Abstract
In plants, reactive oxygen species and, more particularly, hydrogen peroxide (H2O2) play a dual role as toxic by-products of normal cell metabolism and as regulatory molecules in stress perception and signal transduction. Peroxisomal catalases are an important sink for photorespiratory H2O2. Using ATH1 Affymetrix microarrays, expression profiles were compared between control and catalase-deficient Arabidopsis (Arabidopsis thaliana) plants. Reduced catalase levels already provoked differences in nuclear gene expression under ambient growth conditions, and these effects were amplified by high light exposure in a sun simulator for 3 and 8 h. This genome-wide expression analysis allowed us to reveal the expression characteristics of complete pathways and functional categories during H2O2 stress. In total, 349 transcripts were significantly up-regulated by high light in catalase-deficient plants and 88 were down-regulated. From this data set, H2O2 was inferred to play a key role in the transcriptional up-regulation of small heat shock proteins during high light stress. In addition, several transcription factors and candidate regulatory genes involved in H2O2 transcriptional gene networks were identified. Comparisons with other publicly available transcriptome data sets of abiotically stressed Arabidopsis revealed an important intersection with H2O2-deregulated genes, positioning elevated H2O2 levels as an important signal within abiotic stress-induced gene expression. Finally, analysis of transcriptional changes in a combination of a genetic (catalase deficiency) and an environmental (high light) perturbation identified a transcriptional cluster that was strongly and rapidly induced by high light in control plants, but impaired in catalase-deficient plants. This cluster comprises the complete known anthocyanin regulatory and biosynthetic pathway, together with genes encoding unknown proteins.
Catalases are key players within the reactive oxygen-scavenging network that modulates the steady-state levels of reactive oxygen species (ROS) in the different subcellular compartments of plants (Mittler et al., 2004). In the peroxisomes, catalase is the principal scavenging enzyme, acting as an important sink for photorespiratory hydrogen peroxide (H2O2; Dat et al., 2001; Noctor et al., 2002; Mateo et al., 2004). Reduced levels of catalase in mutants and transgenic C3 and C4 plants increase sensitivity toward environmental stresses because of the insufficient removal of photorespiratory H2O2 (Kendall et al., 1983; Willekens et al., 1997). H2O2 diffusing from the peroxisomes into the cytosol can contribute to the interorganellar communication within the cell and lead to nuclear gene activation (Corpas et al., 2001). In accordance, a deficiency in peroxisomal catalase has been shown to deregulate the transcriptome in plants during high light (HL) stress (Vandenabeele et al., 2003, 2004), and transcriptomic reprogrammation has been observed by exogenous application of H2O2 to cell suspensions or by HL stressing of cytosolic ascorbate peroxidase knockout plants (Desikan et al., 2001; Davletova et al., 2005).
With the recent advent of genome-wide microarrays for Arabidopsis (Arabidopsis thaliana), it has become possible to profile dynamic changes in transcript levels of almost the entire genome. Molecular phenotyping of transgenic, mutated, or pathogen-challenged Arabidopsis lines by the use of genome-wide arrays has already significantly expanded the insights into diverse biological processes (Cartieaux et al., 2003; Rohde et al., 2004).
ROS play an important role in the defense of plants against biotic and abiotic stresses and in the induction of plant cell death. However, a clear inventory of downstream responses of ROS in plants is far from complete. Predominantly, parts of ROS signaling have been revealed by mutagenesis and pharmacological studies (Dat et al., 2003; Apel and Hirt, 2004; Laloi et al., 2004; Joo et al., 2005). Analysis of the integrated cellular response to oxidative stress is an area that is particularly suited for the use of genome-wide and proteome-wide approaches. Transcriptomic and proteomic surveys in bacterial, yeast, and mammalian cell lines have provided significant insights into the cellular responses toward elevated levels of oxidative stress (Causton et al., 2001; Mostertz et al., 2004; Murray et al., 2004). Also, in plants, several reports provided partial or genome-wide inventories of oxidative stress-deregulated genes (Desikan et al., 2001; op den Camp et al., 2003; Vandenabeele et al., 2003, 2004). Despite the clear reorientation of the transcriptome by elevated oxidative stress, it remains a challenge to dissect, within these massive deregulations, functional entities that provide a more detailed view into the mechanism. Until recently, published genome-wide studies mainly presented only a snapshot of the transcriptome at a specific moment after stress. In this manner, the dynamics of the transcriptional response could not be visualized and a primary classification of early and late expressed genes was not possible. Such an analysis would permit us to distinguish (early expressed) candidate receptor and signal transducing genes from (later expressed) effector genes, such as those involved in defense response and execution of cell death. Alternatively, when analyzing transcriptome data sets obtained from a combination of different perturbations, it becomes possible to identify more robustly coregulated genes over a wide range of stresses (Chen and Zhu, 2004; Rizhsky et al., 2004), which are to be part of the same cluster and, therefore, to be considered as “brothers in arms” within the studied biological process. From such a guilt-by-association analysis, the function of hitherto unknown genes can be predicted with more certainty.
RESULTS AND DISCUSSION
Molecular Phenotyping of Catalase-Deficient Arabidopsis Plants with ATH1 Affymetrix Microarrays
The transcriptome of catalase-deficient Arabidopsis plants (CAT2HP1, with 20% residual catalase activity) under nonstressed conditions was compared to control plants (PTHW; empty vector control). Plants were grown in Phytotron exposure chambers under strictly controlled environmental conditions, leaf material of 3-week-old plants was harvested, and samples from two biological repeat experiments were hybridized to four ATH1 Genome Arrays (Affymetrix). For details of sampling, hybridization, and data processing, see “Materials and Methods.” To assess transcriptional changes under photorespiratory-promoting conditions, 6-week-old plants were transferred to a sun simulator and exposed to continuous HL irradiation (1,800 μmol m−2 s−1) for 0, 3, 8, and 23 h. The lighting technology of the sun simulator has been especially designed for plant stress research and provides a well-balanced spectrum and environmental parameters, matching natural conditions as closely as possible (Thiel et al., 1996).
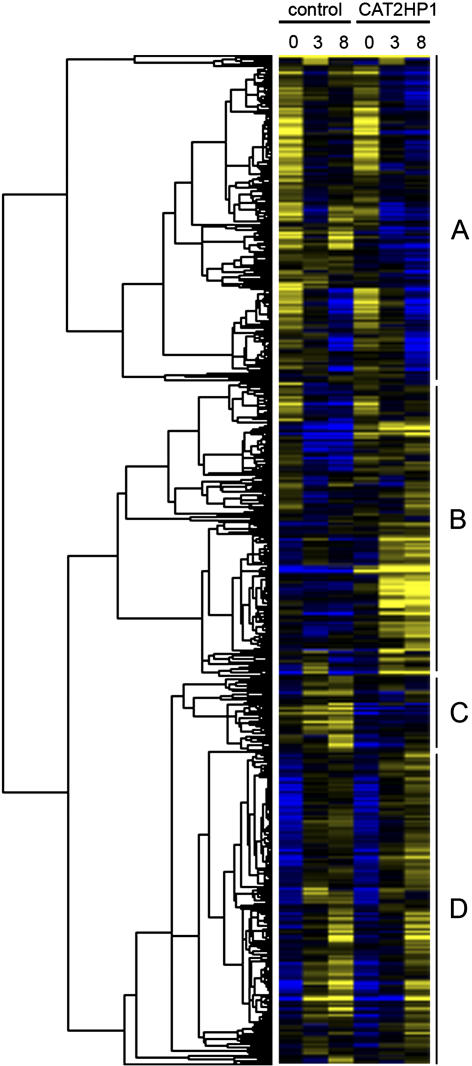
Leaves of catalase-deficient plants developed cell death after 8 h of HL, while in the control plants no cell death was visible even after 23 h of HL. Instead of developing cell death, leaves of control plants colored brown-purple, due to the accumulation of anthocyanins. Cell death in the catalase-deficient plants occurred both in the mesophyll and palisade parenchyma and was spatially correlated with increased H2O2 levels (Vandenabeele et al., 2004). RNA was isolated from pools of leaves of 20 to 30 control and CAT2HP1 plants subjected to 0, 3, and 8 h of HL. Samples from two biological repeat experiments were hybridized to ATH1 Affymetrix Genome Arrays. After data processing and normalization, a coefficient of variation (CV) was used as a selection criterion for differential gene expression during the treatment or between the two genotypes. The CV was calculated as the ratio of the sd on all measurements and the absolute value of the average expression over the time course. A total of 1,495 genes were differentially expressed with a CV > 2 and were subjected to hierarchical average linkage clustering (Fig. 1). Different prominent clusters of transcriptional changes stand out clearly: cluster A (484 genes) represents mainly genotype-independent HL-repressed genes; cluster B groups 437 genes that are exclusively induced by HL in the CAT2HP1 plants; cluster C contains 111 genes that are repressed in the CAT2HP1 plants; and cluster D (463 genes) comprises mainly (genotype-independent) HL-induced genes. For the complete list of genes within these different clusters, see Supplemental Table I. Alternatively, genes were classified according to their fold change in expression. Therefore, the threshold for positive response was set at 3-fold change in expression. Of the 906 genes whose expression was exclusively affected by HL, 379 were up-regulated and 527 were down-regulated. Screening for differentially expressed genes in response to H2O2 revealed 349 and 88 genes induced and repressed by H2O2, respectively. Lists of these genes are available as Supplemental Table II.
Figure 1.
Hierarchical average linkage clustering of 1,495 differentially expressed genes in control and catalase-deficient Arabidopsis plants during HL irradiation. Temporal expression patterns in control and catalase-deficient (CAT2HP1) plants during the HL time course (0, 3, and 8 h) are presented. Yellow and blue correspond to up-regulation and down-regulation, respectively. Four main clusters of transcriptional changes (A, B, C, and D) are indicated.
