Abstract
Histone acetyltransferases (HATs) such as Gcn5 play a role in transcriptional activation. However, the majority of constitutive genes show no requirement for GCN5, and even regulated genes, such as the yeast PHO5 gene, do not seem to be affected significantly by its absence under normal activation conditions. Here we show that even though the steady-state level of activated PHO5 transcription is not affected by deletion of GCN5, the rate of activation following phosphate starvation is significantly decreased. This delay in transcriptional activation is specifically due to slow chromatin remodeling of the PHO5 promoter, whereas the transmission of the phosphate starvation signal to the PHO5 promoter progresses at a normal rate. Chromatin remodeling is equally delayed in a galactose-inducible PHO5 promoter variant in which the Pho4 binding sites have been replaced by Gal4 binding sites. By contrast, activation of the GAL1 gene by galactose addition occurs with normal kinetics. Lack of the histone H4 N-termini leads to a similar delay in activation of the PHO5 promoter. These results indicate that one important contribution of HATs is to increase the rate of gene induction by accelerating chromatin remodeling, rather than to affect the final steady-state expression levels.
Keywords: GCN5/gene regulation/histone acetylation/PHO5
Introduction
The organization of DNA in chromatin provides a major obstacle to gene expression. Much of this occurs at the level of the nucleosome, which plays a highly dynamic role in the regulation of transcription. By controlling the access of the transcription machinery to the DNA template, the nucleosome presents a powerful barrier to transcription. The cell has evolved tools to deal with the nucleosome’s ability to restrict DNA accessibility. Two classes of chromatin modifying activities have been demonstrated to exist in many different systems. These are the ATP-dependent chromatin remodeling machines typified by the Swi/Snf complex (reviewed in Peterson and Workman, 2000; Vignali et al., 2000), and the histone acetyltransferases (HATs) and deacetylases (reviewed in Brown et al., 2000; Ng and Bird, 2000; Sterner and Berger, 2000). In the case of regulated genes, many of those that require remodeling complexes also employ histone acetylation to achieve activation (Pollard and Peterson, 1998). However, there also appears to be a large number of genes whose induction requires none of the currently known chromatin modifying activities.
We have focused our attention on two coregulated phosphatase genes in yeast, the PHO5 gene (Svaren and Hörz, 1997), which encodes an acid phosphatase, and PHO8, the gene for an alkaline phosphatase (Kaneko et al., 1987). Both are activated upon depletion of inorganic phosphate in the medium, and they share the same signal transduction pathway as well as the dedicated transcription factor Pho4, which is directly responsible for the activation of both promoters (Münsterkötter et al., 2000). In phosphate-containing media, i.e. conditions under which both PHO5 and PHO8 are repressed, Pho4 is phosphorylated through the action of Pho80/Pho85, a cyclin/cyclin dependent kinase (CDK) pair (Kaffman et al., 1994). Phosphorylation of Pho4 negatively affects transcriptional activation at three different levels: it promotes export of Pho4 from the nucleus, prevents its reimport into the nucleus, and abolishes Pho4–Pho2 interaction (Komeili and O’Shea, 1999), which is a prerequisite for Pho4 binding to the PHO5 promoter (Barbaric et al., 1998)
Both promoters undergo distinct chromatin remodeling upon activation. At PHO5, four positioned nucleosomes at the repressed promoter undergo a profound structural alteration, resulting in a 600 bp region of the promoter becoming fully accessible (Almer et al., 1986). Upon induction of PHO8, a labile nucleosome located between two hypersensitive regions is disrupted, and a 300 bp hypersensitive region is generated. However, the promoter region downstream of UASp2 acquires only intermediate accessibility to nucleases, consistent with the persistence of unstable, partially remodeled nucleosomes (Barbaric et al., 1992). PHO8 strictly requires histone acetylation by Gcn5 and the Swi/Snf chromatin remodeling activity for induction (Gregory et al., 1999). PHO5, on the other hand, is largely independent of these activities under the standard activation conditions, i.e. phosphate depletion (Gregory et al., 1998). The activity of the promoter in both a gcn5 and a snf2 strain reaches almost wild-type (wt) levels, and nucleosome disruption levels are also very similar to those found in wt strains, suggesting that neither complex plays a significant role in the activation process at this promoter.
