Abstract
CONSTANS (CO) regulates flowering time by positively regulating expression of two floral integrators, FLOWERING LOCUS T (FT) and SUPPRESSOR OF OVEREXPRESSION OF CO 1 (SOC1), in Arabidopsis (Arabidopsis thaliana). FT and SOC1 have been proposed to act in parallel pathways downstream of CO based on genetic analysis using weak ft alleles, since ft soc1 double mutants showed an additive effect in suppressing the early flowering of CO overexpressor plants. However, this genetic analysis was inconsistent with the sequential induction pattern of FT and SOC1 found in inducible CO overexpressor plants. Hence, to identify genetic interactions of CO, FT, and SOC1, we carried out genetic and expression analyses with a newly isolated T-DNA allele of FT, ft-10. We found that ft-10 almost completely suppressed the early flowering phenotype of CO overexpressor plants, whereas soc1-2 partially suppressed the phenotype, suggesting that FT is the major output of CO. Expression of SOC1 was altered in gain- or loss-of-function mutants of FT, whereas expression of FT remained unchanged in gain- or loss-of-function mutants of SOC1, suggesting that FT positively regulates SOC1 to promote flowering. In addition, inactivation of FT caused down-regulation of SOC1 even in plants overexpressing CO, indicating that FT is required for SOC1 induction by CO. Taken together, these data suggest that CO activates SOC1 through FT to promote flowering in Arabidopsis.
The phase transition to flowering in plants is precisely controlled by environmental conditions and endogenous developmental cues so that plants produce their progeny under favorable conditions. The response to multiple factors suggests the existence of a complex network regulating this phase transition in plants (Koornneef et al., 1998). To identify genes that control the transition, mutants that showed accelerated or delayed flowering under different conditions, commonly known as flowering-time mutants, have been isolated (Redei, 1975). These mutants were grouped according to their responses to various physiological conditions and then integrated into genetic pathways to explain the control of flowering time. Four floral promotion pathways have been genetically identified in Arabidopsis (Arabidopsis thaliana): the photoperiod, autonomous, vernalization, and GA pathways (Mouradov et al., 2002). Among these pathways, genes within the photoperiod pathway, or the long-day pathway, play an important role in controlling flowering time (Komeda, 2004), since Arabidopsis is a facultative long-day plant.
One of the central regulators in the photoperiod pathway is CONSTANS (CO), which encodes a nuclear protein that contains a CCT motif and two B-box-type zinc-finger domains (Putterill et al., 1995). Loss of CO function delays the phase transition, whereas gain of function of CO accelerates it, suggesting that CO positively regulates flowering time in Arabidopsis. Furthermore, CO mRNA levels show a circadian rhythm under continuous light, such that CO mRNA levels peak at night and are reduced during the day (Suarez-Lopez et al., 2001). The circadian pattern is altered in circadian clock mutants, such as lhy and elf3-1, but constitutive expression of CO does not alter the circadian rhythm of the CHLOROPHYLL A/B-BINDING PROTEIN gene, suggesting that CO acts as a circadian clock output gene to mediate flowering (Suarez-Lopez et al., 2001; Yanovsky and Kay, 2002).
CO controls flowering time in Arabidopsis by positively regulating two floral integrators, FLOWERING LOCUS T (FT; Kardailsky et al., 1999; Kobayashi et al., 1999) and SUPPRESSOR OF OVEREXPRESSION OF CO 1 (SOC1; Lee et al., 2000; Samach et al., 2000). FT and SOC1 encode a protein similar to the RAF kinase inhibitor protein and a MADS box transcription factor, respectively. These two integrators are early targets of CO within the photoperiod pathway since up-regulation of FT and SOC1 was detected in inducible CO overexpressor plants carrying the 35S∷CO:GR construct, in the presence of cycloheximide (Samach et al., 2000). FT and SOC1 have been confirmed as downstream mediators of CO by a genetic analysis in which FT and SOC1 were identified as strong suppressors of CO overexpressor plants (Onouchi et al., 2000). Furthermore, the early flowering phenotype of CO overexpressor plants was suppressed, albeit weakly, by either ft or soc1 mutations, suggesting that FT and SOC1 are required for the early flowering of 35S∷CO plants. The delay in flowering time in ft soc1 double mutants was additive, compared to the effects of each single mutant. The phenotype of ft soc1 35S∷CO plants was similar to loss-of-function mutants of CO under long-day conditions, implying that inactivation of both FT and SOC1 suppresses signaling from CO. These results, together with the observations that no single mutation that completely suppresses the early flowering of 35S∷CO plants has been isolated, have been used to conclude that CO activates two parallel pathways, which include FT and SOC1, to promote flowering (Samach et al., 2000).
Although genetic studies have inferred that FT and SOC1 act in parallel pathways downstream of CO, up-regulation of FT is detected before SOC1 in CO overexpressor plants (Samach et al., 2000). Furthermore, the genetic studies were performed using weaker ft alleles (Onouchi et al., 2000). We hypothesized that a null allele of FT would produce a stronger suppression of early flowering in CO overexpressor plants. To test this hypothesis, we investigated the genetic interactions between CO and FT with a newly isolated T-DNA-tagged allele of FT, ft-10, in which FT mRNA is almost undetectable. We also examined the expression of FT and SOC1 to determine whether they act independently in parallel pathways.
Here we show that ft-10 almost completely suppressed the early flowering phenotype of CO overexpressor plants, whereas a T-DNA allele of SOC1 only partially suppressed early flowering. These data suggest that FT is the major target of CO. In addition, expression of SOC1 was altered in gain- or loss-of-function mutants of FT, whereas expression of FT remained unchanged in gain- or loss-of-function mutants of SOC1. These data suggest that FT and SOC1 do not act in parallel, but rather that FT positively regulates expression of SOC1. Furthermore, inactivation of FT resulted in down-regulation of SOC1 even in the presence of CO overexpression, suggesting that FT is required for SOC1 induction by CO. These results suggest that FT is the major output of CO and mediates activation of SOC1 by CO to promote flowering in Arabidopsis.
