 5.68E-04 |
| LE15CA07 | AJ785446 | U213060 | 0.0 | 14-3-3 Protein | 8.35 | 3.53E-07 |
| LE08BE05 | AJ784542 | U212855 | 0.0 | Calmodulin | 2.56 | 1.88E-04 |
| LE12DD02 | AJ785238 | U221514 | 0.0 | Protein kinase APK1B | 4.72 | 1.40E-06 |
| LE15DB05 | AJ785520 | U235596 | 0.0 | Phospholipase | 5.96 | 7.09E-06 |
| Classification not yet clear | ||||||
| LE12CB08 | AJ785155 | U213254 | 0.0 | Allergen-like Ole | 2.87 | 1.19E-03 |
| LE12BF02 | AJ785113 | U212552 | 0.0 | Calcium-binding allergen Ole | 3.26 | 7.48E-05 |
| LE15DH09 | AJ785563 | U228352 | 3.3E-37 | Nodulin MtN3 | 6.01 | 1.95E-06 |
| LE12CH08 | AJ785209 | U214871 | 0.0 | Adhesion of calyx edges protein ACE | 1.99 | 4.24E-03 |
| LE08ED10 | AJ784740 | No hit | – | Coat protein | 2.80 | 2.60E-05 |
| LEEBp6-3 | AJ832081 | U216997 | 1.0E-102 | N-acetylethanolamine amidohydrolase | 2.08 | 4.95E-04 |
| cLEF3K1 | AW220963 | U215001 | 0.0 | Dehydration-responsive protein RD22 | 6.80 | 3.35E-05 |
| LE15BE03 | AJ785406 | U219000 | 1.0E-163 | SelT-like protein precursor | 2.03 | 6.40E-04 |
| Control of cellular organization: cell wall | ||||||
| cLES7E1 | AI779074 | U230830 | 0.0 | Extensin HRGP class II | 2.32 | 3.83E-05 |
| LE15DB11 | AJ785525 | No hit | – | Endo-β-1,4-glucanase | 1.73 | 2.82E-03 |
| LE15AG01 | AJ785355 | U222669 | 0.0 | Polygalacturonase inhibitor | 2.35 | 5.14E-06 |
| LE15DA02 | AJ785508 | U216941 | 0.0 | Pectate lyase | 1.95 | 2.34E-04 |
| LE15CA05 | AJ785445 | U213493 | 0.0 | Pectinesterase-like protein | 5.52 | 2.64E-04 |
| Development (systemic): plant development | ||||||
| LE08EF05 | AJ784752 | U215919 | 0.0 | TDR6 transcription factor | 4.97 | 2.64E-05 |
| LE12BF03 | AJ785114 | U220043 | 0.0 | Nam-like protein 2 | 3.72 | 3.13E-05 |
| Energy: electron transport and membrane-associated energy conservation | ||||||
| LE15AC06 | AJ785322 | U225169 | 0.0 | Cytochrome p450 77A2 | 3.80 | 1.46E-04 |
| mo5-10G1-11 | AJ270962 | U220531 | 0.0 | Ferredoxin | 2.23 | 9.68E-04 |
| Energy: fermentation | ||||||
| cLEI5L19 | AW648630 | U220098 | 0.0 | Aldehyde dehydrogenase (NAD+) | 2.13 | 1.59E-03 |
| Energy: glycolysis and gluconeogenesis | ||||||
| cLED19J22 | AI487966 | U228562 | 0.0 | Fructokinase-like protein | 3.78 | 4.46E-05 |
| LE15BB10 | AJ785385 | U213349 | 0.0 | Fructose-1-6-bisphosphate aldolase | 2.42 | 1.32E-03 |
| cLEC35N5 | AW032455 | U213624 | 0.0 | Pyruvate decarboxylase-1 | 5.02 | 1.53E-05 |
| cLET2D18 | AW037934 | U213321 | 0.0 | Ga3PDH-NADP | 2.43 | 1.37E-06 |
| cLES3M17 | AI777819 | U213529 | 0.0 | Triose-P isomerase (cytosolic) | 1.93 | 2.02E-05 |
| LDH1 | Y08887 | U214667 | 0.0 | LDH1 | 1.72 | 1.30E-03 |
| LE12BF10 | AJ785120 | U213348 | 0.0 | Fructose-bisphosphate aldolase (cytosolic) | 2.63 | 6.67E-04 |
| LE08BF04 | AJ784551 | U212825 | 0.0 | Ethylene-responsive enolase | 1.86 | 1.36E-03 |
| Energy: tricarboxylicacid pathway (citrate cycle, Krebs cycle, and tricarboxylic acid cycle) | ||||||
| cLES10I3 | AI780076 | U241915 | 0.0 | 2-Oxoglutarate dehydrogenase E1 component | 37.20 | 3.71E-06 |
| cLEN9B13 | AW222820 | U214866 | 0.0 | Succinyl CoA ligase | 1.63 | 1.67E-05 |
| Metabolism | ||||||
| LE08DC12 | AJ784665 | U213436 | 0.0 | Dehydrogenase | 3.33 | 7.22E-04 |
| Metabolism: amino acid metabolism | ||||||
| cLEC34J20 | AW035240 | U212562 | 0.0 | Glu decarboxylase | 1.69 | 1.47E-03 |
| cLED3A23 | AI484867 | U214486 | 0.0 | His decarboxylase | 2.37 | 6.56E-04 |
| LE08CH07 | AJ784635 | U213162 | 0.0 | Pro oxidase (mitochondrial) | 2.61 | 3.59E-04 |
| LE08DD05 | AJ784669 | U212637 | 0.0 | Adenosylmethionine decarboxylase | 2.93 | 1.35E-05 |
| LE08EC07 | AJ784725 | U215899 | 0.0 | Thr dehydratase/deaminase (chloroplastic) | 1.67 | 1.98E-03 |
| LE15CF04 | AJ785488 | U212948 | 0.0 | Arg decarboxylase | 3.77 | 2.87E-05 |
| LE15DB01 | AJ785516 | U213123 | 0.0 | Arg decarboxylase | 4.72 | 9.66E-05 |
| LE08EB11 | AJ784721 | U215839 | 0.0 | d-3-phosphoglycerate dehydrogenase (chloroplastic) | 2.57 | 1.25E-05 |
| Metabolism: biosynthesis of phenylpropanoids | ||||||
| LE12DH06 | AJ785275 | U212958 | 0.0 | Phe ammonia-lyase | 2.05 | 2.22E-03 |
| cLED8N18 | AI486371 | E213155 | 0.0 | Flavanone 3-hydroxylase | 3.14 | 5.51E-04 |
| LE15DH12 | AJ785566 | U214961 | 0.0 | Flavonol synthase | 12.41 | 1.35E-05 |
| Metabolism: compound and carbohydrate metabolism | ||||||
| LE08EA10 | AJ784713 | U213927 | 0.0 | dTDP-Glc 4 to 6-dehydratase-like protein | 2.92 | 9.78E-04 |
| LE08CG06 | AJ784625 | U222042 | 0.0 | β-Phosphoglucomutase | 1.63 | 2.78E-03 |
| Susy | AJ011319 | U213119 | 0.0 | Sucrose synthase | 4.58 | 6.36E-04 |
| Metabolism: lipid, fatty acid, and isoprenoid metabolism | ||||||
| LE08BH05 | AJ784564 | U212736 | 0.0 | Lipid transfer protein 2 | 28.34 | 1.46E-07 |
| LE15AG03 | AJ785357 | U222218 | 0.0 | Nonspecific lipid transfer protein | 14.93 | 4.55E-06 |
| LE12DB10 | AJ785227 | U217603 | 2.0E-128 | Acetyl-CoA C-acyltransferase | 5.75 | 1.39E-05 |
| LE08BB02 | AJ784516 | U213809 | 0.0 | GDSL-motif lipase/acylhydrolase | 16.64 | 3.03E-06 |
| Metabolism: metabolism of vitamins, cofactors, and prosthetic groups | ||||||
| LE15CH11 | AJ785507 | U212885 | 0.0 | Thiazole biosynthesis enzyme (chloroplastic) | 18.70 | 8.35E-05 |
| LE08BF03 | AJ784550 | U213063 | 0.0 | S-adenosyl-l-homo-Cys hydrolase | 1.86 | 1.07E-03 |
| LE12CF02 | AJ785189 | U213366 | 0.0 | GDP-Man pyrophosphorylase | 3.47 | 2.07E-06 |
| cLED13C20 | AI487366 | U223434 | 0.0 | SRG1-like protein [Fe(II)/ascorbate oxidase] | 3.04 | 9.22E-05 |
| Metabolism: nucleotide metabolism | ||||||
| LE12CB05 | AJ785152 | U214294 | 0.0 | Nucleotide sugar epimerase-like protein | 2.02 | 3.91E-03 |
| LE12BH04 | AJ785132 | U213290 | 0.0 | Guanylate kinase | 2.24 | 4.45E-03 |
| cLED6H5 | AI486997 | U214173 | 0.0 | Nucleoside diphosphate kinase 3 (mitochondrial) | 1.67 | 4.30E-04 |
| Metabolism: secondary metabolism | ||||||
| LE12CH04 | AJ785206 | U214174 | 0.0 | 4-Coumarate-CoA ligase 1 | 2.59 | 2.08E-05 |