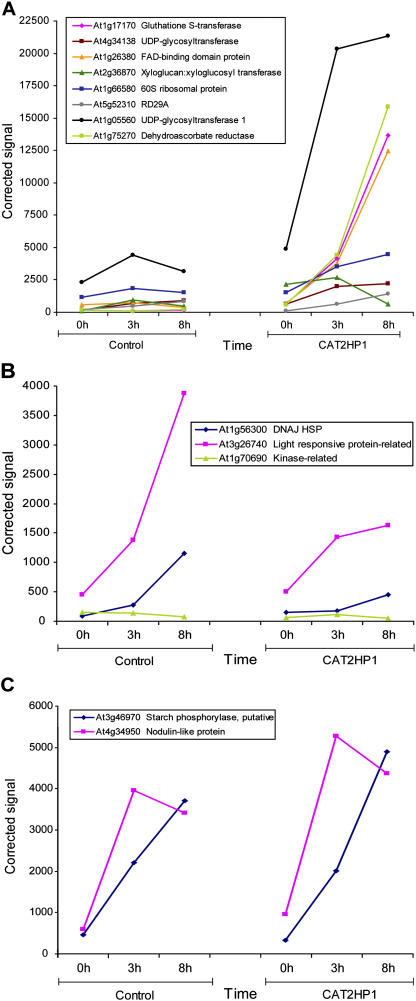
To validate the robustness of the expression data obtained by the microarray analysis, we performed a nonsaturated cDNA-amplified fragment length polymorphism (AFLP) analysis as an alternative strategy to reverse transcription-PCR. cDNA-AFLP is a sensitive and quantitative gel-based transcriptome technology, proven to be a valid alternative to microarrays. Basically, the method consists in the generation of unique restriction fragments from reverse-transcribed messengers, followed by a series of selective PCR amplifications of distinct subsets of transcript tags that are then separated and visualized on high-resolution gels (Breyne et al., 2003). On the same RNA samples used for the microarray hybridizations, 26 randomly chosen AFLP primer combinations with three selective nucleotides were used. For each primer combination, 80 to 100 transcript fragments are visualized. All fragments were quantified, resulting in individual intensities for each time point per transcript fragment. From the differentially expressed fragments, 20 transcripts were excised from the gels, reamplified by PCR, and effectively sequenced. From these sequences, 15 were derived from genes with a unique probe set on the ATH1 microarray, and their expression profiles were compared with those generated by the microarray analysis. The expression kinetics of 13 transcripts, obtained by the cDNA-AFLP analysis, were highly similar to those in the microarray analysis (Fig. 2), except for two genes (At3g11950 and At1g74470). This inconsistency might be explained by a differential sensitivity of the two platforms, but overall the strong correlation between the obtained microarray and cDNA-AFLP data was verified.
Figure 2.
Verification of microarray results by cDNA-AFLP. Expression profiles obtained by cDNA-AFLP are shown for 13 transcripts whose similar expression patterns were found in the microarray analysis. For the sake of clarity, three different graphs (A, B, and C) group transcripts with similar kinetics during the HL treatment. For control and CAT2HP1 plants, each color line connects the different time points at which the samples were collected. Arabidopsis Genome Initiative (AGI) codes and annotations are indicated. Expression data were processed as described in “Materials and Methods.”
Catalase Deficiency under Ambient Growth Conditions Provokes Differences in Gene Expression in 6-Week-Old Plants
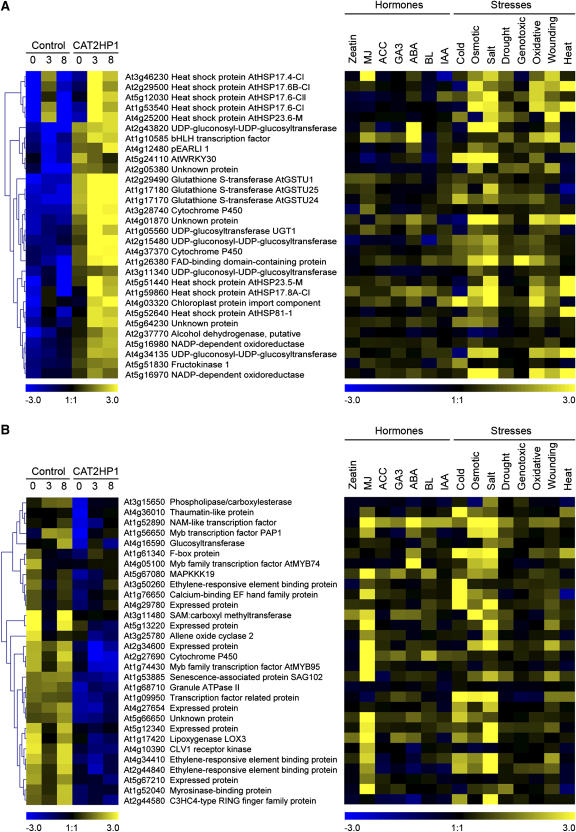
Deficiencies in antioxidative potential are known to affect growth and productivity of plants. Both Arabidopsis knockout ascorbate peroxidase and knockdown Cu/Zn superoxide dismutase plants are characterized by suppressed growth and development (Pnueli et al., 2003; Rizhsky et al., 2003). Although, to our knowledge, no Arabidopsis knockout lines of a catalase gene have been described until now, transgenic catalase RNAi Arabidopsis plants with only 7% residual catalase activity (CAT2HP2) are significantly retarded in growth and undergo spontaneous lesion formation. By contrast, catalase-deficient plants with 20% residual catalase activity (CAT2HP1), grown under controlled conditions, show no apparent phenotypical differences compared to wild-type plants (Vandenabeele et al., 2004). Nevertheless, a lack of any visible effect of this 80% reduction in catalase activity could be accompanied by significant differences at the molecular level. Therefore, we performed a comparative transcriptome analysis on 3- and 6-week-old CAT2HP1 and control plants. At the age of 3 weeks, no drastic differences in gene expression could be observed: Only three genes were more than 3-fold changed in transcript levels, and 30 genes were found by lowering the threshold to 2-fold change in expression (Supplemental Table III). This observation suggests that at this developmental stage and under these conditions, severe deficiencies in peroxisomal catalase do not increase the levels of nonscavenged photorespiratory H2O2 that are able to steer nuclear gene expression. Probably because under these nonstressed conditions and especially at that age, photorespiration activity is minor and the general antioxidant capacity is still high enough to allow the immediate consumption of any photorespiratory H2O2 that might escape. This is in accordance with the suggested age-dependent increases in H2O2 as a result of a programmed repression of antioxidant enzymes (Ye et al., 2000). On the contrary, in leaves of 6-week-old nonstressed CAT2HP1 plants, 179 genes showed at least a 3-fold change in transcript levels compared to control plants. The steady-state levels of 51 transcripts were increased in CAT2HP1 plants under nonstressed conditions (Fig. 3A; Supplemental Table IV). Most genes are stress related, and the prominent groups encode glutathione S-transferases, (small) heat shock proteins (HSPs), and UDP-glycosyltransferases (UGTs). Almost all of these 51 induced transcripts, including a WRKY (At5g24110) and a bHLH (At1g10585) transcription factor, are further highly responsive during the subsequent HL treatment in the sun simulator and show a pronounced responsiveness to various other environmental stresses and hormone treatments. Several members of the bHLH class of transcription factors have been shown to be involved in stress responses in plants and animal systems (Wang et al., 2003; Lorenzo et al., 2004), and the transcription of WRKY genes is strongly and rapidly induced in response to wounding, pathogen infection, or abiotic stress in numerous plant species (Eulgem et al., 2000). By using GENEVESTIGATOR (Zimmermann et al., 2004), we revealed that the 30 highest induced genes in nonstressed CAT2HP1 plants were also consistently induced by wounding, heat, salt, osmotic, and genotoxic stress (Fig. 3A). Therefore, these genes might be involved in primary cellular stress responses mediated by increased levels of H2O2.
Figure 3.
Differentially regulated genes in CAT2HP1 under ambient growth conditions: temporal expression patterns during HL and their responsiveness to various environmental stimuli. The 30 most up-regulated (A) or down-regulated (B) genes in nonchallenged CAT2HP1 plants are presented. Hierarchical average linkage clustering of the genes during 0, 3, and 8 h of HL irradiation in CAT2HP1 and control plants (left) and corresponding response profiles to various stress conditions and treatment with plant hormones (right) are shown. AGI codes and annotations are indicated. Data were obtained with genevestigator, a database and Web browser data mining interface for Affymetrix GeneChip data, allowing a rapid assessment of the expression characteristics of selected genes throughout a wide variety of environmental and developmental conditions (Zimmermann et al., 2004). ACC, 1-Aminocyclopropane-1-carboxylic acid; GA3, gibberellic acid 3; ABA, abscisic acid; BL, brassinolide; IAA, indole-3-acetic acid.
The steady-state levels of over 125 transcripts were decreased in the CAT2HP1 plants under ambient growth conditions (Fig. 3B; Supplemental Table V). Figure 3B lists the 30 most down-regulated genes together with their expression characteristics upon HL and their response profiles to other environmental stresses and stimuli obtained through GENEVESTIGATOR. An inverse responsiveness to methyl jasmonate (MJ) of 23 genes was observed. This antagonistic correlation is significantly higher than expected by chance (P = 0.029; Fisher's exact test), implying that during a stress in which MJ and H2O2 signals act simultaneously, the MJ-driven gene expression will be impaired by H2O2. Such an antagonistic interplay between ROS and jasmonate signaling has recently been reported in chlorophyllase 1-silenced plants. In these plants, jasmonate-induced defense gene expression and associated resistance against Alternaria brassicicola are reduced by ROS (Kariola et al., 2005). Another molecular marker for MJ responsiveness, the vegetative storage protein 1, is 5-fold repressed in CAT2HP1 under ambient growth conditions. MJ-dependent up-regulation of vegetative storage protein 1 is accompanied by the accumulation of anthocyanins in soybean (Glycine max) and Arabidopsis leaves (Franceschi and Grimes, 1991; Ellis and Turner, 2001). Another remarkable trend was that these H2O2-down-regulated genes positively responded to salt, cold, and osmotic stress. Also the H2O2-up-regulated genes were induced upon these stresses (Fig. 3A), illustrating not only the complexity but also the flexibility of the plant's stress response. Looking at a single treatment response of the individual stresses does not necessarily mean that the combination of treatments will be exactly additive. Therefore, it cannot be excluded that one signal pathway is epistatic or dominant over the other, or that the integration of signals is more complicated.
H2O2-Induced Genes and Their Response upon Other Environmental Stresses
Exposure to HL drastically reprograms the transcriptome. In the CAT2HP1 plants, HL irradiation results in elevated levels of H2O2, which also steer nuclear gene expression. Elevated levels of H2O2 positively regulate genes involved in the defense response, hypersensitive response, proteases, transcription and translation, and mitochondrial metabolism (Vandenabeele et al., 2003, 2004). One of the challenges is to organize these transcriptional changes into hierarchical gene networks and to identify the key signal transduction components within these networks that are potentially applicable to the development of (abiotic) stress-resistant plants. To this end, a series of genetic studies can be performed or expression characteristics of stress-induced genes can be compared under a variety of specific stress conditions and mutants (Chen et al., 2002).
