Abstract
Plant genomes are rich in long terminal repeat retrotransposons, and here we describe a plant-specific lineage of Ty1/copia elements called the Sireviruses. The Sireviruses vary greatly in their genomic organization, and many have acquired additional coding information in the form of an envelope-like open reading frame and an extended gag gene. Two-hybrid screens were conducted with the novel domain of Gag (the Gag extension) encoded by a representative Sirevirus from maize (Zea mays) called Hopie. The Hopie Gag extension interacts with a protein related to dynein light chain 8 (LC8). LC8 also interacts with the Gag extension from a Hopie homolog from rice (Oryza sativa). Amino acid motifs were identified in both Hopie Gag and LC8 that are responsible for the interaction. Two amino acids critical for Gag recognition map within the predicted LC8-binding cleft. Two-hybrid screens were also conducted with the Gag extension encoded by the soybean (Glycine max) SIRE1 element, and an interaction was found with light chain 6 (LC6), a member of the LC8 protein family. LC8 and LC6 proteins are components of the dynein microtubule motor, with LC8 being a versatile adapter that can bind many unrelated cellular proteins and viruses. Plant LC8 and LC6 genes are abundant and divergent, yet flowering plants do not encode other components of the dynein motor. Although, to our knowledge, no cellular roles for plant LC8 family members have been proposed, we hypothesize that binding of LC8 proteins to Gag aids in the movement of retrotransposon virus-like particles within the plant cell or possibly induces important conformational changes in the Gag protein.
Retroelements (including retrotransposons and retroviruses) are highly successful biological parasites (Kumar and Bennetzen, 1999; Eickbush and Malik, 2002). Long terminal repeat (LTR) retrotransposons, a specific type of retroelement, are identified by direct sequence repeats that flank a central gag and pol coding region. The life cycle of LTR retrotransposons consists of both nuclear and cytoplasmic stages. Retrotransposons in the genome are transcribed into mRNA, which is then exported to the cytoplasm. In the cytoplasm, the mRNA is translated into Gag and Pol proteins, the former of which form a virus-like particle (VLP). Within the particle, the pol-encoded enzyme reverse transcriptase (RT) makes a cDNA copy of the retrotransposon mRNA. The cDNA is then imported back into the nucleus and integrated into the host's DNA by the pol-encoded integrase. Retroviruses generally undergo the same basic steps in the life cycle as retrotransposons with the exception that retroviruses have an extracellular phase. An additional gene of retroviruses, envelope (env), allows the retroviral particle to escape the cell and infect another cell.
The relationship between LTR retrotransposons and their host is complex and highly evolved (Voytas and Boeke, 2002). These mobile elements require the host for many of the life cycle steps; some examples include a host tRNA that is needed to prime reverse transcription, transcription factors and transcriptional machinery that are needed for mRNA synthesis, and the translation apparatus that is required for protein production. In yeast (Saccharomyces cerevisiae), a model organism for retrotransposon biology, many genes are known to either affect or be required for Ty retrotransposition (Scholes et al., 2001; Griffith et al., 2003; Aye et al., 2004; Irwin et al., 2005). Screens using the yeast gene deletion collection identified approximately 2% to 3% of the yeast genes (involved in many cellular processes) to be involved in Ty1 and Ty3 retrotransposition (Scholes et al., 2001; Griffith et al., 2003). The exact nature of how these genes affect the retroelement life cycle has yet to be determined.
The increase in available DNA sequence data has revealed numerous new retrotransposons, many of which vary greatly from the canonical genomic structure (Havecker et al., 2004). This variability most often takes the form of either a lack of protein-coding genes (nonautonomous elements, for example) or the addition of coding information. The function of additional coding information is often unknown, although it may be that extra retroelement proteins are required for transposition in specific hosts or that extra genes provide an advantage for the retroelements that have acquired them. One of the most frequent additions in coding information is an env-like gene, which is an extra open reading frame (ORF) in the same position and with similar characteristics to the env gene of retroviruses (Malik et al., 2000; Eickbush and Malik, 2002). The most well-studied group of env-containing retrotransposons is the Drosophila Errantiviruses, such as gypsy or ZAM (Leblanc et al., 2000; Pelisson et al., 2002). For the gypsy retrotransposon, the env gene has been implicated in the infection of Drosophila oocytes (Kim et al., 1994; Song et al., 1994). Besides the env-like ORF, there exist other examples of coding information acquisition. For example, some plant retroelements carry multiple antisense ORFs after pol (Martinez-Izquierdo et al., 1997; Ohtsubo et al., 1999). However, no function has been experimentally determined for the retrotransposon env-like ORFs (except gypsy) or for other additional coding sequences.
The Sireviruses are a recently named group of plant retrotransposons that carry additional coding information (Boeke et al., 2004). This group of Ty1/copia retrotransposons, named for the founding member, SIRE1 from soybean (Glycine max), also includes the previously characterized retrotransposons Endovir1-1 from Arabidopsis (Arabidopsis thaliana), Opie-2 and Prem-2 from maize (Zea mays), and ToRTL from tomato (Lycopersicon esculentum; SanMiguel et al., 1996; Laten et al., 1998; Kapitonov and Jurka, 1999; Peterson-Burch et al., 2000; Peterson-Burch and Voytas, 2002). Two striking features characterize the Sireviruses: they too have an env-like ORF after pol, and they also encode a significantly larger Gag protein (Peterson-Burch and Voytas, 2002). To our knowledge, no known functions for either of these additional coding regions have been determined.
Here we report one potential function for the additional information encoded by the gag gene of the Sireviruses. We identified a conserved interaction between Sirevirus Gag proteins and different members of light chain 8 (LC8) protein family (also known as PIN [Jaffrey and Snyder, 1996] or dynein light chain 1 [Dick et al., 1996]). LC8 proteins are highly conserved in all eukaryotic organisms, and LC8 is a known component of the dynein microtubule motor, a complex of proteins that moves cargo toward the minus end of microtubules (King, 2000). In various organisms, one major function of LC8 is to bind cargo (cellular proteins and viruses) for movement along the microtubule (King, 2003). We speculate that the additional coding information in the Sireviruses allows them to proliferate in plant hosts, and we discuss the implications of a Sirevirus Gag-LC8 interaction.
RESULTS
Genomic Organization of the Sireviruses
The Sireviruses were previously reported as a distinct lineage of Ty1/copia retroelements (Pseudoviridae; Peterson-Burch and Voytas, 2002). Because the Sireviruses were unique to plants, they were originally named Agroviruses (Peterson-Burch and Voytas, 2002). More recently, they been designated Sireviruses by the International Committee on the Taxonomy of Viruses (Boeke et al., 2004).
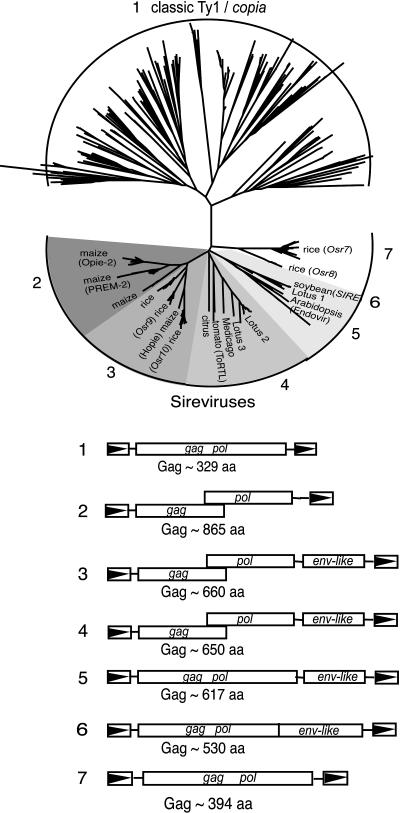
We previously described a phylogenetic analysis of Ty1/copia RT sequences retrieved from GenBank using the Sirevirus Opie-2 RT as an electronic probe (Gao et al., 2003). Two striking features were observed. First, the tree revealed two major lineages of plant Ty1/copia elements, which we refer to as the classical Ty1/copia lineage and the Sirevirus lineage; each represents about one-half of the retrieved RTs (Fig. 1). Second, a striking difference can be seen in the branch lengths between the two clades. In general, longer branch lengths characterize the classical Ty1/copia elements, whereas Sireviruses emanate from the tree with shorter branches (Fig. 1).
Figure 1.