| LE15BG09 | AJ785428 | U219467 | 0.0 | (R)-mandelonitrile lyase | 13.46 | 1.95E-06 |
| LE08AB07 | AJ784451 | U231541 | 1.0E-130 | Cinnamoyl CoA reductase | 3.26 | 2.26E-06 |
| Protein fate (folding, modification, and destination) | ||||||
| PBV1F07 | AJ006376 | U212604 | 0.0 | Subtilisin-like protease | 3.36 | 3.51E-05 |
| cLED19K24 | AI488034 | U215432 | 0.0 | Ubiquitin fusion-degradation protein | 5.56 | 4.55E-06 |
| Protein synthesis: ribosome biogenesis | ||||||
| LE15CG03 | AJ785496 | U213690 | 0.0 | 40S ribosomal protein S16 | 2.63 | 1.13E-03 |
| Protein synthesis: translation | ||||||
| LE12CC10 | AJ785166 | U214373 | 0.0 | Eukaryotic translation initiation factor 5A-3 | 1.77 | 1.36E-03 |
| Subcellular localization: cell wall | ||||||
| LE12AA02 | AJ785006 | U213588 | 0.0 | Pro-rich protein (PRP) | 3.17 | 1.00E-03 |
| LE12CB12 | AJ785159 | U216661 | 0.0 | Expansin precursor | 3.85 | 3.03E-06 |
| LE12DG11 | AJ785269 | U213725 | 0.0 | Expansin precursor | 2.89 | 1.74E-07 |
| cLEX3C8 | AW219178 | U220662 | 0.0 | Fasciclin-like arabinogalactan-protein 9 (FLA) | 2.05 | 6.92E-04 |
| Subcellular localization: cytoskeleton | ||||||
| LE12DG01 | AJ785262 | U212624 | 0.0 | β-tubulin | 1.99 | 1.31E-03 |
| LE15AA04 | AJ785298 | U220529 | 0.0 | Actin-depolymerizing factor | 1.75 | 3.98E-04 |
| cLER5G18 | AI773090 | U214141 | 0.0 | Profilin | 2.02 | 4.23E-05 |
| Systemic regulation of/interaction with environment | ||||||
| LE12CD01 | AJ785169 | U217735 | 0.0 | ERD3 protein | 4.72 | 3.48E-06 |
| LE12CC08 | AJ785164 | U214085 | 0.0 | Auxin growth promotor protein | 2.14 | 1.03E-03 |
| LEEbH9-1 | AJ832104 | U214511 | 2.3E-49 | Ethylene-responsive factor 2 | 3.21 | 6.96E-05 |
| LE15AE01 | AJ785335 | U215501 | 0.0 | Gast1 | 6.13 | 3.88E-07 |
| Transcription | ||||||
| LE15AB07 | AJ785312 | U214908 | 0.0 | Splicing factor | 3.03 | 2.12E-03 |
| LE15AF11 | AJ785354 | U219133 | 0.0 | RNA-binding protein | 1.62 | 2.71E-03 |
| Transport facilitation | ||||||
| cLER2E16 | AI772312 | U229995 | 0.0 | Hexose transporter | 5.50 | 3.57E-06 |
| cLER19B17 | AI776698 | U221086 | 0.0 | Hexose transporter | 9.04 | 1.71E-06 |
| cLER20L10 | AI777293 | U225438 | 0.0 | Sugar transporter | 11.58 | 1.86E-05 |
| LE15CF12 | AJ785494 | U214673 | 0.0 | Vacuolar ATP synthase subunit D | 1.60 | 2.00E-04 |
| LE15CD07 | AJ785470 | U213604 | 0.0 | Vacuolar-type H+-pyrophosphatase | 3.96 | 5.15E-04 |
| cLER16K11 | AI775644 | U222562 | 0.0 | Inorganic pyrophosphatase | 1.83 | 2.11E-03 |
| LE08AH02 | AJ784494 | U216601 | 0.0 | Ca2+-transporting ATPase | 2.16 | 2.93E-04 |
| LE12BD07 | AJ785099 | U213108 | 0.0 | Triose phosphate/phosphate translocator (chloroplastic) | 1.84 | 1.76E-03 |
| LE12AA06 | AJ785010 | U215083 | 0.0 | ABC transporter ATP-binding protein | 2.19 | 8.53E-05 |
| Protein of unknown function | ||||||
| LE15AH03 | AJ785367 | U214454 | 0.0 | Unknown function | 10.72 | 2.94E-06 |
| LE12AD05 | AJ785036 | U222930 | 1.0E-156 | Unknown function | 6.29 | 1.83E-05 |
| LE15DB10 | AJ785524 | U213279 | 1.0E-177 | Unknown function | 5.13 | 1.37E-04 |
| LE12BF04 | AJ785115 | U216056 | 0.0 | Unknown function | 4.44 | 1.00E-05 |
| LE15BB08 | AJ785384 | U213736 | 0.0 | Unknown function | 4.31 | 4.00E-03 |
| LE08DG12 | AJ784697 | U213845 | 0.0 | Unknown function | 4.14 | 1.49E-05 |
| LE08EE06 | AJ784743 | U218676 | 0.0 | Unknown function | 3.42 | 2.14E-05 |
| LE12AH12 | AJ785073 | U217816 | 1.0E-153 | Unknown function | 3.27 | 1.35E-05 |
| LE08BC03 | AJ784524 | U224283 | 0.0 | Unknown function | 3.16 | 2.07E-06 |
| LE08BG02 | AJ784557 | U216056 | 0.0 | Unknown function | 2.95 | 6.50E-06 |
| LE08ED04 | AJ784734 | U218910 | 3.0E-145 | Unknown function | 2.93 | 4.73E-04 |
| LE15BH05 | AJ785435 | U216284 | 0.0 | Unknown function | 2.87 | 3.04E-06 |
| cLED38D5 | AI899252 | U216929 | 0.0 | Similar to Fw2.2 | 2.78 | 2.69E-04 |
| LE08DA11 | AJ784646 | U213772 | 0.0 | Unknown function | 2.41 | 1.96E-04 |
| LE08ED08 | AJ784738 | U216703 | 1.0E-135 | Unknown function | 2.31 | 4.86E-03 |
| LE08EA06 | AJ784710 | U215330 | 0.0 | Unknown function | 2.17 | 6.88E-06 |
| LE15AF07 | AJ785351 | U214933 | 0.0 | Unknown function | 1.79 | 2.04E-04 |
| LE15CB12 | AJ785458 | U218795 | 2.0E-97 | Unknown function | 1.69 | 2.97E-04 |
| LE12CG03 | AJ785197 | U216761 | 0.0 | Unknown function | 1.65 | 8.65E-04 |
| Hypothetical protein | ||||||
| LE12AD06 | AJ785037 | U224304 | 0.0 | Hypothetical protein | 3.40 | 1.81E-04 |
| LE12AE11 | AJ785051 | U217497 | 0.0 | Hypothetical protein | 3.27 | 3.59E-06 |
| LE15CA09 | AJ785448 | U216598 | 0.0 | Hypothetical protein | 3.18 | 1.00E-05 |
| LE15BF08 | AJ785417 | U215214 | 0.0 | Hypothetical protein | 2.88 | 1.37E-04 |
| LE08AE08 | AJ784472 | U219916 | 0.0 | Hypothetical protein | 2.44 | 3.72E-04 |
| LE15DF09 | AJ785548 | U216065 | 0.0 | Hypothetical protein | 2.41 | 1.46E-07 |
| LE15BD01 | AJ785395 | U218211 | 0.0 | Hypothetical protein | 1.94 | 4.12E-03 |
| LE15AE06 | AJ785340 | U222115 | 0.0 | Hypothetical protein | 1.83 | 3.70E-03 |
| LE12AB02 | AJ785014 | U218329 | 0.0 | Hypothetical protein | 1.72 | 1.35E-04 |
| LE15BD04 | AJ785398 | U226333 | 0.0 | Hypothetical protein | 1.70 | 6.71E-04 |
| LE15DD10 | AJ785536 | U231906 | 0.0 | Hypothetical protein | 1.62 | 8.08E-05 |
| LE15CD06 | AJ785469 | U217504 | 0.0 | Hypothetical protein | 1.61 | 2.60E-04 |
| No homology | ||||||
| LE08BA12 | AJ784514 | No hit | – | – | 8.97 | 6.11E-06 |
| LE08DA06 | AJ784641 | No hit | – | – | 2.05 | 4.90E-06 |
SGN identification number of the best BLAST hit in the SGN tomato EST database, if any, and corresponding expect value.
Clone ID in the EMBL or TIGR tomato EST databases.
Putative function and classification into functional categories according to Munich Information Center for Protein Sequences (MIPS; http://www.mips.biochem.mpg.de/).
Mean ratio of the normalized data between exocarp (E) and locular tissue (L).
Probability of the t test.
Table II.
Genes preferentially expressed in the locular tissue
| Clone IDb
|
Accession No.