To present a more justified insight into the generally accepted role of H2O2 as a signal in stress-related gene deregulation, the H2O2-up-regulated genes (349 genes; Supplemental Table III) were compared to those induced by a selection of relevant abiotic stresses, for which Affymetrix ATH1 expression data were publicly available: methyl viologen, cold, heat, and drought. Such a comparison would identify, on one hand, genes that are regulated by all stresses, which therefore are good candidates as key genes within the plant's abiotic stress response, and, on the other hand, obviously, also genes with an expression pattern specific for one of the stresses. The AtGenExpress repository provides several genome-wide transcriptome studies of Arabidopsis seedlings exposed to a variety of environmental stresses within robust time courses (http://web.uni-frankfurt.de/fb15/botanik/mcb/AFGN/atgenextable2.htm). For each of these stresses, the most appropriate time point was selected and fold changes in expression were calculated. For more details on the processing, normalization, time point selection, and size of the data sets, see “Materials and Methods.”
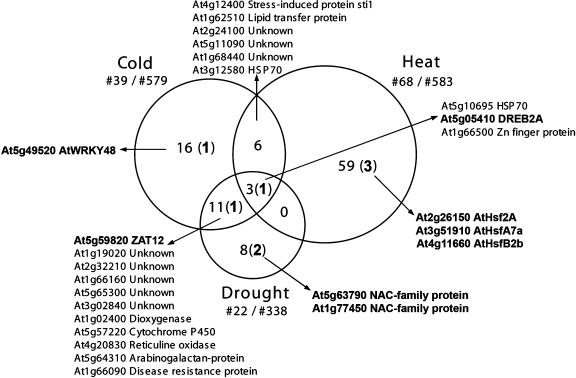
First, we compared the H2O2 data set with methyl viologen up-regulated genes. Methyl viologen is a redox-cycling herbicide that generates superoxide radicals in the chloroplasts and mitochondria. Most of this superoxide subsequently dismutates to H2O2, either spontaneously or enzymatically via superoxide dismutases (Bowler et al., 1992). Within 12 h of methyl viologen treatment (10 μm), 115 genes were at least 3-fold up-regulated, from which 58 (P = 9.3e-18; Fisher's exact test) were also induced by H2O2 (Supplemental Table VI), leaving 291 and 57 genes that responded specifically to H2O2 and superoxide, respectively. Second, we assessed the expression of H2O2-induced genes within three principal environmental stresses: heat, cold, and drought. Figure 4 presents the overlap of the H2O2-induced genes with each of the environmental stresses. Twenty genes were induced in response to H2O2 and under at least two stress conditions. Within these 20 commonly induced genes, two transcription factors, DREB2A and ZAT12, could be identified, which have already been linked to H2O2 responses (Rizhsky et al., 2004). DREB2A is known to be a key regulator of drought response (Shinozaki and Yamaguchi-Shinozaki, 2000), whereas ZAT12 participates in regulation of cold-responsive genes and contributes to an increase in freezing tolerance (Rizhsky et al., 2004; Vogel et al., 2005). The up-regulation of other H2O2-responsive transcription factors was restricted to one specific environmental stress. AtWRKY48 was also induced by cold, whereas two NAC family proteins were responsive to drought. These NAC proteins belong to the ATAF subfamily and have been shown to respond upon abscisic acid, dehydration, and salt treatments (Fujita et al., 2004).
Figure 4.
H2O2-up-regulated genes within three principal environmental stresses. The different stress conditions (cold, heat, and drought) are indicated together with the number of genes within the overlap with the H2O2-responsive genes and the total number of gene input of each of the stresses as well. In the Venn diagram, numbers of genes are given that are unique to that gene set or in the common sections between sets, with the amount of transcription factors present within a specific gene set in parentheses. AGI codes and descriptions are given for genes in the common sections of the stresses. Transcription factors are indicated in bold.
Because deregulated transcription factors in our data set were obviously the primary candidates to steer H2O2-responsive gene expression, we assessed specifically the expression characteristics of all annotated transcription factors during the HL exposure. Of the approximately 1,500 genes annotated as potential transcription factors (Riechmann et al., 2000), 20 were at least 3-fold induced in the CAT2HP1 plants after 3 h of HL. Table I lists these transcription factors and their expression characteristics in the CAT2HP1 plants after 0, 3, and 8 h of HL and in response to the four selected abiotic stresses. Assuming that the increased expression of transcription factors reflects their involvement in steering underlying transcriptional regulons, we can suggest that these 20 different individual transcription factors might be involved directly or indirectly in the up-regulation of the nearly 350 genes during H2O2 stress. Notwithstanding our focus on induced transcription factors, down-regulation of suppressors might as well be a mechanism that regulates H2O2-driven gene expression. Assessing the expression characteristics of the 20 induced transcription factors in the other environmental stresses revealed that some have very similar expression characteristics (Table I), demonstrating the regulatory role of H2O2 in diverse environmental stresses. Future detailed functional analysis, including additional microarray analysis on transgenic plants perturbed in these transcription factors, will reveal more precisely which members of the H2O2-responsive genes are mastered by these individual transcription factors.
Table I.
H2O2-induced transcription factors and their response to other environmental stresses
Relative expression data (CAT2HP1 to control or stress to control) of the 20 H2O2-up-regulated transcription factors are presented. Affymetrix probe set names, AGI codes, and descriptions are given. The induction/suppression of each transcription factor by HL treatment (0, 3, and 8 h) in the CAT2HP1 plants or by the different stressful conditions (heat, cold, drought, and methyl viologen) is indicated. Fold change values marked by asterisks have absent calls (i.e. no detectable expression) in the control or nonstressed condition. Expression data of transcripts with absent calls in both control and stressed condition are indicated with A.
| Probe Set Name | AGI Protein Code | Description | CAT2HP1 0 h | CAT2HP1 3 h | CAT2HP1 8 h | Heat 6 h | Cold 6 h | Drought 1 h | MVa 12 h |
|---|---|---|---|---|---|---|---|---|---|
| 263210_at | At1g10585 | bHLH transcription factor | 7.25 | 85.12 | 16.03 | −1.58 | −1.39 | −4.71 | 1.72 |
| 252081_at | At3g51910 | AtHsfA7a | 1.08 | 28.77* | 4.30* | 20.75* | A | A | −1.28 |
| 266841_at | At2g26150 | AtHsf2A | 1.61 | 27.64 | 112.82 | 1,963.51* | A | A | 18.82* |
| 264202_at | At1g22810 | TINY-like transcription factor | −1.72 | 24.15* | 4.93* | A | A | A | A |
| 249770_at | At5g24110 | AtWRKY30 | 6.00 | 14.85* | 4.84 | A | A | A | A |
| 245173_at | At2g47520 | Putative AP2 domain transcription factor | A | 12.99* | 22.61* | A | A | A | A |
| 257918_at | At3g23230 | Ethylene response factor B-3 | 1.77 | 10.30* | 2.45* | A | A | A | 3.25* |
| 250781_at | At5g05410 | DREB2A | 2.05 | 8.01 | 8.96 | 11.11 | 12.95 | 3.63 | 3.39 |
| 264746_at | At1g62300 | AtWRKY6 | 1.37 | 7.68 | 6.03 | A | A | 2.66 | 4.23* |
| 248611_at | At5g49520 | AtWRKY48 | −1.70 | 6.13 | 3.49 | A | 3.94* | A | A |
| 245976_at | At5g13080 | AtWRKY75 | A | 5.70* | 92.92* | A | A | A | A |
| 264726_at | At1g22985 | Ethylene response factor B-5 | −1.48 | 5.64 | 12.72 | 2.90 | −1.01 | −1.01 | 1.41 |
| 259705_at | At1g77450 | NAM family protein | 1.82 | 5.03 | 3.39 | A | A | 7.30 | 2.19 |
| 267140_at | At2g38250 | GT1-like transcription factor, putative | A | 4.98* | 7.83* | A | A | A | A |
| 254878_at | At4g11660 | AtHsfB2b | 1.07 | 4.38 | 4.06 | 19.42 | A | 1.58* | 1.06 |
| 250024_at | At5g18270 | NAM family protein | 1.60 | 4.10* | 12.99* | A | A | A | A |
| 263823_s_at | At2g40350 | AP2 domain transcription factor | A | 4.03 | 6.55 | −2.81 | 1.49 | A | 1.75 |
| 247655_at | At5g59820 | Zat12 zinc finger protein | 1.04 | 3.27 | 3.41 | 2.49 | 15.73 | 4.91 | 38.62* |
| 247351_at | At5g63790 | NAM family protein | 2.52 | 3.25 | 4.37 | −2.59 | 1.98 | 3.28 | 2.32 |
| 258803_at | At3g04670 | AtWRKY39 | 1.18 | 3.02 | 3.90 | 1.48 | 1.45 | 1.07 | −1.00 |
Methyl viologen.
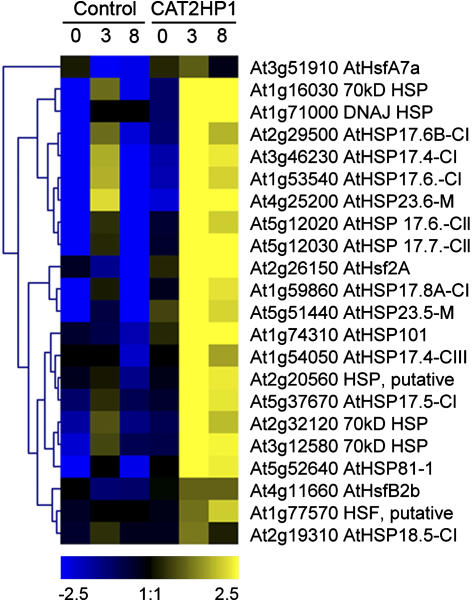
H2O2-Dependent Up-Regulation of Small HSPs during HL Stress
An abundant functional class within the set of H2O2-up-regulated genes (Supplemental Table III) is that of HSPs. Eleven transcripts encoding both cytoplasmic and mitochondrial small HSPs (smHSPs) and four HSP70s were at least 3-fold induced in CAT2HP1 plants within 8 h of HL irradiation. In control plants, a transient induction of the smHSPs is noticed at 3 h of HL, probably reflecting a transient rise in H2O2 that is subsequently scavenged (Fig. 5). Due to a constant electronic monitoring of the growth condition parameters in the sun simulator, artifactual temperature fluctuations can be excluded during the HL treatment (data not shown). Together with the smHSPs, heat shock factors (HSFs) AtHsf2A, AtHsfB2b, and AtHsfA7a showed responsiveness to H2O2. A rapid induction of two other HSFs, AtHsfA4a and AtHsfA8, in response to moderate light stress has been reported in knockout ascorbate peroxidase plants (Pnueli et al., 2003). Surprisingly, the steady-state levels of transcripts encoding these HSFs were not significantly elevated during HL stress in the CAT2HP1 plants. Therefore, our data suggest that, depending on the subcellular origin of the oxidative stress, specific HSFs can trigger the downstream response. Another level of specificity lies in the chemical identity of the ROS. In tomato (Lycopersicon esculentum), an smHSP could only be induced by H2O2 and not by superoxide radicals (Banzet et al., 1998). Further, the complexity of the heat shock response in different organisms is illustrated by its oxidative stress-independent induction in maize (Zea mays) and Caenorhabditis elegans (Kuzmin et al., 2004).
