Abstract
Terfestatin A (TrfA), terphenyl-β-glucoside, was isolated from Streptomyces sp. F40 in a forward screen for compounds that inhibit the expression of auxin-inducible genes in Arabidopsis (Arabidopsis thaliana). TrfA specifically and competitively inhibited the expression of primary auxin-inducible genes in Arabidopsis roots, but did not affect the expression of genes regulated by other plant hormones such as abscisic acid and cytokinin. TrfA also blocked the auxin-enhanced degradation of auxin/indole-3-acetic acid (Aux/IAA) repressor proteins without affecting the auxin-stimulated interaction between Aux/IAAs and the F-box protein TIR1. TrfA treatment antagonized auxin responses in roots, including primary root inhibition, lateral root initiation, root hair promotion, and root gravitropism, but had only limited effects on shoot auxin responses. Taken together, these results indicate that TrfA acts as a modulator of Aux/IAA stability and thus provides a new tool for dissecting auxin signaling.
The plant hormone auxin (indole-3-acetic acid [IAA]) plays a crucial role in most aspects of plant growth and development. At the whole-plant level, auxin regulates tropisms, apical dominance, root development, and ultimately controls the architecture of adult plants (Woodward and Bartel, 2005). Molecular biological and genetic studies on auxin signal transduction have revealed that three major families of genes (Aux/IAAs, GH3s, and SAURs) are induced in response to auxin treatment (Liscum and Reed, 2002). Among the three families, the Aux/IAA family, of which there are 29 members in Arabidopsis (Arabidopsis thaliana), is the best characterized and has a pivotal role in auxin signaling. Aux/IAAs encode short-lived nuclear proteins that are repressors of auxin-responsive gene expression. At least one way in which they impose transcriptional repression is by dimerizing with members of the auxin response factor family of transcription factors that bind the promoters of auxin-regulated genes (Liscum and Reed, 2002; Woodward and Bartel, 2005). Molecular genetic studies and gain-of-function mutants of Aux/IAAs revealed that the proteolysis of Aux/IAA proteins is crucial for primary auxin-responsive gene expression and subsequent developmental processes (Dharmasiri and Estelle, 2002; Liscum and Reed, 2002). TIR1 encodes an F-box protein interacting with Skp1 and Cullin proteins to form an E3 ubiquitin-ligase complex called SCFTIR1 (Ruegger et al., 1998; Gray et al., 1999). SCFTIR1 assembly plays an essential role in the proteolytic pathway regulating auxin-dependant degradation of Aux/IAA repressors. Gain-of-function mutations in domain II of Aux/IAA genes confer resistance to SCFTIR-mediated proteolysis and consequently repress auxin-inducible gene expression (Dharmasiri and Estelle, 2002; Liscum and Reed, 2002). Auxin promotes the interaction between Aux/IAAs and TIR1 protein, thereby enhancing the degradation of Aux/IAA proteins and releasing the repression of auxin-responsive gene expression (Gray et al., 2001; Dharmasiri et al., 2003; Kepinski and Leyser, 2004). Recent reports have demonstrated that the interaction between Aux/IAAs and TIR1 involves the direct binding of IAA and that TIR1 is the auxin receptor for this response (Dharmasiri et al., 2005; Kepinski and Leyser, 2005).
Modification of the cullin CUL1 by the ubiquitin-related protein RUB1/NEDD8 is essential for proper SCFTIR1 function. The components of the RUB-conjugation pathway, including AXR1, ECR1, and RCE1, regulate SCFTIR1 activity, and mutations of these components cause auxin-insensitive phenotypes and severe developmental defects (Dharmasiri and Estelle, 2004; Woodward and Bartel, 2005). Other modulations of SCF activity involve the SCF regulatory proteins CAND1 (cullin-associated and neddylation-dissociated) and SGT1b (Gray et al., 2003; Cheng et al., 2004; Chuang et al., 2004). Loss-of-function mutations in CAND1 and SGT1b also confer auxin resistance in Arabidopsis. However, the molecular mechanism by which SCFTIR1 function is regulated in specific tissue and developmental contexts is still not fully understood.
Bioprobes are very useful tools to elucidate the mechanisms of auxin signal transduction and their role in plant growth and development because they can potentially overcome the functional redundancy of cognate target proteins and accomplish temporal regulation of protein function at specific developmental stages. Previously, we identified a novel auxin signaling inhibitor, Yokonolide B, by a forward screen of microbial extracts using a transgenic auxin-inducible reporter line (Hayashi et al., 2001, 2003). Recent work also demonstrated the identification of synthetic auxin signaling inhibitors from a commercially available chemical library (Armstrong et al., 2004). However, the effects of these inhibitors are not confined to auxin signaling, since they not only repress auxin action but also affect cytokinin-responsive gene expression.
As a result of our ongoing efforts to identify inhibitors specific for auxin signaling, we have found a novel and specific auxin signaling inhibitor, which we designated terfestatin A (TrfA; Fig. 1). TrfA was identified in a screen of Streptomyces sp. F40 extracts for inhibition of expression of an auxin-responsive β-glucuronidase (GUS) reporter construct (Yamazoe et al., 2004). Herein we demonstrate that TrfA specifically blocks auxin-responsive gene expression and other auxin-regulated processes in root. Importantly, TrfA displayed potent antagonistic effects on every known auxin response in the root. This work provides a useful tool for the study of the very early steps of auxin signaling.
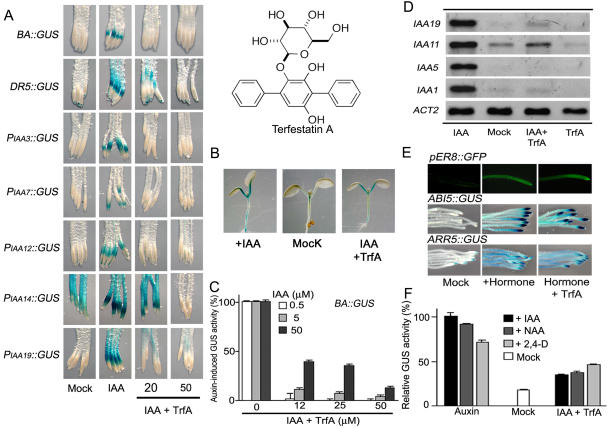
Figure 1.
Effects of TrfA on auxin- and other hormone-responsive reporter gene expression. A, Effects of TrfA on GUS expression in the root elongation zone of the reporter lines. Five-day-old seedlings were treated with 20 μm or 50 μm TrfA and 5 μm IAA for the following times: BA∷GUS, DR5∷GUS, and PIAA14∷GUS, 5 h; PIAA7∷GUS, 6 h; and PIAA3∷GUS, PIAA12∷GUS, and PIAA19∷GUS, 12 h. B, Effects of TrfA on BA∷GUS expression in the shoot. Five-day-old BA∷GUS seedlings were treated with 5 μm IAA and/or 40 μm TrfA for 12 h. C, Competitive inhibition of TrfA on BA∷GUS expression. The roots (5 d old) were treated with TrfA and IAA for 5 h. Treatments with 0.5, 5, or 50 μm IAA are adjusted to 100% value, respectively. Error bars, se. GUS activity was measured fluorometrically. D, TrfA effect on steady-state levels of IAA-induced IAA1, IAA5, IAA11, and IAA19 in Arabidopsis. The roots from 7-d-old light-grown seedlings were treated with 0.5 μm IAA for 45 min after preincubation with or without 50 μm TrfA for 15 min. The transcripts were analyzed by RT-PCR. E, Effect of TrfA on hormone-responsive transgenic reporter gene expression (estrogen-inducible pER8∷GFP, ABA-inducible ABI5∷GUS, and cytokinin-inducible ARR5∷GUS). Seven-day-old pER8∷GFP, 3-d-old ABI5∷GUS, and 5-d-old ARR5∷GUS seedlings were treated with 50 μm TrfA together with 5 μm β-estradiol for 24 h, 5 μm ABA for 12 h, and 5 μm benzyladenine for 5 h, respectively. F, Comparison of TrfA effects on auxin-induced BA∷GUS expression using three diagnostic auxins. The roots (5 d old) were treated with 50 μm TrfA and 10 μm auxin for 5 h. The induced GUS activity by IAA is adjusted to 100% value. Error bars, se.
