Abstract
Rad51 is a homolog of the bacterial RecA recombinase, and a key factor in homologous recombination in eukaryotes. Rad51 paralogs have been identified from yeast to vertebrates. Rad51 paralogs are thought to play an important role in the assembly or stabilization of Rad51 that promotes homologous pairing and strand exchange reactions. We previously characterized two RAD51 paralogous genes in Arabidopsis (Arabidopsis thaliana) named AtRAD51C and AtXRCC3, which are homologs of human RAD51C and XRCC3, respectively, and described the interaction of their products in a yeast two-hybrid system. Recent studies showed the involvement of AtXrcc3 in DNA repair and functional role in meiosis. To determine the role of RAD51C in meiotic and mitotic recombination in higher plants, we characterized a T-DNA insertion mutant of AtRAD51C. Although the atrad51C mutant grew normally during vegetative developmental stage, the mutant produced aborted siliques, and their anthers did not contain mature pollen grains. Crossing of the mutant with wild-type plants showed defective male and female gametogeneses as evidenced by lack of seed production. Furthermore, meiosis was severely disturbed in the mutant. The atrad51C mutant also showed increased sensitivity to γ-irradiation and cisplatin, which are known to induce double-strand DNA breaks. The efficiency of homologous recombination in somatic cells in the mutant was markedly reduced relative to that in wild-type plants.
Chromosomal double-strand DNA breaks (DSBs) are produced by ionizing radiation (IR), oxygen free radicals, DNA cross-linking reagents, and DNA replication failure. There are two major repair pathways for DSBs: nonhomologous end joining (NHEJ) and homologous recombination (HR; Haber, 2000). In yeast, genetic analysis identified a set of genes, called the RAD52 epistasis group genes, which is directly involved in HR (Baumann and West, 1998). This group contains the Mre11, Xrs2, Rad50, Rad51, Rad52, Rad54, Rad55, Rad57, and Rad59 proteins (Paques and Haber, 1999). The eukaryotic Rad51 is structurally and functionally a homolog of the Escherichia coli recombination protein RecA and is involved in both meiotic and mitotic recombination (Shinohara et al., 1992). The yeast genome contains more proteins with homology to RecA and Rad51. Two members of the Rad51 paralogs (Rad55 and Rad57) have been identified (Kans and Mortimer, 1991; Lovett, 1994). In contrast to yeast, five Rad51 paralogs (Rad51B, Rad51C, Rad51D, Xrcc2, and Xrcc3) have been identified in mammals. These proteins share 20% to 30% sequence identity with Rad51 and with each other (Sonoda et al., 2001).
Previous studies reported several mutants of Rad51 paralogous genes. RAD51C, XRCC2, and XRCC3 mutants in Chinese hamster cell lines (CL-V4B, irs1, and irs1SF, respectively) showed extreme sensitivity to cross-linking reagents. The frequencies of spontaneous chromosomal aberrations in these mutants were higher than those in wild-type cell line (Cui et al., 1999; Godthelp et al., 2002). The five knockout mutants of Rad51 paralogs in chicken B-lymphocyte DT40 cell lines were viable, but showed spontaneous chromosomal aberrations, hypersensitivity to DNA-damaging agents, and low levels of HR (Takata et al., 2001). RAD51B, RAD51D, and XRCC2 knockout mutant mice died during embryonic development (Shu et al., 1999; Deans et al., 2000; Pittman and Schimenti, 2000). The RAD51C and XRCC3 mutants in Drosophila (Spn-D and Spn-B, respectively) were partially sterile, defective for meiotic recombination, but not hypersensitive to DSB-inducing agents (Ghabrial et al., 1998; Abdu et al., 2003).
Physical interactions between the five Rad51 paralogs have also been demonstrated using yeast two-hybrid experiments and coimmunoprecipitation (Schild et al., 2000; Masson et al., 2001). Rad51 paralogs form two complexes, Rad51B-Rad51C-Rad51D-Xrcc2 (BCDX2) and Rad51C-Xrcc3 (CX3), both containing Rad51C as a component (Miller et al., 2004). Thus, Rad51C is the only Rad51 paralog that is a common component of distinct Rad51 paralog complexes. These complexes are thought to mediate DNA strand exchange events of Rad51 during the process of HR (Sung et al., 2003; Miller et al., 2004). Recently, Liu et al. (2004) demonstrated that Rad51C was required for resolution of Holliday junctions (HJs). The above studies suggest the involvement of Rad51C in not only the early stages but also the late stages of HR.
In plants, we previously cloned and characterized two RAD51 paralogous genes from Arabidopsis (Arabidopsis thaliana) named AtRAD51C and AtXRCC3, which are homologs of human RAD51C and XRCC3, respectively (Osakabe et al., 2002). Both AtRAD51C and AtXRCC3 transcripts were detected in a variety of tissues, with the highest level of expression in flower buds, and were induced by γ-irradiation. Our previous results suggested that AtRad51C and AtXrcc3 act together and are involved in DNA repair and meiotic recombination. Recently, AtXRCC3 and AtRAD51 knockout mutants have been identified and characterized in Arabidopsis (Bleuyard and White, 2004; Li et al., 2004). atrad51 and atxrcc3 mutants developed normally in vegetative stage but showed male and female sterility, due to chromosome fragmentation after pachytene in both types of gametogenesis. These results suggested that AtRad51 and AtXrcc3 play important roles in both male and female meiosis.
In this study, we characterized an atrad51C mutant of Arabidopsis. Based on the findings, we discuss the possible role of AtRad51C protein in mitotic and meiotic recombination in plant cells.
RESULTS
Identification of an AtRAD51C T-DNA Insertion Mutant in Arabidopsis
To investigate the function of AtRAD51C gene in Arabidopsis, we searched for loss-of-function mutants. A single line, SALK_021960, was found in Salk Institute T-DNA insertion collections (Alonso et al., 2003), and the corresponding allele was named atrad51C. The AtRAD51C/T-DNA junctions were amplified by PCR and sequenced to determine the position of the T-DNA insertion. Figure 1A provides detailed characterization of the T-DNA insertion at the AtRAD51C gene. According to the direct sequencing analysis of PCR products, the T-DNA insertion was surrounded by two left borders in opposite orientations. The insertion caused an 11-bp deletion of the end of the third exon and a 41-bp deletion of the beginning of the third intron in the AtRAD51C locus. We also analyzed the feature of T-DNA insertion at AtRAD51C locus by Southern-blot analysis. Genomic DNA isolated from atrad51C and wild-type plants were digested with either BamHI or XhoI and hybridized with digoxigenin (DIG)-labeled probe A spanning the T-DNA insertion site (Fig. 1, A and B). The results indicated that the length of the insertion was more than 10 kb and composed of several adjacent T-DNAs.
Figure 1.
Molecular analysis of atrad51C T-DNA insertion. A, Genomic organization of the AtRAD51C locus, and the position of the T-DNA insertion at the AtRAD51C locus. Black boxes represent the exons, and white boxes represent 5′ and 3′ untranslated regions. Thick lines indicate probe regions for Southern- and northern-blotting analyses (probe A for Southern blotting and probe B for northern blotting). Arrows indicate PCR primer positions to determine the junction sequences of the T-DNA insertion. B, Southern-blotting analysis of the T-DNA insertion in the AtRAD51C locus. In these experiments, 5 μg of genomic DNA from AtRAD51C+/+ (wild type), AtRAD51C+/− (heterozygous), and atrad51C−/− (homozygous) plants were digested with BamHI or XhoI. The digested DNAs were blotted and probed with the DIG-labeled probe A shown in section A. Asterisks show the nonspecifically hybridized bands because these bands were observed in all lanes regardless of genotypes (+/+, +/−, −/−). C, Northern-blotting analysis of AtRAD51C expression in flower buds of AtRAD51C+/+, and atrad51C−/− plants. In these experiments, 10 μg of total RNAs from flower buds of wild-type and mutant plants were blotted and probed with DIG-labeled probe B shown in section A. RNA Mr markers (0.5–5.0 kb) are shown on the left.
