Abstract
Cells of the marine diatom Phaeodactylum tricornutum Bohlin (UTEX 642) grown in 5% CO2 were transferred to air-level CO2 in the light or dark and allowed to acclimate to air. No accumulation of the transcript of the P. tricornutum β-carbonic anhydrase 1 (ptca1) was detected in 5% CO2-grown cells, but ptca1 mRNA accumulated and reached a peak after 6 h acclimation to air but decreased over the next 18 h. A similar accumulation time course was observed in cells air-acclimated in the dark, except that levels of mRNA were <50% those in the light. These results suggest that air-level [CO2] is required to trigger the transcription of ptca1 and that light affects the extent of acclimation. During acclimation to air for 120 h in the light, levels of ptca1 mRNA exhibited a periodic oscillation with a cycle of about 24 h, which, however, was not reflected in protein accumulation levels. A 5′-upstream region from the transcription-start site toward −1,292 bp of ptca1 was cloned by inverse polymerase chain reaction, and 5′-truncations were carried out on this fragment. The truncated promoter regions were fused with the β-glucuronidase gene (uidA) and introduced into P. tricornutum. The promoter fragments, truncated at positions −1,292, −824, −484, −225, and −70 bp, conferred on transformants clear CO2-responsive β-glucuronidase expressions. In contrast, the CO2-responsive regulation was severely impaired or completely abolished by truncations, respectively, at position −50 or −30 bp. These results indicate that critical cis-elements required for CO2-responsive transcription of ptca1 may be located between −70 and −30 bp relative to the transcription start site.
Marine diatoms are unicellular algae with plastids derived from an ancestral red alga by secondary endosymbiosis and are thought to contribute at least 25% of the global carbon fixation (Werner, 1977; Tréguer et al., 1995; Falkowski et al., 2004). The carbon-concentrating mechanism (CCM) is a system that facilitates an ample flux of CO2 to photosynthesis under CO2 limitation and occurs in cyanobacteria and many eukaryotic algae, including marine diatoms (Badger et al., 1980, 1998; Kaplan et al., 1980; Miller et al., 1990; Colman and Rotatore, 1995; Johnston and Raven, 1996; Tortell et al., 1997; Matsuda et al., 2001). Intracellular carbonic anhydrases (CAs) in microalgae are known to be crucial components for the operation of CCMs in a CO2-limited environment (Funke et al., 1997; Raven, 1997; Karlsson et al., 1998). The activity of the CCM is, in general, suppressed in CO2-enriched air but induced by transferring cells from high CO2 to atmospheric levels of CO2. This CCM induction process has been shown to be accompanied by the activation of a number of gene transcriptions, including those of CAs in Chlamydomonas reinhardtii (Moroney and Somanchi, 1999; Miura et al., 2004). The CCM of C. reinhardtii is induced to a maximum level within 4 h of acclimation to air (Sültemeyer et al., 1991). This acclimation time course corresponded with expression profiles of the genes of periplasmic CA (cah1) and mitochondrial CA (mtca) in C. reinhardtii (Fukuzawa et al., 1990; Eriksson et al., 1996). Similar regulation profiles of CCM expressions in response to changes in the ambient [CO2] have also been reported in two Chlorella species (Matsuda and Colman, 1995a; Bozzo et al., 2000) and in the marine diatom Phaeodactylum tricornutum (Matsuda et al., 2001). However, the time required for the induction of maximum high-affinity photosynthesis in P. tricornutum is relatively longer (24 h) than that of freshwater microalgae (3–6 h; Matsuda et al., 2001). A circadian rhythm is also thought to participate in the regulations of cah1 and mtca in synchronously cultured cells (Rawat and Moroney, 1995; Fujiwara et al., 1996; Eriksson et al., 1998).
In the green algae C. reinhardtii (Bozzo and Colman, 2000) and Chlorella sp. (Matsuda and Colman, 1995b; Bozzo et al., 2000) and in the marine diatom P. tricornutum (Matsuda et al., 2001), it has been shown that the regulation of CCM expression occurs in response to specific CO2 concentrations in the bulk medium rather than concentrations of HCO3− or total dissolved inorganic carbon (DIC). These findings indicate that the ambient [CO2] is the critical species of DIC recognized by these cells.
Light is an additional factor that controls the extent of the algal CCM expression. In C. reinhardtii and P. tricornutum, it has been shown that the CCM expression did not occur under limited CO2 in the dark (Spalding and Ogren, 1982; Matsuda et al., 2002). In contrast, CCM expression was shown to be largely independent of light in Chlorella ellipsoidea (Matsuda and Colman, 1995b). In C. reinhardtii, transcriptional controls of CA genes are found to be more complex than the regulation of high-affinity photosynthesis, and some primary signals seem to interact. That is, trace accumulations of mRNA of CA genes, cah1 and mtca, have been detected during acclimation to limiting CO2 in the dark in synchronously cultured C. reinhardtii (Rawat and Moroney, 1995; Eriksson et al., 1998), indicating that the primary trigger for these expressions is CO2 limitation, but the cell cycle and light also affect this process. In fact, impairment of the cah1 promoter region resulted in a drastic alteration of cah1 regulation in response to light and CO2 in C. reinhardtii (Kucho et al., 1999). The 5′-upstream region of cah1 was shown to contain an enhancer and silencer regions, which control the expression of cah1 positively and negatively, respectively (Kucho et al., 1999), and two consensus DNA sequences that are required for the transcriptional activation of cah1 are included in the enhancer region (Kucho et al., 2003). This consensus sequence is also found in the mtca promoter region (Villand et al., 1997), suggesting that it acts as a CO2-responsive cis-acting element in C. reinhardtii (Kucho et al., 2003).
