Abstract
Phosphatidylethanolamine N-methyltransferase (PEMT) catalyzes phosphatidylcholine synthesis. PEMT knockout mice have fatty livers, and it is possible that, in humans, nonalcoholic fatty liver disease (NAFLD) might be associated with PEMT gene polymorphisms. DNA samples from 59 humans without fatty liver and from 28 humans with NAFLD were genotyped for a single nucleotide polymorphism in exon 8 of PEMT which leads to a V175M substitution. V175M is a loss of function mutation, as determined by transiently transfecting McArdle-RH7777 cells with constructs of wildtype PEMT open reading frame or the V175M mutant. Met/Met at residue 175 (loss of function SNP) occurred in 67.9% of the NAFLD subjects and in only 40.7% of control subjects (p< 0.03). For the first time we report that a polymorphism of the human PEMT gene (V175M) is associated with diminished activity and may confer susceptibility to NAFLD.
Keywords: phosphatidylethanolamine N-methyltransferase (PEMT), choline, liver function, single nucleotide polymorphism (SNP), Nonalcoholic Fatty Liver Disease (NAFLD) pregnancy, neural tube defect (NTD), transfection, site-directed mutagenesis
Introduction
Nonalcoholic Fatty Liver Disease (NAFLD) is the most common reason for abnormal liver function, and may occur in as much as 25% of the population (1). NAFLD can progress to liver cell necrosis, fibrosis and cirrhosis of the liver (1). The mechanisms underlying NAFLD are not well understood. Obesity, diabetes and hypertriglyceridemia are predictive risk factors, but it can appear in humans who are otherwise normal (2, 3). Humans ingesting diets deficient in the nutrient choline develop fatty liver (4, 5) because phosphatidylcholine is required for hepatic secretion of triacylglycerol in very low density lipoproteins (VLDL) (6–8). Phosphatidylethanolamine N -methyltransferase (PEMT; EC 2.1.1.17) catalyzes de novo synthesis of phosphatidylcholine in liver (9, 10) and is responsible for about 30% of phosphatidylcholine formed in liver; the remainder being formed from preexisting choline moiety via an alternative pathway (11). PEMT knockout mice do not express any PEMT activity in liver and completely depend on dietary choline intake to meet daily choline requirements (12, 13). When fed a diet deficient in choline they develop severe fatty liver; a choline supplemented diet prevents this (14) and can reverse hepatic damage if begun early enough (15). The PEMT gene is highly polymorphic; 98 single-nucleotide polymorphisms (SNPs) in PEMT were found in 48 Japanese individuals (16). It is possible that some of these SNPs have functional significance, and if so, could make humans susceptible to fatty liver when dietary intake of choline is low. We identified a variant that resulted in an amino acid substitution (V175M) and report for the first time that this SNP results in partial loss of activity of encoded PEMT and that this SNP occurs 1.7x as frequently in humans with NAFLD as in normal controls.
MATERIALS AND METHODS
Cell culture and transient transfection
McArdle RH-7777 rat hepatoma cells (American Type Culture Collection (ATCC), Manassus, VA) were maintained in Dulbecco’s modified Eagle’s medium/F12 (DMEM/F12; Gibco-BRL Rockville, MD), 10% horse serum, 10% fetal bovine serum, 100 units/ml penicillin (Gibco-BRL) and 100 μg/ml streptomycin sulfate (Gibco-BRL) at 37 °C under 5% CO2. On day 0, cells were plated at a density of 1.5 × 106 cells/60-mm dish. When cells reached 70% confluence they were transfected with test plasmids or mock transfected with empty expression vector using Lipofectamine (Invitrogen, Carlsbad, CA) as per the manufacturer’s protocol. 24 hours post transfection, the expression of enhanced green fluorescent protein (EGFP) was observed under a green fluorescent microscope and the transfection efficiency was calculated based on fluorescence intensity. 48 hours later, cells were harvested and total cellular homogenate and total particulate fraction were prepared for protein assay and enzyme activity analysis.
