Abstract
Our previous research characterized two phosphoenolpyruvate (PEP) carboxylase (PEPC) isoforms (PEPC1 and PEPC2) from developing castor oil seeds (COS). The association of a shared 107-kD subunit (p107) with an immunologically unrelated bacterial PEPC-type 64-kD polypeptide (p64) leads to marked physical and kinetic differences between the PEPC1 p107 homotetramer and PEPC2 p107/p64 heterooctamer. Here, we describe the production of antiphosphorylation site-specific antibodies to the conserved p107 N-terminal serine-6 phosphorylation site. Immunoblotting established that the serine-6 of p107 is phosphorylated in COS PEPC1 and PEPC2. This phosphorylation was reversed in vitro following incubation of clarified COS extracts or purified PEPC1 or PEPC2 with mammalian protein phosphatase type 2A and is not involved in a potential PEPC1 and PEPC2 interconversion. Similar to other plant PEPCs examined to date, p107 phosphorylation increased PEPC1 activity at pH 7.3 by decreasing its Km(PEP) and sensitivity to l-malate inhibition, while enhancing glucose-6-P activation. By contrast, p107 phosphorylation increased PEPC2's Km(PEP) and sensitivity to malate, glutamic acid, and aspartic acid inhibition. Phosphorylation of p107 was promoted during COS development (coincident with a >5-fold increase in the I50 [malate] value for total PEPC activity in desalted extracts) but disappeared during COS desiccation. The p107 of stage VII COS became fully dephosphorylated in planta 48 h following excision of COS pods or following 72 h of dark treatment of intact plants. The in vivo phosphorylation status of p107 appears to be modulated by photosynthate recently translocated from source leaves into developing COS.
Phosphoenolpyruvate (PEP) carboxylase (PEPC; E.C. 4.1.1.31) is a ubiquitous and tightly regulated cytosolic enzyme in vascular plants that is also widely distributed in green algae and bacteria. PEPC catalyzes the irreversible β-carboxylation of PEP to yield oxaloacetate and inorganic phosphate (Pi). The enzyme plays a pivotal photosynthetic role in primary CO2 fixation by C4 and Crassulacean acid metabolism leaves (Chollet et al., 1996; Izui et al., 2004; Nimmo, 2005). PEPC also has a variety of additional important functions in plants, particularly the anaplerotic replenishment of tricarboxylic acid cycle intermediates that are withdrawn for biosynthesis and nitrogen assimilation. Most native plant PEPCs are homotetramers, composed of identical subunits of approximately 100 to 110 kD, with crystal structures having been established for both the maize (Zea mays) and Escherichia coli enzymes (Izui et al., 2004). All PEPCs show allosteric properties: Vascular plant PEPCs are typically inhibited by l-malate and activated by Glc-6-P. In addition, Asp and Glu are allosteric inhibitors of PEPCs in plant tissues active in nitrogen assimilation and/or transamination reactions, thus providing a link between carbon and nitrogen metabolism (Law and Plaxton, 1997; Golombek et al., 1999; Moraes and Plaxton, 2000; Blonde and Plaxton, 2003). It is also well established that the activity of most vascular plant PEPCs is controlled by reversible phosphorylation catalyzed by an endogenous Ca2+-independent PEPC protein kinase, and dephosphorylation by a protein phosphatase type 2A (PP2A) at a highly conserved n-terminal seryl residue (Chollet et al., 1996; Izui et al., 2004; Nimmo, 2005). This usually results in reduced sensitivity of the enzyme to malate inhibition and increased sensitivity to activation by Glc-6-P. The posttranslational control of plant PEPC by reversible phosphorylation has been best characterized in Crassulacean acid metabolism and C4 leaves (Chollet et al., 1996; Izui et al., 2004; Nimmo, 2005), and, to a lesser extent, in C3 leaves and soybean root nodules (Duff and Chollet, 1995; Zhang et al., 1995; Li et al., 1996; Wadham et al., 1996; Zhang and Chollet, 1997; Xu et al., 2003; Sullivan et al., 2004). However, regulatory PEPC phosphorylation in germinated wheat (Triticum aestivum), sorghum (Sorghum bicolor), and barley (Hordeum vulgare) seeds and ripening banana (Musa spp.) fruit has also been reported (Osuna et al., 1996, 1999; Law and Plaxton, 1997; Nhiri et al., 2000).
In developing seeds, the partitioning of imported photosynthate between starch, storage lipid, and storage protein biosynthesis is of great agronomic concern because seeds are a major source of plant-derived nutrients for the worldwide feed and food industry. PEP metabolism via PEPC and cytosolic pyruvate kinase (PKc; E.C. 2.7.1.40) appears to play a prominent role in partitioning seed carbohydrates toward plastidic fatty acid biosynthesis versus the mitochondrial production of ATP and organic acids required for amino acid interconversion in support of storage protein biosynthesis (Ruuska et al., 2002; Schwender et al., 2004). PKc catalyzes the irreversible substrate level of phosphorylation of ADP to ATP with the concomitant conversion of PEP to pyruvate. PEPC and PKc activities are abundant in developing seeds and represent promising targets for metabolic engineering in this tissue (Sangwan et al., 1992; Golombek et al., 1999; Blonde and Plaxton, 2003; Turner et al., 2005; Weber et al., 2005). However, our understanding of the biochemical and molecular properties of PEPC and PKc in developing seeds is relatively sparse. We recently described the physical, immunological, and kinetic/regulatory properties of purified PKc and PEPC from the triglyceride-rich endosperm of developing castor oil seeds (COS; Blonde and Plaxton, 2003; Turner et al., 2005). The combined results suggested an important role for pH and the allosteric effectors Glu and Asp in the coordinate control of the PEP branch point (i.e. PKc versus PEPC) in the developing COS cytosol.
During PEPC purification from developing COS, low- and high-Mr PEPC isoforms, respectively termed PEPC1 and PEPC2, were unexpectedly resolved via gel filtration FPLC (Blonde and Plaxton, 2003). COS PEPC1 and PEPC2 were shown to bear striking physical and kinetic similarities to previously characterized class 1 and class 2 PEPCs from the unicellular green algae Selenastrum minutum and Chlamydomonas reinhardtii (Blonde and Plaxton, 2003; Rivoal et al., 1996, 1998, 2001). The smaller COS PEPC1 exists as a typical 410-kD PEPC homotetramer of 107-kD subunits (p107), whereas the COS PEPC2 exists as an unusual 681-kD heterooctamer that contains the same p107 found in PEPC1 associated with an immunologically unrelated 64-kD polypeptide (p64). Several tryptic peptides from the p64 of COS PEPC2 were sequenced by quadrupole time-of-flight mass spectrometry, and all were shown to be very similar or identical to portions of the deduced sequences of Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa) bacterial-like PEPCs revealed through recent genome sequencing initiatives (Blonde and Plaxton, 2003; Sánchez and Cejudo, 2003). Similarly, the unique green algal high-Mr class 2 PEPCs arise from a tight physical association between immunologically unrelated eukaryotic- and bacterial-like PEPC polypeptides (Rivoal et al., 1996, 1998, 2001; Mamedov et al., 2005). The association of the shared p107 subunit with p64 in COS PEPC2 leads to marked physical and kinetic differences between the PEPC1 p107 homotetramer and the novel PEPC2 p107/p64 heterooctamer. We hypothesized that PEPC1 (activated by Glc-6-P; inhibited by malate, Asp, and Glu) and PEPC2 (relatively insensitive to allosteric effectors compared to PEPC1), respectively, support storage protein versus storage lipid synthesis in developing COS (Blonde and Plaxton, 2003). Exogenous l-malate supported maximal rates of fatty acid synthesis by purified leucoplasts from developing COS (Smith et al., 1992), and malate import from the cytosol into the COS leucoplast stroma is catalyzed by a malate/Pi translocator within the leucoplast envelope (Eastmond et al., 1997).