Next, we analyzed the transcription level of AtRAD51C gene in the atrad51C mutant by northern-blotting analysis. In this experiment, total RNA from flower buds of wild-type and atrad51C mutant were tested because relatively higher levels of AtRAD51C transcripts were detected in flower buds (Osakabe et al., 2002). As shown in Figure 1C, a 1.6-kb mRNA was detected in wild-type flower buds but not in atrad51C (Fig. 1C). Thus, we concluded that the atrad51C mutant could not express a functional Rad51C protein.
AtRad51C Is Not Crucial for Vegetative Development
The atrad51C mutant plant did not show any abnormal phenotypes during the vegetative developmental stage under the standard growth conditions described in “Materials and Methods” (Fig. 2A). Homozygous atrad51C−/− plants were indistinguishable from heterozygous AtRAD51C+/− and wild-type AtRAD51C+/+ plants until the plants started to produce siliques. The numbers of rosette and cauline leaves, size, and the growth rate of atrad51C−/− plants were also similar to those of AtRAD51C+/− and AtRAD51C+/+ plants (data not shown). Thus, the loss of AtRad51C function did not affect vegetative growth. Cytological analysis also showed that mitosis of petals from the atrad51C−/− plant was normal as that of the AtRAD51C+/+ plant (Fig. 3).
Figure 2.
atrad51C mutant plants are sterile. A, Fifteen-day-old AtRAD51C+/+, AtRAD51C+/−, and atrad51C−/− plants on MS agar medium. +/+, +/−, and −/− indicate AtRAD51C+/+, AtRAD51C+/−, and atrad51C−/− plants, respectively. B and C, Open flowers of wild type (B) and atrad51C mutant plants (C). D and E, Siliques of wild-type (D) and atrad51C mutant plants (E). F and G, Siliques resulting from cross-fertilization between wild-type female and wild-type male (F; as a control), and atrad51C mutant and wild-type male (G). H to K, Anthers of wild-type (H and J) and atrad51C mutant plants (I and K). Anthers from 2-mm buds were stained with Alexander's solution (1969; H and I) or stained with I2-KI (J and K). L and M, Tetrad of wild-type (L) and atrad51C mutant plants (M) stained with aniline blue. N, AtRAD51C expression was detected in only PMCs by in situ hybridization. Black arrows indicate signals. O, No hybridization signal was detected when a sense probe was used.
Figure 3.
Mitosis in the petal of wild-type plants (A–E) and atrad51C−/− mutant plants (F–J). A and F, Interphase; B and G, prophase; C and H, metaphase; D and I, anaphase; E and J, telophase. Calibration bar = 10 μm.
The atrad51C Mutant Plant Is Sterile
Next, we determined the sterility phenotypes in the progeny of self-fertilized heterozygous AtRAD51C+/− plants. A total of 188 plants were tested, and 151 (80.3%) were found to be fertile and 37 (19.7%) were sterile, corresponding to a 3:1 segregation expected for a single Mendelian locus (χ2=2.45, 0.1 < P < 0.25 for 3:1 segregation ratio). Using a PCR assay, we identified the genotype of the atrad51C allele in these plants. Thirty-seven of the 188 plants (19.7%) were homozygous for the T-DNA insertion in the AtRAD51C gene (atrad51C−/−), while 103 of 188 (54.8%) were heterozygous (AtRAD51C+/−) and 48 of 188 (25.5%) were wild type (AtRAD51C+/+). This analysis of individual plants confirmed that sterile plants are homozygous for the atrad51C allele.
Heterozygous AtRAD51C+/− plants and homozygous atrad51C−/− mutant plants showed normal flower development similar to the wild-type plants (Fig. 2, B and C). In addition, the fertility of heterozygous plant was similar to that of wild-type plants. However, atrad51C plants produced contractional siliques (Fig. 2E), and almost all these siliques were devoid of seeds. Thus, AtRad51C is required for fertility, but it is not crucial for vegetative development.
atrad51C Mutant Plants Are Defective in Male and Female Gamete Development
To determine the cause(s) of the sterility phenotype in atrad51C plants, we first examined the development of male gametes. To assess pollen grain viability, anthers were dissected from the wild-type and mutant flower buds and stained with Alexander's solution (Alexander, 1969; Fig. 2, H and I) or the iodine potassium iodide solution (I2-KI; Fig. 2, J and K). In Alexander staining, the viable pollen stained red to deep red, while the aborted pollen stained green. On the other hand, the mature pollen, which accumulates starch, stained deep brown by I2-KI. None of the 50 examined atrad51C anthers contained any normal pollen grains (Fig. 2, I and K). Although wild-type plants produced normal tetrad including four spores in meiosis (Fig. 2L), the mutant produced abnormal tetrad-like structures, called polyads, including multiple spores ranging in number from five to seven (Fig. 2M). These findings suggest developmental arrest of the male gamete. In this regard, anthers of heterozygous plants were indistinguishable from those of wild-type plants. Thus, male gametogenesis is severely disturbed in the atrad51C mutant plants. We also determined the expression of AtRAD51C in meiosis by in situ hybridization. Transverse sections of anthers showed strong hybridization signals in only pollen mother cells (PMCs; Fig. 2N); no signals were detected in mature pollen grains (data not shown). This result was consistent with the important function of AtRad51C in meiosis.
Next, to investigate female gametogenesis in atrad51C mutants, atrad51C plants as female parents were cross-pollinated with pollens from wild-type plants. The cross-fertilization resulted in plants with contractional siliques, similar to self-fertilization of atrad51C mutant plants (Fig. 2G). Furthermore, an average of 0.035 seeds per silique (n = 114) was observed. In contrast, when wild-type plants as female parents were cross-pollinated with wild-type pollens, the cross-fertilization resulted in plants produced 22.5 seeds per silique (n = 27). These results suggest that female gametogenesis is also disturbed in the atrad51C mutant plants.
Meiosis Is Severely Disrupted in atrad51C Plants
For further characterization of the atrad51C sterility, we examined meiotic progression in atrad51C and wild-type PMCs by fluorescence microscopy after 4′,6-diamidino-2-phenylindole staining of chromosomes. We observed the progression of normal meiosis in wild-type PMCs as described previously by Ross et al. (1996). Briefly, normal meiosis proceeded by completing synapsis of homologous chromosomes at pachytene (Fig. 4A) and further condensation and separation at diprotene to diakinesis (Fig. 4B), leading to the formation of five condensed bivalents at metaphase I (Fig. 4C). Homologous chromosomes separated from each other and migrated to the opposite poles of the cell at anaphase I (Fig. 4D). Individual chromosomes condensed again and aligned on the metaphase II plate (metaphase II; Fig. 4E). In anaphase II, sister chromatids separated and migrated to the opposite cell poles (Fig. 4F), resulting in decondensation of four groups of five chromatids, with partitioning of the cytoplasm, and finally the formation of a tetrad of haploid nuclei. In atrad51C PMCs, chromosome pairing seemed to occur as in wild-type PMCs during the pachytene stage (Fig. 4, A and G). In diakinesis, chromosomes were further condensed, but the normal bivalent formation was impaired (Fig. 4H). Chromosome fragments and bridges between bivalents were observed in the metaphase I stage (Fig. 4I). These two anomalies were more evident in anaphase I (Fig. 4J). In metaphase II, the majority of the visible chromosome fragments were aligned on the spindle, but some fragments were dispersed throughout the cytoplasm (Fig. 4K). We also found bridges in anaphase II, suggesting that fused chromosomes or chromatid fragments were still present at this stage (Fig. 4L). The result of meiosis in atrad51C PMCs was the aforementioned polyads (Fig. 2M), containing variable number of cells with variable DNA contents (Fig. 4, N–P).
Figure 4.