RESULTS
ft-10 Almost Completely Suppresses the Early Flowering Phenotype of CO Overexpressor Plants
To investigate genetic interactions between CO and FT, we used a new insertional allele of FT (290E08) isolated from a T-DNA library generated by GABI-Kat (http://www.gabi-kat.de/), in which a T-DNA is inserted in the first intron of FT (Fig. 1A; Rosso et al., 2003; Hanzawa et al., 2005). We obtained and plated seeds of the putative insertional mutant of FT and identified a line that was resistant to sulfadiazine. The segregation ratio for sulfadiazine resistance was approximately 3:1, indicating that a single T-DNA is present in the line. This line was PCR genotyped using the JH2295, JH2296, and JH2297 primers to confirm that the line was homozygous for a T-DNA insertion. A single 926-bp PCR product was amplified from the line, confirming homozygosity (Fig. 1B). The line was designated ft-10, and this insertional mutant was used in the following experiments.
Figure 1.
Flowering time of an ft-10 allele and its genetic interaction with 35S∷CO plants. A, Map of a T-DNA insertion 844 bases from the A of the start codon of FT; the allele was named ft-10. Black boxes indicate the four exons of FT. B, PCR genotyping of the ft-10 allele. Genotyping primer sets (JH2295, JH2296, and JH2297) were used to identify a homozygous line. Homozygous ft-10 plants produced a single band of 926 bp in size, whereas wild-type plants produced a band of 1,392 bp in size. M, One-kilobase ladder. C, An RT-PCR experiment showing that ft-10 was a strong knock-down allele. FT mRNA abundance in ft-10 plants was compared to ft-1 (Col), a point mutation allele of FT, and to wild-type Col plants. D and E, Flowering time and phenotypes of 35S∷CO, ft-10, 35S∷CO ft-10, soc1-2, 35S∷CO soc1-2, and wild-type Columbia (WT Col) plants under long-day conditions. ft-10 almost completely suppressed the early flowering phenotype of 35S∷CO plants, whereas soc1-2 partially suppressed the early flowering.
We first monitored the expression levels of FT in ft-10 plants by reverse transcription (RT)-PCR to examine whether ft-10 was an RNA null allele. Total RNA extracted from whole seedlings grown for 10 d under long-day conditions was used to synthesize cDNA. We compared FT expression levels in wild-type and ft-10 plants. Since ft-10 plants were in the Columbia (Col) background, we used an introgressed ft-1 allele in the Col background (ft-1 [Col]) as an additional control. FT mRNA levels are only slightly reduced in ft-1 (Col) plants, indicating that transcription of FT was largely normal in ft-1 (Col) plants. In contrast, FT expression in ft-10 plants was greatly reduced (Fig. 1C), but ft-10 was not completely RNA null.
We measured the flowering time of ft-10 plants under long-day conditions to determine whether a decrease in FT mRNA delayed flowering time. ft-10 plants flowered with 40.6 leaves, whereas wild-type plants flowered with 15.0 leaves, demonstrating that ft-10 plants showed a late-flowering phenotype. In contrast, ft-1 (Col) plants flowered with 33.5 leaves under the same conditions, showing that the T-DNA insertion in ft-10 caused a stronger effect than the point mutation in ft-1 (Fig. 1D).
Since ft-10 was identified as a strong allele showing a late-flowering phenotype, we used ft-10 plants to determine the genetic interactions between FT and CO. To facilitate genetic studies with ft-10 plants, 35S∷CO plants were generated in the Col background. The 35S∷CO plants showed early flowering (6.6 leaves) with formation of a determinate inflorescence under long-day conditions (Fig. 1E). The 35S∷CO plants were crossed with ft-10 plants, and flowering time of the resulting double mutants under long-day conditions was measured. The 35S∷CO ft-10 plants flowered with 37.6 leaves, and ft-10 plants flowered with 40.6 leaves. The flowering times were not significantly different (Fig. 1D). In contrast, 35S∷CO soc1-2 plants flowered with 12.3 leaves, whereas soc1-2 plants flowered with 29.4 leaves, suggesting that inactivation of SOC1 slightly suppresses the early flowering of CO overexpressor plants. This analysis shows that ft-10 strongly suppresses the early flowering of 35S∷CO plants, suggesting that FT is the primary downstream target of CO to promote flowering.
FT Positively Regulates Expression Levels of SOC1
Parallel roles of FT and SOC1 downstream of CO were proposed using ft soc1 double mutants, which largely suppressed the phenotype of the 35S∷CO plants (Samach et al., 2000). However, other data have suggested that FT may act earlier than SOC1 in response to induction of CO (Samach et al., 2000). Therefore, we examined whether inactivation or up-regulation of one gene affected expression of the other gene. We used 35S∷FT and 35S∷SOC1 plants for gain-of-function mutants of FT and SOC1, respectively, and ft-10 and soc1-2 plants (Lee et al., 2000) for loss-of-function mutants of FT and SOC1. All these mutants were in the same genetic background (Col). We extracted total RNA from 6-, 8-, 10-, and 12-d-old plants and examined the time course of SOC1 expression by RT-PCR.