A few CAs have also been identified in marine diatoms (Roberts et al., 1997; Satoh et al., 2001). The intracellular β-carbonic anhydrase in P. tricornutum (PtCA1), which is encoded by the nuclear genome, has been shown to be a relatively abundant species and to be regulated at the transcriptional level in response to the ambient [CO2] (Satoh et al., 2001). Accumulation levels of transcript of the PtCA1 gene (ptca1) in air-grown cells are 5- to 10-fold those in 5% CO2-grown cells (Satoh et al., 2001). PtCA1 could therefore be a suitable model for the elucidation of mechanisms of acclimation to limiting [CO2] in marine diatoms. PtCA1 was recently shown to be localized in the chloroplast of P. tricornutum at the surface of the girdle lamellae as clustered particles (Tanaka et al., 2005). Also recently, a new β-CA isozyme gene has been identified from the expressed sequence tag library of P. tricornutum, and the full-length cDNA was cloned (Harada and Matsuda, 2005). This gene, designated as ptca2, was found to be highly homologous to ptca1 and to be a trace species at the transcript level. However, the function of these diatom CAs is unclear, and the regulation of CA expression in response to [CO2] at the molecular level is not known despite the ecophysiological importance of photosynthetic carbon fixation in marine diatoms.
In this study, accumulation levels of ptca1 mRNA and PtCA1 protein during acclimation of 5% CO2-grown cells of P. tricornutum to atmospheric CO2 were determined under changing light conditions. A putative promoter region of the ptca1 gene was also characterized, and the function of the promoter region was analyzed at the molecular level using a reporter assay of upstream fragments of the ptca1 gene.
RESULTS
Levels of ptca1 mRNA and PtCA1 during Acclimation to Air in the Light or Dark
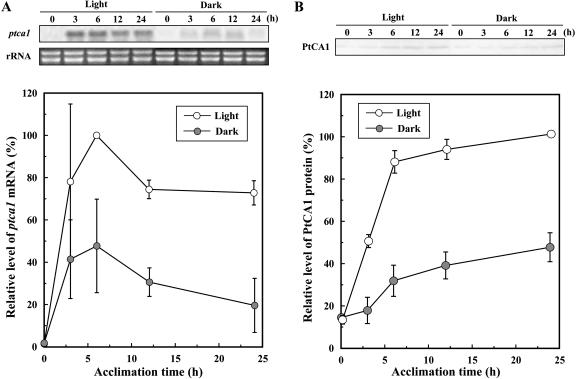
The accumulation of ptca1 mRNA and PtCA1 protein during acclimation of 5% CO2-grown cells of P. tricornutum to air were measured by northern and western blotting. Upon transferring cells from 5% CO2 to air in the light, the transcript of ptca1 started to increase, and the level reached 78% the maximum within 3 h of acclimation (Fig. 1A). Accumulation reached a peak by 6 h of acclimation to air and then gradually decreased over the next 18 h (Fig. 1A). A similar accumulation profile with time was also found when cells were acclimated to air in the dark, except that the accumulation levels of mRNA were 30% to 50% those in the light (Fig. 1A). The accumulation of PtCA1 protein increased to a level approximately 90% the maximum within 6 h of acclimation to air and gradually increased over the next 18 h in the light (Fig. 1B). Changes in accumulation in the dark also exhibited a similar time course, but the levels ranged from 35% to 50% those in the light (Fig. 1B). The levels of PtCA1 protein did not decrease in the last 18 h of acclimation unlike those observed in ptca1 mRNA (Fig. 1).
Figure 1.
Time course of accumulations of ptca1 mRNA and protein during acclimation from 5% CO2 to air in the light or dark. A, Top, Northern blots of ptca1 and ethidium bromide staining of rRNA. RNAs were extracted from air-acclimating cells in the light or dark; bottom, relative intensity of densitometry of northern-blot analysis at different times of acclimation. Open and closed circles represent accumulation profiles in the light and dark, respectively. B, Top, Western blots with anti-PtCA1-rabbit antibody (PtCA1). Proteins were extracted from air-acclimating cells in the light or dark; bottom, relative intensity of densitometry of western-blot analysis at several different times of acclimation. Open and closed circles represent accumulation profiles in the light and dark, respectively. Equal amount of total-soluble protein (20 μg) was loaded in each lane. Densities of bands were expressed as relative values (%) of the maximum signal intensity of each analysis. Densitometry data were quantified using NIH image software. Data are shown as mean values ± sd of three independent measurements.
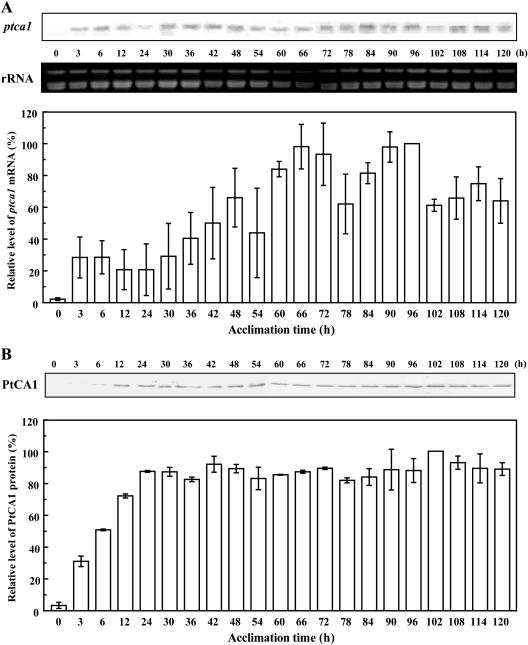
P. tricornutum cells grown under 5% CO2 in the light were transferred and allowed to acclimate to air in the light for 5 d. Northern-blot analysis showed that the ptca1 transcript oscillated in accumulation during prolonged acclimation to air (Fig. 2A). The period of these oscillations was found to be about 24 h (Fig. 2A), reaching peak values at 6, 48, 66, 96, and 114 h and low values at 12, 54, 78, and 102 h of acclimation (Fig. 2A). Peak levels of ptca1 mRNA gradually increased (29% at 6 h, 66% at 48 h, 98% at 66 h, and maximum at 96 h) throughout the acclimation for 5 d. On the other hand, accumulations of PtCA1 protein increased to a level approximately 90% the maximum within 24 h of acclimation to air, and this level was maintained for the next 4 d (Fig. 2B).
Figure 2.
Time course of accumulation of ptca1 mRNA and protein during prolonged acclimation to air in the light. A, Top, Northern blots with partial fragment of ptca1 as probe and ethidium bromide staining of rRNA; bottom, relative intensity of densitometry of northern-blot analysis at different times of acclimation. RNAs were extracted from air-acclimating cells in the light. B, Top, Western blots with anti-PtCA1-rabbit antibody (PtCA1); bottom, relative intensity of densitometry of western-blot analysis at different times of acclimation. Proteins were extracted from air-acclimating cells in the light. Equal amounts of total-soluble protein (20 μg) were loaded in each lane; densities of bands were expressed as relative values (%) of the maximum signal intensity of each analysis (96 h in A, 102 h in B). Densitometry data were quantified using NIH image software. Data are shown as mean values ± sd of three independent measurements.