PEMT activity assay
Liver homogenate or total particulate fraction from McArdle-RH7777 cells were assayed for PEMT activity using a modified method of Ridgway and Vance (17, 18). Briefly, PEMT activity was assayed using 50 μg of protein in 125 mM Tris-HCl (pH 9.2; Mallincrodt, Paris, KY) and 5mM DTT buffer (Sigma, St. Louis, MO) in the presence of 200 μM S-adenosyl-L-methionine containing 0.5 μCi of S-adenosyl-L-methionine (55.70 Ci/mmol; Amersham Biosciences, Piscataway, NJ) and 0.4 mM exogenous phosphatidyldimethylethanolamine (P2, Avanti Polar-lipids, Inc., Alabaster, AL). The reaction was carried out for 30 minutes at 37°C and stopped by addition of chloroform/methanol/hydrochloric acid mixture (100:50:1, v/v). An aliquot of the chloroform phase was applied to a silica gel thin layer chromatography (TLC) plate (Si250-PA (19C)-Silica Gel, Baker, Inc., Phillipsburg, NJ) and was developed in chloroform: methanol: acetic acid: water (50:30:5:2, v/v). Disintegrations per minute from [3H]-PtdCho were determined in bands that co-migrated with authentic standards using liquid scintillation spectrophotometry (Wallac 1410, Pharmacia LKB Nuclear Inc, Gaithersburg, MD).
PEMT protein expression assay
Proteins (25 μg) were separated by SDS-polyacrylamide gel electrophoresis and transferred to a polyvinylidene difluoride membrane (Amersham Biosciences) which was probed with anti-FLAG M2 antibody (Sigma), washed extensively with 1X PBS (Gibco) containing 0.1% Tween 20 (Sigma), and then probed with horseradish peroxidase-conjugated goat anti-mouse IgG (Pierce, Rockford, IL). PEMT-FLAG protein was visualized by a reaction with Supersignal chemiluminescent substrate (Pierce) and exposed to x-ray film (Denville Scientific, Metuchen, NJ). The film was scanned and the integrated optical densities of the bands were measured using the ScionImage software (Scion Corporation, Frederick, MD, USA).
Recombinant plasmid construction /site-directed mutagenesis
Total RNA was extracted from an adult white male human’s liver using Trizol (Life Technologies Inc., Rockville, MD) according to the manufacturer’s instructions. 3μg of total RNA, previously treated with RNase-free DNase I (Life Technologies Inc.), was reverse transcribed using an 18mer oligo(dT) primer and Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA) as per the manufacturer’s instructions. The oligonucleotide (5′MluI PEMT) TAACACGCGTAGTTATGACCCGGCTGCTGGGCTA was used as the 5′ primer, and the oligonucleotide ATCATCATCCTTGTAGTCGAGCACGACGAAGCTGAGAATGTA (3′ PEMT -WT), which adds 19 nucleotides encoding the FLAG epitope (dykddddk), was used as the 3′primer. The PCR product was subcloned into Nhe1 and Mlu1 multicloning sites of mammalian expression vector pBI-EGFP-tet (Clontech, Palo Alto, CA), a bidirectional response plasmid that allows simultaneous expression of both EGFP and PEMT-FLAG under the control of a single tetracycline (doxycycline)-responsive element. With PEMT-WT as the template, site-directed mutagenesis was conducted using GeneTailor™ Site-Directed Mutagenesis System per manufacturer’s instructions (Invitrogen, Carlsbad, CA). Oligonucleotides 5′TGGTGGCCCTCACCTACATAGTGGCTCTCCTATA3′ and 5′ TATGTAGGTGAGGGCCACCAGCACCGTCAG3′ were the forward and reverse primers to introduce the M175V mutation (subsequently we realized that V175M is the SNP and V at 175 is the WT and data analysis was performed accordingly; see discussion).
Human liver specimens
40 human liver samples were obtained from the Liver Procurement and Distribution System (LTPADS) (University of Minnesota, Minneapolis MN 55455, USA; funded by NIH contract N01 DK92310). Of these 40 subjects, 28 had fatty liver and 12 had normal livers. Synopses of tissue donors’ medical histories, including pathologist’s impression diagnosis on liver fat content, were also obtained. The specimens were snap-frozen once removed from the organ donors, delivered on dry ice and stored at −80°C until analysis.
Normal human blood collection
47 healthy volunteers were recruited for a protocol approved by the Institutional Review Board at the University of North Carolina at Chapel Hill. These individuals consumed a diet adequate in choline content and had no liver disease by review of medical history, serum liver function tests (bilirubin, alanine aminotransferase, aspartate aminotransferase, creatine phosphokinase, γ-glutamyl transpeptidase, lactic dehydrogenase, alkaline phosphatase, prothrombin time, partial thromboplastin time, and albumin) and did not have fatty liver as assessed by magnetic resonance imaging of liver (see below). Blood samples were obtained by venipuncture and peripheral lymphocytes isolated by Ficoll-Hypaque gradient using Vacutainer® CPT™ tubes with sodium citrate (Becton Dickinson, Franklin Lakes, NJ) (19, 20) and prepared for SNP analyses as described below.