Genomic organization of the Sireviruses. A phylogenetic analysis of the plant Pseudoviridae (Ty1/copia elements) was previously described (Gao et al., 2003). The neighbor-joining tree resulting from this analysis revealed two major groups, the classical Ty1/copia elements and the Sireviruses. The Sirevirus clade contains elements with unusually large Gag proteins and varied genomic organization with respect to env-like ORFs and breaks in translation between gag and pol.
We wanted to superimpose on the phylogenetic tree the variety observed in the genomic organization of Sirevirus genomes. Many retrotransposon sequences are degenerate; that is, they have accumulated mutations in the form of stop codons and frameshifts that disrupt their ORFs and render them nonfunctional. Two methods were used to assess the organization of ORFs in what are likely functional elements. For families with multiple, highly homogenous members, the full-length elements were aligned and a strict consensus nucleotide sequence was determined (referred to as Osr9, Osr10, and Lotus2). For other more highly degenerate Sireviruses, multiple elements were compared to determine whether or not a frameshift separated gag and pol and whether or not an env-like ORF was present. The variation in the genomic organization of the Sireviruses can be seen in Figure 1.
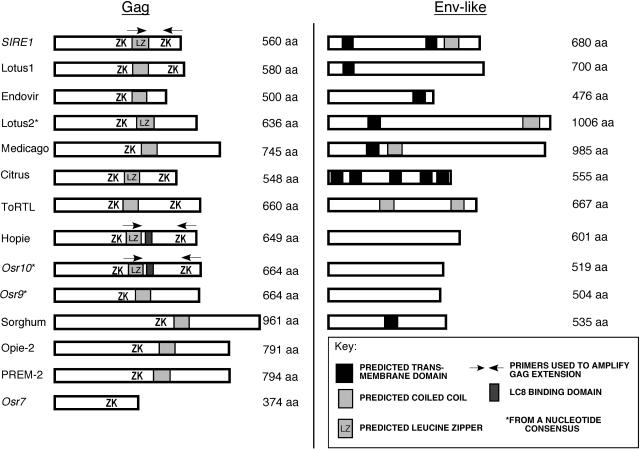
Considerable variation was observed among Sireviruses with respect to coding sequences downstream of pol. In addition to previously reported Sireviruses, we identified a number of new elements with env-like ORFs in both monocots (maize and sorghum) and dicots (lotus, medicago, and citrus). Some rice (Oryza sativa) elements were previously named (Osr7, Osr8, Osr9, and Osr10; McCarthy et al., 2002; Fig. 1). Not all Sireviruses, however, encode an obvious env-like ORF. Among these are the Opie-2 and Prem-2 elements of maize as well as some members from rice (Osr7 and Osr8). Little amino acid similarity (if any) is present among predicted consensus Env-like proteins. Furthermore, the consensus elements generated in this study and the newly identified elements reveal great variation in the length of the predicted Env-like proteins (from 504–1,006 amino acids). Based on an analysis of the Endovir1-1, SIRE1, and ToRTL Env sequences, Peterson-Burch and Voytas (2002) had noted that some secondary structural features are conserved, such as the presence of a transmembrane domain. However, neither transmembrane nor coiled-coil domains are consistently predicted in the new Env-like ORFs we describe (Fig. 2).
Figure 2.
Cartoon depicting the structural features of the Gag and Env-like proteins of individual Sirevirus elements. Boxes represent ORFs followed by their approximate length in amino acids. Arrows represent primers that delimit the regions of the Gag extension that were cloned and used for two-hybrid assays. The asterisks identify those ORFs predicted from a nucleotide consensus. ZK, CX2CX3HX4C zinc knuckle.
In an initial characterization of the Sireviruses, it was noted that they encode unusually large gag genes (Peterson-Burch and Voytas, 2002). The lengths of Gag for each of the Sirevirus groups in Figure 1 were estimated by averaging the number of amino acids in Gag for all elements having a given genomic structure. Gag lengths range from about 548 to 961 amino acids. Figure 2 depicts some of the variation in individual gag genes. The gag genes are most conserved in their N-terminal halves, and all encode a central CCHC zinc knuckle. With the exception of Osr7 and Osr8, the central zinc knuckle is followed by a predicted coiled-coil domain. Throughout this manuscript we use the term Gag extension, which refers to this coiled-coil domain and sequences downstream. The Gag extensions are mainly responsible for variations in the lengths of the entire gag genes. In addition, some Sireviruses have a second CCHC zinc knuckle at the Gag C terminus. Finally, and as previously reported, some Sireviruses that have a shift in reading frame between gag and pol also have a conserved amino acid sequence domain at the very C terminus of gag (Gao et al., 2003).
The Gag Extension Interacts with Proteins Related to the Dynein Light Chain Family LC8
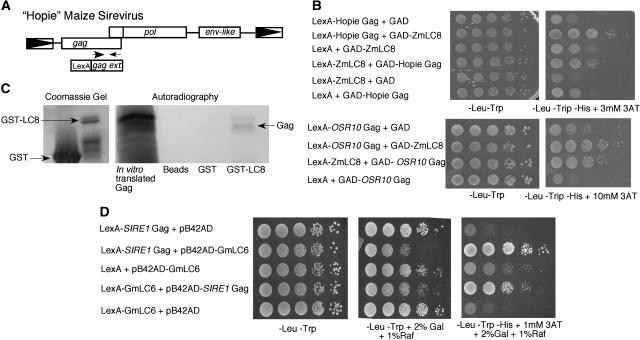
To help understand the biological roles of the additional coding information present in Sireviruses, we sought to establish a model element for study. Ideally, the element should be functional and typify the diversity in coding and structural features seen for the Sirevirus group. We settled on a maize element we call Hopie, whose sequence was obtained from bacterial artificial chromosome ZM15C05 (GenBank accession no. AC116033; Nagaki et al., 2003). Hopie has the Gag extension (containing the coiled-coil domain and second zinc knuckle), an env-like ORF, and a shift in reading frame between gag and pol (Gao et al., 2003; Fig. 3A). Thus, Hopie contains all the unique features of Sireviruses thus far identified.
Figure 3.
Interaction of some Sireviruses with LC8 family members. A, Hopie has all features associated with Sireviruses, namely an extended gag gene, a break in reading frame between gag and pol, and an env-like ORF. Primers (represented by arrows) amplify the gag extension, which was fused to the LexA DNA-binding domain for use in a two-hybrid screen. Boxed arrows represent LTRs; open rectangles represent coding regions. B, Ten-fold serial dilutions of yeast cells expressing either the Hopie (maize) or Osr10 (rice) Gag extensions as bait and ZmLC8 as the prey. Yeast grown on −Leu −Trp serve as a control for cell number. Yeast grown on media lacking His identify two-hybrid interactions. C, ZmLC8 and the Hopie Gag extension interact in vitro. A Coomassie-stained polyacrylamide gel shows purification of GST-LC8. The autoradiograph demonstrates that GST-LC8 can immunoprecipitate radiolabeled Gag translated in vitro. D, Ten-fold serial dilutions of yeast cells expressing the SIRE1-4 Gag extension as a bait and GmLC6 as the prey. The plasmid containing the B42p transcription activation domain is under the control of the galactose (Gal) promoter. Yeast grown on −Leu −Trp 2% dextrose (Dex) serve as a loading control. Yeast grown on −Leu −Trp 2% Gal 1% raffinose (Raf) show a growth defect when both the Gag and GmLC6 proteins are present.
To understand a potential function of the additional information encoded by the Sireviruses, yeast two-hybrid screens were completed using the Hopie Gag extension and Env-like proteins as bait (Fig. 3A). Both screens were carried out using a library made from juvenile shoot expressed sequence tags (ESTs) fused to the Gal4p transcriptional activation domain (GAD; Moose and Sisco, 1996). Growth of yeast on media lacking His indicated a two-hybrid interaction. LexA fusions to both a full-length Env-like protein and its C terminus were used as bait. However, no ESTs were identified that interacted with any portion of the Hopie Env-like protein.