|
SGNa
|
Annotationc
|
FoldChange L/Ed
|
Adjusted P Valuee
|
|
|---|---|---|---|---|---|---|
| Identifier | E Value | |||||
| Cell cycle and DNA processing: cell cycle | ||||||
| CDKB2 | AJ297917 | U225483 | 0.0 | B2-type cyclin dependent kinase | 2.33 | 1.36E-03 |
| Cell cycle and DNA processing: DNA processing | ||||||
| cLEX11N17 | AW621342 | U224740 | 0.0 | Minichromosome maintenance 7 | 1.68 | 2.00E-04 |
| PBV1F01 | AW429173 | U214722 | 0.0 | Histone H4 | 2.39 | 1.84E-03 |
| Cell fate: growth regulators/regulation of cell size | ||||||
| cLED4L11 | AI485554 | U218137 | 0.0 | Spindly petunia | 1.84 | 4.55E-04 |
| LE12DH08 | AJ785277 | U239606 | 0.0 | IAA-amino acid hydrolase 1 | 2.01 | 1.90E-04 |
| cLER7E24 | AI773528 | U222172 | 0.0 | IAA-amino acid hydrolase | 6.56 | 1.37E-04 |
| LE12BA10 | AJ832000 | U214745 | 3.0E-130 | Zeatin O-glucosyltransferase | 1.74 | 1.59E-03 |
| cLES14P11 | AI781149 | U226448 | 1.2E-63 | Gibberellin 20-oxidase-1 | 4.46 | 7.07E-05 |
| Cell rescue defense and virulence: disease, virulence, and defense | ||||||
| LE08CD10 | AJ784602 | U236605 | 6.2E-63 | Meloidogyne-induced giant cell protein | 12.52 | 6.23E-05 |
| Cell rescue defense and virulence: stress response | ||||||
| LE08BH07 | AJ784566 | U212640 | 0.0 | Heat shock cognate protein 80 | 2.15 | 2.48E-03 |
| LE12BD08 | AJ785100 | U216966 | 0.0 | Wound-induced protein Sn-1 | 2.97 | 1.63E-04 |
| LE15CB10 | AJ785456 | U213892 | 0.0 | Aluminium induced protein | 3.17 | 1.35E-03 |
| LE08DE07 | AJ784680 | U217567 | 0.0 | Osmotin-like protein | 8.98 | 3.99E-06 |
| LE12BA07 | AJ785076 | U234306 | 0.0 | Calmodulin-binding heat shock protein | 1.69 | 3.45E-04 |
| Cellular communication/signal transduction mechanism: intracellular signaling | ||||||
| cLED6E18 | AI486576 | U217063 | 0.0 | MYB8 protein | 2.45 | 3.99E-05 |
| cLED26G15 | AI897007 | U235872 | 0.0 | bZIP50 protein | 2.70 | 1.37E-04 |
| LE15AC09 | AJ785325 | U213708 | 0.0 | bZIP DNA-binding protein | 4.43 | 9.49E-05 |
| LE08EE01 | AJ784741 | U213473 | 0.0 | Leu zipper-containing protein | 4.54 | 5.87E-05 |
| LE12CD07 | AJ785174 | U229573 | 0.0 | ZFHD-homeobox protein | 3.59 | 4.15E-03 |
| MO5_3G16_6 | AJ831478 | U213820 | 1.0E-136 | Mitogen-activated protein kinase MMK2 | 2.66 | 4.79E-03 |
| Classification not yet clear | ||||||
| cLED30N18 | AI897870 | U213316 | 0.0 | Pro-rich protein | 4.21 | 6.88E-06 |
| LE12AC05 | AJ785026 | U215643 | 0.0 | Nucleoid DNA binding protein (chloroplastic) | 8.06 | 1.84E-04 |
| Control of cellular organization: cell wall | ||||||
| LE08DB03 | AJ784648 | U221840 | 0.0 | Polygalacturonase | 2.43 | 1.00E-05 |
| Control of cellular organization: cytoskeleton | ||||||
| cLED5D5 | AI486189 | U218801 | 0.0 | Microtubule-associated protein MAP65 | 1.81 | 1.03E-03 |
| Control of cellular organization: endoplasmic reticulum | ||||||
| LE12AD12 | AJ785042 | U222488 | 0.0 | Endoplasmic reticulum lumen protein retaining receptor | 1.92 | 5.77E-05 |
| Energy: electron transport and membrane-associated energy conservation | ||||||
| LE12BE11 | AJ785110 | U212982 | 0.0 | Cytochrome P450 | 3.28 | 2.19E-03 |
| LE12CA10 | AJ785145 | U233399 | 1.0E-144 | Cytochrome P450 | 2.79 | 7.74E-05 |
| LE08DA02 | AJ784638 | U212714 | 0.0 | Ferredoxin-I | 2.04 | 2.07E-06 |
| Energy: fermentation | ||||||
| LE08BC01 | AJ784523 | U232963 | 1.9E-29 | Alcohol dehydrogenase | 2.44 | 9.88E-05 |
| LEECp10-3 | AJ832084 | U214593 | 1.0E-114 | Alcohol dehydrogenase | 3.59 | 3.83E-05 |
| Energy: glycolysis and gluconeogenesis | ||||||
| LePPC1 | AJ243416 | U214321 | 0.0 | Phosphoenolpyruvate carboxylase 1 | 3.56 | 2.07E-06 |
| LE15BH07 | AJ785437 | U214321 | 3.0E-125 | Phosphoenolpyruvate carboxylase 2 | 2.95 | 4.00E-05 |
| cLED24K5 | AI490644 | U219468 | 0.0 | PEP carboxylase-related protein 3 | 2.86 | 4.35E-04 |
| Energy: photosynthesis | ||||||
| LE08EH04 | AJ784768 | U212743 | 0.0 | Oxygen-evolving enhancer protein 1 (chloroplastic) | 1.88 | 1.90E-04 |
| cLEI6J23 | AW648964 | U215938 | 0.0 | Pyruvate phosphate dikinase (chloroplastic) | 2.71 | 9.68E-04 |
| LE08CC01 | AJ784584 | U213031 | 0.0 | Plastocyanin (chloroplastic) | 1.96 | 2.66E-07 |
| LE08AE04 | AJ784469 | U213039 | 0.0 | Chlorophyll a/b-binding protein CP29 | 1.98 | 7.51E-05 |
| LE15AA07 | AJ785300 | U212697 | 0.0 | Chlorophyll a/b-binding protein | 2.54 | 4.93E-04 |
| LE08AF03 | AJ784476 | U212742 | 0.0 | Oxygen-evolving enhancer protein (chloroplastic) | 2.50 | 2.14E-05 |
| LE08DA10 | AJ784645 | U213105 | 0.0 | Chlorophyll a/b-binding protein | 3.50 | 1.47E-05 |
| LE15AG07 | AJ785360 | U213041 | 0.0 | Chlorophyll a/b-binding protein | 3.91 | 3.88E-07 |
| LEEaA1-4 | AJ832091 | U212863 | 3.0E-109 | Chlorophyll a/b-binding protein of light-harvesting complex II type I | 5.55 | 2.10E-05 |
| LE15AE04 | AJ785338 | U218914 | 0.0 | Chlorophyll a/b-binding protein | 10.08 | 1.45E-06 |
| Metabolism: amino acid metabolism | ||||||
| LE08CB02 | AJ784576 | U212860 | 0.0 | Thr deaminase chloroplastic | 2.10 | 3.46E-05 |
| LE15BH02 | AJ785432 | U216858 | 0.0 | Dehydroquinate synthase | 1.80 | 1.03E-03 |
| cLED2M7 | AI486042 | U221579 | 0.0 | Anthranilate phosphoribosyltransferase | 1.66 | 1.14E-03 |
| Metabolism: nitrogen and sulfur metabolism | ||||||
| LE08AF10 | AJ784483 | U215649 | 0.0 | Steroid sulfotransferase | 2.61 | 8.15E-06 |
| Metabolism: compound and carbohydrate metabolism | ||||||
| LE08AF02 | AJ784475 | U225512 | 1.0E-135 | Rubisco, small subunit | 1.66 | 2.69E-04 |
| LEEaA3-1 | AJ832092 | U225534 | 1.0E-104 | Rubisco, small subunit | 1.74 | 4.22E-03 |
| cLED21P10 | AI490453 | U216862 | 0.0 | LeLIN5 cell wall invertase | 1.73 | 2.54E-05 |
| MO7_10T2_4 | AJ831483 | U212903 | 2.0E-119 | Vacuolar invertase | 9.82 | 1.13E-05 |
| cLED24i21 | AI483400 | U216715 | 0.0 | Invertase Inhibitor | 1.98 | 2.45E-04 |
| cLED5N5 | AI486229 | U218220 | 0.0 | Starch phosphorylase | 1.75 | 6.73E-04 |
| Metabolism: lipid, fatty acid, and isoprenoid metabolism | ||||||
| LEEA3-1 | AJ832060 | U214114 | 0.0 | Epoxide hydrolase | 2.09 | 1.98E-05 |
| cLEN15E17 | AW441298 | U212777 | 0.0 | Lipoxygenase | 6.08 | 1.01E-04 |
| cLPT8I1 | AW399592 | U217409 | 0.0 | HMGR CoA reductase | 8.28 | 1.46E-07 |
| PTOX | AF177980 | U218630 | 0.0 | Plastid terminal oxidase | 1.81 | 1.40E-03 |
| Metabolism: metabolism of vitamins, cofactors, and prosthetic groups | ||||||
| LE08CE12 | AJ784611 | U213738 | 0.0 | GSH-dependent dehydroascorbate reductase | 2.29 | 2.82E-04 |
| Metabolism: secondary metabolism | ||||||
| LE08AD10 | AJ784465 | U230863 | 0.0 | 1,4-benzoquinone reductase | 2.95 | 6.88E-06 |
| Protein fate (folding, modification, and destination) | ||||||
| LE12BH02 | AJ785130 | U213337 | 0.0 | Metallocarboxypeptidase inhibitor | 3.53 | 7.04E-05 |
| LeVPE1-7 | AJ243876 | U217676 | 0.0 | Vacuolar processing enzyme (VPEγ) | 7.05 | 2.14E-05 |
| LE08DE04 | AJ784677 | No hit | – | Ser carboxypeptidase II-like | 2.43 | 2.15E-05 |
| LEEbB6-1 | AJ832099 | U224513 | 0.0 | Cyclophilin | 2.29 | 3.77E-04 |
| LE15CC04 | AJ785460 | U242133 | 0.0 | Subtilisin-like Ser protease | 7.33 | 4.73E-04 |
| LE12DA07 | AJ785217 | U213211 | 0.0 | Cys endopeptidase | 4.41 | 2.82E-04 |
| Storage protein | ||||||
| mo6-MgT2-1 | AJ270964 | U215172 | 1.0E-108 | Vicilin | 25.53 | 7.72E-05 |
| LE15BG07 | AJ785426 | U214940 | 0.0 | Legumin precursor | 69.59 | 3.01E-06 |
| Subcellular localization | ||||||
| LE12AD09 | AJ785040 | U215827 | 0.0 | Outer membrane lipoprotein | 4.61 | 4.73E-04 |
| Systemic regulation of/interaction with environment | ||||||
| LE08AE10 | AJ784474 | U213616 | 0.0 | Auxin repressed protein | 2.73 | 1.53E-05 |
| LE08EG11 | AJ784766 | U214220 | 0.0 | Auxin responsive protein | 4.39 | 2.33E-05 |
| cLEI2G22 | AW647801 | U226470 | 0.0 | PIN7-like Auxin transport protein | 4.20 | 4.73E-04 |
| cLED19H14 | AI487976 | U220722 | 0.0 | AXR1 RUB1-E1-activating | 1.95 | 6.49E-04 |
| cLEN17N15 | AW441576 | U220580 | 0.0 | AXR1 RUB1-E1-activating | 1.62 | 6.49E-04 |
| cLED16G4 | AI489353 | U214220 | 0.0 | Aux/IAA protein | 4.30 | 3.40E-06 |