Activation or inactivation of FT altered expression of SOC1; however, gain or loss of SOC1 function did not alter expression of FT (Fig. 2). Expression of SOC1 is greatly up-regulated in 35S∷FT plants, such that expression levels of SOC1 in 35S∷FT plants were similar to those of 35S∷SOC1 plants, as previously suggested (Moon et al., 2005). Up-regulation of SOC1 was delayed in ft-10 plants, indicating that changes in FT expression affected expression of SOC1. In contrast, expression levels of FT in 35S∷SOC1 plants did not increase over the time tested; rather, the levels were similar to those of wild-type plants. Furthermore, expression levels of FT in soc1-2 plants were comparable to those of wild-type plants. These results demonstrate that expression of FT is not affected by SOC1 and that FT is likely to be upstream of SOC1. This experiment further suggests that FT and SOC1 are not independent, but rather that FT positively regulates SOC1. These data contradict a recent model suggesting parallel roles for FT and SOC1 (Onouchi et al., 2000; Samach et al., 2000). To confirm whether FT positively regulates SOC1, we employed a β-glucuronidase (GUS) reporter assay. We used FT∷GUS (Takada and Goto, 2003) and SOC1∷GUS (I. Lee, personal communication) plants to visualize expression of FT and SOC1, respectively. We generated 35S∷FT/+ SOC1∷GUS/+ plants to examine whether SOC1 promoter-driven expression of GUS was affected by overexpression of FT. The 35S∷SOC1/+ FT∷GUS/+ plants were generated to test whether FT promoter-driven expression of GUS was altered by overexpression of SOC1.
Figure 2.
Time-course expression of FT, SOC1, and AP1 in 35S∷FT, 35S∷SOC1, ft-10, soc1-2, and wild-type plants grown under long-day conditions. soc1-2 is a T-DNA-tagged allele of SOC1 (Lee et al., 2000). Gene expression levels of FT and SOC1 in 6-, 8-, 10-, and 12-d-old seedlings were measured. AP1 and UBQ10 were used as a molecular marker of floral transition and an internal positive control, respectively.
A histochemical GUS assay showed that expression of SOC1 was dependent on the mRNA level of FT. In SOC1∷GUS plants, GUS staining was faintly observed in the shoot apex and vascular bundles in cotyledons (Fig. 3A). In contrast, strong GUS staining was observed in the shoot apex and roots in 35S∷FT/+ SOC1∷GUS/+ plants (Fig. 3, B and E). In addition, increased GUS staining was seen in vascular bundles of cotyledons (Fig. 3C). This suggests that SOC1 expression is induced by FT and that the strong SOC1 expression at the apical region during the vegetative phase is associated with the early flowering of 35S∷FT plants. In contrast, similar localization patterns of GUS staining with similar staining intensities were observed in 35S∷SOC1/+ FT∷GUS/+ plants (Fig. 3F). The vascular tissue-specific GUS expression of FT∷GUS was not altered by activation of SOC1 (Fig. 3H). Furthermore, overexpression of SOC1 did not cause ectopic staining in hypocotyls and roots of 35S∷SOC1/+ FT∷GUS/+ plants (Fig. 3, I and J). This indicates that expression of FT was not affected by SOC1. Taken together, the results from the GUS reporter assays were consistent with those of the RT-PCR expression analysis (Fig. 2), suggesting that expression of SOC1 is positively regulated by FT, which is consistent with the previous findings (Michaels et al., 2005).
Figure 3.
Histochemical GUS analysis to examine genetic interactions between FT and SOC1. A, Comparison of GUS-staining pattern of 5-d-old seedlings of SOC1∷GUS (left) and 35S∷FT/+ SOC1∷GUS/+ plants (right). B to E, Close-up views of GUS-staining patterns in different tissues of SOC1∷GUS (left) and 35S∷FT/+ SOC1∷GUS/+ plants (right): apical region (B), cotyledon (C), hypocotyl (D), and root (E). An arrowhead indicates faint staining in the shoot apex of a SOC1∷GUS/+ seedling. F, Comparison of the staining pattern of 10-d-old seedlings of FT∷GUS (left) and 35S∷SOC1/+ FT∷GUS/+ plants (right). G to J, Close-up views of staining patterns observed in different tissues of FT∷GUS (left) and 35S∷SOC1/+ FT∷GUS/+ plants (right); apical region (G), cotyledon (H), hypocotyl (I), and root (J).
Double Overexpression of FT and SOC1 Does Not Have an Additive Effect
To confirm the results obtained from the RT-PCR analysis and GUS reporter assay, we investigated the genetic interactions between FT and SOC1. We measured the flowering time of plants in which both FT and SOC1 are overexpressed to examine whether the mutations had an additive effect. The single overexpressor plants, 35S∷FT and 35S∷SOC1, flowered with 4.0 and 8.6 leaves under long-day conditions (Fig. 4A), respectively. Homozygous 35S∷FT 35S∷SOC1 plants flowered with 3.9 leaves under the same conditions, indicating that the phenotype of the double-overexpressor plants was similar to the 35S∷FT plants, implying that FT and SOC1 do not act in parallel. This result is not consistent with previous genetic results that showed an additive effect of FT and SOC1 overexpression (Moon et al., 2005). This raises the possibility that the difference in flowering time was due to the 35S promoter, which may cause overexpression of SOC1 not reflective of the normal role of SOC1. Thus, to confirm our genetic data, we generated additional double-overexpressor plants using an activation-tagged allele of SOC1, soc1-101D (Lee et al., 2000). Flowering time of the resulting 35S∷FT/+ soc1-101D/+ plants, in which both FT and SOC1 were activation tagged, was measured. The 35S∷FT/+ and soc1-101D/+ plants that were used for controls flowered with 4.2 and 5.5 leaves, respectively. In contrast, 35S∷FT/+ soc1-101D/+ plants exhibited an intermediate flowering time (5.0 leaves; Fig. 4). These results using the activation-tagged lines and the 35S promoter-driven overexpressor lines indicate that overexpression of FT and SOC1 did not have an additive effect. However, soc1-2 only partially suppressed the early flowering of 35S∷FT plants (8.4 leaves), suggesting that SOC1 is not the only gene downstream of FT. Interestingly, the ft-10 soc1-2 double mutant flowered with 52.0 leaves, showing an additive delay in flowering time.