RESULTS
TrfA Inhibits the Expression of Primary Auxin-Responsive Genes
We first investigated the effects of TrfA on the expression of two GUS reporter gene constructs driven by different synthetic auxin-responsive promoters. The transgenic Arabidopsis BA∷GUS line contains a GUS reporter gene under the control of a promoter composed of the primary auxin-responsive elements derived from the pea (Pisum sativum) Aux/IAA promoter PS-IAA 4/5 (Oono et al., 1998). The DR5 promoter is comprised of tandem elements taken from the primary auxin-responsive GH3 promoter (Ulmasov et al., 1997). TrfA completely inhibited IAA-induced reporter gene expression in the roots of BA∷GUS and DR5∷GUS lines at 20 and 50 μm, respectively (Fig. 1, A and C). Furthermore, TrfA also blocked GUS expression driven by the native auxin-inducible promoters PIAA3∷GUS, PIAA7∷GUS, PIAA12∷GUS, PIAA14∷GUS, and PIAA19∷GUS (Fig. 1A; Fukaki et al., 2002; Tatematsu et al., 2004; Weijers et al., 2005). Interestingly, TrfA had no effect on IAA-induced BA∷GUS expression in shoots (Fig. 1B). The inhibitory mode of TrfA on BA∷GUS expression was examined by the quantitative measurement for GUS activity of roots treated with various concentrations of TrfA and IAA. Figure 1C clearly indicates that TrfA competitively inhibits IAA-induced reporter gene expression. To confirm the results of the reporter line experiments, we carried out reverse transcription (RT)-PCR on selected primary auxin-responsive genes. The roots or shoots excised from 7-d-old seedling were treated with IAA and TrfA for 45 min after preincubation with TrfA for 15 min. TrfA blocked auxin-induced Aux/IAA transcription in roots (Fig. 1D) but did not inhibit their auxin-induction in the shoot (data not shown).
To address issues of the specificity of TrfA on auxin signaling, we investigated the effects of TrfA on abscisic acid (ABA)- and cytokinin-responsive gene expression, in addition to estrogen-inducible green fluorescent protein (GFP) reporter gene expression. TrfA had no effect on estrogen-induced GFP expression at 50 μm in a pER8∷GFP line (Zuo et al., 2000), indicating TrfA is not a general inhibitor of transcription or translation (Fig. 1E). The effects of TrfA on ABA and cytokinin responses were examined by using cytokinin-inducible ARR5∷GUS (D'Agostino et al., 2000) and ABA-inducible ABI5∷GUS (Lopez-Molina et al., 2002) reporter lines. In response to exogenous hormones, these two reporter lines strongly express the GUS reporter gene in roots. TrfA (50 μm) had no effect on ABA- and cytokinin-responsive gene expression in the root (Fig. 1E). These results indicate the inhibitory effects of TrfA appear to be specific to auxin. Moreover, TrfA itself was incapable of inducing auxin-responsive gene expression and inhibiting GUS enzymatic activity (supplemental data).
To test whether the inhibitory effects of TrfA are caused by disrupting auxin transport, we studied the effects of TrfA on responses to the synthetic auxins 2,4-dichlorophenoxyacetic acid (2,4-D) and 1-naphthaleneacetic acid (NAA). It is believed that auxin influx carriers transport IAA and 2,4-D into the cell, whereas NAA can move into the cell by passive diffusion. In contrast, IAA and NAA are good substrates for auxin efflux carriers, but 2,4-D is not (Delbarre et al., 1996). If TrfA modulates auxin transport, then there should be differences in the antagonistic effect of TrfA on responses elicited by the three auxins. TrfA blocked BA∷GUS and DR5∷GUS expression induced by all three auxins (Fig. 1F; supplemental data for NAA- or 2,4-D-induced DR5∷GUS expression), suggesting TrfA does not act by directly affecting auxin transport.
TrfA Inhibits Auxin-Enhanced Aux/IAA Protein Degradation But Not 26S Proteasome Activity
Auxin promotes the degradation of Aux/IAA repressor proteins via the ubiquitin-proteasome pathway and thereby induces primary auxin-responsive gene expression (Dharmasiri and Estelle, 2004). To address the effect of TrfA on auxin-enhanced degradation of Aux/IAA proteins, we examined the effect of TrfA on Aux/IAA stability using the Arabidopsis HS∷AXR3NT-GUS line, in which a translational fusion between domains I and II of AXR3 and the GUS reporter protein is expressed under the control of a heat shock promoter (Gray et al., 2001). After heat induction of the HS∷AXR3NT-GUS line, the seedlings were treated with IAA in the presence or absence of inhibitors. After a 60-min incubation without IAA, the AXR3NT-GUS fusion protein was broken down as described previously (Gray et al., 2001). Treatment with 50 μm TrfA for 60 min caused the accumulation of the fusion protein to the same extent as treatment with 50 μm MG132, a potent proteasome inhibitor (Fig. 2A). Treatment with IAA alone for 20 min enhanced the degradation of the fusion protein. TrfA blocked this IAA-enhanced degradation, stabilizing the fusion protein to a similar extent as that observed with MG132 (Fig. 2B). To confirm that AXR3NT-GUS protein accumulated as a consequence of TrfA effects on the protein's stability, the line HS∷axr3-1NT-GUS, in which a mutation in domain II of AXR3 confers increased stability on the fusion protein, was used as a control. Figure 2B shows that TrfA did not cause the accumulation of the axr3-1NT-GUS fusion protein in the presence of IAA, implying that TrfA specifically affects the degradation of Aux/IAA proteins.
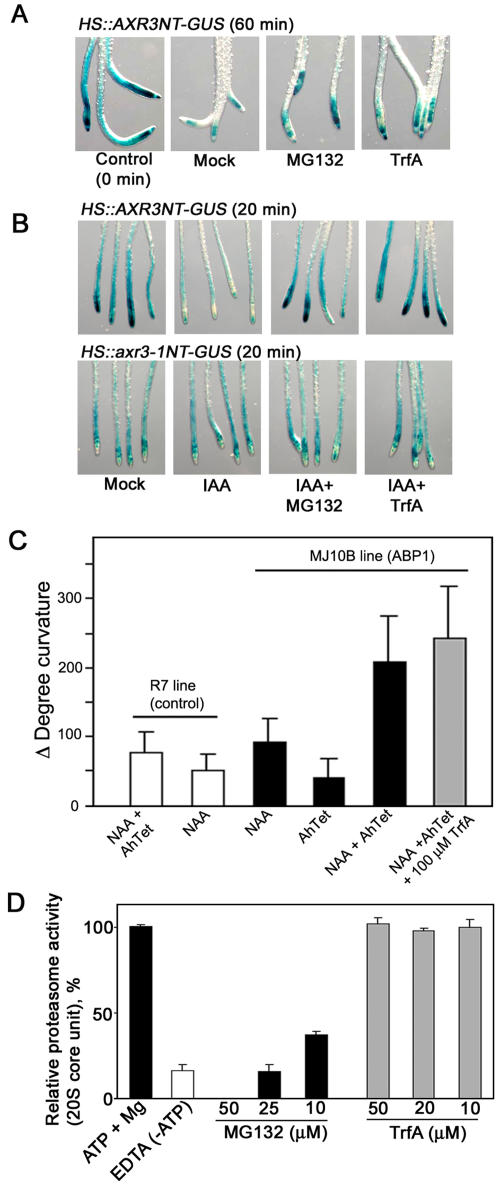
Figure 2.