| LE15AC12 | AJ785327 | U213952 | 0.0 | Cell differentiation protein RCD1 | 1.68 | 2.27E-03 |
| LE15DE02 | AJ785537 | U217589 | 0.0 | Rho GDP-dissociation inhibitor 1 | 1.63 | 2.04E-04 |
| Transcription: mRNA transcription | ||||||
| LE08BF01 | AJ784548 | No hit | – | RNA polymerase II largest subunit | 1.73 | 5.51E-04 |
| Transport facilitation | ||||||
| LE08AC10 | AJ784459 | U212568 | 4.0E-152 | Aquaporin PIP-type TRAMP | 2.88 | 5.94E-06 |
| LE12DE07 | AJ785248 | U214295 | 0.0 | Tonoplast intrinsic protein (δTIP) | 2.76 | 2.34E-04 |
| LE12DH11 | AJ785280 | U213272 | 0.0 | γTIP | 7.58 | 6.65E-07 |
| LE12AE02 | AJ785044 | U215949 | 0.0 | Plasma membrane intrinsic protein | 2.02 | 9.45E-05 |
| LE12DC03 | AJ785232 | No hit | – | Ycf1 | 1.67 | 2.69E-03 |
| Protein of unknown function | ||||||
| LE15BG11 | AJ785430 | U224778 | 0.0 | Unknown function | 34.99 | 2.85E-05 |
| LE15CG06 | AJ785499 | U225059 | 0.0 | Unknown function | 10.17 | 3.37E-05 |
| LE08CF01 | AJ784612 | U215776 | 0.0 | Unknown function | 9.30 | 1.46E-07 |
| LE15DB03 | AJ785518 | U231609 | 0.0 | Unknown function | 5.79 | 4.11E-04 |
| LE12AB10 | AJ785020 | U239000 | 0.0 | Unknown function | 5.74 | 4.06E-05 |
| LEEBp4-1 | AJ832080 | U215120 | 1.3E-73 | Unknown function | 4.05 | 1.31E-04 |
| LEEDp3-1 | AJ832087 | U214474 | 2.1E-81 | Unknown function | 3.66 | 1.35E-05 |
| LE12AA03 | AJ785007 | U218722 | 0.0 | Unknown function | 3.64 | 2.18E-06 |
| LE15AF02 | AJ785347 | U214236 | 0.0 | Unknown function | 3.55 | 1.48E-05 |
| LED83 | AJ831585 | U218518 | 0.0 | Unknown function | 2.87 | 3.63E-04 |
| LE08DF10 | AJ784689 | U235233 | 1.5E-08 | Unknown function | 2.82 | 5.25E-04 |
| LE12DC05 | AJ785233 | U218142 | 7.0E-176 | Unknown function | 2.20 | 2.26E-03 |
| LE08DH03 | AJ784699 | No hit | – | Unknown function | 1.69 | 1.45E-03 |
| LE08EA02 | AJ784706 | U224380 | 0.0 | Unknown function | 1.68 | 5.67E-04 |
| LE12DB03 | AJ785222 | U217461 | 0.0 | Unknown function | 1.64 | 2.00E-04 |
| Hypothetical protein | ||||||
| LE15BB12 | AJ785386 | U227260 | 0.0 | Hypothetical protein | 2.72 | 9.82E-04 |
| LE08AG05 | AJ784489 | U220976 | 0.0 | Hypothetical protein | 2.28 | 6.14E-06 |
| LE08AB03 | AJ784448 | U220987 | 5.0E-175 | Hypothetical protein | 2.17 | 2.56E-05 |
| LE08CG01 | AJ784620 | No hit | – | Hypothetical protein | 1.76 | 5.67E-04 |
| LE08CD02 | AJ784596 | U218544 | 0.0 | Hypothetical protein | 1.70 | 8.84E-04 |
| LE15AF03 | AJ785348 | U231894 | 0.0 | Hypothetical protein | 1.70 | 1.98E-03 |
| LE15DC03 | AJ785527 | U236020 | 1.0E-127 | Hypothetical protein | 1.64 | 3.91E-04 |
| No homology | ||||||
| LE08BA02 | AJ784504 | No hit | – | – | 27.00 | 3.15E-06 |
| LE15CF01 | AJ785485 | No hit | – | – | 1.87 | 9.31E-05 |
| LE15BB07 | AJ785383 | No hit | – | – | 1.75 | 3.33E-03 |
SGN identification number of the best BLAST hit in the SGN tomato EST database, if any, and corresponding expect value.
Clone ID in the EMBL or TIGR tomato EST databases.
Putative function and classification into functional categories according to MIPS (http://www.mips.biochem.mpg.de/).
Mean ratio of the normalized data between exocarp (E) and locular tissue (L).
Probability of the t test.
RT-PCR Confirms Preferential Gene Expression in Exocarp and Locular Tissue
To gain additional information on the tissue specificity of the genes detected by microarray analysis (Tables I and II), we performed RT-PCR analyses on 21 genes preferentially expressed in exocarp (7 genes) or in locular tissue (14 genes). The RT-PCR analyses confirmed the preferential expression of the selected genes in the exocarp (Fig. 5B) or in the locular tissue (Fig. 5C). Few genes are specifically expressed in a single tissue (e.g. the nonspecific transfer protein; Fig. 5B). In contrast, the detailed analysis in the different fruit tissues (exocarp, mesocarp, columella, locular tissue, and seeds) of several genes fulfilling different functions in the fruit (e.g. water transport for γ-tonoplast intrinsic protein [TIP], hormone synthesis for GA20 oxidase, or transcriptional regulation for leucine zipper protein) indicates that most of the genes selected from the locular tissue (Fig. 5C) follow a gradient of expression from the central part of the fruit (columella and locular tissue) to the outer part of the fruit (exocarp). Another interesting feature is that most of the genes analyzed also showed strong expression in the developing seed, suggesting common developmental programs between developing seed and fleshy parts of the developing fruit.
Figure 5.
RT-PCR analyses confirm differential gene expression in exocarp and locular tissue. The expression of 21 genes preferentially expressed in fruit exocarp or locular tissue according to the microarray data was assayed by semiquantitative RT-PCR in different tissues dissected from 22-DPA fruit (cv Ferum) using gene-specific primers. E, Exocarp; M, mesocarp; C, columella; L, locular tissue; F, fruit without seeds; S, seed. A, LeActin (U60480) was used as control. For each gene and tissue, the relative abundance of mRNA was normalized toward that of tomato actin in the corresponding tissue. The results are presented as percentage of the highest relative expression for the considered gene. B, Transcripts showing preferential expression in the exocarp. C, Transcripts showing preferential expression in the locular tissue.
DISCUSSION
To date, studies on tomato fruit development have focused primarily on the ripening process (for review, see Giovannoni, 2001, 2004; Seymour et al., 2002), and few genes from early developing fruit have been studied until now. Early fruit growth conforms to a complex developmental program that involves cell division followed by cell expansion (Gillaspy et al., 1993). Functionally distinct tissues, each exhibiting a specific pattern of development, are apparent in the developing tomato fruit after 6 to 10 DPA (e.g. locular, mesocarp, or columella tissues; Fig. 4). While some tissues are differentiating (e.g. locular tissue), others (e.g. exocarp) retain mitotic activity late during fruit development (Joubès et al., 1999; Cong et al., 2002). The relative development of the various fruit tissues will affect both fruit size and anatomy but also fruit quality. Our aim was to identify genes linked with the differentiation of specialized tissues in early developing tomato fruit through global analysis of gene expression. These data will help in elucidating the molecular mechanisms underlying fleshy fruit development and their regulation.
Identification of Novel Genes Associated with the Early Stages of Fruit Development in Tomato
Our present data indicate that the sequencing of new fruit cDNA libraries targeted to specific stages of early fruit development (8-, 12-, and 15-DPA fruit) may reveal new tomato genes not present in EST databases and a whole range of new functions not previously described in the developing fruit. The higher proportion of genes with no known functions is found in the 8-DPA cherry tomato fruit library (Fig. 4), suggesting that future studies on early tomato fruit development should preferentially focus on this stage, which is concomitant with the transition from cell division to cell expansion in most tomato fruit tissues (Fig. 4; Joubès et al., 1999). However, the most important finding is perhaps that the comparative transcriptome analyses of developing fruit and of other tomato organs failed to identify large sets of genes specifically expressed in the fruit. Instead, the majority of the growing fruit-related genes are also expressed in additional organs (Fig. 3), reflecting either ontogenic relationships (e.g. between fruit and leaf; see Gillaspy et al., 1993) or processes common to fruit and other organs (e.g. cell division and cell elongation in fruit and roots). One likely explanation is that the acquisition of specialized functions in fleshy fruit cells is achieved through the coordinated temporal and spatial expression in the distinct fruit tissues of genes that also participate in other developmental programs and in other plant tissues.
