Abstract
To identify transcription factors (TFs) involved in jasmonate (JA) signaling and plant defense, we screened 1,534 Arabidopsis (Arabidopsis thaliana) TFs by real-time quantitative reverse transcription-PCR for their altered transcript at 6 h following either methyl JA treatment or inoculation with the incompatible pathogen Alternaria brassicicola. We identified 134 TFs that showed a significant change in expression, including many APETALA2/ethylene response factor (AP2/ERF), MYB, WRKY, and NAC TF genes with unknown functions. Twenty TF genes were induced by both the pathogen and methyl JA and these included 10 members of the AP2/ERF TF family, primarily from the B1a and B3 subclusters. Functional analysis of the B1a TF AtERF4 revealed that AtERF4 acts as a novel negative regulator of JA-responsive defense gene expression and resistance to the necrotrophic fungal pathogen Fusarium oxysporum and antagonizes JA inhibition of root elongation. In contrast, functional analysis of the B3 TF AtERF2 showed that AtERF2 is a positive regulator of JA-responsive defense genes and resistance to F. oxysporum and enhances JA inhibition of root elongation. Our results suggest that plants coordinately express multiple repressor- and activator-type AP2/ERFs during pathogen challenge to modulate defense gene expression and disease resistance.
Plants grown in the natural environment are continually exposed to a variety of potential pathogens without becoming diseased. This is because plants can recognize and respond to pathogenic organisms and activate multiple defenses, including the production of diverse antimicrobial metabolites and proteins. The key to understanding plant defense responses lies in the elucidation of the signaling pathways involved in their regulation. A large amount of research has identified several important defense-signaling pathways regulated by low-molecular weight signal molecules, such as salicylic acid (SA), jasmonic acid (JA), abscisic acid (ABA), and ethylene (ETH). In the model plant Arabidopsis (Arabidopsis thaliana), it has been proposed that the SA pathway primarily regulates resistance to biotrophic pathogens, as mutants defective in SA biosynthesis or signaling show increased susceptibility to pathogens such as Peronospora parasitica, Erysiphe cichoracearum, and Pseudomonas syringae (Thomma et al., 1998; McDowell et al., 2000; Lu et al., 2001; Liu et al., 2005). In addition, transgenic plants overexpressing the NPR1 gene, a positive regulator of a subset of SA responses, showed increased resistance to biotrophic pathogens (Cao et al., 1998). Conversely, jasmonate (JA) and ETH signaling has been proposed to be more effective against necrotrophic pathogens such as Botrytis cinerea, Pythium irregulare, Plectosphaerella cucumerina, and Fusarium oxysporum. Arabidopsis mutants that are unable to activate JA-dependent defense gene expression showed compromised resistance to necrotrophic fungal pathogens (Staswick et al., 1998; Thomma et al., 1998), and plants that overexpress transcription factors (TFs) involved in the ETH and JA pathways show an increased resistance to several necrotrophs (Berrocal-Lobo et al., 2002; Berrocal-Lobo and Molina, 2004).
In pathogen-challenged plants, a complex cross-talk among the various defense-signaling pathways occurs, resulting in the activation of the most appropriate defense responses for the type of threat. For instance, mutually antagonistic interactions between the SA and JA pathways, as well as between the JA/ETH and ABA pathways, act to fine tune plant defense responses (Spoel et al., 2003; Anderson et al., 2004; Takahashi et al., 2004; Kariola et al., 2005). However, the antagonistic nature of these pathways is not absolute, and many plant defense genes show coordinated induction patterns utilizing multiple signaling pathways during the defense responses (Schenk et al., 2000; Campbell et al., 2003).
TFs are believed to play a crucial role in the transmission of pathogen-derived defense signals to either activate or suppress downstream defense gene expression as well as in the regulation of cross-talk between different signaling pathways (Lorenzo et al., 2003; Anderson et al., 2004). The Arabidopsis genome contains more than 1,500 TFs (Ratcliffe and Riechmann, 2002), and, within this large group of TFs, several gene families that share similarity in their binding domains exist. Virtually every major TF gene family harbors members that have been implicated in some aspect of plant defense (for review, see Singh et al., 2002). Examples of these include the APETALA2/ethylene response factors (AP2/ERFs; Solano et al., 1998), WRKYs (Dong et al., 2003), MYBs (Vailleau et al., 2002; Mengiste et al., 2003), NACs (Delessert et al., 2005), and basic helix-loop-helices (Anderson et al., 2004; Lorenzo et al., 2004), as well as the recently described whirly-domain TFs (Desveaux et al., 2004). In many instances, genes encoding TFs that are involved in plant defense signaling are also transcriptionally regulated by pathogen challenge and treatment with defense elicitors. Examples of these are the JA-activated ERF1 and AtMYC2 TFs that regulate JA responses (Anderson et al., 2004; Lorenzo et al., 2004). This suggests that a possible strategy to identify TFs with roles in plant defense may be to first identify TF genes that show altered transcript levels during the early stages of the defense response, followed by a more focused functional analysis of these candidate genes.
With the advent of genomic technologies, it is possible to analyze the expression of a large number of Arabidopsis genes, including many TF genes, in parallel (Chen et al., 2002; Zimmerli et al., 2004). However, hybridization-based assays may not be very suitable for the measurement of small, but biologically significant, changes in the low-abundance transcripts, such as those of many TFs. Reverse transcription (RT)-quantitative (Q)-PCR is known to be more sensitive than hybridization-based methods for the quantification of low-abundance transcripts (Horak and Snyder, 2002). Although RT-Q-PCR is often utilized for validating and extending the results of microarray experiments (Schenk et al., 2003), until recently the use of this technique for large-scale gene expression analyses has been limited. In an attempt to overcome the problems associated with array-based methods of expression analysis, Czechowski et al. (2004) developed a genome-wide RT-Q-PCR-based resource for quantitative measurements of transcripts of 1,465 Arabidopsis TF genes. In these experiments, RT-Q-PCR profiling of TF gene expression successfully identified a substantial number of novel root- and shoot-specific TF genes. The same resource was also used in combination with array-based methods to identify processes affected by long-term nitrogen deprivation or short-term nitrate nutrition in Arabidopsis (Scheible et al., 2004). To date, this resource has not been applied to the analysis of defense-associated TF genes in Arabidopsis.
We report here the screening of the 1,534-member TF complement of Arabidopsis during defense responses triggered by either the incompatible necrotrophic pathogen Alternaria brassicicola or the signaling compound methyl jasmonate (MeJA) as a means of identifying TFs involved in the JA-regulated biotic defense-signaling pathway. The sensitive RT-Q-PCR screen initially developed by Czechowski et al. (2004) identified several TF genes that have not been previously implicated in plant defense, and particularly demonstrated that the AP2/ERF family of TF genes predominated among the spectrum of gene families activated by both MeJA and pathogen challenge. Functional characterization of two MeJA- and A. brassicicola-responsive AP2/ERF TF genes by insertional inactivation and/or constitutive overexpression demonstrated roles for these genes as negative or positive regulators of resistance to the necrotrophic fungal pathogen F. oxysporum as well as defense gene expression and MeJA sensitivity. These results suggest that the extent of the JA-activated defense response is modulated by the coordinated expression of both repressor- and activator-type AP2/ERF TF genes.