To carry out the Gag extension two-hybrid screens, the C-terminal portion of the Hopie gag gene, which encodes the coiled-coil region and the second zinc knuckle (see Fig. 2), was fused to the LexA DNA-binding domain (Fig. 3A). A yeast two-hybrid screen was then performed using the same juvenile shoot EST library. Four different ESTs were recovered; however, only one retained the interaction when the bait (Gag) and prey (EST) reading frames were switched (Fig. 3B). This EST sequence was retrieved numerous times and encoded a homolog of a dynein light chain gene (known as LC8 [King et al., 1996], PIN [Jaffrey and Snyder, 1996], and dynein light chain 1 [Dick et al., 1996]) and is hereafter referred to as ZmLC8. LC8 in animals is most often identified as a component of the dynein microtubule motor complex but has other functions that will be discussed in depth in the discussion (King, 2003). To confirm the interaction between ZmLC8 and the maize Gag extension, an in vitro binding assay was conducted. A glutathione S-transferase (GST)-ZmLC8 fusion protein was purified from Escherichia coli and found to pull down a S35-labeled in vitro-translated Gag extension. No interaction was observed when Gag was incubated with GST or the agarose beads alone (Fig. 3C).
To assess the universality of the Gag/LC8 interaction, the Gag extension was cloned from a related rice Sirevirus (Osr10). The rice and maize Gag extensions are 40% similar. A strong two-hybrid interaction was observed between the Osr10 Gag extension and ZmLC8 (Fig. 3B). Because the maize and rice Gag extensions shared sequence similarity, we also tested a Gag extension from the previously characterized soybean SIRE1-4 element (Laten et al., 2003). The SIRE1-4 Gag and the Hopie Gag extension do not share obvious amino acid similarity, but they do share some structural features such as the coiled-coil region and the second zinc knuckle. However, the SIRE1-4 Gag extension did not interact with ZmLC8 in a directed two-hybrid screen. We then used the soybean SIRE1-4 element for another yeast two-hybrid screen with a library of soybean ESTs from 2- to 4-d-old hypocotyls (H. Gao and M. Bhattacharyya, unpublished data). In this screen, an EST with some sequence similarity to ZmLC8 was identified (Fig. 3D). This EST was more closely related to another dynein light chain, light chain 6 (LC6) from Chlamydomonas reinhardtii (King and Patel-King, 1995). Approximately 30% of the His+ colonies obtained in the screen contained the EST encoding a soybean homolog of LC6 (referred to now as GmLC6).
Sequences Critical for the Gag/LC8 Interaction
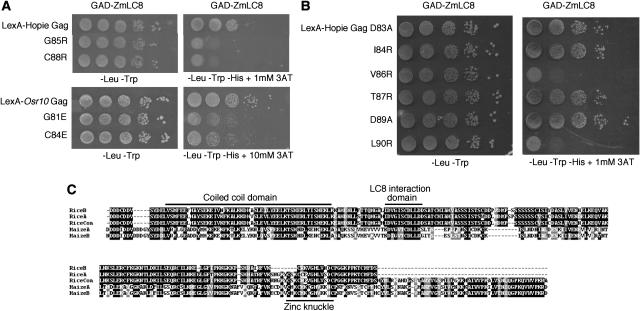
Experiments were conducted to characterize regions of the maize Gag extension and ZmLC8 responsible for interaction. In initial experiments, portions of the N and C termini of the Hopie Gag extension were deleted, and the region between the coiled-coil domain and the CCHC motif was found necessary and sufficient for the interaction (data not shown). To further define the Gag interaction domain, a reverse-yeast two-hybrid assay was employed (Vidal et al., 1996). PCR mutagenesis of the Gag extension was followed by selection of yeast that failed to grow on media lacking His, thereby identifying candidate Gag mutants that no longer could sustain a productive interaction. Two critical residues were identified, G85 and C88 (Fig. 4A), which are located in a region conserved between the maize and rice Osr10 Gag extensions (Fig. 4B). The corresponding residues in the Osr10 Gag extension were also found to be important for interactions with ZmLC8 (Fig. 4A). To determine if any of the amino acids surrounding G85 and C88 were necessary for the interaction, maize Gag extension mutants (D83A, I84R, V86R, T87R, D89A, and L90R) were made by site-directed PCR mutagenesis. The mutant analysis indicated that both V86 and L90 were necessary for the Gag/ZmLC8 interaction (Fig. 4B). Of these two amino acids, L90 is also conserved in the rice Gag extension, but V86 has been conservatively replaced by an Ile in Osr10 (Fig. 4C).
Figure 4.
Mutations in Gly and Cys residues of the Hopie and Osr10 Sirevirus Gag extension abrogate binding to ZmLC8. A, The first section depicts 10-fold serial dilutions of yeast cells expressing the Hopie Gag or the G85R or C88R mutants along with ZmLC8. The second section depicts cells expressing either the cloned Osr10 Gag or the G81E or C84E mutants. B, Hopie Gag mutants made by site-directed mutagenesis. Yeast grown on −Leu −Trp serve as a loading control. All Gag mutant constructs are fused to the LexA DNA-binding domain, and two-hybrid interactions are measured against GAD-ZmLC8. C, ClustalX alignment of maize and rice Sirevirus gag sequences. RiceA is the sequence used as bait. RiceCon is the Osr10 consensus sequence, whereas RiceB is another Osr10 sequence cloned during our experiments. MaizeA is the Hopie sequence used for these experiments, whereas MaizeB is from the sequence present in GenBank for the first-described maize Sirevirus (Hopie) element. The LC8 interaction domain is located between a predicted coiled-coil domain and a CX2CX3HX4C zinc knuckle.
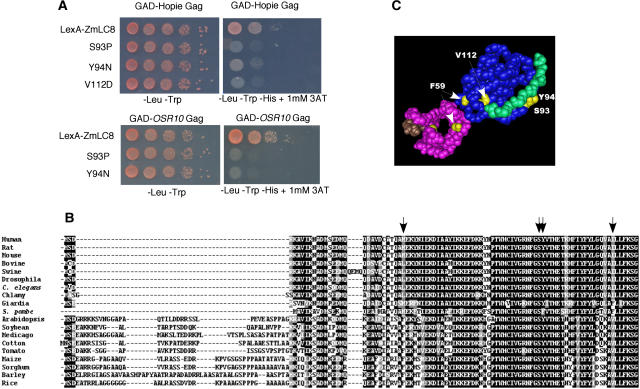
To determine which amino acids in ZmLC8 were responsible for recognizing the Hopie Gag extension, a reverse two-hybrid assay was again employed, this time using a PCR-mutagenized ZmLC8 library. Residues in ZmLC8 important for the interaction included F58, S93, Y94, and V112 (Fig. 5A). Also, disruption of the last three amino acids of the C terminus, either by removal or addition of extra amino acids, resulted in a loss of interaction with the Gag extensions (data not shown). All of the ZmLC8 mutants failed to interact with the cloned Osr10 Gag extension (Fig. 5A).
Figure 5.
ZmLC8 mutants that abrogate binding to the Hopie Gag extension. A, Ten-fold serial dilutions are shown of cells expressing three of the four ZmLC8 mutants: S93P, Y94N, and V112D. All of these mutants fail to grow on media lacking His when either the Hopie or Osr10 Sirevirus Gag extension is expressed as a GAD fusion. B, Alignment of some LC8 protein sequences from animals, fungi, and plants. Arrows represent amino acid positions, which, when mutated, abrogate binding of ZmLC8 to the Hopie and Osr10 Gag extensions. C, The amino acids important in binding the maize and rice Gag extensions were mapped onto the crystal structure of human LC8 bound to a peptide. LC8 was crystallized as a dimer, and the blue and pink domains represent a monomer of that dimer. The green and brown residues represent the bound peptide. Yellow amino acids were identified in this study as being important for binding the Gag extension.
The crystal structure of human LC8 has been determined (Protein Data Bank accession: 1CMI; Liang et al., 1999). The threading program Cn3D (Hogue, 1997) was used to map the location of the amino acids necessary for Gag extension interaction onto the structure of human LC8. This was possible because human LC8 and plant LC8 sequences are approximately 65% similar (Fig. 5B). S93 and Y94 are not only conserved with animal LC8 sequences (Fig. 5B), but they also map to the peptide-binding cleft of human LC8 (Liang et al., 1999; Fig. 5C). The crystal structure shows that V112 and the C terminus of the protein are located near the peptide-binding cleft, whereas F58 is located away from the binding region.
