Abstract
Activation of proMMP-2 by MT1-MMP is considered to be a critical event in cancer cell invasion. In the activation step, TIMP-2 bound to MT1-MMP on the cell surface acts as a receptor for proMMP-2. Subsequently, adjacent TIMP-2-free MT1-MMP activates the proMMP-2 in the ternary complex. In this study, we demonstrate that MT1-MMP forms a homophilic complex through the hemopexin-like (PEX) domain that acts as a mechanism to keep MT1-MMP molecules close together to facilitate proMMP-2 activation. Deletion of the PEX domain in MT1-MMP, or swapping the domain with the one derived from MT4-MMP, abolished the ability to activate proMMP-2 on the cell surface without affecting the proteolytic activities. In addition, expression of the mutant MT1-MMP lacking the catalytic domain (MT1PEX-F) efficiently inhibited complex formation of the full-length enzymes and activation of pro MMP-2. Furthermore, expression of MT1PEX-F inhibited proMMP-2 activation and Matrigel invasion activity of invasive human fibrosarcoma HT1080 cells. These findings elucidate a new function of the PEX domain: regulating MT1-MMP activity on the cell surface, which accelerates cellular invasiveness in the tissue.
Keywords: activation/homophilic complex/invasion/MT1-MMP/proMMP-2
Introduction
The extracellular matrix (ECM) is a physical barrier for cells migrating in tissue; therefore, it has to be degraded in the direction that the cells are migrating. Matrix metalloproteinases (MMPs) are a group of enzymes that are thought to be critical for this process (Nagase and Woessner, 1999). To date, 21 MMP genes have been cloned in mammals (Nagase and Woessner, 1999; Pei, 1999; Benoit De Coignac et al., 2000; Park et al., 2000; Velasco et al., 2000; Lohi et al., 2001) and their products can be subgrouped into soluble MMPs and membrane-anchored MMPs (membrane-type MMPs, MT-MMPs) (Seiki, 1999). Soluble MMPs are thought to be responsible for ECM degradation over broad areas of tissue, while MT-MMPs are thought to participate in pericellular ECM degradation, since they are tethered to the plasma membrane (Nagase and Woessner, 1999; Seiki, 1999; Hotary et al., 2000).
MMPs are composed of the conserved domain structures of a pre/propeptide, a catalytic domain, a hinge and a hemopexin-like domain (PEX) (Nagase and Woessner, 1999). In MT-MMPs, an additional transmembrane and cytoplasmic domain (Sato et al., 1994) or a glycosylphosphatidyl inositol (GPI) anchor signal (Itoh et al., 1999; Kojima et al., 2000) are present. Among these domains, PEX is uniquely present in the MMP family and is not found in the other metzincin family of metalloprotein ases. All MMPs have this domain, except for MMP-7, MMP-23 (Nagase and Woessner, 1999) and MMP-26 (Benoit De Coignac et al., 2000; Park et al., 2000). Thus, it is conceivable that MMPs might have acquired certain unique functions during evolution by having PEX. Thus, understanding the role of the PEX domain may help in understanding their unique biological activities.
MT1-MMP was the first MT-MMP to be discovered, and has been shown to promote cancer invasion and metastasis (Sato et al., 1994; Seiki, 1999). It degrades collagen types I, II and III, fibronectin, laminin 1 and 5, vitronectin, fibrin and aggrecan (d’Ortho et al., 1997; Ohuchi et al., 1997; Buttner et al., 1998; Fosang et al., 1998; Hiraoka et al., 1998; Hotary et al., 2000; Koshikawa et al., 2000). MT1-MMP also activates other MMPs, such as proMMP-2 (progelatinase A) (Sato et al., 1994) and proMMP-13 (procollagenase 3) (Knäuper et al., 1996). Thus, expression of MT1-MMP is thought to initiate multiple proteinase cascades on the cell surface. MT1-MMP has been shown to participate not only in cancer cell invasion and metastasis (Sato et al., 1994; Seiki, 1999), but also in other physiological processes such as angiogenesis (Hiraoka et al., 1998; Zhou et al., 2000), wound healing (Okada et al., 1997) and skeletal development (Holmbeck et al., 1999; Zhou et al., 2000). Thus, MT1-MMP is likely to be essential for both physiological and pathological processes of invasion.
The cell surface activation of proMMP-2 by MT1-MMP has been considered to be a particularly important step in tumor invasion because MMP-2 degrades collagen type IV and laminin, which are the major components of basement membrane (Stetler-Stevenson et al., 1993). The appearance of the active MMP-2 is well correlated with tumor invasiveness (Nomura et al., 1995; Tokuraku et al., 1995). The activation steps include the tri-molecular complex formation of MT1-MMP, TIMP-2 and proMMP-2 (Strongin et al., 1995; Butler et al., 1998; Kinoshita et al., 1998). The interaction of MT1-MMP and TIMP-2 is through the active site of MT1-MMP and the inhibitory site of TIMP-2; thus, the activity of MT1-MMP is inhibited in the complex (Imai et al., 1996; Itoh et al., 1998). This MT1-MMP–TIMP-2 complex acts as a receptor to bind proMMP-2 to the cell surface through the interaction of the exposed TIMP-2 C-terminal domain and the PEX of proMMP-2 (Butler et al., 1998; Kinoshita et al., 1998). Since the ability of MT1-MMP in the complex to process the propeptide of MMP-2 proteolytically is inhibited by TIMP-2, MT1-MMP that is adjacent to the complex but not interacting with TIMP-2 is required for the processing of proMMP-2 (Butler et al., 1998; Kinoshita et al., 1998). However, it is unclear whether the molecular arrangement of the tri-molecular complex and the adjacent MT1-MMP on the cell surface occurs randomly or in a regulated manner.
Here, we report that MT1-MMP forms a homophilic complex on the cell surface through the PEX domain, and that this complex facilitates proMMP-2 activation and regulates cellular invasiveness. These results suggest the importance of the PEX domain of MT1-MMP in proMMP-2 activation, as well as in promoting tumor cell invasion.
Results
MT1-MMP forms a homophilic complex through the PEX domain
The activation of proMMP-2 by MT1-MMP on the cell surface requires the ternary complex formation of proMMP-2, TIMP-2 and MT1-MMP, and also the presence of TIMP-2-free MT1-MMP near the ternary complex (Strongin et al., 1995; Butler et al., 1998; Kinoshita et al., 1998). Therefore, if TIMP-2-free MT1-MMP and the ternary complex are far apart, activation may not occur efficiently. We thus hypothesized that MT1-MMP would form a homophilic complex on the cell surface to arrange the effective activation complex for MMP-2. To test this possibility, FLAG-tagged MT1-MMP (MT1-F) and c-Myc-tagged MT1-MMP (MT1-Myc) (Figure 1A) were co-expressed in COS1 cells, and MT1-F was immunoprecipitated with anti-FLAG M2 antibody-conjugated beads. As shown in Figure 1B, MT1-Myc was co-immunoprecipitated with MT1-F (lane 6, Anti-FLAG and Anti-Myc), whereas MT1-Myc alone did not immunoprecipitate (lane 7, Anti-Myc). This indicates that the MT1-MMP molecules with different tags formed a complex with each other. Next, to test the domain requirement for complex formation, MT1-F was co-expressed with MT1-MMP lacking either the PEX domain (MT1Cat) or the catalytic domain (MT1PEX) (Figure 1A), and subjected to immunoprecipitation with anti-FLAG beads. The samples were then subjected to western blotting using anti-FLAG M2, anti-MT1PEX and anti-MT1Cat to examine the species co-immunoprecipitated with MT1-F. As shown in Figure 1C, MT1PEX, but not MT1Cat, was co-immunoprecipitated with MT1-F (Anti-PEX and Anti-Cat, lanes 8 and 9, respectively). This indicates that MT1-MMP forms a homophilic complex through the PEX domain.
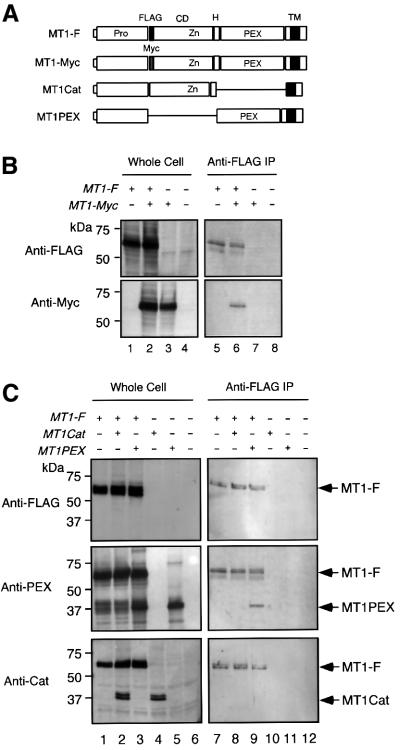
Fig. 1. MT1-MMP forms a homophilic complex through the PEX domain. (A) A schematic representation of MT1-MMP mutants used in the experiments. Pro, propeptide; FLAG, FLAG epitope; Myc, c-Myc epitope; CD, catalytic domain; H, hinge; PEX, hemopexin-like domain; TM, transmembrane domain. (B) COS1 cells were transfected with the expression plasmid for MT1-F and/or MT1-Myc. The cells were lysed and subjected to immunoprecipitation using anti-FLAG M2 antibody-conjugated beads as described in Materials and methods. The whole-cell lysates (Whole Cell) and immunoprecipitated materials (Anti-FLAG IP) were subjected to western blotting using an anti-FLAG M2 antibody (Anti-FLAG) or anti-c-Myc antibody (Anti-Myc). (C) COS1 cells were transfected with the expression plasmids for MT1-F, MT1Cat and/or MT1PEX. The cells were lysed and subjected to immunoprecipitation using anti-FLAG beads as above. The samples were analyzed by western blotting using an anti-FLAG M2 antibody (Anti-FLAG), anti-MT1-MMP PEX (Anti-PEX) or anti-MT1Cat (Anti-Cat).
