Abstract
Plant acclimation to environmental stress is controlled by a complex network of regulatory genes that compose distinct stress-response regulons. In contrast to many signaling and regulatory genes that are stress specific, the zinc-finger protein Zat12 responds to a large number of biotic and abiotic stresses. Zat12 is thought to be involved in cold and oxidative stress signaling in Arabidopsis (Arabidopsis thaliana); however, its mode of action and regulation are largely unknown. Using a fusion between the Zat12 promoter and the reporter gene luciferase, we demonstrate that Zat12 expression is activated at the transcriptional level during different abiotic stresses and in response to a wound-induced systemic signal. Using Zat12 gain- and loss-of-function lines, we assign a function for Zat12 during oxidative, osmotic, salinity, high light, and heat stresses. Transcriptional profiling of Zat12-overexpressing plants and wild-type plants subjected to H2O2 stress revealed that constitutive expression of Zat12 in Arabidopsis results in the enhanced expression of oxidative- and light stress-response transcripts. Under specific growth conditions, Zat12 may therefore regulate a collection of transcripts involved in the response of Arabidopsis to high light and oxidative stress. Our results suggest that Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis.
Plants are sessile organisms that evolved complex regulatory networks to control their response to changes in environmental conditions (Arabidopsis Genome Initiative, 2000). Interestingly and in contrast to prior belief, little overlap in transcript expression was found between the response of plants to different environmental stress conditions (Kreps et al., 2002; Rizhsky et al., 2004b). Thus, transcriptome profiling of plants subjected to heat, drought, cold, salt, high light, or mechanical stress revealed that very few genes respond in a similar manner to all of these stresses (Cheong et al., 2002; Fowler and Thomashow, 2002; Kreps et al., 2002; Rossel et al., 2002; Rizhsky et al., 2004b). Moreover, although reactive oxygen species (ROS) were implicated as signals and/or byproducts of many different biotic and abiotic stress conditions, different genes of the ROS gene network of Arabidopsis (Arabidopsis thaliana) were found to respond differently to different stress treatments (Mittler et al., 2004).
A complex network of transcription factors orchestrates the response of plants to changes in environmental conditions (Chen et al., 2002). These include WRKY and other zinc-finger proteins (72 WRKY genes and more than 600 zinc-finger proteins in Arabidopsis; Eulgem et al., 2000), MYB transcription factors (133 genes in Arabidopsis; Stracke et al., 2001), and heat shock transcription factors (21 genes in Arabidopsis; Nover et al., 2001). However, only a few of these transcription factors appear to respond in a similar manner to all or most of the different environmental stress conditions tested in Arabidopsis. One representative of the small group of genes that responds similarly to many different environmental stress conditions is the zinc-finger protein Zat12 (At5g59820). Zat12 was found to respond at the steady-state transcript level to ozone fumigation; wounding; bacterial, fungal, or nematode infection; H2O2; heat; cold; drought; elicitor; heavy metal; methanol; fumonisin; or UV application (Iida et al., 2000; Cheong et al., 2002; Fowler and Thomashow, 2002; Kreps et al., 2002; Rizhsky et al., 2004b; Davletova et al., 2005; a search of 1,809 ATH1 Affymetrix chips in https://www.genevestigator.ethz.ch/). In addition, Zat12 is expressed in roots, flowers, and developing seeds, tissues associated with expression of stress-response genes (https://www.genevestigator.ethz.ch/).
Zat12 was originally isolated as a light stress-response cDNA (rhl41) by Iida et al. (2000), and was later identified by transcriptome analyses of plants subjected to different biotic and abiotic stress conditions. Recent analysis of Zat12 using transgenic plants suggested a role for Zat12 in cold acclimation and in the response of plants to oxidative stress (Rizhsky et al., 2004a; Vogel et al., 2005). However, the three different studies that used transgenic plants overexpressing Zat12 to study Zat12 function were very different in their description of plant phenotype (Iida et al., 2000; Rizhsky et al., 2004a; Vogel et al., 2005). In addition, very little overlap in transcript expression was found between the transcriptome of mature transgenic plants expressing Zat12 in experiments performed by Vogel et al. (2005) and Rizhsky et al. (2004a). Perhaps the involvement of Zat12 in many different stress-response pathways, the differences in experimental conditions used for the assays, and the differences in genetic background, age, and physical condition of plants might explain the variations observed in the effects of Zat12 overexpression in these experiments.
The enhanced expression of Zat12 in response to many different biotic and abiotic stress conditions makes Zat12 an interesting subject for analysis. What is the function of Zat12 in Arabidopsis? Is it involved in all of the stresses its expression is associated with? Is it related to ROS signaling as previously suggested (Rizhsky et al., 2004a)? To address these questions, we functionally characterized Zat12 in Arabidopsis. Using transgenic plants expressing the reporter gene luciferase under the control of the Zat12 promoter, we studied the transcriptional activation of Zat12 during different abiotic stresses. Using gain- and loss-of-function plants for Zat12, we studied the function of Zat12 during different stresses, and using microarray analysis we compared the transcriptome of Zat12-expressing plants to that of plants subjected to H2O2 stress. Our results identify a function for Zat12 during oxidative, osmotic, salinity, high light, and heat stresses. In addition, our analysis suggests that under certain growth conditions, Zat12 may control a collection of transcripts involved in plant acclimation to high light and oxidative stress.
RESULTS
Activation of Zat12 Transcription in Arabidopsis
The response of Zat12 to different stress treatments is typically measured as a transient increase in steady-state transcript level (mainly by RNA blots or Affymetrix chips; Iida et al., 2000; Cheong et al., 2002; Fowler and Thomashow, 2002; Kreps et al., 2002; Rizhsky et al., 2004a; Davletova et al., 2005). To examine whether the increase in Zat12 steady-state transcript level results from an increase in Zat12 transcription or from a change in the stability of the Zat12 transcript, we generated a fusion between the Zat12 promoter and the reporter gene luciferase. For this analysis, a 1,000-bp fragment corresponding to position 1 to −1,000 of the Zat12 gene (At5g59820) was amplified by PCR from genomic DNA, sequenced, and cloned into a binary vector that contained the luciferase reporter gene. Three independent transgenic lines were chosen for analysis.