Cellular Processes and Metabolic Pathways in the Exocarp from Developing Tomato Fruit
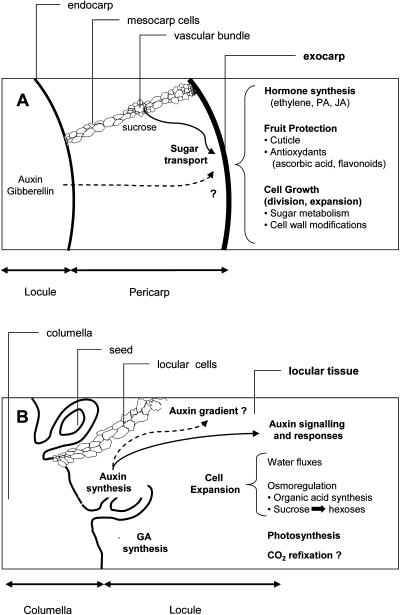
The sample referred to as exocarp in this study (Fig. 4) is composed of the skin and of several additional layers of outer pericarp cells that include mitotically active cells and enlarging cells (Joubès et al., 2000). The skin comprises the cuticle, one epidermal cell layer, and about four subepidermal layers of small thick-walled cells with few chloroplasts. Genes preferentially expressed in the exocarp (Table I) encode a wide range of proteins, reflecting the complexity of this tissue. However, as summarized in Figure 6A, two broad categories based on the biological functions of exocarp in the fruit can be defined.
Figure 6.
Schematic representation of the major mechanisms and regulations in early developing fruit tissues. A, Exocarp; B, locular tissue.
Fruit Protection
This category includes proteins possibly involved in the formation of the skin, which provides a physical barrier to the fruit. Among them are lipid transfer proteins with suggested roles in transfer of wax or cutin monomers to the cuticle (Kader, 1996) and various cell-wall-related proteins that can participate in the thickening of subepidermal cell walls. Other exocarp-expressed proteins can be involved in fruit defense against pathogens (glucanase and chitinase) or in stress tolerance (UV radiation and heat). These include key enzymes required for the synthesis of flavonoids (Phe ammonia-lyase, flavanone hydroxylase, and flavonol synthase; Muir et al., 2001) and of ascorbic acid (GDP Man pyrophosphorylase; Smirnoff et al., 2001), two fruit antioxidants with high nutritional value.
Fruit Growth
The second category contains genes encoding proteins required for sustaining the growth of the dividing and enlarging cells of the exocarp. Among them are cell-wall-related proteins involved in polysaccharide synthesis and modifications (pectate lyase, pectinesterase, endo-β-1,4 glucanase [EGase], and polygalacturonase inhibitor), cell wall structure (PRP and extensin), cell adhesion (FLA; Johnson et al., 2003), and cell wall relaxation (expansins; Cosgrove, 1997). In particular, EGase and expansin are cell wall loosening factors expressed during the stages of highest growth in the expanding tomato fruit (Catala et al., 2000), where they are presumed to allow fruit growth by modifying epidermal cell wall properties, as previously suggested for xyloglucan endotransglycosylase (Thompson et al., 1998). GAST1, another putative cell wall protein of unknown function expressed in the exocarp (Table I; Fig. 5B), belongs to a family of GA-inducible genes functioning in cell expansion mechanisms necessary for the regulation of cell and organ shape in Gerbera hybrida flower (Kotilainen et al., 1999). Consistent with the growth activity of the exocarp, sugar transport proteins (Suc transporter and hexose transporters), Suc synthase, and various glycolytic enzymes (Table I; Fig. 5B) are highly expressed in this tissue. Indeed, high sugar import and metabolic activities are necessary to support the strong utilization sink activity of the young growing fruit (Ho, 1996; Barker et al., 2000; Koch, 2004). In addition, Suc synthase may also act as a metabolic channel for cell wall synthesis (Cosgrove, 1997), together with its role in symplastic unloading of sugar in the young developing fruit (Nguyen-Quoc and Foyer, 2001).
Regulation of Exocarp-Expressed Genes
Besides sugars, auxin, and gibberellins, which are known regulators of sugar- and cell-growth-related genes such as expansin, EGase, and GAST1 (Kotilainen et al., 1999; Catala et al., 2000), other phytohormones may also serve as signaling molecules in the developing exocarp tissue (Gillaspy et al., 1993). As shown in Table I, several enzymes involved in the synthesis of ethylene or polyamines (SAM synthetase, ACC oxidase, and Arg decarboxylase) and jasmonic acid (12-oxophytodienoate reductase) are indeed expressed in the exocarp, and at least two proteins, EREBP-3 (Leubner-Metzger et al., 1998) and JERF1 (Zhang et al., 2004), are ethylene-responsive transcription factors that may participate in the regulation of stress tolerance and fruit growth (Jones et al., 2002; Balbi and Lomax, 2003). In addition, a wide range of regulatory proteins can modulate gene expression in the exocarp from growing fruit (Table I). They include transcription factors controlling flower and fruit development, such as TDR6 (Busi et al., 2003; Favaro et al., 2003; Giovannoni, 2004), various WRKY-DNA binding proteins belonging to a large group of zinc-finger proteins implicated primarily in defense responses but also in plant development (Eulgem et al., 2000), and 14-3-3 proteins that function in regulation of carbon metabolism and in signaling for plant defense and development (Roberts, 2003).
Molecular Mechanisms in the Differentiating Locular Tissue
The ontogenic origin of the locular cells remains unclear since they differentiate from a central axial placenta, to which the seeds are attached and which is formed by the fusion of the carpels with a column extending from the central part of the floral meristem (Gasser and Robinson-Beers, 1993). As shown in Figure 4, locular tissue is homogeneous and is composed of thin-walled and highly vacuolated cells undergoing cell expansion in the young growing fruit (Fig. 4), concomitant with DNA endoreduplication (Joubès et al., 1999). Its physiological functions in the fruit, if any, are not obvious. However, locular tissue constitutes a well-defined model for studying cell expansion and its regulation in the fruit, a process crucial for the acquisition of the fleshy fruit trait. Gene expression profiling of locular tissue (Table II) allowed the identification of major functional categories of genes underlying the molecular mechanisms involved in locular tissue differentiation (summarized in Fig. 6B).
Cell Expansion
Cell enlargement depends on both cell wall loosening and increase in turgor pressure (Cosgrove, 1997), which is itself driven by osmolyte (sugars, organic acids, and K+) and water accumulation inside the vacuole of fruit cells. Several genes expressed in locular tissue (Table II) encode known proteins controlling the flux of water from the symplast and across the vacuolar membrane (the plasma membrane intrinsic protein [PIP] and TIP aquaporins; Maurel and Chrispeels, 2001; Ozga et al., 2002), the accumulation of soluble sugars in the vacuole (the vacuolar invertase that is controlled by the vacuolar processing enzyme VPEγ, targeting to the vacuole, and the invertase inhibitor; Koch, 2004), and the accumulation of organic acids (citric and malic acids) used as counterions in the expanding vacuoles (the phosphoenolpyruvate carboxylase that controls a key step in organic acid synthesis and the zinc-finger homeodomain transcription factor, a putative regulator of its expression; Windhovel et al., 2001; Guillet et al., 2002). More complex roles may be fulfilled by TRAMP, a fruit-expressed PIP found in the locular tissue, since its inhibition by antisense technology altered the organic-acid-to-sugar ratio in the fruit (Chen et al., 2001). Interestingly, detailed analysis of the distribution of locular tissue-expressed genes in the different fruit tissues indicates that, in most cases, genes controlling the cell expansion process (γTIP and PRP) or its regulation (hormone synthesis, signaling, and response) are expressed along a gradient from the inner part to the outer part of the fruit (Fig. 5C). These results are consistent with the existence of a gradient in cell size observed in the distinct tissues of the expanding fruit (Fig. 4).
Photosynthesis
A surprisingly high number of genes found in the locular tissue (15) are related to chloroplastic functions (Table II). The locular tissue is indeed capable of photosynthetic activity (Laval-Martin et al., 1977) and shows high promoter activity of Rubisco genes (Meier et al., 1995), despite its localization in locules surrounded by carpel walls (Fig. 4). One possible explanation is that the CO2 arising from fruit respiration, which may account for as much as 25% of the imported carbon in the growing fruit (Ho, 1996), is retained by the cuticle barrier and is recaptured in the central part of the fruit via photosynthesis or via dark fixation with PEP carboxylase. In contrast, the exocarp tissue, which is localized on the external side of the fruit and corresponds to the abaxial side of the leaf, is poor in chloroplasts (Gillaspy et al., 1993), which may explain the stronger expression of photosynthesis-related genes in the locular tissue.
Is Locular Tissue Differentiation and Development Controlled by Auxins and Gibberellins?
In addition to 3-hydroxy-3-methylglutaryl CoA reductase controlling a key step in isoprenoid metabolism and previously shown to control early fruit growth (Gillaspy et al., 1993; Rodriguez-Conception and Gruissem, 1999), key candidate genes functioning in auxin synthesis (indole-3-acetic acid [IAA] amidohydrolases), transport (PIN7-like protein), and responses (auxin-repressed protein, Aux-IAA protein, and AXR1) showed a preferential expression in locular tissue. Some other candidate genes encoding proteins with roles in auxin perception and signaling (RCE1, ARF, and FIN219), not included in Table II (P value > 0.005), could also play a role in locular tissue (expression ratio >1.6). The use of more sensitive analytical tools, such as RT-PCR, may confirm their differential expression in the locular tissue, as was done for FIN219, which links auxin to light regulation (Hsieh et al., 2000; Fig. 5C).