Meiosis is severely disrupted in atrad51C mutant plants. A to F, Wild type; G to P, atrad51C. A and G, Pachytene stage; B and H, diakinesis; C and I, metaphase I; D and J, anaphase I; E and K, metaphase II; F and L, anaphase II; M to P, telophase II. Arrows with b in sections I, J, and L indicate bridges. Arrows indicate abnormal chromosomal fragments. Calibration bar = 10 μm.
atrad51C Plants Are Hypersensitive to a Cross-Linking Reagent, Cisplatin
We performed the sensitivity test to a cross-linking reagent, cisplatin, in atrad51C and wild-type plants. Cross-linking reagents produce intra- and interstrand DNA cross-links. Both intra- and interstrand cross-links induce DSBs during DNA synthesis, and these DNA lesions are thought to be repaired predominantly by HR as one of the major repair pathways during S phase. Especially, interstrand DNA cross-links are exclusively repaired by HR (De Silva et al., 2002; Sasaki et al., 2004). Therefore, this test aims to provide evidence for the role of AtRad51C in HR processes. Cisplatin sensitivity was scored based on the number of true leaves produced on cisplatin-containing Murashige and Skoog (MS) agar medium. In cisplatin-free medium, both the wild-type and mutant plants produced six true leaves at 15 d after germination. Wild-type plants also produced six true leaves on 15 μm cisplatin medium and four to five true leaves on 30 μm cisplatin medium at 15 d after germination on 15 μm cisplatin medium (Fig. 5A). In mutant plants, the average numbers of true leaves were reduced to 50% and 60% on 15 μm and 30 μm cisplatin medium, respectively, compared to those in wild-type plants. In addition, the mutant plants produced two true leaves on 30 μm and 50 μm cisplatin medium, but these true leaves were not fully opened (Fig. 5B, center of 30 μm and 50 μm sections). After 11 d treatment with cisplatin, the plants were transferred onto cisplatin-free MS agar medium and allowed further growing. Wild-type plants first treated with 30 μm and 50 μm cisplatin produced additional true leaves, although wild-type plants treated with 50 μm cisplatin produced abnormal leaves with a slender leaf shape (Fig. 5C, top of the 50 μm section). On the other hand, the mutant plants that were first treated with cisplatin did not produce any additional leaves when allowed to grow in cisplatin-free medium (Fig. 5C, center of each section), and these plants later died (data not shown). We also tested the sensitivity of atrad51B null mutant plants (Osakabe et al., 2005) to cisplatin by the sensitivity test described in this article. When 3-d-old seedlings of atrad51B were treated with 30 μm cisplatin, atrad51B plants produced three to four true leaves (Fig. 5, A and B, bottom of the 30 μm sections). In contrast to the case of atrad51C, true leaves of atrad51B were expanded, and additional true leaves were also produced after transfer of the plants to cisplatin-free medium, although atrad51B plants produced abnormal leaves with a slender leaf shape (Fig. 5C, bottom of 30 μm section). These results indicated that atrad51C was more sensitive to cisplatin than atrad51B. Next, we tested the sensitivity to cisplatin by root growth assay on MS agar medium containing 15 or 30 μm cisplatin. Unexpectedly, the root growth of atrad51C−/− plants was not different from that of AtRAD51C+/+ and AtRAD51C+/− plants by 3-d cisplatin treatment (Fig. 5D). We also found that the root growth atrad51B was not different from that of wild type (data not shown).
Figure 5.
Cisplatin sensitivity of wild-type, atrad51C, and atrad51B plants. A, Production of true leaves in wild-type, atrad51C, and atrad51B plants at 12 d after treatment with 15 to 75 μm cisplatin. Asterisks indicate true leaf production with small and slender leaves. These leaves were not fully opened. Data represent mean ± sd of 50 plants. B, Fifteen-day-old wild-type, atrad51C, and atrad51B plants treated or untreated with cisplatin. C, Three-week-old wild-type, atrad51C, and atrad51B plants treated or untreated with cisplatin. Fifteen-day-old plants untreated and treated with 30 μm and 50 μm cisplatin were transferred on MS agar medium without cisplatin, and cultured for additional 6 to 7 d. Top of each section, wild-type plants; center of each section, atrad51C plants; bottom of each section, atrad51B plants. D, Comparison of the root growth of AtRAD51C+/+, AtRAD51C+/−, and atrad51C−/− plants after 3-d cisplatin treatment. Six-day-old seedlings were transferred to cisplatin-containing MS agar medium and incubated for another 3 d. Root length of each plant was measured using the public domain NIH Image program (developed at the United States National Institutes of Health and available on the Internet at http://rsb.info.nih.gov/nih-image). Data represent mean ± sd of 30 to 40 measurements.
We also tested the sensitivity to cisplatin in cultured cells derived from wild type and atrad51C. Wild-type and mutant calli were transferred to callus-inducing medium (CIM) containing 0 to 30 μm of cisplatin, and the growth of these calli was scored visually after 3 weeks. As shown in Figure 6A, 15 μm cisplatin severely affected the growth of atrad51C cells, while wild-type cells grew normally similar to wild-type cells grown on cisplatin-free CIM. Thirty millimolar cisplatin reduced the growth of both mutant and wild-type cells. These results suggested that AtRAD51C functions as a factor of DSB repair in the somatic cells of Arabidopsis.
Figure 6.
A, Cisplatin sensitivity of wild-type and atrad51C-cultured cells. Calli were obtained from excised root segments of wild-type and atrad51C plants. These calli were maintained on CIM and subcultured every 3 weeks. Seven-day-old calli were transferred and grown on CIM containing 0 to 50 μm cisplatin. The sensitivity to cisplatin was scored visually 3 weeks later. B, Complementation of atrad51C. Calli from wild-type roots transformed with pZH1 (wild type [pZH1]), calli from the atrad51C roots transformed with pZH1 (atrad51C [pZH1]), and calli from the atrad51C roots transformed with pZHg51C (atrad51C [pZHg51C]) were maintained on CIM containing hygromycin. Sensitivity to cisplatin of transgenic callus was scored as mentioned above. The expression of AtRAD51C mRNA in calli from the atrad51C roots transformed with pZHg51C was similar to that in wild-type callus based on reverse transcription-PCR analysis (data not shown).
To confirm that the hypersensitive phenotype of the atrad51C mutant to cisplatin is indeed due to the loss of function of AtRAD51C locus, a 4.2-kb XbaI-SalI genomic AtRAD51C DNA fragment containing 1.3-kp 5′-upstream, 2.0-kb coding, and 0.9-kb 3′-downstream regions was cloned into the binary vector pZH1 (M. Kuroda and S. Toki, unpublished data) and used for transformation with root segments from wild type and the atrad51C mutant plants. Transgenic calli derived form wild-type and the mutant plants were selected on CIM containing hygromycin and further used for the sensitivity test to cisplatin. As shown in Figure 6B, complementation of atrad51C with the wild-type AtRAD51C resulted in wild-type sensitivity to cisplatin, demonstrating that the hypersensitivity of atrad51C to cisplatin was indeed because of the mutated AtRAD51C gene.
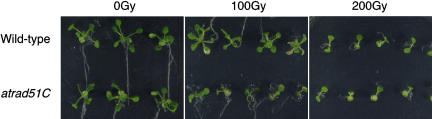
atrad51C Plants Are Hypersensitive to γ-Irradiation
We performed the sensitivity test to γ-irradiation in order to confirm the involvement of AtRad51C in DSB repair induced by another genotoxic stresses other than cisplatin. We irradiated 3-d-old seedlings of wild type and atrad51C to γ-irradiation. A dose of 100 Grey (Gy) strongly suppressed development of true leaves (Fig. 7, bottom of the center section). Mutant plants produced one or two thin and small true leaves at 12 d postirradiation and did not produce additional true leaves after two more weeks. These sensitive plants turned yellow and died (data not shown). In contrast, wild-type plants irradiated with 100 Gy showed only minimal growth defects (Fig. 7, top of the center section). However, 200 Gy strongly suppressed the development of true leaves in both wild-type and mutant plants (Fig. 7, right section). Considered together with the result of the hypersensitivity of atrad51C to cisplatin, we thought that AtRad51C is involved in efficient repair of the DSBs.
Figure 7.
Sensitivity of wild-type and atrad51C−/− plants to γ-irradiation. In these experiments, 3-d-old seedlings of wild-type and mutant plants were either left untreated (as 0 Gy) or irradiated with a dose of 100 Gy or 200 Gy. The photographs were taken at 12 d after irradiation. Top of each section, wild-type plants; bottom of each section, atrad51C plants.