The Plant LC8 Family and Their Interactions with Gag
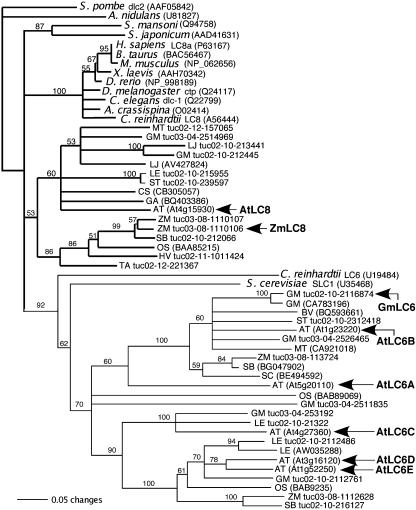
We have shown that multiple Sirevirus Gag extensions interact with two different LC8 family members (ZmLC8 and GmLC6). To further characterize the LC8 gene family, LC8 and LC6 sequences were retrieved using the BLAST program against the Plant Genome Database (http://www.plantgdb.org; Dong et al., 2004). This database contains EST consensus sequences from a variety of plant species. The plant LC8 and LC6 sequences as well as some sequences from other organisms were aligned, and a neighbor-joining tree was created (Fig. 6). The LC8 and LC6 sequences cluster in distinct clades. In addition, members of the LC8 family are more closely related, especially within animals, than are LC6 family members. Plants harbor clear LC8 homologs and multiple, divergent LC6 sequences. The LC6 sequences appear to bifurcate into two lineages.
Figure 6.
Neighbor-joining phylogenetic tree of LC8 (represented by bold-line branches) and LC6 sequences (represented by thin-line branches) with emphasis placed on those genes found in plants. Arrows identify the LC8/LC6 sequences discussed in this study. Numbers represent bootstrap values (100 replicates). GenBank or PlantGDB identifiers follow each entry. PlantGDB identifiers contain “tuc” and can be retrieved from www.plantgdb.org. For entries found in GenBank, the genus initial followed by the species name is given. For plant sequences, hosts can be identified by the following abbreviations: (AT, Arabidopsis; BV, Beta vulgaris; CS, Citrus sinesis; GA, Gossypium arboretum; GM, soybean; HV, Hordeum vulgare; LE, tomato; LJ, Lotus japonicus; MT, Medicago truncatula; OS, rice; SB, Sorghum bicolor; SC, Secale cereale; ST, Solanum tuberosum; TA, Triticum aestivum; ZM, maize).
In Arabidopsis, there are six members in the LC8/LC6 gene family (At1g52250, At1g23220, At3g16120, At4g15930, At4g27360, and At5g20110). To our knowledge, nothing is known about the function of any of these genes, although they are annotated as being part of the “microtubule-based process” (http://www.arabidopsis.org), inferred from their structural similarity to other LC8 and LC6 proteins known to associate with the dynein microtubule motor. At4g15930 is the clear Arabidopsis LC8 ortholog (Fig. 6) and it interacts with the Hopie Gag extension in a yeast two-hybrid assay (data not shown; Fig. 7). To differentiate the LC6 paralogs, letter designations have been given to various family members (AtLC6A At5g20110, AtLC6B At1g23220, AtLC6C At4g27360, AtLC6D At3g16120, and AtLC6E At1g52250).
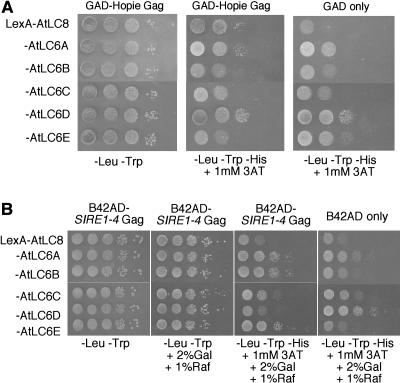
Figure 7.
Interactions between the various Arabidopsis LC8/LC6 proteins and either the Hopie Gag extension (A) or the SIRE1-4 Gag extension (B). Ten-fold serial dilutions were plated onto either control media or media lacking His to identify protein-protein interactions. In these experiments, the baits were the different Arabidopsis dynein light chains expressed as LexA fusions. In B, the LexA plasmid is under the control of the GAL promoter. The preys are indicated above each section. The last sections in both A and B serve as negative controls for autoactivation of the HIS3 marker gene by the bait. Cell number controls for these sections were consistent with the other sections (data not shown).
Because the Hopie Gag extension interacted with ZmLC8, and the soybean SIRE1-4 Gag extension interacted with GmLC6 but not ZmLC8, we wanted to determine which of the Arabidopsis LC6 genes and/or LC8 would interact with the various Gag extensions in directed yeast two-hybrid assays. The cDNAs for all the corresponding genes (AtLC8, AtLC6A–E) were fused to the LexA DNA-binding domain and tested for a two-hybrid interaction with the Hopie Gag extension (Fig. 7A) or the SIRE1-4 Gag extension (Fig. 7B). The Hopie Gag extension, which had originally been identified as interacting with ZmLC8, interacted with AtLC8 but not any of the Arabidopsis LC6 proteins (Fig. 7A). The SIRE1-4 Gag extension was found to initially interact with GmLC6, and we did not observe an interaction with AtLC8. However, the SIRE1-4 Gag extension interacted with three of the five Arabidopsis LC6 homologs (AtLC6A, AtLC6B, and AtLC6E; Fig. 7B). AtLC6C did not interact with the SIRE1-4 Gag in the yeast two-hybrid assay, and AtLC6D showed autoactivation in the absence of SIRE1 Gag (Fig. 7B), and so no conclusion could be drawn as to whether the SIRE1-4 Gag extension interacts with AtLC6D. Figure 6 maps the location of these Arabidopsis, maize, and soybean LC8 genes onto the LC8/LC6 phylogenetic tree.
If LC8 and LC6 interact in the same complex in plants, the Sirevirus Gags may use these mediators to associate with this complex. It is known that LC8 and LC6 work together as part of the dynein microtubule motor, and preliminary data suggest that LC8 and LC6 interact in plants. A yeast two-hybrid assay was conducted using ZmLC8 as bait. Of 18 clones recovered, 14 were for a maize LC6 homolog (GenBank accession no. CN844329) most similar to AtLC6D, and two were for another LC6 (GenBank accession no. CF627955) most similar to AtLC6B. Initial subcellular localization studies indicate that both ZmLC8 and a ZmLC6 (CN844329) are found in the cytoplasm and nucleus (Supplemental Fig. 1A). Retrotransposon VLPs are normally cytoplasmic, and therefore retrotransposon proteins and LC8 or LC6 can come into contact. RT-PCR experiments were also completed with Hopie and with ZmLC8 to assess expression patterns in maize plants. Overlapping RNA expression was observed in maize roots and juvenile shoots (Supplemental Fig. 1B). These results indicate that there can be spatial and temporal overlap of expression between Hopie and ZmLC8.
DISCUSSION
Diversity in the Genomic Organization of the Sireviruses
The sequencing of various eukaryotic genomes, particularly those of plants, has revealed the magnitude to which transposable elements have colonized their hosts. Numerous studies have documented the number, diversity, and distribution of transposable elements within genomes (for review, see Kumar and Bennetzen, 1999; Feschotte et al., 2002; Deininger et al., 2003; Havecker et al., 2004). However, outside of yeast, relatively little information is available about specific relationships between transposable elements and host cellular processes.
The Sireviruses are not canonical retrotransposons. Phylogenetic analyses indicate Sireviruses are Ty1/copia LTR retrotransposons, yet their genomic organization appears more similar to the close cousins of the retrotransposons, the retroviruses. This is principally because some Sireviruses encode an extra ORF after the pol gene, referred to as an env-like gene. Here we show that many plants harbor the Sireviruses and that the Sirevirus genomic structure itself can be very variable. Many, but not all Sireviruses have env-like ORFs, and they share little amino acid similarity. Nonetheless, the Env-like protein probably plays a similar role in each of its hosts, as several of its secondary structural characteristics are broadly shared, specifically the presence of coiled-coil domains and transmembrane domains. Although we did complete a two-hybrid screen with both the Hopie and the SIRE1-4 Env-like proteins, no interacting partners could be found (data not shown). Thus, clues as to the function of the Env-like protein still escape us.
In contrast, a two-hybrid screen with both the Hopie and SIRE1-4 Gag extension revealed an interaction with two proteins belonging to the LC8 protein family, ZmLC8 and GmLC6, respectively. Peterson-Burch and Voytas (2002) first described the Sirevirus Gag extensions in their analysis of the Pseudoviridae. However, no known function for the extra coding capacity of the gag gene was proposed. As Figure 1 demonstrates, the extended gag gene is broadly found among Sireviruses, even among those that lack the env-like ORF. All Gag extensions have a coiled-coil domain, whereas the presence of a second zinc knuckle and the length of the Gag extension are not uniform. It is possible the Gag extension has evolved in the Sireviruses because an interaction with LC8 family members facilitates replication of these mobile elements (see discussion below).