RESULTS
AP2/ERF Genes Are the Predominant TF Family Responsive to Both JA and A. brassicicola
To identify TF genes with potential roles in JA signaling and defense against necrotrophic pathogens, we undertook a global screen of Arabidopsis TF genes by RT-Q-PCR following either challenge by A. brassicicola or treatment with MeJA. Three independent replicate experiments were performed, and the samples were used to produce the cDNA used in RT-Q-PCR analysis of the expression of 1,534 known or putative Arabidopsis TF genes, using an updated version of the specific primer set devised and used by Czechowski et al. (2004). The complete dataset from these experiments is available as Supplemental Table I. A total of 134 genes, representing approximately 9% of the estimated Arabidopsis TF suite, showed a significant (P < 0.05) and 2-fold or greater change in expression at 6 h following exposure to either MeJA or A. brassicicola. The number of TFs showing altered expression following treatment was similar for both MeJA (75 TF genes) and A. brassicicola (84 TF genes). Classification of the inducible TF genes into their respective families revealed that the AP2/ERF, MYB, WRKY, and NAC TF families showed the greatest number of inducible members. A total of 24 genes showed coregulation by both treatments, and 20 of these genes were induced by both treatments with the remaining four repressed. Interestingly, members of the AP2/ERF TF family showed the highest number of coregulated genes, with 10 members (7% of total AP2/ERFs) being significantly induced by both MeJA and A. brassicicola, suggesting that this family of TFs may play a particularly important role in the regulation of JA-dependent defense response pathways. We further focused only on the 10 AP2/ERF genes that were significantly induced following 6-h exposure to both MeJA and A. brassicicola. We performed three further independent time-course experiments and assayed gene expression at 1, 3, 6, and 24 h after either inoculation with A. brassicicola or exposure to MeJA. In these experiments, all 10 of the 10 AP2/ERFs identified in the initial screen were significantly (P < 0.05) induced more than 2-fold (Table I) following both treatments.
Table I.
Effect of A. brassicicola inoculation and MeJA treatment on the expression of AP2/ERF genes
Results obtained in the initial genome-wide RT-Q-PCR screen were verified in a secondary independent time-course expression analysis for each ERF/AP2 gene initially identified as differentially expressed at 6 h after either MeJA treatment or fungal inoculation. Ab, A. brassicicola. Values shown indicate average relative expression ratio to control (data from three independent experiments for both initial screening and secondary time course). Bold text indicates statistically significant induction (P < 0.05).
| Locus No.
|
Name
|
Initial Screening
|
Secondary Time Course
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
Ab
|
MeJA
|
Ab
|
MeJA
|
||||||||
| 6 h | 6 h | 1 h | 3 h | 6 h | 24 h | 1 h | 3 h | 6 h | 24 h | ||
| At1g04370 | AtERF14 | 3.5 | 8.3 | 3.3 | 1.0 | 0.41 | 13.9 | 0.7 | 4.7 | 2.7 | 4.4 |
| At1g06160 | 2.7 | 5.7 | 1.0 | 1.2 | 1.3 | 41.8 | 1.1 | 6.3 | 3.2 | 22.6 | |
| At1g28370 | AtERF11 | 2.6 | 2.7 | 1.8 | 2.8 | 4.7 | 8.7 | 1.7 | 6.8 | 3.7 | 3.0 |
| At1g33760 | Tiny-like | 2.7 | 3.0 | 1.6 | 0.86 | 9.8 | 1.5 | 1.6 | 2.2 | 1.1 | 0.9 |
| At2g44840 | AtERF13 | 4.2 | 4.3 | 2.5 | 0.4 | 4.7 | 4.4 | 4.1 | 19.7 | 99.6 | 4.7 |
| At3g15210 | AtERF4 | 2.1 | 2.6 | 1.4 | 1.6 | 1.9 | 2.4 | 2.0 | 2.9 | 2.5 | 3.0 |
| At3g23230 | TDR1 | 14.2 | 2.8 | 1.0 | 1.9 | 30.8 | 5.4 | 1.0 | 1.8 | 0.68 | 3.0 |
| At3g23240 | ERF1 | 4.0 | 5.9 | 1.1 | 2.8 | 1.8 | 11.8 | 6.4 | 4.2 | 13.7 | 24.4 |
| At4g17500 | AtERF1 | 2.3 | 3.7 | 1.3 | 0.79 | 1.0 | 2.5 | 2.4 | 9.1 | 5.6 | 8.8 |
| At5g47220 | AtERF2 | 2.2 | 3.5 | 0.74 | 1.0 | 1.3 | 3.3 | 6.7 | 7.7 | 7.4 | 9.4 |
AP2 Domain Sequence Similarity of JA- and Pathogen-Responsive AP2/ERFs
A sequence similarity analysis of the AP2/ERF gene family conducted using the 59 to 60 amino acids of the conserved AP2 domain (Pfam, PF00847; Bateman et al., 2004) showed that the AP2/ERF genes that are coinduced by MeJA and A. brassicicola form distinct groups within the B3 and B1a clusters of the gene family (Fig. 1). Seven of the 10 MeJA- and pathogen-regulated ERFs are located within the B3 subcluster, which includes TFs such as ERF1 and AtERF2, which have been shown to act as positive regulators of defense-related transcription (Solano et al., 1998; Brown et al., 2003). Additionally, two more genes within this cluster, AtERF14 and AtERF15, were previously reported to be responsive to the bacterial pathogen P. syringae (Onate-Sanchez and Singh, 2002). Subsequently, we analyzed all 17 members of the B3 subcluster in the full time-course experiments that showed that the expression of each gene was significantly altered more than 2-fold by either MeJA or A. brassicicola in at least one of the time points analyzed, with the exception of At4g18450 (data not shown).
Figure 1.
Phylogenetic tree of the AP2/ERF TF family formed by sequence analysis of 155 Arabidopsis AP2 domains. Bar equals 0.2 substitutions per site. TFs induced by both A. brassicicola and MeJA indicated by squares (▪). Thirteen AP2/ERFs contain two AP2 domains and these genes were named as locus number 1 or 2 (e.g. At1g78080-1 and At1g78080-2; see A6 subcluster).
A cluster of eight genes within the B1 subcluster included two genes that also showed altered expression from the treatments, and we have termed this the B1a cluster (Fig. 1). These genes all contain a sequence that encodes for a protein motif identified as the ERF-associated amphiphilic repression (EAR) motif (Ohta et al., 2001), which has previously been shown to function as an active repressor of transcription (Fujimoto et al., 2000; Hiratsu et al., 2003). Full time-course analysis of the MeJA and A. brassicicola inducibility of the remaining members of the B1 cluster failed to reveal any significant change in their expression (data not shown). The other AP2/ERF (At1g33760) identified in the initial screen was located in the drought-responsive element-binding subfamily (Fig. 1), members of which are typically involved in drought and cold stress (Liu et al., 1998), and this suggests a potential interaction point between biotic and abiotic stress signaling.