Isolation of the 5′-Upstream Region of the ptca1 Gene by Inverse PCR
To isolate the 5′-upstream region of the ptca1 gene, two pairs of PCR primers, which are oriented in the reverse directions toward genomic regions flanking the inserted ptca1, were designed (Fig. 3, underlines). The inverse PCR amplifications of the adjacent sequences of ptca1 were carried out on self-ligated circular DNAs of PstI-digested fragments of the genomic DNA as templates. Approximately 3 kb of the inverse PCR product was obtained after the nested PCR (data not shown). The size of the fragment agreed well with that predicted from genomic southern-blot analysis of PstI-digested genomic DNA. A 1.3-kb 5′-upstream region of ptca1 was cloned, and the sequence was determined (Fig. 3). This 1.3-kb region was characterized for known basic transcriptional elements in eukarya. There were no obvious TATA and CAAT boxes found between −200 bp and the transcription start site of ptca1 (Fig. 3), although the sequence from −2 to +2 (CACA) relative to the transcription start site (Fig. 3, closed box) was identical to the initiator-like sequence of the promoter region of fucoxanthin-chlorophyll a/c binding protein genes (fcp), which was previously reported in P. tricornutum (Bhaya and Grossman, 1993). Two highly conserved sequences of the cAMP-response element (CRE; 5′-TGACGTCA-3′) were found at positions from −70 to −63 and from −21 to −14 (Fig. 3, open boxes). In addition, the position from −52 to −46 (5′-GGGAGTG-3′) was identical to the DNA-binding element, E1A-associated 300-kD protein (p300), which is a functional homolog of the binding protein of the CRE-binding protein (CBP), CREB. However, there was little sequence similarity to the known CO2-responsive gene promoters of cah1 and mtca found in the green alga C. reinhardtii.
Figure 3.
Nucleotide sequence of 5′-upstream region of ptca1. Underline indicates sequences at which primers were designed for inverse PCR. The initiator-like sequence is shown as white letters on the black box. The coding sequence of ptca1 is indicated with lowercase letters. An intron is indicated with uppercase italic letters. Arrowheads show the initial base of each truncated promoter. Open boxes show CRE-like elements found in the isolated 5′-upstream region. Double underline indicates putative p300-binding element.
β-Glucuronidase Reporter Assay of the Promoter Region of ptca1
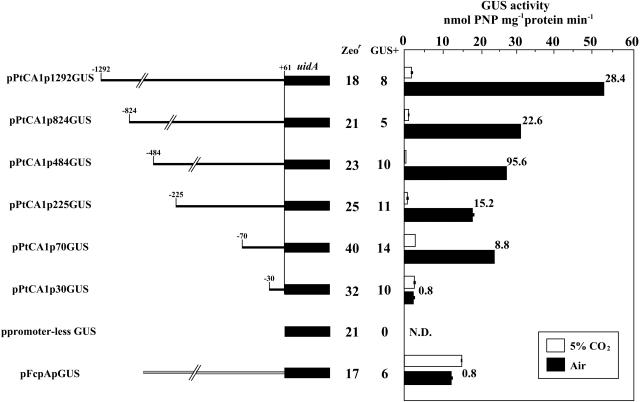
In order to investigate the CO2-responsive regulatory mechanisms of the ptca1 gene, a series of 5′-deletions of the ptca1 promoter region were carried out, and the resulting truncated fragments were ligated with the β-glucuronidase (GUS) reporter gene (uidA) at the position of +61 from the transcription start site (Fig. 4). Each fusion construct was packaged into the transformation vector pPha-T1 and introduced into P. tricornutum by microprojectile bombardment. Twenty to forty of Zeocin-resistant clones were obtained out of 108 possible transformants, and approximately 50% of the Zeocin-resistant clones were found to be GUS positive (Fig. 4). No transformants expressing GUS activity were obtained with the introduction of promoterless uidA (Fig. 4, ppromoter-less GUS). Genomic southern-blot analysis with uidA gene probe of 350 bp indicated that these transformants possess one to four independent inserts of uidA genes, which would be integrated randomly into the nuclear DNA (data not shown). Transformants with a single insertion of uidA were selected for further experiments.
Figure 4.
The 5′-deletion constructs with fused GUS reporter gene, uidA, and GUS reporter assays of transformants with each 5′-deletion construct of ptca1 promoter. Left, Schematics of 5′-deletion constructs of ptca1 promoter. The initial base of each construct is indicated in base pairs from the transcription start site (arrowheads in Fig. 3). The 5′-truncated ptca1 promoters were fused to uidA (accession no. S69414). Each construct was introduced into cells by microprojectile bombardment. Numbers of Zeocin-resistant (Zeor) clones and GUS-positive clones (GUS+) are indicated. Right, Levels of GUS activity measured in each GUS+ transformant grown in 5% CO2 or air. GUS activities in lysates of 5% CO2-grown transformants (white bars) and air-grown transformants (black bars) were determined per total protein bases. Ratios of GUS activity in air-grown cells to that in 5% CO2-grown cells are indicated at the right of each black bar. N.D., Not detected; error bars, sd of three independent experiments.
GUS activities of each transformant, grown in 5% CO2 or air, were measured (Fig. 4). uidA driven with promoters truncated at positions −1,292, −824, −484, −225, and −70 exhibited a clear CO2-responsive regulation of GUS expression (Fig. 4). In these transformants, extremely low basal expressions of GUS activity of 0.5 to 3.0 nmol p-nitrophenol (PNP) mg−1 protein min−1 were detected in 5% CO2-grown cells; thus, uidA was found to be almost totally repressed in 5% CO2 (Fig. 4). The induced levels of GUS activities in the transformants grown in air ranged from 18 to 53 nmol PNP mg−1 protein min−1, which were 9- to 96-fold those in 5% CO2-grown transformants (Fig. 4). A significant drop in the induced level of GUS activity under air conditions was observed in the PtCA1p824GUS transformant as compared to that in the PtCA1p1292GUS transformant (Fig. 4). In PtCA1p824GUS transformants grown in air, GUS activity was found to be 31 nmol PNP mg−1 protein min−1, which was about 58.5% of that in air-grown PtCA1p1292GUS (Fig. 4). The induced GUS levels in air-grown transformants seemed to be relatively stable with truncations at −824 throughout −70 (Fig. 4). Interestingly, a further 5′-deletion toward −31 bp completely abolished the CO2-responsive expression of uidA (Fig. 4), and the levels of GUS activity were found to be basal (3.0 nmol PNP mg−1 protein min−1) irrespective of growth [CO2].