Sequences Critical for Gag/LC8 Interaction
A number of proteins are now known to bind nonplant LC8 proteins, and in general two amino acid motifs allow LC8 binding: (K/R)XTQT and GIQVDR (for review, see Wu and King, 2003). We identified four amino acids in Hopie Gag responsible for the interaction with ZmLC8 and suggest G(I/V)XCXL as a possible LC8 interaction motif in plants. The LC8-binding motif of Hopie Gag is also present in the Osr10 Gag, but these specific amino acids were not identified in any of the other Sirevirus Gag extensions. In these other Gag proteins, binding to an LC8 family member could occur through other related motifs. For example, GmLC6 interacts with the SIRE1-4 Gag extension through a yet uncharacterized sequence motif.
Four ZmLC8 amino acids were identified, which when mutated, abrogated the interaction with the Hopie Gag extension. Interestingly, two of the residues, S93 and Y94, are highly conserved among LC8 proteins; in the crystallized human LC8 protein, these residues abut the target protein in the LC8-binding cleft. As human LC8 and ZmLC8 are about 65% similar, it is likely that the same or similar binding clefts as in the human protein exist in the plant LC8 molecules. Further analysis would be needed to confirm such a hypothesis. Based on the crystal structure of the human protein, the other ZmLC8 amino acids important for Gag binding do not contact the peptide directly, but could influence binding in another way. For example, they may be important for the overall structural integrity of the LC8 protein.
The LC8 Protein Family and Its Possible Role in the Sirevirus Life Cycle
The dynein light chains LC8 and LC6 were originally identified in the outer arm of C. reinhardtii flagella and are associated with the dynein complex (King and Patel-King, 1995). These light chains were subsequently identified in EST libraries from organisms/tissues that do not contain motile cilia or flagella (King and Patel-King, 1995). Soon thereafter LC8 was shown to be part of cytoplasmic dynein (as opposed to flagellar dynein; Dick et al., 1996; King et al., 1996) and a stoichiometric component of the myosin-Va complex, an actin motor, which carries LC8 in its tail domain (Espindola et al., 2000).
As a component of cytoskeletal machinery, many viruses bind LC8 and thus hijack the cytoskeleton to move throughout the cell. Viral proteins implicated in binding LC8 for intracellular movement include the African swine fever virus protein p54 (Alonso et al., 2001), two different Lyssavirus (rabies) P proteins (Jacob et al., 2000; Raux et al., 2000), and the Gag protein of human foamy retrovirus (Petit et al., 2003). In each of these cases, the virus/LC8 interaction is proposed to allow the viral particle movement along either the actin skeleton (via myosin) or toward the nucleus via LC8 in the dynein microtuble motor. For example, in the case of the Gag protein of the human foamy retrovirus, the association with LC8 is thought to enable retroviral particles to traffic toward the microtubule organizing center and ultimately enter the nucleus to continue their life cycle (Petit et al., 2003).
In addition to its role in the cytoskeleton, LC8 binds a number of cellular proteins, including neuronal nitric oxide synthetase, IκBα, Bim, a Bcl-2 family member, the Drosophila swallow protein, a regulator of the bicoid RNA, and p53 Binding protein1 (for review, see King, 2000, 2003; Vallee et al., 2003). In these cases, other functions of LC8 have been proposed, including the sequestering of proteins within a cell (such as the proapoptitic factor Bim [Puthalakath et al., 1999]) or inhibiting protein function (Kaiser et al., 2003). LC8 binding to targets may also facilitate protein folding. For two proteins—IC74, an intermediate chain of the dynein complex, and swallow—conformational changes were demonstrated after binding to LC8 (Nyarko et al., 2004; Wang et al., 2004).
In contrast to animals, flowering plants lack all components of the dynein microtubule motor except LC8 and the related LC6 sequences (Lawrence et al., 2001). The lack of a plant dynein motor and the fact that animal LC8 proteins can bind numerous targets suggests that plant LC8 proteins likely do not carry out all of the same functions as their animal counterparts (King, 2003). Nevertheless, the sequence conservation of LC8 across kingdoms is high; plant and animal LC8 sequences share approximately 65% similarity (89% similarity is observed between the human, Drosophila, Caenorhabditis, elegans, and C. reinhardtii orthologs; King and Patel-King, 1995; King et al., 1996). The N terminus of the plant LC8 proteins is the most variable in terms of length and sequence composition. Programs that predict protein subcellular localization signals fail to identify any such motifs in the LC8 amino terminus (data not shown). To our knowledge, for LC6, no role outside of the dynein microtubule motor has been described (King and Patel-King, 1995). BLAST searches of the plant genome database (www.plantgdb.org) identified a number of LC6-related sequences that are members of large gene families exhibiting considerable sequence diversity. This suggests that, like LC8, LC6 proteins may have assumed some unique functions in plants.
We tested for specificity in the binding of Sirevirus Gags to both LC8 and LC6. Hopie Gag interacted with both ZmLC8 and AtLC8, but no binding was observed to any of the AtLC6 proteins. In contrast, the soybean SIRE1-4 Gag, which interacted with a soybean LC6 protein, bound to multiple AtLC6 proteins but not AtLC8. Humans have three highly conserved isoforms of LC8, and it is unclear if they are able to discriminate targets (Wilson et al., 2001). Our data suggests that plant LC8 and LC6 proteins distinguish targets, but among LC6 proteins in Arabidopsis, some overlap in binding occurs.
The reason why the Sirevirus Gags bind to members of the LC8 gene family remains unclear. It is possible that both LC8 and LC6 associate with other microtubule motor complexes, such as kinesin. The kinesin gene family has greatly expanded in plants and some kinesins translocate toward the minus end of microtubules similar to dynein (Reddy and Day, 2001; Lee and Liu, 2004). Because animal LC8 proteins associate with Myosin-V (Espindola et al., 2000), it may be that the Gag proteins hijack LC8 (or LC6) to allow the VLP to move within the cell using the actin cytoskeleton. It is not known how retrotransposon particles navigate within a cell; nonetheless the cytoplasmic VLP needs to reach a nucleus to complete transposition. An alternative hypothesis is that interactions between Gag and LC8 or LC6 lead to conformational changes that facilitate VLP assembly. Ultimately, understanding the function of the Gag extension in the Sireviruses will require an active element and a further understanding of the role of the dynein light chains in plants.
MATERIALS AND METHODS
Sirevirus Sequence Analysis
The Opie-2 (U68408) RT amino acid sequence was used to query the tBLASTn nonredundant database of GenBank (November, 2002) as previously described (Gao et al., 2003). Individual full-length Sireviruses were retrieved from various bacterial artificial chromosomes manually or with the software package RetroMap (Peterson-Burch et al., 2004). Representative elements include the maize (Zea mays) Sirevirus Hopie (AC116033), sorghum Sirevirus (AF503433), medicago Sirevirus (AC122723 and AC130810), citrus Sirevirus (AF506028), and SIRE1-4 (Laten et al., 2003). Consensus sequences were generated for other full-length elements. Because of high sequence divergence in noncoding regions, we were unable to build a complete consensus sequence for some elements. Also, for some nucleotide positions in coding regions, it was impossible to obtain a consensus nucleotide. In these cases, a consensus sequence was made by one of three methods: (1) a nucleotide consensus was made incorporating any available EST sequences, (2) a protein consensus was made if the ambiguous nucleotide fell into the wobble base, or (3) the consensus genomic structure was determined in cases where a nucleotide or protein consensus failed. In all cases, a genomic structure (the presence or absence of a continuous ORF) could be discerned. Consensus elements for Osr9, Osr10, and Lotus2 are available at http://www.public.iastate.edu/∼voytas/MSsupplementary/Gag_LC8.
Average Sirevirus Gag lengths were estimated for either representative or consensus sequences for each of the genomic structures outlined in Figure 1. The Gag lengths from classical Ty1/copia retrotransposons were previously reported and averaged from 27 diverse Ty1/copia elements (Peterson-Burch and Voytas, 2002). Gag protein length was estimated in either of two ways: if a conserved break in frame was observed between the putative Gag and Pol proteins, the Gag protein was assumed to be the first reading frame; if Gag and Pol were predicted to be encoded in the same reading frame, then Gag was arbitrarily estimated to end approximately 20 amino acids upstream of the predicted protease-active site (Peterson-Burch and Voytas, 2002).