It would be expected that coregulated genes would share similar regulatory motifs in their promoter regions. Surprisingly, a search of the 1-kb region upstream of the coregulated AP2/ERFs using commonly available motif-scanning software (Toufighi et al., 2005) failed to reveal motifs that were significantly enriched in our inducible gene set compared with a random sample of Arabidopsis promoters. Despite this, the identification of the B3 ERFs as potential activators in this study is consistent with the proposal of Gutterson and Reuber (2004) that these genes play important roles in plant defense, although an increased disease resistance phenotype has so far only been demonstrated for ERF1 (Berrocal-Lobo and Molina, 2004). Importantly, direct involvement of the B1a subcluster containing potential repressor-type AP2/ERFs in plant defense has not been previously reported in Arabidopsis.
AtERF4 Negatively Regulates JA-Responsive Expression of PDF1.2 and Disease Resistance
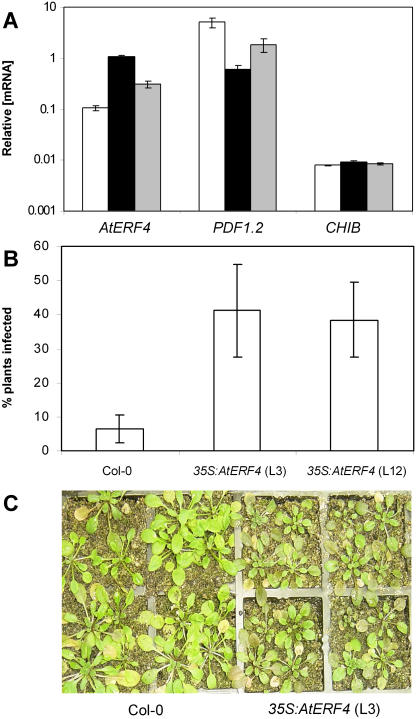
To test for a role of the B1a-type TF, AtERF4, in defense gene regulation and disease resistance, we analyzed Arabidopsis plants constitutively expressing AtERF4 under the control of the cauliflower mosaic virus 35S promoter. RT-Q-PCR analysis of homozygous 35S:AtERF4 lines revealed between 3- and 10-fold increases in AtERF4 transcript levels relative to wild-type plants (Fig. 2A). In addition, we examined the expression of two defense-associated genes, PDF1.2 and CHIB. Both genes are known to contain a GCC-box motif in their promoters (imperfect in the case of CHIB), and are regulated by the JA pathway (Penninckx et al., 1998; Anderson et al., 2004). In AtERF4-overexpressing plants, basal (e.g. not induced by MeJA) transcript levels of PDF1.2 and CHIB remained at levels similar to those observed in wild-type plants (data not shown). However, treatment of AtERF4-overexpressing plants with MeJA resulted in PDF1.2 induction ratios that were 2.7- and 10-fold lower than those observed in MeJA-treated wild-type plants (Fig. 2A). Interestingly, we did not observe any significant change in the induction profile of CHIB between MeJA-treated wild-type and AtERF4-overexpressing lines. These results suggest that AtERF4 negatively regulates the MeJA-responsive expression of PDF1.2 preferentially to that of CHIB.
Figure 2.
The 35S:ERF4 lines show reduced MeJA-responsive PDF1.2 expression and increased disease susceptibility to F. oxysporum. A, RT-Q-PCR analysis of AtERF4, PDF1.2, and CHIB mRNA levels in wild-type (white bars) and two independent 35S:AtERF4 transgenic lines, line 7 (black bars) and line 12 (gray bars), after 24-h exposure to MeJA. Average data with standard errors from three independent biological experiments are shown. B, Percentage of wild-type and 35S:AtERF4 plants showing disease symptoms 3 d after inoculation with F. oxysporum. Average data with standard errors from four biological replicates are shown. C, Wild-type (Col-0) and 35S:AtERF4 plants (line 3) 3 d after inoculation with F. oxysporum, with 35S:AtERF4 plants showing enhanced disease symptoms (see “Materials and Methods” for description).
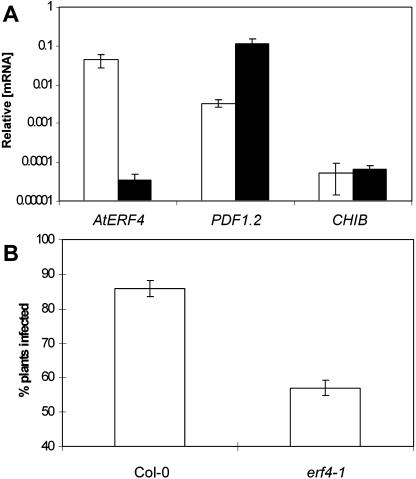
To further examine the role of AtERF4 as a negative regulator of PDF1.2, we studied PDF1.2 and CHIB basal transcript levels in untreated plants of an Arabidopsis line containing a homozygous T-DNA insertion in the coding region of AtERF4 (termed erf4-1). The T-DNA insertion in erf4-1 truncates the EAR domain that is necessary for repressor activity. RT-Q-PCR analysis of these plants using primers designed to amplify the native AtERF4 transcript detected very low levels in the erf4-1 mutant (Fig. 3A). As expected from a negative regulator, we found a more than 30-fold increase in PDF1.2 basal transcript levels in untreated erf4-1 plants compared to those in untreated wild-type plants, whereas CHIB levels remain unchanged (Fig. 3A). These results further suggest that AtERF4 acts as a negative regulator of PDF1.2 expression.
Figure 3.
The erf4-1 mutant shows increased defense gene expression and disease resistance to F. oxysporum. A, RT-Q-PCR analysis of PDF1.2 and CHIB mRNA levels in wild-type (white bars) and erf4-1 mutant line (black bars). Average data with standard errors from three independent biological replicates are shown. B, Percentage of wild-type and erf4-1 mutant plants showing disease symptoms 8 d after inoculation with F. oxysporum. Average data with standard errors from four biological replicates are shown.