Magnetic Resonance Imaging of liver
Changes in relative hepatic fat levels were determined utilizing a modified “In and Out of Phase” magnetic resonance imaging (MRI) technique of Dixon (21–23) using a Siemens Vision 1.5T clinical MR system. Briefly, quantification of fat within the liver using MRI is possible because of the resonant frequency differences between fat and water. The resonant frequency differences in fat and water is reflected in the transverse magnetization changes and signal intensity changes at particular time intervals. Utilizing a “breath-hold” fast field echo sequence (FLASH; TE=2.2 msec and 4.5 msec, with a flip angle of 80°, and TR=140 msec) at an echo time of 2.2 msec (TE) MRI signals from water and fat will be 180° out-of-phase with each other while at a TE=4.5 msec the signals from fat and water will be in-phase with each other. MR images obtained from the in-phase and the out-of phase can then be processed to derive the fat fraction from the differences in the MR image pixel intensity values. Serial FLASH MRI studies utilizing a TE= 2.2 and 4.5 msec were performed on subjects. The MR image sets were processed to determine the fat fraction in the liver utilizing software provided by Siemens Medical Solutions (Malvern, PA). Five liver slices in each subject were analyzed and compared to the fat fraction found in the spleen. Additionally, the relative changes in the levels of fat and water were monitored utilizing a localized single volume proton magnetic resonance spectroscopy technique (PRESS; TE=135, TR=1500 msec ) (24).
Genomic DNA extraction
Genomic DNA was extracted from liver tissues using TriZol (Life Technologies Inc.) and from peripheral lymphocytes using PureGene (Gentra Systems, Minneapolis, MN) according to the manufacturer’s instructions.
SNPs detection
DNA sequencing was performed on double stranded DNA templates obtained from genomic DNA by PCR amplification. Exon 8 of PEMT was amplified with the oligonucleotides 5′GGAGCACTTTGCCCCAGAATC3′ and 5′GACTTGGAGCCTTCAGAGCG3′ as forward and reverse primers, respectively. PCR products were purified with QIAquick® PCR Purification Kit 250 (QIAGEN Inc, Valencia CA) according to the manufacturer’s instructions. Sequencing reactions were performed by the University of North Carolina at Chapel Hill Genome Analysis Facility, using a capillary sequencing machine (model 3100, Applied Biosystems, Foster City, CA). The sequences obtained were compared with ones stored in the NCBI database (http://www.ncbi.nlm.nih.gov/entrez/, accession number AF294467) using ClustalW multiple sequence alignment software (http://www.ebi.ac.uk/Tools/sequence.html). Sequence homology identity was determined in accordance with criteria as previously described (25).
Statistics
All data are presented as mean ± standard error of the mean. Differences in the prevalences of V175M polymorphic genotypes in normal controls and patients with NAFLD were tested with Fisher’s Exact Test (26). PEMT activity data were analyzed using one way analysis of variance (27).
RESULTS
In Vitro study
Wildtype and V175M PEMT plasmids were transiently transfected into McArdle-RH7777 cells, which does not show endogenous PEMT activity (Figure 1). Transfection efficiency as assessed by the expression of EGFP was not different in the three groups (vector, wildtype or V175M; data not shown). The amount of PEMT protein expressed was assessed by measuring the FLAG epitope expressed. Whether PEMT activity was expressed per mg protein or per expressed FLAG, the V175M form of PEMT had significantly diminished activity (Figure 1). Thus, V175M is a loss of function polymorphism. We analyzed enzyme activity using saturating concentrations of substrates; it is possible that activity differences between the polymorphic proteins might be different using other incubation conditions.
Figure 1.
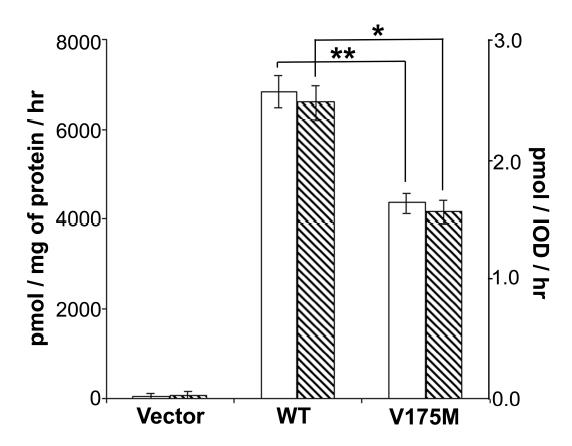
Val to Met substitution at residue 175 of PEMT leads to expression of PEMT with reduced activity.