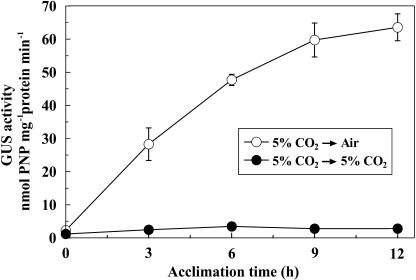
The time course of induction of GUS activity during acclimation from 5% CO2 to air was examined in the transformant PtCA1p1292GUS (Fig. 5). About half maximum GUS activity was induced within 3 h of acclimation to air, which was followed by a steady increase over the next 9 h, resembling the changes in the induction profiles of endogenous PtCA1 (Fig. 5).
Figure 5.
Time course of the GUS activity in the PtCA1p1292GUS transformant during acclimation from 5% CO2 to air. White circles indicate the GUS activity of transformants during the acclimation from 5% CO2 to air. Black circles indicate control experiment in which 5% CO2-grown cells were resuspended in new F/2ASW and were maintained in 5% CO2. Error bars indicate sd of three independent experiments.
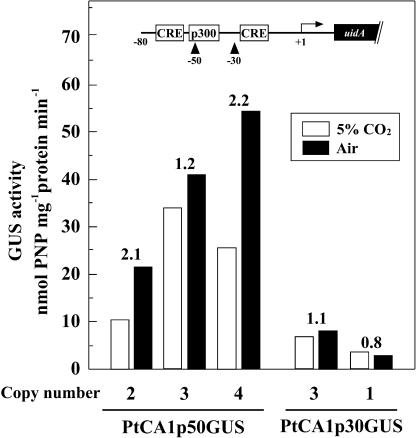
In order to focus on the critical CO2-responsive region in between −70 and −30 of the ptca1 promoter, a short upstream sequence was prepared by deleting the 5′-end at −50 (PtCA1p50GUS) relative to the transcription start site. Transformants obtained with the introduction of pPtCA1p50GUS were found to contain two to four copies of the uidA sequence, but no transformant with a single insertion of uidA was obtained in this strain. Although the basal expression levels varied among transformants, presumably due to the variation of copy number of uidA, the ratio of GUS activities in cells grown in air over that in 5% CO2-grown cells was relatively stable among clones irrespective of copy number (Fig. 6). GUS activity of three PtCA1p50GUS transformants containing two to four copies of uidA showed CO2-responsive expressions of GUS, but GUS activities under 5% CO2 were found to range from 10.7 to 34.5 nmol PNP mg−1 protein min−1 (Fig. 6), and the repression of uidA was apparently incomplete (Fig. 6). GUS activities in air-grown cells of PtCA1p50GUS transformants were found to be 1.2- to 2.2-fold those in 5% CO2-grown PtCA1p50GUS (Fig. 6). The PtCA1p30GUS transformants containing one or three copies of uidA showed no CO2-responsive expression of GUS irrespective of copy number of uidA (Fig. 6).
Figure 6.
GUS reporter assays of two types of transformants, PtCA1p50GUS and PtCA1p30GUS. PtCA1p50GUS and PtCA1p30GUS were generated by introducing pPtCA1p50GUS and pPtCA1p30GUS, respectively. GUS activity in lysates of 5% CO2-grown transformants (white bars) or air-grown transformants (black bars) were determined on total protein bases. Copy numbers were determined by genomic southern analysis. Ratios of GUS activity in air-grown cells to those in 5% CO2-grown cells were indicated at the top of the each column. The structure of the ptca1 promoter region (−80 to uidA) with putative cis-elements is depicted at the top part of the diagram box. Two truncation sites at −50 and −30 relative to the transcription start site are indicated by arrowheads.
DISCUSSION
The rapid accumulation of ptca1 mRNA in the dark after a few hours acclimation to air (Fig. 1A) implies that the absence of light did not alter the apparent time course of the induction. Similar induction profiles in the dark were also observed in changes in the relative accumulation of PtCA1 protein (Fig. 1B). These data indicate that the primary trigger for ptca1 induction is a decrease in the ambient [CO2] and that light increases the expression of PtCA1 presumably by enhancing the transcription of ptca1. It should be noted that the effect of light on CCM induction is still significant, since CCM in P. tricornutum did not show a measurable increase in the dark under air in the previous study (Matsuda et al., 2001). It is probable that the CCM as a whole cell event requires activations of multiple components; thus, trace increase in PtCA1 protein alone in the dark has little physiological contribution to it.
Light-independent inductions of CA under limiting [CO2] has been previously reported for the genes of periplasmic and mitochondrial CAs, cah1 and mtca, respectively, in C. reinhardtii when cells were synchronously cultured on a light/dark cycle (Rawat and Moroney, 1995; Eriksson et al., 1998). Given the culture conditions employed in these studies, the expression of CAs in the dark in C. reinhardtii was thought to be regulated by the stage in the cell cycle as well as CO2 limitation. In fact, induction of these CAs in the dark was not observed in nonsynchronous cultures (Dionisio-Sese et al., 1990; Rawat and Moroney, 1995; Eriksson et al., 1998). In contrast, the culture of 5% CO2-grown P. tricornutum, in this study, was not synchronous, and the induction time course in the dark was comparable to that in the light.
It is possible that an abrupt change in the concentration of DIC in the bulk medium, upon transferring cells from 5% CO2 to air, might reset the cell cycle of P. tricornutum. Supporting this consideration, accumulation of ptca1 mRNA exhibited an apparent peak after 6 h of acclimation to air (Fig. 1A), and this tendency became more evident by prolonged acclimation under air in continuous light (Fig. 2A). This interpretation in turn suggests that the initial cells of P. tricornutum exhibited an intrinsic circadian rhythm with a period of about 1 d even under continuous light for 5 d. Nonetheless, this oscillation of mRNA accumulation during acclimation to air seems to have a negligible physiological effect since fluctuations were not reflected in levels of protein accumulation (Fig. 2B), presumably due to the stability of PtCA1 protein in vivo.