The Defect of Rad51C Reduced Frequencies of HR in Somatic Cells
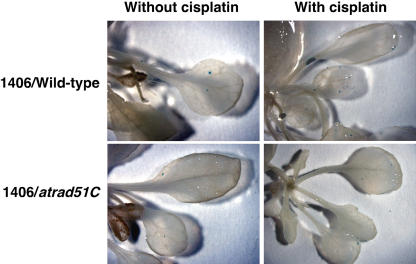
For further characterization of AtRad51 function in HR, the atrad51C plants were monitored for somatic HR using an in planta recombination assay with a reporter gene consisting of two overlapping fragments of the β-glucuronidase (GUS) gene separated by a hygromycin-resistant marker (Swoboda et al., 1994, Gherbi et al., 2001). The GUS sequences have an overlap of 618 bp, and recombination event between the two overlapping sequences can produce a functional GUS gene resulting in blue-colored GUS spots in cells, which are detected by histochemical staining with 5-bromo-4-chloro-3-indoryl glucuronide. We used the two GUS recombination reporter lines, 1406 (direct-repeat recombination reporter line) and 1415 (inverted-repeat recombination reporter line), for in planta recombination assay (Gherbi et al., 2001). The GUS recombination reporter line 1406 or 1415 was crossed with the heterozygous AtRAD51C+/− plant. The frequency of somatic HR (HRF) was monitored in F3 progeny plants homozygous for the GUS recombination substrate and homozygous for the atrad51C mutation allele or the wild-type AtRAD51C allele. The results are summarized in Table I and are presented as frequency distribution histogram in Figure 8. Spontaneous HRF of the atrad51C−/− plants was reduced 2-fold compared to that of the wild-type plants (Table I; Fig. 8, A and B). Somatic HRF in wild-type plants was enhanced with approximately 4- to 5-fold induction after the treatment with 5 μm cisplatin (Table I; Fig. 8, C and D, and images, top). We also measured somatic HRF for bleomycin-treated plants. Bleomycin is a γ-ray mimetic agent and is known to induce DSBs (Favaudon, 1982). After bleomycin treatment, somatic HRF in wild-type plants was also enhanced with 4- to 6-fold induction (Table I; Fig. 8, E and F). In contrast, somatic HRF in the atrad51C mutant plants increased by only 2- to 3-fold after cisplatin or bleomycin treatment (Table I; Fig. 8, C–F, and images, bottom). Thus, atrad51C showed hyporecombination phenotype for somatic chromosomal recombination.
Table I.
Somatic HR in atrad51C and wild-type plants
Data are numbers of plants tested (n), total blue-stained recombination spots (N), and the mean number of spots per plant (M) of chromosomal recombination assays of wild-type and atrad51C plants under three different treatments.
| Wild-Type
|
atrad51C
|
Fold Reduction
|
|||||
|---|---|---|---|---|---|---|---|
| n | N | M (Enhancement) | n | N | M (Enhancement) | ||
| No genotoxic stresses | |||||||
| DRa | 50 | 241 | 4.82 | 50 | 117 | 2.34 | 0.49 |
| IRb | 50 | 357 | 7.14 | 50 | 176 | 3.92 | 0.55 |
| Cisplatin treatment | |||||||
| DR | 50 | 1285 | 26.3 (5.46) | 50 | 368 | 7.36 (3.15) | 0.28 |
| IR | 50 | 1562 | 31.2 (4.38) | 50 | 396 | 7.92 (2.02) | 0.25 |
| Bleomycin treatment | |||||||
| DR | 50 | 1521 | 30.4 (6.31) | 50 | 378 | 7.56 (3.23) | 0.25 |
| IR | 50 | 1671 | 33.4 (4.68) | 50 | 371 | 7.42 (1.89) | 0.22 |
Results with the direct-repeat reporter line 1406.
Results with the inverted-repeat reporter line 1415.
Figure 8.
Frequencies of intrachromosomal HR in atrad51C and wild-type plants. A to F, Frequency distribution histogram shows the proportions of plants with a given number of blue GUS spots in the direct repeat (A, without genotoxic stresses; C, with cisplatin treatment; E, with bleomycin treatment) and inverted repeat (B, without genotoxic stresses; D, with cisplatin treatment; F, with bleomycin treatment) populations. atrad51C mutant and wild-type plants are shown as black and white bars, respectively. Images, Visualization by histochemical staining of recombination events in the direct-repeat line 1406. Top left, Wild-type plant without genotoxic stress; top right, wild-type plant with cisplatin treatment; bottom left, atrad51C plant without genotoxic stress; bottom right D, atrad51C plant with cisplatin treatment.
DISCUSSION
We reported previously the isolation and characterization of Arabidopsis RAD51 paralogous genes AtRAD51C and AtXRCC3 (Osakabe et al., 2002). Transcription of AtRAD51C and AtXRCC3 was detected in various tissues, with the highest level of expression in flower buds, and was induced by γ-irradiation. In this report, we characterized the atrad51C mutant caused by T-DNA insertion in the Arabidopsis RAD51C gene.
Phenotypic analysis of the atrad51C plant indicated that AtRad51C does not play a crucial role in vegetative development under normal growth conditions, but it is essential for male and female meiosis (Figs. 2 and 4). The lack of AtRad51C resulted in severely disrupted meiosis with chromosome fragmentations and appearance of bridges between chromosomes during meiosis I, and these anomalies were also present during meiosis II (Fig. 4), similar to the case of the atxrcc3 mutant (Bleuyard and White, 2004; Bleuyard et al., 2005; Li et al., 2005; this study). It is noted that other Rad51 family proteins, including AtXrcc3, cannot fully substitute the function of AtRad51C in meiosis because the sterility of atrad51C is quite severe. The atdmc1 mutant was deficient for meiotic homologous chromosome synapsis, arrested by the 10 univalents observed at metaphase I (Couteau et al., 1999). The atrad51 mutant also failed to synapse homologous chromosomes and formed synaptonemal complexes (Li et al., 2004). Chromosome fragmentations were also observed at early stage of meiosis I in atrad51 (Li et al., 2004). In contrast to AtDmc1 and AtRad51, AtRad51C and AtXrcc3 are not required for synapsis but essential for postsynaptic events in meiosis (Bleuyard and White, 2004; this study). Thus, the functions of Arabidopsis Dmc1, Rad51, and Rad51 paralogs in meiosis are different, but AtRad51C acts in concert with AtXrcc3 to ensure the correct progression of meiosis. Recently, Bleuyard et al. (2004) reported that the atspo11-1 mutation suppressed the fragmentation phenotype that arose from meiosis I of the atxrcc3 mutant, but chromosome fragmentations are still observed in anaphase II of the atxrcc3/spo11-1 double mutant. In contrast to the case of atxrcc3/spo11-1, no chromosome fragmentation is observed in anaphase II of the atrad51/atspo11-1 double mutant (Li et al., 2004). Grelon et al. (2001) reported the presence of bridges in anaphase I of the spo11-1 mutant but no bridges in anaphase II. These observations suggest that AtXrcc3, and presumably also AtRad51C, functions in a post-AtRad51 role in the repair Spo11-induced DSBs or DSBs in homologous chromosomes and sister chromatids damaged spontaneously or by a minor meiotic endonuclease other than Spo11. Recent studies of Rad51 paralogs in mammals suggest the involvement of Rad51 paralogs in modification of HJ (Braybrooke et al., 2003; Yokoyama et al., 2003; Liu et al., 2004) and that Rad51C and Xrcc3 contribute to HJ resolution (Liu et al., 2004). These findings support the hypothesis that the defect of meiosis in atrad51C with chromosome fragmentations and bridges is due to the absence of HJ resolution activity as thought in the case of atxrcc3 (Bleuyard and White, 2004). Furthermore, the fragmentations in anaphase II of atrad51C mutants seem to arise from unresolved sister chromatid.
On the other hand, earlier studies suggested that the Rad51 paralogs also act in early steps of the HR process (Bishop et al., 1998; Godthelp et al., 2002). Recently, Forget et al. (2004) showed the localization of Xrcc3 at sites of DNA breaks within 10 min after exposure to γ-irradiation and that such localization is independent of Rad51. In plants, meiosis is initiated by the formation of DSB, which is introduced by Spo11, in a manner similar to that in yeast (Grelon et al., 2001; Li et al., 2004). Further studies should be performed to determine the localization of AtRad51C and AtXrcc3 following DSB formation by Spo11 in meiosis to understand the function of AtRad51C and AtXrcc3 in HR.