The possibility that developing COS PEPC1 and/or PEPC2 might also be controlled by reversible phosphorylation was indicated by (1) the presence of the highly conserved n-terminal regulatory seryl phosphorylation site in p107 that is characteristic of most plant PEPCs studied to date (Blonde and Plaxton, 2003); and (2) the report that the PEPC activity of Vicia faba cotyledons showed a progressive diminished sensitivity to malate inhibition as seed development proceeded (Golombek et al., 1999). However, direct evidence for PEPC phosphorylation in maturing seeds has not been obtained. This study addresses this deficiency through the use of antiphosphorylation site-specific antibodies (APS-IgG), which have been an invaluable tool for investigation of eukaryotic protein kinase cascades and associated signaling pathways, including regulatory phosphorylation of the C4 leaf (Pacquit et al., 1995; Ueno et al., 2000) and germinating wheat, barley, and sorghum seed PEPCs (Osuna et al., 1996, 1999; Nhiri et al., 2000). Here, we describe the production of affinity-purified APS-IgG to the conserved n-terminal phosphorylation site (Ser-6) of COS p107. The APS-IgG was shown to be useful for monitoring the phosphorylation status of p107 in COS PEPC1 and PEPC2 under various developmental and physiological conditions. Our results indicate that in vivo phosphorylation of p107 in COS PEPC1 and PEPC2 is likely modulated by the supply of photosynthate recently translocated from leaves.
RESULTS
Specificity of APS-IgG
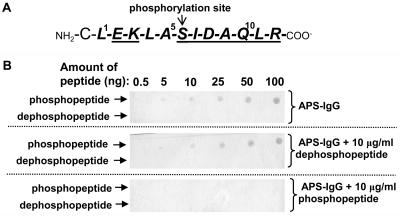
To determine whether Ser-6 is phosphorylated in COS p107, we generated a phosphospecific antibody using a synthetic phosphopeptide corresponding to the conserved n-terminal sequence of p107 (Fig. 1A). As shown in Fig. 1B, the affinity-purified APS-IgG detected as little as 10 ng of the phosphopeptide, but failed to cross-react with up to 100 ng of the corresponding dephosphopeptide. Moreover, the cross-reaction between the APS-IgG and the phosphopeptide was abolished when the dot blot was incubated with APS-IgG in the presence of the blocking phosphopeptide. By contrast, the addition of the dephosphopeptide exerted no influence on the cross-reactivity of the APS-IgG with the phosphopeptide (Fig. 1B).
Figure 1.
Specificity of APS-IgG. A, Sequence of phosphorylated synthetic peptide that was covalently coupled to KLH and used for rabbit immunization. The sequence numbering represents amino acid position relative to the N terminus of the shared p107 subunit of purified COS PEPC1 and PEPC2 (indicated in bold; Blonde and Plaxton, 2003). The peptide was synthesized with an extra N-terminal Cys residue to facilitate its conjugation with KLH prior to rabbit immunization. Underlined letters denote the highly conserved consensus target sequence for plant PEPC kinase (Chollet et al., 1996). The seryl phosphorylation site is indicated. B, Dot blots of varying amounts of the synthetic phosphopeptide and corresponding dephosphopeptide were probed with 10 μg/mL of the affinity-purified APS-IgG in the presence and absence of 10 μg/mL of dephosphopeptide or corresponding phosphopeptide.
p107 Phosphorylation in PEPC1 and PEPC2 from Stage VII Developing COS
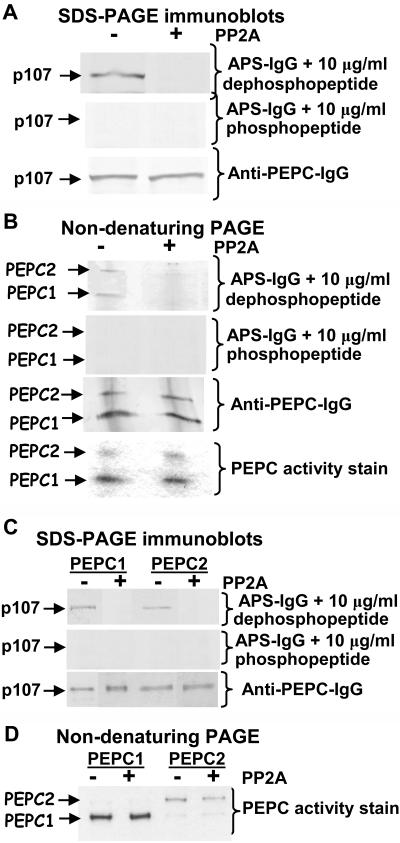
The use of the APS-IgG for examining p107 phosphorylation in COS PEPC1 and PEPC2 was complemented with an affinity-purified polyclonal antibody raised against purified PEPC from Brassica napus suspension cell cultures (Moraes and Plaxton, 2000). This latter antibody, which detects both phosphorylated and dephosphorylated forms of PEPC, allowed for the normalization of total PEPC on immunoblots. The 410-kD PEPC1 homotetramer and 681-kD PEPC2 heterooctamer of stage VII (midcotyledon) developing COS can be readily separated via nondenaturing PAGE (Blonde and Plaxton, 2003). Thus, both SDS-PAGE and nondenaturing PAGE immunoblots of clarified extracts from stage VII developing COS were incubated with the APS-IgG in the presence of the corresponding dephosphopeptide or blocking phosphopeptide (Fig. 2, A and B). Results presented in Figure 2A show that COS p107 is phosphorylated at Ser-6 and that the APS-IgG is indeed specific for the phosphorylated p107 as the signal was eliminated by PP2A pretreatment or incubation with the blocking phosphopeptide. Nondenaturing PAGE of a clarified extract from stage VII COS followed by immunoblotting demonstrated that p107 is phosphorylated in both PEPC1 and PEPC2 (Fig. 2B). Laser densitometry of the immunoblots indicated that the p107 of PEPC1 is significantly (up to 50%) more phosphorylated relative to the p107 of PEPC2. Although PP2A treatment (20 min) fully dephosphorylated the p107 of both PEPC isoforms present in the COS extract, it did not influence the proportion of PEPC1 relative to PEPC2 as determined by nondenaturing PAGE followed by immunoblotting with the anti-(B. napus PEPC)-IgG or in-gel PEPC activity staining (Fig. 2B).
Figure 2.
p107 exists as a phosphoprotein in the PEPC1 homotetramer or PEPC2 heterooctamer present in clarified extracts or purified from endosperm of stage VII developing COS. A clarified COS extract (A and B) or purified COS PEPC1 and PEPC2 (C and D) were subjected to SDS-PAGE (10% gel; A and C) or nondenaturing PAGE (7% gel; B and D) before and following a 20-min incubation with 5 milliunits/mL of bovine heart PP2A as indicated. Gels were electroblotted onto a PVDF membrane. The immunoblots (A–C) were probed with either 10 μg/mL APS-IgG in the presence of 10 μg/mL of the corresponding dephosphopeptide or phosphopeptide, or affinity-purified anti-(B. napus PEPC)-IgG (labeled as anti-PEPC-IgG; Moraes and Plaxton, 2000). In-gel PEPC activity staining was also performed following nondenaturing PAGE (B and D) using a fast violet B method (Blonde and Plaxton, 2003). Protein loading was as follows: clarified extracts, 40 μg/lane (A and B); purified PEPC1 and PEPC2, 50 and 300 ng/lane for SDS-PAGE immunoblots probed with anti-PEPC-IgG and APS-IgG, respectively (C); or 1 μg purified PEPC1 or 0.5 μg purified PEPC2 for in-gel PEPC activity staining (D).