The Env-like proteins were analyzed with HMMTop to predict transmembrane domains (Tusnady and Simon, 1998, 2001). Coiled-coil predictions were generated by Paircoil (Berger et al., 1995), and a coiled-coil region was considered significant if it had a probability of 0.5 or greater.
Plasmid Construction
The Hopie Gag extension was PCR amplified from B73 maize genomic DNA with primers DVO2657 5′-GAATTCATCACCGATTTAAATGATATAAAAG-3′ and DVO2658 5′-GGATCCCTCAATCTCTTTTAGGTACCCAAAC-3′. The rice (Oryza sativa) Gag C terminus was amplified from O. sativa cv japonicus DNA using primers DVO2854 5′-GAATTCGATGATGATTGTGATGATGTTTCC-3′ and DVO2855 5′-GGATCCCTAAGAATCAAACATGCATGTCTTAGGAGG-3′. These PCR products were digested with EcoRI and BamHI and cloned into pBTM116 (Bartel and Fields, 1995). Three rice and maize Gags were sequenced, and one was chosen for the two-hybrid assays based on the presence of complete ORFs and sequences that most closely conformed to a consensus sequence. The SIRE1-4 C terminus of gag was cloned from a previously characterized SIRE1-4 element (AY205608; Laten et al., 2003). The gag gene was digested with EcoRI and XhoI and treated with Klenow (Promega). The insert was cloned into the SmaI site of pYZ275, a LexA DNA-binding plasmid identical to pBTM116 except that is has a LEU2-selectable marker instead of TRP1.
ZmLC8, GmLC6, ZmLC6 (CN844329), and ZmLC6 (CF627955) were isolated from the yeast (Saccharomyces cerevisiae) two-hybrid libraries (see below). Five of the six LC8/LC6 family cDNAs from Arabidopsis (Arabidopsis thaliana) were obtained from the Arabidopsis Biological Resource Center (http://www.arabidopsis.org). These included At1g23220 (AY096447), At1g52250 (BT010683), At4g27360 (AY096707), At4g15930 (BT004785), and At5g20110 (BT011208). A cDNA clone of At3g16120 was obtained using RT-PCR. First-strand cDNA synthesis was completed using a polyT primer to amplify Arabidopsis ecotype Columbia 7-d seedling RNA. Gene-specific PCR was then completed with the gene-specific primers DVO3425 5′-GAATTCATGTTGGAAGGGAAAGCGAAGGT-3′ and DVO3426 5′-CTCGAGTAAAGAGTGGCGCCTTTGAAG-3′. The PCR product was cloned into the pGEM-T Easy vector (Promega). All cloned cDNAs were PCR amplified with high-fidelity polymerase from the putative start codon to the stop codon with primers containing restriction enzyme sites EcoRI and XhoI. The products were digested with EcoRI and XhoI, and cloned into the LexA plasmids pBTM116 or pYZ275 between EcoRI and SalI. For the subcellular localization studies, the cDNAs were cloned using EcoRI and XhoI into a vector containing the 35S promoter, enhanced green fluorescent protein, a seven-amino acid polyalanine linker followed by a multiple cloning site containing EcoRI and XhoI and the NOS terminator (pEH375; E. Havecker, unpublished data).
PCR mutagenesis of the maize gag was completed with primers DVO2657 and DVO2658 using Taq polymerase under standard thermocycling conditions. PCR mutagenesis of ZmLC8 was completed using primers DVO1157 5′-CTATTCGATGATGAAGATACCCCACC-3′ and DVO1158 5′-GCGGGGTTTTTCAGTATCTACGATTC-3′ that anneal to the GAD and the ADH1 terminator, respectively. The mutagenesis rate for each library approximated one mutation every 500 bp. Single-point mutations were introduced into the various clones using overlapping primers. All clones were sequenced to ensure that only the desired mutations were present.
Yeast Two-Hybrid Screens
All two-hybrid assays were performed in the yeast strain L40 (Hollenberg et al., 1995), which has a HIS3 gene with upstream LexA operators. The library used to screen interacting clones for the Hopie Gag was a juvenile shoot EST library (Moose and Sisco, 1996) fused to the GAD. Yeast containing the Hopie Gag were transformed at 10-fold coverage and plated on media selecting for a two-hybrid interaction: synthetic complete (SC)-Trp-Leu-His-Ura plus 1 mm 3-amino-1,2,4-triazole and 2% dextrose.
The soybean (Glycine max) two-hybrid screen was performed with the SIRE1-4 Gag extension fused to LexA. The library used for screening was made from poly (A+) RNAs from etiolated hypocotyls (H. Gao and M. Bhattacharya, unpublished data). The ESTs from this library were cloned into the pB42AD plasmid (Clonetech). The activating domain B42p is under the control of a Gal promoter. A 10-fold coverage of this library was transformed into yeast, and colonies were first allowed to grow on nonselective media. Colonies were scraped from the plate, titered, and then plated onto selective media (SC)-Trp-Leu-His-Ura plus 1mm 3-amino-1,2,4-triazole media with 2% Gal, 1% raffinose at a further 10-fold coverage.
For individual two-hybrid assays, yeast were grown in 5 mL of selective media and shaken at 30°C for approximately 24 h. Ten-fold serial dilutions of the cultures (OD600 1.0) were made, and 10 μL of each dilution was plated onto control plates (SC)-Trp-Leu-Ura or two-hybrid interaction selective media. Expression of various constructs was tested by western-blot analysis.
In Vitro Binding Assay
To determine whether the maize Sirevirus Gag binds in vitro to ZmLC8, a plasmid expressing Hopie Gag under the T7 promoter was constructed (pEH293). To do this, the original Hopie Gag was digested from the LexA DNA-binding domain vector with EcoRI and SalI and inserted into pCite-2a(+) (Novagen). A coupled transcription-translation reaction in rabbit reticulocyte lysate was completed with 20 μCi of [S35]Met according to manufacturer's recommendations (Promega).
ZmLC8 was fused to GST in the pgex4t-1 plasmid (Amersham Biosciences) to give rise to pEH277. Methods for expressing and purifying the GST-ZmLC8 fusion were done in accordance with Smith and Corcoran (1987) with induction of GST-ZmLC8 in with 0.1 mm isopropylthio-β-galactoside at 30°C for 4 to 6 h. Pull-down assays with GST-ZmLC8 were similar to those described (Xie et al., 2001). Radiolabeled Gag pulled down by ZmLC8 was visualized by autoradiography.
Plant LC8/LC6 Sequences
All LC8/ LC6 sequences were obtained from GenBank or the plant genome database (www.plantgdb.org; Dong et al., 2004). The identifiers for each are provided in Figure 6. All amino acid sequences were aligned using ClustalX (Higgins and Sharp, 1988), and the neighbor-joining tree was constructed using Phylogenetic Analysis Using Parsimony, version 4.0 (Sinaur Associates; Saitou and Nei, 1987; Swofford, 2001).
ZmLC8 was fused to the C terminus of YFP (pEH441; E. Havecker and D. Voytas, unpublished data) and ZmLC6 (most similar to GenBank accession no. CN844329) was fused to the C terminus of CFP (pEH442; E. Havecker and D. Voytas, unpublished data) to determine their subcellular localization in plant cells. These constructs, driven by the cauliflower mosaic virus 35S promoter and terminated with NOS terminator, were transiently expressed in tobacco (Nicotiana tabacum) protoplasts using a modified protocol based on Sheen (2001) and Locatelli et al. (2003). Briefly, tobacco protoplasts were collected according to Havecker and Voytas (2003) and resuspended in 0.4 m mannitol, 15 mm MgCl2, 5 mm MES, pH 5.7, to a concentration of 3 × 106 protoplasts/mL. 1 × 106 protoplasts in 0.4 m mannitol, 15 mm MgCl2, 5 mm MES, pH 5.7 (approximately 300 μL) were then aliquoted to individual 15-mL tubes. Subsequently, 30 μg plasmid DNA (Qiagen Midi Prep) and 300 μL 40% polyethylene glycol solution [40% polyethylene glycol (Fluka), 100 mm Ca(NO3)2, 0.4 m Mannitol] were added. Protoplasts were incubated at room temperature for 30 min with occasional gentle shaking, diluted with 1:15 (v/v) W5 (154 mm NaCl, 5 mm KCl, 125 mm CaCl2, 5 mm Glc, pH 5.8), centrifuged at 300 rpm for 5 min, and collected by centrifugation (600 rpm, 5 min). Transformed protoplasts were resuspended in 1.5 mL K3/G1 (Havecker and Voytas, 2003) and incubated in the dark at 25°C.