MT1-MMP forms a homophilic complex directly through the PEX domain
To test whether the MT1-MMP complex formation is a result of direct interaction or not, MT1-MMP without the propeptide and transmembrane/cytoplasmic domains (MT1EAΔTM) (Figure 2A) was expressed in Escherichia coli and refolded. The active site was mutated to make it inactive (Glu248 to Ala) to avoid autodegradation during experiments. This mutation should not create an overall structural alteration, as has been shown in other MMPs (Atkinson et al., 1995). The expressed protein appeared to be >98% pure, as shown in the inset in Figure 2B. MT1EAΔTM had a different mobility under reducing (Red) and non-reducing (NR) SDS–PAGE, suggesting that the disulfide bond in the PEX domain was correctly formed and there is no intermolecular disulfide bond formation. Also, purified MT1EAΔTM formed a complex with TIMP-2 (data not shown), suggesting that the catalytic domain was also folded correctly.
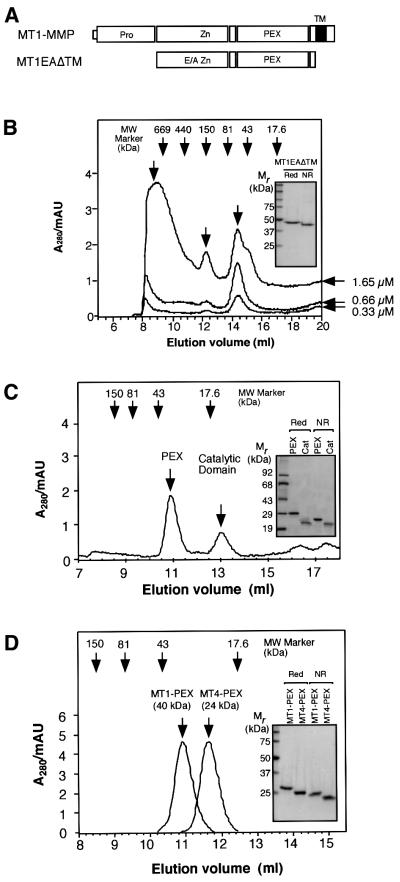
Fig. 2. MT1-MMP directly forms a homophilic complex. (A) A schematic representation of MT1EAΔTM expressed in E.coli. E/A, Glu240 to Ala mutation. (B) Gel filtration analysis of MT1EAΔTM at different concentrations. Three different concentrations of MT1EAΔTM (0.33, 0.66 and 1.65 µM) were subjected to gel-permeation column chromatography on Superdex 200. The inset shows SDS–PAGE analysis of MT1EAΔTM under reducing (Red) and non-reducing (NR) conditions. Bands were visualized by Coomassie Blue. (C) MT1EAΔTM (50 µg/ml) was treated with trypsin (0.1 µg/ml) at 37°C for 1 h to separate the catalytic domain and PEX. Trypsin was inactivated by the addition of 2 mM phenylmethylsulfonyl fluoride to the mixture. The sample was then subjected to gel-permeation column chromatography on Superdex 75. The inset shows the SDS–PAGE analysis of the purified PEX and catalytic domains on the column under reducing and non-reducing conditions; the bands were visualized by Coomassie Blue. (D) Escherichia coli-expressed MT1-MMP PEX and MT4-MMP PEX were subjected to gel-permeation column chromatography on Superdex 75. The inset shows the SDS–PAGE analysis of MT4-MMP PEX under reducing and non-reducing conditions; the bands were visualized by Coomassie Blue.
MT1EAΔTM did not elute from the gel-permeation column where expected for its size of 49 kDa, but rather at a volume suggesting a larger molecular mass (Figure 2B). This suggested that MT1-MMP formed a homophilic complex. The formation of the complex was concentration dependent (Figure 2B). At a concentration of 0.33 µM, there was a monomer peak around 50 kDa, a dimer peak around 130 kDa and a higher oligomer peak above 600 kDa. The ratio of the peak heights was 1:0.2:0.78, respectively. Increasing the concentration to 0.66 µM resulted in a similar ratio of 1:0.3:0.8. However, at 1.65 µM, the ratio was 1:0.75:1.6, representing the increased relative amounts of the higher molecular mass complex.
Next, MT1EAΔTM was treated with trypsin to separate the catalytic and PEX domains, since trypsin was found to cleave the middle of the hinge region, which contains several Arg and Lys. As shown in Figure 2C, these domains were separated as expected, and the catalytic domain was eluted at around 17 kDa, but the PEX domain was eluted at a volume corresponding to ∼40 kDa. It is notable that trypsin treatment resulted in loss of all the high molecular mass peaks as well as monomer peaks, suggesting that the PEX domain alone has tighter affinity to form a dimer complex. Since the catalytic domain and PEX migrate in SDS–PAGE under non-reducing conditions to 21 and 24 kDa, respectively (Figure 2C, inset, NR), it is suggested that PEX, but not the catalytic domain, forms a homophilic complex in solution.
To test whether the complex formation of PEX is specific to MT1-MMP, the PEX of MT4-MMP (MT4- PEX) was also expressed in E.coli. The calculated molecular weight based on amino acid composition is 26 714 for MT4-PEX and 26 304 for MT1-PEX. The overall topological structure of PEX is also expected to be similar. As shown in Figure 2D, the apparent molecular mass of MT4-PEX is almost identical to that of MT1-PEX on SDS–PAGE under reducing and non-reducing conditions. However, on gel-permeation chromatography, MT4-PEX eluted at ∼24 kDa (Figure 2D), whereas MT1-PEX eluted at ∼40 kDa, suggesting that dimer formation was specific for MT1-PEX (Figure 2D, inset). These results indicate that MT1-MMP forms a homophilic complex through the direct interaction of the PEX domains.
Activation of proMMP-2 by MT1-MMP on the cell surface requires PEX
To test the importance of the MT1-MMP PEX domain in activation of proMMP-2, the PEX domain was exchanged with one derived from MT4-MMP (Figure 3A, MT1-MT4PEX). As shown in Figure 3B, COS1 cells that express MT1-MT4PEX did not activate proMMP-2 (top panel), although the expression level is similar to that of the wild-type enzyme (second panel, Anti-Cat). MT1-MT4PEX is expressed on the cell surface, as the molecule was sensitive to surface biotinylation (bottom panel, Surface Biotinylation). MT1-MT4PEX can form a ternary complex of MT1-MT4PEX–TIMP-2–pro MMP-2 on the cell surface as the proMMP-2 bound to MT1-MT4PEX-expressing cells (Figure 3C, lanes 2 and 3). To examine whether MT1-MT4PEX is proteolytically active on the cell surface, the transfected cells were cultured on a DQ gelatin substrate. DQ gelatin is a fluorescence-quenched substrate that fluoresces upon proteolytic cleavage. As shown in Figure 3D, MT1-MMP- as well as MT1-MT4PEX-expressing cells showed gelatinolytic activity, whereas mock-transfected cells did not. This activity was also completely inhibited by BB94 (data not shown). We have also confirmed that the PEX domain-deleted MT1-MMP (MT1Cat) cannot activate proMMP-2, although it retains its proteolytic activity as well as its ability to form the ternary complex of MT1Cat–TIMP-2–proMMP-2 on the cell surface, as does MT1-MT4PEX (data not shown). Taken together, these data show that the PEX domain plays an important role in proMMP-2 activation, presumably by forming a homophilic complex.
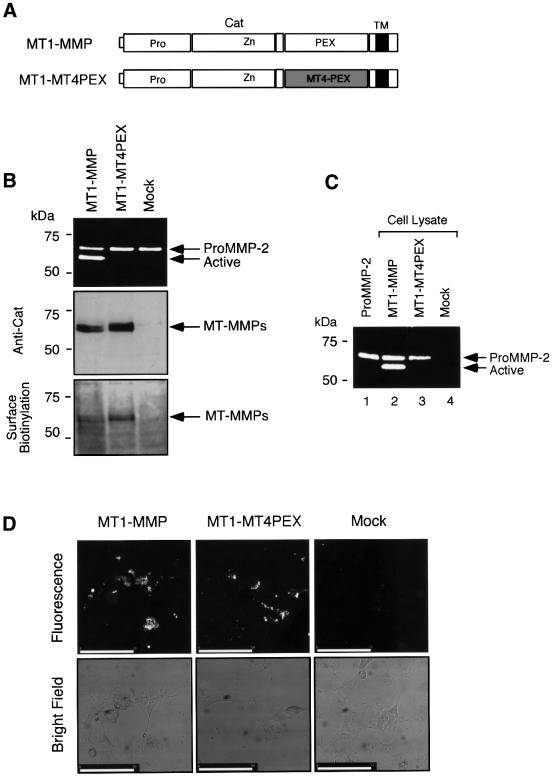
Fig. 3. Examination of MT1-MT4PEX for the activation of proMMP-2 on the cell surface. (A) A schematic representation of MT1-MT4PEX. MT4-PEX, PEX domain derived from MT4-MMP. (B) Activation of proMMP-2 by MT1-MMPs. COS1 cells were transfected with the expression plasmids for MT1-MMP, MT1-MT4PEX or vector alone (Mock), and these cells were reacted with purified proMMP-2 (0.25 µg/ml) in the culture medium without serum at 37°C for 18 h. The medium was analyzed by zymography for proMMP-2 processing (top panel), and the cell lysate by western blotting using anti-MT1Cat (middle panel, Anti-Cat). Transfected cells were also subjected to surface biotinylation as described in Materials and methods, and analyzed by western blotting using anti-MT1Cat (bottom panel, Surface Biotinylation). (C) Binding of proMMP-2 to the MT1-MT4PEX-expressing cells tested as described in Materials and methods. Samples were analyzed by gelatin zymography. (D) In situ gelatin degradation activities of MT1-MT4PEX-expressing cells were studied by culturing cells on DQ gelatin-coated coverslips as described in Materials and methods. Gelatin degradation activity was visualized as the fluorescent area. A bright field image is also shown. Bars: 50 µm.