Immunoblot analyses were also employed to assess whether the Ser-6 of p107 is phosphorylated in either PEPC isoform purified to homogeneity from stage VII developing COS. The results corroborate those of Figure 2, A and B, and demonstrate that, although p107 is phosphorylated in both purified PEPCs (Fig. 2C), the p107 of PEPC2 was about 50% less phosphorylated relative to the p107 of PEPC1 (as indicated by laser densitometry).
Influence of p107 Phosphorylation Status on Activity and Oligomeric Structure of Purified PEPC1 and PEPC2 from Stage VII Developing COS
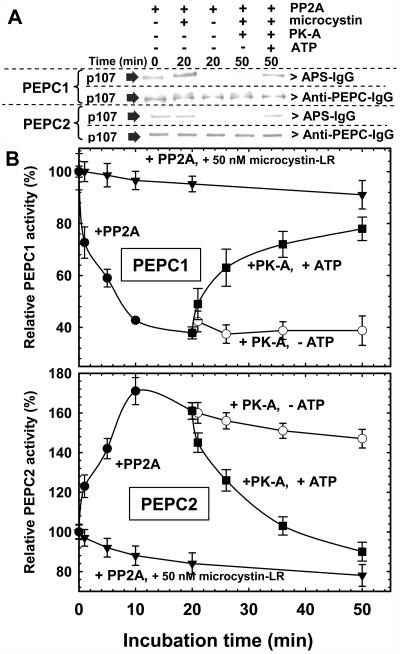
The p107 subunit of the COS PEPC1 homotetramer and PEPC2 heterooctamer was dephosphorylated and subsequently rephosphorylated when either purified enzyme from stage VII COS was incubated with bovine heart PP2A for 20 min, followed by the catalytic subunit of porcine heart cAMP-dependent protein kinase (PK-A) and MgATP for an additional 30 min (Fig. 3A). No detectable change in p107 phosphorylation occurred when either PEPC isoform was incubated in the presence of PP2A and 50 nm microcystin-LR (a specific and potent inhibitor of animal and plant protein phosphatase type 1 and PP2A activities; Fig. 3A), or when the non-PP2A-treated PEPC1 or PEPC2 preparations were incubated with PK-A and MgATP for up to 50 min (data not shown).
Figure 3.
Dephosphorylation and subsequent rephosphorylation of purified PEPC1 and PEPC2 from stage VII developing COS by exogenous mammalian PP2A and PK-A, respectively. A, Immunoblot analysis was performed using APS-IgG or anti-(B. napus PEPC)-IgG (300 and 50 ng PEPC1 or PEPC2 loaded/lane, respectively) and chromogenic staining as in Blonde and Plaxton (2003). B, Time course for PP2A-dependent inhibition, and PK-A and ATP-dependent reactivation of PEPC1 and PEPC2. PEPC activity was determined using suboptimal conditions (pH 7.3 and 0.2 mm PEP, but 0.125 and 1.25 mm malate for PEPC1 and PEPC2, respectively). All values represent mean ± se for n = 3 separate determinations.
No influence of PP2A-mediated dephosphorylation on enzymatic activity occurred when PEPC1 or PEPC2 were assayed under optimal conditions (i.e. pH 8.0, 2 mm PEP; data not shown). However, PEPC1 dephosphorylation caused an approximate 60% decrease in PEPC1 activity when assayed under suboptimal conditions (i.e. pH 7.3, 0.2 mm PEP, 0.125 mm malate; Fig. 3B), without influencing the oligomeric structure of the native homotetrameric enzyme (Fig. 2D). PEPC1 activity recovered to about 75% of its initial activity when the dephosphorylated enzyme was rephosphorylated with the catalytic subunit of PK-A (Fig. 3B). The results suggest that one kinetic effect of COS PEPC1 phosphorylation, shared by all vascular plant PEPCs examined to date (Duff and Chollet, 1995; Zhang et al., 1995; Chollet et al., 1996; Li et al., 1996; Law and Plaxton, 1997; Izui et al., 2004; Nimmo, 2005), is to decrease enzyme sensitivity to inhibition by l-malate. This is supported by the observation that, when assayed at pH 7.3 with subsaturating PEP, the PEPC1 I50(malate) value was reduced by over 250% following its dephosphorylation by PP2A (Table I). In addition, dephosphorylated PEPC1 displayed reduced activation by Glc-6-P and a significantly higher Km(PEP) at pH 7.3 relative to phosphorylated PEPC1 (Table I).
Table I.
Effect of PP2A-mediated p107 dephosphorylation on kinetic constants of PEPC1 and PEPC2 from stage VII developing COS
Purified PEPC1 and PEPC2 were incubated for 20 min with 5 milliunits/mL of PP2A in the standard dephosphorylation assay buffer. Assays were conducted at pH 7.3. I50 and Ka values were determined with subsaturating PEP (0.6 and 0.2 mm for PEPC1 and PEPC2, respectively). All values represent the mean of at least three separate experiments and are reproducible to within ±10% SE of the mean value.
| Kinetic Parameter
|
PEPC1
|
PEPC2
|
||
|---|---|---|---|---|
| Phospho | Dephospho | Phospho | Dephospho | |
| mm | ||||
| Km(PEP) | 0.55 | 0.92 | 0.28 | 0.18 |
| I50(malate) | 0.075 | 0.029 | 0.57 | 1.47 |
| I50(glutamate) | 2.1 | 2.2 | 4.1 | 7.0 |
| I50(aspartate) | 0.35 | 0.32 | 2.6 | 4.5 |
| Ka(Glc-6-P) | 0.22 (2.0)a | 0.25 (1.5) | n.d.b | n.d. |
Values in parenthesis indicate maximal fold activation of PEPC1 by saturating Glc-6-P.
n.d., Not determined (activity of phosphorylated [phospho] or dephosphorylated [dephospho] PEPC2 is essentially unresponsive to Glc-6-P).
In agreement with the results of Figure 2B, PP2A-catalyzed p107 dephosphorylation did not influence the native Mr of purified COS PEPC2 (approximately 680 kD; Blonde and Plaxton, 2003) as shown by nondenaturing PAGE followed by in-gel PEPC2 activity staining (Fig. 2D), as well as analytical gel filtration FPLC on a calibrated Superose 6 HR10/30 column (data not shown). However, the kinetic studies unexpectedly revealed that, when assayed under suboptimal conditions (i.e. pH 7.3, 0.2 mm PEP, 1.25 mm malate), p107 dephosphorylation exerted a reciprocal influence on the activity of purified PEPC2 relative to PEPC1 (Fig. 3B). In contrast to PEPC1, PP2A-dependent p107 dephosphorylation caused an approximate 60% increase in PEPC2 activity (Fig. 3B). Rephosphorylation of dephosphorylated p107 in PEPC2 with the catalytic subunit of PK-A caused a time-dependent reduction of PEPC2 activity to its initial state. Dephosphorylated PEPC2 displayed 2- to 2.5-fold greater I50 values for malate, Glu, and Asp, and a significantly lower Km(PEP) value (Table I). Similar findings were obtained following PP2A-catalyzed dephosphorylation of an independent preparation of purified PEPC2 from stage VII COS (data not shown).