The truncation of 5′-upstream sequences of ptca1 to −824 resulted in a significant decrease in the expression levels of GUS under air conditions (Fig. 4). Such changes in expression levels, depending on the length of upstream sequences relatively distant from the minimal promoter region, are often observed in plants and algae probably due to the existence of cis-elements, which participate in controlling the extent of transcript levels. In C. reinhardtii, deletions several hundreds of base pairs upstream of the basal promoter region of cah1 resulted in a moderate stimulation or an abrupt decrease in transcription in response to changes in the ambient [CO2] and/or light, and these changes in transcription were more evident when a foreign minimal promoter was ligated with these putative enhancer or silencer regions (Kucho et al., 1999). Given these examples, the upstream sequence between −1,292 and −824 of ptca1 might comprise regulatory sequences that control the extent of transcript in response to CO2. The sequence between −1,292 and −824 contains a CCAAT-box (5′-CCAAT-3′) and a Myc element (5′-CANNTG-3′), although the functions of these elements in marine diatoms are not known. Another important controlling factor of ptca1 expression is light; it was shown in this study that light apparently affects the extent of ptca1 expression. However, truncations carried out in this study did not alter the apparent responses of uidA expression to light or dark conditions, irrespective of growth [CO2] (data not shown), suggesting that the repressive effect of darkness on ptca1 expression is not directly due to the upstream sequence of up to −1.3 kb. Alternatively, it is also possible that the effect of light on the ptca1 promoter interacts with other signal-response systems; thus, clear changes in the light response were not observed in wide-range truncations of 5′-upstream regions employed in this study. Considering that the CO2-responsive promoter regions in cah1 in C. reinhardtii, in sharp contrast to the case of ptca1, were affected by light drastically, it is strongly suggested that marine diatoms might have developed a unique system of CO2 responses that are apparently different from those in green algae.
It is clear from this study that the critical sequences required for the CO2-responsive regulations of ptca1 are present between −70 and −30 relative to the transcription start site of ptca1. Although the minimal promoter region of ptca1 is not clear, it is not likely that the truncation toward −30 severely damaged the binding of RNA polymerase, that is, trace GUS expression was clearly observed both in 5% CO2 and air in the transformant PtCA1p30GUS (Fig. 6), whereas GUS activity was absent in cells that were transformed with promoterless uidA. Nevertheless, it is still possible that the minimal promoter sequence was partially impaired in pPtCA1p30GUS. It is therefore not clear at the moment whether the region between −70 and −30 activates a repressive or an inducible response to changes in the ambient [CO2]. Truncations toward −50 might give important clues on this issue. In transformants with pPtCA1p50GUS, CO2-responsive expressions of GUS were apparently disturbed (Fig. 6). Although influences of multiple insertions of uidA in these transformants should be addressed carefully, the expression levels of GUS in 5% CO2-grown cells of PtCA1p50GUS tend to be significantly higher than those in other transformants with longer promoter regions when grown in 5% CO2 (Fig. 6). The ratios of GUS levels in air-grown cells to those in 5% CO2-grown cells were also significantly reduced in these transformants (Fig. 6); thus, the phenotype of PtCA1p50GUS became weakly responsive to the ambient [CO2]. The upstream region between −70 and −50 therefore appears to include elements that participate in the repression of ptca1 under 5% CO2 conditions. Interestingly, the promoter region of ptca1 between −70 and +61 comprises two highly conserved sequences of CRE at −70 to −63 and −21 to −14 (Fig. 3, open boxes). The 20 bp between −70 and −50 thus include one of the CREs. It should also be noted that the weak response to [CO2] of GUS expression was completely abolished by the deletion of 20 bp between −50 and −30 (Fig. 6). It is therefore possible that one of the crucial sequences for the CO2-responsive regulations of ptca1 might also be present between −50 and −30 relative to the transcription start site of ptca1. Relating to this, a p300-binding element was found at −52 to −46 relative to the transcription start site (Fig. 3, double underline), suggesting a possible involvement of p300 in CO2-responsive regulation of ptca1. p300 is structurally similar to CBP and functionally redundant protein essential for proper development of mammals (Rikitake and Moran, 1992; Tanaka et al., 1997; Oike et al., 1999).
CRE is found in many eukaryotic promoters, which are regulated by the cytosolic concentration of cAMP. cAMP is known to be a multifunctional second messenger in a wide variety of living organisms from prokaryotes to eukaryotes, although the function of cAMP in higher plants and eukaryotic algae has been controversial. It has been reported that soluble-adenylyl cyclase might be a HCO3−- and pH-sensing protein that has been evolutionally conserved from cyanobacteria (Chen et al., 2000; Pastor-Soler et al., 2003). A possible function of cAMP in the regulation of CCMs in response to changes in the ambient [CO2] in cyanobacteria and the green alga C. reinhardtii has been proposed but with little direct evidence. The data obtained in this study strongly suggest a possible participation of CRE-like elements and, hence, cAMP in the CO2-responsive regulation of a part of the CO2-acquisition systems in the marine diatom P. tricornutum. The existence of a p300-binding element (at −52 to −46) shortly downstream of the CRE (at −70 to −63) would also support this assumption, since p300 has been shown to interact with CREB as a transcriptional coactivator and is known to be a functional homolog of CBP (Chrivia et al., 1993; Kwok et al., 1994). It is of particular interest to know whether any functional relationship exists between CRE and p300 in the CO2-responsive regulation of ptca1.
This study also showed that there is little sequence similarity in the ptca1 promoter region in P. tricornutum with the known CO2-responsive gene promoters found in the green alga C. reinhardtii. It is probable that the mechanism of CO2-responsive transcriptional controls in P. tricornutum might have evolved independently and thus is distinct from that in the green alga C. reinhardtii. The function of the ptca1 promoter region awaits detailed study and is crucial to a better understanding of acclimation mechanisms of marine diatoms to the changing [CO2] at the ocean surface.