The role of auxins in early fruit growth, known for a long time (see Varga and Bruinsma, 1986), has been emphasized by recent studies on growing tomato fruit (Bohner and Bangerth, 1988; Catala et al., 2000), tomato mutants (diageotropica; Balbi and Lomax, 2003), and transgenic lines altered for auxin biosynthesis or signaling genes (Pandolfini et al., 2002; Carmi et al., 2003). In particular, auxin signaling appears as a prerequisite for the development of the locular tissue and for the enlargement of the locular cells. According to our gene expression data (Table II; Fig. 5C), in agreement with the established notion that auxins produced by the seeds and/or surrounding fruit tissues are released to trigger or accelerate fruit growth by cell expansion (see Gillaspy et al., 1993), the following picture can be drawn (Fig. 6B). Free IAA hydrolyzed from its conjugates by IAA amino acid hydrolase in the central parts of tomato fruit (seeds and/or locules) could in turn activate and coordinate the expression of cell-expansion-related proteins, such as aquaporins, vacuolar invertase, and PEP carboxylase, thus enabling cell enlargement in locular tissue. The auxin-transport protein (PIN7-like) could well play a crucial role in the differentiation of the locular tissue, and more largely in fruit morphogenesis, by establishing auxin gradients in the fruit tissues (Benkova et al., 2003). Several putative transcription factors expressed in the locular tissue, including Aux/IAA proteins (Jones et al., 2002) or bZIP DNA-binding proteins (Strathmann et al., 2001; Heinekamp et al., 2004), may link hormonal signals by auxin or other phytohormones (Jones et al., 2002; Balbi and Lomax, 2003) to cell growth. Cross-talk between auxins, gibberellins, and cytokinins for the stimulation of cell growth, already demonstrated in developing pea (Pisum sativum) fruit for auxin and gibberellin (Ozga et al., 2002, 2003), is also likely to occur in developing tomato locules where GA20 oxidase (gibberellin synthesis), SPINDLY GA response inhibitor, and zeatin O-glucosyltransferase (cytokinin biosynthesis) are also expressed (Table II; Rebers et al., 1999).
In conclusion, transcriptome analysis of early developmental stages and of distinct fruit tissues allowed the identification of new candidate genes controlling early fruit development and provided new insights into the mechanisms involved in the acquisition of the fleshy fruit trait in tomato. Future studies using new generations of tomato microarrays generated from the nonredundant tomato EST collections (Giovannoni, 2004) and focused on specific fruit tissues, or even single fruit cells, should allow more comprehensive analysis of the mechanisms and regulations controlling fleshy fruit development.
MATERIALS AND METHODS
Plant Material and RNA Isolation
Tomato (Solanum lycopersicum) plants were grown either in a growth chamber (cherry tomato, cv West Virginia 106) with a 15-h-day (25°C)/9-h-night (20°C) cycle with an irradiance of 400 μmol m−2 s−1 and 75% to 80% humidity or in a greenhouse during the spring season (medium-sized tomato, cv Ferum). Whole fruits (8 and 15 DPA), seeds (15 DPA), young leaves, and roots were collected from WVa106 cultivar. Fruit tissues (exocarp including epiderm and outer pericarp cell layers, mesocarp + endocarp, locular tissue, columella, and seeds) were collected from Ferum cultivar (22-DPA fruit). Individual flowers were tagged on the day of anthesis (flower opening). The fruits were further selected according to size and/or color and collected at the indicated times (expressed in DPA). All plant materials were frozen in liquid nitrogen and stored at −80°C until use. Samples were composed of at least 6 to 100 fruit or flowers according to the developmental stages and the tissues analyzed. Total RNA was isolated as described previously (Lemaire-Chamley et al., 2000), then treated with RNase-free DNase (RQ1; Promega) for 15 min at 37°C, extracted with phenol:chloroform (1:1) and chloroform, precipitated with ethanol, and finally resuspended in diethyl pyrocarbonate-treated water. For microarray probe synthesis, total RNA quality was further checked using Agilent Microchips (Agilent Bionalyzer 2100). Poly(A)+ mRNAs were isolated using the Oligotex Midi kit (Qiagen) according to the manufacturer's instructions.
cDNA Library Construction and Sequence Analysis
Three cDNA libraries were constructed from WVa106 cherry tomato fruits at 8 (Le08), 12 (Le12), and 15 (Le15) DPA using the λ UniZap cDNA library construction kit (Stratagene), which allows directional cloning between the EcoRI and XhoI sites of pBluescript. A total of 1,248 randomly selected cDNA clones were sequenced from the 5′ end. After vector clipping and sequence trimming (Ewing et al., 1998), clustering and assembly of the sequences were achieved to provide a nonredundant set of ESTs using StackPack v2.2 (Miller et al., 1999). The resulting consensus and singleton sequences were sequentially compared to the Swiss-Prot database, TrEMBL, EMBL, and dbEST using the appropriate BLAST programs as described hereafter. Sequences showing a hit with E value < 10−20 with a sequence present in the first database (Swiss-Prot) were retrieved, visually inspected, and assigned a putative function if (1) the entire EST sequence showed an E value < 10−50 or if (2) the sequence showed an E value < 10−20 with a lack of homology in the 3′ or 5′ ends of the sequence, which was considered as indicative of the presence of 3′ or 5′ untranslated regions in these regions. The same flowchart was applied to the remaining sequences with the next database (i.e. TrEMBL) and so on. In parallel, consensus and singleton sequences were systematically compared to the tomato TIGR EST database (http://www.tigr.org) and to SGN database (http://www.sgn.cornell.edu). Functional categories were assigned following the MIPS (http://mips.gsf.de/) role characterization. The EST sequences have been deposited in the EMBL database and in the SGN tomato EST database (http://www.sgn.cornell.edu/). Clones are available by request to the authors.
Microarray Design and Construction
The expanding fruit tomato cDNA microarray was produced using 923 nonredundant cDNAs from Le08, Le12, and Le15 libraries, 165 cDNAs related to early fruit development previously isolated in our group (Lemaire-Chamley et al., 2000), and 305 cDNAs involved in a range of growth-related processes (regulation, hormones, metabolism, etc.) selected from the TIGR tomato EST database. In addition, negative controls (yeast [Saccharomyces cerevisiae] and wheat [Triticum aestivum] cDNAs) and positive controls (full-length and partial luciferase fragments) were included in the microarray. A detailed list of the cDNAs spotted is available as supplemental data. Purified PCR products were spotted in duplicate on amino silane slides (CMT Gaps-II; Corning) using a PixSys 7500 arrayer (Cartesian Technologies).
Preparation of Labeled cDNA Probes
mRNA [1 μg poly(A)+ mRNA] was reverse transcribed using 400 units of Superscript II reverse transcriptase (Life Technologies) in the presence of aminoallyl-dUTP (100 μM), 2 μg oligo(dT)21-mer, 2 μg random hexamer (Promega), 250 μm each dATP, dCTP, and dGTP, 150 μm dTTP, 15 units RNasin (Promega), and 10 μm dithiothreitol in the provided buffer (final volume 30 μL). After incubation at 42°C for 90 min, 400 units of Superscript II reverse transcriptase were added, and incubation was continued for 90 min at 42°C. Sample tubes were treated with 0.25 n NaOH, incubated 10 min at 70°C, and neutralized by addition of HCl (0.25 n). Aminoallyl-labeled cDNA was ethanol-precipitated overnight at −20°C in the presence of glycogen (20 μg). After centrifugation, the pellet was rinsed with 70% ethanol, air dried, and resuspended in 10 μL NaHCO3, pH 9.3. Post-labeling was performed by incubation of the aminoallyl cDNA 90 min in the dark at room temperature after addition of the 3′ Cy3 N-hydroxysuccinimide ester (Cy3) or 3′ Cy5 N-hydroxysuccinimide ester (Cy5) resuspended in dimethyl sulfoxide (10 μL). Cy dye-labeled cDNA was purified using a Qiaquick PCR purification kit (Qiagen) according to the manufacturer's instructions. Each sample was concentrated on Microcon YM-30 columns (Amicon Bioseparations, Millipore) and quantified by spectrophotometry.
Microarray Hybridization and Data Acquisition
Slides were prehybridized in the hybridization solution containing 1:1 (v/v) formamide (5× SSC, 0.25% SDS and 5× Denhardt's solution, with 1 μg mL−1 denaturated salmon sperm DNA [Stratagene] during 2 h at 42°C) then washed sequentially in water and isopropanol and dried by centrifugation at 450g for 1 min. A gene frame (ABgene; Epsom) was applied around the spotted area and closed by a coverslip. The pooled Cy3- and Cy5-labeled cDNAs were mixed with 75 μL of hybridization solution and 0.5 μg mL−1 denaturated salmon sperm DNA. The probe solution was boiled for 1 min and injected into the frame. Slides were incubated at 42°C for 16 h in a glass beaker placed in a water bath, then washed sequentially at room temperature in 1× SSC, 0.1% SDS for 5 min, 0.1× SSC, 0.1% SDS for 5 min, and 0.1× SSC for 1 min. Slides were dried by centrifugation at 450g for 3 min. Microarray slides were then scanned with a Genepix 4000 B fluorescence reader (Axon Instruments) using Genepix 3.0 image acquisition software with photomultiplier tube voltage adjusted to 620 V for Cy3 and 700 V for Cy5. Each of the microarray experiments was performed in duplicate with the dyes reversed (i.e. four slides or eight subarrays per experiment). All slides were visually inspected and nonhomogeneous and aberrant spots were flagged.