Phosphorylation of p107 Is Promoted during COS Development But Absent in the Mature Seed
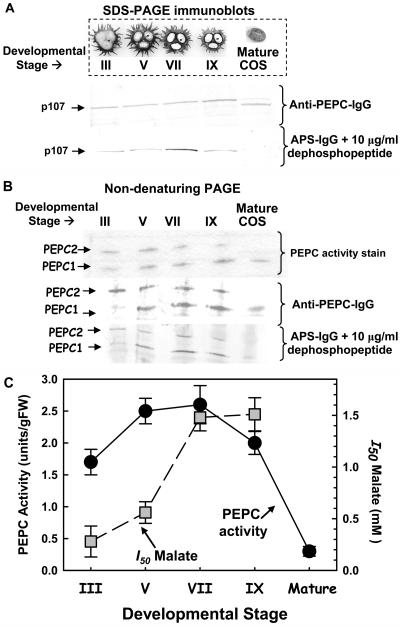
SDS-PAGE followed by immunoblotting of clarified endosperm extracts with the APS-IgG indicated that, although p107 is phosphorylated in vivo throughout seed development, it appears to be maximally phosphorylated in stage VII (full cotyledon) COS, and then becomes undetectable in the dry (fully mature) COS (Fig. 4A). Parallel immunoblots probed with anti-(B. napus PEPC)-IgG (Fig. 4A) agreed with our previous study (Blonde and Plaxton, 2003), suggesting that, during the desiccation phase, a polypeptide of about 120 amino acids is proteolytically cleaved from the n-terminal end of p107 in vivo to yield a truncated 98-kD polypeptide. Nondenaturing PAGE of clarified extracts followed by in-gel PEPC activity staining or immunoblotting with the anti-(B. napus PEPC)-IgG also corroborated our earlier study (Blonde and Plaxton, 2003) and indicated that the ratio of PEPC1 to PEPC2 progressively increases during COS development. PEPC2 was mainly detected in the younger stage III (heart-shaped embryo) COS, whereas only PEPC1 was present at maturity (Fig. 4B). Nondenaturing PAGE immunoblots of clarified extracts probed with the anti-APS-IgG suggested that p107 phosphorylation in PEPC2 is relatively constant in stage III to IX developing COS, whereas in PEPC1 it increases to a maximum at stages VII to IX (Fig. 4, A and B). Enhanced p107 phosphorylation during COS development was coincident with a marked increase in the I50(malate) for total PEPC activity present in desalted extracts (Fig. 4C).
Figure 4.
Developmental profiles of relative p107 phosphorylation (A and B), and extractable PEPC activity and sensitivity to malate inhibition (C) during COS development. A and B, Clarified COS endosperm extracts were subjected to SDS-PAGE (10% gel; A) or nondenaturing PAGE (7% gel; B) and immunoblotting with anti-(B. napus PEPC)-IgG or anti-APS-IgG (40 μg protein/lane). In-gel PEPC activity staining was also performed following nondenaturing PAGE (B). Inset in A shows a photograph of cross-sections of COS pods at developmental stages III, V, VII, and IX that respectively correspond to the heart-shaped embryo, midcotyledon, full cotyledon, and maturation phases of development (Greenwood and Bewley, 1982). C, Relationship between maximal PEPC activity (measured under optimal assay conditions at pH 8.0) and corresponding I50(malate) (measured at pH 7.3, 0.2 mm PEP) for total PEPC activity in the respective desalted clarified extracts (means ± se of n = 3 separate extracts). PEPC activity of mature COS was too low to allow accurate I50(malate) determinations for this tissue.
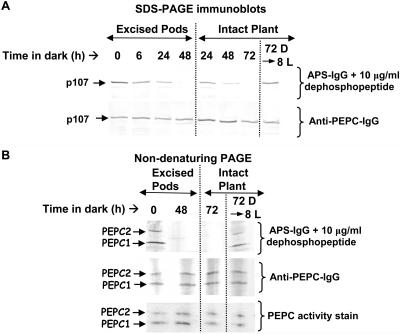
COS Pod Excision or Extended Darkness of Intact Plants Provokes in Vivo p107 Dephosphorylation in Stage VII Developing COS
Shoot decapitation, stem girdling, or prolonged darkness increased the malate sensitivity (reduced phosphorylation) of soybean root nodule PEPC due to the down-regulation of PEPC kinase activity caused by the elimination of photosynthate supply (Zhang et al., 1995; Wadham et al., 1996; Zhang and Chollet, 1997; Xu et al., 2003; Sullivan et al., 2004). Similarly, the p107 of PEPC1 and PEPC2 from stage VII COS progressively became fully dephosphorylated in planta 48 h following pod excision or 72 h following dark treatment of intact castor plants (Fig. 5, A and B). However, neither pretreatment influenced the amount or the proportion of PEPC1 to PEPC2 in stage VII developing COS as assessed by nondenaturing PAGE followed by immunoblotting with the anti-(B. napus PEPC)-IgG or in-gel PEPC activity staining (Fig. 5B). Likewise, optimal PEPC activity, which is not influenced by the phosphorylation status of p107 in PEPC1 or PEPC2 (see above), and levels of total soluble protein demonstrated no change in stage VII endosperms from these pretreated plants (data not shown). Thus, these various pretreatments do not cause global effects on COS PEPC isoforms, but appear to specifically perturb the phosphorylation status of their mutual p107 subunit.
Figure 5.
Influence of seed pod excision or prolonged dark treatment of intact plants on p107 phosphorylation in stage VII developing COS. Stems containing intact pods of developing COS were excised and placed in water in the dark at 24°C. Alternately, intact plants cultivated under a 16-h-light/8-h-dark regime in a growth chamber as described in “Materials and Methods” were subjected to darkness for 72 h and then illuminated for an additional 8 h (72 D → 8 L). At the indicated times, endosperm tissue from stage VII COS was harvested, extracted, and subjected to SDS-PAGE (A) or nondenaturing PAGE (B) and immunoblotting with anti-(B. napus PEPC)-IgG or APS-IgG (40 μg protein/lane). In-gel PEPC activity staining was also performed following nondenaturing-PAGE (B).
DISCUSSION
This study builds upon our previous examination of the molecular and kinetic properties of unusual high- and low-molecular-mass PEPC isoforms in developing COS and unicellular green algae (Blonde and Plaxton, 2003; Rivoal et al., 1996, 1998, 2001, 2002b). The production of APS-IgG against the conserved n-terminal seryl phosphorylation site of COS p107 (Fig. 1A) has provided a direct and relatively straightforward means to assess the in vivo phosphorylation status of the shared p107 subunit of the PEPC1 homotetramer and novel PEPC2 heterooctamer of developing COS. The results clearly establish the existence of the regulatory phosphorylation of a C3 plant PEPC by a protein Ser/Thr kinase in developing COS.