McArdle-RH7777 cells were transiently transfected with plasmids containing sequence for wild type PEMT (WT) or V175M mutated form (V175M) or vector only (Vector). 50 μg of total particulate protein from cells harvested 48 hr after transfection was used for PEMT activity assay in the presence of 200 μM AdoMet. Results are expressed as mean ± standard error for n=3/group. Open bars: Activity expressed as pmol phosphatidylcholine formed per mg protein per hour. Hatched bars: Activity expressed as pmol phosphatidylcholine formed per integrated optical density of FLAG epitope per hour. The experiment was duplicated with similar results.
* = p<0.01 by one way ANOVA; ** = p<0.005 by one way ANOVA.
Subject demographics
In patients with NAFLD (n=28; all confirmed as NAFLD from liver biopsy specimens with appropriate history), there were 12 males and 16 females, age range was 5–77 years (mean 51 ± 3 years), with 23 Caucasians, 2 African Americans, 1 Hispanic and 1 Asian (ethnicity not known for 1 subject) and with body mass indices ranging from 18.4 to 48.5 (mean 30 ± 1.4 units; BMI unknown for 2 subjects). In control subjects (n=59; 12 confirmed normal from liver biopsy specimens and 47 confirmed as having normal liver by MRI), there were 30 males and 29 females, age range was 5–72 years (mean 37.1 ± 2.1 years), with 37 Caucasians and 14 African Americans, 3 Hispanics, 3 Asians, 1 Native American, and 1 Trinidadian. Body mass indices ranged from 15.3 to 33 (mean 24.7 ± 0.5 units) in the controls.
SNP detection
The V175M (G to A substitution) polymorphism in PEMT was differentially distributed in controls and NAFLD patients. The allele frequency for G and A were 0.19 and 0.81 respectively in 28 NAFLD patients. Among 59 normal controls, they are 0.39 and 0.61 respectively. The frequencies of the three genotypes among patients were GG (Val/Val) 7.1%, GA (Val/Met) 25%, and AA (Met/Met) 67.9%. The frequencies of the three genotypes among normal controls were GG, 18.6%, GA, 40.7%, and AA, 40.7%. (Table 1). Almost twice as many controls had the GG and GA genotype than did the NAFLD patients, while the AA genotype was over represented in the NAFLD patients compared with the normal controls (p=0.02 by two tailed Fischer’s Exact Test). We observed no significant sexual dimorphism in this distribution. The observed distribution of the V175 and M175 alleles is similar to that previously reported in the public SNP databases.
Table 1.
PEMT Exon 8 SNP distribution in human subjects.
| Percent of Population | |||
|---|---|---|---|
| GG Val/Val | GA Val/Met | AA Met/Met | |
| NAFLD (n=28) | 7.1 | 25.0 | 67.9 * |
| Controls (n=59) | 18.6 | 40.7 | 40.7 |
DNA was isolated from subjects with nonalcoholic fatty liver disease (NAFLD) and from controls with normal liver fat and was analyzed for the V175M SNP. Data are presented as percent of group (n/group indicated in parentheses).
= p<0.03 that AA genotype (loss of function SNP) occurred more frequently in NAFLD patients than in control subjects by two-tailed Fisher’s Exact Test.
Discussion
Nonalcoholic fatty liver disease is the most common reason for liver test abnormalities in the general population (1). While it is usually identified in adults with obesity, it can also occur in people with normal body weight without underlying diabetes and hyperlipidemia and has been described in children as well (2, 3). We identified a loss of function SNP in PEMT (G→A in exon 8 of the PEMT gene in humans; results in a V175M substitution in the encoded protein) that occurs more frequently in patients with NAFLD. This SNP was previously detected in humans in a Japanese population but no functional significance was attributed to it (16). Currently, there are no noninvasive tests to diagnose and stage NAFLD (1). Liver biopsy remains the most sensitive diagnostic test but cannot distinguish NAFLD from other causes of fatty liver disease, such as alcohol abuse (28). Identification of genetic predisposition factors to this disease would help to identify individuals who are at risk for NAFLD and who warrant evaluation for pre-symptomatic prediction and prevention.
The mechanism whereby a loss of function SNP in PEMT might be associated with NAFLD involves the role of this enzyme in lipoprotein secretion from liver. Triacylglycerol is formed in the liver and then secreted in very low density lipoprotein (VLDL). Synthesis of new phosphatidylcholine molecules is required for VLDL formation, and when they are not available fat droplets accumulate in the cytosol of liver cells (6, 7, 29). When diet is deficient in choline (4, 8, 30) or when PEMT activity is inhibited or deleted (12, 13, 15, 29), fatty liver ensues.