MT1-MMP forms a homophilic complex on the cell surface
To monitor the dimer formation of the enzyme in the cells, we took another approach to detect the molecular interactions. Growth factor receptors, including nerve growth factor receptor (NGFR), form a dimer or oligomer complex through the ectodomain upon binding to their specific ligands (Jing et al., 1992; Lemmon and Schlessinger, 1994). Subsequently, the cytoplasmic domains containing tyrosine kinase (TK) are brought into close proximity, resulting in trans-phosphorylation at the tyrosine residues (Jing et al., 1992; Lemmon and Schlessinger, 1994). It is thus possible to determine the specific homo-dimer interactions of a protein by making a chimeric protein with a receptor molecule. For example, the fusion protein between the cytoplasmic domain of NGFR and BCR of the oncoprotein BCR-ABL resulted in constitutive phosphorylation of NGFR’s tyrosine residues since BCR forms a tetrameric complex (Maru et al., 1998). We thus constructed a chimeric protein consisting of the ectodomain of MT1-MMP and the transmembrane and cytoplasmic domains of NGFR (Figure 4A, MT1-F/NGFR). MT1-F/NGFR was expressed in COS1 cells, as detected by the anti-FLAG M2 antibody (Figure 4B, top panel, Anti-FLAG, lane 1). The same sample was analyzed by anti-PY, and phosphorylation was detected (Figure 4B, bottom panel, Anti-PY, lane 1). This suggested that the MT1-F/NGFR tyrosine residues were phosphorylated and that the ectodomain of MT1-MMP formed a homophilic dimer on the cell surface.
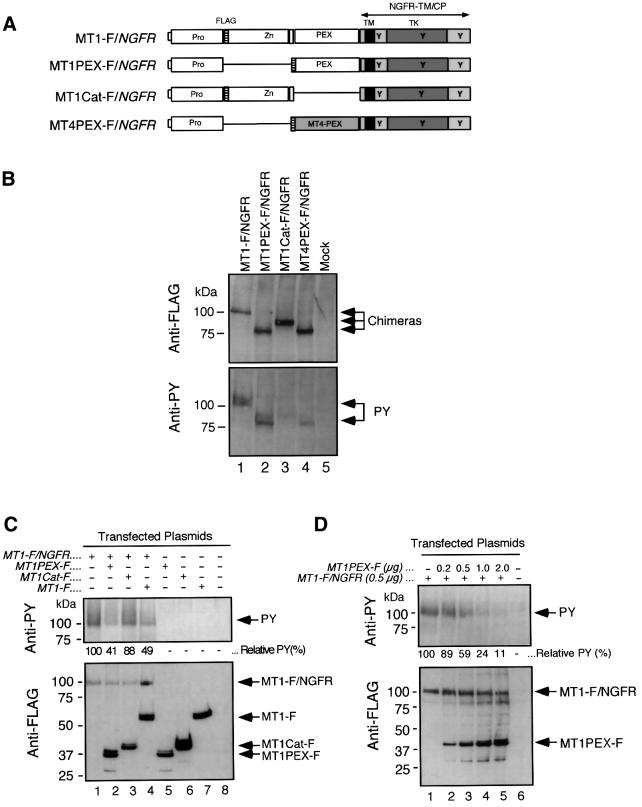
Fig. 4. Examination of dimer formation of the MT1-MMP ectodomain on the cell surface. (A) A schematic representation of the mutant MT1-MMP/NGFR chimeras. NGFR-TM/CP, NGFR-derived transmembrane and cytoplasmic domains; TK, tyrosine kinase domain; Y, possible tyrosine residue to be phosphorylated; MT4-PEX, PEX domain derived from MT4-MMP. (B) COS1 cells were transfected with expression plasmids for MT1-F/NGFR, MT1PEX-F/NGFR, MT1Cat-F/NGFR, MT4PEX-F/NGFR or vector alone (Mock). Cell lysates were subjected to western blot analysis using the anti-FLAG M2 monoclonal antibody (Anti-FLAG, upper panel) or anti-phosphotyrosine PY20 monoclonal antibody (Anti-PY). (C) COS1 cells were co-transfected with MT1-F/NGFR and/or MT1PEX-F, MT1Cat-F, or MT1-F. Cells were then subjected to western blotting using anti-FLAG M2 (Anti-FLAG) or PY20 (Anti-PY). The relative intensity of the bands detected by PY20 was analyzed with NIH Image, and is indicated. (D) COS1 cells were co-transfected with MT1-F/NGFR and MT1PEX-F at the indicated amounts of DNA. Cells were then subjected to western blotting using anti-FLAG M2 (Anti-FLAG) or PY20 (Anti-PY). The relative intensity of the bands detected by PY20 is indicated.
To test for domain-specific interactions in this sys tem, the following chimeras were also constructed: MT1PEX-F/NGFR, MT1Cat-F/NGFR and MT4PEX-F/NGFR (Figure 4A). These constructs were expressed in COS1 cells at levels similar to MT1-F/NGFR expression, as detected by the anti-FLAG M2 antibody (Figure 4B, top panel, Anti-FLAG, lanes 2–4). When tyrosine phosphorylation in these samples was detected by the anti-PY antibody, only MT1PEX-F/NGFR was shown to be strongly phosphorylated (Figure 4B, bottom panel, Anti-PY, lane 2). Phosphorylation levels of MT1Cat-F/NGFR and MT4PEX-F/NGFR were very low (lanes 3 and 4, respectively). These results indicate that the chimera formed a homophilic complex through PEX on the cell surface, supporting the earlier data (Figures 1 and 2).
Next, the expression plasmids for MT1-F/NGFR and FLAG-tagged MT1PEX (MT1PEX-F), MT1Cat (MT1Cat-F) or MT1-F were co-transfected to investigate whether the dimer formation of the chimera can be disrupted or not. As shown in Figure 4C, co-expression of MT1PEX-F and MT1-F effectively competed in the dimer formation of MT1-F/NGFR (lanes 2 and 4). On the other hand, the expression of MT1Cat-F did not decrease the intensity of the band very much (lane 3). Furthermore, up to 90% of the inhibition of phosphorylation was observed by increasing levels of MT1PEX-F expression (Figure 4D, lanes 1–5). This further strengthened the idea that MT1-MMP molecules interact through their PEX domains.
Rac1 augments dimer formation of MT1-MMP and proMMP-2 activation at lamellipodia
Using the chimera construct, we next investigated whether there are any factors that might regulate the dimer formation of MT1-MMP on the cell surface. Since it has been reported that MT1-MMP is localized at the lamellipodia structure in walking osteoclasts (Sato et al., 1997), we looked at the effect of a constitutively active form of the small GTPase Rac1 (V12Rac1, Rac1DA) on dimer formation as a GTP-bound form of Rac1 stimulates the generation of lamellipodia in cells (Michiels et al., 1995). MT1-F/NGFR was co-expressed with Rac1DA in COS1 cells and the phosphotyrosine signal was monitored. As shown in Figure 5A, co-transfection of Rac1DA and MT1-F/NGFR resulted in an enhanced phosphotyrosine signal compared with MT1-F/NGFR alone (Anti-PY, lanes 1 and 3). When MT1PEX-F was co-expressed, the signal decreased from 182 to 120%, suggesting that the increased phosphotyrosine signal was likely to be the result of enhanced dimer formation (Figure 5A, Anti-PY, lanes 2 and 4, see also Figure 4). Immunostaining of MT1-F/NGFR-expressing cells for phosphotyrosine revealed that distinct signals were detected, especially where the F-actin was concentrated (Figure 5B, MT1-F/NGFR). Expression of Rac1DA generated ruffled membranes at the edge of the cell (Figure 5B, MT1-F/NGFR⋅Rac1DA and Rac1DA), and phosphotyrosine signals were detected extensively at the edge of the membrane structure (MT1-F/NGFR⋅Rac1DA). These signals were generated by MT1-F/NGFR as they were co-localized with the staining pattern with the anti- FLAG M2 antibody (data not shown), and no such signals were detected in Rac1DA- or mock-transfected cells (Rac1DA and Mock). These results suggest that accelerated dimer formation occurs at the ruffling membrane edge. We then tested the effect of Rac1DA expression on proMMP-2 activation by MT1-MMP. As shown in Figure 5C, co-expression of MT1-MMP and Rac1DA clearly enhanced the processing of proMMP-2 to the active form. Immunolocalization of MT1-F in the transfected cells showed that MT1-F is distributed on the cell surface and is relatively concentrated at the lamellipodia structure where F-actin is concentrated (Figure 5D, Anti-FLAG, MT1-F). On the other hand, co-expression of Rac1DA with MT1-F reinforced the localization of MT1-F at the ruffled membrane edge (Anti-FLAG, MT1-F/RacDA). Taken together, these data suggest that the expression of Rac1DA generates ruffling membrane and forces MT1-MMP to localize at the site. Subsequently, the local concentration of MT1-MMP is likely to increase at the site of the membrane, and it promotes homophilic complex formation and activation of proMMP-2.