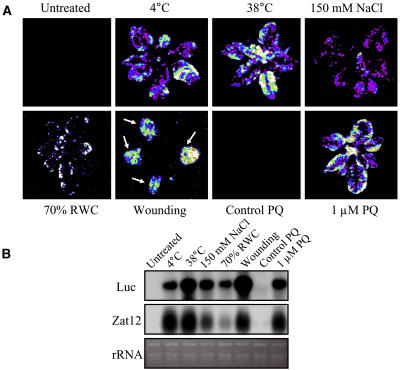
As shown in Figure 1A, activation of Zat12 transcription occurred in response to cold (4°C, 2 h), heat (38°C, 1 h), salinity (150 mm NaCl, 4 h), drought (75% relative water content), wounding (arrows indicate leaves that were wounded 10 times each with a needle, 30 min), and application of the superoxide-generating agent methyl viologen (10−6 m, 1 h). These stresses were applied to soil-grown 3-week-old plants as described (Mittler and Zilinskas, 1992; Kreps et al., 2002; Rizhsky et al., 2004a, 2004b). As shown in Figure 1B, RNA-blot analysis of transgenic plants expressing the Zat12 promoter::luciferase construct confirmed that the expression of luciferase correlated with the steady-state level of Zat12 transcripts, the stress treatment, and the intensity of luciferase activity (compare the steady-state level of Zat12 and luciferase in Fig. 1B to luciferase activity in Fig. 1A). Luciferase transcription was also activated in response to light stress (1,000 μmol m−2 s−1; 1 h), the application of H2O2 (1, 2, 5, or 20 mm, 1 h), ferric-citrate (1 mm, 5 h), dark treatment (4 h), or the application of the plant hormones auxin or abscisic acid (data not shown).
Figure 1.
Promoter∷reporter analysis of Zat12 expression in Arabidopsis. A, Luciferase imaging in transgenic Arabidopsis plants expressing luciferase under the control of the Zat12 promoter in response to different environmental stresses. B, RNA-blot analysis of luciferase and Zat12 steady-state transcript level in the plants shown in A in response to stress. Construction of transgenic plants, stress assays, and luciferase imaging were performed as described in “Materials and Methods.” Abbreviations: Luc, luciferase; PQ, methyl viologen (paraquat); rRNA, ribosomal RNA; RWC, relative water content.
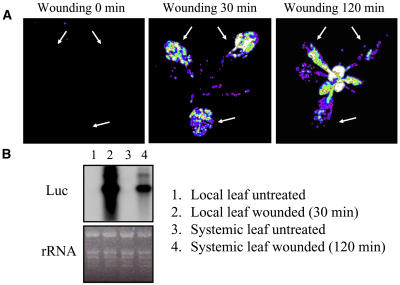
Several stresses, such as pathogen infection, high light, and wounding, induce a systemic response in plants (Alvarez et al., 1998; Karpinski et al., 1999; Orozco-Cardenas et al., 2001). Typically, this response requires a few hours to develop and is thought to involve salicylic acid, jasmonic acid, systemin, and/or H2O2. To test whether Zat12 expression is induced systemically in Arabidopsis, we wounded plants on three different leaves and followed the activation of luciferase transcription in the entire plant. As shown in Figure 2A, 30 min following wounding (middle section), luciferase activity was detected in the wounded leaves (wounded leaves are indicated by arrows in all sections). Two hours following wounding, luciferase activity in the wounded leaves subsided and strong luciferase activity was observed in upper young leaves that were not treated (right section). As shown in Figure 2B, RNA-blot analysis of treated and untreated local and systemic leaves confirmed that expression of the luciferase transcript is enhanced in wounded (local) and unwounded systemic (upper young leaves of wounded plants) tissues. The results presented in Figures 1 and 2 indicate that transcription of the Zat12 gene is activated in response to different stimuli. These may be directly related to stress or wounding, or may occur as a result of a systemic signal that is generated in the wounded leaves and transferred to untreated parts of the plant.
Figure 2.
Systemic induction of the Zat12 promoter in response to wounding. A, Luciferase imaging in a transgenic Arabidopsis plant expressing luciferase under the control of the Zat12 promoter in response to wounding. Arrows indicate the wounded leaves. Luciferase imaging was performed at 0, 30, and 120 min following wounding. B, RNA-blot analysis of luciferase steady-state transcript level in transgenic Arabidopsis plant expressing luciferase under the control of the Zat12 promoter in response to wounding. Luciferase transcripts are shown to accumulate in local wounded leaves (lane 2) and systemic leaves of wounded plants (lane 4). Construction of transgenic plants, wounding, and luciferase imaging were performed as described in “Materials and Methods.” Abbreviations: Luc, luciferase; rRNA, ribosomal RNA.
Tolerance of Gain- and Loss-of-Function Zat12 Lines to Abiotic Stress
The response of Zat12 to many different stresses is intriguing. Is Zat12 essential for plant tolerance to all of these stresses? To address this question, we subjected seedlings of knockout Zat12 plants (KO-Zat12; SALK_037357 and SAIL 792_F04) and seedlings of plants that overexpress Zat12 (OE-Zat12; three independent lines) to different abiotic stresses and scored them for root growth and percentage of germination, parameters that reflect overall health and stress tolerance of plants (Zhu, 2002). The following stresses were tested: heat (38°C; 1, 6, 24, and 48 h), cold (10°C; 1, 2, and 3 d), osmotic stress (50, 100, 200, and 300 mm sorbitol), salinity (50, 100, 150, and 200 mm NaCl), ferric-citrate (0.01, 0.1, 0.5, and 1 mm), high light (1,000 μmol m−2 s−1 for 24, 48, and 72 h), and methyl viologen (0.01, 0.1, 0.5, 1, and 10 μm).
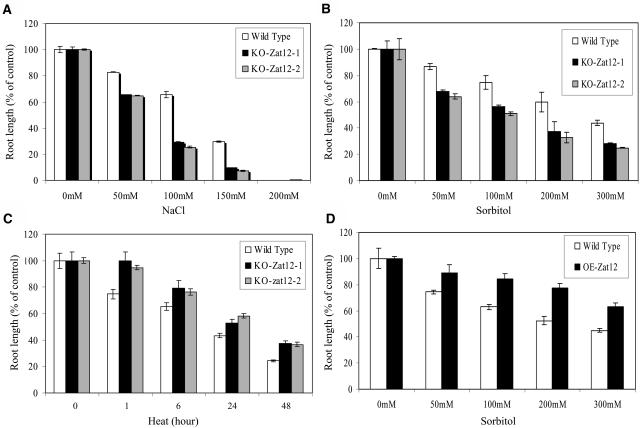
As shown in Figure 3 and Supplemental Figures 1 and 2 (that represent different sets of experiments and include measurements of root growth, as well as data collected for percentage of germination), knockout Zat12 plants were more sensitive than wild-type plants to salinity stress (Fig. 3A) and osmotic stress (Fig. 3B). In contrast, knockout Zat12 plants were more tolerant than wild-type plants to heat stress (Fig. 3C). Compared to wild-type plants, transgenic plants overexpressing Zat12 were more tolerant to osmotic stress (Fig. 3D), light stress, and methyl viologen (data not shown; see Iida et al. [2000] for enhanced tolerance of Zat12-overexpressing lines to light stress, and Rizhsky et al. [2004a] for enhanced tolerance of transgenic Zat12 plants to methyl viologen). These results suggest that, at least when tested with 5-d-old seedlings, Zat12 is required for plant tolerance to osmotic, oxidative, and salinity stresses. In contrast, Zat12 may have a negative effect on plant tolerance to heat stress.