In the literature, the sequence for human PEMT was originally reported to contain a methionine at residue 175 (GenBank™ accession number AAK19172); however in later reports on human PEMT and in the reported sequence for mouse, rat and cow (GenBank™ accession numbers are NP_009100, AAH26796, Q08388, and AAQ01191, respectively) PEMT contains a valine at this residue (Table 2). Therefore, we argue that in evolution the earliest sequence for PEMT contains a valine at this position, and that the mutation to encode a methionine is the genetic polymorphism.
Table 2.
Amino acid sequence alignment of mammalian PEMT orthologs.
| RAT | LDNPMYWGSTANYLGWALMHASPTGLLLTVLVALVYVVALLFE 180 |
| MOUSE | LDNPMYWGSTANYLGWALMHASPTGLLLTVLVAIVYVVALLYE 180 |
| HUMAN | LDNPMYWGSTANYLGWAIMHASPTGLLLTVLVALTYIVALLYE 180 |
| COW | LDNPMYWGSTAIYLGWAIVHASPTGLLLTALVALIYMVAIVYE 180 |
| ***********:***** :**********:***: * **:: * |
We recognize that our observation is derived from a small study and that the group of NAFLD patients was drawn from a national pool, while many of our control subjects were recruited in North Carolina. The demographics of the two groups were similar, though average BMI was higher in the NAFLD group. Since high BMI can increase incidence of fatty liver, it is possible that the PEMT SNP we report is somehow associated with factors that increase BMI. However, it is unlikely that this is the sole reason that this SNP is associated with risk for NAFLD.
The relatively common occurrence of this loss of function SNP (81% of controls and 93% of NAFLD have at least one allele) suggests that it provides some evolutionary advantage to humans. We previously reported (31) that mice in which this gene is deleted have excess S-adenosylmethionine available for methylation reactions because PEMT activity uses an appreciable portion of available methyl groups for formation of choline. Perhaps, when humans eat enough choline in their diet, the V175M SNP is beneficial because it reduces waste of methyl-groups for making choline moiety. Sometimes a SNP is selected for because it protects against disease (32). Human erythrocytes infected with Plasmodium develop new pathways for accumulating choline from plasma (33). Up to 45% of the malaria parasite is composed of phosphatidylcholine and the malaria parasite actively accumulates choline from its host (34). Finally, there is a clear correlation between antimalarial activity of some drugs and their ability to inhibit choline uptake into the parasite (35). Perhaps the V175M SNP that we describe diminishes the availability of choline in the human host and thereby impairs the replication of the malaria parasite. Whatever the reason, it is interesting that the V175M SNP is so prevalent in humans.
It is interesting that the incidence of NAFLD is lower in premenopausal women than in men or postmenopausal women (36). This would be consistent with our hypothesis that low PEMT activity is a risk factor for developing NAFLD. Female rats are less likely to develop fatty liver when fed choline deficient diets than are male rats (37), because females have greater capacity to form the choline moiety de novo via PEMT pathway in liver. It is estimated that female rats have 10–50% more PEMT activity than do males (38, 39). A woman’s capacity to form the choline moiety de novo may be highest before menopause because estrogens increase PEMT activity in humans (40) and in castrated-rats (41). Thus, premenopausal women may be less sensitive to a loss of function SNP in PEMT because they have excess PEMT activity as compared to men or postmenopausal women.
The requirement for choline (from diet or from PEMT synthesis) is spared, in part, by the availability of methyl groups from 1-carbon metabolism (via methyltetrahydrofolate) (42). It is possible that the PEMT SNP we describe will interact with other commonly known SNPs in humans. For example, the thermolabile variant (677C→T) of 5,10-methylenetetrahydrofolate reductase (MTHFR, E.C. 1.5.1.20) occurs in 15–30% of humans (43). We found that mice in which MTHFR was deleted develop fatty liver that resolves when mice are fed the choline metabolite betaine (43). These mice require more choline or betaine because homocysteine remethylation to methionine, in the absence of 1-carbon units via the folate pathway shifts to a pathway that uses choline as a precursor. Homocysteine can be remethylated to methionine by methionine synthase, using 5-methylfolate which is supplied by MTHFR (43). Alternatively, betaine:homocysteine methyltransferase (BHMT, EC 2.1.1.5) catalyzes a methyl transfer from betaine to homocysteine (43). When 5-methylfolate is not available, more betaine is required. Thus, humans who have diminished capacity to synthesize choline moiety via PEMT activity, and who have diminished capacity to form 5-methylfolate will have difficulty producing increased betaine from choline when it is needed for homocysteine methylation.