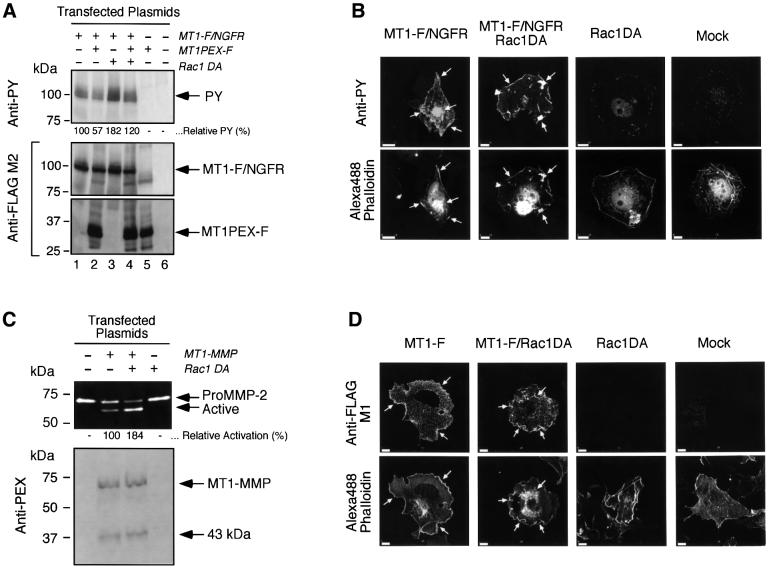
Fig. 5. Effect of the constitutively active form of Rac1 (Rac1DA) on the dimer formation of MT1-MMP and its ability to activate proMMP-2. (A) Effect of Rac1DA on the dimer formation of MT1-F/NGFR. COS1 cells were transfected with the expression plasmids for MT1-F/NGFR, MT1PEX-F and/or Rac1DA. Cell lysates were subjected to western blot analysis using PY20 (Anti-PY) and anti-FLAG M2 antibodies (Anti-FLAG). The relative intensity of the bands detected by PY20 was analyzed with NIH Image, normalized by the relative intensity of the bands of MT1-F/NGFR detected by Anti-FLAG M2 antibody, and is indicated. (B) Transfected COS1 cells were stained with PY20 (Anti-PY) as described in Materials and methods. F-actin was also stained with Alexa488-conjugated Phalloidin (Alexa488 Phalloidin). The white arrows indicate the sites where F-actin and PY signals were co-localized. Bars: 10 µm. (C) COS1 cells were transfected with the expression plasmids for MT1-MMP and Rac1DA, and these cells were reacted with purified proMMP-2 (0.25 µg/ml). The medium was analyzed by zymography for proMMP-2 processing (top panel), and the cell lysate was analyzed by western blotting using the anti-MT1PEX antibody (bottom panel). The relative activation of proMMP-2 to the active form was calculated by measuring the intensity of the bands of proMMP-2 with NIH Image, and is indicated. (D) Transfected COS1 cells were stained with anti-FLAG M1 antibody as described in Materials and methods. F-actin was also stained with Alexa488-conjugated phalloidin (Alexa488 Phalloidin). The white arrows indicate the sites where F-actin and FLAG signals were co-localized. Bars: 10 µm.
Biological outcome by MT1-MMP depends on its ability to form a homophilic complex on the cell surface
We next examined the effect of MT1PEX-F on the activation of proMMP-2 since MT1PEX-F effectively inhibited the dimer formation of MT1-F/NGFR (Figure 4C and D). COS1 cells were transfected with the plasmids for MT1-MMP and MT1PEX-F in different ratios. As shown in Figure 6A, MT1PEX-F expression inhibited the cell surface activation of proMMP-2 by MT1-MMP (top and middle panels). The expression of MT1PEX-F did not affect the appearance of MT1-MMP on the surface of the cells, since a similar amount of MT1-MMP was biotinylated (Figure 6A, bottom panel, Surface Biotinylation). As shown in Figure 6B, proMMP-2 binding to the cell surface was also not affected by MT1PEX-F expression. Co-expression of MT1PEX-F did not decrease the amount of proMMP-2 bound to the cell surface, and a significantly lower amount of active species was detected. MT1PEX-F also did not affect the proteolytic activity of MT1-MMP on the cell surface, as shown in Figure 6C, showing a similar gelatin degradation pattern and area. Therefore, MT1PEX-F inhibits the dimer formation of MT1-MMP and acts as a dominant-negative form of MT1-MMP in activation of proMMP-2 on the cell surface.
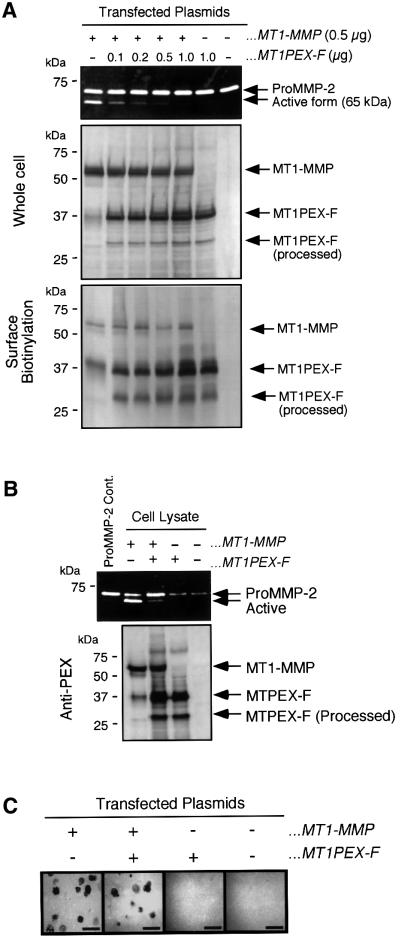
Fig. 6. Effect of MT1PEX-F on the proMMP-2 activation by MT1-MMP on the cell surface. (A) COS1 cells were co-transfected with the expression plasmids for MT1-MMP and MT1PEX-F at the amounts of DNA indicated. Purified proMMP-2 (0.25 µg/ml) was then reacted with the cells at 37°C for 18 h. MMP-2 in the culture medium was analyzed by gelatin zymography (top panel), and the cell lysate was subjected to western blotting using anti-MT1PEX antibody (middle panel, Whole cell). Transfected cells were also subjected to surface biotinylation as described in Materials and methods and analyzed by western blotting using anti-MT1PEX (bottom panel, Surface Biotinylation). (B) Binding of proMMP-2 to the transfected cells was tested as described in Materials and methods. Samples were analyzed by gelatin zymography. (C) In situ gelatin degradation activity of transfected cells was studied by culturing cells on the glass slide coated with Alexa488-labeled gelatin as described in Materials and methods. Gelatin degradation activity was visualized as dark, non-fluorescent areas. Bars: 50 µm.
HT1080 cells are known to express MT1-MMP and MMP-2 endogenously (Strongin et al., 1995). Thus, we next looked at the effect of the expression of MT1PEX-F on the activation of endogenous proMMP-2 in HT1080 cells. The cells were transfected with MT1PEX-F plasmid or the empty vector, which bears the hygromycin-resistant gene. MMP-2 produced in the culture supernatant by whole hygromycin-resistant populations of the cells was then analyzed by zymography. As shown in Figure 7A, mock-transfected cells spontaneously activate endogenous MMP-2 in a manner sensitive to BB94 (top panel, lanes 1 and 2). On the other hand, the cells expressing MT1PEX-F did not effectively activate proMMP-2 (top panel, lane 3). Western blot analysis of the cell lysates indicated that HT1080 cells spontaneously produce MT1-MMP, and it is partially processed to a 43 kDa fragment, as reported previously (Lehti et al., 1998; Stanton et al., 1998). MT1PEX-F did not affect the expression and processing of endogenous MT1-MMP. Co-immunolocalization of endogenous MT1-MMP and MT1-PEX-F clearly indicated the co-localization of both molecules at the edge of the cells (Figure 7B). Finally, the effect of MT1PEX-F on the invasive activity of HT1080 cells was tested (Figure 7C). Exogenously added TIMP-2 inhibited the invasion by mock-transfected cells by 50%. By transfecting the expression plasmid for MT1PEX-F, invasion was suppressed by 70%. Adding TIMP-2 to the MT1PEX-F-expressing cells produced no further reduction in the amount of invasion.