Figure 3.
Tolerance of gain- and loss-of-function Zat12 Arabidopsis lines to abiotic stress. A, Root growth of wild-type and KO-Zat12 seedlings subjected to salinity stress. B, Root growth of wild-type and KO-Zat12 seedlings subjected to osmotic stress. C, Root growth of wild-type and KO-Zat12 seedlings subjected to heat stress. D, Root growth of wild-type and OE-Zat12 seedlings subjected to osmotic stress. Stress assays were performed as described in “Materials and Methods” using independent knockout and overexpression lines. Additional assays are shown in Supplemental Figures 1 and 2. Abbreviations: KO-Zat12-1, knockout Zat12, SALK_037357; KO-Zat12-2, knockout Zat12, SAIL 792_F04; OE-Zat12, transgenic seedlings overexpressing Zat12.
Transcriptome Profiling of Transgenic Plants Overexpressing Zat12
To complement the characterization of abiotic stress tolerance performed with seedlings of gain- and loss-of-function lines (Fig. 3) and to address the question of Zat12 association with H2O2 (Rizhsky et al., 2004a), we performed transcriptome profiling of transgenic plants overexpressing Zat12 (obtained from a pool of three independent lines). For this analysis, we used 5-d-old seedlings, comparable in age and growth conditions to the seedlings used for all stress assays in this study (Fig. 3; Supplemental Figs. 1 and 2) and in Rizhsky et al. (2004a). Our profiling analysis was, however, not comparable to that reported by Rizhsky et al. (2004a) and Vogel et al. (2005) that used older or more mature plants grown in soil. In parallel to the profiling of Zat12-overexpressing seedlings, we performed profiling of wild-type seedlings subjected to H2O2 stress. This design enabled us to check for overlap between transcripts elevated or suppressed in transgenic seedlings overexpressing Zat12 and in wild-type seedlings treated with H2O2. Four different treatments were used: wild-type untreated (WT), wild-type treated with 20 mm H2O2 for 1 h (WT + H2O2), transgenic plants overexpressing Zat12 (Zat12), and transgenic plants overexpressing Zat12 treated with 20 mm H2O2 for 1 h (Zat12 + H2O2). All treatments were sampled at the same time and the experiment was repeated three times (total of 12 samples representing three biological repeats). We did not use loss-of-function (KO-Zat12) lines for transcriptome profiling because the quality of RNA extracted from these lines following the H2O2 treatment was insufficient for microarray analysis (data not shown).
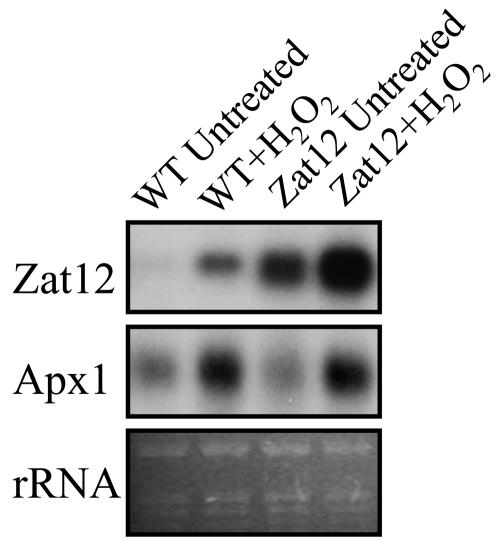
As shown in Figure 4, expression of Zat12 and ascorbate peroxidase 1 (Apx1), a H2O2-response transcript (Mittler and Zilinskas, 1992; Davletova et al., 2005), is elevated in wild-type plants following the application of H2O2 (WT + H2O2). The steady-state level of Zat12 transcripts found in transgenic plants overexpressing Zat12 (Zat12 untreated) was higher than that of treated wild-type plants (compare Zat12 untreated to WT + H2O2). The level of Zat12 found in untreated transgenic plants overexpressing Zat12 was elevated further following H2O2 application (Zat12 + H2O2).
Figure 4.
RNA blots showing the expression of Zat12 and Apx1 in H2O2-treated and untreated wild-type plants and transgenic plants overexpressing Zat12. RNA blots were performed as described in “Materials and Methods.” Abbreviations: rRNA, ribosomal RNA; WT, wild type; Zat12, transgenic plants overexpressing Zat12.
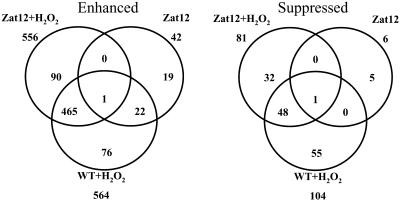
Treatment of wild-type plants with H2O2 resulted in the significant elevation of 564 transcripts (more than 2-fold; Fig. 5; Supplemental Table II). To validate the results obtained with this treatment (i.e. H2O2 stress), we compared the transcripts enhanced in 5-d-old wild-type seedlings in response to H2O2 application with those enhanced in mature knockout plants deficient in Apx1 (knockout-Apx1) plants in response to light stress, a stress that results in the accumulation of H2O2 in knockout-Apx1 plants (Davletova et al., 2005). Overall, 32% of the transcripts elevated in wild-type plants in response to H2O2 application were also elevated in knockout-Apx1 plants in response to a light stress treatment. Signal transduction, ROS-related, and defense-related transcripts significantly elevated in wild-type plants treated with H2O2 as well as in knockout-Apx1 plants subjected to light stress (Davletova et al., 2005) are shown in Table I. They include heat shock transcription factor 21 (HSF21), NADPH oxidase D (RbohD), mitochondrial alternative oxidase, mitogen-activated protein kinase MAPK3, several WRKY transcription factors (40, 33, 18, and 70), several zinc-finger proteins including Zat10 and Zat12, and a number of transcripts associated with calcium signaling. We previously used dominant-negative lines for HSF21 and knockout lines for RbohD to show that these two proteins are directly involved in H2O2 sensing in Arabidopsis (Davletova et al., 2005). The transcripts shown in Table I might represent a collection of genes directly associated with H2O2 signaling and stress in Arabidopsis.