The vast majority of experiments in which the requirement for a coactivator was investigated have focused on steady-state expression of a given gene in the presence or absence of the coactivator. In the case of regulated genes, the final level of activation with and without coactivator is usually compared. Work on Elp3, the HAT subunit of Elongator, a multisubunit complex associated with elongating RNA polymerase II, has recently revealed that the rate of induction rather than final transcription level for a variety of genes can be affected by this acetyltransferase (Otero et al., 1999; Wittschieben et al., 1999). In its absence, gene induction proceeded significantly more slowly. It seemed important to know whether this feature of Elp3 is a more general property of HATs, especially since a more rapid response of the cell to a signal may actually be just as important or even more important physiologically than achieving a high final level of induction. Gcn5 is the paradigm HAT in yeast and has been investigated more extensively than any other HAT. We decided, therefore, to look for possible kinetic effects of the Gcn5 HAT on induction of the PHO5 gene upon phosphate depletion. We find a striking effect of Gcn5 on the rate of chromatin remodeling at the PHO5 promoter and the kinetics of PHO5 induction. Both are strongly delayed if Gcn5 is deleted or its acetylase activity destroyed by targeted mutation. We provide evidence that this delay is related to the chromatin structure of the PHO5 promoter, that it is not a genome-wide phenomenon, and that it is not an effect on the act of transcription itself.
Results
PHO5 induction is strongly delayed in the absence of Gcn5
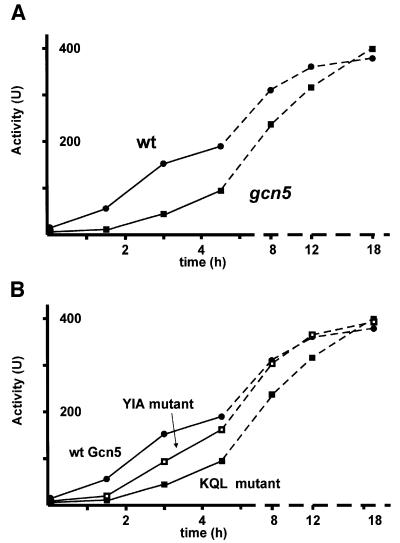
We have previously shown that Gcn5 is not required for induction of the PHO5 promoter and chromatin remodeling. Under phosphate starvation conditions, acid phosphatase activity reaches levels very similar to those of wt cells. Moreover, chromatin opening at the PHO5 promoter is indistinguishable in a gcn5 and a wt strain (Gregory et al., 1998). However, when we looked at the time course of activation we found that acid phosphatase activity accumulated at a strikingly reduced rate in a gcn5 strain compared with the isogenic wt strain (Figure 1A). The wt strain took 4–5 h to reach 50% of full activity (200 U). After the same period of induction, a gcn5 strain attained only 20–25% of final activity.
Fig. 1. The kinetics of PHO5 induction is strongly dependent on Gcn5. (A) The time course of PHO5 induction was followed by measuring acid phosphatase activity at the indicated time points after transferring cells from phosphate-containing media (+Pi) to phosphate-free media (–Pi) in a wt strain (circles) and a Δgcn5 strain (squares). The scale of the time axes is different for the first 5 h (solid lines) and the remaining 13 h (broken lines). (B) Identical measurements were carried out with the Gcn5 HAT domain mutant strains KQL (solid squares) and YIA (open squares) (Gregory et al., 1998; Wang et al., 1998). The solid circles denote the wt strain.