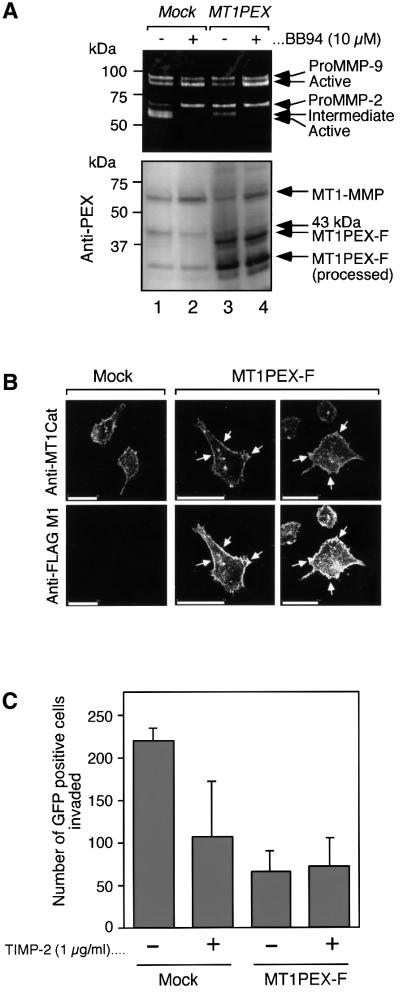
Fig. 7. Effect of MT1PEX-F on the activation of proMMP-2 and the Matrigel invasion activity of HT1080 cells. (A) HT1080 cells were transfected with the stable expression plasmid for MT1PEX-F and an empty vector, and the whole population of hygromycin-resistant cells was pooled and reseeded. After 24 h, the medium was exchanged for serum-free DMEM in the presence or absence of BB94 (10 µM) and the cells were further cultured for 24 h. The culture medium and cell lysate were analyzed by gelatin zymography and western blotting using anti-MT1PEX. (B) HT1080 cells were transfected with the expression plasmid for MT1PEX-F or empty vector. Cells were co-immunostained for endogenous MT1-MMP and MT1PEX-F using rabbit anti-MT1Cat antibody and anti-FLAG M1 antibody, respectively. The white arrows indicate the sites where endogenous MT1-MMP (Anti-MT1Cat) and MT1PEX-F (Anti-FLAG M1) signals were co-localized. Bars: 50 µm. (C) HT1080 cells were transfected with the expression plasmid for MT1PEX-F or empty vector together with the plasmid for GFP. Cells were then subjected to a Matrigel invasion assay using the modified Boiden chamber method as described in Materials and methods. TIMP-2 at 0.5 µg/ml was added to both the upper and lower chamber. GFP-positive cells on the lower chamber surface were counted under fluorescence microscopy. The total numbers of GFP-positive cells in mock and MT1PEX-F-transfected cells were 1.31 × 104 and 1.12 × 104, respectively. The mean and standard deviation of four in each group are indicated as a graph.
Discussion
Homophilic complex formation of MT1-MMP facilitates the activation of proMMP-2 on the cell surface
In the present study, we have shown that MT1-MMP forms a homophilic complex through the PEX domain on the cell surface. The complex formation of MT1-MMP on the cell surface allows a closer molecular arrangement between enzyme molecules. Thus, it is conceivable that one of the MT1-MMP molecules in the complex binds to TIMP-2 to act as a proMMP-2 receptor, and the other TIMP-2-free MT1-MMP in the complex cleaves the propeptide of proMMP-2 bound to the receptor. Co-expression of MT1PEX-F with full-length MT1-MMP results in the formation of a hetero-complex of MT1PEX-F–MT1-MMP (see Figure 1), and creates a certain distance between the receptor-MT1-MMP–TIMP-2 and the catalytic MT1- MMP. This disables the catalytic enzyme, preventing it from reacting with the proMMP-2 in the ternary complex. Thus, the interaction of MT1-MMP molecules through PEX is most likely a mechanism for assembling the activation complex by bringing together the receptor for proMMP-2 (i.e. MT1-MMP–TIMP-2) and catalytic MT1- MMP on the cell surface. It is not clear at present whether dimer formation is sufficient for effective activation, or if oligomerization would enhance its activity further. Nonetheless, PEX domain interaction in MT1-MMP is critical for proMMP-2 activation.
Regulation of complex formation on the cell surface
The expression of Rac1DA induced ruffled membrane that is rich in F-actin, and MT1-MMP on the cell surface was concentrated at the membrane structure. Our results indicated that the homophilic complex was formed exclusively at the ruffling membrane structure generated by Rac1DA. Presumably, increased local concentration of MT1-MMP at the membrane structure enhanced complex formation and resulted in efficient proMMP-2 activation, although the exact mechanism underlying the phenomenon is not known at present. While this manuscript was in preparation, it was reported that Rac1 mediates type I collagen-dependent proMMP-2 activation in HT1080 cells (Zhuge and Xu, 2001). Zhuge and Xu (2001) showed that expression of Rac1DA up-regulated MT1-MMP expression, thereby enhancing proMMP-2 activation. In our experiments, however, Rac1DA did not affect MT1-MMP expression levels and, therefore, the increased activation of proMMP-2 we observed was attributed to a post-translational event: complex formation. It is possible, however, that proMMP-2 activation can be stimulated synergistically by both mechanisms when cells are in a type I collagen-rich environment.
A mode of dimer formation
Homophilic complex formation of E.coli-expressed MT1EAΔTM was concentration dependent. MT1-MMP expressed on the cell surface is likely to be in the same manner as described above. However, an excess of exogenously added soluble MT1PEX fragment failed to inhibit proMMP-2 activation on the cell surface (data not shown). This is presumably due to the tighter complex formation of purified PEX without the catalytic domain, and it would not be able to exchange efficiently with cell surface-located PEX in an intact MT1-MMP. It is also possible that soluble MT1PEX may not be able to compete the binding with membrane-anchored MT1-MMP, and it may require to be anchored to the plasma membrane to concentrate both molecules at the same site effectively.
Of interest are previous reports in which an MT1-MMP with the catalytic domain deleted was generated in HT1080 cells (Lehti et al., 1998; Stanton et al., 1998). This product is the result of a proteolytic cleavage at an Ala255–Ile bond located at the end of the catalytic domain of MT1-MMP by the action of either MT1-MMP itself or MMP-2 activated by MT1-MMP (Lehti et al., 1998; Stanton et al., 1998). Since this product retains the PEX domain, it may have a similar effect to MT1PEX-F. Thus, this processing not only removes the activity of MT1-MMP, but may also inhibit homophilic complex formation of the intact MT1-MMP molecules on the cell surface.
Overall et al. (2000) have recently suggested that MT1-MMP may not form homophilic dimer or oligomer complexes, which they speculated from gel-permeation column chromatography of an E.coli-expressed recombinant MT1-MMP PEX fragment. However, we were able to detect dimer formation of E.coli-expressed MT1PEX, as well as of MT1-MMP expressed in mammalian cells through the PEX domain. It is most likely that the discrepancy comes from different experimental conditions or folding of the recombinant PEX domain.
Role of the PEX domain in the biological activities of MMPs
Since the PEX domain is found in most of the MMPs, but not in the other metzincin metalloproteinase family members, this domain is likely to determine the unique properties of MMPs. The crystal structure of the PEX domain from MMP-2 and MMP-13 revealed an ellipsoidal disk shape with a four-bladed β-propeller structure (Gohlke et al., 1996; Gomis-Rüth et al., 1996). It has been shown that collagenases require the PEX domain to cleave triple helical collagens (Murphy et al., 1992; Knäuper et al., 1997), and probably that the combination of PEX and the catalytic domain unwinds the triple helical structure to cleave each peptide bond one by one (Gomis-Rüth et al., 1996). Another role of PEX is shown in the progelatinases proMMP-9 and proMMP-2. They form a complex with TIMP-1 and TIMP-2, respectively, through the interactions of their PEX domain and the C-terminal domain of the TIMPs (Goldberg et al., 1989; Wilhelm et al., 1989). In the case of the proMMP-9–TIMP-1 complex, TIMP-1 is thought to regulate the activation of proMMP-9 by inhibiting its activators, such as MMP-3 (Ogata et al., 1995) and MMP-7 (Imai et al., 1995; Ogata, 1995). In the case of the proMMP-2–TIMP-2 complex, it is now clear that complex formation is rather essential for the activation of proMMP-2 by MT1-MMP on the cell surface (Will et al., 1996; Butler et al., 1998; Kinoshita et al., 1998). It has also been reported that MMP-2 binds to αvβ3 integrin through the PEX domain (Brooks et al., 1996), although its relationship to MT1-MMP is not clear. In addition to these previous findings, the present study provides the novel example of the role of the PEX domain in MT1-MMP homophilic complex formation and in the activation of proMMP-2 on the cell surface. Thus, PEX, a uniquely conserved domain in MMPs, is thought to be a general device for interaction with molecules that determine the fate of the enzyme in biological systems.
A new strategy to test the biological function of MT1-MMP
Using MT1PEX-F, one can test the role of proMMP-2 activation or dimer formation of MT1-MMP in certain biological systems. Our data indicate that the invasive activity of HT1080 cells in the Matrigel system was effectively inhibited by MT1PEX-F. The major components of the Matrigel are laminin and type IV collagen, and HT1080 cells need to degrade these components to penetrate the membrane efficiently. Since MT1-MMP does not degrade type IV collagen by itself (Ohuchi et al., 1997), activation of proMMP-2 is likely to play an important role in this system. Therefore, it is likely that one of the major causes of the inhibition of the invasion by MT1PEX-F is inhibition of the activation of proMMP-2 by endogenous MT1-MMP, although it still cannot be ruled out that PEX binds another partner molecule on the cell surface to diminish unknown MT1-MMP activity. Nevertheless, MT1PEX-F is a useful tool to test the role of MT1-MMP in certain biological systems.
Evidence is accumulating as to the importance of MT1-MMP activity in many biological systems. There fore, the dominant-negative MT1-MMP, MT1PEX-F, would help to elucidate the biological significance of MT1-MMP in tissue and to develop new strategies for treating MT1-MMP-related diseases, including cancer.