Figure 5.
Venn diagrams showing the overlap between transcript expression in wild-type plants subjected to H2O2 stress (WT + H2O2), transgenic plants overexpressing Zat12 (Zat12), and transgenic plants overexpressing Zat12 subjected to H2O2 stress (Zat12 + H2O2). Transcriptome profiling and data analysis were performed as described in “Materials and Methods.”
Table I.
Signal transduction, ROS-related, and defense-related transcripts significantly elevated in wild-type plants treated with H2O2 as well as in knockout-Apx1 plants subjected to light stress
Signal transduction, ROS-related, and defense-related transcripts significantly elevated in wild-type plants treated with H2O2 and in knockout-Apx1 plants subjected to light stress are shown. Affymetrix ATH1 accessions are given on left. Locus identification numbers are given in the second column from left. Gene annotation and fold change in log2 in response to H2O2 application to wild-type plants are given in the third and fourth columns from left, respectively. Transcriptome profiling and data analysis were performed as described in “Materials and Methods.”
| Chip Annotation | Locus Identifier | Gene Annotation | Fold (log2) |
|---|---|---|---|
| 261892_at | AT1G80840 | WRKY40 transcription factor | 5.310 |
| 261648_at | AT1G27730 | Zinc-finger (C2H2-type) family protein (ZAT10) | 4.550 |
| 267028_at | AT2G38470 | WRKY33 transcription factor | 3.724 |
| 248657_at | AT5G48570 | Peptidyl-prolyl cis-trans isomerase | 3.415 |
| 263379_at | AT2G40140 | Zinc finger (CCCH type), Ankyrin repeat | 3.081 |
| 251745_at | AT3G55980 | Zinc finger (CCCH type), Ankyrin repeat | 2.955 |
| 254926_at | AT4G11280 | Aminocyclopropane carboxylic acid synthase 6 | 2.923 |
| 262119_s_at | AT1G02920 | Glutathione S-transferase | 2.922 |
| 267083_at | AT2G41100 | Touch-responsive/calmodulin-related protein 3 | 2.791 |
| 253414_at | AT4G33050 | Calmodulin-binding family protein | 2.537 |
| 248164_at | AT5G54490 | Calcium-binding EF-hand protein | 2.330 |
| 247655_at | AT5G59820 | Zinc-finger (C2H2-type) family (ZAT12) | 2.313 |
| 252592_at | AT3G45640 | Mitogen-activated protein kinase (MPK3) | 2.276 |
| 253915_at | AT4G27280 | Calcium-binding EF-hand protein | 2.245 |
| 259879_at | AT1G76650 | Calcium-binding EF protein | 2.169 |
| 246777_at | AT5G27420 | Zinc finger (C3HC4-type RING finger) | 2.114 |
| 254549_at | AT4G19880 | Glutathione S-transferase | 2.105 |
| 248719_at | AT5G47910 | Respiratory burst oxidase protein D (RbohD) | 2.000 |
| 252474_at | AT3G46620 | Zinc finger (C3HC4-type RING finger) | 1.996 |
| 253485_at | AT4G31800 | WRKY18 transcription factor | 1.906 |
| 254592_at | AT4G18880 | HSF21 | 1.904 |
| 247137_at | AT5G66210 | Calcium-dependent protein kinase/CDPK | 1.858 |
| 259272_at | AT3G01290 | Band 7 protein, hypersensitive induced | 1.800 |
| 249417_at | AT5G39670 | Calcium-binding EF-hand protein | 1.663 |
| 248327_at | AT5G52750 | Heavy-metal-associated protein | 1.630 |
| 250676_at | AT5G06320 | Harpin-induced family protein/HIN1 | 1.620 |
| 258921_at | AT3G10500 | No apical meristem (NAM) protein | 1.595 |
| 260399_at | AT1G72520 | Lipoxygenase | 1.509 |
| 253125_at | AT4G36040 | DNAJ heat shock N-terminal domain containing | 1.493 |
| 258452_at | AT3G22370 | Alternative oxidase 1a | 1.490 |
| 267293_at | AT2G23810 | Senescence-associated protein 5 | 1.438 |
| 251705_at | AT3G56400 | WRKY70 | 1.261 |
| 266746_s_at | AT4G02520 | Glutathione S-transferase | 1.238 |
| 267381_at | AT2G26190 | Calmodulin-binding protein | 1.217 |
| 260648_at | AT1G08050 | Zinc finger (C3HC4-type RING finger) | 1.166 |
As shown in Figure 5, 42 transcripts were elevated and six transcripts suppressed (more than 2-fold) in transgenic plants overexpressing Zat12 in the absence of stress. Of the 42 transcripts elevated in Zat12-overexpressing lines, 23 were also elevated in wild-type plants following H2O2 application. As shown in Table II, the majority of these transcripts are encoded by chloroplastic genes (the 23 transcripts elevated in Zat12-overexpressing plants and in wild-type plants treated with H2O2 are indicated in bold in Table II). The transcriptome of transgenic plants overexpressing Zat12 resembled the transcriptome of plants subjected to light stress (Rossel et al., 2002; Rizhsky et al., 2003). Thus, chloroplastic transcripts elevated in Zat12-overexpressing lines included psbA, B and H, psaA, B and I, small ribosomal proteins, and clpP protease. The detection of chloroplastic transcripts by Affymetrix chips (Table II) indicates that these transcripts were not removed by the oligo(dT) purification step (http://www.affymetrix.com/support/technical/manual/expression_manual.affx). Because polyadenylation of chloroplastic transcripts is associated with increased turnover (Kudla et al., 1996), it is possible that the turnover of the chloroplastic transcripts shown in Table II is enhanced in Zat12-overexpressing plants and in wild-type plants subjected to a H2O2 treatment. This possibility is in accordance with the enhanced turnover of many chloroplastic transcripts during light stress (Kudla et al., 1996). Further studies are required to address this possibility and to elucidate the possible role of Zat12 in regulating light stress responses in Arabidopsis. Nuclear genes elevated in Zat12-overexpressing lines included chloroplastic and cytosolic copper/zinc superoxide dismutases and the superoxide dismutase copper chaperone (Table II). In addition to these transcripts, transcripts encoding six different Lea (late embryogenesis abundant) proteins were elevated in transgenic plants overexpressing Zat12. The elevated expression of these transcripts might explain the enhanced tolerance of Zat12-overexpressing lines to osmotic stress (Fig. 3D). The six transcripts suppressed in transgenic plants overexpressing Zat12 include the following: copper amine oxidase (At3g43670), copper homeostasis factor (At3g56240), Cys proteinase (At4g11310), harpin-induced protein (At2g35960), GDSL-lipase/hydrolase (At4g28780), and iron superoxide dismutase (At4g25100).