MATERIALS AND METHODS
Cells and Culture Conditions
The marine diatom Phaeodactylum tricornutum Bohlin (UTEX 642) was obtained from the University of Texas Culture Collection and was grown in artificial seawater, which was supplemented with half-strength Guillard's “f” solution (F/2ASW; Guillard and Ryther, 1962; Harrison et al., 1980) under continuous illumination (50 μmol m−2 s−1) at 20°C with constant aeration of 5% CO2 or atmospheric air.
Two liters of 5% CO2-grown cells were centrifuged at 3,500g at 20°C for 5 min, washed twice in 10 mL of CO2-free F/2ASW, resuspended in 2 L of CO2-free F/2ASW, and allowed to acclimate to air by aeration with atmospheric air. Prior to acclimation to air in the dark, 5% CO2-grown cells were dark adapted for 1 h in 5% CO2.
Extraction of Total RNA and Northern-Blot Analysis
Total RNA was extracted as described by Chomczynski and Sacchi (1987). Total RNA (12 μg) was separated by formaldehyde agarose gel (1%) electrophoresis and blotted onto a nylon membrane (Hybond N+; Amersham Biosciences). The 573 bp of ptca1 cDNA amplified by PCR was used as a probe. The labeling of the probe, hybridization, and signal detection were carried out as described previously (Satoh et al., 2001) using Gene Images random prime labeling module and CDPstar (Amersham Biosciences). The hybridization signal was captured as a digital image and was subjected to the quantitative densitometry using the NIH image program (National Institutes of Health). Densities of bands were converted into relative values (%) of the maximum signal intensity measured in each analysis.
Extraction of Protein and Western-Blot Analysis
Total soluble proteins were extracted as described by Guy and Haskell (1987) with some modifications. Protein concentration was determined by the Bradford assay (Bradford, 1976) using the Bio-Rad Protein Assay Kit. Protein samples (20 μg) were separated by SDS-PAGE (12%) as described by Laemmli (1970) and electroblotted onto Immobilon PVDF membrane (Millipore). The purified protein of PtCA1 (Satoh et al., 2001) was used as the antigen in a custom protocol for rabbit immunization (Asahi Techno Glass). The protein-blotted PVDF membrane was incubated with PtCA1 antibody and detected by biotin-streptavidin system labeled with horseradish peroxidase using the VECTASTAIN Elite ABC kit (Vector Laboratories) and the TMB substrate kit (Vector Laboratories) according to the manufacturer's protocols. Chromogenic signal data were quantitatively analyzed using the NIH image program as described in northern-blot analysis.
Isolation of 5′-Upstream Region of ptca1
The genomic DNA of P. tricornutum was extracted as described by Sambrook and Russell (2001). Five hundred nanograms of genomic DNA was digested with PstI at 37°C for 16 h and then self-ligated with T4 DNA ligase (TaKaRa Bio). The resulting circular genomic DNAs were used as templates for inverse PCR. The primary and the nested PCR were done using two primer pairs (5′-GCGATGGAGGCTGACAAGAAC-3′ and 5′-TAACGTCCTCAAGATCCCGCA-3′ for the primary PCR and 5′-CAGTTTCAAGATGGTCGGTGAG-3′ and 5′-AATGTGGTGGTGTCCGTGCTTC-3′ for the nested PCR) as indicated in Figure 3 using high-fidelity DNA polymerase (KOD-Plus; TOYOBO). The 3-kb fragment amplified by the inverse PCR was cloned into pBluescript II SK+ (Stratagene) and sequenced.
Construction of Chimeric Genes
The full-length uidA gene, which encodes GUS of Escherichia coli, was amplified by PCR using two primers with an attached EcoRI site at the 5′-terminus of the forward primer and a HindIII site at 3′-terminus of the reverse primer. The PCR product was packaged into EcoRI and HindIII sites of the transformation vector pPha-T1 (Zaslavskaia et al., 2000). This plasmid was designated as pFcpApGUS as described in Figure 4. To create a series of 5′-deletions of ptca1 promoter (Pptca1)-uidA constructs, pFcpApGUS plasmid was digested with EcoRI and NdeI to remove the promoter of fcpA and then blunt-ended by T4 DNA polymerase. A series of the 5′-deleted fragments of Pptca1 region (−1,292 to +61, −824 to +61, −484 to +61, −225 to +61, −70 to +61, −50 to +61, and −30 to +61) was amplified by PCR, phosphorylated, and inserted into the blunt-ended site of pFcpApGUS to produce constructs as indicated in Figure 4.
Transformation of P. tricornutum
P. tricornutum cells grown in 5% CO2 under continuous illumination were harvested at mid-logarithmic phase (optical density at 730 nm of 0.3–0.4). Approximately 5×107 cells were spotted as a plaque of 2.5 cm diameter on the surface of the F/2ASW agar plate. Five hundred micrograms of tungsten microcarriers (1.1-μm particle size) were coated with approximately 1 μg of plasmid DNA containing 1 m CaCl2 and 16 mm spermidine. The PDS-1000/He biolistic particle-delivery system (Bio-Rad) was used for microprojectile bombardments of microcarriers. The bombardment was done at 1,550 psi under a negative pressure of 27 inches mercury with a target distance of 6 cm. Bombarded cells were cultured for 1 d under illumination and suspended in 5 mL of F/2ASW. After centrifugation at 3,000 rpm at 20°C for 5 min, cells were resuspended in 0.3 mL of F/2ASW, plated onto F/2ASW agar plates containing 100 μg mL−1 Zeocin, and allowed to grow for 3 to 4 weeks under continuous light.
GUS Assays
The transformants were cultured in 100 mL of F/2ASW in 5% CO2 or air. Five to ten milliliters of cells were harvested and disrupted by a sonicator (Ultrasonic disruptor model UD-201; TOMY Seiko) at output level 3 in 0.5 mL GUS extraction buffer (50 mm sodium phosphate, pH 7.0, 10 mm β-mercaptoethanol, 0.1% [w/v] sodium lauryl sarcosine, 0.1% [v/v] Triton X-100) and then centrifuged at 13,000 rpm for 5 min at 4°C. Twenty microliters of lysate was added to 980 μL of GUS assay buffer (10 mm p-nitrophenyl β-d-glucuronide in the GUS extraction buffer) and incubated at 37°C for 1 h. The reaction was terminated by the addition of 400 μL of 0.5 m Na2CO3 at every 10 min after starting the reaction, and the optical density of PNP released from the substrate was measured at 405 nm.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers S69414 and AF414191 for uidA and ptca1, respectively.