Based on our previous findings of the interaction between AtRad51C and AtXrcc3, and AtRad51B and AtRad51C (Osakabe et al., 2002, 2005), and of the inducibility of AtRAD51B, AtRAD51C, and AtXRCC3 genes in response to genotoxic stress inducing DSBs (Osakabe et al., 2002, 2005), we hypothesized the involvement of AtRad51C in DSB repair and the increased sensitivity of atrad51C to genotoxic stresses compared to wild-type plants, although the atrad51C mutants plant did not show any abnormal growth in vegetative development under the standard growth condition used in our experiment. As seen in Figure 5, atrad51C showed hypersensitivity to the cross-linking reagent as reported in chicken DT40 and Chinese hamster CL-V4B cells with impaired Rad51C function (Takata et al., 2001; Godthelp et al., 2002; Drexler et al., 2004; Sasaki et al., 2004). Cisplatin forms DNA cross-links, which are intrastrand cross-links and interstrand cross-links (Zamble and Lippard, 1995). Intra- and interstrand DNA cross-links disturb DNA synthesis and lead to DSBs during DNA replication (Dronkert and Kanaar, 2001; De Silva et al., 2002). In this regard, HR seems to repair this type of DNA damage efficiently during S to G2 phase. Indeed, hypersensitivity to cross-linking reagents is a consistent feature of HR repair in deficient mutants described in vertebrate cell lines (Liu et al., 1998; Takata et al., 2000; Sasaki et al., 2004). Furthermore, a recent study reported that the Arabidopsis mutant with impaired Xrcc3, which is the interaction partner of AtRad51C (Osakabe et al., 2002), also shows hypersensitivity to the cross-linking reagent (Bleuyard et al., 2004). Unexpectedly, the root growth of atrad51C and atrad51B was not affected by cisplatin treatment in contrast to the production of true leaves (Fig. 2B). According to studies on yeast and mammals, damaged DNA by cross-linking reagents such as cisplatin and mitomycin C (MMC) is repaired through three potential repair pathways: nucleotide excision repair (NER), postreplication repair/translesion DNA synthesis, and HR repair (Dronkert and Kanaar, 2001). Other than mutants of HR factor, two mutants and antisense-suppressed plants show higher sensitivity to the cross-linking reagents: antisense AtRAD1 (NER factor; Gallego et al., 2000) and atrev3 (translesion DNA synthesis factor; Sakamoto et al., 2002). MMC inhibits both the production of true leaves and growth of roots of antisense AtRAD1 plants (Gallego et al., 2000). MMC also inhibits the root growth of the rev3 mutant plant, although the effect of MMC on the production of true leaves has not been determined (Sakamoto et al., 2002). These results suggest that the contribution of HR repair to the cross-linked DNA repair might be tissue specific, although the cooperation of several DNA repair pathways must be operational for the efficient repair of cross-linked DNAs in plant cells.
As shown in Figure 7, atrad51C seedlings also showed hypersensitivity to γ-irradiation, which is a direct DSB inducer (Dizdaroglu and Bergtold, 1986; Thacker, 1999). In contrast to our result, Bleuyard et al. (2005) reported that atrad51C does not show any increased sensitivity to γ-irradiation compared to wild-type plants. However, they used imbibed seeds in their sensitivity tests to γ-irradiation. When Arabidopsis embryo becomes fully mature, the cells stop division, and it is thought that the cells of Arabidopsis embryo are completely arrested in the G1 phase for long-term quiescence (Laufs et al., 1998; Preuss and Britt, 2003). In vertebrates and higher plants, NHEJ is though to be the predominant repair pathway, especially in mature organisms and during the G1 and early S phases of the cell cycle. On the other hand, HR is important during the late S and G2 phases of the cell cycle when the sister chromatid is available as a template and during early development (Essers et al., 2000; Richardson and Jasin, 2000). Thus, NHEJ repair of the DSBs in seeds was predominant during germination after γ-ray irradiation, and the role of AtRad51C in HR is thought to be masked by NHEJ activity in the range of lower dose of γ-irradiation.
It is interesting to note that atrad51C mutant plants were more sensitive to cisplatin than atrad51B, when we tested the sensitivity to cisplatin by the method used in this study (Fig. 5, B and C). Previous studies of mammals suggested that Rad51 paralogs form two complexes, BCDX2 and CX3, both containing Rad51C (Miller et al., 2004). Moreover, Lio et al. (2004) reported that inhibition of Rad51C reduced the protein levels of Xrcc3 and Rad51B in four other Rad51 paralogs, although the reduction of Rad51B was less and more transient. Thus, it is thought that Rad51C is a central component of Rad51 paralog complexes and that it may regulate the amount of Rad51 paralog complexes. We also showed the interaction between AtRad51C and AtXrcc3, and AtRad51B and AtRad51C (Osakabe et al., 2002, 2005). Several mechanisms could explain the higher sensitivity of atrad51C than atrad51B. First, Arabidopsis might possess two different types of Rad51 paralog complexes similar to vertebrates, and, if CX3 and BCDX2 complexes participate in DSB repair in a different manner, the sensitivity to cisplatin of the atrad51C mutant should be more severe than that of other Rad51 paralog mutants. The other possibility is that AtRad51C possesses multiple functions in HR processes, such as the assembling step of Rad51-nucleoprotein filament (Sung et al., 2003; Miller et al., 2004) and the resolution step of HJs (Liu et al., 2004). Our results clearly showed a higher sensitivity of atrad51C compared to atrad51B, but the hypothesis of different functions for Arabidopsis Rad51 paralogs and Rad51 paralog complexes remains to be confirmed.
In addition to the hypersensitivity to genotoxic stresses, Rad51C deficiency causes impaired sister chromatid cohesion and lack of induction of sister chromatid exchange after genotoxic treatment, as well as increased levels of chromosome aberrations (Godthelp et al., 2002). Previous studies using chicken DT40 cells also indicated that the deficiency of Rad51C resulted in low levels of both spontaneous and DNA cross-linking reagents-induced sister chromatid exchange (Takata et al., 2001; Sasaki et al., 2004). Rad51C deficiency also results in reduced frequencies of somatic recombination in Rad51C knockout DT40 and CL-V4B cells (Takata et al., 2001; Drexler et al., 2004). To determine the direct involvement of AtRad51C in HR, we measured the frequencies of somatic recombination in the atrad51C mutant with GUS recombination reporter system developed by the group of B. Hohn (Swoboda et al., 1994; Gherbi et al., 2001). Using this system, we confirmed that AtRad51C required the efficient recombination in somatic cells (Table I; Fig. 8). We used both direct- and inverted-repeat recombination reporter lines (1406 and 1415) for the evaluation of somatic HRF in the atrad51C mutant. In the case of the direct repeat, the production of a functional recombined GUS gene occurs by gene conversion/crossover or single-strand annealing mechanisms; on the other hand, only a gene conversion/crossover produced a functional recombined GUS gene for the inverted repeat (Gherbi et al., 2001; Dubest et al., 2004). Somatic HRF in atrad51C reduced in both direct- and inverted-repeat reporter lines at similar rates under or without genotoxic stresses (Table I). This finding suggested that AtRad51C participates in the processes of single-strand annealing as well as that of gene conversion. Dubest et al. (2004) reported that the deficiency of AtErcc1 shows hyporecombination phenotype as similar to atrad51C. In this regard, Ercc1 (Rad10) is a key component of NER as well as recombination as part of a structure-specific endonuclease (Davies et al., 1995; Niedernhofer et al., 2001; Motycka et al., 2004). As seen in the case of atrad51C, the somatic HRF is reduced in atercc1 compared to wild type, but some stimulation is observed after bleomycin treatment. This indicates that AtRad51C and AtErcc1 play a cooperative role in somatic recombination and also in DSB repair, especially in repair of DNA cross-links as discussed above.