Table II.
Transcripts significantly elevated (more than 2-fold) in transgenic Arabidopsis seedlings overexpressing Zat12
Transcripts that are significantly elevated in transgenic plants overexpressing Zat12 compared to wild-type plants (more than 2-fold) are shown. Affymetrix ATH1 accessions are given on left. Locus identification numbers are given in the second column from left. Gene annotation and fold change in log2 are given in the third and fourth columns from left, respectively. Transcripts that are elevated in transgenic plants overexpressing Zat12, as well as in wild-type plants subjected to H2O2 stress, are indicated in bold. Locus identifiers that begin with an ATC denote chloroplastic genes. Transcriptome profiling and data analysis were performed as described in “Materials and Methods.”
| Chip Annotation | Locus Identifier | Gene Annotation | Fold (log2) |
|---|---|---|---|
| 247655_at | AT5G59820 | Zinc-finger (C2H2-type) protein (ZAT12) | 3.444 |
| 245007_at | ATCG00350 | psaA PSI P700 apoprotein | 2.895 |
| 244965_at | ATCG00590 | orf31 hypothetical protein | 2.632 |
| 244940_at | ATCG00900 | rps7.1 ribosomal protein S12 | 2.563 |
| 244992_s_at | ATCG01240 | rps7.2 ribosomal protein S7 | 2.372 |
| 245003_at | ATCG00280 | psbC PSII 43-kD protein | 2.226 |
| 245025_at | ATCG00130 | atpF ATPase I subunit | 2.151 |
| 259511_at | AT1G12520 | Superoxide dismutase copper chaperone | 2.141 |
| 245006_at | ATCG00340 | psaB PSI P700 apoprotein A2 | 2.119 |
| 266165_at | AT2G28190 | Copper/zinc superoxide dismutase (CSD2) | 1.983 |
| 245024_at | ATCG00120 | atpA ATPase α-subunit | 1.919 |
| 262128_at | AT1G52690 | Lea group 3 | 1.903 |
| 251838_at | AT3G54940 | Cys proteinase | 1.893 |
| 244975_at | ATCG00710 | psbH PSII 10-kD phosphoprotein | 1.887 |
| 245026_at | ATCG00140 | atpH ATPase III subunit | 1.865 |
| 245047_at | ATCG00020 | psbA PSII 32-kD protein | 1.793 |
| 258224_at | AT3G15670 | Lea group 3 | 1.736 |
| 254805_at | AT4G12480 | Protease inhibitor/seed storage/lipid transfer protein | 1.711 |
| 266544_at | AT2G35300 | Lea group 3 | 1.689 |
| 246099_at | AT5G20230 | Plastocyanin like | 1.686 |
| 254550_at | AT4G19690 | Iron-responsive transporter (IRT1) | 1.651 |
| 244976_at | ATCG00720 | petB cytochrome B6 | 1.619 |
| 244936_at | ATCG01100 | ndhA NADH dehydrogenase ND1 | 1.538 |
| 244972_at | ATCG00680 | psbB PSII 47-kD protein | 1.504 |
| 244971_at | ATCG00670 | clpP ATP-dependent protease | 1.503 |
| 258498_at | AT3G02480 | Abscisic acid-responsive, cold-induced protein kin1 | 1.497 |
| 264809_at | AT1G08830 | Copper/zinc superoxide dismutase (CSD1) | 1.495 |
| 250780_at | AT5G05290 | Expansin At-EXP2 | 1.485 |
| 245009_at | ATCG00380 | rps4 ribosomal protein S4 | 1.447 |
| 244961_at | ATCG01040 | ycf5 hypothetical protein | 1.399 |
| 244935_at | ATCG01090 | ndhI NADH dehydrogenase subunit | 1.322 |
| 254818_at | AT4G12470 | Protease inhibitor/seed storage/lipid transfer protein | 1.319 |
| 245002_at | ATCG00270 | psbD PSII D2 protein | 1.263 |
| 250648_at | AT5G06760 | Lea group 5 | 1.263 |
| 260556_at | AT2G43620 | Basic endochitinase CHB4 | 1.216 |
| 245017_at | ATCG00510 | psaI PSII protein | 1.208 |
| 245335_at | AT4G16160 | Mitochondrial inner membrane translocase | 1.179 |
| 265211_at | AT2G36640 | Lea group 3 | 1.106 |
| 246242_at | AT4G36600 | Lea group 3 | 1.058 |
| 265208_at | AT2G36690 | Oxidoreductase, 2OG-Fe(II) oxygenase | 1.050 |
| 249353_at | AT5G40420 | Gly-rich protein/oleosin | 1.030 |
| 250624_at | AT5G07330 | Expressed protein | 1.005 |
As shown in Figure 5, 465 transcripts were elevated in wild-type or Zat12-overexpressing plants treated with H2O2. Of these, only eight transcripts were elevated to a higher degree in Zat12-overexpressing lines compared to wild-type plants (HSP17.4-CIII, At1g54050; HSP17.6A-CI, At1g59860; HSP17.6B-CI, At2g29500; stress-inducible protein, At4g12400; and four expressed proteins of unknown function, At5g24660, At5g10695, At4g36500, and At3g10930; Supplemental Table III). In addition, as shown in Figure 5, 90 transcripts were specifically elevated in Zat12-overexpressing plants in response to H2O2 application. These transcripts are summarized in Supplemental Table IV. Interestingly, a number of ROS-response genes are found in this group of genes, suggesting that Zat12 expression during H2O2 stress might activate parts of the ROS-response pathway in Arabidopsis (Mittler et al., 2004). They include glutathione S-transferases, DNAJ heat shock proteins, heat shock transcription factors 4 and 6, thioredoxin, calmodulin-9, ubiquitin, and glutathione peroxidase. Supplemental Table V summarizes all transcripts significantly elevated in Zat12-overexpressing plants in response to H2O2 stress.