Materials and methods
Cell culture and transfection
COS1 and HT1080 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Sigma Chemical Co. Ltd, St Louis, MO) containing 10% fetal bovine serum and kanamycin in a humidified 37°C incubator. At 16 h before the transfection was performed, the cells were seeded in six-well plates at 1 × 105/well. Expression plasmids for each protein were transfected using FuGENE6™ (Roche Molecular Biochemicals, Basel, Switzerland) according to the manufacturer’s instructions.
Construction of MT1-MMP mutants and chimeras
The cDNA of MT1-MMP was used as a template to generate DNA fragments for MT1Cat (ΔIle318-Gly535) and MT1PEX (ΔTyr112-Pro312) by the PCR extension method of Ho et al. (1989). A FLAG epitope (Asp-Tyr-Lys-Asp-Asp-Asp-Asp-Lys)-tagged MT1-MMP (MT1-F) expression construct was generated as described previously (Itoh et al., 1999) and used as a template to generate cDNA for FLAG-tagged MT1Cat (MT1Cat-F) and MT1PEX (MT1PEX-F). The cDNA was subcloned into pSG5 (Stratagene, CA). For stable expression, the cDNA of MT1PEX-F was subcloned into pCEP4 (Invitrogen, Groningen, The Netherlands), which bears EBNA-1 and hygromycin resistance gene expression cassettes.
The DNA corresponding to Cys319–Cys508 in MT1-MMP and MT1PEX-F was replaced with Cys336–Cys527 of mouse MT4-MMP to generate MT1-MT4PEX and MT4PEX-F, respectively.
A c-Myc epitope (Glu-Gln-Lys-Leu-Ile-Ser-Glu-Glu-Asp-Leu)-tagged MT1-MMP (MT1-Myc) expression construct was generated by PCR in a manner similar to that described for MT1-F (Itoh et al., 1999). A c-Myc tag was inserted between Arg111–Tyr, which is the same site for insertion of a FLAG tag in MT1-F.
Chimera mutants of the ectodomain of MT1-F, MT1Cat-F, MT1PEX-F or MT4PEX-F and the transmembrane/cytoplasmic domain of NGFR (MT1-F/NGFR, MT1Cat-F/NGFR, MT1PEX-F/NGFR or MT4PEX-F/NGFR, respectively) were also generated by PCR and subcloned into pSG5. The mutant is derived from sequences corresponding to Met1–Asp515 of MT1-MMP and Glu384–Gly790 of NGFR. The other chimera mutants were also generated at the corresponding sites. All the PCR-generated fragments were confirmed by DNA sequencing.
The expression plasmid for the constitutively active form of Rac1 (V17, Rac1DA) was kindly provided by Dr Yoshimi Takai at Osaka University.
Antibodies
A polyclonal anti-(MT1-MMP catalytic domain) antibody (anti-Cat) was generated by injecting the E.coli-expressed purified MT1-MMP catalytic domain into rabbits. The antibody was confirmed to recognize MT1-MMP specifically as it did not recognize other MT-MMPs, including MT2-, MT3-, MT4-, MT5-, MT6-MMPs. A monoclonal mouse anti-(human MT1-MMP PEX domain) antibody (anti-PEX, 222-1D8) was generated by injecting purified E.coli-expressed human MT1-MMP PEX domain into mice. This antibody was confirmed to be specific to MT1-MMP.
We also used commercially available antibodies as follows: a monoclonal mouse anti-FLAG epitope M2 antibody (Sigma), a mouse monoclonal anti-c-Myc antibody (Ab-1) (Oncogene, MA; 1 µg/ml), a mouse monoclonal anti-phosphotyrosine antibody (PY20) (ICN Biochemicals, Inc., OH), alkaline phosphatase-conjugated goat anti-mouse IgG and anti-rabbit IgG (Sigma).
Immunoprecipitation and western blotting
Preparation of cell lysate and immunoprecipitation with anti-FLAG M2 antibody-conjugated beads (Sigma) were carried out as described in Itoh et al. (1999). To detect FLAG-tagged proteins and associating molecules, the bound protein was eluted by the FLAG peptide (200 µg/ml; Sigma) in Tris-buffered saline (TBS), and subjected to western blot analysis.
Western blotting was also carried out as described previously (Itoh et al., 1999) and proteins reacted with probe antibodies were visualized using alkaline phosphatase-conjugated second antibodies.
Purification and folding of MT1EAΔTM, MT1-PEX and MT4-PEX expressed in E.coli
The cDNA fragment encoding Tyr112–Gly535 of MT1-MMP with the point mutation of Glu240–Ala (MT1EAΔTM) was generated by PCR and subcloned into a pET3a. Constructs of Cys319–Gly535 (MT1PEX) of MT1-MMP and Cys336–Gly550 in mouse MT4-MMP were generated in the same manner. The sequence encoding Met was included at the 5′ end of all constructs. All the PCR-generated DNAs were confirmed by DNA sequencing. BL21(DE3)pLysS cells were transformed with these pET3a constructs, and protein expression was induced by 0.4 mM isopropyl- β-d-thiogalactopyranoside. Proteins were purified and folded according to the method of Huang et al. (1996).
Purification of proMMP-2 and TIMP-2
Recombinant human proMMP-2 and TIMP-2 were expressed in HighFive insect cells (Invitrogen, CA) infected with recombinant baculoviruses engineered to express human proMMP-2 and TIMP-2. Recombinant viruses were made using Bac-To-Bac™ Baculovirus Expression Systems (Life Technologies, MD). ProMMP-2 was purified from the culture medium of infected cells by gelatin–Sepharose (Amersham Pharmacia Biotech Inc., Uppsala, Sweden) and a gel-permeation column on Sephacryl S-200 (Amersham Pharmacia Biotech, Inc.) as described previously (Itoh et al., 1995). TIMP-2 was purified from the medium by Green A Dyematrex (Millipore, MA) and a gel-permeation column on S-200.
Gel-permeation column chromatography
Purified MT1EAΔTM, MT1PEX and MT4PEX were subjected to gel-permeation column chromatography on Superdex 200 or Superdex 75 equilibrated with TNC buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 10 mM CaCl2, 0.02% NaN3) containing 0.05% Brij35 and analyzed under the ÄKTA explorer 10S system (Amersham Pharmacia Biotech, Inc.). The calibration was performed using thyroglobulin (mol. wt 669 000), ferritin (mol. wt 440 000), gammaglobulins (mol. wt 150 000), transferrin (mol. wt 81 000), ovalbumin (mol. wt 43 000) and myoglobin (mol. wt 17 600) as standards.
Gelatin zymography
Gelatin zymography was conducted with an SDS–polyacrylamide gel containing gelatin (0.8 mg/ml) as described previously (Morodomi, 1992; Itoh et al., 1998). The samples were mixed with SDS–PAGE loading buffer without a reducing agent and subjected to electrophoretic analysis at room temperature. Enzyme activity was visualized as negative staining with Coomassie Blue.
Surface biotinylation and subsequent immunoprecipitation
Transfected COS1 cells were washed three times with chilled phosphate-buffered saline (PBS) containing 1 mM MgCl2 and 0.1 mM CaCl2. Cells were then incubated with sulfo-NHS-biotin (Pierce) in the same buffer (2 mg/ml) at 4°C for 30 min. The reaction was terminated by further incubating the cells with 25 mM lysine in PBS. The cells were lysed in RIPA buffer, and the biotinylated proteins were precipitated with streptavidin–Sepharose beads (Amersham-Pharmacia). The samples were analyzed by western blotting using anti-MT1Cat or anti-PEX antibody.
In situ gelatin degradation assay
We utilized two different in situ gelatin degradation assays. One used DQ™ gelatin (Molecular Probes, Inc., OR). DQ gelatin is a fluorescence-quenched gelatin substrate and will fluoresce upon proteolytic cleavage. Glass coverslips were coated with 5% DQ gelatin in 10 mg/ml unlabeled gelatin solution, and dried. Transfected cells were cultured on the coverslip in the humidified chamber for 16 h. Cells were then fixed with 3% paraformaldehyde and the degraded areas were visualized as those areas showing fluorescence.
The other assay used Alexa488-conjugated gelatin made with an Alexa488-labeling kit (Molecular Probes). The bottom surface of four-well chamber slides (Nunc) was coated with Alexa488-conjugated gelatin according to the method of Yana and Weiss (2000). Transfected COS1 cells were cultured in the chamber slides for 16 h in the humidified culture incubator. Cells were then fixed with 3% paraformaldehyde in PBS and the degraded area was analyzed under a fluorescence microscope. The degraded area was visualized as a dark, non-fluorescent area. Fluorescence images were analyzed using confocal microscopy (Bio-Rad).
The activities of MT1-MMPs detected by the above methods are confirmed to be inhibited by the peptidyl hydroxamate MMP inhibitor BB94, which was kindly provided by Dr Peter D.Brown at British Biotech Pharmaceuticals Ltd (Oxford, UK).
Indirect immunofluorescence staining
Cells cultured on coverslips coated with gelatin were fixed with 3% paraformaldehyde in PBS containing 75 mM lysine. After blocking with 5% goat serum and 3% bovine serum albumin in TBS for 1 h at room temperature, cells were reacted with an anti-FLAG M1 antibody (5 µg/ml), PY20 (2 µg/ml) or anti-Cat serum (1:1000) at room temperature for 2 h. To detect the phosphotyrosine signal, cells were permeabilized with 0.1% Triton X-100 in TBS after fixation. CaCl2 (1 mM) was included throughout the procedure of washing and incubation for the staining with the anti-FLAG M1 antibody. Cy3- or Alexa488-conjugated goat anti-mouse IgG was used to visualize the antigen signal. To visualize F-actin, cells were further incubated with Alexa488- or Alexa594-conjugated phalloidin (Molecular Probes). The signals were analyzed by confocal microscopy (Bio-Rad).