We studied a relatively small number of subjects (28 with NAFLD and 59 controls); it would be valuable to characterize this SNP in larger populations. It would also be useful to determine whether diets high in choline reduce hepatic steatosis in humans with this SNP. We recently published data on choline content of foods (44), and the US Department of Agriculture maintains an updated food composition table online (http://www.nal.usda.gov/fnic/foodcomp/Data/Choline/Choline.html). We are currently examining whether there are other loss of function SNPs in the PEMT gene.
Acknowledgments
We thank Olga G. Kozyreva, PhD and Umadevi Veluvolu, MS for their technical assistance and Lawrence L. Kupper, PhD for his assistance with statistical analyses. This work was supported by grants from the NIH to SZ (DK55865, AG09525). JS is a recipient of the Royster Fellowship from UNC-Chapel Hill. Support for this work was also provided by grants from the NIH to the UNC Clinical Nutrition Research Unit (DK56350) and the Center for Environmental Health and Susceptibility (ES10126).
References
- 1.Browning JD, Horton JD. Molecular mediators of hepatic steatosis and liver injury. J Clin Invest. 2004;114:147–152. doi: 10.1172/JCI22422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brunt EM. Nonalcoholic steatohepatitis: definition and pathology. Semin Liver Dis. 2001;21:3–16. doi: 10.1055/s-2001-12925. [DOI] [PubMed] [Google Scholar]
- 3.Falck-Ytter Y, Younossi ZM, Marchesini G, McCullough AJ. Clinical features and natural history of nonalcoholic steatosis syndromes. Semin Liver Dis. 2001;21:17–26. doi: 10.1055/s-2001-12926. [DOI] [PubMed] [Google Scholar]
- 4.Zeisel SH, daCosta KA, Franklin PD, Alexander EA, Lamont JT, Sheard NF, Beiser A. Choline, an essential nutrient for humans. FASEB J. 1991;5:2093–2098. [PubMed] [Google Scholar]
- 5.Buchman A, Dubin M, Moukarzel A, Jenden D, Roch M, Rice K, Gornbein J, Ament M. Choline deficiency: a cause of hepatic steatosis during parenteral nutrition that can be reversed with intravenous choline supplementation. Hepatology. 1995;22:1399–1403. [PubMed] [Google Scholar]
- 6.Yao ZM, Vance DE. The active synthesis of phosphatidylcholine is required for very low density lipoprotein secretion from rat hepatocytes. J Biol Chem. 1988;263:2998–3004. [PubMed] [Google Scholar]
- 7.Yao ZM, Vance DE.1989Head group specificity in the requirement of phosphatidylcholine biosynthesis for very low density lipoprotein secretion from cultured hepatocytes J Biol Chem 26411373–11380. [PubMed] [Google Scholar]
- 8.Yao ZM, Vance DE. Reduction in VLDL, but not HDL, in plasma of rats deficient in choline. Biochem Cell Biol. 1990;68:552–558. doi: 10.1139/o90-079. [DOI] [PubMed] [Google Scholar]
- 9.Bremer J, Greenberg D. Methyl transfering enzyme system of microsomes in the biosynthesis of lecithin (phosphatidylcholine) Biochim Biophys Acta. 1961;46:205–216. [Google Scholar]
- 10.Vance DE, Walkey CJ, Cui Z. Phosphatidylethanolamine N-methyltransferase from liver. Biochim Biophys Acta. 1997;1348:142–150. doi: 10.1016/s0005-2760(97)00108-2. [DOI] [PubMed] [Google Scholar]
- 11.Reo NV, Adinehzadeh M, Foy BD. Kinetic analyses of liver phosphatidylcholine and phosphatidylethanolamine biosynthesis using (13)C NMR spectroscopy. Biochim Biophys Acta. 2002;1580:171–188. doi: 10.1016/s1388-1981(01)00202-5. [DOI] [PubMed] [Google Scholar]