DISCUSSION
The enhanced expression of Zat12 in response to many different abiotic stress conditions (Figs. 1 and 2; Iida et al., 2000; Cheong et al., 2002; Fowler and Thomashow, 2002; Kreps et al., 2002; Rizhsky et al., 2004b; Davletova et al., 2005) suggests that Zat12 is involved in the response of plants to all of these stresses. However, functional characterization of Zat12 demonstrated that Zat12 is essential for plant tolerance to only a few of the stresses its expression is associated with (Fig. 3; Supplemental Figs. 1 and 2). How could this result be explained? We see at least two different possibilities. (1) Zat12 performs different functions as a transcriptional regulator during different stresses, and our functional characterization could not have detected all of the cellular and stress-response functions associated with Zat12. (2) Zat12 expression is not directly linked to its function, and although in many stress situations Zat12 expression is enhanced, the Zat12 protein might not be required for stress tolerance to all stresses. The broad response of the Zat12 gene to different stresses might result from the activation of Zat12 expression by ROS such as H2O2 that accumulate in cells in response to almost all stress conditions (Dat et al., 2000; Apel and Hirt, 2004; Mittler et al., 2004).
The transcriptome profiling experiments performed in this study suggest that, under specific growth conditions, Zat12 regulates a collection of transcripts involved in light and oxidative stress responses (Table II; Fig. 5). Our profile analyses results are also in good agreement with the functional characterization of gain-of-function lines for Zat12 (Fig. 3). Thus, the six different Lea proteins expressed in Zat12-overexpressing lines (Table II) might explain the high tolerance of these lines to osmotic stress (Fig. 3D), and the copper/zinc superoxide dismutases (CSD1, CSD2, and the copper chaperone) expressed in Zat12-overexpressing lines (Table II) might explain the high tolerance of these lines to methyl viologen (data not shown; Rizhsky et al., 2004a) and high light stress (data not shown; Iida et al., 2000). If indeed the expression of the same Lea proteins enhanced in Zat12-overexpressing lines is linked to Zat12 function, then this result might also explain the sensitivity of KO-Zat12 lines to salinity (Fig. 3A) and osmotic stress (Fig. 3B). The similarity between the transcriptome of Zat12-overexpressing lines and plants subjected to light stress (Table II; Rizhsky et al., 2003) might explain the high tolerance of Zat12-overexpressing lines to light stress (Iida et al., 2000; D. Davletova, J. Coutu, and R. Mittler, unpublished data).
Zat12 contains an EAR-motif-like sequence that may function as a repression domain (Ohta et al., 2001; Hiratsu et al., 2002; Vogel et al., 2005). In response to cold stress, the Zat12 protein was suggested to act as a suppressor of CBF transcription factors (Vogel et al., 2005). Our transcriptional profiling analysis revealed several transcripts suppressed in Zat12-overexpressing plants in the absence of stress and in response to H2O2 application (Supplemental Table II). However, additional studies are required to confirm a possible role for Zat12 as a repressor of these transcripts. Our analysis of knockout Zat12 plants subjected to heat stress revealed that the absence of Zat12 might have resulted in an enhanced tolerance of knockout plants to this stress (Fig. 3C). Thus, at least in response to heat stress, Zat12 might function as a repressor. It should however be noted that transgenic plants overexpressing Zat12 were not found to be more sensitive to heat stress than wild-type plants (Supplemental Fig. 1). Further studies, including transcriptome profiling of wild-type and Zat12 knockout plants subjected to heat stress, are required to address the possibility that Zat12 functions as a repressor during stress.
We previously proposed that Zat12 expression is directly linked to reactive oxygen metabolism in Arabidopsis (Rizhsky et al., 2004a; see also Table I). This hypothesis was based on the enhanced expression of Zat12 in knockout-Apx1 plants (Pnueli et al., 2003; Davletova et al., 2005), the lack of Apx1 expression in knockout Zat12 plants subjected to oxidative stress (Rizhsky et al., 2004a), the high sensitivity of knockout Zat12 plants to oxidative stress, and the enhanced tolerance of Zat12-overexpressing lines to methyl viologen application (Rizhsky et al., 2004a). Our current analysis reveals an overlap between transcript expression in Zat12-overexpressing seedlings grown under controlled conditions and transcript expression in wild-type seedlings subjected to H2O2 stress (Fig. 5; Table II). Surprisingly, the majority of transcripts that were expressed in Zat12-overexpressing seedlings, as well as in wild-type seedlings subjected to H2O2 stress, were encoded by the chloroplast genome and associated with the response of plants to high light stress (Rossel et al., 2002; Rizhsky et al., 2003). Reactive oxygen species such as H2O2, O2−, and 1O2 are thought to function as early signals for high light stress in plants (Pnueli et al., 2003; Rizhsky et al., 2003; Apel and Hirt, 2004; Mittler et al., 2004). Because Zat12 expression is enhanced in response to light stress (Iida et al., 2000; Davletova et al., 2005) and its expression in transgenic plants enhances their tolerance to high light stress (Iida et al., 2000; D. Davletova, J. Coutu, and R. Mittler, unpublished data), it is possible that Zat12 functions as part of a signal transduction pathway that directly or indirectly alters chloroplast gene expression in response to an H2O2 signal generated during light stress.
We previously reported that Zat12 is required for Apx1 expression during oxidative stress (Rizhsky et al., 2004a). However, Zat12 overexpression did not result in the enhanced expression of Apx1 (more than wild type) in the presence or absence of oxidative stress (Fig. 4). This result suggests that the relationship between Zat12 and Apx1 expression is complex and that further studies are required to resolve it. It is possible, for example, that Zat12 functions as part of a transcriptional or signal transduction complex required for Apx1 expression. In its absence, Apx1 expression is prevented during oxidative stress (Rizhsky et al., 2004a). However, simply increasing the level of Zat12 in cells might not be sufficient to increase Apx1 expression because other components or subunits of the complex might be rate limiting.