Figure 4.
Genetic interactions of FT and SOC1. Flowering time (A) and phenotypes (B) of single overexpressors, double overexpressors, and double mutants under long-day conditions are shown. All plants were in the Col background. soc1-101D is an activation-tagged line of SOC1 (Lee et al., 2000). The total number of primary leaves formed was scored in at least 10 plants to measure flowering time. The horizontal T-bars indicate the sd.
Inactivation of FT Causes Down-Regulation of SOC1 in the Presence of CO Overexpression
Genetic interaction studies using ft-10 together with expression analysis indicated that FT, which primarily mediates signaling inputs from CO, positively regulates SOC1. These results strongly suggest that FT acts downstream of CO and SOC1 in turn acts downstream of FT. Therefore, to determine the signaling network downstream of CO, we monitored the expression level of SOC1 in 6-, 8-, 10-, and 12-d-old 35S∷CO, ft-10, 35S∷CO ft-10, and wild-type plants. If FT mediates SOC1 activation by CO, inactivation of FT would cause down-regulation of SOC1 even in the presence of abundant CO mRNA.
Indeed, inactivation of FT caused down-regulation of SOC1 in 35S∷CO ft-10 plants. Although the CO mRNA level was greatly up-regulated in 35S∷CO ft-10 plants, expression of SOC1 was significantly down-regulated over the time tested (Fig. 5). The SOC1 expression level in 35S∷CO ft-10 plants was similar to that of wild-type plants. Considering that expression of SOC1 was greatly up-regulated in 35S∷CO plants, these data suggest that loss of FT function altered expression of SOC1 even in the presence of abundant CO mRNA, which is a strong activator of SOC1 in wild-type plants. This result suggests that FT is required for SOC1 activation by CO to promote flowering.
Figure 5.
Expression of SOC1 in 6-, 8-, 10-, and 12-d-old 35S∷CO ft-10, ft-10, 35S∷CO, and wild-type Col plants grown under long-day conditions. Semiquantitative RT-PCR was used to measure expression of SOC1. UBQ10 and CO were used for internal positive controls.
DISCUSSION
FT Is the Major Target of CO in the Photoperiod Pathway
FT is one of the early targets of CO because inactivation of FT delayed early flowering resulting from the constitutive expression of CO (Onouchi et al., 2000). However, since previously identified ft alleles only partially suppressed the early flowering phenotype of 35S∷CO plants, the existence of additional genes downstream of CO was inferred and led to the identification of SOC1 (Samach et al., 2000). Indeed, ft soc1 double mutants proposed an additive effect in suppressing the early flowering phenotype of 35S∷CO plants. Therefore, parallel flowering-time pathways downstream of CO have been proposed (Samach et al., 2000). However, our data suggest that FT is the major target of CO, as suggested by observation that ft-10 was almost completely epistatic to 35S∷CO (Fig. 1).
This genetic result revisits the molecular data obtained by Samach et al. (2000) using a dexamethasone (DEX)-inducible CO construct. The authors showed that FT is rapidly up-regulated within 2 h of DEX treatment, whereas SOC1 was up-regulated 4 h after DEX induction. According to this sequential induction pattern, it appeared that FT is an earlier acting gene in response to CO activation. However, since this sequential induction pattern is largely incompatible with the genetic analysis, in which preexisting ft soc1 double mutants showed additive effects on flowering time (Onouchi et al., 2000), a model of parallel pathways was proposed instead. In contrast, the almost complete suppression of the early flowering of 35S∷CO plants by ft-10 (Fig. 1) is consistent with the sequential induction model. In addition, incomplete suppression by soc1-2 also supports the interpretation that FT is the major target of CO. Our interpretation is also consistent with the previous finding that showed the genomic response of co and ft mutants using microarray analysis (Schmid et al., 2003). The authors showed that co and ft had very similar expression profiles and, therefore, suggested that FT is the major output of CO at the shoot apex. Our genetic interaction data using ft-10 strongly supports this interpretation. Considering that CO is a B-type zinc-finger protein, it remains to be determined whether CO directly binds to FT or recruits additional components to promote flowering, as previously suggested (Hepworth et al., 2002).