Acknowledgments
We thank Dr. Brian Colman (York University, Toronto, Canada) for critical reading and comments on this manuscript, Dr. Kirk E. Apt (Martek Biosciences, Columbia, MD) for providing the transformation vector, pPha-T1, Ms. Nobuko Higashiuchi for her technical assistance, and Ms. Miyabi Inoue for her skillful secretarial aid.
This research was supported in part by the Kato-Memorial-Bioscience Foundation to Y.M., in part by the Grant of Salt Science Foundation to Y.M., and in part by the University-Industry Joint Research Project of the Ministry of Education, Culture, Sports, Science, and Technology.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.065185.
References
- Badger MR, Andrews TJ, Whitney SM, Ludwig M, Yellowlees DC, Leggat W, Price GD (1998) The diversity and co-evolution of Rubisco, plastids, pyrenoids and chloroplast-based CCMs in the algae. Can J Bot 76: 1052–1071 [Google Scholar]
- Badger MR, Kaplan A, Berry JA (1980) Internal inorganic carbon pool of Chlamydomonas reinhardtii: evidence for a carbon dioxide concentrating mechanism. Plant Physiol 66: 407–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaya D, Grossman AR (1993) Characterization of gene clusters encoding the fucoxanthin chlorophyll proteins of the diatom Phaeodactylum tricornutum. Nucleic Acids Res 21: 4458–4466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozzo GG, Colman B (2000) The induction of inorganic carbon and external carbonic anhydrase in Chlamydomonas reinhardtii is regulated by external CO2 concentration. Plant Cell Environ 23: 1137–1144 [Google Scholar]
- Bozzo GG, Colman B, Matsuda Y (2000) Active transport of CO2 and bicarbonate is induced in response to external CO2 concentration in the green alga Chlorella kessleri. J Exp Bot 349: 1341–1348 [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Chen Y, Cann MJ, Litvin TN, Iourgenko V, Sinclair ML, Levin LR, Buck J (2000) Soluble adenylyl cyclase as an evolutionarily conserved bicarbonate sensor. Science 289: 625–628 [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162: 156–159 [DOI] [PubMed] [Google Scholar]
- Chrivia JC, Kwok RP, Lamb N, Hagiwara M, Montminy MR, Gooman RH (1993) Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature 365: 855–859 [DOI] [PubMed] [Google Scholar]
- Colman B, Rotatore C (1995) Photosynthetic inorganic carbon uptake and accumulation in two marine diatoms. Plant Cell Environ 18: 919–924 [Google Scholar]
- Dionisio-Sese ML, Fukuzawa H, Miyachi S (1990) Light-induced carbonic anhydrase expression in Chlamydomonas reinhardtii. Plant Physiol 94: 1103–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson M, Karlsson J, Ramazanov Z, Garderström P, Samuelsson G (1996) Discovery of an algal mitochondrial carbonic anhydrase: molecular cloning and characterization of a low-CO2-induced polypeptide in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 93: 12031–12034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson M, Villand P, Garderström P, Samuelsson G (1998) Induction and regulation of expression of a low-CO2-induced mitochondrial carbonic anhydrase in Chlamydomonas reinhardtii. Plant Physiol 116: 637–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkowski PG, Katz ME, Knoll AH, Quigg A, Raven JA, Schofield O, Taylor FJR (2004) The evolution of modern eukaryotic phytoplankton. Science 305: 354–360 [DOI] [PubMed] [Google Scholar]
- Fujiwara S, Ishida N, Tsuzuki M (1996) Circadian expression of the carbonic anhydrase gene, Cah1, in Chlamydomonas reinhardtii. Plant Mol Biol 32: 745–749 [DOI] [PubMed] [Google Scholar]
- Fukuzawa H, Fujiwara S, Yamamoto Y, Dionisio-Sese ML, Miyachi S (1990) cDNA cloning, sequence, and expression of carbonic anhydrase in Chlamydomonas reinhardtii: regulation by environmental CO2 concentration. Proc Natl Acad Sci USA 87: 4383–4387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funke RP, Kovar JL, Weeks DP (1997) Intracellular carbonic anhydrase is essential to photosynthesis in Chlamydomonas reinhardtii at atmospheric levels of CO2: demonstration via genomic complementation of the high-CO2-requiring mutant ca-1. Plant Physiol 114: 237–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillard RRL, Ryther JH (1962) Studies of marine planktonic diatoms. I. Cyclotella nana Hustedt and Detonula confervacea (Cleve) Grun. Can J Microbiol 8: 229–239 [DOI] [PubMed] [Google Scholar]
- Guy CL, Haskell D (1987) Induction of freezing tolerance in spinach is associated with the synthesis of cold acclimation induced proteins. Plant Physiol 84: 872–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada H, Matsuda Y (2005) Identification and characterization of a new carbonic anhydrase in the marine diatom Phaeodactylum tricornutum. Can J Bot (in press)
- Harrison PJ, Waters RE, Taylor FJR (1980) A broad spectrum artificial seawater medium for coastal and open ocean phytoplankton. J Phycol 16: 28–35 [Google Scholar]
- Johnston AM, Raven JA (1996) Inorganic carbon accumulation by the marine diatom Phaeodactylum tricornutum. Eur J Phycol 31: 285–290 [Google Scholar]
- Kaplan A, Badger MR, Berry JA (1980) Photosynthesis and intracellular inorganic carbon pool in the blue-green algae Anabaena variabilis: response to external CO2 concentration. Planta 149: 219–226 [DOI] [PubMed] [Google Scholar]