Confocal images were obtained using a Leica TCS-NT confocal microscope (Leica Microsystems) at the Iowa State University confocal microscopy facility using a 63× lens with a zoom factor of 2. This YFP protein has an absorbance/emission of 516 nm/529 nm. The CFP protein has an absorbance/emission of 452 nm/476 nm.
The RNA from bis-(2-mercaptoethylsulfone) suspension cells and Mo17 10-d-old seedlings dissected into endosperm, root, and shoot tissues (meristem included) was isolated using PUREscript RNA isolation kit (Gentra Systems). RT-PCR was completed as previously described (Wright and Voytas, 2002). The gene-specific primers for second-round PCR were DVO2665 5′-CACATTGAGCTCATTTGTGATC-3′ for Hopie (located in the LTR) and DVO2809 5′-AGTGCCGACATGAAGGAGGAGATG-3′ for ZmLC8. A primer that anneals to an adapter at the 3′ end of the polyT primer was used as the reverse strand primer for the second round of PCR. All RT-PCR products were cloned (pGEM-T Easy; Promega) and sequenced to ensure that they were Hopie and ZmLC8. No RT-PCR products were detected in the absence of the RT enzyme.
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requester.
Supplementary Material
Acknowledgments
We would like to thank Steve Moose for giving us the juvenile shoot library, and Hongyu Gao and Madan Bhattacharya for allowing us to use their soybean two-hybrid library. We would also like to acknowledge Sarah Tucker and Kent Doolittle for their help with the yeast two-hybrid screens, and Kevin Geiken for help with the figures. Finally, Qunfeng Dong helped with the plant genome database and wrote a nucleotide consensus-building computer program.
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.065680.
References
- Alonso C, Miskin J, Hernaez B, Fernandez-Zapatero P, Soto L, Canto C, Rodriguez-Crespo I, Dixon L, Escribano JM (2001) African swine fever virus protein p54 interacts with the microtubular motor complex through direct binding to light-chain dynein. J Virol 75: 9819–9827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aye M, Irwin B, Beliakova-Bethell N, Chen E, Garrus J, Sandmeyer S (2004) Host factors that affect Ty3 retrotransposition in Saccharomyces cerevisiae. Genetics 168: 1159–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel PL, Fields S (1995) Analyzing protein-protein interactions using two-hybrid system. Methods Enzymol 254: 241–263 [DOI] [PubMed] [Google Scholar]
- Berger B, Wilson DB, Wolf E, Tonchev T, Milla M, Kim PS (1995) Predicting coiled coils by use of pairwise residue correlations. Proc Natl Acad Sci USA 92: 8259–8263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke JD, Eickbush TH, Sandmeyer SB, Voytas DF (2004) Pseudoviridae. In CM Fauquet, ed, Virus Taxonomy: Eighth Report of the International Committee on Taxonomy of Viruses. Academic Press, New York, pp 663–672
- Deininger PL, Moran JV, Batzer MA, Kazazian HH Jr (2003) Mobile elements and mammalian genome evolution. Curr Opin Genet Dev 13: 651–658 [DOI] [PubMed] [Google Scholar]
- Dick T, Ray K, Salz HK, Chia W (1996) Cytoplasmic dynein (ddlc1) mutations cause morphogenetic defects and apoptotic cell death in Drosophila melanogaster. Mol Cell Biol 16: 1966–1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Q, Schlueter SD, Brendel V (2004) PlantGDB, plant genome database and analysis tools. Nucleic Acids Res 32: D354–D359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickbush TH, Malik HS (2002) Origins and evolution of retrotransposons. In NL Craig, R Craigie, M Gellert, AL Lambowitz, eds, Mobile DNA II. ASM Press, Washington, DC, pp 1111–1144
- Espindola FS, Suter DM, Partata LB, Cao T, Wolenski JS, Cheney RE, King SM, Mooseker MS (2000) The light chain composition of chicken brain myosin-Va: calmodulin, myosin-II essential light chains, and 8-kDa dynein light chain/PIN. Cell Motil Cytoskeleton 47: 269–281 [DOI] [PubMed] [Google Scholar]
- Feschotte C, Jiang N, Wessler SR (2002) Plant transposable elements: where genetics meets genomics. Nat Rev Genet 3: 329–341 [DOI] [PubMed] [Google Scholar]
- Gao X, Havecker ER, Baranov PV, Atkins JF, Voytas DF (2003) Translational recoding signals between gag and pol in diverse LTR retrotransposons. RNA 9: 1422–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith JL, Coleman LE, Raymond AS, Goodson SG, Pittard WS, Tsui C, Devine SE (2003) Functional genomics reveals relationships between the retrovirus-like Ty1 element and its host Saccharomyces cerevisiae. Genetics 164: 867–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havecker ER, Gao X, Voytas DF (2004) The diversity of LTR retrotransposons. Genome Biol 5: 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havecker ER, Voytas DF (2003) The soybean retroelement SIRE1 uses stop codon suppression to express its envelope-like protein. EMBO Rep 4: 274–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins DG, Sharp PM (1988) CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene 73: 237–244 [DOI] [PubMed] [Google Scholar]
- Hogue CW (1997) Cn3D: a new generation of three-dimensional molecular structure viewer. Trends Biochem Sci 22: 314–316 [DOI] [PubMed] [Google Scholar]
- Hollenberg SM, Sternglanz R, Cheng PF, Weintraub H (1995) Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol Cell Biol 15: 3813–3822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin B, Aye M, Baldi P, Beliakova-Bethell N, Cheng H, Dou Y, Liou W, Sandmeyer S (2005) Retroviruses and yeast retrotransposons use overlapping sets of host genes. Genome Res 15: 641–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob Y, Badrane H, Ceccaldi PE, Tordo N (2000) Cytoplasmic dynein LC8 interacts with lyssavirus phosphoprotein. J Virol 74: 10217–10222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffrey SR, Snyder SH (1996) PIN: an associated protein inhibitor of neuronal nitric oxide synthase. Science 274: 774–777 [DOI] [PubMed] [Google Scholar]
- Kaiser FJ, Tavassoli K, Van den Bemd GJ, Chang GT, Horsthemke B, Moroy T, Ludecke HJ (2003) Nuclear interaction of the dynein light chain LC8a with the TRPS1 transcription factor suppresses the transcriptional repression activity of TRPS1. Hum Mol Genet 12: 1349–1358 [DOI] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J (1999) Molecular paleontology of transposable elements from Arabidopsis thaliana. Genetica 107: 27–37 [PubMed] [Google Scholar]
- Kim A, Terzian C, Santamaria P, Pelisson A, Purd'homme N, Bucheton A (1994) Retroviruses in invertebrates: the gypsy retrotransposon is apparently an infectious retrovirus of Drosophila melanogaster. Proc Natl Acad Sci USA 91: 1285–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King SM (2000) The dynein microtubule motor. Biochim Biophys Acta 1496: 60–75 [DOI] [PubMed] [Google Scholar]
- King SM (2003) Dynein motors: structure, mechanochemistry and regulation. In M Schliwa, ed, Molecular Motors. Wiley-VCH Verlag GmbH & Co., Viernheim, Germany, pp 45–78
- King SM, Barbarese E, Dillman JF III, Patel-King RS, Carson JH, Pfister KK (1996) Brain cytoplasmic and flagellar outer arm dyneins share a highly conserved Mr 8,000 light chain. J Biol Chem 271: 19358–19366 [DOI] [PubMed] [Google Scholar]
- King SM, Patel-King RS (1995) The M(r) = 8,000 and 11,000 outer arm dynein light chains from Chlamydomonas flagella have cytoplasmic homologues. J Biol Chem 270: 11445–11452 [DOI] [PubMed] [Google Scholar]
- Kumar A, Bennetzen JL (1999) Plant retrotransposons. Annu Rev Genet 33: 479–532 [DOI] [PubMed] [Google Scholar]