Our analyses (Figs. 1–3; Table II; Rizhsky et al., 2004a) and that of others (Iida et al., 2000; Vogel et al., 2005) suggest that Zat12 plays an important role in abiotic stress signaling in Arabidopsis. The enhanced tolerance to different stress conditions and the presumed activation of a collection of transcripts involved in the defense response of plants against light and oxidative stress in Zat12-expressing plants suggest that Zat12, or its orthologs, could be used for the development of stress-tolerant crops. In addition, the Zat12 promoter::luciferase fusion plants (Figs. 1 and 2) generated during our study could be used as a valuable tool for the isolation of stress- and ROS-response signaling mutants (Zhu, 2002). These could be used in future studies to identify pathways and receptors involved in stress and reactive oxygen perception and defense in plants.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Five-day-old Arabidopsis (Arabidopsis thaliana cv Columbia) seedlings were used for all stress assays described in this study. Seedlings were grown on 1%, 0.5× Murashige and Skoog (MS) agar plates or in sterile 0.5× MS media on a shaker in growth chambers (Percival E-30) under controlled conditions (21°C–22°C, 18 h or constant light cycle, 100 μmol m−2 s−1, and a relative humidity of 70%; Rizhsky et al., 2004a; Davletova et al., 2005). Two independent homozygous knockout Arabidopsis lines containing T-DNA inserts in the Zat12 gene (KO-Zat12; SALK_037357 and SAIL 792_F04) and three independent homozygous lines of transgenic plants overexpressing Zat12 (OE-Zat12; lines 4, 12, and 17, all with similar expression level) were obtained as described previously (Rizhsky et al., 2004a). Transformation of Arabidopsis plants with the Zat12 promoter (1,000-bp fragment)::luciferase fusion was performed as described (Bent and Clough, 2000; Rizhsky et al., 2003), and transgenic plants were screened by RNA blots (Rizhsky et al., 2004a). Three independent lines expressing the Zat12 promoter (1,000-bp fragment)::luciferase fusion construct were used for Zat12 expression analysis (lines 4-4, 5-4, and 12-1). These were similar to wild-type plants in all aspects of plant growth and development (data not shown). All experiments were performed with three to five technical replications and repeated at least three times.
Molecular and Biochemical Analysis
RNA was isolated and analyzed by RNA blots as described previously (Rizhsky et al., 2004a; Davletova et al., 2005). RNA staining or a ribosomal 18S rRNA probe was used to control for RNA loading. Luciferase imaging was performed with a Kodak 2000MM image station. Plants were sprayed with 1 mm luciferine (Promega) prepared in water, incubated for 30 min, placed on the imager, and exposed for 3 to 5 min using the luminescence setting with a fully opened aperture.
Stress Assays
For the analysis of stress tolerance, seeds of wild-type and Zat12-perturbed lines (two independent Zat12 knockout and three independent Zat12-overexpressing lines) were surface sterilized with bleach and placed in rows on 1% agar plates (0.5× MS medium), containing different concentrations of methyl viologen, NaCl, sorbitol, or ferric-citrate (Sigma; Rizhsky et al., 2003, 2004a). Each row of seeds placed on a plate was divided into two parts: wild-type seeds, and seeds of transgenic plants overexpressing Zat12 or seeds of knockout Zat12 plants. Thus, the different seeds were placed side by side on the same plate. Plates were maintained vertically in a growth chamber (21°C–22°C, constant light, 100 μmol m−2 s−1), and percentage of germination and root length were scored 5 d after seed plating. Four- or five-day-old seedlings grown on 0.5× MS agar plates were also subjected to heat stress (38°C), cold stress (10°C), or light stress (1,000 μmol m−2 s−1). For DNA chip analysis, 5-d-old seedlings grown in 0.5× MS media were subjected to H2O2 (1, 5, 10, or 20 mm) stress (Rizhsky et al., 2003, 2004a). Wounding was performed as described previously (Rizhsky et al., 2004a). All stress experiments were performed with three to five technical replications, each containing 15 to 30 seeds per line, and repeated at least three times.
DNA Chip Analysis
In three independent experiments, RNA was isolated from control and H2O2 (20 mm, 1 h)-treated 5-d-old wild-type and Zat12-overexpressing seedlings (pooled from three independent lines) grown in 0.5× MS medium as described above. This treatment was not lethal to wild-type or Zat12-overexpressing lines as determined by continued growth of seedlings in culture (data not shown). A total of 12 RNA samples were used: three wild type untreated, three wild type treated with H2O2, three Zat12 overexpressing untreated, and three Zat12 overexpressing treated with H2O2. At least 150 seedlings were used per RNA sample, and RNA was isolated using Trizol (Davletova et al., 2005). RNA samples were used to perform chip hybridization analyses (Arabidopsis ATH1 chips; Affymetrix) at the Virginia Bioinformatics Institute Core Laboratory Gene Expression Facility (https://www.vbi.vt.edu/). Conditions for RNA isolation, labeling, hybridization, and data analysis are described (Davletova et al., 2005). Data visualization and analysis were performed with Silicon Genetics GeneSpring Version 5.1 and ArraAassit (IobionLab). Some of the results were confirmed by RNA blots. See details for data analysis below.
GeneChip Data Processing and Statistical Analysis
All GeneChip arrays were processed first by Robust Multi-Array Average (RMA; Irizarry et al., 2003) using the R package affy (Gautier et al., 2004). Expression values were computed from raw CEL files by first applying the RMA model of probe-specific correction of PM (perfect match) probes. These corrected probe values were then normalized via quantile normalization, and a median polish was applied to compute one expression measure from all probe values. Resulting RMA expression values were log2 transformed. These are standard methods for processing Affymetrix data (Davletova et al., 2005). Please see the affy manual at www.bioconductor.org/repository/devel/vignette/affy.pdf for details. A visual inspection of the distributions of raw PM probes values for all 12 arrays showed no outlying arrays; similarly, both density plots and boxplots of RMA expression value distributions of all arrays were very similar with no apparent outlying arrays. Curves describing trends in RNA degradation between the 5′ end and the 3′ end in each probe set were generated, and all 12 proved very similar, with an evident downward trend at the 5′ end. Pearson correlation coefficients and Spearman rank coefficients were computed on the RMA expression values (log base 2) for each set of biological triplicates. Spearman coefficients ranged from 0.985 to 0.997; Pearson coefficients ranged between 0.989 and 0.998 (for quality control measures, see Supplemental Figs. 3–8 and Supplemental Table I).
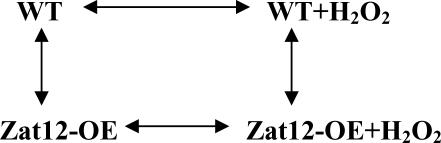
To determine whether genes were differentially expressed between genotypes across the temporal states, an ANOVA was performed on the RMA expression values. For an overview on the application of ANOVA to microarray data, see Kerr et al. (2000). The following model was used for this analysis: yijk = Vi + (VT)ij + ɛijk, where yijk denotes the log2 signal measured for variety i, treatment j, and biological replicate k, with 1 ≤ i ≤ 2, 1 ≤ j ≤ 2, and 1 ≤ k ≤ 3. The terms Vi and Tj measure the effect of the variety and treatment, respectively, and the interaction term (VT)ij accounts for the interaction between variety and treatment. An ANOVA was performed on each gene using the linear model above, with four contrasts based on the comparisons between varieties and treatments as outlined in the diagram below.
Figure .