An important question is why previously identified ft alleles, for example, ft-1 and ft-7, did not suppress the early flowering phenotype of 35S∷CO plants. The partial suppression of the CO overexpressor phenotype by ft-1 (Onouchi et al., 2000) can be explained by different levels of FT transcripts between ft-1 and ft-10. Unlike ft-10, transcription of FT is largely normal in ft-1 plants (Fig. 1); thus, a mutant form of FT protein is likely produced. This mutant ft-1 protein appeared to be partially functional since ft-1 showed a weaker phenotype than did ft-10. Moreover, the mutant ft-1 protein is probably overproduced in 35S∷CO ft-1 plants because ft-1 mRNA was up-regulated by overexpression of CO (K.S. Chung and J.H. Ahn, unpublished data). The overproduced ft-1 proteins in 35S∷CO ft-1 plants may weakly promote flowering, for instance, by increasing expression of SOC1. The ft-7 (=ft-2) mutant, in which a point mutation changes Trp-138 to a stop codon (TGA), most likely produced a truncated FT protein, which is predicted to show a stronger effect. Indeed, ft-7 plants showed a later flowering phenotype than did ft-1 plants, indicating that the ft-7 mutation has a more severe effect. The difference between ft-1 and ft-7 is seemingly comparable to that between ft-1 and ft-10, suggesting that ft-7 may also be a null or almost null. However, ft-7 partially suppressed the early flowering phenotype of 35S∷CO plants (Onouchi et al., 2000). There are two possibilities to explain the discrepancy. First, the truncated FT protein produced in ft-7 plants is likely to be partially functional. This is inferred by the fact that the truncated FT protein contains a ligand-binding pocket that is important for FT function (Banfield and Brady, 2000; Hanzawa et al., 2005) and an important domain in the C terminus that determines the functional specificity of FT (J.H. Ahn and D. Weigel, unpublished data). Secondly, using different genetic backgrounds for ft-7 may lead to discrepancies in phenotype. Previous genetic analysis with 35S∷CO ft-7 plants used the ecotype Landsberg erecta background (Onouchi et al., 2000), whereas our genetic studies with 35S∷CO ft-10 plants used the Col background. The functional FLOWERING LOCUS C (FLC), which encodes a MADS domain-containing transcription factor that acts as a floral repressor (Michaels and Amasino, 1999; Sheldon et al., 1999), presented in the Col ecotype may further suppress expression of SOC1, causing an additional delay in flowering time in 35S∷CO ft-10 plants. Genetic analysis using an introgressed allele of ft-7 in the Col background may provide a clue to explain this discrepancy.
Given that FT is the major mediator of floral inductive inputs from CO, an important question is whether two other genes induced in 35S∷CO:GR plants, ACS10 and AtP5CS2 (Samach et al., 2000), play a redundant role in flower development. Some evidence suggests that ACS10 and AtP5CS2 do not play a role in reproductive development. First, a null mutation of ACS10 does not have a phenotype associated with flowering time, but the gravitropic curvature of hypocotyls, which is related to auxin distribution and ethylene synthesis, is increased in ACS10 T-DNA mutants (Porter and Harrison, 2003). Second, expression of ACS10 is not affected in co-2 mutants (Schmid et al., 2003) or in other flowering-time mutants such as leafy (lfy) and ft. Third, ACS10 is located between the EMB173 and FHA markers on chromosome 1, which have not been reported to contain flowering-time mutants. Rather, ACS10 encodes an aminotransferase with broad specificity for Asp and aromatic amino acids (Bernier, 2004). Another gene activated in 35S∷CO:GR plants is AtP5CS2, but AtP5CS2 does not appear to be involved in regulation of flowering time. To the best of our knowledge, no loss-of-function mutant of AtP5CS2 has been reported. The loss-of-function phenotype of AtP5CS2 may be inferred by transgenic plants expressing antisense AtP5CS1 (Nanjo et al., 1999), since AtP5CS1 and AtP5CS2 share 85% sequence homology. Transgenic plants expressing antisense AtP5CS1 exhibited severe defects in vegetative tissues, but flowering time was largely normal. Thus, it is likely that down-regulation of AtP5CS2 does not have a flowering-time phenotype. Rather, AtP5CS2 is likely to be involved in stress signaling and low-temperature regulation of gene expression (Gilmour et al., 2000; Fabro et al., 2004).
Nonetheless, why ACS10 and AtP5CS2 are induced in 35S∷CO:GR plants remains unanswered. One possibility is that the inducible version of CO does not precisely duplicate the normal role of CO. For instance, 35S promoter-driven expression of SEP3, a floral organ identity gene, caused early flowering (Honma and Goto, 2001), and overexpression of CO resulted in pleiotropic carpel defects (Jeong and Clark, 2005), suggesting that overexpressor phenotypes are not necessarily correlated with the wild-type function of the genes tested. An alternative explanation is that CO may play a role in vegetative growth. Microarray results showed that CO expression is also detected in vegetative tissues, and both spatial and developmental expression patterns of ACS10 and AtP5CS2 mRNA abundance largely overlap with CO during vegetative development (Schmid et al., 2003). Colocalization of ACS10 and AtP5CS2 with CO suggests that vegetative tissue-specific functions of CO may require ACS10 and AtP5CS2.
Genetic Framework of Flowering-Time Pathways Downstream of CO
Expression analyses in this study showed that FT positively regulates expression of SOC1 (Figs. 2 and 3). Our results are consistent with a previous result obtained from an experiment using an activation-tagged mutant of FT (Michaels et al., 2005). The authors showed that activation of FT or TWIN SISTER OF FT, which acts as a floral pathway integrator redundantly with FT (Yamaguchi et al., 2005), leads to up-regulation of SOC1, even in the presence of functional FRIGIDA and FLC. They further showed that FT activation causes increased SOC1∷GUS expression throughout the plant, similar to our results. In this study, we additionally showed that SOC1∷GUS expression is strongly induced in the shoot apex during vegetative phase by FT activation and, furthermore, that loss of FT function delays expression of SOC1. All these results suggest that SOC1 expression is regulated by FT in the photoperiod pathway.
If FT positively regulates SOC1, then the question of how CO regulates expression of SOC1 arises. Two possible scenarios have been suggested, based on the expression patterns of FT and SOC1 (Michaels et al., 2005). CO may positively regulate SOC1 through indirect mechanisms, such that CO promotes expression of FT, which in turn promotes expression of SOC1. Alternatively, CO could regulate SOC1 by binding to the SOC1 promoter as part of a regulatory complex, as suggested in previous binding assays (Hepworth et al., 2002). Our results support the indirect mechanism of CO function to activate SOC1, since FT is the major target of CO and FT is required for SOC1 activation by CO (Fig. 5). However, it is still possible that genes other than FT contribute, albeit weakly, to mediate floral inductive inputs from CO, since FT is not completely epistatic to CO (Fig. 1).