- Laten HM, Havecker ER, Farmer LM, Voytas DF (2003) SIRE1, an endogenous retrovirus family from Glycine max, is highly homogeneous and evolutionarily young. Mol Biol Evol 20: 1222–1230 [DOI] [PubMed] [Google Scholar]
- Laten HM, Majumdar A, Gaucher EA (1998) SIRE-1, a copia/Ty1-like retroelement from soybean, encodes a retroviral envelope-like protein. Proc Natl Acad Sci USA 95: 6897–6902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence CJ, Morris NR, Meagher RB, Dawe RK (2001) Dyneins have run their course in plant lineage. Traffic 2: 362–363 [DOI] [PubMed] [Google Scholar]
- Leblanc P, Desset S, Giorgi F, Taddei AR, Fausto AM, Mazzini M, Dastugue B, Vaury C (2000) Life cycle of an endogenous retrovirus, ZAM, in Drosophila melanogaster. J Virol 74: 10658–10669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YR, Liu B (2004) Cytoskeletal motors in Arabidopsis: sixty-one kinesins and seventeen myosins. Plant Physiol 136: 3877–3883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Jaffrey SR, Guo W, Snyder SH, Clardy J (1999) Structure of the PIN/LC8 dimer with a bound peptide. Nat Struct Biol 6: 735–740 [DOI] [PubMed] [Google Scholar]
- Locatelli F, Vannini C, Magnani E, Coraggio I, Bracale M (2003) Efficiency of transient transformation in tobacco protoplasts is independent of plasmid amount. Plant Cell Rep 21: 865–871 [DOI] [PubMed] [Google Scholar]
- Malik HS, Henikoff S, Eickbush TH (2000) Poised for contagion: evolutionary origins of the infectious abilities of invertebrate retroviruses. Genome Res 10: 1307–1318 [DOI] [PubMed] [Google Scholar]
- Martinez-Izquierdo JA, Garcia-Martinez J, Vicient CM (1997) What makes Grande1 retrotransposon different? Genetica 100: 15–28 [PubMed] [Google Scholar]
- McCarthy EM, Liu J, Lizhi G, McDonald JF (2002) Long terminal repeat retrotransposons of Oryza sativa. Genome Biol 3: RESEARCH0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moose SP, Sisco PH (1996) Glossy15, an APETALA2-like gene from maize that regulates leaf epidermal cell identity. Genes Dev 10: 3018–3027 [DOI] [PubMed] [Google Scholar]
- Nagaki K, Song J, Stupar RM, Parokonny AS, Yuan Q, Ouyang S, Liu J, Hsiao J, Jones KM, Dawe RK, et al (2003) Molecular and cytological analyses of large tracks of centromeric DNA reveal the structure and evolutionary dynamics of maize centromeres. Genetics 163: 759–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyarko A, Hare M, Hays TS, Barbar E (2004) The intermediate chain of cytoplasmic dynein is partially disordered and gains structure upon binding to light-chain LC8. Biochemistry 43: 15595–15603 [DOI] [PubMed] [Google Scholar]
- Ohtsubo H, Kumekawa N, Ohtsubo E (1999) RIRE2, a novel gypsy-type retrotransposon from rice. Genes Genet Syst 74: 83–91 [DOI] [PubMed] [Google Scholar]
- Pelisson A, Mejlumian L, Robert V, Terzian C, Bucheton A (2002) Drosophila germline invasion by the endogenous retrovirus gypsy: involvement of the viral env gene. Insect Biochem Mol Biol 32: 1249–1256 [DOI] [PubMed] [Google Scholar]
- Peterson-Burch BD, Nettleton D, Voytas DF (2004) Genomic neighborhoods for Arabidopsis retrotransposons: a role for targeted integration in the distribution of the Metaviridae. Genome Biol 5: R78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson-Burch BD, Voytas DF (2002) Genes of the Pseudoviridae (Ty1/copia retrotransposons). Mol Biol Evol 19: 1832–1845 [DOI] [PubMed] [Google Scholar]
- Peterson-Burch BD, Wright DA, Laten HM, Voytas DF (2000) Retroviruses in plants? Trends Genet 16: 151–152 [DOI] [PubMed] [Google Scholar]
- Petit C, Giron ML, Tobaly-Tapiero J, Bittoun P, Real E, Jacob Y, Tordo N, De The H, Saib A (2003) Targeting of incoming retroviral Gag to the centrosome involves a direct interaction with the dynein light chain 8. J Cell Sci 116: 3433–3442 [DOI] [PubMed] [Google Scholar]
- Puthalakath H, Huang DC, O'Reilly LA, King SM, Strasser A (1999) The proapoptotic activity of the Bcl-2 family member Bim is regulated by interaction with the dynein motor complex. Mol Cell 3: 287–296 [DOI] [PubMed] [Google Scholar]
- Raux H, Flamand A, Blondel D (2000) Interaction of the rabies virus P protein with the LC8 dynein light chain. J Virol 74: 10212–10216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy AS, Day IS (2001) Kinesins in the Arabidopsis genome: a comparative analysis among eukaryotes. BMC Genomics 2: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425 [DOI] [PubMed] [Google Scholar]
- SanMiguel P, Tikhonov A, Jin YK, Motchoulskaia N, Zakharov D, Melake-Berhan A, Springer PS, Edwards KJ, Lee M, Avramova Z, et al (1996) Nested retrotransposons in the intergenic regions of the maize genome. Science 274: 765–768 [DOI] [PubMed] [Google Scholar]
- Scholes DT, Banerjee M, Bowen B, Curcio MJ (2001) Multiple regulators of Ty1 transposition in Saccharomyces cerevisiae have conserved roles in genome maintenance. Genetics 159: 1449–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J (2001) Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol 127: 1466–1475 [PMC free article] [PubMed] [Google Scholar]
- Smith DB, Corcoran LM (1987) Expression and purification of glutathione-S-transferase fusion proteins. In FM Ausubel, R Brent, RE Kingston, DD Moore, JG Seidman, JA Smith, K Struhl, eds, Current Protocols in Molecular Biology, Vol 3. Greene/Wiley Interscience, New York, pp 16.17.11–16.17.17 [DOI] [PubMed]
- Song SU, Gerasimova T, Kurkulos M, Boeke JD, Corces VG (1994) An env-like protein encoded by a Drosophila retroelement: evidence that gypsy is an infectious retrovirus. Genes Dev 8: 2046–2057 [DOI] [PubMed] [Google Scholar]
- Swofford DL (2001) PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. Sinauer Associates, Sunderland, MA
- Tusnady GE, Simon I (1998) Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J Mol Biol 283: 489–506 [DOI] [PubMed] [Google Scholar]
- Tusnady GE, Simon I (2001) The HMMTOP transmembrane topology prediction server. Bioinformatics 17: 849–850 [DOI] [PubMed] [Google Scholar]
- Vallee RB, Williams JC, Varma D, Barnhart LE (2003) Dynein: an ancient motor protein involved in multiple modes of transport. J Neurobiol 58: 189–200 [DOI] [PubMed] [Google Scholar]
- Vidal M, Brachmann RK, Fattaey A, Harlow E, Boeke JD (1996) Reverse two-hybrid and one-hybrid systems to detect dissociation of protein-protein and DNA-protein interactions. Proc Natl Acad Sci USA 93: 10315–10320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voytas DF, Boeke JD (2002) Ty1 and Ty5 of Saccharomyces cerevisiae. In NL Craig, R Craigie, M Gellert, AL Lambowitz, eds, Mobile DNA II. ASM Press, Washington, DC, pp 631–662
- Wang L, Hare M, Hays TS, Barbar E (2004) Dynein light chain LC8 promotes assembly of the coiled-coil domain of swallow protein. Biochemistry 43: 4611–4620 [DOI] [PubMed] [Google Scholar]
- Wilson MJ, Salata MW, Susalka SJ, Pfister KK (2001) Light chains of mammalian cytoplasmic dynein: identification and characterization of a family of LC8 light chains. Cell Motil Cytoskeleton 49: 229–240 [DOI] [PubMed] [Google Scholar]
- Wright DA, Voytas DF (2002) Athila4 of Arabidopsis and Calypso of soybean define a lineage of endogenous plant retroviruses. Genome Res 12: 122–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, King SM (2003) Backbone dynamics of dynein light chains. Cell Motil Cytoskeleton 54: 267–273 [DOI] [PubMed] [Google Scholar]
- Xie W, Gai X, Zhu Y, Zappulla DC, Sternglanz R, Voytas DF (2001) Targeting of the yeast Ty5 retrotransposon to silent chromatin is mediated by interactions between integrase and Sir4p. Mol Cell Biol 21: 6606–6614 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.