Specificity of the affinity-purified APS-IgG for the corresponding phosphopeptide was demonstrated by probing dot blots of various amounts of the phosphopeptide and equivalent dephosphopeptide (Fig. 1B). Subsequent results demonstrated that p107 in clarified extracts from stage VII COS is phosphorylated at Ser-6 and that the APS-IgG is indeed specific for the phosphorylated form of p107, as the signal was nullified following mammalian PP2A pretreatment or by incubation with the blocking phosphopeptide (Fig. 2, A and B). PP2A catalyzes the dephosphorylation of several cytosolic regulatory enzymes of plant carbon and nitrogen metabolism, including phosphorylated PEPCs from various green and non-green plant tissues (Chollet et al., 1996; Law and Plaxton, 1997; Nimmo, 2005). Nondenaturing PAGE immunoblots of stage VII COS extracts revealed that p107 is phosphorylated in both the 410-kD PEPC1 homotetramer and the 680-kD PEPC2 heterooctamer (Fig. 2B). Analogous results were obtained with PEPC1 and PEPC2 purified to homogeneity from stage VII COS (Figs. 2C and 3A). As the native Mr of PEPC1 or PEPC2 or proportion of PEPC1 to PEPC2 was not influenced following in vitro or in vivo p107 dephosphorylation (Figs. 2D and 5B), reversible p107 phosphorylation does not appear to be involved in a potential interconversion of PEPC1 and PEPC2.
The p107 of both purified COS PEPC isoforms was dephosphorylated and subsequently rephosphorylated in vitro when the final preparations were incubated with mammalian PP2A for 20 min, followed by the catalytic subunit of mammalian PK-A and ATP (Fig. 3A). Densitometric analysis of the immunoblots indicated that the p107 polypeptide of the COS PEPC2 heterooctamer contains a lower (approximately 50%) stoichiometric incorporation of Pi relative to that of the PEPC1 homotetramer. Nevertheless, the p64 subunit in the PEPC2 heterooctamer must interact with p107 so as to maintain the p107 n-terminal Ser-6 phosphorylation site accessible to the solvent. No kinetic effect of p107 dephosphorylation/rephosphorylation was discernible when either COS PEPC isoform was assayed under optimal conditions (i.e. pH 8.0, 2 mm PEP). However, PEPC1 dephosphorylation resulted in a time-dependent 60% reduction in its activity within 20 min following the addition of mammalian PP2A when assayed under suboptimal conditions (Fig. 3B). Additional kinetic studies revealed that phosphorylated PEPC1 displayed a 2.5-fold greater I50(malate) value, enhanced activation by Glc-6-P, and a significantly lower Km(PEP) at pH 7.3 (Table I). Comparable kinetic results have been reported for phosphorylated PEPCs from various green and non-green plant tissues (Duff and Chollet, 1995; Zhang et al., 1995; Chollet et al., 1996; Li et al., 1996; Law and Plaxton, 1997; Izui et al., 2004; Nimmo, 2005). By contrast, PP2A-mediated dephosphorylation of p107 in PEPC2 resulted in a marked time-dependent increase in PEPC activity, which was reversed following subsequent p107 rephosphorylation by PK-A (Fig. 3B). These results are unprecedented for vascular plant PEPC. However, they are quite reminiscent of the analogous high-molecular-mass class 2 PEPC isoforms of the unicellular green algae S. minutum, which were also reported to be less active in their phosphorylated state (Rivoal et al., 2002b). It is intriguing that the p64-associating polypeptide of the COS PEPC2 heterooctamer not only desensitizes the enzyme to allosteric effectors (relative to PEPC1; Table I; Blonde and Plaxton, 2003), but also appears to mediate a reciprocal kinetic effect to PEPC1 when their shared p107 subunit is phosphorylated on its conserved n-terminal Ser-6 residue.
The in vivo physiological significance of the aforementioned results is difficult to assess without knowledge of (1) possible PEPC1 and PEPC2 microcompartmentation and protein-protein interactions within the cytosol of developing COS; (2) in vivo cytosolic concentrations of PEP and PEPC allosteric effectors in the PEPC1 and PEPC2 respective microenvironments; and (3) how these parameters may be modulated during COS development or in response to treatments that disrupt photosynthate import into this tissue. The concentration of malate within the developing endosperm of midcotyledon (stage V) developing COS was estimated by Smith and coworkers (1992) to be about 5 mm. Although the subcellular compartmentation of this metabolite is unknown, it seems likely that cytosolic malate levels in developing COS could be sufficient to exert feedback inhibition on both PEPC isoforms.
Time-course immunoblots revealed that, although the p107 of PEPC1 and PEPC2 is phosphorylated in vivo throughout COS development, it appears to become maximally phosphorylated in PEPC1 in full cotyledon (stage VII) COS (Fig. 4, A and B). This pattern parallels triglyceride and storage protein synthesis in this tissue, which peak at stages VII and IX, respectively (Simcox et al., 1979). Enhanced p107 phosphorylation during COS development coincided with a significant increase in the I50(malate) for total PEPC activity present in desalted extracts (Fig. 4C). This result parallels that of Golombek et al. (1999), who hypothesized that PEPC becomes increasingly phosphorylated during the development of V. faba seeds as its activity in clarified cotyledon extracts showed a progressive diminished sensitivity to malate inhibition as seed development proceeded.
The results from the pod excision and extended darkness experiments established that p107 phosphorylation status of developing COS PEPC1 and PEPC2 is modulated by photosynthate supply recently translocated from the source leaves. As reported for soybean root nodule PEPC (Zhang et al., 1995), in vivo dephosphorylation of p107 in stage VII COS was faster in excised pods compared to intact plants that remained in darkness; 48 h following pod excision, the p107 of stage VII COS was completely dephosphorylated, whereas it retained some degree of phosphorylation after the same period of subjecting the intact plants to prolonged darkness (Fig. 5). This observation could reflect the existence of Suc and starch stores in the leaves and other parts of the plant to which seeds might have access, but that become unavailable when the pods are excised from the plant. Developing seed respiration, storage product synthesis, and carbon-nitrogen interactions are highly dependent upon the supply of recent photosynthate (Weber et al., 2005). The in vivo phosphorylation of PEPC1 and PEPC2 may be one of the regulatory components involved in photosynthate partitioning in developing COS. Interestingly, Suc synthase, a major sink-activity-related enzyme of developing seeds, is induced by Suc at the transcriptional level through a signaling pathway that appears to involve protein phosphorylation (Weber et al., 2005). The likely role of protein kinase-mediated enzyme phosphorylation in the metabolic control of developing legume seed carbon metabolism was recently emphasized (Weber et al., 2005). This has been corroborated by a cDNA microarray study that identified numerous putative protein kinase and phosphoprotein phosphatase genes whose expression was significantly modified during Arabidopsis embryogenesis (Ruuska et al., 2002). Clearly, additional research is warranted on the role of enzyme phosphorylation and the identification and characterization of the responsible protein kinases and phosphatases in the integration and control of cytosolic carbon metabolism and photosynthate partitioning during seed maturation.