Effects of TrfA on auxin-dependent degradation of Aux/IAA protein. A and B, Effects of TrfA and MG132 on IAA17/AXR3-GUS fusion protein degradation. Six-day-old HS∷AXR3NT-GUS and HS∷axr3-1NT-GUS transgenic seedlings were heat shocked for 2 h at 37°C to induce the fusion protein expression. After incubation for 20 min at 23°C, the seedlings were incubated in 50 μm TrfA or 50 μm MG132 with/without 1 μm IAA for 60 min (A) or 20 min (B). C, Effect of TrfA on auxin-induced cell elongation mediated by ABP1. The MJ10B line expresses ABP1 under the control of a tetracycline-inducible promoter. The R7 line harbors a corresponding empty vector. Leaf strips from each line were incubated in 5 μm NAA and/or TrfA together with AhTet (inducer) for 12 h after preincubation with AhTet for 4 h. Error bars, se. D, Arabidopsis ATP-dependent proteasome inhibition by TrfA and MG132 in T-87 suspension cells. Error bars, se.
To explore further the specificity of TrfA action on auxin signaling, we examined its effect on ABP1-mediated cell elongation. Although ABP1 is reported to be involved in the pathway leading to epinastic cell elongation of young tobacco (Nicotiana tabacum) leaves (Jones et al., 1998; Chen et al., 2001), there is no demonstrated link between ABP1 and the TIR1-Aux/IAA pathway. In the transgenic tobacco line MJ10B, which expresses ABP1 under the control of a tetracycline-inducible promoter, overexpressed ABP1 mediates epinastic leaf curvature in response to auxin in young leaves (Jones et al., 1998). Strips of interveinal leaf excised from the base of young MJ10B leaves expressing ABP1 (plus anhydrotetracycline [AhTet]) exhibited NAA-dependent curvature resulting from epinastic cell elongation. The control R7 line harboring the empty vector did not respond to NAA in the presence of AhTet (Fig. 2C). TrfA had no effect on NAA-induced curvature in AhTet-treated MJ10B leaf strips at 100 μm (Fig. 2C), further supporting the idea that TrfA acts specifically on the TIR1-Aux/IAA pathway.
In order to address whether TrfA is a general proteasome inhibitor, we examined the effect of TrfA on ATP-dependent proteasome activity using Arabidopsis T-87 cultured cells. Proteolytic activity of the 20S core unit of the 26S proteasome in cell homogenates was directly measured with a fluorogenic peptide substrate. As shown in Figure 2D, while MG132 completely repressed the proteolytic activity in 26S proteasome, TrfA had no effect.
TrfA Does Not Inhibit Auxin-Enhanced Interaction between TIR1 and Aux/IAAs
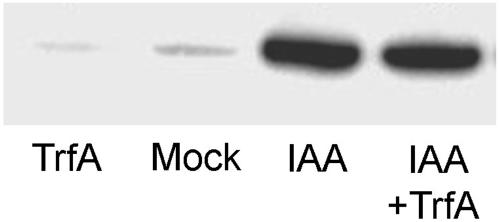
The binding of auxin and Aux/IAAs to the ubiquitin-ligase component TIR1 is an important step in the proteolysis of Aux/IAAs (Dharmasiri et al., 2005; Kepinski and Leyser, 2005). To address whether TrfA affects the binding of auxin and/or Aux/IAAs to TIR1, we carried out pull-down assays using a biotinylated peptide corresponding to the core residues of the Aux/IAA domain II degron and myc-tagged TIR1 (Kepinski and Leyser, 2004) in the presence and absence of both IAA and TrfA. Figure 3 shows that even at high concentrations (100 μm) TrfA did not inhibit the auxin-enhanced interaction between TIR1 and domain II Aux/IAA peptides.
Figure 3.
Effects of TrfA on auxin-enhanced interaction between SCFTIR1 and Aux/IAA. Anti-c-myc immunoblot showing pull-down assays using a synthetic Aux/IAA domain II peptide, and extracts of tir1-1[TIR1myc] plants in the presence or absence of both 0.5 μm IAA and 100 μm TrfA as indicated. The recovery of TIR1myc on domain II peptide beads was assessed by immunoblotting with anti-c-myc antibody.
TrfA Inhibits Auxin-Dependent Cell Division
Auxin is a positive regulator in cell division and promotes lateral root initiation (Hellmann and Estelle, 2002). We first assessed TrfA effects on cell division with tobacco suspension-cultured BY2 cells. Cell division of BY2 cultured cells is critically dependent on auxin and is interrupted by auxin deprivation. Readdition of auxin stimulates division in a semisynchronous manner (Winicur et al., 1998). As shown in Figure 4A, the cell division in auxin-starved cells was initiated by the addition of 2,4-D within 3 h as described previously (Winicur et al., 1998). Application of 20 μm TrfA completely inhibited cell division induced by 2,4-D and cells entered stasis (Fig. 4A) without any effect on viability (data not shown). Auxin deprivation also altered the cell shape, resulting in larger cells compared to the chains of small cells typical of cells cultured in auxin. The cells grown in 2,4-D with TrfA swelled and phenocopied auxin-starved cells (Fig. 4B).
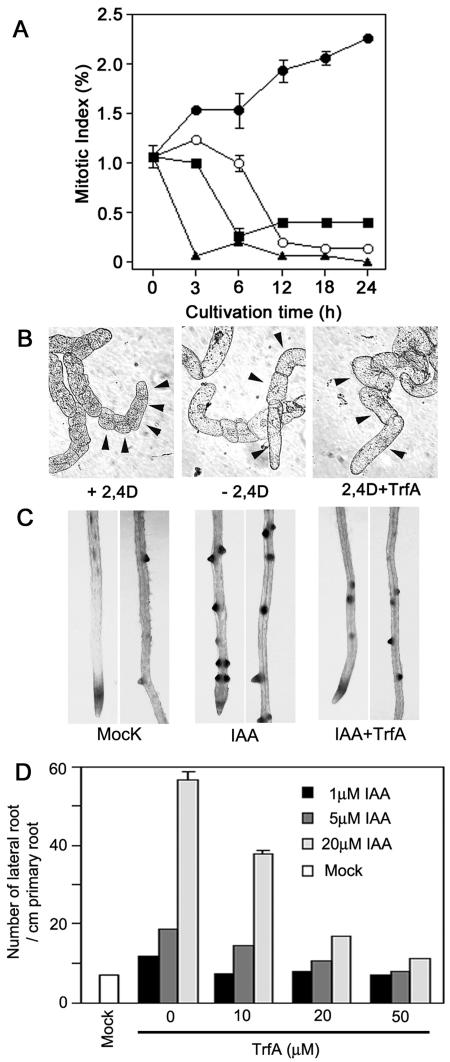
Figure 4.