Statistical Analysis
Raw data, corresponding to the median spot intensities with no background substraction, were submitted to R/MAANOVA v0.91 to 3 software (R package for the analysis of microarray) for data visualization, preprocessing, normalization, and statistical analysis. Raw data were transformed (Cui et al., 2003) using an intensity-based LOWESS function (using a smoother span of 20%). After visual validation of the preprocessing step on ratio versus intensity plot for each slide, data were submitted to a two-stage ANOVA (Wolfinger et al., 2001). Replicates were averaged, and variance of systematic effects was evaluated by the normalization ANOVA with the following model: ygi,jk = μ + Ai + Dj + ɛgijk, where ygi,jk is the base-2 logarithm of measurement from gene g, μ is the overall mean signal, Ai is the main effect for arrays, Dj is the main effect for dyes, and ɛ is the error. Normalized values for each gene (ygi,jk corrected by the substraction of the fitted values for the main effects Ai + Dj) were used as input for the gene by gene model: rgijk = Gg + (GA)gi + (GD)gj + (GS)gk + egijk. For each gene, differences between the gene-specific sample effects (GS)gk were tested using F-tests. Corresponding P values are based upon the hybrid test (F2) of R/MAANOVA v0.91 to 3 (Cui et al., 2003) that uses a combination of global and gene-specific variance estimates in the denominator of the statistics. To avoid distributional assumptions, the F-test was performed with restricted residual shuffling and 1,000 iterations. The calculated P values were adjusted with the linear step-up procedure included in R/MAANOVA in order to take into account the false discovery rate. In an additional step, the mean and sd of the normalized values of the negative controls were calculated, and the genes whose normalized values were less than mean plus 2 sd of negative controls (cutoff value) for all tissue or organ comparisons were excluded from further analysis. Expression profile data clustering was done on the log2-based relative expression values of the genes (available online at http://cbi.labri.fr/outils/data/Tomato/Sup_Tomato.html) using EPCLUST (http://ep.ebi.ac.uk/EP/EPCLUST/) with the correlation-based distance measure and the average linkage clustering method. The relative expression value represents the expression level of a gene g in a sample k relative to the weighted average expression of that gene over all of the samples in the experiment, as described by Wu et al. (2003).
RT-PCR Analyses
For the RT-PCR experiments, 2 μg of DNase-treated total RNA were denatured and reverse transcribed using 0.5 μm oligo(dT)20 to prime the reaction. The RT product was diluted 10-fold in water, and aliquots of 4 μL were used in PCR reactions (40 μL final volume) in the presence of 0.4 μm clone-specific primers. The primers used for the 24 clones tested are presented as supplemental data online (primer table). Tomato actin (accession no. U60480) was used as a constitutive control. PCR reaction parameters were as follows: 5 min at 95°C, 20 cycles of 30 s at 95°C, 45 s at 60°C, and 45 s at 72°C, and 5 min at 72°C. The PCR products (10 μL) were separated by electrophoresis on a 1% (w/v) agarose 1× Tris-acetate EDTA gel. After denaturation with 0.4 n NaOH, DNA was blotted onto Hybond N+ membrane (Amersham Biosciences) and hybridized at 65°C with radiolabeled specific probes as described previously (Lemaire-Chamley et al., 2000).
Cytological Studies
Ovaries at anthesis and fruit samples (6, 12, and 25 DPA) were collected, cut (approximately 0.3–0.6-mm-thick pieces), and immersed in 2.5% (p/v) glutaraldehyde in 100 mm phosphate buffer, pH 7.2, for 3 to 4 h at room temperature under partial vacuum. The samples were rinsed, dehydrated by an ethanol series, and embedded in Technovit 7100 (Kulzer). Sections (3 μm) were obtained with glass knives, stained with 0.04% (p/v) toluidine blue, and photographed on a Zeiss Axiophot microscope with a Spot digital camera (Diagnostic instruments). Measurements were made using Image Pro Plus software (Media Cybernetics).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AJ784436 to AJ784774, AJ785005 to AJ785280, AJ785296 to AJ785566, and AJ831846 to AJ832104.
Supplementary Material
Acknowledgments
We thank Christian Chevalier for valuable comments and Bérénice Ricard for English editing of the manuscript. We also thank Philippe Gallusci and Jean-Pierre Renaudin for in silico screening of the tomato EST database for genes related to signaling processes, Mathilde Causse for access to Ferum cultures, and Patricia Coat for greenhouse experiments.
This work was supported by the Région Aquitaine, the Fonds Européen de Développement Régional, the Groupement d'Intérêt Économique “Fruits et Légumes,” and the French Tomato Genome Program from the Institut National de la Recherche Agronomique.
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.063719.
References
- Alba R, Fei Z, Payton P, Liu Y, Moore SL, Debbie P, Cohn J, D'Ascenzo M, Gordon JS, Rose JK, et al (2004) ESTs, cDNA microarrays, and gene expression profiling: tools for dissecting plant physiology and development. Plant J 39: 697–714 [DOI] [PubMed] [Google Scholar]
- Balbi V, Lomax TL (2003) Regulation of early tomato fruit development by the Diageotropica gene. Plant Physiol 131: 186–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker L, Kühn C, Weise A, Schulz A, Gebhardt C, Hirner B, Hellman H, Schulze W, Ward JM, Frommer WB (2000) SUT2, a putative sucrose sensor in sieve elements. Plant Cell 12: 1153–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter CJ, Carrari F, Bauke A, Overy S, Hill SA, Quick P, Fernie AR, Sweetlove LJ (2005) Fruit carbohydrate metabolism in an introgression line of tomato with increased fruit soluble solids. Plant Cell Physiol 46: 425–437 [DOI] [PubMed] [Google Scholar]
- Benkova E, Michniewicz M, Sauer M, Teichmann T, Seifertova D, Jurgens G, Friml J (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 26: 591–602 [DOI] [PubMed] [Google Scholar]
- Bohner J, Bangerth F (1988) Effects of fruit set sequence and defoliation on cell number, cell size and hormone levels of tomato fruits (Lycopersicon esculentum Mill.) within a truss. Plant Growth Regul 7: 141–155 [Google Scholar]
- Busi MV, Bustamante C, D'Angelo C, Hidlago-Cuevas M, Boggio SB, Valle EM, Zabaleta E (2003) MADS-box genes expressed during tomato seed and fruit development. Plant Mol Biol 2003: 801–815 [DOI] [PubMed] [Google Scholar]
- Carmi N, Salts Y, Dedicova B, Shabtai S, Barg R (2003) Induction of parthenocarpy in tomato via specific expression of the rolB gene in the ovary. Planta 217: 726–735 [DOI] [PubMed] [Google Scholar]
- Catala C, Rose JKC, Benett AB (2000) Auxin-regulated genes encoding cell wall-modifying proteins that are expressed during tomato fruit growth. Plant Physiol 122: 527–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causse M, Duffe P, Gomez MC, Buret M, Damidaux R, Zamir D, Gur A, Chevalier C, Lemaire-Chamley M, Rothan C (2004) A genetic map of candidate genes and QTLs involved in tomato fruit size and composition. J Exp Bot 55: 1671–1685 [DOI] [PubMed] [Google Scholar]
- Chen GP, Wilson ID, Kim SH, Grierson D (2001) Inhibiting expression of a tomato ripening-associated membrane protein increases organic acids and reduces sugar levels of fruit. Planta 212: 799–807 [DOI] [PubMed] [Google Scholar]
- Cheng GW, Huber DJ (1996) Alterations in structural polysaccharides during liquefaction of tomato locule tissue. Plant Physiol 111: 447–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong B, Liu J, Tanksley SD (2002) Natural alleles at a tomato fruit size quantitative trait locus differ by heterochronic regulatory mutations. Proc Natl Acad Sci USA 99: 13606–13611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ (1997) Assembly and enlargement of the primary cell wall in plants. Annu Rev Cell Dev Biol 13: 171–201 [DOI] [PubMed] [Google Scholar]
- Cui X, Kerr MK, Churchill GA (2003) Transformations for cDNA microarray data. Stat Appl Genet Mol Biol 2: Art4 [DOI] [PubMed]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5: 199–206 [DOI] [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P (1998) Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res 8: 175–185 [DOI] [PubMed] [Google Scholar]
- Favaro R, Pinyopich A, Battaglia R, Kooiker M, Borghi L, Ditta G, Yanofsky MF, Kater MM, Colombo L (2003) MADS-box protein complexes control carpel and ovule development in Arabidopsis. Plant Cell 15: 2603–2611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrandiz C, Pelaz S, Yanofsky MF (1999) Control of carpel and fruit development in Arabidopsis. Annu Rev Biochem 68: 321–354 [DOI] [PubMed] [Google Scholar]
- Frary A, Nesbitt TC, Grandillo S, Knaap E, Cong B, Liu J, Meller J, Elber R, Alpert KB, Tanksley SD (2000) fw2.2: a quantitative trait locus key to the evolution of tomato fruit size. Science 289: 85–88 [DOI] [PubMed] [Google Scholar]
- Fridman E, Pleban T, Zamir D (2000) A recombination hotspot delimits a wild-species quantitative trait locus for tomato sugar content to 484 bp within an invertase gene. Proc Natl Acad Sci USA 97: 4718–4723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser CS, Robinson-Beers K (1993) Pistil development. Plant Cell 5: 1231–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillaspy G, Ben-David H, Gruissem W (1993) Fruits: a developmental perspective. Plant Cell 5: 1439–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni JJ (2001) Molecular biology of fruit maturation and ripening. Annu Rev Plant Physiol Plant Mol Biol 52: 725–749 [DOI] [PubMed] [Google Scholar]
- Giovannoni JJ (2004) Genetic regulation of fruit development and ripening. Plant Cell (Suppl) 16: S170–S180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillet C, Just D, Benard N, Destrac-Irvine A, Baldet P, Hernould M, Causse M, Raymond P, Rothan C (2002) A fruit-specific phosphoenolpyruvate carboxylase is related to rapid growth of tomato fruit. Planta 214: 717–726 [DOI] [PubMed] [Google Scholar]