Although we suggest that FT and SOC1 do not act in parallel, our results with the ft-10 soc1-2 double mutants (Fig. 4) are not consistent with a linear relationship since ft-10 soc1-2 double mutants showed an additive delay. One reason for this discrepancy may be that the additive delay in flowering time is the result of repression of multiple pathways by the soc1 mutation. Although FT most likely mediates flowering signals in the photoperiod pathway, SOC1 is downstream of multiple pathways including the autonomous, GA, and vernalization pathways (Moon et al., 2003). These data suggest that inactivation of SOC1 inhibits signaling in each of the floral promotion pathways (Mouradov et al., 2002). Thus, although FT is sufficient to induce expression of SOC1, suppression of signaling inputs from multiple pathways, due to inactivation of SOC1, may cause an additional delay under long-day conditions. An alternative explanation is that the synergistic delay simply resulted from greater suppression of SOC1 expression in ft-10 plants. Expression of SOC1 is detectable, albeit down-regulated, in ft-10 plants. However, expression of SOC1 is further down-regulated in ft-10 soc1-2 plants, due to a T-DNA insertion in SOC1. The discrepancy can be resolved by a genetic study using null alleles of both FT and SOC1. Nevertheless, we favor the first explanation since SOC1 is involved in more floral promotion pathways than FT (Mouradov et al., 2002; Michaels et al., 2005).
The partial suppressions of the phenotype of 35S∷FT plants by soc1-2 (Fig. 4) and the phenotype of ft-1 plants by 35S∷SOC1 (S.K. Yoo and J.H. Ahn, unpublished data) are not consistent with a simple linear-pathway model. The partial suppression suggests that the floral promotion signal from FT is not solely mediated by SOC1, but that FT activates several downstream mediators. One such candidate is APETALA1 (AP1), which encodes a MADS box protein and functions both as a flowering-time gene and a floral meristem identity gene. AP1 is an early target of FT since the AP1 level is precociously elevated before LFY up-regulation in FT overexpressor plants (Kardailsky et al., 1999). Additionally, ft lfy double mutants have a phenotype associated with loss of function of AP1 (Ruiz-Garcia et al., 1997). Thus, AP1 may be required to promote flowering in response to FT activation. However, it is not clear why ap1 loss-of-function mutants did not suppress early flowering of FT overexpressor plants (Kardailsky et al., 1999). One possibility is that AP1 and SOC1 have redundant roles. We are currently investigating whether ap1-10 soc1-2 double mutants can completely block the accelerated flowering phenotype of 35S∷FT plants. Another candidate for a downstream mediator of FT is FD, a bZIP transcription factor expressed in the shoot apex (Abe et al., 2005; Wigge et al., 2005). Some previous studies suggest that FD is downstream of FT. First, fd loss-of-function mutants strongly repressed the early flowering of 35S∷FT plants, although the loss-of-function phenotype on its own is very weak (Abe et al., 2005; Wigge et al., 2005). Secondly, the FT protein interacts with FD protein by two-hybrid analysis in yeast, as well as genetically (Abe et al., 2005; Wigge et al., 2005). These results indicate that floral promotion by FT requires the activity of FD and suggest that FD is downstream of FT. However, FD does not simply act downstream of FT; rather, FT and FD are interdependent, since overexpression of FD did not rescue the late-flowering phenotype of ft mutants (T. Araki, personal communication) and activation of AP1 by FD correlates with the level of FT mRNA (P.A. Wigge, M.C. Kim, and D. Weigel, personal communication).
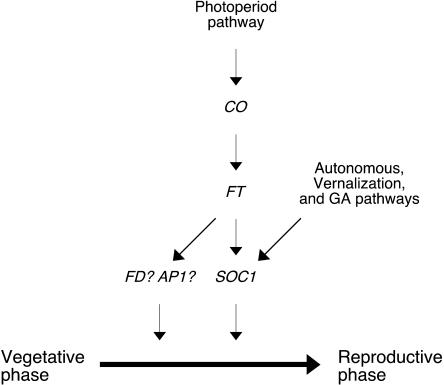
A model of the roles of CO, FT, and SOC1 in the regulation of flowering is presented in Figure 6. It is likely that FT is the major downstream target of CO and SOC1 is in turn activated downstream of FT. Although FT is sufficient to induce expression of SOC1, it appears that these proteins integrate different floral promotion pathways. For instance, SOC1 is mainly regulated by the autonomous pathway through FLC (Moon et al., 2005), whereas FT is strongly dependent on the photoperiod pathway through CO. The effects of photoperiod and vernalization are thus additive and overlap on these floral integrators. FT most likely has additional downstream mediators other than SOC1. Possible candidates for downstream mediators of FT include AP1 and FD.
Figure 6.
Model for genetic interactions of CO, FT, and SOC1 in determination of flowering time in Arabidopsis. FT primarily mediates floral inductive inputs from CO. Although FT positively regulates SOC1, FT appears to have other downstream mediators in parallel with SOC1. Putative candidates for downstream mediators are FD and AP1.