In conclusion, AtRad51C is essential for meiosis and is required for efficient repair of damaged DNA induced by cisplatin and γ-irradiation in somatic cells. In addition, we provided direct evidence for the involvement of AtRad51C in HR in somatic cells of higher plants.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia was used in this study. The atrad51C T-DNA insertion line (Salk_021960) was found by searching the T-DNA express database (http://signal.salk.edu) established by the Salk Institute Genomic Analysis Laboratory (Alonso et al., 2003). Plants were grown on a soil mixture of equal parts of vermiculite and commercial soil (Sakata Super Mix; Sakata Seed) in a growth chamber at 22°C under 16/8-h photoperiod at 65 μmol m−2 s−1 with cool-white fluorescent lamps. Sterile plants were incubated on MS medium (Murashige and Skoog, 1962) solidified with 0.25% gelrite (Wako; MS gelrite plate) in a growth chamber at 22°C under 16/8-h photoperiod at 65 μmol m−2 s−1 with cool-white fluorescent lamps.
Isolation of DNA and RNA
Genomic DNA was isolated from 2- to 4-week-old sterile plants with the DNeasy Plant Maxi kit (Qiagen). Total RNA was isolated from flower buds of 4-week-old plants using the RNeasy Plant Mini kit (Qiagen) according to the instructions provided by the supplier.
DNA Sequencing
Sequencing reactions were performed with the BigDye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems). The reaction products were analyzed with an automatic DNA sequencer (ABI PRISM 3100-Avant Genetic Analyzer; Applied Biosystems).
PCR Genotyping Assay
Because the kanamycin-resistance gene present in atrad51C T-DNA insertion is silenced, the plant genotypes of SALK_021960 for the atrad51C mutation were identified by PCR. The wild-type AtRAD51C locus was identified by PCR with primers 3ex-F (5′-TTGCCGCGTATT ACTACATCTTGCTCTGA-3′) and 8ex-R (5′-ATGCGATTTGCTCGGTGTAACTGCAGAC-3′). The mutant locus was identified by PCR with LBa-1 (5′-GCGTGGACCGCTTGCTGCAACT-3′) and 8ex-R. The left and right sides of the T-DNA border sequences were amplified with primers 1ex-F (5′-ATGCGATTTGCTCGGTGTAA CTGCAGAC-3′) and LB-a1, and primers 8ex-R1 and LB-a1, respectively (Fig. 1A). PCR products were determined by direct sequencing.
Hybridization
Southern- and northern-blotting analyses were performed according to the standard protocols. DNA probes were prepared by PCR DIG probe synthesis kit (Roche) according to the manufacturer's instruction. For southern-blotting analysis, 3ex-F and 6ex-R (5′-ATGCGATTTGCTCGGTGTAACTGCAGAC-3′) were used as primers. For northern-blotting analysis, 1ex-F and 3ex-R (5′-GCAAAGACTCCTCCTCGTGAAGC-3′) were used as primers. Hybridization and washing were performed under high-stringency conditions with 0.1× SSC.
Cytological Analysis
Viability of pollen grains was examined by staining with Alexander's solution (Alexander, 1969) or the I2-KI (Jefferies, 1977). Tetrads were dissected and stained with aniline blue according to Smith and McCully (1978). Mitotic chromosome preparations were obtained from petals of young floral buds, and meiotic chromosomes were prepared from anthers of young floral buds by enzyme maceration method (Murata and Motoyoshi, 1995; Ross et al., 1996; Armstrong et al., 2001; Nakayama et al., 2004). Whole inflorescences were fixed in freshly prepared fixatives (3:1 ethanol-acetic acid mixture) at 4°C. After rinsing in deionized water for 30 min twice, the floral buds were macerated with an enzyme mixture, which contained 0.8% Cellulase Onozuka RS (Yakult), 0.3% Macerozyme R200 (Yakult), 0.14% Cytohelicase (Sigma-Aldrich), 0.14% Pectolyase Y23 (Seishin Pharmaceutical), and 0.1 mm ethylenediaminetetraacetic acid at pH 4.2 at 37°C for 1 h. After dissection of petals or anthers, the chromosomes were spread by tapping with forceps and dried in air. The chromosomes were counter stained with 1.0 μg/mL of 4′,6-diamidino-2-phenylindole in Vectashield (Vector Laboratories) and observed with a Zeiss Axioplan II using Cascade blue filter set. Fluorescent images were captured using a cooled CCD camera (PentaMax, Photometrics) and analyzed using IPLab (Scanalytics) and Adobe Photoshop. Meiotic stages of wild type were detected as described previously (Ross et al., 1996).
RNA in Situ Hybridization
Plant materials were fixed in 4% (w/v) paraformaldehyde in 100 mm sodium phosphate buffer, pH 7.4, overnight at 4°C, dehydrated through a graded ethanol series and t-butanol series, and finally embedded in Paraplast Plus (Sherwood Medical). Microtome sections (6–10 μm thick) were placed onto silane-treated glass slides. The antisense and sense RNAs of AtRAD51C were labeled with DIG through in vitro transcription of linearized pBluescript KS(+) carrying a fragment of the entire coding sequence of the AtRAD51C cDNA. Hybridization and immunological detection were conducted according to the methods of Kouchi and Hata (1993).
Cisplatin Treatment and γ-Ray Irradiation
To study the effect of cross-linking reagents on Arabidopsis plants, we used cisplatin (cis-diamminedichloroplatinum [II]; Wako Pure Chemical Industries). For assay of cisplatin sensitivity based on production of true leaves, 3-d-old seedlings germinated on MS agar medium were transferred onto MS agar medium containing appropriate concentrations of cisplatin. Plants were scored at 12 d posttreatment of cisplatin for the formation of true leaves. To test the effects of cisplatin on root growth, 3-d-old seedlings were transferred onto MS agar medium containing appropriate concentrations of cisplatin and set vertically for 3 d. After 3-d cisplatin treatment, root length was measured. To test the effect of cisplatin on callus growth, calli were obtained from root segments of 2-week-old wild-type and atrad51C plants. Roots were cut into 3- to 5-mm segments and placed onto callus-inducing medium (3.3 g/L B5 basal minimal salts [Wako], 0.5g/L MES, pH 5.7, 1 mL/L vitamin stock solution [0.5 mg/mL nicotinic acid, 0.5 mg/mL pyridoxine, and 0.5 mg/mL thiamine-HCl], 100 mg/L myo-inositol, 20 g/L Glc, 5 mg/L indoleacetic acid, 0.5 mg/L 2,4-dichlorophenoxyacetic acid, 0.06 mg/L kinetin, and 0.75% bactoagar). Wild-type and atrad51C calli were subcultured on CIM every 2 weeks. Five-day-old calli were transferred onto CIM containing appropriate concentration of cisplatin. Sensitivity to cisplatin was scored visually at 2 weeks posttreatment of cisplatin for callus growth. For assay of sensitivity to γ-ray irradiation, 3-d-old seedlings germinated on MS agar medium were irradiated using a 60Co source at a dose rate of 14 Gy/min. Plants were further cultured on MS agar medium and scored at 12 d postirradiation for formation of true leaves. After scoring the sensitivity, genomic DNAs were isolated from the tested plants to determine their genotypes.
Complementation Test
For complementation of atrad51C, 4.2-kb XbaI-SalI genomic AtRAD51C DNA fragment containing 1.3-kb 5′-upstream, 2.0-kb coding, and 0.9-kb 3′-downstream regions was subcloned from bacterial artificial chromosome DNA F4L23 (GenBank accession no. AC002387) into binary vector pZH1 (M. Kuroda and S. Toki, unpublished data) to yield pZHg51C. pZH1 and pZHg51C vectors carry the hygromycin-resistant gene as a selectable marker. The binary vectors were introduced in Agrobacterium tumefaciens strain EHA105. Wild-type and atrad51C plants were transformed by the root transformation method described by Nam et al. (1997). Transgenic calli were selected and maintained on CIM containing 20μg/L hygromycin (Wako). These calli were later analyzed for AtRAD51C mRNA expression and the sensitivity to cisplatin.