ProMMP-2 binding assay
Transfected COS1 cells were cultured in a 24-well plate (0.2 × 104/well). Forty-eight hours after transfection, the cells were washed three times with serum-free DMEM and incubated with 2 µg/ml purified proMMP-2 in the serum-free DMEM at 25°C for 1 h. Cells were then washed three times with 5% dimethyl sulfoxide (DMSO) containing serum-free DMEM to remove free proMMP-2, and lysed in 50 µl of non-reducing SDS loading buffer. MMP-2 bound to the cells was analyzed by gelatin zymography.
Matrigel invasion assay
Matrigel invasion chambers were prepared according to the manufacturer’s instructions (Becton Dickinson Labware, MA) by coating 10 µg of Matrigel on the 8 µm pore membrane of the culture inserts for 24-well plates (Falcon). A 0.5 ml aliquot of transfected HT1080 cell suspension at 1 × 105/ml was seeded on the upper chamber and incubated at 37°C in a humidified chamber. Hepatocyte growth factor (Toyobo; 10 ng/ml) was added to the lower chamber as a chemoattractant. After 14 h of incubation, cells remaining on the upper surface of the membrane were removed, and cells on the lower surface were fixed with 3% paraformaldehyde in PBS. Green fluorescent protein (GFP)-positive cells that had invaded the lower surface were counted using fluorescence microscopy (×40).
Acknowledgments
Acknowledgements
We thank Dr Kirsten J.Lampi at Oregon Health Science University for critical reading of the manuscript. This work was supported by the Special Coordination Fund for promoting Science and Technology from the Ministry of Science and Technology of Japan, and by a grant-in-aid for Cancer Research from the Ministry of Education, Science and Culture of Japan.
References
- Atkinson S.J., Crabbe,T., Cowell,S., Ward,R.V., Butler,M.J., Sato,H., Seiki,M., Reynolds,J.J. and Murphy,G. (1995) Intermolecular autolytic cleavage can contribute to the activation of progelatinase A by cell membranes. J. Biol. Chem., 270, 30479–30485. [DOI] [PubMed] [Google Scholar]
- Benoit De Coignac A. et al. (2000) Cloning of MMP-26 a novel matrilysin-like proteinase. Eur. J. Biochem., 267, 3323–3329. [DOI] [PubMed] [Google Scholar]
- Brooks P.C., Stromblad,S., Sanders,L.C., von Schalscha,T.L., Aimes,R.T., Stetler-Stevenson,W.G., Quigley,J.P. and Cheresh,D.A. (1996) Localization of matrix metalloproteinase MMP-2 to the surface of invasive cells by interaction with integrin α v β 3. Cell, 85, 683–693. [DOI] [PubMed] [Google Scholar]
- Butler G.S. et al. (1998) The TIMP2 membrane type 1 metalloproteinase ‘Receptor’ regulates the concentration and efficient activation of progelatinase A. A kinetic study. J. Biol. Chem., 273, 871–880. [DOI] [PubMed] [Google Scholar]
- Buttner F.H., Hughes,C.E., Margerie,D., Lichte,A., Tschesche,H., Caterson,B. and Bartnik,E. (1998) Membrane type 1 matrix metalloproteinase (MT1-MMP) cleaves the recombinant aggrecan substrate rAgg1mut at the ‘aggrecanase’ and the MMP sites. Characterization of MT1-MMP catabolic activities on the interglobular domain of aggrecan. Biochem. J., 333, 159–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Ortho M.P. et al. (1997) Membrane-type matrix metalloproteinases 1 and 2 exhibit broad-spectrum proteolytic capacities comparable to many matrix metalloproteinases. Eur. J. Biochem., 250, 751–757. [DOI] [PubMed] [Google Scholar]
- Fosang A.J., Last,K., Fujii,Y., Seiki,M. and Okada,Y. (1998) Membrane-type 1 MMP (MMP-14) cleaves at three sites in the aggrecan interglobular domain. FEBS Lett., 430, 186–190. [DOI] [PubMed] [Google Scholar]
- Gohlke U., Gomis-Rüth,F.X., Crabbe,T., Murphy,G., Docherty,A.J. and Bode,W. (1996) The C-terminal (haemopexin-like) domain structure of human gelatinase A (MMP2): structural implications for its function. FEBS Lett., 378, 126–130. [DOI] [PubMed] [Google Scholar]
- Goldberg G.I., Marmer,B.L., Grant,G.A., Eisen,A.Z., Wilhelm,S. and He,C. (1989) Human 72-kilodalton type IV collagenase forms a complex with a tissue inhibitor of metalloproteinases designated TIMP-2. Proc. Natl Acad. Sci. USA, 86, 8207–8211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomis-Rüth F.X., Gohlke,U., Betz,M., Knäuper,V., Murphy,G., López-Otín,C. and Bode,W. (1996) The helping hand of collagenase-3 (MMP-13): 2.7 Å crystal structure of its C-terminal haemopexin-like domain. J. Mol. Biol., 264, 556–566. [DOI] [PubMed] [Google Scholar]
- Hiraoka N., Allen,E., Apel,I.J., Gyetko,M.R. and Weiss,S.J. (1998) Matrix metalloproteinases regulate neovascularization by acting as pericellular fibrinolysins. Cell, 95, 365–377. [DOI] [PubMed] [Google Scholar]
- Ho S.N., Hunt,H.D., Horton,R.M., Pullen,J.K. and Pease,L.R. (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene, 77, 51–59. [DOI] [PubMed] [Google Scholar]
- Holmbeck K. et al. (1999) MT1-MMP-deficient mice develop dwarfism, osteopenia, arthritis and connective tissue disease due to inadequate collagen turnover. Cell, 99, 81–92. [DOI] [PubMed] [Google Scholar]
- Hotary K., Allen,E., Punturieri,A., Yana,I. and Weiss,S.J. (2000) Regulation of cell invasion and morphogenesis in a three-dimensional type I collagen matrix by membrane-type matrix metalloproteinases 1, 2 and 3. J. Cell Biol., 149, 1309–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Suzuki,K., Nagase,H., Arumugam,S., Van Doren,S.R. and Brew,K. (1996) Folding and characterization of the amino-terminal domain of human tissue inhibitor of metalloproteinases-1 (TIMP-1) expressed at high yield in E.coli. FEBS Lett., 384, 155–161. [DOI] [PubMed] [Google Scholar]
- Imai K., Ohuchi,E., Aoki,T., Nomura,H., Fujii,Y., Sato,H., Seiki,M. and Okada,Y. (1996) Membrane-type matrix metalloproteinase 1 is a gelatinolytic enzyme and is secreted in a complex with tissue inhibitor of metalloproteinases 2. Cancer Res., 56, 2707–2710. [PubMed] [Google Scholar]
- Imai K., Yokohama,Y., Nakanishi,I., Ohuchi,E., Fujii,Y., Nakai,N. and Okada,Y. (1995) Matrix metalloproteinase 7 (matrilysin) from human rectal carcinoma cells. Activation of the precursor, interaction with other matrix metalloproteinases and enzymic properties. J. Biol. Chem., 270, 6691–6697. [DOI] [PubMed] [Google Scholar]
- Itoh Y., Binner,S. and Nagase,H. (1995) Steps involved in activation of pro-matrix metalloproteinase 2 (progelatinase A) and tissue inhibitor of metalloproteinases (TIMP)-2 by 4-aminophenylmercuric acetate. Biochem. J., 308, 645–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh Y., Ito,A., Iwata,K., Tanzawa,K., Mori,Y. and Nagase,H. (1998) Plasma membrane-bound tissue inhibitor of metalloproteinases (TIMP)-2 specifically inhibits matrix metalloproteinase 2 (gelatinase A) activated on the cell surface. J. Biol. Chem., 273, 24360–24367. [DOI] [PubMed] [Google Scholar]
- Itoh Y., Kajita,M., Kinoh,H., Mori,H., Okada,A. and Seiki,M. (1999) Membrane type 4 matrix metalloproteinase (MT4-MMP, MMP-17) is a glycosylphosphatidylinositol-anchored proteinase. J. Biol. Chem., 274, 34260–34266. [DOI] [PubMed] [Google Scholar]
- Jing S., Tapley,P. and Barbacid,M. (1992) Nerve growth factor mediates signal transduction through trk homodimer receptors. Neuron, 9, 1067–1079. [DOI] [PubMed] [Google Scholar]
- Kinoshita T., Sato,H., Okada,A., Ohuchi,E., Imai,K., Okada,Y. and Seiki,M. (1998) TIMP-2 promotes activation of progelatinase A by membrane-type 1 matrix metalloproteinase immobilized on agarose beads. J. Biol. Chem., 273, 16098–16103. [DOI] [PubMed] [Google Scholar]
- Knäuper V., Cowell,S., Smith,B., López-Otín,C., Oshea,M., Morris,H., Zardi,L. and Murphy,G. (1997) The role of the C terminal domain of human collagenase 3 (MMP 13) in the activation of procollagenase 3, substrate specificity and tissue inhibitor of metalloproteinase interaction. J. Biol. Chem., 272, 7608–7616. [DOI] [PubMed] [Google Scholar]