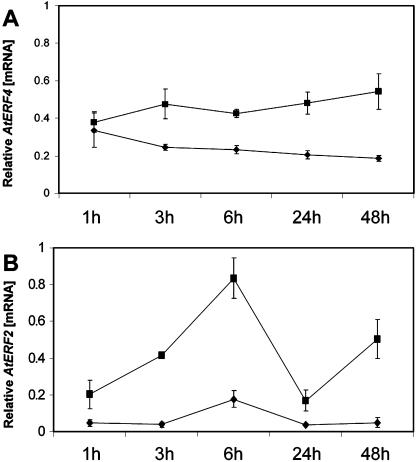
To test whether AtERF4 is involved in regulating disease resistance, AtERF4-overexpressing lines and erf4-1 mutant plants were inoculated with the pathogen F. oxysporum, and disease development was followed over 15 d and compared to that in similarly inoculated wild-type plants. F. oxysporum was used in these assays because it is a compatible pathogen of Arabidopsis (Anderson et al., 2004). In addition, F. oxysporum was shown to induce AtERF4 expression in wild-type Arabidopsis plants (Fig. 4A). No phenotypic differences were observed between the ecotype Columbia (Col-0), 35S:AtERF4 lines, and erf4-1 under normal growing conditions. However, when inoculated with F. oxysporum, we found that only 5% of inoculated wild-type plants showed disease symptoms 3 d after treatment, whereas the percentage of both AtERF4-overexpressing lines displaying disease symptoms was approximately 40% (P < 0.05; Fig. 2B). The difference in F. oxysporum resistance between the transgenic and Col-0 lines was visually apparent, with the 35S:AtERF4 lines showing postinoculation stunting and disease symptoms typical of F. oxysporum infection (Fig. 2C). Complementing these data, the percentage of plants of the erf4-1 mutant that displayed typical disease symptoms 8 d after treatment was approximately 57% of total plants, which was markedly reduced (P < 0.05) from that of wild-type plants in which 85% of plants displayed symptoms (Fig. 3B). Together, these results indicate that AtERF4 is a negative regulator of resistance to necrotrophic pathogens.
Figure 4.
RT-Q-PCR analysis of gene expression in Arabidopsis following inoculation with F. oxysporum. Data come from three independent replicates, with standard error shown. A, AtERF4 expression in F. oxysporum-treated plants (square) compared with mock-inoculated plants (diamond). B, AtERF2 expression in F. oxysporum-treated plants (square) compared with mock-inoculated plants (diamond).
AtERF4 Negatively Regulates Root Sensitivity to JA
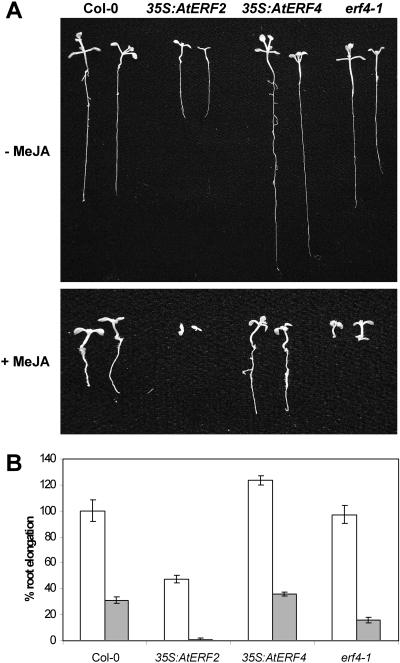
To determine whether AtERF4 functions as a negative regulator of other JA-related responses, we germinated seeds from Col-0, 35S:AtERF4 lines, and the erf4-1 mutant in the presence or absence of MeJA. We found that root elongation in the presence of MeJA was significantly enhanced by 15% to 50% in 35S:AtERF4 lines, while significantly reduced by 50% in the erf4-1 mutant when compared to wild-type plants (Fig. 5, A and B). These results further support the notion that AtERF4 negatively regulates JA-dependent responses in Arabidopsis.
Figure 5.
JA sensitivity in Arabidopsis is positively and negatively influenced by AtERF2 and AtERF4, respectively. Shown is JA-mediated root elongation inhibition 7 d following germination. A, Seed from Col-0, 35S:AtERF2, 35S:AtERF4, and erf4-1 mutant germinated in the presence or absence of MeJA. B, Average root length for plants grown on control (white bars) or media containing MeJA (gray bars), shown as a percentage relative to Col-0 (without MeJA). Standard errors are indicated.
AtERF2 Functions as a Positive Regulator of Disease Resistance and JA-Dependent Responses
To test whether JA- and pathogen-inducible members of the B3 subcluster of AP2/ERF genes identified here may have a role in disease resistance, we analyzed the effect of overexpression of AtERF2 on resistance to F. oxysporum. AtERF2 was identified here as inducible by both A. brassicicola and MeJA, supporting previous results showing its induction by MeJA, ETH, wounding, and P. syringae inoculation (Chen et al., 2002; Brown et al., 2003). In addition, AtERF2 has previously been implicated in the regulation of PDF1.2 and CHIB (Brown et al., 2003), but a role in disease resistance has not been demonstrated.
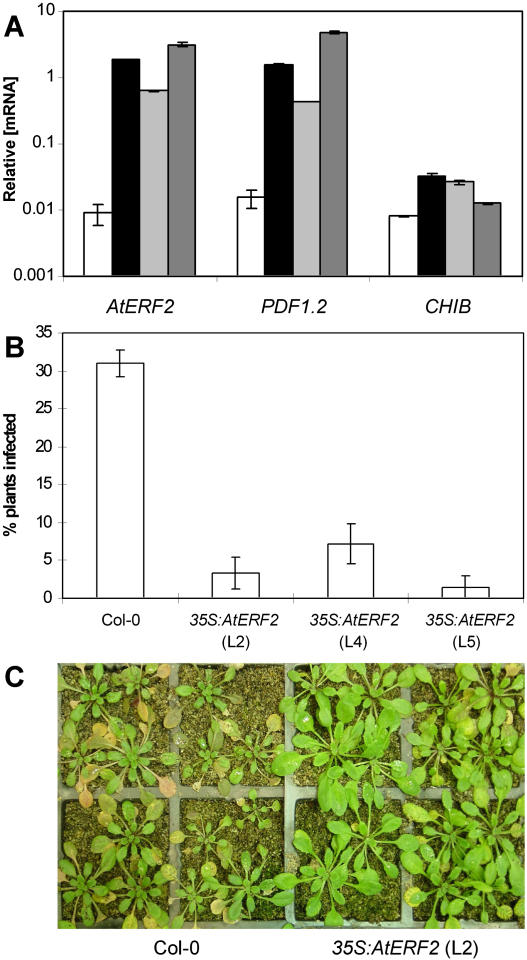
Analysis of three independent homozygous lines transformed with a 35S:AtERF2 construct showed a 70- to 350-fold increase in AtERF2 expression over wild type in these lines (Fig. 6A). Correspondingly, transcript levels of PDF1.2 were elevated between 28- and 310-fold that of wild-type plants. In addition, CHIB was shown to be elevated in the AtERF2-overexpressing lines but only between 1.5- and 4-fold. These results confirm the function of AtERF2 as a positive regulator of defense gene expression (Brown et al., 2003). In addition, AtERF2 expression was observed to increase following inoculation with F. oxysporum (Fig. 4B). For each of the AtERF2-overexpressing lines, we observed a significant reduction in the development of disease symptoms in independent inoculation experiments. Six days following inoculation with F. oxysporum, 30% of wild-type plants showed symptoms of infection, whereas only between 1.5% and 7.5% of plants from AtERF2-overexpressing lines were symptomatic (Fig. 6, B and C).
Figure 6.