In Planta Recombination Assay
The GUS recombination substrate lines 1406 (direct-repeat recombination substrate line) and 1415 (inverted-repeat recombination substrate line) were crossed with the heterozygous AtRAD51C+/− plant. F3 seeds from plants homozygous for the GUS recombination reporter and heterozygous for the mutant atrad51C-1 allele were germinated on MS medium. The aerial part from each of 3-week-old plants was incubated at 37°C for 48 h in sterile staining buffer (100 mm sodium phosphate buffer [pH 7.0], 0.4 mg/mL 5-bromo-4-chloro-3-indoryl glucuronide, 0.25 mm potassium ferricyanide, 0.25 mm potassium ferrocyanide, and 0.1% Triton X-100]. Genomic DNA was isolated from the remaining root of each plant for genotyping. The number of blue spots, resulting from a recombination event, on each plant was determined visually under a dissecting microscope. For the DNA damage-induced recombination assay, 2-week-old plants were transferred onto MS agar medium containing 5 μm cisplatin or 0.05 μg/L bleomycin for 7 d, and used for GUS histochemical staining.
Acknowledgments
We thank J.M. Lucht and B. Hohn for providing GUS recombination reporter lines. We thank the SALK Institute Genomic Analysis Laboratory and the Arabidopsis Biological Resource Center for providing the T-DNA insertion line. We also thank the Arabidopsis Biological Resource Center for providing the bacterial artificial chromosome DNA. We thank R. Aoto, E. Ozawa, A. Nagashii, Y. Nomura, and F. Suzuki for their technical help.
This work was supported by Grants-in-Aid from the Program for Promotion of Basic Research Activities for Innovative Biosciences (grant to H.I. and S.T.), by the Ministry of Agriculture, Forestry and Fisheries of Japan, and by a Grant-in-Aid from the Japan Society for the Promotion of Science (fellowship to M.E.). Part of this work was also supported by the Budget for Nuclear Research of the Ministry of Education, Culture, Sports, Science and Technology, based on the screening and counseling by the Atomic Energy Commission.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.065243.
References
- Abdu U, Gonzalez-Reyes A, Ghabrial A, Schupbach T (2003) The Drosophila spn-D gene encodes a RAD51C-like protein that is required exclusively during meiosis. Genetics 165: 197–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander MP (1969) Differential staining of aborted and nonaborted pollen. Stain Technol 44: 117–122 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Armstrong SJ, Christopher F, Franklin H, Jones GH (2001) Nucleolus-associated telomere clustering and pairing precede meiotic chromosome synapsis in Arabidopsis thaliana. J Cell Sci 114: 4207–4217 [DOI] [PubMed] [Google Scholar]
- Baumann P, West SC (1998) Role of human Rad51 protein in homologous recombination and double-stranded-break repair. Trends Biochem Sci 23: 247–251 [DOI] [PubMed] [Google Scholar]
- Bishop DK, Ear U, Bhattacharyya A, Calderone C, Beckett M, Weichselbaum RR, Shinohara A (1998) Xrcc3 is required for assembly of Rad51 complexes in vivo. J Biol Chem 273: 21482–21488 [DOI] [PubMed] [Google Scholar]
- Bleuyard JY, Gallego ME, Savigny F, White CI (2005) Differing requirements for the Arabidopsis Rad51 paralogs in meiosis and DNA repair. Plant J 41: 533–545 [DOI] [PubMed] [Google Scholar]
- Bleuyard JY, Gallego ME, White CI (2004) The atspo11–1 mutation rescues atxrcc3 meiotic chromosome fragmentation. Plant Mol Biol 56: 217–224 [DOI] [PubMed] [Google Scholar]
- Bleuyard JY, White CI (2004) The Arabidopsis homologue of Xrcc3 plays an essential role in meiosis. EMBO J 23: 439–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braybrooke JP, Li JL, Wu L, Caple F, Benson FE, Hickson ID (2003) Functional interaction between the Bloom's syndrome helicase and the RAD51 paralog, RAD51L3 (RAD51D). J Biol Chem 278: 48357–48366 [DOI] [PubMed] [Google Scholar]
- Couteau F, Belzile F, Horlow C, Grandjean O, Vezon D, Doutriaux MP (1999) Random chromosome segregation without meiotic arrest in both male and female meiocytes of a dmc1 mutant of Arabidopsis. Plant Cell 11: 1623–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Brenneman M, Meyne J, Oshimura M, Goodwin EH, Chen DJ (1999) The XRCC2 and XRCC3 repair genes are required for chromosome stability in mammalian cells. Mutat Res 434: 75–88 [DOI] [PubMed] [Google Scholar]
- Davies AA, Friedberg EC, Tomkinson AE, Wood RD, West SC (1995) Role of the Rad1 and Rad10 proteins in nucleotide excision repair and recombination. J Biol Chem 270: 24638–24641 [DOI] [PubMed] [Google Scholar]
- Deans B, Griffin CS, Maconochie M, Thacker J (2000) Xrcc2 is required for genetic stability, embryonic neurogenesis and viability in mice. EMBO J 19: 6675–6685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva IU, McHugh PJ, Clingen PH, Hartley JA (2002) Defects in interstrand cross-link uncoupling do not account for the extreme sensitivity of ERCC1 and XPF cells to cisplatin. Nucleic Acids Res 30: 3848–3856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dizdaroglu M, Bergtold DS (1986) Characterization of free radical-induced base damage in DNA at biologically relevant levels. Anal Biochem 156: 182–188 [DOI] [PubMed] [Google Scholar]
- Drexler GA, Rogge S, Beisker W, Eckardt-Schupp F, Zdzienicka MZ, Fritz E (2004) Spontaneous homologous recombination is decreased in Rad51C-deficient hamster cells. DNA Repair (Amst) 3: 1335–1343 [DOI] [PubMed] [Google Scholar]
- Dronkert MLG, Kanaar R (2001) Repair of DNA interstrand cross-links. Mutat Res 486: 217–247 [DOI] [PubMed] [Google Scholar]
- Dubest S, Gallego ME, White CI (2004) Roles of the AtErcc1 protein in recombination. Plant J 39: 334–342 [DOI] [PubMed] [Google Scholar]
- Essers J, van Steeg H, de Wit J, Swagemakers SM, Vermeij M, Hoeijmakers JH, Kanaar R (2000) Homologous and non-homologous recombination differentially affect DNA damage repair in mice. EMBO J 19: 1703–1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaudon V (1982) On the mechanism of reductive activation in the mode of action of some anticancer drugs. Biochimie 64: 457–475 [DOI] [PubMed] [Google Scholar]
- Forget AL, Bennett BT, Knight KL (2004) Xrcc3 is recruited to DNA double strand breaks early and independent of Rad51. J Cell Biochem 93: 429–436 [DOI] [PubMed] [Google Scholar]
- Gallego F, Fleck O, Li A, Wyrzykowska J, Tinland B (2000) AtRAD1, a plant homologue of human and yeast nucleotide excision repair endonucleases, is involved in dark repair of UV damages and recombination. Plant J 6: 507–518 [DOI] [PubMed] [Google Scholar]
- Ghabrial A, Ray RP, Schupbach T (1998) okra and spindle-B encode components of the RAD52 DNA repair pathway and affect meiosis and patterning in Drosophila oogenesis. Genes Dev 12: 2711–2723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gherbi H, Gallego ME, Jalut N, Lucht JM, Hohn B, White CI (2001) Homologous recombination in planta is stimulated in the absence of Rad50. EMBO Rep 2: 287–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godthelp BC, Wiegant WW, van Duijin-Goedhart A, Schärer OD, van Buul PPW, Kanaar R, Zdzienicka MZ (2002) Mammalian Rad51C contributes to DNA cross-link resistance, sister chromatid cohesion and genomic stability. Nucleic Acids Res 30: 2172–2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grelon M, Vezon D, Gendrot G, Pelletier G (2001) AtSPO11-1 is necessary for efficient meiotic recombination in plants. EMBO J 20: 589–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber JE (2000) Partners and pathways repairing a double-strand break. Trends Genet 16: 259–264 [DOI] [PubMed] [Google Scholar]
- Jefferies CJ (1977) Sequential staining to assess viability and starch content in individual pollen grains. Stain Technol 52: 277–283 [DOI] [PubMed] [Google Scholar]