- Knäuper V., Will,H., López-Otín,C., Smith,B., Atkinson,S.J., Stanton,H., Hembry,R.M. and Murphy,G. (1996) Cellular mechanisms for human procollagenase-3 (MMP-13) activation. Evidence that MT1-MMP (MMP-14) and gelatinase A (MMP-2) are able to generate active enzyme. J. Biol. Chem., 271, 17124–17131. [DOI] [PubMed] [Google Scholar]
- Kojima S., Itoh,Y., Matsumoto,S., Masuho,Y. and Seiki,M. (2000) Membrane-type 6 matrix metalloproteinase (MT6-MMP, MMP-25) is the second glycosyl-phosphatidyl inositol (GPI)-anchored MMP. FEBS Lett., 480, 142–146. [DOI] [PubMed] [Google Scholar]
- Koshikawa N., Giannelli,G., Cirulli,V., Miyazaki,K. and Quaranta,V. (2000) Role of cell surface metalloprotease MT1-MMP in epithelial cell migration over laminin-5. J. Cell Biol., 148, 615–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehti K., Lohi,J., Valtanen,H. and Keski-Oja,J. (1998) Proteolytic processing of membrane-type-1 matrix metalloproteinase is associated with gelatinase A activation at the cell surface. Biochem. J., 334, 345–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon M.A. and Schlessinger,J. (1994) Regulation of signal transduction and signal diversity by receptor oligomerization. Trends Biochem. Sci., 19, 459–463. [DOI] [PubMed] [Google Scholar]
- Lohi J., Wilson,C.L., Roby,J.D. and Parks,W.C. (2001) Epilysin, a novel human matrix metalloproteinase (MMP-28) expressed in testis and keratinocytes and in response to injury. J. Biol. Chem., 276, 10134–10144. [DOI] [PubMed] [Google Scholar]
- Maru Y., Yamaguchi,S. and Shibuya,M. (1998) Flt-1, a receptor for vascular endothelial growth factor, has transforming and morphogenic potentials. Oncogene, 16, 2585–2595. [DOI] [PubMed] [Google Scholar]
- Michiels F., Habets,G.G., Stam,J.C., van der Kammen,R.A. and Collard,J.G. (1995) A role for Rac in Tiam1-induced membrane ruffling and invasion. Nature, 375, 338–340. [DOI] [PubMed] [Google Scholar]
- Morodomi T., Ogata,Y., Sasaguri,Y., Morimatsu,M. and Nagase,H. (1992) Purification and characterization of matrix metalloprotein ase 9 from U937 monocytic leukaemia and HT-1080 fibrosarcoma cells. Biochem. J., 285, 603–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy G., Allan,J.A., Willenbrock,F., Cockett,M.I., O’Connell,J.P. and Docherty,A.J.P. (1992) The role of the C-terminal domain in collagenase and stromelysin specificity. J. Biol. Chem., 267, 9612–9618. [PubMed] [Google Scholar]
- Nagase H. and Woessner,J.F.,Jr (1999) Matrix metalloproteinases. J. Biol. Chem., 274, 21491–21494. [DOI] [PubMed] [Google Scholar]
- Nomura H., Sato,H., Seiki,M., Mai,M. and Okada,Y. (1995) Expression of membrane-type matrix metalloproteinase in human gastric carcinomas. Cancer Res., 55, 3263–3266. [PubMed] [Google Scholar]
- Ogata Y., Itoh,Y. and Nagase,H. (1995) Steps involved in activation of the pro-matrix metalloproteinase 9 (progelatinase B)–tissue inhibitor of metalloproteinase-1 complex by 4-aminophenylmercuric acetate and proteinases. J. Biol. Chem., 270, 18506–18511. [DOI] [PubMed] [Google Scholar]
- Ohuchi E., Imai,K., Fujii,Y., Sato,H., Seiki,M. and Okada,Y. (1997) Membrane type 1 matrix metalloproteinase digests interstitial collagens and other extracellular matrix macromolecules. J. Biol. Chem., 272, 2446–2451. [DOI] [PubMed] [Google Scholar]
- Okada A., Tomasetto,C., Lutz,Y., Bellocq,J.P., Rio,M.C. and Basset,P. (1997) Expression of matrix metalloproteinases during rat skin wound healing: evidence that membrane type-1 matrix metalloproteinase is a stromal activator of pro-gelatinase A. J. Cell Biol., 137, 67–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overall C.M., Tam,E., McQuibban,G.A., Morrison,C., Wallon,U.M., Bigg,H.F., King,A.E. and Roberts,C.R. (2000) Domain interactions in the gelatinase A.TIMP-2.MT1-MMP activation complex. The ectodomain of the 44-kDa form of membrane type-1 matrix metalloproteinase does not modulate gelatinase A activation. J. Biol. Chem., 275, 39497–39506. [DOI] [PubMed] [Google Scholar]
- Park H.I., Ni,J., Gerkema,F.E., Liu,D., Belozerov,V.E. and Sang,Q.X. (2000) Identification and characterization of human endometase (matrix metalloproteinase-26) from endometrial tumor. J. Biol. Chem., 275, 20540–20544. [DOI] [PubMed] [Google Scholar]
- Pei D. (1999) Leukolysin/MMP25/MT6-MMP: a novel matrix metalloproteinase specifically expressed in the leukocyte lineage. Cell Res., 9, 291–303. [DOI] [PubMed] [Google Scholar]
- Sato H., Takino,T., Okada,Y., Cao,J., Shinagawa,A., Yamamoto,E. and Seiki,M. (1994) A matrix metalloproteinase expressed on the surface of invasive tumour cells. Nature, 370, 61–65. [DOI] [PubMed] [Google Scholar]
- Sato T., del Carmen Ovejero,M., Hou,P., Heegaard,A.M., Kumegawa,M., Foged,N.T. and Delaisse,J.M. (1997) Identification of the membrane-type matrix metalloproteinase MT1-MMP in osteoclasts. J. Cell Sci., 110, 589–596. [DOI] [PubMed] [Google Scholar]
- Seiki M. (1999) Membrane-type matrix metalloproteinases. Apmis, 107, 137–143. [DOI] [PubMed] [Google Scholar]
- Stanton H., Gavrilovic,J., Atkinson,S.J., d’Ortho,M.P., Yamada,K.M., Zardi,L. and Murphy,G. (1998) The activation of ProMMP-2 (gelatinase A) by HT1080 fibrosarcoma cells is promoted by culture on a fibronectin substrate and is concomitant with an increase in processing of MT1-MMP (MMP-14) to a 45 kDa form. J. Cell Sci., 111, 2789–2798. [DOI] [PubMed] [Google Scholar]
- Stetler-Stevenson W.G., Aznavoorian,S. and Liotta,L.A. (1993) Tumor cell interactions with the extracellular matrix during invasion and metastasis. Annu. Rev. Cell Biol., 9, 541–573. [DOI] [PubMed] [Google Scholar]
- Strongin A.Y., Collier,I., Bannikov,G., Marmer,B.L., Grant,G.A. and Goldberg,G.I. (1995) Mechanism of cell surface activation of 72-kDa type IV collagenase. Isolation of the activated form of the membrane metalloprotease. J. Biol. Chem., 270, 5331–5338. [DOI] [PubMed] [Google Scholar]
- Tokuraku M., Sato,H., Murakami,S., Okada,Y., Watanabe,Y. and Seiki,M. (1995) Activation of the precursor of gelatinase A/72 kDa type IV collagenase/MMP-2 in lung carcinomas correlates with the expression of membrane-type matrix metalloproteinase (MT-MMP) and with lymph node metastasis. Int. J. Cancer, 64, 355–359. [DOI] [PubMed] [Google Scholar]
- Velasco G., Cal,S., Merlos-Suarez,A., Ferrando,A.A., Alvarez,S., Nakano,A., Arribas,J. and López-Otín,C. (2000) Human MT6-matrix metalloproteinase: identification, progelatinase A activation and expression in brain tumors. Cancer Res., 60, 877–882. [PubMed] [Google Scholar]
- Wilhelm S.M., Collier,I.E., Marmer,B.L., Eisen,A.Z., Grant,G.A. and Goldberg,G.I. (1989) SV40-transformed human lung fibroblasts secrete a 92-kDa type IV collagenase which is identical to that secreted by normal human macrophages. J. Biol. Chem., 264, 17213–17221. [PubMed] [Google Scholar]
- Will H., Atkinson,S.J., Butler,G.S., Smith,B. and Murphy,G. (1996) The soluble catalytic domain of membrane type 1 matrix metalloproteinase cleaves the propeptide of progelatinase A and initiates autoproteolytic activation. Regulation by TIMP-2 and TIMP-3. J. Biol. Chem., 271, 17119–17123. [DOI] [PubMed] [Google Scholar]
- Yana I. and Weiss,S.J. (2000) Regulation of membrane type-1 matrix metalloproteinase activation by proprotein convertases. Mol. Biol. Cell, 11, 2387–2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Apte,S.S., Soininen,R., Cao,R., Baaklini,G.Y., Rauser,R.W., Wang,J., Cao,Y. and Tryggvason,K. (2000) Impaired endochondral ossification and angiogenesis in mice deficient in membrane-type matrix metalloproteinase I. Proc. Natl Acad. Sci. USA, 97, 4052–4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuge Y. and Xu,J. (2001) Rac1 mediates type I collagen-dependent MMP-2 activation: role in cell invasion across collagen barrier. J. Biol. Chem., 276, 16248–16256. [DOI] [PubMed] [Google Scholar]