- 12.Walkey CJ, Donohue LR, Bronson R, Agellon LB, Vance DE. Disruption of the murine gene encoding phosphatidylethanolamine N-methyltransferase. Proc Natl Acad Sci U S A. 1997;94:12880–12885. doi: 10.1073/pnas.94.24.12880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu X, Song J, Mar MH, Edwards LJ, Zeisel SH. Phosphatidylethanolamine N-methyltransferase (PEMT) knockout mice have hepatic steatosis and abnormal hepatic choline metabolite concentrations despite ingesting a recommended dietary intake of choline. Biochem J. 2003;370:987–993. doi: 10.1042/BJ20021523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walkey CJ, Yu L, Agellon LB, Vance DE. Biochemical and evolutionary significance of phospholipid methylation. J Biol Chem. 1998;273:27043–27046. doi: 10.1074/jbc.273.42.27043. [DOI] [PubMed] [Google Scholar]
- 15.Waite KA, Cabilio NR, Vance DE. Choline deficiency-induced liver damage is reversible in Pemt(−/−) mice. J Nutr. 2002;132:68–71. doi: 10.1093/jn/132.1.68. [DOI] [PubMed] [Google Scholar]
- 16.Saito S, Iida A, Sekine A, Miura Y, Sakamoto T, Ogawa C, Kawauchi S, Higuchi S, Nakamura Y. Identification of 197 genetic variations in six human methyltransferase genes in the Japanese population. J Hum Genet. 2001;46:529–537. doi: 10.1007/s100380170035. [DOI] [PubMed] [Google Scholar]
- 17.Ridgway ND, Vance DE. Kinetic mechanism of phosphatidylethanolamine N-methyltransferase. J Biol Chem. 1988;263:16864–16871. [PubMed] [Google Scholar]
- 18.Ridgway ND, Vance DE. Specificity of rat hepatic phosphatidylethanolamine N-methyltransferase for molecular species of diacyl phosphatidylethanolamine. J Biol Chem. 1988;263:16856–16863. [PubMed] [Google Scholar]
- 19.Fotino M, Merson EJ, Allen FH., Jr Micromethod for rapid separation of lymphocytes from peripheral blood. Ann Clin Lab Sci. 1971;1:131–133. [PubMed] [Google Scholar]
- 20.Ting A, Morris PJ. A technique for lymphocyte preparation from stored heparinized blood. Vox Sang. 1971;20:561–563. doi: 10.1111/j.1423-0410.1971.tb00469.x. [DOI] [PubMed] [Google Scholar]
- 21.Dixon WT. Simple proton spectroscopic imaging. Radiology. 1984;153:189–194. doi: 10.1148/radiology.153.1.6089263. [DOI] [PubMed] [Google Scholar]
- 22.Fishbein MH, Gardner KG, Potter CJ, Schmalbrock P, Smith MA. Introduction of fast MR imaging in the assessment of hepatic steatosis. Magn Reson Imaging. 1997;15:287–293. doi: 10.1016/s0730-725x(96)00224-x. [DOI] [PubMed] [Google Scholar]
- 23.Lee JK, Dixon WT, Ling D, Levitt RG, Murphy WA., Jr Fatty infiltration of the liver: demonstration by proton spectroscopic imaging. Preliminary observations. Radiology. 1984;153:195–201. doi: 10.1148/radiology.153.1.6089264. [DOI] [PubMed] [Google Scholar]
- 24.Bottomly PA. Spatial localization in NMR spectroscopy in vivo. . Ann NY Acad Sci. 1987;508:376–385. doi: 10.1111/j.1749-6632.1987.tb32915.x. [DOI] [PubMed] [Google Scholar]
- 25.Goldenberger D, Kunzli A, Vogt P, Zbinden R, Altwegg M. Molecular diagnosis of bacterial endocarditis by broad-range PCR amplification and direct sequencing. J Clin Microbiol. 1997;35:2733–2739. doi: 10.1128/jcm.35.11.2733-2739.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Analyze-It for Microsoft Excel (2004) Analyze-It for Microsoft Excel, Ltd., Leeds, UK
- 27.SAS (1999) SAS/STAT® User’s Guide, Version 8. SAS Institute, Inc., Cary, NC
- 28.Bravo AA, Sheth SG, Chopra S. Liver biopsy. N Engl J Med. 2001;344:495–500. doi: 10.1056/NEJM200102153440706. [DOI] [PubMed] [Google Scholar]
- 29.Noga AA, Zhao Y, Vance DE. An unexpected requirement for phosphatidylethanolamine N-methyltransferase in the secretion of very low density lipoproteins. J Biol Chem. 2002;277:42358–42365. doi: 10.1074/jbc.M204542200. [DOI] [PubMed] [Google Scholar]
- 30.da Costa KA, Garner SC, Chang J, Zeisel SH. Effects of prolonged (1 year) choline deficiency and subsequent refeeding of choline on 1,2,-sn-diradylglycerol, fatty acids and protein kinase C in rat liver. Carcinogenesis. 1995;16:327–334. doi: 10.1093/carcin/16.2.327. [DOI] [PubMed] [Google Scholar]