- Heinekamp T, Strathmann A, Kuhlmann M, Froissard M, Muller A, Perrot-Rechenmann C, Droge-Laser W (2004) The tobacco bZIP transcription factor BZI-1 binds the GH3 promoter in vivo and modulates auxin-induced transcription. Plant J 38: 298–309 [DOI] [PubMed] [Google Scholar]
- Ho LC (1996) Tomato. In E Zomstein, AA Schaffer, eds, Photoassimilate Distribution in Plants. Marcel Dekker, New York, pp 709–728
- Hsieh HL, Okamoto H, Wang M, Ang LH, Matsui M, Goodman H, Deng XW (2000) FIN219, an auxin-regulated gene, defines a link between phytochrome A and the downstream regulator COP1 in light control of Arabidopsis development. Genes Dev 14: 1958–1970 [PMC free article] [PubMed] [Google Scholar]
- Johnson KL, Jones BJ, Bacic A, Schultz CJ (2003) The fasciclin-like arabinogalactan proteins of Arabidopsis. A multigene family of putative cell adhesion molecules. Plant Physiol 133: 1911–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B, Frasse P, Olmos E, Zegzouti H, Guo Li Z, Latché A, Pech JC, Bouzayen M (2002) Down-regulation of DR12, an auxin-response-factor homolog, in the tomato results in a pleiotropic phenotype including dark green and blotchy ripening fruit. Plant J 32: 603–613 [DOI] [PubMed] [Google Scholar]
- Joubès J, Phan TH, Just D, Rothan C, Bergounioux C, Raymond P, Chevalier C (1999) Molecular and biochemical characterization of the involvement of cyclin-dependent kinase A during the early development of tomato fruit. Plant Physiol 121: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubès J, Walsh D, Raymond P, Chevalier C (2000) Molecular characterization of the expression of distinct classes of cyclins during the early development of tomato fruit. Planta 211: 430–439 [DOI] [PubMed] [Google Scholar]
- Kader JC (1996) Lipid-transfer proteins in plants. Annu Rev Plant Physiol Plant Mol Biol 47: 627–654 [DOI] [PubMed] [Google Scholar]
- Knapp S (2002) Tobacco to tomatoes: a phylogenetic perspective on fruit diversity on the Solanaceae. J Exp Bot 53: 2001–2022 [DOI] [PubMed] [Google Scholar]
- Koch K (2004) Sucrose metabolism: regulatory mechanisms and pivotal roles in sugar sensing and plant development. Curr Opin Plant Biol 7: 235–246 [DOI] [PubMed] [Google Scholar]
- Kotilainen M, Helaruttia Y, Mehto M, Pöllänen E, Albert VA, Elomaa P, Teeri TH (1999) GEG participates in the regulation of cell and organ shape during corolla and carpel development in Gerbera hybrida. Plant Cell 11: 1093–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laval-Martin D, Farineau J, Diamond J (1977) Light versus dark carbon metabolism in cherry tomato fruits. I. Occurence of photosynthesis: studies of the intermediates. Plant Physiol 60: 872–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire-Chamley M, Petit J, Causse M, Raymond P, Chevalier C (2000) Identification of differentially expressed genes during early tomato fruit development. Characterization of a novel cDNA coding for a RAD23 protein. Aust J Plant Physiol 27: 911–920 [Google Scholar]
- Leonhardt N, Kwak JM, Robert N, Waner D, Leonhardt G, Schroeder JI (2004) Microarray expression analyses of Arabidopsis guard cells and isolation of a recessive abscissic acid hypersensitive protein phosphatase 2 C mutant. Plant Cell 16: 596–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leubner-Metzger G, Petruzzelli L, Waldvogel R, Vogeli-Lange R, Meins F, Jr. (1998) Ethylene-responsive element binding protein (EREBP) expression and the transcriptional regulation of class I beta-1,3-glucanase during tobacco seed germination. Plant Mol Biol 38: 785–795 [DOI] [PubMed] [Google Scholar]
- Liu J, Van Eck J, Cong B, Tanksley SD (2002) A new class of regulatory genes underlying the cause of pear-shaped tomato fruit. Proc Natl Acad Sci USA 99: 13302–13306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurel C, Chrispeels MJ (2001) Aquaporins. A molecular entry into plant water relation. Plant Physiol 125: 135–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier I, Callan KL, Fleming AJ, Gruissem W (1995) Organ-specific differential regulation of a promoter subfamily for the ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit genes in tomato. Plant Physiol 107: 1105–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menda N, Semel Y, Peled D, Eshed Y, Zamir D (2004) In silico screening of a saturated mutation library of tomato. Plant J 38: 861–872 [DOI] [PubMed] [Google Scholar]
- Miller RT, Christoffels AG, Gopalakrishnan C, Burke J, Ptitsyn AA, Broveak TR, Hide WA (1999) A comprehensive approach to clustering of expressed human gene sequence: the sequence tag alignment and consensus knowledge base. Genome Res 9: 1143–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr WP, Stein M (1969) Fine structure of fruit development in tomato. Can J Plant Sci 49: 549–553 [Google Scholar]
- Muir SR, Collins GF, Robinson S, Hughes S, Bovy A, Ric De Vos CH, van Tunen AJ, Verhoeyen M (2001) Overexpression of petunia chalcone isomerase in tomato results in fruit containing increased levels of flavonols. Nat Biotechnol 18: 666–669 [DOI] [PubMed] [Google Scholar]
- Nguyen-Quoc B, Foyer CH (2001) A role for “futile cycles” involving invertase and sucrose synthase in sucrose metabolism of tomato fruit. J Exp Bot 52: 881–889 [DOI] [PubMed] [Google Scholar]
- Obiadalla-ali H, Fernie AF, Kossman J, Lloyd JR (2004) Developmental analysis of carbohydrate metabolism in tomato (Lycopersicon esculentum cv. Micro-Tom) fruits. Physiol Plant 120: 196–204 [DOI] [PubMed] [Google Scholar]
- Ozga JA, van Huizen R, Reinecke DM (2002) Hormone and seed-specific regulation of pea fruit growth. Plant Physiol 128: 1379–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozga JA, Yu J, Reinecke DM (2003) Pollination-, development-, and auxin-specific regulation of gibberellin 3beta-hydroxylase gene expression in pea fruit and seeds. Plant Physiol 131: 1137–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandolfini T, Rotino GL, Camerini S, Defez R, Spena A (2002) Optimisation of transgene action at the post-transcriptional level: high quality parthenocarpic fruits in industrial tomatoes. BMC Biotechnol 2: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebers M, Kaneta T, Kawaide H, Yamaguchi S, Yang YY, Imai R, Sekimoto H, Kamiya Y (1999) Regulation of gibberellin biosynthesis genes during flower and early fruit development of tomato. Plant J 17: 241–250 [DOI] [PubMed] [Google Scholar]
- Roberts MR (2003) 14-3-3 proteins find new partners in plant cell signalling. Trends Plant Sci 8: 218–223 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Conception M, Gruissem W (1999) Arachidonic acid alters tomato HMG expression and fruit growth and induces 3-hydroxy-3-methylglutaryl coenzyme A reductase-independent lycopene accumulation. Plant Physiol 119: 41–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer AA, Petreikov M (1997) Sucrose-to-starch metabolism in tomato fruit undergoing transient starch accumulation. Plant Physiol 113: 739–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable PS, Hochholdinger F, Nakazono M (2004) Global expression profiling applied to plant development. Curr Opin Plant Biol 7: 50–56 [DOI] [PubMed] [Google Scholar]
- Seymour GB, Manning K, Eriksson EM, Popovich AH, King GJ (2002) Genetic identification and genomic organization of factors affecting fruit texture. J Exp Bot 53: 2065–2071 [DOI] [PubMed] [Google Scholar]
- Smirnoff N, Conklin PL, Loewus FA (2001) Biosynthesis of ascorbic acid in plants: a renaissance. Annu Rev Plant Physiol Plant Mol Biol 52: 437–467 [DOI] [PubMed] [Google Scholar]
- Strathmann A, Kuhlmann M, Heinekamp T, Droge-Laser W (2001) BZI-1 specifically heterodimerises with the tobacco bZIP transcription factors BZI-2, BZI-3/TBZF and BZI-4, and is functionally involved in flower development. Plant J 28: 397–408 [DOI] [PubMed] [Google Scholar]
- Tanksley SD (2004) The genetic, developmental, and molecular bases of fruit size and shape variation in tomato. Plant Cell (Suppl) 16: S181–S189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DS, Davies WJ, Ho LC (1998) Regulation of tomato fruit growth by epidermal cell wall enzymes. Plant Cell Environ 21: 589–599 [Google Scholar]
- Van der Hoeven R, Ronning C, Giovannoni J, Martin G, Tanksley S (2002) Deductions about the number, organization, and evolution of genes in the tomato genome based on analysis of a large expressed sequence tag collection and selective genomic sequencing. Plant Cell 14: 1441–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga A, Bruinsma J (1986) Tomato. In SP Monselise, ed, CRC Handbook of Fruit Set and Development. CRC Press, Boca Raton, FL, pp 461–491
- Windhovel A, Hein I, Dabrowa R, Stockhaus J (2001) Characterization of a novel class of plant homeodomain proteins that bind to the C4 phosphoenolpyruvate carboxylase gene of Flaveria trinervia. Plant Mol Biol 45: 201–214 [DOI] [PubMed] [Google Scholar]
- Wolfinger RD, Gibson G, Wolfinger ED, Bennett L, Hamadeh H, Bushel P, Afshari C, Paules RS (2001) Assessing gene significance from cDNA microarray expression data via mixed models. J Comput Biol 8: 625–637 [DOI] [PubMed] [Google Scholar]
- Wu H, Kerr K, Cui X, Churchill GA (2003) MAANOVA: A software package for the analysis of spotted cDNA microarray experiments. In G Parmigiani, ES Garett, RA Irizarry, SL Zeger, eds, The Analysis of Gene Expression Data. Methods and Software. Springer-Verlag, Heidelberg, pp 313–341
- Zhang H, Uang Z, Chen Q, Tian X, Zhang X, Zhang H, Lu X, Huang D, Huang R (2004) The ethylene-, jasmonate-, abscisic acid- and NaCl-responsive tomato transcription factor JERF1 modulates expression of GCC box-containing genes and salt tolerance in tobacco. Planta 220: 262–270 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.