Effect of TrfA on auxin-dependent cell division. A, Inhibition of auxin-induced cell division of auxin-deprived BY2 cells by TrfA. Auxin-starved cells cultured without 2,4-D for 24 h were then incubated with 2,4-D and/or TrfA. Black circle, 1 μm 2,4-D; white circle, without 2,4-D; black square, 1 μm 2,4-D plus 20 μm TrfA; black triangle, 20 μm TrfA. Error bars, se. B, Auxin-starved cells treated with or without 1 μm 2,4-D, and with 10 μm TrfA plus 1 μm 2,4-D for 3 d. Arrow indicates each cell. The photographs are of the same magnification. C, Effects of TrfA on IAA-induced lateral root formation in CycB1;1∷GUS line. Six-day-old seedlings grown in 0.5 μm NPA were treated with 5 μm IAA and/or 50 μm TrfA for 2 d. The expression of CycB1;1 was visualized by histochemical staining. D, Antagonistic effects of TrfA on auxin-induced lateral root formation. Six-day-old seedlings grown in 0.5 μm NPA were treated with various concentrations of IAA and/or TrfA and for 2 d. The number of lateral roots per centimeter of primary root was calculated in each seedling (n = 15). Error bars, se.
The effect of TrfA on cell division was also examined by auxin-induced CycB1;1 expression and subsequent lateral root initiation using the transgenic Arabidopsis CycB1;1∷GUS line (Ferreira et al., 1994). CycB1;1 gene expression is closely associated with the G2 and M phases of cell division and can be used as an indicator for early mitotic events leading to a lateral root initiation (Himanen et al., 2002). Seedlings were grown vertically for 6 d on agar plates containing an auxin efflux transport inhibitor (0.5 μm 1-naphthylphthalamic acid [NPA]) to repress a lateral root formation, and then treated with IAA in the presence or absence of TrfA for 48 h. CycB1;1∷GUS expression and lateral root initiation were induced by IAA (Fig. 4C) as reported previously (Himanen et al., 2002). TrfA repressed IAA-induced CycB1;1∷GUS expression and lateral root initiation (Fig. 4C). Consistent with the antagonistic effects of TrfA on auxin-responsive gene expression (Fig. 1C), TrfA counteracted auxin-induced lateral root initiation (Fig. 4D). In addition, TrfA displayed antagonistic effects on lateral root initiation induced by 2,4-D and NAA (supplemental data). MG132 also inhibited lateral root initiation, but MG132 displayed toxic effects on the root growth (Hayashi et al., 2003) and no antagonistic activity on auxin responses (data not shown). These data suggest TrfA blocks the very early events of auxin-dependent lateral root initiation.
TrfA Suppresses Root Auxin Responses
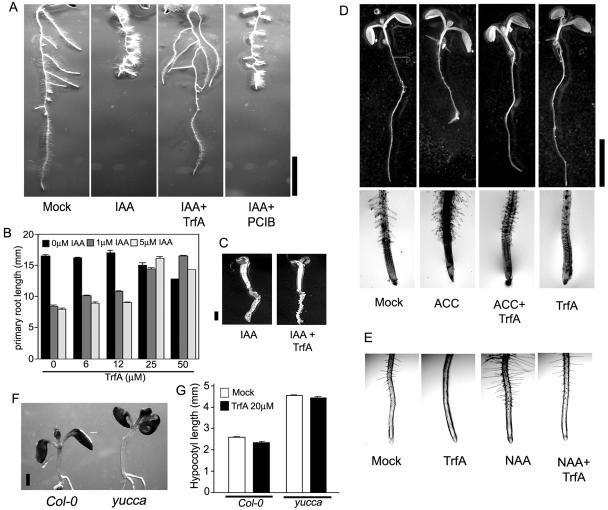
In addition to stimulating lateral root growth, it is known that auxin inhibits primary root elongation and promotes root hair formation. We therefore assessed the physiological effects of TrfA on root auxin responses in Arabidopsis plants. Treatment with 2 μm IAA for 3 d inhibited the primary root elongation and promoted the formation of root hairs and lateral roots (Fig. 5A). These IAA-induced root responses were completely suppressed by 20 μm TrfA (Fig. 5A). A quantitative analysis of primary root elongation (Fig. 5B) illustrated further the antagonistic effect of TrfA on IAA-induced primary root inhibition, although TrfA could no longer counteract auxin responses in roots treated with very high levels of exogenous IAA (100 μm; Fig. 5C). TrfA also suppressed NAA- or 2,4-D-inhibited primary root elongation and lateral root promotion (supplemental data). Unlike TrfA, the classic anti-auxin p-chlorophenoxyisobutyric acid (PCIB) could not counteract IAA-inhibited primary root elongation (Fig. 5A) as described previously (Oono et al., 2003). TrfA itself had a slight inhibitory effect on primary root elongation at 50 μm (Fig. 5B). Interestingly, this small effect on primary root elongation was reduced by 1 μm IAA (Fig. 5B). With respect to NAA-induced root hair formation, TrfA itself displayed complete inhibition of root hair formation at 20 μm, and this inhibition was recovered to normal levels with NAA application (Fig. 5E), indicating TrfA also competed with auxin in root hair formation.
Figure 5.
Effect of TrfA on physiological action in Arabidopsis plant. A, Seven-day-old roots (Columbia [Col-0]) treated with 2 μm IAA in the presence of 20 μm TrfA or 20 μm PCIB for 3 d. Bar represents 5 mm. B, Effect of TrfA on IAA-induced primary root inhibition. Three-day-old seedlings were cultured in the presence of TrfA and IAA. The primary root length was measured after another 4 d culture. Error bars, se. C, Seven-day-old seedlings treated with 100 μm IAA in the presence of 25 μm TrfA for 3 d. Bar, 1 mm. D, Six-day-old seedlings treated with or without 2 μm ACC in the presence of 20 μm TrfA for 2 d. Bar, 5 mm. E, Effect of TrfA on auxin-induced root hair promotion. Five-day-old seedlings grown vertically were transferred on GM plate containing 20 μm TrfA and/or 0.2 μm NAA and incubated vertically for additional 3 d. F, Seven-day-old yucca, an auxin overproduction mutant, and wild-type seedlings (Col-0) treated with 20 μm TrfA for 3 d. Bar, 1 mm. G, Effect of TrfA on hypocotyl elongation in yucca and wild-type (Col-0). Four-day-old seedlings were treated with 20 μm TrfA for 3 d. The hypocotyl length of 7-d-old seedlings was measured. Error bars, se.
Many auxin-insensitive mutants are also resistant to ethylene, and cross-talk between auxin and ethylene has been shown to play an essential role in root hair formation (Swarup et al., 2002). Ethylene also inhibits primary root elongation. To clarify TrfA action on these ethylene responses, seedlings were incubated in medium containing the ethylene precursor, 1-aminocyclopropane-1-carboxylic acid (ACC), with or without 20 μm TrfA for 2 d. ACC treatment inhibited root elongation and promoted root hair formation (Fig. 5D). Twenty micromolar TrfA significantly suppressed these ethylene responses in root (Fig. 5D).