- 31.Zhu X, Mar MH, Song J, Zeisel SH. Deletion of the Pemt gene increases progenitor cell mitosis, DNA and protein methylation and decreases calretinin expression in embryonic day 17 mouse hippocampus. Brain Res Dev Brain Res. 2004;149:121–129. doi: 10.1016/j.devbrainres.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 32.Yates Z, Lucock M. Methionine synthase polymorphism A2756G is associated with susceptibility for thromboembolic events and altered B vitamin/thiol metabolism. Haematologica. 2002;87:751–756. discussion 756. [PubMed] [Google Scholar]
- 33.Kirk K, Wong HY, Elford BC, Newbold CI, Ellory JC. Enhanced choline and Rb+ transport in human erythrocytes infected with the malaria parasite Plasmodium falciparum. Biochem J. 1991;278 (Pt 2):521–525. doi: 10.1042/bj2780521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lehane AM, Saliba KJ, Allen RJ, Kirk K. Choline uptake into the malaria parasite is energized by the membrane potential. Biochem Biophys Res Commun. 2004;320:311–317. doi: 10.1016/j.bbrc.2004.05.164. [DOI] [PubMed] [Google Scholar]
- 35.Ancelin ML, Calas M, Bompart J, Cordina G, Martin D, Ben Bari M, Jei T, Druilhe P, Vial HJ. Antimalarial activity of 77 phospholipid polar head analogs: close correlation between inhibition of phospholipid metabolism and in vitro Plasmodium falciparum growth. Blood. 1998;91:1426–1437. [PubMed] [Google Scholar]
- 36.Shen L, Fan JG, Shao Y, Zeng MD, Wang JR, Luo GH, Li JQ, Chen SY. Prevalence of nonalcoholic fatty liver among administrative officers in Shanghai: an epidemiological survey. World J Gastroenterol. 2003;9:1106–1110. doi: 10.3748/wjg.v9.i5.1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tessitore L, Sesca E, Greco M, Pani P, Dianzani M. Sexually differentiated response to choline in choline deficiency and ethionine intoxication. Int J Exper Path. 1995;76:125–129. [PMC free article] [PubMed] [Google Scholar]
- 38.Bjornstad P, Bremer J. In vivo studies on pathways for the biosynthesis of lecithin in the rat. J Lipid Res. 1966;7:38–45. [PubMed] [Google Scholar]
- 39.Lyman RL, Sheehan G, Tinoco J. Diet and 14CH3-methionine incorporation into liver phosphatidylcholine fractions of male and female rats. Can J Biochem. 1971;49:71–79. doi: 10.1139/o71-011. [DOI] [PubMed] [Google Scholar]
- 40.Drouva SV, LaPlante E, Leblanc P, Bechet JJ, Clauser H, Kordon C. Estradiol activates methylating enzyme(s) involved in the conversion of phosphatidylethanolamine to phosphatidylcholine in rat pituitary membranes. Endocrinol. 1986;119:2611–2622. doi: 10.1210/endo-119-6-2611. [DOI] [PubMed] [Google Scholar]
- 41.Young DL. Estradiol- and testosterone-induced alterations in phosphatidylcholine and triglyceride synthesis in hepatic endoplasmic reticulum. J Lipid Res. 1971;12:590–595. [PubMed] [Google Scholar]
- 42.Zeisel SH, Blusztajn JK. Choline and human nutrition. Ann Rev Nutr. 1994;14:269–296. doi: 10.1146/annurev.nu.14.070194.001413. [DOI] [PubMed] [Google Scholar]
- 43.Schwahn BC, Chen Z, Laryea MD, Wendel U, Lussier-Cacan S, Genest J, Jr, Mar MH, Zeisel SH, Castro C, Garrow T, Rozen R. Homocysteine-betaine interactions in a murine model of 5,10-methylenetetrahydrofolate reductase deficiency. FASEB J. 2003;17:512–514. doi: 10.1096/fj.02-0456fje. [DOI] [PubMed] [Google Scholar]
- 44.Zeisel SH, Mar MH, Howe JC, Holden JM. Concentrations of choline-containing compounds and betaine in common foods. J Nutr. 2003;133:1302–1307. doi: 10.1093/jn/133.5.1302. [DOI] [PubMed] [Google Scholar]