- Kans JA, Mortimer RK (1991) Nucleotide sequence of the RAD57 gene of Saccharomyces cerevisiae. Gene 105: 139–140 [DOI] [PubMed] [Google Scholar]
- Kouchi H, Hata S (1993) Isolation and characterization of novel nodulin cDNAs representing genes expressed at early stages of soybean nodule development. Mol Gen Genet 238: 106–119 [DOI] [PubMed] [Google Scholar]
- Laufs P, Grandjean O, Jonak C, Kieu K, Traas J (1998) Cellular parameters of the shoot apical meristem in Arabidopsis. Plant Cell 1998: 1375–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Chen C, Markmann-Mulisch U, Timofejeva L, Schmelzer E, Ma H, Reiss B (2004) The Arabidopsis AtRAD51 gene is dispensable for vegetative development but required for meiosis. Proc Natl Acad Sci USA 101: 10596–10601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Yang X, Lin Z, Timofejeva L, Xiao R, Makaroff CA, Ma H (2005) The AtRAD51C gene is required for normal meiotic chromosome synapsis and double-stranded break repair in Arabidopsis. Plant Physiol 138: 965–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lio Y-C, Schild D, Brenneman MA, Redpath JL, Chen DJ (2004) Human Rad51C deficiency destabilizes XRCC3 impairs recombination, and radiosensitizes S/G2-phase cells. J Biol Chem 279: 42313–42320 [DOI] [PubMed] [Google Scholar]
- Liu N, Lamerdin JE, Tebbs RS, Schild D, Tucker JD, Shen MR, Brookman KW, Siciliano MJ, Walter CA, Fan W, et al (1998) XRCC2 and XRCC3, new human Rad51-family members, promote chromosome stability and protect against DNA cross-links and other damages. Mol Cell 1: 783–793 [DOI] [PubMed] [Google Scholar]
- Liu Y, Masson JY, Shah R, O'Regan P, West SC (2004) Rad51C is required for Holliday junction processing in mammalian cells. Science 303: 243–246 [DOI] [PubMed] [Google Scholar]
- Lovett ST (1994) Sequence of the RAD55 gene of Saccharomyces cerevisiae: similar to prokaryotic RecA and other RecA-like proteins. Gene 142: 103–106 [DOI] [PubMed] [Google Scholar]
- Masson JY, Tarsounas MC, Stasiak AZ, Stasiak A, Shah R, McIlwraith MJ, Benson FE, West SC (2001) Identification and purification of two distinct complexes containing the five RAD51 paralogs. Genes Dev 15: 3296–3307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KA, Sawicka D, Barsky D, Albala JS (2004) Domain mapping of the Rad51 paralog protein complexes. Nucleic Acids Res 32: 169–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motycka TA, Bessho T, Post SM, Sung P, Tomkinson AE (2004) Physical and functional interaction between the XPF/ERCC1 endonuclease and hRad52. J Biol Chem 279: 13634–13639 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant 15: 473–497 [Google Scholar]
- Murata M, Motoyoshi F (1995) Floral chromosomes of Arabidopsis thaliana for detecting low-copy DNA sequences by fluorescence in situ hybridization. Chromosoma 104: 39–43 [DOI] [PubMed] [Google Scholar]
- Nakayama S, Fujishita M, Ohyama K (2004) FISH shows structural differentiation between liverwort sex chromosomes. In AK Sharma, A Sharma, eds, Plant Genome: Biodiversity and Evolution, Lower Groups, Vol 2A. Science Publishers, Enfield, NH, pp 235–246
- Nam J, Matthysse AG, Gelvin SB (1997) Differences in susceptibility of Arabidopsis ecotypes to crown gall disease may result from a deficiency in T-DNA integration. Plant Cell 9: 317–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedernhofer LJ, Essers J, Weeda G, Beverloo B, de Wit J, Muijtjens M, Odijk H, Hoeijmakers JH, Kanaar R (2001) The structure-specific endonuclease Ercc1-Xpf is required for targeted gene replacement in embryonic stem cells. EMBO J 20: 6540–6549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakabe K, Abe K, Yamanouchi H, Takyuu T, Yoshioka T, Ito Y, Kato T, Tabata S, Kurei S, Yoshioka Y, et al (2005) Arabidopsis Rad51B is important for double-strand DNA breaks repair in somatic cells. Plant Mol Biol 57: 819–833 [DOI] [PubMed] [Google Scholar]
- Osakabe K, Yoshioka T, Ichikawa H, Toki S (2002) Molecular cloning and characterization of RAD51-like genes from Arabidopsis thaliana. Plant Mol Biol 50: 71–81 [DOI] [PubMed] [Google Scholar]
- Paques F, Haber JE (1999) Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 63: 349–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittman DL, Schimenti JC (2000) Midgestation lethality in mice deficient for the RecA-related gene, Rad51d/Rad51l3. Genesis 26: 167–173 [DOI] [PubMed] [Google Scholar]
- Preuss SB, Britt AB (2003) A DNA-damage-induced cell cycle checkpoint in Arabidopsis. Genetics 164: 323–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson C, Jasin M (2000) Coupled homologous and nonhomologous repair of a double-strand break preserves genomic integrity in mammalian cells. Mol Cell Biol 20: 9068–9075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross KJ, Franz P, Jones GH (1996) A light microscopic atlas of meiosis in Arabidopsis thaliana. Chromosome Res 4: 507–516 [DOI] [PubMed] [Google Scholar]
- Sakamoto A, Lan VT, Hase Y, Shikazono N, Matsunaga T, Tanaka A (2002) Disruption of the AtREV3 gene causes hypersensitivity to ultraviolet B light and gamma-rays in Arabidopsis: implication of the presence of a translesion synthesis mechanism in plants. Plant Cell 15: 2042–2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki MS, Takata M, Sonoda E, Tachibana A, Takeda S (2004) Recombination repair pathway in the maintenance of chromosomal integrity against DNA interstrand crosslinks. Cytogenet Genome Res 104: 28–34 [DOI] [PubMed] [Google Scholar]
- Schild D, Lio YC, Collins DW, Tsomondo T, Chen DJ (2000) Evidence for simultaneous protein interactions between human Rad51 paralogs. J Biol Chem 275: 16443–16449 [DOI] [PubMed] [Google Scholar]
- Shinohara A, Ogawa H, Ogawa T (1992) Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell 69: 457–470 [DOI] [PubMed] [Google Scholar]
- Shu Z, Smith S, Wang L, Rice MC, Kmiec EB (1999) Disruption of muREC2/RAD51L1 in mice results in early embryonic lethality which can be partially rescued in a p53(−/−) background. Mol Cell Biol 19: 8686–8693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MM, McCully ME (1978) A critical evaluation of the specificity of aniline blue induced fluorescence. Protoplasma 95: 229–254 [Google Scholar]
- Sonoda E, Takata M, Yamashita YM, Morrison C, Takeda S (2001) Homologous DNA recombination in vertebrate cells. Proc Natl Acad Sci USA 98: 8388–8394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung P, Krejci L, Van Komen S, Sehorn MG (2003) Rad51 recombinase and recombination mediators. J Biol Chem 278: 42729–42732 [DOI] [PubMed] [Google Scholar]
- Swoboda P, Gal S, Hohn B, Puchta H (1994) Intrachromosomal homologous recombination in whole plants. EMBO J 13: 484–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata M, Sasaki MS, Sonoda E, Fukushima T, Morrison C, Albala J, Swagemakers MA, Kanaar R, Thompson LH, Takeda S (2000) The Rad51 paralogs Rad51B promotes homologous recombinational repair. Mol Cell Biol 20: 6476–6482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata M, Sasaki MS, Tachiiri S, Fukushima T, Sonoda E, Schild D, Thompson LH, Takeda S (2001) Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Mol Cell Biol 21: 2858–2866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thacker J (1999) Repair of ionizing radiation damage in mammalian cells: alternative pathways and their fidelity. C R Acad Sci III 322: 103–108 [DOI] [PubMed] [Google Scholar]
- Yokoyama H, Kurumizaka H, Ikawa S, Yokoyama S, Shibata T (2003) Holliday junction binding activity of the human Rad51B protein. J Biol Chem 278: 2767–2772 [DOI] [PubMed] [Google Scholar]
- Zamble DB, Lippard SJ (1995) Cisplatin and DNA in cancer chemotherapy. Trends Biochem Sci 20: 435–439 [DOI] [PubMed] [Google Scholar]