As mentioned above, TrfA had no apparent effects on auxin-responsive gene expression in aerial parts of Arabidopsis plants. To assess the effect of TrfA on shoot growth, we performed assays of auxin-induced hypocotyl elongation and auxin-induced epinastic cell elongation. The Arabidopsis mutant yucca overexpresses a flavin monooxygenase involved in a Trp-dependant auxin biosynthetic pathway and thus has elevated auxin biosynthesis (Zhao et al., 2001). Four-day-old yucca and wild-type seedlings were treated in the presence or absence of 20 μm TrfA for 3 d. Light-grown yucca seedling showed longer hypocotyl than wild type (Fig. 5F), an aerial phenotype indicative of high endogenous auxin levels (Zhao et al., 2001). TrfA had no effects on hypocotyl elongation of light-grown yucca and wild-type seedlings (Fig. 5G). Interestingly, TrfA antagonistically suppressed IAA-inhibited hypocotyl growth in etiolated seedlings (supplemental data). This different effect on etiolated hypocotyl may reflect distinct signaling pathways for hypocotyl auxin response between light-grown and etiolated seedlings (Gray et al., 1998).
TrfA Inhibits Gravitropic Response of Arabidopsis Roots
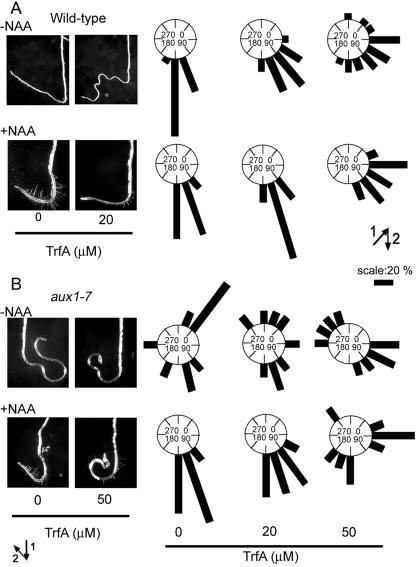
Auxin is an essential regulator of the root gravitropic response. Wild-type seedlings were grown on vertically oriented agar plates for 5 d in light and then transferred onto agar plates with or without both NAA and TrfA. The plate was then rotated by 135° and cultured for another 3 d in the dark. As shown in Figure 6A, the roots of wild-type seedlings treated with TrfA did not properly realign to the new gravity vector. This disruption of gravitropic response by 20 μm TrfA was dramatically restored by the addition of 0.2 μm NAA. The restored response by NAA was repressed again at 50 μm TrfA (Fig. 6A). The Arabidopsis mutant aux1-7 has mutation in a putative auxin influx carrier and has agravitropic roots because of poor uptake of endogenous IAA into cells (Yamamoto and Yamamoto, 1998; Marchant et al., 1999). The agravitropic phenotype of aux1-7 is rescued by treatment with 0.2 μm NAA (Fig. 6B). TrfA disrupted the NAA rescue of gravitropic response in aux1-7 roots at 50 μm (Fig. 6B). These observations suggest that TrfA can also antagonize auxin-mediated root gravitropism.
Figure 6.
Effect of TrfA on root gravitropic response. A and B, Wild-type or aux1-7 seedlings (5 d old) were transferred onto GM agar plate containing various concentrations of TrfA in the presence or absence of 0.2 μm NAA and grown in the dark for 3 d after rotating plates at 135° angle against vertical direction. The arrows indicate the vector of gravity before (1) and after (2) the commencement of gravistimulation. In right section, zero degrees represents roots that showed nil response. The angles were grouped into 22° classes and plotted as circular histograms.
DISCUSSION
TrfA Inhibits the Expression of Primary Auxin-Responsive Genes via the SCFTIR1 Pathway
SCFTIR and Aux/IAAs play a central role in the regulation of auxin-responsive gene expression. Aux/IAA repressors interact with auxin response factors to repress the expression of numerous auxin-responsive genes (Tiwari et al., 2004). Auxin relieves this repression by accelerating the degradation of Aux/IAA repressors via the ubiquitin-proteolytic pathway mediated by the SCFTIR complex (Gray et al., 2001). TrfA inhibited these processes, blocking the auxin-dependent turnover of Aux/IAA analogs (AXR3NT-GUS) in vivo and, hence, the induction of primary auxin-responsive genes by auxin (Figs. 1, A, C, and D, and 2A). We showed that TrfA is not a general inhibitor of the ubiquitin-proteolytic pathway and does not affect cytokinin and ABA signaling pathways. We also found no effect of TrfA on ABP1-mediated cell elongation, a response that does not appear to involve Aux/IAA turnover, further supporting the idea that TrfA affects specifically the TIR1-Aux/IAA pathway.
To distinguish between effects on auxin transport or signaling, we also examined the influence of TrfA on responses induced by auxins with differing transport properties. IAA and 2,4-D are good substrates for auxin influx carriers, but membrane-permeable NAA is not (Delbarre et al., 1996; Marchant et al., 1999). 1-Naphthoxyacetic acid, an auxin-influx inhibitor, represses IAA-induced BA∷GUS expression but failed to inhibit NAA-induced expression (Rahman et al., 2002). TrfA displayed similar effects on auxin responses induced by all three types of auxins, suggesting that TrfA does not perturb auxin transport machinery. Together, these data suggest that TrfA is auxin specific and acts on a component(s) involved in the SCFTIR1-mediated degradation of Aux/IAAs.
TrfA Inhibits Several Typical Auxin Responses
Consistent with its effect of auxin-induced gene expression, TrfA antagonized several classic auxin responses. In the root, TrfA suppressed the inhibition of primary root growth and the stimulation of lateral rooting and root hair formation in response to exogenous auxin. TrfA (50 μm) also completely inhibited root hair formation and gravitropic response in the absence of exogenous auxin. In contrast, primary root elongation was only slightly repressed by 50 μm TrfA treatment without additional auxin (Fig. 5B). Under certain conditions, very low concentrations of exogenous auxin can stimulate elongation of the primary root, but, as concentrations increase, auxin quickly becomes inhibitory to root growth (Lincoln et al., 1990; Mussig et al., 2003). Auxin-resistant mutants such as axr1-12, tir1-1, and aux1-7 (Lincoln et al., 1990; Ruegger et al., 1998; Rahman et al., 2002) produce longer roots in the absence of exogenous auxin, suggesting that the endogenous auxin concentration is often supraoptimal for maximum primary root growth. Thus, it might be expected that lower concentrations of TrfA would phenocopy the longer root phenotype of auxin-resistant mutants while higher concentrations would eventually inhibit primary root growth. Since TrfA clearly and specifically antagonizes several important auxin responses, this minor paradox may just reflect the complexity of auxin's contribution to growth of the primary root.
TrfA treatment also conferred ethylene resistance in roots. Most auxin-insensitive mutants also display ethylene-resistant phenotypes in the root (Swarup et al., 2002). The ethylene insensitivity of the auxin influx and efflux facilitator mutants aux1-7 and eir1/pin2 is rescued by exogenous NAA (Rahman et al., 2001). These observations indicate that ethylene response in the root requires auxin signaling. TrfA also significantly suppressed ethylene-induced root phenotypes, indicating that ethylene response in root requires TrfA-targeted auxin signaling.