- Karlsson J, Clarke AK, Chen ZY, Hugghins SY, Park YI, Husic HD, Moroney JV, Samuelsson G (1998) A novel α-type carbonic anhydrase associated with the thylakoid membrane in Chlamydomonas reinhardtii is required for growth at ambient CO2. EMBO J 17: 1208–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucho K, Ohyama K, Fukuzawa H (1999) CO2-responsive transcriptional regulation of CAH1 encoding carbonic anhydrase is mediated by enhancer and silencer regions in Chlamydomonas reinhardtii. Plant Physiol 121: 1329–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucho K, Yoshioka S, Taniguchi F, Ohyama K, Fukuzawa H (2003) Cis-acting elements and DNA-binding proteins involved in CO2-responsive transcriptional activation of Cah1 encoding a periplasmic carbonic anhydrase in Chlamydomonas reinhardtii. Plant Physiol 133: 783–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok RP, Lundblad JR, Chrivia JC, Richards JP, Bachinger HP, Brennan RG, Roberts SG, Green MR, Goodman RH (1994) Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature 370: 223–226 [DOI] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Matsuda Y, Colman B (1995. a) Induction of CO2 and bicarbonate transport in green alga Chlorella ellipsoidea. Time course of induction of the two systems. Plant Physiol 108: 247–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda Y, Colman B (1995. b) Induction of CO2 and bicarbonate transport in green alga Chlorella ellipsoidea. Evidence for induction in response to external CO2 concentration. Plant Physiol 108: 253–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda Y, Hara T, Colman B (2001) Regulation of the induction of bicarbonate uptake by dissolved CO2 in the marine diatom Phaeodactylum tricornutum. Plant Cell Environ 24: 611–620 [Google Scholar]
- Matsuda Y, Satoh K, Harada H, Satoh D, Hiraoka Y, Hara T (2002) Regulation of the expressions of HCO3− uptake and intracellular carbonic anhydrase in response to CO2 concentration in the marine diatom Phaeodactylum sp. Funct Plant Biol 29: 279–287 [DOI] [PubMed] [Google Scholar]
- Miller AG, Espie GS, Canvin DT (1990) Physiological aspects of CO2 and HCO3− transport by cyanobacteria: a review. Can J Bot 68: 1291–1302 [Google Scholar]
- Miura K, Yamano T, Yoshioka S, Kohinata T, Inoue Y, Taniguchi F, Asamizu E, Nakamura Y, Tabata S, Yamato KT, Ohyama K, Fukuzawa H (2004) Expression profiling-based identification of CO2-responsive genes regulated by CCM1 controlling a carbon-concentrating mechanism in Chlamydomonas reinhardtii. Plant Physiol 135: 1595–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroney JV, Somanchi A (1999) How do algae concentrate CO2 to increase the efficiency of photosynthetic carbon fixation? Plant Physiol 119: 9–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oike Y, Takakura N, Hata A, Kaname T, Akizuki M, Yamaguchi Y, Yasue H, Araki K, Yamamura K, Suda T (1999) Mice homozygous for a truncated form of CREB-binding protein exhibit defects in hematopoiesis and vasculo-angiogenesis. Blood 93: 2771–2779 [PubMed] [Google Scholar]
- Pastor-Soler N, Beaulieu V, Litvin TN, Da Silva N, Chen Y, Brown D, Buck J, Levin LR, Breton S (2003) Bicarbonate-regulated adenylyl cyclase (sAC) is a sensor that regulates pH-dependent V-ATPase recycling. J Biol Chem 278: 49523–49529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raven JA (1997) CO2-concentrating mechanism: a direct role for thylakoid lumen acidification? Plant Cell Environ 20: 147–154 [Google Scholar]
- Rawat M, Moroney JV (1995) The regulation of carbonic anhydrase and ribulose-1,5-bisphosphate carboxylase/oxygenase activase by light and CO2 in Chlamydomonas reinhardtii. Plant Physiol 109: 937–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rikitake Y, Moran E (1992) DNA-binding properties of the ElA-associated 300-kilodalton protein. Mol Cell Biol 12: 2826–2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SB, Lane TW, Morel FMM (1997) Carbonic anhydrase in the marine diatom Thalassiosira weissflogii (Bacillariophyceae). J Phycol 33: 845–850 [Google Scholar]
- Sambrook J, Russell DW (2001) Molecular Cloning: A Laboratory Manual, Ed 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 6.4–6.11
- Satoh D, Hiraoka Y, Colman B, Matsuda Y (2001) Physiological and molecular biological characterization of intracellular carbonic anhydrase from the marine diatom Phaeodactylum tricornutum. Plant Physiol 126: 1459–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalding MH, Ogren WL (1982) Photosynthesis is required for induction of the CO2-concentrating system in Chlamydomonas reinhardtii. FEBS Lett 145: 41–44 [Google Scholar]
- Sültemeyer DF, Fock HP, Canvin DT (1991) Active uptake of inorganic carbon by Chlamydomonas reinhardtii: evidence for simultaneous transport of HCO3− and CO2 and characterization of active CO2 transport. Can J Bot 69: 995–1002 [Google Scholar]
- Tanaka Y, Nakatsuma D, Harada H, Ishida M, Matsuda Y (2005) Localization of soluble β-carbonic anhydrase in the marine diatom Phaeodactylum tricornutum. Sorting to the chloroplast and cluster formation on the girdle lamellae. Plant Physiol 138: 207–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Naruse I, Maekawa T, Masuya H, Shiroishi T, Ishii S (1997) Abnormal skeletal patterning in embryos lacking a single Cbp allele: a partial similarity with Rubinstein-Taybi syndrome. Proc Natl Acad Sci USA 94: 10215–10220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortell PD, Reinfelder JR, Morel FMM (1997) Active uptake of bicarbonate by diatoms. Nature 390: 243–2449384376 [Google Scholar]
- Tréguer P, Nelson D, Van Bennekom A, DeMaster D, Leynaert A, Quéguiner B (1995) The silica balance in the world ocean: a reestimate. Science 268: 375–379 [DOI] [PubMed] [Google Scholar]
- Villand P, Eriksson M, Samuelsson G (1997) Carbon dioxide and light regulation of promoters controlling the expression of mitochondrial carbonic anhydrase in Chlamydomonas reinhardtii. Biochem J 327: 51–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner D (1977) Introduction with a note on taxonomy. In D. Werner, ed, The Diatoms. Blackwell Scientific Publications, Oxford, pp 1–17
- Zaslavskaia LA, Lippmeier JC, Kroth PG, Grossman AR, Apt KE (2000) Transformation of the diatom Phaeodactylum tricornutum (Bacillariophyceae) with a variety of selectable marker and reporter genes. J Phycol 36: 379–386 [Google Scholar]