Concluding Remarks
This study provides definitive evidence that a developing seed PEPC is subject to regulatory seryl phosphorylation in vivo. Our results have implications not only for the control of cytosolic carbon metabolism in developing oilseeds but also for the control of this ubiquitous plant cytosolic enzyme by reversible phosphorylation in non-green tissues. Furthermore, the results from the experiments in which pod excision or prolonged darkness pretreatments were imposed on castor plants indicate that, similar to soybean root nodule PEPC (Zhang et al., 1995), the in vivo phosphorylation status of p107 in COS PEPC1 and PEPC2 is reversibly modulated in some manner by the supply of photosynthate recently translocated from the leaves. Our results also demonstrate that the PEPC (p107) kinase and corresponding phosphatase respectively required to catalyze the in vivo phosphorylation and dephosphorylation of p107 are present and functional during COS development. Preliminary studies using dephosphorylated maize leaf PEPC and [γ-32P]ATP as COS PEPC kinase substrates suggested that the phosphorylation status of COS p107 is controlled by factors other than the turnover of COS (p107) PEPC kinase (Tripodi and Plaxton, 2005). This is similar to PEPC kinase from germinated barley seeds (Osuna et al., 1999), but contrasts with leaf and root nodule PEPC kinases that appear to be largely controlled by their synthesis/degradation (Chollet et al., 1996; Zhang and Chollet, 1997; Xu et al., 2003; Izui et al., 2004; Sullivan et al., 2004; Nimmo, 2005). Our present efforts are focused on validating the COS PEPC (p107) kinase activity assay previously described (Tripodi and Plaxton, 2005), and verifying the influence that COS development or perturbation of photosynthate supply to intact pods has on its activity. We are also examining the possibility that the p64 of COS PEPC2 might represent an in vivo or in vitro proteolytically truncated form of a larger (i.e. approximately 120 kD) bacterial-type PEPC polypeptide (Blonde and Plaxton, 2003; Sánchez and Cejudo, 2003), as well as the potential role that reversible phosphorylation of this polypeptide may play in the metabolic control and structural organization of COS PEPC2. Nevertheless, we hypothesize that p107 phosphorylation may help PEPC1 to compete with the allosterically desensitized PEPC2 to facilitate the partial redirection of cytosolic PEP flux away from storage lipids and toward storage protein biosynthesis during the latter stages of COS endosperm development.
MATERIALS AND METHODS
Chemicals and Plant Material
Acrylamide, bisacrylamide, and dithiothreitol (DTT) were from ICN Pharmaceuticals, whereas microcystin-LR was from Alexis Biochemicals (Cedarlane Laboratories). Ribi adjuvant was obtained from Corixa, polyvinylidene difluoride (PVDF) membranes (0.45-μm pore size; Immobilon) were from Millipore, and alkaline phosphatase-tagged goat anti-rabbit IgG was from Promega (Fisher Scientific). Purified bovine heart PP2A (7.5 units/mL) was a gift from Prof. Greg Moorhead (University of Calgary; 1 unit of PP2A dephosphorylates 1 μmol of muscle glycogen phosphorylase/min at 30°C). The catalytic subunit of porcine heart PK-A and other biochemical reagents were from Sigma-Aldrich Canada. All other chemicals were of analytical grade and obtained from BDH Chemicals. Affinity-purified rabbit anti-(Brassica napus suspension cell PEPC)-IgG was obtained as previously described (Moraes and Plaxton, 2000). Homogeneous PEPC1 and PEPC2 were isolated from stage VII (full cotyledon) developing COS as previously described (Blonde and Plaxton, 2003), with the exception that the final Superose 6 HR10/30 gel filtration FPLC step was replaced by ion-exchange FPLC on a Mono-Q HR5/5 column (final PEPC1- and PEPC2-specific activities were 22.6 and 31.2 units/mg protein, respectively). All solutions were prepared using Milli-Q-processed water (Millipore). Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes.
Castor bean (Ricinus communis L. var. Baker 296) plants were routinely cultivated in Promix BX general-purpose potting mixture (Premier Horticulture) in a greenhouse (10-inch-diameter pots) at 24°C and 70% relative humidity under natural light supplemented with 16 h of artificial light (high-pressure sodium lamps). Fertilizer (20:20:20 at 1.5 g/L; Plant Products) was administered every 7 d. Pods containing developing COS at heart-shaped embryo, midcotyledon, full cotyledon, and maturation stages of development (corresponding to 15, 25, 35, and 50 d after pollination, respectively; Greenwood and Bewley, 1982) were harvested at midday. Endosperm (free of cotyledon) was rapidly dissected, frozen under liquid N2, and stored at −80°C until used. Plants destined for prolonged darkness pretreatments were first transferred to a controlled environment Conviron PGR-15 growth chamber and cultivated for 5 to 7 d at 70% relative humidity with a 16-h-light (800 μE s−1 m−2 at canopy level; 24°C)/8-h-dark (22°C) photoperiod.
Preparation of Clarified Extracts
Quick-frozen COS endosperm was homogenized (1:2, w/v) using a Brinkmann PT-3100 Polytron in 50 mm HEPES-KOH (pH 7.5), containing 1 mm EDTA, 1 mm EGTA, 25 mm NaF, 0.1% (v/v) Triton X-100, 20% (v/v) glycerol, 10 mm MgCl2, 5 mm thiourea, 2 mm DTT, 50 nm microcystin-LR, 10 μg/mL chymostatin, 5 μg/mL leupeptin, 2 mm phenylmethylsulfonyl fluoride, 2 mm 2,2′-dipyridyl disulfide, 5 mm malate, and 1% (w/v) polyvinyl polypyrrolidone. Homogenates were centrifuged at 15,000g for 15 min at 4°C. Supernatants were either immediately (1) analyzed via nondenaturing PAGE; (2) boiled for 3 min in SDS sample buffer prior to SDS-PAGE; or (3) 0.5-mL aliquots desalted through 3-mL Sephadex G-50 spin columns (Penefsky, 1977) equilibrated in extraction buffer (minus phenylmethylsulfonyl fluoride and polyvinyl polypyrrolidone) for subsequent kinetic studies.
Enzyme and Protein Assays and Kinetic Studies
The standard PEPC assay mix contained 50 mm HEPES-KOH (pH 8.0), 5 mm MgCl2, 2.5 mm PEP, 0.15 mm NADH, 2 mm NaHCO3, 15% (v/v) glycerol, and 5 units/mL malate dehydrogenase (250 μL final volume). PEP-dependent NADH oxidation was monitored at 340 nm using a Molecular Devices SpectraMax kinetics microplate reader. All assays were linear with respect to time and concentration of enzyme assayed. One unit of activity is defined as the amount of PEPC resulting in the formation of 1 μmol of oxaloacetate/min at 25°C. Protein concentrations were determined with the Coomassie Blue G-250 dye-binding method (Bollag et al., 1996) using bovine γ-globulin as the protein standard.
Apparent Vmax, Km, and I50 and Ka values (concentrations of inhibitors and activators producing 50% inhibition and activation, respectively) were calculated using the Brooks (1992) computer kinetics program. All kinetic parameters are the means of a minimum of three separate experiments and are reproducible within ±10% se of the mean value. Stock solutions of PEP, amino acids, and organic acids were made equimolar with MgCl2 and adjusted to pH 7.5.
Preparation of APS-IgG Antibodies
Antiserum against the conserved n-terminal phosphorylation domain of plant PEPC was generated using a synthetic phosphopeptide corresponding to residues 1 through 12 in COS p107 (Blonde and Plaxton, 2003). The parent 12-mer PEPC peptide was synthesized as a C-terminal amide, and an additional Cys residue was introduced at the extreme n terminus. This base peptide was synthesized in two forms, with or without a phosphate group at the target Ser-6 residue (Fig. 1A), using a solid-phase peptide synthesis strategy and an Applied Biosystems 433A peptide synthesizer at the Peptide Synthesis Laboratory of the Protein Function Discovery Facility of Queen's University (Kingston, Ontario, Canada). Both peptides were purified by reverse-phase HPLC on a C18 Vydac column, and their sequences verified by MALDI-TOF mass spectrometry (Waters). The purified phosphopeptide was coupled to the carrier keyhole limpet hemocyanin (KLH) through the heterobifunctional reagent m-maleimidobenzoyl-N-hydroxysulfosuccinimide ester (Pierce; purchased from Biolynx), according to the manufacturer's protocols. The phosphopeptide-KLH conjugate (200 μg) was dialyzed overnight against Pi-buffered saline (100 mm NaPi, pH 6.8, containing 500 mm NaCl; PBS), filtered through an 0.2-μm membrane, and emulsified in Ribi adjuvant (1 mL total volume). After collection of preimmune serum, the conjugate was injected (0.6 mL subcutaneously, 0.4 intramuscularly) into an approximately 2-kg New Zealand rabbit. A booster injection (200 μg) was administered subcutaneously after 28 d. At 10 d following the final injection, blood was collected into Vacutainers (Beckton Dickinson) by cardiac puncture. After removal of clotted blood cells by centrifugation at 1,000g, the antiserum was frozen in liquid N2 and stored at −80°C.