Root gravitropic response is mediated by asymmetric auxin distribution and subsequent differential root elongation. The aux1-7 mutant displays agravitropic phenotype that is fully rescued by NAA, but not by IAA or 2,4-D (Yamamoto and Yamamoto, 1998; Marchant et al., 1999). TrfA conferred an agravitropic root phenotype in wild type that was antagonized by NAA (Fig. 6A). In addition, TrfA also repressed NAA-restored gravitropic response of aux1-7 (Fig. 6B). These data provide yet more support for the idea that TrfA acts by affecting intracellular auxin signaling rather than auxin transport.
Interestingly, many of the effects of TrfA seem to be confined to the root. However, TrfA did suppress the auxin-induced inhibition of hypocotyl growth in dark-grown seedlings (supplemental data). In the absence of additional auxin, TrfA also slightly inhibited the elongation of etiolated hypocotyls, similar to the effects of the axr1-3 mutation on hypocotyl growth (Lincoln et al., 1990). In light-grown seedlings, TrfA did not affect hypocotyl elongation, which again is similar to the effect of the auxin-resistant axr1 mutations on hypocotyl growth (Lincoln et al., 1990). It is also interesting to note that TrfA had a significant effect on auxin-stimulated cell division in tobacco BY2 cultures, cell types with no specific root or shoot identity. Thus, it remains to be determined whether the apparent largely root-specific effect of TrfA represents a true root specificity or is an artifact of differential metabolism or pharmacokinetics of TrfA in the plant.
TrfA May Act by Regulating Aux/IAA Turnover or Distribution
Our molecular and physiological investigation of TrfA clearly indicates that it can act as a specific auxin antagonist that competes with auxin in root auxin responses. However, the fact that TrfA did not block the auxin-enhanced interaction of the auxin receptor TIR1 and Aux/IAA peptides in pull-down assays suggests that TrfA does not compete directly with auxin at the receptor binding site.
This prompts the following question: How does TrfA exert its effects? There are several possibilities. One possible mechanism for the repression of auxin action by TrfA could be the enhanced accumulation of Aux/IAA proteins in the nucleus. A recent report demonstrates a role for Rac GTPases in Aux/IAA degradation because they are involved in the recruitment of nucleoplasmic Aux/IAAs into proteolytically active nuclear protein bodies (Tao et al., 2005). If TrfA blocked these or any components involved in the translocation of AUX/IAAs to nuclear protein bodies, the resulting accumulation of Aux/IAA proteins would be consistent with the observed effects of TrfA.
A second possible mechanism is that TrfA blocks (root-localized) positive regulators of SCFTIR1 assembly. Previous studies have identified two regulators of SCF assembly. ETA3/SGT1b is a positive regulator of R gene-mediated defense against certain pathogens and was found to be a positive regulator of SCF activity. Defects in ETA3/SGT1b result in auxin resistance, and ETA3/SGT1b is not required for SCFTIR1 binding to its Aux/IAA substrates (Gray et al., 2003). Regulation of SCF activity is also mediated by a plant ortholog of the human CAND protein. Mutations in CAND1 also result in auxin resistance. The expression of CAND1 was reported to be very high in the root and weaker in the shoot of the seedling (Cheng et al., 2004; Chuang et al., 2004). Further investigation is required to examine the effects of TrfA on these modes of SCF regulation.
Lastly, it is possible that TrfA activates negative regulator(s) of SCF activity in root. Identifying the target(s) of TrfA and the mechanism by which it exerts its effects will be the subject of future work. We anticipate that a combination of chemical and genetic approaches using TrfA will enable the identification of new components in auxin signal transduction.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia was used for all experiments. Arabidopsis mutant yucca was a gift from Dr. Y. Zhao (University of California at San Diego). Arabidopsis aux1-7 was obtained from the Arabidopsis Biological Resource Center. Transgenic Arabidopsis reporter lines PIAA7∷GUS, DR5∷GUS, and ARR5∷GUS were provided by Drs. J. Reed, A.M. Jones, and J. Kieber (University of North Carolina). Arabidopsis CycB1;1∷GUS and pER8∷GFP lines were provided by Dr. D. Inzé (Ghent University, Belgium) and N.-H. Chua (Rockefeller University). Arabidopsis PIAA3∷GUS, PIAA12∷GUS, PIAA14∷GUS, and PIAA19∷GUS lines were gifts from Drs. D. Weijers (ZMBP, University of Tuebingen, Germany), H. Fukaki (Nara Institute of Science and Technology, Japan), and K.T. Yamamoto (Hokkaido University, Japan). The HS∷AXR3NT-GUS and HS∷axr3-1NT-GUS lines were provided by Dr. M. Estelle (University of Indiana). Arabidopsis suspension T-87 cells and tobacco (Nicotiana tabacum) BY2 cells were obtained from the Riken Bioresource Center, Japan. Suspension-cultured tobacco cells (cv BY2) were maintained in a modified Murashige and Skoog medium as described previously (Winicur et al., 1998) on a rotary shaker (100 rpm) at 25°C in the dark. Auxin deprivation was carried out by washing a 7-d culture twice with the same medium lacking 2,4-D and then culturing in auxin-free medium for 24 h before auxin addition and determination of mitotic indices over time. Mitotic index of BY2 cells was determined as described previously (Winicur et al., 1998). Arabidopsis suspension T-87 cells were cultured as described (Axelos et al., 1992). Tobacco MJ10B line carrying a tetracycline-inducible ABP1 transgene and control R7 line, the corresponding empty vector, were provided by Dr. A.M. Jones (University of North Carolina) and were grown in soil under continuous light at 23°C.
Chemicals
TrfA used in this work is a synthetic sample as described (Yamazoe et al., 2004). The sample of TrfA is available for academic research from us. MG132 was purchased from The Peptide Institute. PCIB and NPA were obtained from Sigma-Aldrich and Tokyo Kasei Kogyo, respectively.
Hormone Induction
The seedlings (n = 10–15) grown vertically in continuous light were transferred to a 12- or 24-well microtiter plate containing 1 or 0.5 mL of a germination medium (GM; 0.5× Murashige and Skoog salts [Gibco-BRL], 1% Suc, 1× B5 vitamins, and 0.2 g/L MES, pH 5.8) containing the indicated hormone and/or chemicals and then incubated for the indicated time to induce each responsive gene.
Histochemical and Quantitative GUS Measurements
For GUS histochemical analysis, the seedlings were washed with a GUS-staining buffer (100 mm sodium phosphate, pH 7.0, 10 mm EDTA, 0.5 mm K4Fe(CN)6, 0.5 mm K3Fe(CN)6, and 0.1% Triton X-100) and transferred to the GUS-staining buffer containing 1mm 5-bromo-4-chloro-3-indolyl-β-d-glucuronide (X-Gluc), the substrate for histochemical staining, and incubated at 37°C until sufficient staining developed. For quantitative measurement, seedlings or the excised roots (n = 15–20) were homogenized in an extraction buffer as described previously (Oono et al., 2003). After centrifugation to remove cell debris, GUS activity was measured with 1 mm 4-methyl umbelliferyl-β-d-glucuronide as a fluorogenic substrate at 37°C. Protein concentration was determined by Bradford protein assay (Bio-Rad Japan). The experiments were repeated at least three times with four replications.