Figure 5.
Induction of HSPs in catalase-deficient plants during HL stress. Hierarchical average linkage clustering of the HSPs and HSFs induced during 0, 3, and 8 h of HL irradiation in the control and CAT2HP1 plants. Only HSPs/HSFs with a more than 3-fold increase in expression are included. AGI codes and annotations are indicated.
A more general comparison between H2O2- and heat shock-responsive genes (see Fig. 4) indicated that of the 583 heat shock-responsive genes, 68 were also up-regulated by H2O2 in the CAT2HP1 plants (Supplemental Table VII). Such an overlap is highly significant (P = 1.77e-14; Fisher's exact test). Together with the previously mentioned HSPs and HSFs, several unknown and defense-related genes were present within this overlap. Because temperature-independent induction of smHSPs has been observed in response to HL (Pnueli et al., 2003; Yamamoto et al., 2004) and to various other abiotic stress conditions (genevestigator; Zimmermann et al., 2004), a subset of genes within the heat shock response might be triggered by increased levels of H2O2, implicating the presence of two transcriptional regulons during heat stress: one regulated by the increase in temperature and a smaller one triggered by bursts of H2O2 generated during heat stress (Larkindale and Knight, 2002). H2O2 is clearly able to induce the smHSPs of the Hsp17.6 class. In the different assessed abiotic stresses, these smHSPs are coexpressed with AtHsf2A; recently, this class of cytoplasmic smHSPs has been shown not to be under transcriptional control of HsfA1a/HsfA1b during heat shock (Busch et al., 2005), suggesting that AtHsf2A might be the driving force for the transcriptional up-regulation of smHSPs under abiotic stress conditions, even in the absence of a heat stress. Interdependence of Hsf2A and smHSPs has also been observed at the posttranscriptional level: in tomato, the activity of Hsf2A is controlled by a network of nucleocytoplasmic smHSPs, influencing its solubility, intercellular distribution, and functional state (Port et al., 2004). In fruit fly (Drosophila melanogaster) and mammalian cells, HSFs might act as direct sensors of H2O2 (Zhong et al., 1998; Ahn and Thiele, 2003), and recently a role for AtHsfA4a in the early sensing of H2O2 stress has been demonstrated in Arabidopsis (Davletova et al., 2005).
The Reactive Oxygen Gene Network
A tight regulation of the steady-state levels of ROS in plants is necessary to avoid cellular injury and to maintain a base level of ROS on which different environmental and developmental signals can be registered. Oxidative stress management happens at both production and scavenging levels by a network of at least 152 genes in Arabidopsis (Mittler et al., 2004). Genetic analyses of some key antioxidant components clearly showed a high degree of flexibility and redundancy in this reactive oxygen gene network. Changing the balance of ROS scavenging by knocking out or decreasing the expression of one ROS gene is mostly accompanied by increased transcript levels of another ROS gene (Mittler et al., 2004). Here, almost one-third of the reactive oxygen gene network was found to be transcriptionally deregulated (at least 2-fold) by the catalase 2 deficiency or the HL irradiation (Supplemental Table VIII). Peroxiredoxin D (At1g60740), alternative oxidase a At3g22370), and a putative mitochondrial alternative oxidase (At1g32350), which were also induced by other abiotic stresses, such as drought and salt (Mittler et al., 2004), were very strongly elevated by H2O2. Surprisingly, except for dehydroascorbate reductase 3, from which the subcellular localization is not known exactly, no chloroplastic ROS genes were differentially regulated in CAT2HP1 plants within 8 h of HL. In contrast, several other chloroplastic ROS genes, such as iron superoxide dismutases (FSD2 and FSD3), glutathione peroxidases (GPX6 and GPX7), ferritin 2, and stromal/mitochondrial ascorbate peroxidase, were induced by HL irradiation itself both in CAT2HP1 and control plants, showing their independency of H2O2 during this induction. Besides the apparent down-regulation of the two other catalase genes, CAT1 and CAT3, after 3 and 8 h of HL, no other members of the reactive oxygen gene network were more than 3-fold down-regulated by H2O2.
H2O2 Impairs the HL Induction of a Transcriptional Cluster Involved in Anthocyanin Biosynthesis
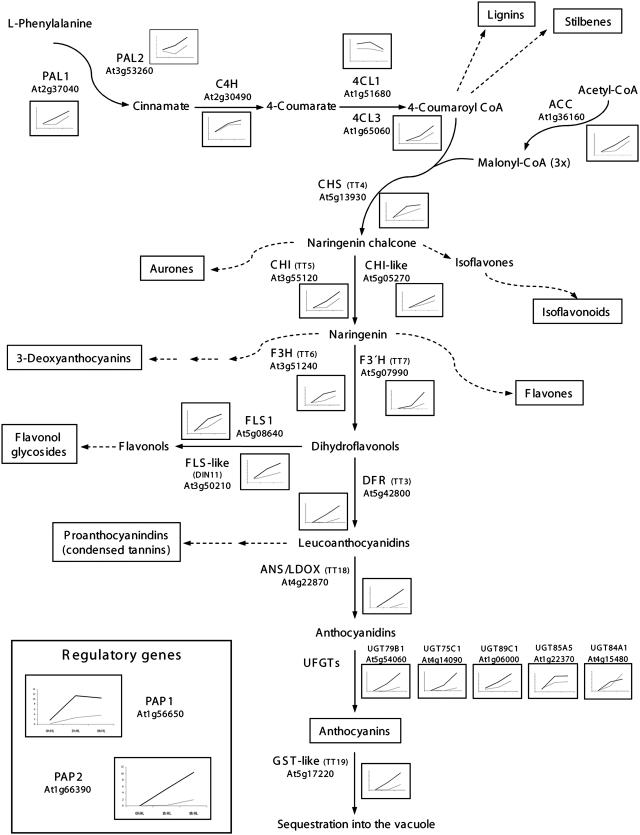
Global changes in gene expression in plants are steered by HL. Previously reported transcriptome studies in Arabidopsis revealed a differential expression of at least 185 and 110 genes in response to 1 and 3 h of HL, respectively (Rossel et al., 2002; Kimura et al., 2003). In our analysis, after 3 h, HL drives the up-regulation of nearly 380 genes in control plants (Supplemental Table III). When assessing the expression profiles of these genes in both HL-exposed CAT2HP1 and control plants, a clear subcluster could be recognized in which the induction by HL was significantly delayed in the CAT2HP1 plants. Whereas in control plants transcripts levels increased rapidly within 3 h of HL, they only reached their highest expression levels after 8 h in the CAT2HP1 plants. Within this subcluster, genes known to be involved in the regulation, biosynthesis, and sequestration of anthocyanins were predominantly present. To explore this aspect in more detail, we first assessed the individual expression levels of all known genes of the flavonoid biosynthetic pathway in plants (Winkel-Shirley, 2002; Kitamura et al., 2004). This flavonoid pathway is part of the larger phenylpropanoid pathway that also produces a range of other secondary metabolites, such as lignins and stilbenes. The biochemistry of the core steps in both the general phenylpropanoid and flavonoid pathways has been completely elucidated (Schwinn and Davies, 2004). Figure 6 depicts the expression profiles in response to HL in both control and CAT2HP1 plants for (1) genes annotated to the general phenylpropanoid pathway, which delivers 4-coumaroyl-CoA to chalcone synthase (CHS) for the biosynthesis of anthocyanins and other flavonoids. In addition, a gene encoding acetyl-CoA carboxylase is present. Acetyl-CoA carboxylase is needed for the production of malonyl-CoA, the second start product for the biosynthesis of chalcone; (2) genes encoding the enzymatic steps from chalcone to anthocyanins; (3) genes involved in glycosylation and sequestration; and (4) two known regulatory MYB transcription factors, production of anthocyanin pigment 1 (PAP1) and PAP2 (Borevitz et al., 2000; Winkel-Shirley, 2002; Springob et al., 2003).
Figure 6.
Schematic overview of the flavonoid biosynthetic pathway in Arabidopsis (based on Winkel-Shirley, 2002; Kitamura et al., 2004). Catalyzing enzymes, AGI codes, and corresponding genetic loci in parentheses are indicated. Graphs represent the expression profiles of the corresponding genes in response to 0, 3, and 8 h of HL irradiation: solid line, control; dashed, CAT2HP1. Dashed arrows indicate different branches of the flavonoid pathway involved in the biosynthesis of other flavonoids or secondary metabolites that were not activated in the experiment. The names of the major classes of (flavonoid) end products are boxed. C4H, Cinnamate-4-hydroxylase; ACC, acetyl-CoA carboxylase; CHI, chalcone isomerase; F3H, flavonone 3-hydroxylase; F3′H, flavonoid 3′ hydroxylase; DFR, dihydroflavonol reductase; ANS/LDOX, anthocyanidin synthase/leucoanthocyanidin dioxygenase; UFGT, UDP-Glc:flavonoid glycosyltransferase; GST, glutathione S-transferase; TT, testa transparent; DIN, dark-inducible.
Some components of the phenylpropanoid pathway, such as Phe ammonia-lyase (PAL) and 4-coumarate:CoA ligase (4CL), are encoded by small gene families (Raes et al., 2003). From the four PAL gene family members, only PAL1 and PAL2 have similar expression profiles during HL stress. These most prominent stress-responsive PAL genes are also the best candidates for a function in monolignol biosynthesis (Raes et al., 2003). Similarly, from the four 4CL genes, only 4CL1 and more pronounced 4CL3 respond upon exposure to HL (Fig. 6). Interestingly, 4CL3 has been suggested to be the most important substrate deliverer for CHS and, subsequently, to flavonoid biosynthesis (Ehlting et al., 1999). 4CL1 has a rather distinct expression pattern in our analysis and has been shown to be involved in lignin formation and production of additional phenolic compounds other than flavonoids (Ehlting et al., 1999; Raes et al., 2003). Flavonol synthase (FLS), which is part of the flavonol glycoside branch, is also represented by a multigene family. Only FLS1 and an FLS-like protein, encoded by dark-inducible 11 (Fujiki et al., 2001), are responsive to HL and are present in the proposed anthocyanin cluster. Therefore, out of the six members of the FLS family, FLS1 is the primary candidate to participate in the flavonol biosynthesis pathway in Arabidopsis. Tissue-specific regulatory genes, such as TT2, TT8, and the biosynthetic key enzyme anthocyanidin reductase (BANYULS) involved in branched anthocyanin pathways, such as proanthocyanidins, are absent in this cluster (Xie et al., 2003; Baudry et al., 2004). This is not unexpected because earlier biochemical analyses have revealed that flavones and anthocyanin derivatives accumulate in vegetative parts, whereas flavonols and proanthocyanidins predominantly accumulate in the endothelium layer of the seed coat (Chapple et al., 1994).