The 35S:AtERF2 lines show increased defense gene expression and disease resistance. A, RT-Q-PCR analysis of AtERF2, PDF1.2, and CHIB mRNA levels in wild-type (white bars) and the three independent 35S:AtERF2 transgenic lines, line 2 (black bars), line 4 (light-gray bars), and line 5 (dark-gray bars). Average data with standard errors from three independent biological experiments are shown. B, Percentage of wild-type and 35S:AtERF2 lines showing disease symptoms 6 d after inoculation with F. oxysporum. Average data with standard errors from four biological replicates are shown. C, Col-0 and 35S:AtERF2 (line 2), 6 d after inoculation with F. oxysporum, with 35S:AtERF2 plants showing reduced disease symptoms (see “Materials and Methods” for description).
The AtERF2-overexpressing lines were also analyzed for MeJA-dependent root elongation inhibition. We found that AtERF2-overexpressing lines showed increased sensitivity to MeJA, with the highest expressing line showing complete inhibition of root elongation by MeJA (Fig. 5, A and B). These results support the function of AtERF2 as a positive regulator of MeJA responses, including plant defense gene expression (Brown et al., 2003) and disease resistance, and, taken together with previous work on ERF1 (Berrocal-Lobo et al., 2002; Lorenzo et al., 2003), suggest that multiple activator-type AP2/ERF genes in the B3 subcluster have regulatory roles in disease resistance.
DISCUSSION
In this article, we describe the screening of the entire Arabidopsis TF transcriptome for altered expression following treatment with the defense-signaling regulator MeJA and the incompatible pathogen A. brassicicola using RT-Q-PCR. This analysis identified many novel TF genes as candidates for further functional studies. We demonstrated the utility of this approach by characterizing functions of two AP2/ERFs in JA signaling and plant defense. These studies showed that multiple AP2/ERF genes, including both activator and repressor types, are induced during the plant defense response and that both of these types of AP2/ERFs have important functions in regulating disease resistance.
A Repressor-Type AP2/ERF Regulates Disease Resistance in Arabidopsis
In contrast to the activator-type ERF TFs located in the B3 subcluster, in planta functional roles of repressor-type ERFs from the B1a subcluster have not been described previously. Here we demonstrated that the erf4-1 mutant was more resistant to F. oxysporum, while transgenic lines overexpressing AtERF4 were more susceptible, and therefore AtERF4 negatively regulates resistance to this necrotrophic pathogen. Overexpression of AtERF4 had no effect on basal PDF1.2 transcript levels in untreated plants, but, following MeJA treatment, the AtERF4-overexpressing lines showed significantly lower induction of PDF1.2 than that observed in MeJA-treated wild-type plants (Fig. 2A) and reduced sensitivity to JA inhibition of root elongation (Fig. 5A). The role of AtERF4 as a repressor of JA-dependent plant responses was further strengthened by the increased PDF1.2 levels observed in the erf4-1 mutant as well as the increased sensitivity of root growth to MeJA. The AtERF4 protein has previously been shown to bind to the GCC-box motif and act as an active repressor of transcription in vitro (Fujimoto et al., 2000). This GCC-box motif is essential for MeJA induction of PDF1.2 (Brown et al., 2003). Therefore, our results provide in planta evidence to support the notion that AtERF4 is an active repressor of GCC-box-mediated transcription.
During review of this article, Yang et al. (2005) reported that AtERF4 acts as a repressor of transcription in transient assays. In addition, these authors demonstrated that overexpression of an AtERF4∷GFP fusion construct acts as a repressor of both ETH and ABA responses in Arabidopsis. AtERF4∷GFP is localized to nuclear bodies within the nucleus, suggesting that protein degradation could be a potential mechanism in the regulation of gene expression by this TF (Yang et al., 2005).
The role of AtERF4 in the negative regulation of JA-responsive defense gene expression and disease resistance is also reminiscent of the role of the recently characterized NIMIN1, which also contains the EAR motif and negatively regulates SA-responsive defense gene expression (Weigel et al., 2005). The role of pathogen-inducible repressor-type TFs may be to regulate the activation of plant defense responses in a finely controlled manner in coordination with other cellular processes. Negative regulators may also be necessary to modulate antagonistic interactions between signaling pathways. To this end, a possible function for AtERF4 is to mediate antagonistic interactions between SA- and JA-signaling pathways (Schenk et al., 2000; Spoel et al., 2003). It is plausible that SA suppression of the JA inducibility of PDF1.2 is partly mediated by AtERF4. Our initial experiments have shown SA inducibility of AtERF4. Previously, AtWRKY70 has been implicated in regulating the SA-mediated suppression of PDF1.2, acting downstream of NPR1 (Li et al., 2004). However, the absence of any W-box in the PDF1.2 promoter argues against a direct suppression of PDF1.2 by AtWRKY70. In contrast, the AtERF4 promoter contains five putative W-boxes (core motif TGAC) in the 1-kb region upstream of the start codon, suggesting the potential for WRKY factors such as AtWRKY70 to play some role in its regulation. The hypothesis that AtERF4 acts downstream from NPR1 and AtWRKY70 in SA-mediated suppression of JA-inducible PDF1.2 expression is currently being tested in our laboratory. The inducibility of AtERF4 by ABA (Yang et al., 2005) also indicates the possibility that AtERF4 plays an integral role in both biotic and abiotic signaling pathways and may be responsible for regulating the antagonism between them (Anderson et al., 2004).
The repression-associated EAR motif found in AtERF4 [consensus sequence L/FDLNL/F(x)P] was first identified in the C-terminal region of the class II ERFs (e.g. NtERF3, AtERF3, and AtERF4) and TFIIIA-type zinc finger proteins (e.g. ZAT10 and ZAT11). More recently, NIMIN1 (Weigel et al., 2005) and HSI proteins (Tsukagoshi et al., 2005) have also been shown to contain the EAR motif in their C-terminal region. Furthermore, the repression domains in Aux/indole acetic acid (IAA)-responsive IAA proteins (e.g. IAA17) contain an EAR-like motif [L(x)L(x)L; Tiwari et al., 2004]. Although the mechanism by which EAR or EAR-like motifs inhibit transcription is currently unknown, these class II ERFs were shown to down-regulate both basal transcription levels of a reporter gene as well as the transactivation activity of other TFs (Ohta et al., 2001). The presence of this motif across a range of TF families suggests it is a distinct regulatory motif utilized in the repression of a number of genes in different signaling pathways.
Members of the B3 Subcluster of AP2/ERFs Share Similar Functions
In addition to the seven members of the AP2/ERF B3 subcluster that were identified as being both MeJA and A. brassicicola inducible by the screening process, we also established that nine of the remaining 10 members of the B3 subcluster were significantly altered by one of these treatments in at least one of the time points analyzed here. This implicates almost every member of the B3 subgroup of AP2/ERF genes in defense responses in Arabidopsis. The B3 subcluster contains ERF1, which is known to provide enhanced disease resistance when overexpressed in transgenic plants, including resistance to F. oxysporum (Berrocal-Lobo et al., 2002; Berrocal-Lobo and Molina, 2004). Our results show that another B3 gene of Arabidopsis, AtERF2, can also result in increased resistance to F. oxysporum when overexpressed.