RT-PCR
Total RNA was isolated from the excised root of 7-d-old seedlings using RNeasy Plant Mini kit (Qiagen) and treated with on-column DNase digestion according to the manufacturer's instruction. The corresponding cDNAs were synthesized and amplified by the PCR using primers for the indicated genes as follows: IAA1, 5′-ggattacccggagcacaag and 5′-ggagctccgtccatactcac; IAA5, 5′-agatatcgtcgtctccggtg and 5′-gccgaagcaagatcttggta; IAA11, 5′-ggtcttacgttgagccttgg and 5′-gtggctgaagccttagcttg; IAA19, 5′-gagcatggatggtgtgccttat and 5′-ttcgcagttgtcaccatctttc; RAB18, 5′-ttgggaggaatgcttcacc and 5′-ttgttcgaagcttaacggc; and ACTIN (ACT2), 5′-aacattgtgctcagtggtgg and 5′-tcatcatactcggccttgg. The amplified products (IAA1, 208 bp after 24 cycles; IAA5, 251 bp after 27 cycles; IAA11, 229 bp after 27 cycles; IAA19, 141 bp after 27 cycles ; RAB18, 269 bp after 27 cycles; ACT2, 206 bp after 25 cycles) were analyzed by 3% agarose gel electrophoresis.
Aux/IAA Protein Degradation Assay
Seven-day-old HS∷AXR3NT-GUS and HS∷axr3-1NT-GUS transgenic seedlings were incubated in liquid GM medium for 2 h at 37°C, followed by transfer of the seedlings into liquid GM medium at 23°C. After 10 min at 23°C, the indicated inhibitors were added into the medium. IAA was added after additional 10 min incubation with inhibitors. The seedlings were immersed with 70% cold acetone and washed with water after incubation for another 20 min. GUS activity was histochemically stained for 12 h for the HS∷AXR3NT-GUS or 3 h for the HS∷axr3-1NT-GUS lines. For proteasome assay, ATP-dependant 20S core unit activity of 26S proteasome in Arabidopsis T-87 suspension cells was measured by peptide-hydrolysis activity using succinyl-Leu-Leu-Val-Tyr-4-methyl-coumaryl-7-amide as the substrate with or without ATP and Mg2+, as described previously (Fujinami et al., 1994). The experiments were repeated three times with three replications.
ABP1-Mediated Tobacco Leaf Curvature Assay
Measurement of NAA-induced tobacco leaf curvature was carried out as described previously (Chen et al., 2001) with slight modification. The leaf strips (n = 10–15) were excised from the sixth leaf of seven-leaf-staged transgenic tobacco MJ10B (tetracycline-inducible ABP1) or R7 (corresponding empty vector) lines. One end of each strip was clamped by a small rubber block. Blocks of 12 interveinal leaf strips were placed in petri dishes containing 5 mL of buffer (10 mm Suc, 10 mm KCl, and 0.5 mm MES, pH 6.0) with or without the indicated concentrations of the chemicals. To induce ectopic expression of ABP1, MJ10B and R7 strips were incubated in solution containing 4 μg/mL AhTet for 4 h prior to NAA and TrfA addition. Photographs of the strips were taken and the degree of curvature for each strip was measured with NIH Image software (developed at the United States National Institutes of Health and available on the Internet at http://rsb.info.nih.gov/nih-image).
Pull-Down Assay with Aux/IAA Domain II Peptide and c-Myc-Tagged TIR1
Pull-down assays with the biotinylated domain II peptide were performed as described previously (Kepinski and Leyser, 2004). Briefly, where indicated, auxin (IAA 0.5 μm) and inhibitors (TrfA 100 μm) were added directly to the extracts of tir1-1[TIR1myc] plants with 6.5 μg of biotinylated domain II peptide immobilized on streptavidin agarose (Novagen). After 45 min incubation at 4°C, streptavidin beads were collected by brief centrifugation, washed three times, resuspended in SDS-PAGE sample buffer, and subjected to SDS-PAGE electrophoresis and immunoblotting with anti-c-myc antibody.
Measurements for Growth of Hypocotyls, and Primary and Lateral Roots
For hypocotyl and primary root growth, the seedlings (3 or 4 d old) were transferred into liquid GM medium containing the indicated concentration of TrfA with or without auxin. The seedlings were then cultivated under continuous light for indicated period at 24°C. The length of hypocotyl and primary root was recorded by digital camera and analyzed by NIH Image software.
For lateral root growth, Arabidopsis CycB1;1∷GUS seedlings were vertically grown for 6 d in continuous light on GM agar plate containing 0.5 μm NPA, auxin efflux inhibitor. The seedlings were transferred to liquid GM medium containing the indicated concentration of IAA and TrfA. The seedlings (n = 15) were cultivated under continuous light for another 2 d at 24°C. The expression of GUS was stained by X-Gluc, and the number of lateral roots was counted by eye. The experiments were repeated three times with three replications.
Root Gravitropic Response Assay
Five-day-old seedlings grown vertically on GM agar plate under continuous light were transferred on GM agar plates containing the indicated concentration of TrfA. The plate was rotated to 135° angle against vertical direction and incubated for 3 d in dark. The angle between root tip orientation and vertical direction was recorded by digital camera and analyzed by the NIH Image software. The experiments were carried out two times (n = 20–25) with three replications.
Supplementary Material
Acknowledgments
We thank Drs. J. Reed, A.M. Jones, J. Kieber, D. Inzé, A. Theologis, H. Fukaki, K.T. Yamamoto, J. Callis, N.-H. Chua, M. Estelle, Y. Takahashi, Y. Oono, and Y. Zhao for providing materials; Drs. Y. Oono, H. Fukaki, A. Kuboki, and S. Ohira for useful suggestions; and Drs. J.-G. Chen and Y. Zhao for critical reading of the manuscript.
This work was supported by a Grant-in-Aid for Scientific Research (grant no. KAKEN16710162) to K.H. from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, and in part by the Biotechnology and Biological Sciences Research Council (grant no. 87/G14634 to O.L.).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.068924.