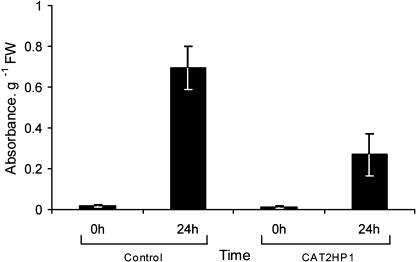
By combining a genetic (catalase deficiency) and an environmental (HL) perturbation, we were able to dissect this transcriptional cluster involved in anthocyanin biosynthesis out of the general light stress response. This molecular phenotype also correlated with the phenology of the CAT2HP1 plants under HL stress. Control plants accumulated much more anthocyanins than CAT2HP1 plants after exposure to HL (Fig. 7). In contrast to the control plants, CAT2HP1 plants developed cell death within 8 h of HL, suggesting a protective role of anthocyanins against HL stress. Anthocyanins have the ability to reduce the potential to oxidative damage via light attenuation (Steyn et al., 2002). Failure of anthocyanin accumulation (sunscreen) might contribute to the increased sensitivity of CAT2HP1 plants under HL exposure (Hoch et al., 2003). Under low-light conditions or in covered leaves of control plants, no or only a strongly reduced coloration was observed (Fig. 7).
Figure 7.
Quantification of anthocyanin content. Bars represent the mean anthocyanin content (expressed as absorbance per g FW) in control and CAT2HP1 plants exposed to 0 and 24 h of HL irradiation. Error bars correspond to ±se.
Identification of Candidate Genes Involved in Anthocyanin Biosynthesis and Regulation
Anthocyanin pigments are predominantly present in glycosylated forms. This glycosylation contributes to the increased stability of anthocyanins and influences their color variation (Schwinn and Davies, 2004). Glycosylation steps are catalyzed by UDP-Glc:anthocyanidin 3-O-glycosyltransferases and 5-O-glycosyltransferases (Springob et al., 2003) that belong to a larger protein family of UGTs. In Arabidopsis, 107 gene sequences have been annotated as glycosyltransferases, which can be divided into 14 major evolutionary groups (Ross et al., 2001). However, the knowledge on the functionality of the individual genes remains scarce because the corresponding substrate and the associated biological processes are mostly unknown. Until now, only two flavonoid glycosyltransferases have been functionally characterized in Arabidopsis: UGT73C6 (At2g36790) and UGT78D1 (At1g30530; Jones et al., 2003). These UGTs are involved in flavonol glycoside biosynthesis, which is a side branch away from anthocyanidin production. UGT73C6 was induced upon H2O2 stress in the CAT2HP1, but both UGTs were not present within the identified transcriptional regulon. Using a genomic approach in which most UGT sequences were cloned, recombinantly expressed, and subsequently tested on a large scale for their catalytic activity, a few UGTs involved in monolignol, auxin, and salicylic acid have been characterized (Jackson et al., 2001; Li et al., 2001; Lim et al., 2002). An alternative strategy to address their potential functionality is to identify UGTs in transcriptional regulons associated with a specific biological process. By assuming the guilt-by-association principle, these UGTs would most probably be involved in this process. Whereas 11 UGTs were up-regulated by H2O2 stress in the CAT2HP1 plants, several other UGTs (Fig. 6) were found in the anthocyanin cluster. Hence, these UGTs would be very good candidates for the anthocyanin modification during HL stress. Very recently, two of these UGTs have been functionally validated as genuine flavonoid glycosyltransferases (Tohge et al., 2005). Similarly, other genes that are part of this cluster are most probably also involved in the production of anthocyanins or derivative products. To enable a more robust identification of UGTs and other genes in the cluster, we selected all genes whose expression levels were at least 3-fold induced after 3 h of HL stress but had at least a 1.5-fold lower expression in the CAT2HP1 compared to control plants. The expression characteristics of 98 genes matched these criteria (Supplemental Table IX). Within this list, we recognized, in addition to the earlier identified complete anthocyanin biosynthesis pathway, several unknown genes and genes involved in signal transduction and carbohydrate metabolism, such as glycosyltransferases and glycosyl hydrolases. Of the UGTs present (UGT89C1, UGT85A5, UGT75C1, UGT79B1, and UGT84A1; Fig. 6), three (UGT79B1, UGT75C1, and UGT84A1) were assigned to branches of the phylogenetic tree of the Arabidopsis UGT superfamily that are homologous to other plant UGTs involved in the 3-O-glycosylation of anthocyanins (Ross et al., 2001). Further functional analysis of these potential anthocyanin-related UGTs will now allow a more targeted validation toward their involvement in anthocyanin modifications. One can presume that within this selection of 98 genes, the genes with a hitherto unknown function or that had not been previously associated with anthocyanin biosynthesis are either part of the regulatory network, biosynthetic pathway(s), and associated processes involved in anthocyanin metabolism, or alternatively their regulation is triggered by the increased levels. Together with known regulators (PAP1-AtMYB75 and PAP2-AtMYB90), also several novel candidate signal transducers are present in this transcriptional cluster.
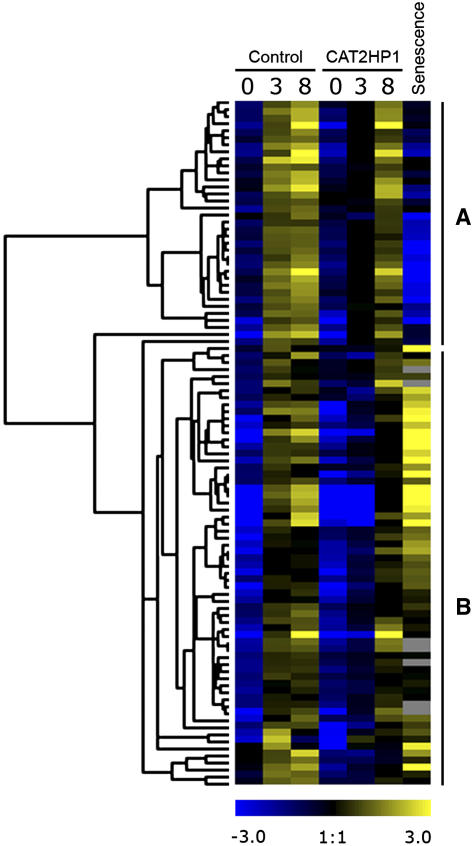
To validate the robustness of the selected genes, we assessed their expression during leaf senescence. Senescence is a well-characterized process in which anthocyanin levels are elevated (Hoch et al., 2001). To this end, we used the publicly available microarray data on senescing leaves (Schmid et al., 2005). For more details on the experiment, see “Materials and Methods.” Expression profiles of the 98 genes during the HL treatment and their behavior during senescence were clustered together. Unexpectedly, a division into two clusters was obtained (Fig. 8). Despite a similar H2O2-impaired HL induction of the genes in both clusters, the transcripts in cluster A were clearly down-regulated during senescence, whereas those in cluster B were up-regulated. Closer inspection of the functional categories within both clusters revealed also a functional divergence. Besides the genes of the general phenylpropanoid pathway, CHS, chalcone isomerase, flavonone 3-hydroxylase, and FLS1, cluster A contained mainly genes involved in starch metabolism: SEX1, starch phosphorylase, starch debranching enzymes, and glycoside hydrolases (Smith et al., 2003). Cluster B (65 genes) grouped, in addition to multiple unknown genes, the genes involved in the final anthocyanin biosynthesis and regulation. The apparently less efficient HL-induced up-regulation of transcripts involved in starch metabolism (Fig. 8) could be explained by the lower photosynthetic capacities that had been previously observed in catalase-deficient plants (Vandenabeele et al., 2003).
Figure 8.
Hierarchical average linkage clustering of the identified anthocyanin cluster and its behavior during senescence. Temporal expression patterns in control and CAT2HP1 plants during the HL time course (0, 3, and 8 h) are presented together with their expression profile during senescence. Two main clusters of transcriptional changes (A and B) are indicated.
Tissue-specific regulation of anthocyanin biosynthetic genes is controlled by a combination of MYB-related and bHLH-related transcription factors (Mol et al., 1998; Vom Endt et al., 2002). In Arabidopsis, two MYB transcription factors have been described to positively regulate anthocyanin accumulation via activation tagging (Borevitz et al., 2000). The resulting pap-1D mutant is characterized by an enhanced pigmentation and a massive activation of genes encoding PAL, CHS, dihydroflavonol reductase, and a glutathione S-transferase. Accordingly, expression of a chimeric PAP1 repressor results in a reduced accumulation of anthocyanins and suppressed expression of PAL, CHS, and dihydroflavonol reductase, suggesting that these genes are regulated by PAP1 (Hiratsu et al., 2003). Both PAP1 and PAP2 interact synergistically with the bHLH transcription factors GL3 and EGL3, which are in turn regulated by the WD repeat-containing protein TTG1 (Zhang et al., 2003). Both regulatory bHLH proteins and TTG1 were not transcriptionally deregulated upon HL stress in our analysis, suggesting that the induction of PAP1 and PAP2 might be sufficient to switch on the anthocyanin biosynthetic pathway.
Very recently and independently, a genome-wide transcriptome analysis on transgenic plants with ectopic PAP1 overexpression has been reported (Tohge et al., 2005) that led to the induction of 39 genes, among which are most of the known genes involved in anthocyanin production. In addition, 12 genes were down-regulated that in our analysis all have higher expression levels in CAT2HP1 plants compared to control plants. This independent analysis surely consolidates the robustness of our in silico-identified regulon, pointing out PAP1 as a key regulator within the HL-driven induction of the anthocyanin biosynthesis pathway. Whereas assessing downstream gene expression within a transgenic line overexpressing a transcription factor strictly identifies the underlying transcriptional gene network of this specific transcription factor, our (in silico) expression analysis allows the identification of a similar regulon together with genes that have a high degree of coregulation but whose transcription cannot be steered by the sole overproduction of PAP1. Among these genes, we anticipate to identify potential upstream regulators, potential coregulators, and even parallel pathways that are governed by a common upstream regulator. The advantage of identifying coregulated genes, through the analysis of transcriptomic changes provoked by multiple challenges and perturbations, is that the selection of identified genes is probably more confined and avoids the identification of coincidentally coregulated genes because they are not solely selected on the basis of an artificial overproduction of one regulatory gene (Bruce et al., 2000; Broun, 2004). In addition, even further upstream regulators might be identified, assuming that within a transcriptional cascade these upstream factors have similar expression characteristics (Vom Endt et al., 2002).