The shared function of B3-like TFs is evident from studies of orthologs from other species. For example, overexpression of Pti4 and Pti5, AP2/ERF TFs from tomato (Lycopersicon esculentum) that show substantial similarity within their AP2 domain to B3 ERFs, also induced defense genes and provided resistance to Erysiphe orontii and P. syringae when overexpressed in transgenic Arabidopsis (He et al., 2001; Gu et al., 2002). Transgenic tobacco (Nicotiana tabacum) plants overexpressing NtERF5, a wound- and pathogen-responsive tobacco gene related to ERF1, showed enhanced resistance to Tobacco mosaic virus (Fischer and Dröge-Laser, 2004). These results are consistent with the view that diverse B3-like TFs, both within Arabidopsis and in other species, have conserved functions in activating plant defenses. It must be noted, however, that the effects from overexpression studies could be the result of nonspecific activation of GCC-box-containing genes due to elevated levels of the overexpressed TFs. Further studies using gene silencing or insertional inactivation (such as that shown here for AtERF4) techniques would allow more specific conclusions to be made about the functions of these genes.
The effects on disease resistance through the activation of JA-responsive defense genes suggest that substantial functional redundancy may exist in the B3 subcluster of genes. Previously, ERF1 was shown to integrate signals from both JA and ET signaling, as mutations in either pathway (i.e. coi1 and ein3) abolish JA- and ET-inducible expression of ERF1 (Lorenzo et al., 2003). Interestingly, through searching a publicly available DNA chip database (Zimmermann et al., 2004), we found that, similar to ERF1, expression of AtERF2 was abolished in the coi1 and ein3 mutants. Furthermore, overexpression of AtERF2 increases sensitivity to JA as measured by root inhibition assays (Fig. 5, A and B), which is also observed in plants overexpressing ERF1 (Lorenzo et al., 2003). This suggests that the expression of both genes is regulated by the same upstream signaling pathways and that both genes activate the same or overlapping patterns of downstream gene expression. It is possible, however, that a certain hierarchy may exist in the signaling pathway leading to the induction of these and other AP2/ERF genes in response to pathogen attack, and that some B3 AP2/ERFs act as the activators of others. In addition, variations in the binding affinities of ERF1 and AtERF2 to different GCC-box motifs (Fujimoto et al., 2000) may allow for some degree of target gene specificity.
Coordination of AP2/ERF Gene Expression Is a Key Aspect of the Plant Defense Response
The coordinated induction of both activator- and repressor-type AP2/ERFs may at first seem to be paradoxical, considering that B3 activators, such as ERF1 and AtERF2, and the B1a repressors, such as AtERF4, most probably compete for the same binding site (i.e. GCC-box). It is not known how the expression and interaction of opposing regulators is coordinated to regulate the wide spectrum of downstream defense genes that are induced by MeJA or pathogen challenge. In tobacco, ETH and fungal elicitors similarly induce expression from both activator- and repressor-type AP2/ERFs (Ohme-Takagi and Shinshi, 1995; Yamamoto et al., 1999). It is possible that the repressor activity of TFs might be down-regulated at a posttranscriptional level, such as the control of protein stability. Supporting this view, an interaction between a repressor-type AP2/ERF (ERF3) and the ubiquitin-conjugating enzyme NtUBC2 has recently been identified in tobacco (Koyama et al., 2003). Therefore, it would be interesting to examine the repressor- and activator-type AP2/ERFs at the protein level, not only to correlate transcript expression with protein abundance but also to test for the possibility of posttranscriptional regulation. Studies at the protein level could also investigate whether the multiple activators and repressors differ in their binding affinities for the GCC-box motif. The preferential binding affinities that some ERF members display for mutant GCC-box motifs has already been analyzed in vitro (Fujimoto et al., 2000).
To summarize, in this study, we undertook a global analysis of TF gene expression to identify candidate genes that may regulate disease resistance. Our functional analysis of selected AP2/ERF genes has highlighted the value of this approach, demonstrating roles for both repressor- and activator-type ERFs in disease resistance and JA signaling. Additionally, several genes identified in our initial screen have also previously been implicated in plant defense (Berrocal-Lobo et al., 2002; Chen and Chen, 2002; Brown et al., 2003; Dong et al., 2003), while many other inducible genes (Supplemental Table I) do not yet have known functions. Further work on these candidate TF genes is likely to reveal other new aspects of the regulation of plant defense and related signaling pathways.
MATERIALS AND METHODS
Plant Growth Conditions, Chemical Treatments, and Pathogen Inoculations
Plant growth conditions, fungal inoculations, and chemical treatments were described previously (Schenk et al., 2000; Campbell et al., 2003; Anderson et al., 2004). Tissue samples were taken after the required times by removing the entire above-ground plant and stored at −80°C.
RT-Q-PCR Analysis
Initial RT-Q-PCR of the 6-h cDNA samples was carried out according to Czechowski et al. (2004), with the primer sequences of the original and additional genes available in Supplemental Table I. All subsequent RT-Q-PCR analysis used for the follow-up time-course experiments was performed at the CSIRO, Brisbane. The RT-Q-PCR parameters used in the initial screening were replicated.
For all data analysis, the PCR primer efficiency (E value) of each primer pair in each individual reaction was calculated from the ΔRn values of each amplification plot using linear regression analysis. The primer efficiencies for each gene were averaged across all samples, except those that showed primer efficiency values with an R2 value of less than 0.999, which were ignored. Amplification plots were analyzed using an Rn threshold of 0.3 to give a cycle threshold value for each gene-cDNA combination. Absolute gene expression levels relative to the housekeeping gene actin-2 (At3g18780) were calculated for each cDNA sample using the equation: relative ratiogene/actin = (Egene−(Ct gene))/(Eactin-2−(Ct actin-2)). The values of the three control and three treated samples were used in a Student's t test to calculate probabilities of distinct induction or repression, and the average ratio of these values was used to determine the fold change in transcript level in treatment samples compared with control.
Sequence Analysis of 155 Arabidopsis AP2 Domains
The amino acid sequences of the AP2 domains (defined by PF00847) were obtained from the genome repository of the Pfam database (Bateman et al., 2004). All sequences were aligned to the AP2 protein family hidden Markov model using hmmalign of the HMMER software package (Eddy, 1998). Neighbor-joining trees of 1,000 bootstrapped samples were constructed (Kimura distance correction method, including gaps). Nomenclature of the AP2/ERF clustering was done according to Sakuma et al. (2002).
Characterization of the erf4-1 Mutant
The location of the T-DNA insertion in the AtERF4 gene (SALK_073394) was verified by using a nested PCR approach (Alonso et al., 2003), and homozygous plants were used in all subsequent experiments.