These four comparisons were performed simultaneously, and a multiple testing correction (Benjamini and Hochberg, 1995) was applied to the P values of the test statistics to adjust the false discovery rate. Genes with adjusted P values < 0.05 were extracted for further analysis. This resulted in 5,417 genes with significant differential expression in at least one of the comparisons performed based on the ANOVA above. Expression values of this selection were then inverse-log transformed, and genes with differential expression in each of the four comparisons of more than 2-fold were selected. Specifically, 668 genes were found to be significantly differentially expressed by more than 2-fold in wild type compared to wild type treated with H2O2: 637 between Zat12 overexpressing and Zat12 overexpressing treated with H2O2; 48 between wild type and Zat12 overexpressing; and 54 between wild type treated with H2O2 and Zat12 overexpressing treated with H2O2. The R package limma was used for ANOVA methods (www.bioconductor.org/repository/devel/vignette/affy.pdf). Microarray data from this experiment were submitted to NASCArrays at http://affymetrix.arabidopsis.info/.
Supplementary Material
This work was supported by the National Science Foundation (grant nos. NSF–0431327 and NSF–0420033).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.068254.
References
- Alvarez ME, Pennell RI, Meijer PJ, Ishikawa A, Dixon RA, Lamb C (1998) Reactive oxygen intermediates mediate a systemic signal network in the establishment of plant immunity. Cell 92: 773–784 [DOI] [PubMed] [Google Scholar]
- Apel K, Hirt H (2004) reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55: 373–399 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Statist Soc Ser B Methodological 57: 289–300 [Google Scholar]
- Bent AF, Clough SJ (2000) Plant transformation. In SB Gelvin, RA Schilperoort, eds, Plant Molecular Biology Manual, Ed 2. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 1–14
- Chen W, Provart NJ, Glazebrook J, Katagiri F, Chang HS, Eulgem T, Mauch F, Luan S, Zou G, Whitham SA, et al (2002) Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell 14: 559–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong YH, Chang HS, Gupta R, Wang X, Zhu T, Luan S (2002) Transcriptional profiling reveals novel interactions between wounding, pathogen, abiotic stress, and hormonal responses in Arabidopsis. Plant Physiol 129: 661–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dat J, Vandenabeele S, Vranova E, Van Montagu M, Inze D, Van Breusegem F (2000) Dual action of the active oxygen species during plant stress responses. Cell Mol Life Sci 57: 779–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davletova S, Rizhsky L, Liang H, Shengqiang Z, Oliver DJ, Coutu J, Shulaev V, Schlauch K, Mittler R (2005) Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. Plant Cell 17: 268–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5: 199–206 [DOI] [PubMed] [Google Scholar]
- Fowler S, Thomashow MF (2002) Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14: 1675–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier L, Cope L, Bolstad B, Irizarry RA (2004) Affy: analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20: 307–315 [DOI] [PubMed] [Google Scholar]
- Hiratsu K, Ohta M, Matsui K, Ohme-Takagi M (2002) The SUPERMAN protein is an active repressor whose carboxy-terminal repression domain is required for the development of normal flowers. FEBS Lett 514: 351–354 [DOI] [PubMed] [Google Scholar]
- Iida A, Kazuoka T, Torikai S, Kikuchi H, Oeda K (2000) A zinc finger protein RHL41 mediates the light acclimatization response in Arabidopsis. Plant J 24: 191–203 [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs R, Collin R, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264 [DOI] [PubMed] [Google Scholar]
- Karpinski S, Reynolds H, Karpinska B, Wingsle G, Creissen G, Mullineaux P (1999) Systemic signaling and acclimation in response to excess excitation energy in Arabidopsis. Science 284: 654–657 [DOI] [PubMed] [Google Scholar]
- Kerr MK, Martin M, Churchill GA (2000) Analysis of variance for gene expression microarray data. J Comput Biol 7: 819–837 (R) [DOI] [PubMed] [Google Scholar]
- Kreps JA, Wu Y, Chang H-S, Zhu T, Wang X, Harper JF (2002) Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol 130: 2129–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla J, Hayes R, Gruissem W (1996) Polyadenylation accelerates degradation of chloroplast mRNA. EMBO J 15: 7137–7146 [PMC free article] [PubMed] [Google Scholar]
- Mittler R, Zilinskas B (1992) Molecular cloning and characterization of a gene encoding pea cytosolic ascorbate peroxidase. J Biol Chem 267: 21802–21807 [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Gollery M, Van Breusegem F (2004) The reactive oxygen gene network of plants. Trends Plant Sci 9: 490–498 [DOI] [PubMed] [Google Scholar]
- Nover L, Bharti K, Doring P, Mishra SK, Ganguli A, Scharf KD (2001) Arabidopsis and the heat stress transcription factor world: How many heat stress transcription factors do we need? Cell Stress Chaperones 6: 177–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M (2001) Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13: 1959–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco-Cardenas ML, Narvaez-Vasquez J, Ryan CA (2001) Hydrogen peroxide acts as a second messenger for the induction of defense genes in tomato plants in response to wounding, systemin, and methyl jasmonate. Plant Cell 13: 179–191 [PMC free article] [PubMed] [Google Scholar]
- Pnueli L, Hongjian L, Mittler R (2003) Growth suppression, altered stomatal responses, and augmented induction of heat shock proteins in cytosolic ascorbate peroxidase (Apx1)-deficient Arabidopsis plants. Plant J 34: 187–203 [DOI] [PubMed] [Google Scholar]
- Rizhsky L, Liang H, Mittler R (2003) The water-water cycle is essential for chloroplast protection in the absence of stress. J Biol Chem 278: 38921–38925 [DOI] [PubMed] [Google Scholar]
- Rizhsky L, Davletova S, Liang H, Mittler R (2004. a) The zinc-finger protein Zat12 is required for cytosolic ascorbate peroxidase 1 expression during oxidative stress in Arabidopsis. J Biol Chem 279: 11736–11743 [DOI] [PubMed] [Google Scholar]
- Rizhsky L, Liang H, Shuman J, Shulaev V, Davletova S, Mittler R (2004. b) When defense pathways collide: the response of Arabidopsis to a combination of drought and heat stress. Plant Physiol 134: 1683–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossel JB, Wilson IW, Pogson BJ (2002) Global changes in gene expression in response to high light in Arabidopsis. Plant Physiol 130: 1109–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R, Werber M, Weisshaar B (2001) The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 4: 447–456 [DOI] [PubMed] [Google Scholar]
- Vogel JT, Zarka DG, Van Buskirk HA, Fowler SG, Thomashow MF (2005) Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J 41: 195–211 [DOI] [PubMed] [Google Scholar]
- Zhu J-K (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Physiol Plant Mol Biol 53: 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.