The Spatial Regulation of CO, FT, and SOC1
The spatial pattern of flowering-time gene activities has recently been studied, since little is known about how a flowering stimulus is transmitted to the shoot apex. Classical grafting experiments showed that day length is perceived in leaves, which generate a systemic, graft-transmissible signal. Recently, CO was shown to act noncell autonomously in leaves to induce flowering (An et al., 2004), since phloem-specific expression of CO is sufficient to induce early flowering. FT, the major target of CO, is also expressed in vascular bundles of leaves. Interestingly, phloem-specific expression of CO causes increased FT expression, specifically in the phloem, suggesting that CO activates FT in a cell-autonomous manner in leaves (An et al., 2004). However, SOC1 is expressed in the shoot apex and leaves (Lee et al., 2000; Samach et al., 2000). Thus, to activate SOC1 in the shoot apex to induce flowering, the flowering stimulus generated by FT must be transmitted to the shoot apex. One possibility is that FT itself, either the FT RNA or the FT protein, physically moves to the apex to trigger flowering. FT protein movement is the more likely hypothesis, since microarray results have shown that FT RNA is not detected at the shoot apex (Schmid et al., 2003). The possibility of FT protein movement is further supported by the molecular size of the FT protein (23 kD), which is below the size exclusion limit of plasmodesmata (Imlau et al., 1999). The FT protein may thus freely move to the shoot apex. However, this FT protein movement should be precisely coordinated to induce flowering since the size exclusion limit can change during organ development (Imlau et al., 1999). Nevertheless, we cannot exclude the movement of FT RNA since FT RNA may be present at the shoot apex below the level of detection. An alternative explanation is that FT signaling requires an intermediate component, which generates a downstream mobile signal. A possible candidate for this component is a gene that is genetically downstream of FT but is not expressed in the shoot apex. However, it is still possible that our interpretation on the spatial regulation of CO, FT, and SOC1 based on our expression studies can be misleading because we used the constitutive 35S promoter for overexpression. Taken together, the intercellular signaling processes to transmit a flowering stimulus are largely unknown and await further investigation.
MATERIALS AND METHODS
Plant Materials and Measurement of Flowering Time
Wild-type Arabidopsis (Arabidopsis thaliana ecotype Col) was used to generate transgenic plants. Plants were grown in Sunshine Mix 5 (Sungro Horticulture) under long-day conditions (16/8-h photoperiod at 120 μmol m−2 s−1) at 23°C. SKI083 plants (Kardailsky et al., 1999), in which transcription of FT is increased by four copies of the 35S enhancers, are referred to as 35S∷FT plants in this study. SK231 and pJA1148 plants were generated using the 35S promoter-fused SOC1 cDNA and CO cDNA constructs, respectively, which were kindly provided by Dr. George Coupland (Max Planck Institute, Germany). These SK231 plants and pJA1148 plants were referred to as 35S∷SOC1 and 35S∷CO plants, respectively, in this study. The original ft-1 allele, which is in the ecotype Landsberg erecta background, was introgressed seven times into the Col background to generate the ft-1 (Col) plants used in this study. FT∷GUS, LFY∷GUS, and SOC1∷GUS seeds were kind gifts from Dr. Goji Koto (Research Institute for Biological Science, Japan), Dr. Detlef Weigel (Max Planck Institute), and Dr. Ilha Lee (Seoul National University, Korea), respectively. soc1-2 is a T-DNA insertional mutant of SOC1 that was described previously (Lee et al., 2000). The flowering time of the plants used in this study was measured by scoring the total number of primary leaves of at least 10 plants grown under long-day conditions.
RT-PCR and PCR-Genotyping
Sequence information of the oligonucleotides used in this study is available in Supplemental Table I. The reverse transcriptase-mediated PCR procedure has been described previously (Kardailsky et al., 1999). Total RNA was isolated from whole seedlings using Trizol reagent (Invitrogen Life Technologies) according to the manufacturer's instructions. cDNA was synthesized from 1 μg of total RNA treated with DNaseI. The expression levels were normalized against UBQ10. The JH1061 and JH1063 primers were used to detect FT mRNA. The JH1145 and JH1146 primers were used to measure transcript levels of SOC1. The JH1015 and JH1016 primers were used to monitor the mRNA levels of CO. To genotype soc1-2 plants, a new set of primers, JH2537, JH2538, and JH2539, was used. The JH2295, JH2296, and JH2297 primers were used to genotype ft-10 plants.
Histochemical GUS Assay and Microscopy
GUS staining of whole seedlings and tissue sections were performed as described previously (Sessions et al., 2000). GUS-stained seedlings were photographed using a Nikon SMZ1000 dissection microscope. Stained primary inflorescences were fixed and embedded in paraffin. After deparaffinization, sections of primary inflorescences were examined using a Nikon Optiphot-2 microscope.
Supplementary Material
Acknowledgments
We thank X. Wu, P.A. Wigge, and D. Weigel for critical reading of our manuscript.
This work was supported by the Plant Signaling Network Research Center, Plant Metabolism Research Center, and the Korea Research Foundation (grant no. KRF–2002–015–CS0038 to J.H.A.), and by a grant from the Crop Functional Genomics Center to J.S.L. S.Y.Y., S.K.Y., S.M.H., and S.J.Y. were supported by the BK21 program, and J.H.L. was supported by the Plant Signaling Network Research Center.
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.066928.