Construction of the Binary Vectors and Plant Transformation
The construction of the AtERF2 (At5g47220) overexpression construct has been described previously (Brown et al., 2003). The AtERF4 gene (At3g15210; also known as RAP2.5) of Arabidopsis (Arabidopsis thaliana) was PCR amplified from the Col-0 ecotype using the primers AtERF4-F (5′-CGAGAATGGCCAAGATGGGC-3′) and AtERF4-R (5′-GCTCAGGCCTGTTCCGATGG-3′). The amplification product for each gene was cloned into the binary vector pKEN containing Basta resistance in the T-DNA region (Brown et al., 2003). The recombinant plasmid was transformed into Agrobacterium tumefaciens (strain AGL-1) cells containing the pSoup plasmid (Hellens et al., 2000). Arabidopsis Col-0 plants were transformed using the floral-dip transformation procedure. The transgenic plants were selected based on their resistance to Basta. Segregation analysis of the resulting T1 generation using Basta selection allowed for selection of homozygous T2 lines.
RT-Q-PCR Analysis of Gene Expression in erf4-1 and AtERF2- and AtERF4-Overexpressing Lines
cDNAs obtained from untreated or MeJA-treated wild-type and transgenic plants were used in quantification of gene expression. Three independent samples of each control and transgenic line were taken and analyzed. The RT-Q-PCR reaction, as well as the primer sequences for AtERF2, PDF1.2 (At5g44420), and CHIB (At3g12500), and the internal standardization and calculations were performed as described previously (Brown et al., 2003). The primers used to detect transcript levels of AtERF4 were 5′-GACTCTGATTCGTCATCGGTCG-3′ and 5′-AGGCCTGTTCCGATGGAGG-3′.
Fusarium oxysporum Disease Assays
Wild-type and transgenic Arabidopsis (Col-0) plants were grown in soil to the eight- to 10-leaf stage. Inoculation with F. oxysporum was carried out as described by Campbell et al. (2003). Disease development was observed over the following 15 d. Inoculated plants were scored based on the presence or absence of any disease symptoms, including reduced growth or wilting, chlorosis of the leaf vasculature, curling and necrosis of the leaves, or collapse of the petioles. For each of the wild-type and transgenic plant lines, four biological replicates were performed in parallel, with each replicate containing between 24 and 36 plants. In addition, the entire experiment was repeated for confirmation of the original results.
MeJA Root Elongation Inhibition Assays
Seeds of Col-0, AtERF2, and AtERF4 overexpressors, as well as erf4-1, were surface sterilized and plated onto Murashige and Skoog media in either the presence or absence of 50 μm MeJA. At least 40 seeds were used for each plant line. Root length was measured for all germinating seeds over a 2-week period.
Supplementary Material
Acknowledgments
We acknowledge the University of Queensland's Graduate School for a Research Travel Award to K.C.M. We thank Thomas Czechowski and Yves Gibon for assistance with RT-Q-PCR procedures, Anca Rusu and Christina Bakker for technical assistance, and Gang-Ping Xue for critical reading of the manuscript.
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.068544.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Anderson JP, Badruzsaufari E, Schenk PM, Manners JM, Desmond OJ, Ehlert C, Maclean DJ, Ebert PR, Kazan K (2004) Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell 16: 3460–3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer ELL, et al (2004) The Pfam protein families database. Nucleic Acids Res 32: D138–D141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrocal-Lobo M, Molina A (2004) Ethylene response factor 1 mediates Arabidopsis resistance to the soilborne fungus Fusarium oxysporum. Mol Plant Microbe Interact 17: 763–770 [DOI] [PubMed] [Google Scholar]
- Berrocal-Lobo M, Molina A, Solano R (2002) Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J 29: 23–32 [DOI] [PubMed] [Google Scholar]
- Brown RL, Kazan K, McGrath KC, Maclean DJ, Manners JM (2003) A role for the GCC-box in jasmonate-mediated activation of the PDF1.2 gene of Arabidopsis. Plant Physiol 132: 1020–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell EJ, Schenk PM, Kazan K, Penninckx I, Anderson JP, Maclean DJ, Cammue BPA, Ebert PR, Manners JM (2003) Pathogen-responsive expression of a putative ATP-binding cassette transporter gene conferring resistance to the diterpenoid sclareol is regulated by multiple defense signaling pathways in Arabidopsis. Plant Physiol 133: 1272–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Li X, Dong XN (1998) Generation of broad-spectrum disease resistance by overexpression of an essential regulatory gene in systemic acquired resistance. Proc Natl Acad Sci USA 95: 6531–6536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CH, Chen ZX (2002) Potentiation of developmentally regulated plant defense response by AtWRKY18, a pathogen-induced Arabidopsis transcription factor. Plant Physiol 129: 706–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WQ, Provart NJ, Glazebrook J, Katagiri F, Chang HS, Eulgem T, Mauch F, Luan S, Zou GZ, Whitham SA, et al (2002) Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell 14: 559–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Bari RP, Stitt M, Scheible WR, Udvardi MK (2004) Real-time RT-PCR profiling of over 1400 Arabidopsis transcription factors: unprecedented sensitivity reveals novel root- and shoot-specific genes. Plant J 38: 366–379 [DOI] [PubMed] [Google Scholar]
- Delessert C, Kazan K, Wilson IW, Van Der Straeten D, Manners J, Dennis ES, Dolferus R (2005) The transcription factor ATAF2 represses the expression of pathogenesis-related genes in Arabidopsis. Plant J 43: 745–757 [DOI] [PubMed] [Google Scholar]
- Desveaux D, Subramaniam R, Despres C, Mess JN, Levesque C, Fobert PR, Dangl JL, Brisson N (2004) A “whirly” transcription factor is required for salicylic acid-dependent disease resistance in Arabidopsis. Dev Cell 6: 229–240 [DOI] [PubMed] [Google Scholar]
- Dong JX, Chen CH, Chen ZX (2003) Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol Biol 51: 21–37 [DOI] [PubMed] [Google Scholar]
- Eddy S (1998) Profile hidden Markov models. Bioinformatics 14: 755–763 [DOI] [PubMed] [Google Scholar]
- Fischer U, Dröge-Laser W (2004) Overexpression of NtERF5, a new member of the tobacco ethylene response transcription factor family enhances resistance to tobacco mosaic virus. Mol Plant Microbe Interact 17: 1162–1171 [DOI] [PubMed] [Google Scholar]
- Fujimoto SY, Ohta M, Usui A, Shinshi H, Ohme-Takagi M (2000) Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 12: 393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu YQ, Wildermuth MC, Chakravarthy S, Loh YT, Yang CM, He XH, Han Y, Martin GB (2002) Tomato transcription factors Pti4, Pti5, and Pti6 activate defense responses when expressed in Arabidopsis. Plant Cell 14: 817–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutterson N, Reuber TL (2004) Regulation of disease resistance pathways by AP2/ERF transcription factors. Curr Opin Plant Biol 7: 465–471 [DOI] [PubMed] [Google Scholar]
- He P, Warren RF, Zhao TH, Shan LB, Zhu LH, Tang XY, Zhou JM (2001) Overexpression of Pti5 in tomato potentiates pathogen-induced defense gene expression and enhances disease resistance to Pseudomonas syringae pv. tomato. Mol Plant Microbe Interact 14: 1453–1457 [DOI] [PubMed] [Google Scholar]