How H2O2 impinges negatively on this HL-induced anthocyanin regulon remains elusive. H2O2 might impair the transcriptional induction or activity of PAP1 and PAP2 directly, thereby blocking a putative autoregulatory transcriptional activation. However, because both PAP1 and PAP2 are already completely repressed under ambient growth conditions in CAT2HP1 plants, the elevated H2O2 levels more probably affect an upstream regulator, which steers the transcript levels of PAP1 directly or via a transcriptional cascade. An upstream regulator of PAP1 regulation might be present among genes whose expression is already down-regulated in nonstressed CAT2HP1 plants. Within this list, we can recognize several transcription factors and unknown proteins as primary candidates (Supplemental Table V). Alternatively, interplay with an H2O2-induced/activated inhibitor might occur. The Arabidopsis homolog AtMYB4 (At4g38620) of a reported anthocyanin biosynthesis repressor from strawberry (Fragaria ananassa; FaMYB1) is, at least transcriptionally, not induced by H2O2 (Jin et al., 2000; Aharoni et al., 2001). This inhibitor might be present in the list of genes that are up-regulated in the CAT2HP1 plants grown under ambient growth conditions. Obviously, a posttranscriptional modification (such as oxidation) of an existing protein cannot be excluded from modulating the activity of such an upstream regulator (Johansson et al., 2004). Future functional analysis of these signal transduction components and unknown genes will be necessary to assess whether and in which branches of the flavonoid biosynthesis they play a regulatory role.
MATERIALS AND METHODS
Plant Material, Growth Conditions, and Stress Treatments
Catalase-deficient (CAT2HP1) and control (PTHW) plants were obtained as described by Vandenabeele et al. (2004). Unless mentioned otherwise, the plants were grown under controlled conditions in phytotron exposure chambers, which had been especially designed for plant stress research (Thiel et al., 1996). The light regime was 12 h/12 h at 100 to 140 μmol m−2 s−1, the climate adjusted to a relative humidity of 70%, and 22°C/18°C day/night temperatures. For HL treatments, 6-week-old plants were transferred to a sun simulator with identical growth conditions and exposed to continuous HL irradiation (photosynthetically active radiation 400 to 700 nm at approximately 1,600 to 1,800 μmol m−2 s−1). Middle-aged leaves of 20 to 30 plants per line were sampled 0, 3, and 8 h after the onset of HL stress and pooled for RNA analysis. The two biological repeat experiments were done with a temporal interval of 1 year.
Microarray Analysis
In two independent experiments, RNA was isolated from leaves of 20 to 30 control or catalase-deficient plants with the TRIzol Reagent (Invitrogen). The concentration of total RNA was determined with a Nanodrop ND-1000 spectrophotometer, and the quality was examined with the RNA 6000 Nano Assay (Agilent Technologies, 2100 Bioanalyzer). Each of the different pools of control and CAT2HP1 plants, subjected for 0, 3, and 8 h of HL irradiation, was hybridized to one chip (Genechip Arabidopsis ATH1 Genome Array; Affymetrix). For each hybridization, 15 μg of total RNA was used. Affymetrix chip analyses were performed at the Functional Genomics Center Zurich and the VIB Microarray Facility (Leuven, Belgium). Conditions for reverse transcription, RNA labeling, hybridization, and scanning were according to the manufacturer's instructions (Affymetrix; https://www.affymetrix.com/). Raw data were processed with the statistical algorithm of the Affymetrix Microarray Suite 5.0 as described by Liu et al. (2002) and subsequently normalized by dividing all measurements on each chip by the 50th percentile value (median). To calculate the median, only measurements flagged as present (i.e. detectable expression) were used. Genes with at least four present calls over the 12 different data points were retained for further analysis. Expression values were obtained by averaging the normalized values of the two independent repeats. As a selection criterion for differential expression, a CV was used, namely the ratio of the sd on all measurements and the (absolute value of the) average expression over the time course. Expression values of genes with CV > 2 were taken for hierarchical cluster analysis with the Cluster and TreeView software (Eisen et al., 1998) to obtain a global view of the transcriptional changes. For the in-pair comparison at different time points, fold changes were calculated by using the average expression value of the two independent experiments. Only fold changes with at least two present calls over the four data points were used. Analyses were based on annotations compiled by The Arabidopsis Information Resource (http://www.arabidopsis.org/; Rhee et al., 2003), and the affy25k_array_elements-2003-05-30 version was used for ATH1 Affymetrix array element information.
cDNA-AFLP
RNA samples of the second biological repeat experiment were used and templates for cDNA-AFLP were prepared as described by Breyne et al. (2003). Twenty-six randomly chosen primer combinations (BstT1-Mse14, BstT1-Mse21, BstT1-Mse23, BstT1-Mse24, BstT2-Mse34, BstT2-Mse42, BstT3-Mse14, BstT3-Mse21, BstT3-Mse42, BstT4-Mse11, BstT4-Mse12, BstT4-Mse13, BstT4-Mse14, BstT4-Mse21, BstT4-Mse31, BstT4-Mse32, BstT4-Mse33, BstT4-Mse34, BstT4-Mse41, BstT4-Mse42, BstT4-Mse43, BstT4-Mse44, BstT4-Mse23, BstT4-Mse24, BstC4-Mse23, and BstC4-Mse13 [with 1, 2, 3, and 4 representing the selective nucleotides A, C, G, and T, respectively]) were used, and amplification products were separated on 5% polyacrylamide gels with the Sequigel system (Bio-Rad). Sodium acetate (22 mg/L) was added to the buffer in the bottom tank to avoid run-off of the smallest fragments. Gels were dried on 3MM Whatman paper, exposed to Kodak BioMax MS films, and scanned in a PhosphorImager 445 SI (GE-Biosciences). The gel images were processed with AFLP-QuantarPRO (Keygene Products), a software application that allows an accurate quantification of band intensities in DNA and RNA fingerprints. Intensity of individual bands was determined and normalized semiautomatically for variations, such as running or loading differences and image artifacts. The obtained raw expression data were further processed via the ARRAY-AN application as described by Vandenabeele et al. (2003). Bands corresponding to differentially expressed transcript tags were excised from the gels and reamplified by PCR with BstYI+2/MseI+0 primers. PCR fragments were sequenced directly.
Publicly Available Affymetrix GeneChip Data
The GeneChip data were retrieved from the international AtGenExpress repository (from The Arabidopsis Functional Genomics Network, http://www.uni-frankfurt.de/fb15/botanik/mcb/AFGN/AFGNHome.html) and downloaded from The Arabidopsis Information Resource (http://www.arabidopsis.org/servlets/Search?type=expr&search_action=new_search). Raw data were processed with the statistical algorithm of Affymetrix Microarray Suite 5.0 (Liu et al., 2002), and each chip was normalized as described above. Growth stage annotations were based on Boyes et al. (2001).
Abiotic Stresses
The (publicly available) cold and drought stress experiments were performed by J. Kudla's group (Ulm, Germany), the heat experiment by the group of L. Nover and P. von Koskull-Döring (Frankfurt, Germany), and the methyl viologen treatment by the group of D. Bartels (Bonn, Germany). For more information on the exact conditions of the different stress treatments, see http://web.uni-frankfurt.de/fb15/botanik/mcb/AFGN/atgenextable2.htm.
All abiotic stress treatments and sample preparations considered in this study were done in the laboratory of J. Kudla. Seeds of Arabidopsis (Arabidopsis thaliana L.) Heynh. wild type (ecotype Columbia-0) were sown on rafts in agar with Murashige and Skoog medium and grown under a 16 h/8 h light/dark regime, 24°C, 50% relative humidity, and 150 μE cm−2 s−1 light intensity. At day 11, the rafts were transferred to liquid media. At day 16, stress treatments were started at 3 h of light and treated with a specific stress for 0.5, 1, 3, 6, 12, and 24 h. Samples of all stress treatments were harvested in duplicate at the growth stage 1.04 and hybridized to Affymetrix Genechip arrays (see below). After processing and normalization of the data, genes with at least six present calls over the 24 different data points were retained for further analysis. Expression values were obtained by averaging the normalized values of the two biological repeats.
For the comparative analysis, only one time point was used to represent the stress condition: A CV was applied on the whole time course experiment to get insight into the general response profiles upon the stress treatment and to select the most appropriate time point. For heat and cold, the 6-h time point was selected; for drought and methyl viologen, the 1- and 12-h time points, respectively. Fold changes for these specific time points were calculated as described above, and data sets were created of genes with at least a 3-fold induction at the specific time point. The cold, heat, drought, and methyl viologen data sets at the previously indicated time points contained 579, 583, 338, and 115 up-regulated genes, respectively. Statistical significance of the different positive correlations was evaluated with the Fisher's exact test. A probability value (P) of less than 0.05 was considered to indicate statistically significant overlaps between compared groups of genes.
Senescence
The senescence experiment is part of the larger Development Baseline experiment from the group of D. Weigel. (Tübingen, Germany; Schmid et al., 2005). Arabidopsis wild-type (ecotype Columbia-0) plants were grown under continuous light at 23°C, 65% relative humidity, and harvested at different growth stages. For the senescence samples, leaf material was harvested in triplicate at the growth stage 6.20 and corresponds to the AtGenExpress Array data ATGE_25_A, ATGE_25_B, and ATGE_25_C. For the control samples, whole-plant material was harvested in triplicate at the growth stage 3.20 and corresponds to the AtGenExpress Array data ATGE_26_A, ATGE_26_B, and ATGE_26_C. After processing and normalization of the data, genes with at least three present calls over the six different data points were retained for further analysis. Expression values were obtained by averaging the normalized values of the three independent repeats. Finally, senescent leaf expression data were compared with whole-plant expression data.
Data Obtained from genevestigator
Within the different tools of GENEVESTIGATOR, the Response Viewer was used to reveal the response profiles of genes to different stimuli (Zimmermann et al., 2004). Out of the different conditions annotated, the abiotic stresses (Kudla's Laboratory, Ulm, Germany) and hormone treatments (Yoshida's Laboratory, Saitama, Japan) were selected for comparison with our data or selected genes. For more information, see https://www.genevestigator.ethz.ch.