The IgG fraction from the antiserum was partially purified by ammonium sulfate fractionation (0%–50% saturation) and dialyzed against 50 mm Tris-HCl, pH 7.5, containing 500 mm NaCl. APS-IgG were purified via chromatography of the dialyzed IgG fraction on a phosphopeptide affinity column. Affi-Gel 15 (2.0 mL; Bio-Rad), an N-hydroxysuccinimide ester of a derivatized cross-linked agarose, was washed with coupling buffer (100 mm MOPS-NaOH, pH 7.7) twice and centrifuged at 500g for 2 min. Phosphopeptide (20 mg) was dissolved in 1 mL of coupling buffer and incubated with the resin end-over-end for 4 h at room temperature. Resin was pelleted by centrifugation as above, and remaining amino reactive groups blocked by incubation with 100 mm Tris-HCl (pH 8.0) for 1 h at room temperature. The resin was equilibrated with Tris-buffered saline containing 20 mm NaF, and the dialyzed IgG fraction (3 mL containing approximately 30 mg of protein) made 20 mm NaF and absorbed batchwise onto the resin. After end-over-end incubation for 4 h at 24°C, the resin was packed into a column (9 mm i.d.) and washed with Tris-buffered saline until the A280 decreased to zero. The APS-IgG was eluted with 50 mm Gly-HCl (pH 2.5), and 0.5-mL fractions were received into 50 μL of 1 m Tris-HCl (pH 8.0). IgG-containing fractions were pooled (1.5 mL at about 0.3 mg protein/mL), adjusted to contain 20% (v/v) glycerol, and stored in 50-μL aliquots at −80°C. APS-IgG specificity was tested by immunoblotting. Nonphosphorylated peptide was used to block any nonspecific antibodies raised against the nonphosphorylated sequence, whereas the corresponding phosphopeptide was used as a blocking peptide to confirm the specific affinity of the APS-IgG for the phosphopeptide as well as phosphorylated p107.
Protein Electrophoresis and Immunoblotting
Nondenaturing and SDS-PAGE using a Bio-Rad minigel apparatus (7% and 10% separating gels, respectively), in-gel PEPC activity staining, and immunoblotting were as previously described (Rivoal et al., 2002a; Blonde and Plaxton, 2003). Densitometric analyses of immunoblots were performed using an LKB Ultroscan XL laser densitometer and GELSCAN software (version 2.1; Pharmacia LKB Biotech). Derived A663 values were linear with respect to the amount of the immunoblotted extract. All immunoblot results were replicated a minimum of three times. Representative results are shown in the various figures.
In Vitro Dephosphorylation and Rephosphorylation of Purified COS PEPC1 and PEPC2
Aliquots (50 μL) of purified PEPC1 or PEPC2 were desalted into the standard dephosphorylation buffer (50 mm Tris-HCl, pH 7.5, containing 5 mm MgCl2, 1 mm DTT, and 20% [v/v] glycerol) using Micro Spin-OUT GT-1200 desalting columns (Geno Technology) according to the manufacturer's instructions. Desalted COS PEPC1 or PEPC2 (50 μg each) were incubated at 30°C with 5 milliunits/mL of bovine PP2A in 50 μL of 50 mm Tris-HCl (pH 7.5) containing 5 mm MgCl2, 1 mm DTT, and 20% (v/v) glycerol. Aliquots of PP2A-treated PEPC1 and PEPC2 were subsequently incubated at 30°C in the presence of 20 units/mL of the catalytic subunit of porcine heart PK-A and 50 nm microcystin-LR ±1 mm ATP. Aliquots were withdrawn at the specified times and assayed for PEPC activity or analyzed by nondenaturing and SDS-PAGE and immunoblotting as described above.
Acknowledgments
We are very grateful to Dr. Pauline Douglas (University of Calgary) and Dr. Raymond Chollet (University of Nebraska-Lincoln) for helpful discussions, and to the Protein Function Discovery Research and Training Program (Queen's University) for peptide synthesis, purification, and sequence verification. We also thank Dr. Greg Moorhead (University of Calgary) for helpful discussions, as well as for supplying bovine heart PP2Ac.
This work was supported by research and equipment grants from the Natural Sciences and Engineering Research Council of Canada.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.066647.
References
- Blonde JD, Plaxton WC (2003) Structural and kinetic properties of high and low molecular mass phosphoenolpyruvate carboxylase isoforms from the endosperm of developing castor oil seeds. J Biol Chem 278: 11867–11873 [DOI] [PubMed] [Google Scholar]
- Bollag DM, Rozycki MD, Edelstein SJ (1996) Protein Methods. Wiley-Liss, New York, pp 62–70
- Brooks SPG (1992) A program for analyzing enzyme rate data obtained from a microplate reader. Biotechniques 17: 1154–1161 [PubMed] [Google Scholar]
- Chollet R, Vidal J, O'Leary MH (1996) Phosphoenolpyruvate carboxylase: a ubiquitous, highly regulated enzyme in plants. Annu Rev Plant Physiol Plant Mol Biol 47: 273–298 [DOI] [PubMed] [Google Scholar]
- Duff SMG, Chollet R (1995) In vivo regulation of wheat-leaf phosphoenolpyruvate carboxylase by reversible phosphorylation. Plant Physiol 107: 775–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastmond PJ, Dennis DT, Rawsthorne S (1997) Evidence that a malate/inorganic phosphate exchange translocator imports carbon across the leucoplast envelope for fatty acid synthesis in developing castor seed endosperm. Plant Physiol 114: 851–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golombek S, Heim U, Horstmann C, Wobus U, Weber H (1999) Phosphoenolpyruvate carboxylase in developing seeds of Vicia faba L.: gene expression and metabolic regulation. Planta 208: 66–72 [DOI] [PubMed] [Google Scholar]
- Greenwood JS, Bewley JD (1982) Seed development in Ricinus communis (castor bean). I. Descriptive morphology. Can J Bot 60: 1751–1760 [Google Scholar]
- Izui K, Matsumura H, Furumoto T, Kai Y (2004) Phosphoenolpyruvate carboxylase: a new era of structural biology. Annu Rev Plant Biol 55: 69–84 [DOI] [PubMed] [Google Scholar]
- Law RD, Plaxton WC (1997) Regulatory phosphorylation of banana fruit phosphoenolpyruvate carboxylase by a copurifying phosphoenolpyruvate carboxylase-kinase. Eur J Biochem 247: 642–651 [DOI] [PubMed] [Google Scholar]
- Li B, Zhang X-Q, Chollet R (1996) Phosphoenolpyruvate carboxylase kinase in tobacco leaves is activated by light in a similar but not identical way as in maize. Plant Physiol 111: 497–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamedov TG, Moellering ER, Chollet R (2005) Identification and expression analysis of two inorganic C- and N-responsive genes encoding novel and distinct molecular forms of eukaryotic phosphoenolpyruvate carboxylase in the green microalga Chlamydomonas reinhardtii. Plant J 42: 832–843 [DOI] [PubMed] [Google Scholar]