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM (2000) pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M (2003) Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J 34: 733–739 [DOI] [PubMed] [Google Scholar]
- Horak CE, Snyder M (2002) Global analysis of gene expression in yeast. Funct Integr Genomics 2: 171–180 [DOI] [PubMed] [Google Scholar]
- Kariola T, Brader G, Li J, Palva ET (2005) Chlorophyllase 1, a damage control enzyme, affects the balance between defense pathways in plants. Plant Cell 17: 282–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T, Okada T, Kitajima S, Ohme-Takagi M, Shinshi H, Sato F (2003) Isolation of tobacco ubiquitin-conjugating enzyme cDNA in a yeast two-hybrid system with tobacco ERF3 as bait and its characterization of specific interaction. J Exp Bot 54: 1175–1181 [DOI] [PubMed] [Google Scholar]
- Li J, Brader G, Palva ET (2004) The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 16: 319–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu GS, Holub EB, Alonso JM, Ecker JR, Fobert PR (2005) An Arabidopsis NPR1-like gene, NPR4, is required for disease resistance. Plant J 41: 304–318 [DOI] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1998) Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10: 1391–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O, Chico JM, Sanchez-Serrano JJ, Solano R (2004) Jasmonate-insensitive1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16: 1938–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O, Piqueras R, Sanchez-Serrano JJ, Solano R (2003) ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15: 165–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M, Tang X, Zhou J-M (2001) Arabidopsis NHO1 is required for general resistance against Pseudomonas bacteria. Plant Cell 13: 437–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell JM, Cuzick A, Can C, Beynon J, Dangl JL, Holub EB (2000) Downy mildew (Peronospora parasitica) resistance genes in Arabidopsis vary in functional requirements for NDR1, EDS1, NPR1 and salicylic acid accumulation. Plant J 22: 523–529 [DOI] [PubMed] [Google Scholar]
- Mengiste T, Chen X, Salmeron J, Dietrich R (2003) The BOTRYTIS SUSCEPTIBLE1 gene encodes an R2R3MYB transcription factor protein that is required for biotic and abiotic stress responses in Arabidopsis. Plant Cell 15: 2551–2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohme-Takagi M, Shinshi H (1995) Ethylene-inducible DNA-binding proteins that interact with an ethylene-responsive element. Plant Cell 7: 173–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M (2001) Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13: 1959–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onate-Sanchez L, Singh KB (2002) Identification of Arabidopsis ethylene-responsive element binding factors with distinct induction kinetics after pathogen infection. Plant Physiol 128: 1313–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx I, Thomma B, Buchala A, Metraux JP, Broekaert WF (1998) Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 10: 2103–2113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe OJ, Riechmann JL (2002) Arabidopsis transcription factors and the regulation of flowering time: a genomic perspective. Curr Issues Mol Biol 4: 77–91 [PubMed] [Google Scholar]
- Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K (2002) DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem Biophys Res Commun 290: 998–1009 [DOI] [PubMed] [Google Scholar]
- Scheible WR, Morcuende R, Czechowski T, Fritz C, Osuna D, Palacios-Rojas N, Schindelasch D, Thimm O, Udvardi MK, Stitt M (2004) Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol 136: 2483–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk PM, Kazan K, Manners JM, Anderson JP, Simpson RS, Wilson IW, Somerville SC, Maclean DJ (2003) Systemic gene expression in Arabidopsis during an incompatible interaction with Alternaria brassicicola. Plant Physiol 132: 999–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk PM, Kazan K, Wilson I, Anderson JP, Richmond T, Somerville SC, Manners JM (2000) Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc Natl Acad Sci USA 97: 11655–11660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh KB, Foley RC, Onate-Sanchez L (2002) Transcription factors in plant defense and stress responses. Curr Opin Plant Biol 5: 430–436 [DOI] [PubMed] [Google Scholar]
- Solano R, Stepanova A, Chao QM, Ecker JR (1998) Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev 12: 3703–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel SH, Koornneef A, Claessens SMC, Korzelius JP, Van Pelt JA, Mueller MJ, Buchala AJ, Metraux JP, Brown R, Kazan K, et al (2003) NPR1 modulates cross-talk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell 15: 760–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Yuen GY, Lehman CC (1998) Jasmonate signaling mutants of Arabidopsis are susceptible to the soil fungus Pythium irregulare. Plant J 15: 747–754 [DOI] [PubMed] [Google Scholar]
- Takahashi H, Kanayama Y, Zheng MS, Kusano T, Hase S, Ikegami M, Shah J (2004) Antagonistic interactions between the SA and JA signaling pathways in Arabidopsis modulate expression of defense genes and gene-for-gene resistance to cucumber mosaic virus. Plant Cell Physiol 45: 803–809 [DOI] [PubMed] [Google Scholar]
- Thomma B, Eggermont K, Penninckx I, Mauch-Mani B, Vogelsang R, Cammue BPA, Broekaert WF (1998) Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc Natl Acad Sci USA 95: 15107–15111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle TJ (2004) Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 16: 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toufighi K, Brady SM, Austin R, Ly E, Provart NJ (2005) The botany array resource: e-northerns, expression angling, and promoter analyses. Plant J 43: 153–163 [DOI] [PubMed] [Google Scholar]
- Tsukagoshi H, Saijo T, Shibata D, Morikami A, Nakamura K (2005) Analysis of a sugar response mutant of Arabidopsis identified a novel B3 domain protein that functions as an active transcriptional repressor. Plant Physiol 138: 675–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vailleau F, Daniel X, Tronchet M, Montillet J-L, Triantaphylides C, Roby D (2002) A R2R3-MYB gene, AtMYB30, acts as a positive regulator of the hypersensitive cell death program in plants in response to pathogen attack. Proc Natl Acad Sci USA 99: 10179–10184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel RR, Pfitzner UM, Gatz C (2005) Interaction of NIMIN1 with NPR1 modulates PR gene expression in Arabidopsis. Plant Cell 17: 1279–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S, Suzuki K, Shinshi H (1999) Elicitor-responsive, ethylene-independent activation of GCC box-mediated transcription that is regulated by both protein phosphorylation and dephosphorylation in cultured tobacco cells. Plant J 20: 571–579 [DOI] [PubMed] [Google Scholar]
- Yang Z, Tian L, Latoszek-Green M, Brown D, Wu K (2005) Arabidopsis ERF4 is a transcriptional repressor capable of modulating ethylene and abscisic acid responses. Plant Mol Biol 58: 585–596 [DOI] [PubMed] [Google Scholar]
- Zimmerli L, Stein M, Lipka V, Schulze-Lefert P, Somerville S (2004) Host and non-host pathogens elicit different jasmonate/ethylene responses in Arabidopsis. Plant J 40: 633–646 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.