References
- Armstrong JI, Yuan S, Dale JM, Tanner VN, Theologis A (2004) Identification of inhibitors of auxin transcriptional activation by means of chemical genetics in Arabidopsis. Proc Natl Acad Sci USA 101: 14978–14983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelos M, Curie C, Mazzolini L, Bardet C, Lescure B (1992) A protocol for transient gene expression in Arabidopsis thaliana protoplasts isolated from cell suspension cultures. Plant Physiol Biochem 30: 123–128 [Google Scholar]
- Chen JG, Shimomura S, Sitbon F, Sandberg G, Jones AM (2001) The role of auxin-binding protein 1 in the expansion of tobacco leaf cells. Plant J 28: 607–617 [DOI] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y (2004) AtCAND1, a HEAT-repeat protein that participates in auxin signaling in Arabidopsis. Plant Physiol 135: 1020–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang HW, Zhang W, Gray WM (2004) Arabidopsis ETA2, an apparent ortholog of the human cullin-interacting protein CAND1, is required for auxin responses mediated by the SCF(TIR1) ubiquitin ligase. Plant Cell 16: 1883–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Agostino IB, Deruere J, Kieber JJ (2000) Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol 124: 1706–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delbarre A, Muller P, Imhoff V, Guern J (1996) Comparison of mechanisms controlling uptake and accumulation of 2,4-dichlorophenoxy-acetic acid, naphthalene-1-acetic acid, and indole-3-acetic acid in suspension-cultured tobacco cells. Planta 198: 532–541 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M (2005) The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Jones AM, Estelle M (2003) Auxin action in a cell-free system. Curr Biol 13: 1418–1422 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Estelle M (2004) Auxin signaling and regulated protein degradation. Trends Plant Sci 9: 302–308 [DOI] [PubMed] [Google Scholar]
- Dharmasiri S, Estelle M (2002) The role of regulated protein degradation in auxin response. Plant Mol Biol 49: 401–409 [PubMed] [Google Scholar]
- Ferreira PC, Hemerly AS, Engler JD, van Montagu M, Engler G, Inzé D (1994) Developmental expression of the Arabidopsis cyclin gene cyc1At. Plant Cell 6: 1763–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujinami K, Tanahashi N, Tanaka K, Ichihara A, Cejka Z, Baumeister W, Miyawaki M, Sato T, Nakagawa H (1994) Purification and characterization of the 26 S proteasome from spinach leaves. J Biol Chem 269: 25905–25910 [PubMed] [Google Scholar]
- Fukaki H, Tameda S, Masuda H, Tasaka M (2002) Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J 29: 153–168 [DOI] [PubMed] [Google Scholar]
- Gray WM, del Pozo JC, Walker L, Hobbie L, Risseeuw E, Banks T, Crosby WL, Yang M, Ma H, Estelle M (1999) Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev 13: 1678–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M (2001) Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature 414: 271–276 [DOI] [PubMed] [Google Scholar]
- Gray WM, Muskett PR, Chuang HW, Parker JE (2003) Arabidopsis SGT1b is required for SCF(TIR1)-mediated auxin response. Plant Cell 15: 1310–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Ostin A, Sandberg G, Romano CP, Estelle M (1998) High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc Natl Acad Sci USA 95: 7197–7202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Jones AM, Ogino K, Yamazoe A, Oono Y, Inoguchi M, Kondo H, Nozaki H (2003) Yokonolide B, a novel inhibitor of auxin action, blocks degradation of AUX/IAA factors. J Biol Chem 278: 23797–23806 [DOI] [PubMed] [Google Scholar]
- Hayashi K, Ogino K, Oono Y, Uchimiya H, Nozaki H (2001) Yokonolide A, a new inhibitor of auxin signal transduction, from Streptomyces diastatochromogenes B59. J Antibiot (Tokyo) 54: 573–581 [DOI] [PubMed] [Google Scholar]
- Hellmann H, Estelle M (2002) Plant development: regulation by protein degradation. Science 297: 793–797 [DOI] [PubMed] [Google Scholar]
- Himanen K, Boucheron E, Vanneste S, de Almeida Engler J, Inzé D, Beeckman T (2002) Auxin-mediated cell cycle activation during early lateral root initiation. Plant Cell 14: 2339–2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AM, Im KH, Savka MA, Wu MJ, DeWitt NG, Shillito R, Binns AN (1998) Auxin-dependent cell expansion mediated by overexpressed auxin-binding protein 1. Science 282: 1114–1117 [DOI] [PubMed] [Google Scholar]
- Kepinski S, Leyser O (2004) Auxin-induced SCFTIR1-Aux/IAA interaction involves stable modification of the SCFTIR1 complex. Proc Natl Acad Sci USA 101: 12381–12386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski S, Leyser O (2005) The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435: 446–456 [DOI] [PubMed] [Google Scholar]
- Lincoln C, Britton JH, Estelle M (1990) Growth and development of the axr1 mutants of Arabidopsis. Plant Cell 2: 1071–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum E, Reed JW (2002) Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol Biol 49: 387–400 [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, McLachlin DT, Chait BT, Chua NH (2002) ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J 32: 317–328 [DOI] [PubMed] [Google Scholar]
- Marchant A, Kargul J, May ST, Muller P, Delbarre A, Perrot-Rechenmann C, Bennett MJ (1999) AUX1 regulates root gravitropism in Arabidopsis by facilitating auxin uptake within root apical tissues. EMBO J 18: 2066–2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mussig C, Shin GH, Altmann T (2003) Brassinosteroids promote root growth in Arabidopsis. Plant Physiol 133: 1261–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oono Y, Chen QG, Overvoorde PJ, Kohler C, Theologis A (1998) age mutants of Arabidopsis exhibit altered auxin-regulated gene expression. Plant Cell 10: 1649–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oono Y, Ooura C, Rahman A, Aspuria ET, Hayashi K, Tanaka A, Uchimiya H (2003) p-Chlorophenoxyisobutyric acid impairs auxin response in Arabidopsis root. Plant Physiol 133: 1135–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman A, Amakawa T, Goto N, Tsurumi S (2001) Auxin is a positive regulator for ethylene-mediated response in the growth of Arabidopsis roots. Plant Cell Physiol 42: 301–307 [DOI] [PubMed] [Google Scholar]
- Rahman A, Hosokawa S, Oono Y, Amakawa T, Goto N, Tsurumi S (2002) Auxin and ethylene response interactions during Arabidopsis root hair development dissected by auxin influx modulators. Plant Physiol 130: 1908–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruegger M, Dewey E, Gray WM, Hobbie L, Turner J, Estelle M (1998) The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast grr1p. Genes Dev 12: 198–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R, Parry G, Graham N, Allen T, Bennett M (2002) Auxin cross-talk: integration of signalling pathways to control plant development. Plant Mol Biol 49: 409–424 [DOI] [PubMed] [Google Scholar]
- Tao LZ, Cheung AY, Nibau C, Wu HM (2005) RAC GTPases in tobacco and Arabidopsis mediate auxin-induced formation of proteolytically active nuclear protein bodies that contain Aux/IAA proteins. Plant Cell 17: 2369–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatematsu K, Kumagai S, Muto H, Sato A, Watahiki MK, Harper RM, Liscum E, Yamamoto KT (2004) MASSUGU2 encodes Aux/IAA19, an auxin-regulated protein that functions together with the transcriptional activator NPH4/ARF7 to regulate differential growth responses of hypocotyl and formation of lateral roots in Arabidopsis thaliana. Plant Cell 16: 379–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle TJ (2004) Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 16: 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D, Benkova E, Jager KE, Schlereth A, Hamann T, Kientz M, Wilmoth JC, Reed JW, Jurgens G (2005) Developmental specificity of auxin response by pairs of ARF and Aux/IAA transcriptional regulators. EMBO J 24: 1874–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winicur ZM, Zhang GF, Staehelin LA (1998) Auxin deprivation induces synchronous Golgi differentiation in suspension-cultured tobacco BY-2 cells. Plant Physiol 117: 501–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward AW, Bartel B (2005) Auxin: regulation, action, and interaction. Ann Bot (Lond) 95: 707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Yamamoto KT (1998) Differential effects of 1-naphthaleneacetic acid, indole-3-acetic acid and 2,4-dichlorophenoxyacetic acid on the gravitropic response of roots in an auxin-resistant mutant of arabidopsis, aux1. Plant Cell Physiol 39: 660–664 [DOI] [PubMed] [Google Scholar]
- Yamazoe A, Hayashi K-I, Kuboki A, Ohira S, Nozaki H (2004) The isolation, structural determination, and total synthesis of terfestatin A, a novel auxin signaling inhibitor from Streptomyces sp. Tetrahedron Lett 45: 8359–8362 [Google Scholar]
- Zhao Y, Christensen SK, Fankhauser C, Cashman JR, Cohen JD, Weigel D, Chory J (2001) A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291: 306–309 [DOI] [PubMed] [Google Scholar]
- Zuo J, Niu QW, Chua NH (2000) Technical advance: an estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J 24: 265–273 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.