Anthocyanin Measurements
Catalase-deficient (CAT2HP1) and control (PTHW) plants were grown under controlled conditions: 21°C to 22°C, 12 h/12 h photoperiod, 100 μmol m−2 s−1, and 70% relative humidity. For the light stress treatment, plants were exposed to continuous HL irradiation (approximately 1,000 μmol m−2 s−1) for 24 h. Fresh weight (FW) was recorded for each sample, and ranged from 0.04 to 0.12 g per sample. Samples were frozen in liquid nitrogen and ground with mortar and pestle. Anthocyanins were measured according to a procedure based on the methods of Rabino and Mancinelli (1986) and Feinbaum and Ausubel (1988). Total plant pigments were extracted in 0.75 mL of 1% HCl/methanol, and 0.5 mL of distilled water was added. Chlorophyll was separated from the anthocyanins by back extraction with chloroform. The quantity of anthocyanin pigments was determined by spectrophotometric measurements of the aqueous/methanol phase. The absorbance at 530 nm minus the absorbance at 657 nm was used as a measure of anthocyanin content, and values were normalized to the FW of each sample. Results are expressed as absorbance per g FW and represent the mean ± se of three repeat experiments.
Supplementary Material
Acknowledgments
We thank Dr. Paul Van Hummelen, Dr. Lars Hennig, Mari Ikonen, Dr. Antje Rohde, and Dr. Marnik Vuylsteke for helpful contribution and discussions; Lucia Gössl, Claudia Knappe, and Stijn Morsa for excellent technical assistance; Dr. Martine De Cock for help in preparing the manuscript; and Karel Spruyt for artwork.
This work was supported by the Research Fund of the Ghent University (Geconcerteerde Onderzoeksacties grant no. 12051403), the Institute for the Promotion of Innovation by Science and Technology in Flanders (grant no. 040134/Bayer), and the Commission of the European Communities (Marie Curie Fellowship MOIF–CT–2004 to S.V.).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.065896.
References
- Aharoni A, De Vos CHR, Wein M, Sun Z, Greco R, Kroon A, Mol JNM, O'Connell AP (2001) The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J 28: 319–332 [DOI] [PubMed] [Google Scholar]
- Ahn S-G, Thiele DJ (2003) Redox regulation of mammalian heat shock factor 1 is essential for Hsp gene activation and protection from stress. Genes Dev 17: 516–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apel K, Hirt H (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55: 373–399 [DOI] [PubMed] [Google Scholar]
- Banzet N, Richaud C, Deveaux Y, Kazmaizer M, Gagnon J, Triantaphylidès C (1998) Accumulation of small heat shock proteins, including mitochondrial HSP22, induced by oxidative stress and adaptive response in tomato cells. Plant J 13: 519–527 [DOI] [PubMed] [Google Scholar]
- Baudry A, Heim MA, Dubreucq B, Caboche M, Weisshaar B, Lepiniec L (2004) TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J 39: 366–380 [DOI] [PubMed] [Google Scholar]
- Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C (2000) Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12: 2383–2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler C, Van Montagu M, Inzé D (1992) Superoxide dismutase and stress tolerance. Annu Rev Plant Physiol Plant Mol Biol 43: 83–116 [Google Scholar]
- Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Görlach J (2001) Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13: 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breyne P, Dreesen R, Cannoot B, Rombaut D, Vandepoele K, Rombauts S, Vanderhaeghen R, Inzé D, Zabeau M (2003) Quantitative cDNA-AFLP analysis for genome-wide expression studies. Mol Genet Genomics 269: 173–179 [DOI] [PubMed] [Google Scholar]
- Broun P (2004) Transcription factors as tools for metabolic engineering in plants. Curr Opin Plant Biol 7: 202–209 [DOI] [PubMed] [Google Scholar]
- Bruce W, Folkerts O, Garnaat C, Crasta O, Roth B, Bowen B (2000) Expression profiling of the maize flavonoid pathway genes controlled by estradiol-inducible transcription factors CRC and P. Plant Cell 12: 65–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch W, Wunderlich M, Schöffl F (2005) Identification of novel heat shock factor-dependent genes and biochemical pathways in Arabidopsis thaliana. Plant J 41: 1–14 [DOI] [PubMed] [Google Scholar]
- Cartieaux F, Thibaud M-C, Zimmerli L, Lessard P, Sarrobert C, David P, Gerbaud A, Robaglia C, Somerville S, Nussaume L (2003) Transcriptome analysis of Arabidopsis colonized by a plant-growth promoting rhizobacterium reveals a general effect on disease resistance. Plant J 36: 177–188 [DOI] [PubMed] [Google Scholar]
- Causton HC, Ren B, Koh SS, Harbison CT, Kanin E, Jennings EG, Lee TI, True HL, Lander ES, Young RA (2001) Remodeling of yeast genome expression in response to environmental changes. Mol Biol Cell 12: 323–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapple CCS, Shirley BW, Zook M, Hammerschmidt R, Somerville SC (1994) Secondary metabolism in Arabidopsis. In EM Meyerowitz, CR Somerville, eds, Arabidopsis, Cold Spring Harbor Monograph Series, Vol 27. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 989–1030
- Chen W, Provart NJ, Glazebrook J, Katagiri F, Chang H-S, Eulgem T, Mauch F, Luan S, Zou G, Whitham SA, et al (2002) Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell 14: 559–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WJ, Zhu T (2004) Networks of transcription factors with roles in environmental stress response. Trends Plant Sci 9: 591–596 [DOI] [PubMed] [Google Scholar]
- Corpas FJ, Barroso JB, del Río LA (2001) Peroxisomes as a source of reactive oxygen species and nitric oxide signal molecules in plant cells. Trends Plant Sci 6: 145–150 [DOI] [PubMed] [Google Scholar]
- Dat JF, Inzé D, Van Breusegem F (2001) Catalase-deficient tobacco plants: tools for in planta studies on the role of hydrogen peroxide. Redox Rep 6: 37–42 [DOI] [PubMed] [Google Scholar]
- Dat JF, Pellinen R, Beeckman T, Kangasjärvi J, Langebartels C, Inzé D, Van Breusegem F (2003) Changes in hydrogen peroxide homeostasis trigger an active cell death process in tobacco. Plant J 33: 621–632 [DOI] [PubMed] [Google Scholar]
- Davletova S, Rizhsky L, Liang H, Zhong S, Oliver DJ, Coutu J, Shulaev V, Schlauch K, Mittler R (2005) Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. Plant Cell 17: 268–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan R, Mackerness SA-H, Hancock JT, Neill SJ (2001) Regulation of the Arabidopsis transcriptome by oxidative stress. Plant Physiol 127: 159–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlting J, Büttner D, Wang Q, Douglas CJ, Somssich IE, Kombrink E (1999) Three 4-coumarate:coenzyme A ligases in Arabidopsis thaliana represent two evolutionarily divergent classes in angiosperms. Plant J 19: 9–20 [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis C, Turner JG (2001) The Arabidopsis mutant cev1 has constitutively active jasmonate and ethylene signal pathways and enhanced resistance to pathogens. Plant Cell 13: 1025–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5: 199–206 [DOI] [PubMed] [Google Scholar]
- Feinbaum RL, Ausubel FM (1988) Transcriptional regulation of the Arabidopsis thaliana chalcone synthase gene. Mol Cell Biol 8: 1985–1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschi VR, Grimes HD (1991) Induction of soybean vegetative storage proteins and anthocyanins by low-level atmospheric methyl jasmonate. Proc Natl Acad Sci USA 88: 6745–6749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki Y, Yoshikawa Y, Sato T, Inada N, Ito M, Nishida I, Watanabe A (2001) Dark-inducible genes from Arabidopsis thaliana are associated with leaf senescence and repressed by genes. Physiol Plant 111: 345–352 [DOI] [PubMed] [Google Scholar]
- Fujita M, Fujita Y, Maruyama K, Seki M, Hiratsu K, Ohme-Takagi M, Tran L-SP, Yamaguchi-Shinozaki K, Shinozaki K (2004) A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J 39: 863–876 [DOI] [PubMed] [Google Scholar]
- Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M (2003) Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J 34: 733–739 [DOI] [PubMed] [Google Scholar]
- Hoch WA, Singsaas EL, McCown BH (2003) Resorption protection. Anthocyanins facilitate nutrient recovery in autumn by shielding leaves from potentially damaging light levels. Plant Physiol 133: 1296–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch WA, Zeldin EL, McCown BH (2001) Physiological significance of anthocyanins during autumnal leaf senescence. Tree Physiol 21: 1–8 [DOI] [PubMed] [Google Scholar]
- Jackson RG, Lim E-K, Li Y, Kowalczyk M, Sandberg G, Hoggett J, Ashford DA, Bowles DJ (2001) Identification and biochemical characterization of an Arabidopsis indole-3-acetic acid glucosyltransferase. J Biol Chem 276: 4350–4356 [DOI] [PubMed] [Google Scholar]
- Jin H, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, Tonelli C, Weisshaar B, Martin C (2000) Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J 19: 6150–6161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson E, Olsson O, Nyström T (2004) Progression and specificity of protein oxidation in the life cycle of Arabidopsis thaliana. J Biol Chem 279: 22204–22208 [DOI] [PubMed] [Google Scholar]
- Jones P, Messner B, Nakajima J-I, Schäffner AR, Saito K (2003) UGT73C6 and UGT78D1, glycosyltransferases involved in flavonol glycoside biosynthesis in Arabidopsis thaliana. J Biol Chem 278: 43910–43918 [DOI] [PubMed] [Google Scholar]
- Joo JH, Yoo HJ, Hwang I, Lee JS, Nam KH, Bae YS (2005) Auxin-induced reactive oxygen species production requires the activation of phosphatidylinositol 3-kinase. FEBS Lett 579: 1243–1248 [DOI] [PubMed] [Google Scholar]
- Kariola T, Brader G, Li J, Palva ET (2005) Chlorophyllase 1, a damage control enzyme, affects the balance between defense pathways in plants. Plant Cell 17: 282–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall AC, Keys AJ, Turner JC, Lea PJ, Miflin BJ (1983) The isolation and characterization of a catalase-deficient mutant of barley (Hordeum vulgare). Planta 159: 505–511 [DOI] [PubMed] [Google Scholar]