- Moraes TF, Plaxton WC (2000) Purification and characterization of phosphoenolpyruvate carboxylase from Brassica napus (rapeseed) suspension cell cultures. Implications for phosphoenolpyruvate carboxylase regulation during phosphate starvation and the integration of glycolysis with nitrogen assimilation. Eur J Biochem 267: 4465–4476 [DOI] [PubMed] [Google Scholar]
- Nhiri M, Bakrim N, Bakrim N, El Hachimi-Messouak Z, Echevarria C, Vidal J (2000) Posttranslational regulation of phosphoenolpyruvate carboxylase during germination of Sorghum seeds: influence of NaCl and L-malate. Plant Sci 151: 29–37 [Google Scholar]
- Nimmo HG (2005) Control of phosphoenolpyruvate carboxylase in plants. In WC Plaxton, MT McManus, eds, Control of Primary Metabolism in Plants. Blackwell Scientific Publishing, Oxford, UK (in press)
- Osuna L, González M-C, Cejudo FJ, Vidal J, Echevarría C (1996) In vivo and in vitro phosphorylation of the phosphoenolpyruvate carboxylase from wheat seeds during germination. Plant Physiol 111: 551–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osuna L, Pierre J-N, González M-C, Alvarez R, Cejudo FJ, Echevarría C, Vidal J (1999) Evidence for a slow-turnover form of the Ca2+-independent phosphoenolpyruvate carboxylase kinase in the aleurone-endosperm tissue of germinating barley seeds. Plant Physiol 119: 511–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacquit V, Giglioli N, Crétin C, Pierre JN, Vidal J, Echevarria C (1995) Regulatory phosphorylation of C4 phosphoenolpyruvate carboxylase from Sorghum: an immunological study using specific anti-phosphorylation site-antibodies. Photosynth Res 43: 283–288 [DOI] [PubMed] [Google Scholar]
- Penefsky HS (1977) Reversible binding of Pi by beef heart mitochondrial adenosine triphosphatase. J Biol Chem 252: 2891–2899 [PubMed] [Google Scholar]
- Rivoal J, Dunford R, Plaxton WC, Turpin DH (1996) Purification and properties of four phosphoenolpyruvate carboxylase isoforms from the green alga Selenastrum minutum. Evidence that association of the 102 kD catalytic subunit with unrelated polypeptides modifies the physical and kinetic properties of the enzyme. Arch Biochem Biophys 332: 47–57 [DOI] [PubMed] [Google Scholar]
- Rivoal J, Plaxton WC, Turpin DH (1998) Purification and characterization of high and low molecular mass isoforms of phosphoenolpyruvate carboxylase from Chlamydomonas reinhardtii: kinetic, structural and immunological evidence suggest that the green algal enzyme is distinct from the prokaryotic and higher plant enzymes. Biochem J 331: 201–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivoal J, Smith CR, Moraes TF, Turpin DH, Plaxton WC (2002. a) Enzyme activity staining after native polyacrylamide gel electrophoresis using fluorescence detection. Anal Biochem 300: 94–99 [DOI] [PubMed] [Google Scholar]
- Rivoal J, Trzos S, Gage DA, Plaxton WC, Turpin DH (2001) Two unrelated phosphoenolpyruvate carboxylase polypeptides physically interact in the high molecular mass isoform of this enzyme in the unicellular green alga Selenastrum minutum. J Biol Chem 276: 12588–12597 [DOI] [PubMed] [Google Scholar]
- Rivoal J, Turpin DH, Plaxton WC (2002. b) In vitro phosphorylation of phosphoenolpyruvate carboxylase from the green alga Selenastrum minutum. Plant Cell Physiol 43: 785–792 [DOI] [PubMed] [Google Scholar]
- Ruuska SA, Girke T, Benning C, Ohlrogge JB (2002) Contrapuntal networks of gene expression during Arabidopsis seed filling. Plant Cell 14: 1191–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez S, Cejudo FJ (2003) Identification and expression analysis of a gene encoding a bacterial-type phosphoenolpyruvate carboxylase from Arabidopsis and rice. Plant Physiol 132: 949–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangwan RS, Singh N, Plaxton WC (1992) Phosphoenolpyruvate carboxylase activity and concentration in the endosperm of developing and germinating castor oil seeds. Plant Physiol 99: 445–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwender J, Ohlrogge JB, Shachar-Hill Y (2004) Understanding flux in plant metabolic networks. Curr Opin Plant Biol 7: 309–317 [DOI] [PubMed] [Google Scholar]
- Simcox PD, Garland W, DeLuca V, Canvin DT, Dennis DT (1979) Respiratory pathways and fat synthesis in the developing castor oil seed. Can J Bot 57: 1008–1014 [Google Scholar]
- Smith RG, Gauthier DA, Dennis DT, Turpin DH (1992) Malate and pyruvate dependent fatty acid synthesis in leucoplasts from developing castor endosperm. Plant Physiol 98: 1233–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan S, Jenkins GI, Nimmo HG (2004) Roots, cycles and leaves. Expression of the phosphoenolpyruvate carboxylase gene family in soybean. Plant Physiol 135: 2078–2087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripodi KE, Plaxton WC (2005) In vivo regulatory phosphorylation of phosphoenolpyruvate carboxylase in developing castor oil seeds. In A van der Est, D Bruce, eds, Photosynthesis: Fundamental Aspects to Global Perspectives. Allen Press, Lawrence, KS, pp 915–917
- Turner WL, Knowles VL, Plaxton WC (2005) Cytosolic pyruvate kinase subunit composition, activity, and amount in developing castor and soybean seeds, and biochemical characterization of the purified castor seed enzyme. Planta doi/10.1007/s00425-005-0044-8 [DOI] [PubMed]
- Ueno Y, Imanari E, Emura J, Yoshizawa-Kumagaye K, Nakajima K, Inami K, Shiba T, Sakakibara H, Sugiyama T, Izui K (2000) Immunological analysis of the phosphorylation state of maize C4-form phosphoenolpyruvate carboxylase with specific antibodies raised against a synthetic phosphorylated peptide. Plant J 21: 17–26 [DOI] [PubMed] [Google Scholar]
- Wadham C, Winter H, Schuller KA (1996) Regulation of soybean nodule phosphoenolpyruvate carboxylase in vivo. Physiol Plant 97: 531–535 [Google Scholar]
- Weber H, Borisjuk L, Wobus U (2005) Molecular physiology of legume seed development. Annu Rev Plant Biol 56: 252–279 [DOI] [PubMed] [Google Scholar]
- Xu W, Zhou Y, Chollet R (2003) Identification and expression of a soybean nodule-enhanced PEP-carboxylase kinase gene (NE-PpcK) that shows striking up-/down-regulation in vivo. Plant J 34: 441–452 [DOI] [PubMed] [Google Scholar]
- Zhang X-Q, Chollet R (1997) Phosphoenolpyruvate carboxylase protein kinase from soybean root nodules: partial purification, characterization, and up/down-regulation by photosynthate supply from the shoots. Arch Biochem Biophys 343: 260–269 [DOI] [PubMed] [Google Scholar]
- Zhang X-Q, Li B, Chollet R (1995) In vivo regulatory phosphorylation of soybean nodule phosphoenolpyruvate carboxylase. Plant Physiol 108: 1561–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]