References
- Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T (2005) FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309: 1052–1056 [DOI] [PubMed] [Google Scholar]
- An H, Roussot C, Suarez-Lopez P, Corbesier L, Vincent C, Pineiro M, Hepworth S, Mouradov A, Justin S, Turnbull C, et al (2004) CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131: 3615–3626 [DOI] [PubMed] [Google Scholar]
- Banfield MJ, Brady RL (2000) The structure of Antirrhinum centroradialis protein (CEN) suggests a role as a kinase regulator. J Mol Biol 297: 1159–1170 [DOI] [PubMed] [Google Scholar]
- Bernier G (2004) Increased ethylene production in long days does not arise from activation of the ACS10 gene. Flowering Newsletter 38: 49–50 [Google Scholar]
- Fabro G, Kovacs I, Pavet V, Szabados L, Alvarez ME (2004) Proline accumulation and AtP5CS2 gene activation are induced by plant-pathogen incompatible interactions in Arabidopsis. Mol Plant Microbe Interact 17: 343–350 [DOI] [PubMed] [Google Scholar]
- Gilmour SJ, Sebolt AM, Salazar MP, Everard JD, Thomashow MF (2000) Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol 124: 1854–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanzawa Y, Money T, Bradley D (2005) A single amino acid converts a repressor to an activator of flowering. Proc Natl Acad Sci USA 102: 7748–7753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth SR, Valverde F, Ravenscroft D, Mouradov A, Coupland G (2002) Antagonistic regulation of flowering-time gene SOC1 by CONSTANS and FLC via separate promoter motifs. EMBO J 21: 4327–4337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honma T, Goto K (2001) Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409: 525–529 [DOI] [PubMed] [Google Scholar]
- Imlau A, Truernit E, Sauer N (1999) Cell-to-cell and long-distance trafficking of the green fluorescent protein in the phloem and symplastic unloading of the protein into sink tissues. Plant Cell 11: 309–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong S, Clark SE (2005) Photoperiod regulates flower meristem development in Arabidopsis thaliana. Genetics 169: 907–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D (1999) Activation tagging of the floral inducer FT. Science 286: 1962–1965 [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T (1999) A pair of related genes with antagonistic roles in mediating flowering signals. Science 286: 1960–1962 [DOI] [PubMed] [Google Scholar]
- Komeda Y (2004) Genetic regulation of time to flower in Arabidopsis thaliana. Annu Rev Plant Biol 55: 521–535 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Alonso-Blanco C, Peeters AJ, Soppe W (1998) Genetic control of flowering time in Arabidopsis. Annu Rev Plant Physiol Plant Mol Biol 49: 345–370 [DOI] [PubMed] [Google Scholar]
- Lee H, Suh SS, Park E, Cho E, Ahn JH, Kim SG, Lee JS, Kwon YM, Lee I (2000) The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev 14: 2366–2376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM (1999) FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11: 949–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Himelblau E, Kim SY, Schomburg FM, Amasino RM (2005) Integration of flowering signals in winter-annual Arabidopsis. Plant Physiol 137: 149–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J, Lee H, Kim M, Lee I (2005) Analysis of flowering pathway integrators in Arabidopsis. Plant Cell Physiol 46: 292–299 [DOI] [PubMed] [Google Scholar]
- Moon J, Suh SS, Lee H, Choi KR, Hong CB, Paek NC, Kim SG, Lee I (2003) The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J 35: 613–623 [DOI] [PubMed] [Google Scholar]
- Mouradov A, Cremer F, Coupland G (2002) Control of flowering time: interacting pathways as a basis for diversity. Plant Cell (Suppl) 14: S111–S130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanjo T, Kobayashi M, Yoshiba Y, Kakubari Y, Yamaguchi-Shinozaki K, Shinozaki K (1999) Antisense suppression of proline degradation improves tolerance to freezing and salinity in Arabidopsis thaliana. FEBS Lett 461: 205–210 [DOI] [PubMed] [Google Scholar]
- Onouchi H, Igeno MI, Perilleux C, Graves K, Coupland G (2000) Mutagenesis of plants overexpressing CONSTANS demonstrates novel interactions among Arabidopsis flowering-time genes. Plant Cell 12: 885–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter JE, Harrison M (2003) Role of ethylene biosynthesis in hypocotyl gravitropism. In 14th International Conference on Arabidopsis Research, June 20–24, 2003, Madison, WI
- Putterill J, Robson F, Lee K, Simon R, Coupland G (1995) The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80: 847–857 [DOI] [PubMed] [Google Scholar]
- Redei GP (1975) Arabidopsis as a genetic tool. Annu Rev Genet 9: 111–127 [DOI] [PubMed] [Google Scholar]
- Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B (2003) An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol Biol 53: 247–259 [DOI] [PubMed] [Google Scholar]
- Ruiz-Garcia L, Madueno F, Wilkinson M, Haughn G, Salinas J, Martinez-Zapater JM (1997) Different roles of flowering-time genes in the activation of floral initiation genes in Arabidopsis. Plant Cell 9: 1921–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, Coupland G (2000) Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288: 1613–1616 [DOI] [PubMed] [Google Scholar]
- Schmid M, Uhlenhaut NH, Godard F, Demar M, Bressan R, Weigel D, Lohmann JU (2003) Dissection of floral induction pathways using global expression analysis. Development 130: 6001–6012 [DOI] [PubMed] [Google Scholar]
- Sessions A, Yanofsky MF, Weigel D (2000) Cell-cell signaling and movement by the floral transcription factors LEAFY and APETALA1. Science 289: 779–782 [DOI] [PubMed] [Google Scholar]
- Sheldon CC, Burn JE, Perez PP, Metzger J, Edwards JA, Peacock WJ, Dennis ES (1999) The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11: 445–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez-Lopez P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G (2001) CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410: 1116–1120 [DOI] [PubMed] [Google Scholar]
- Takada S, Goto K (2003) Terminal flower2, an Arabidopsis homolog of heterochromatin protein1, counteracts the activation of flowering locus T by constans in the vascular tissues of leaves to regulate flowering time. Plant Cell 15: 2856–2865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D (2005) Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309: 1056–1059 [DOI] [PubMed] [Google Scholar]
- Yamaguchi A, Kobayashi Y, Goto K, Abe M, Araki T (2005) TWIN SISTER OF FT (TSF) acts as a floral pathway integrator redundantly with FT. Plant Cell Physiol 46: 1175–1189 [DOI] [PubMed] [Google Scholar]
- Yanovsky MJ, Kay SA (2002) Molecular basis of seasonal time measurement in Arabidopsis. Nature 419: 308–312 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.