- Kimura M, Yamamoto YY, Seki M, Sakurai T, Sato M, Abe T, Yoshida S, Manabe K, Shinozaki K, Matsui M (2003) Identification of Arabidopsis genes regulated by high light-stress using cDNA microarray. Photochem Photobiol 77: 226–233 [DOI] [PubMed] [Google Scholar]
- Kitamura S, Shikazono N, Tanaka A (2004) TRANSPARENT TESTA 19 is involved in the accumulation of both anthocyanins and proanthocyanidins in Arabidopsis. Plant J 37: 104–114 [DOI] [PubMed] [Google Scholar]
- Kuzmin EV, Karpova OV, Elthon TE, Newton KJ (2004) Mitochondrial respiratory deficiencies signal up-regulation of genes for heat shock proteins. J Biol Chem 279: 20672–20677 [DOI] [PubMed] [Google Scholar]
- Laloi C, Apel K, Danon A (2004) Reactive oxygen signalling: the latest news. Curr Opin Plant Biol 7: 323–328 [DOI] [PubMed] [Google Scholar]
- Larkindale J, Knight MR (2002) Protection against heat stress-induced oxidative damage in Arabidopsis involves calcium, abscisic acid, ethylene, and salicylic acid. Plant Physiol 128: 682–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Baldauf S, Lim E-K, Bowles DJ (2001) Phylogenetic analysis of the UDP-glycosyltransferase multigene family of Arabidopsis thaliana. J Biol Chem 276: 4338–4343 [DOI] [PubMed] [Google Scholar]
- Lim E-K, Doucet CJ, Li Y, Elias L, Worrall D, Spencer SP, Ross J, Bowles DJ (2002) The activity of Arabidopsis glycosyltransferases toward salicylic acid, 4-hydroxybenzoic acid, and other benzoates. J Biol Chem 277: 586–592 [DOI] [PubMed] [Google Scholar]
- Liu W-m, Mei R, Di X, Ryder TB, Hubbell E, Dee S, Webster TA, Harrington CA, Ho M-h, Baid J, et al (2002) Analysis of high density expression microarrays with signed-rank call algorithms. Bioinformatics 18: 1593–1599 [DOI] [PubMed] [Google Scholar]
- Lorenzo O, Chico JM, Sánchez-Serrano JJ, Solano R (2004) JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16: 1938–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateo A, Mühlenbock P, Rustérucci C, Chi-Chen Chang C, Miszalski Z, Karpinska B, Parker JE, Mullineaux PM, Karpinski S (2004) LESION SIMULATING DISEASE 1 is required for acclimation to conditions that promote excess excitation energy. Plant Physiol 136: 2818–2830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Gollery M, Van Breusegem F (2004) The reactive oxygen gene network in plants. Trends Plant Sci 9: 490–498 [DOI] [PubMed] [Google Scholar]
- Mol J, Grotewold E, Koes R (1998) How genes paint flowers and seeds. Trends Plant Sci 3: 212–217 [Google Scholar]
- Mostertz J, Scharf C, Hecker M, Homuth G (2004) Transcriptome and proteome analysis of Bacillus subtilis gene expression in response to superoxide and peroxide stress. Microbiology 150: 497–512 [DOI] [PubMed] [Google Scholar]
- Murray JI, Whitfield ML, Trinklein ND, Myers RM, Brown PO, Botstein D (2004) Diverse and specific gene expression responses to stresses in cultured human cells. Mol Biol Cell 15: 2361–2374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G, Veljovic-Jovanovic S, Driscoll S, Novitskaya L, Foyer CH (2002) Drought and oxidative load in the leaves of C3 plants: a predominant role for photorespiration? Ann Bot (Lond) 89: 841–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- op den Camp RGL, Przybyla D, Ochsenbein C, Laloi C, Kim C, Danon A, Wagner D, Hideg E, Göbel C, Feussner I, et al (2003) Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis. Plant Cell 15: 2320–2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pnueli L, Liang H, Rozenberg M, Mittler R (2003) Growth suppression, altered stomatal responses, and augmented induction of heat shock proteins in cytosolic ascorbate peroxidase (Apx1)-deficient Arabidopsis plants. Plant J 34: 187–203 [DOI] [PubMed] [Google Scholar]
- Port M, Tripp J, Zielinski D, Weber C, Heerklotz D, Winkelhaus S, Bublak D, Scharf K-D (2004) Role of Hsp17.4-CII as coregulator and cytoplasmic retention factor of tomato heat stress transcription factor HsfA2. Plant Physiol 135: 1457–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabino I, Mancinelli AL (1986) Light, temperature and anthocyanin production. Plant Physiol 81: 922–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raes J, Rohde A, Christensen JH, Van de Peer Y, Boerjan W (2003) Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol 133: 1051–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee SY, Beavis W, Berardini TZ, Chen G, Dixon D, Doyle A, Garcia-Hernandez M, Huala E, Lander G, Montoya M, et al (2003) The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res 31: 224–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Heard J, Martin G, Reuber L, Jiang C-Z, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, et al (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290: 2105–2110 [DOI] [PubMed] [Google Scholar]
- Rizhsky L, Liang H, Mittler R (2003) The water-water cycle is essential for chloroplast protection in the absence of stress. J Biol Chem 278: 38921–38925 [DOI] [PubMed] [Google Scholar]
- Rizhsky L, Liang H, Shuman J, Shulaev V, Davletova S, Mittler R (2004) When defense pathways collide: the response of Arabidopsis to a combination of drought and heat stress. Plant Physiol 134: 1683–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde A, Morreel K, Ralph J, De Rycke R, Kushnir S, Van Doorsselaere J, Goeminne G, Joseleau J-P, Vuylsteke M, Van Driessche G, et al (2004) Molecular phenotyping of the pal1 and pal2 mutants of Arabidopsis thaliana reveals far-reaching consequences on phenylpropanoid, amino acid, and carbohydrate metabolism. Plant Cell 16: 2749–2771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J, Li Y, Lim E-K, Bowles DJ (2001) Higher plant glycosyltransferases. Genome Biol 2: REVIEWS3004 [DOI] [PMC free article] [PubMed]
- Rossel JB, Wilson IW, Pogson BJ (2002) Global changes in gene expression in response to high light in Arabidopsis. Plant Physiol 130: 1109–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 35: 501–506 [DOI] [PubMed] [Google Scholar]
- Schwinn KE, Davies KM (2004) Flavonoids. In KM Davies, ed, Plant Pigments and Their Manipulation, Annual Plant Reviews, Vol 14. Blackwell Publishing, Oxford, pp 92–149
- Shinozaki K, Yamaguchi-Shinozaki K (2000) Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Curr Opin Plant Biol 3: 217–223 [PubMed] [Google Scholar]
- Smith AM, Zeeman SC, Thorneycroft D, Smith SM (2003) Starch mobilization in leaves. J Exp Bot 54: 577–583 [DOI] [PubMed] [Google Scholar]
- Springob K, Nakajima J-i, Yamazaki M, Saito K (2003) Recent advances in the biosynthesis and accumulation of anthocyanins. Nat Prod Rep 20: 288–303 [DOI] [PubMed] [Google Scholar]
- Steyn WJ, Wand SJE, Holcroft DM, Jacobs G (2002) Anthocyanins in vegetative tissues: a proposed unified function in photoprotection. New Phytol 155: 349–361 [DOI] [PubMed] [Google Scholar]
- Thiel S, Döhring T, Köfferlein M, Kosak A, Martin P, Seidlitz HK (1996) A phytotron for plant stress research: How far can artificial lighting compare with natural sunlight? J Plant Physiol 148: 456–463 [Google Scholar]
- Tohge T, Nishiyama Y, Hirai MY, Yano M, Nakajima J-i, Awazuhara M, Inoue E, Takahashi H, Goodenowe DB, Kitayama M, et al (2005) Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J 42: 218–235 [DOI] [PubMed] [Google Scholar]
- Vandenabeele S, Van Der Kelen K, Dat J, Gadjev I, Boonefaes T, Morsa S, Rottiers P, Slooten L, Van Montagu M, Zabeau M, et al (2003) A comprehensive analysis of hydrogen peroxide-induced gene expression in tobacco. Proc Natl Acad Sci USA 100: 16113–16118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenabeele S, Vanderauwera S, Vuylsteke M, Rombauts S, Langebartels C, Seidlitz HK, Zabeau M, Van Montagu M, Inzé D, Van Breusegem F (2004) Catalase deficiency drastically affects gene expression induced by high light in Arabidopsis thaliana. Plant J 39: 45–58 [DOI] [PubMed] [Google Scholar]
- Vogel JT, Zarka DG, Van Buskirk HA, Fowler SG, Thomashow MF (2005) Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J 41: 195–211 [DOI] [PubMed] [Google Scholar]
- Vom Endt D, Kijne JW, Memelink J (2002) Transcription factors controlling plant secondary metabolism: What regulates the regulators? Phytochemistry 61: 107–114 [DOI] [PubMed] [Google Scholar]
- Wang Y-J, Zhang Z-G, He X-J, Zhou H-L, Wen Y-X, Dai J-X, Zhang J-S, Chen S-Y (2003) A rice transcription factor OsbHLH1 is involved in cold stress response. Theor Appl Genet 107: 1402–1409 [DOI] [PubMed] [Google Scholar]
- Willekens H, Chamnongpol S, Davey M, Schraudner M, Langebartels C, Van Montagu M, Inzé D, Van Camp W (1997) Catalase is a sink for H2O2 and is indispensable for stress defence in C3 plants. EMBO J 16: 4806–4816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley B (2002) Biosynthesis of flavonoids and effects of stress. Curr Opin Plant Biol 5: 218–223 [DOI] [PubMed] [Google Scholar]
- Xie D-Y, Sharma SB, Paiva NL, Ferreira D, Dixon RA (2003) Role of anthocyanidin reductase, encoded by BANYULS in plant flavonoid biosynthesis. Science 299: 396–399 [DOI] [PubMed] [Google Scholar]
- Yamamoto YY, Shimada Y, Kimura M, Manabe K, Sekine Y, Matsui M, Ryuto H, Fukunishi N, Abe T, Yoshida S (2004) Global classification of transcriptional responses to light stress in Arabidopsis thaliana. Endocytobiosis Cell Res 15: 438–452 [Google Scholar]
- Ye Z, Rodriguez R, Tran A, Hoang H, de los Santos D, Brown S, Vellanoweth RL (2000) The developmental transition to flowering represses ascorbate peroxidase activity and induces enzymatic lipid peroxidation in leaf tissue in Arabidopsis thaliana. Plant Sci 158: 115–127 [DOI] [PubMed] [Google Scholar]
- Zhang F, Gonzalez A, Zhao M, Payne CT, Lloyd A (2003) A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130: 4859–4869 [DOI] [PubMed] [Google Scholar]
- Zhong M, Orosz A, Wu C (1998) Direct sensing of heat and oxidation by Drosophila heat shock transcription factor. Mol Cell 2: 101–108 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.