Gcn5 is a member of two multiprotein complexes termed SAGA and ADA within which it serves as a nucleosomal HAT. If Gcn5 is absent from the cell through deletion of its gene, the composition or stability of these complexes is likely to change. The observed kinetic difference could therefore be an indirect effect via other components of SAGA/ADA and/or the result of partial decomposition of these complexes. To address this possibility, we made use of mutants of Gcn5 that abolish the catalytic HAT activity of the protein while leaving the complexes intact (Gregory et al., 1998; Wang et al., 1998). The results for this experiment are shown in Figure 1B. The KQL mutant strain, which possesses HAT activity equivalent to that of the Δgcn5 strain, gave the same delay in activation as the strain disrupted for GCN5. Furthermore, the YIA mutant (which is intermediate in HAT activity) was also intermediate in activation kinetics. These results provide a strong correlation between the histone acetylation activity of Gcn5 and the rate at which PHO5 is activated in response to phosphate starvation.
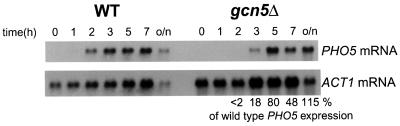
The delay in PHO5 induction as measured through the levels of acid phosphatase accumulation could occur at several stages in the process of gene expression. The data shown in Figure 2 demonstrate, however, that the production of mRNA was similarly delayed, ruling out the possibility that the kinetic effect was due to a delay in post-transcriptional processes like translation and/or phosphatase export to the periplasmic space. In order to determine whether the process of transcription itself proceeds at a lower rate in the absence of Gcn5, or whether Gcn5 affects prior steps, we compared the rate of PHO5 chromatin remodeling [which has been shown to occur independently of transcription (Fascher et al., 1993)] in the presence and absence of Gcn5.
Fig. 2. The Gcn5-dependent delay in PHO5 expression occurs at the level of transcription. Total RNA was extracted at the indicated time points after transferring cells from phosphate-containing media (+Pi) to phosphate-free media (–Pi) in a wt and a Δgcn5 strain, blotted and hybridized with a PHO5 and an ACT1 coding region specific probe (load control). Autoradiographs are shown. The amounts of PHO5 mRNA in the Δgcn5 relative to the wt cells at the different time points (normalized for ACT1 expression) were calculated and are shown at the bottom. Expression of the highly expressed, constitutive ACT1 gene is not affected by a GCN5 deletion (Lee et al., 2000).
Gcn5 affects the rate of chromatin remodeling at the PHO5 promoter
Measuring the accessibility of restriction sites in the PHO5 promoter has been shown to be a reliable way of following chromatin opening in a quantitative way. Notably, the accessibility of a ClaI site located in the precisely positioned nucleosome –2 differs dramatically between repressed and active nuclei. Two other sites, a BamHI and a BstEII site, are 50% accessible in repressed nuclei, consistent with their presence in nucleosomal linker regions. Like ClaI, they reach close to 100% accessibility when the PHO5 promoter is fully activated.
We previously reported that these sites became almost fully accessible also in a gcn5 strain after the usual overnight induction (Gregory et al., 1998). However, when we determined the accessibility of restriction sites at earlier time points of PHO5 induction, the presence or absence of Gcn5 made a clear difference. The accessibility data after 3 h of induction are shown in Figure 3. The ClaI site in the wt strain was highly accessible, while the same site in a Δgcn5 strain was only ∼20% accessible. A similar difference was observed for the BstEII site, which was 70–80% accessible in the wt strain and remained at 50% in the Δgcn5 strain. This is the value found also under repressing conditions, and it reflects the location of this site between two positioned nucleosomes in the promoter. These data clearly demonstrate that Gcn5 is necessary for the normal kinetics of chromatin remodeling at the PHO5 promoter. This delay is independent of the coding sequence and chromosomal location of the promoter since an identical delay in chromatin remodeling was seen when PHO5–lacZ fusion constructs were analyzed on centromeric plasmids (data not shown), indicating that the delay is specifically due to the structure of the PHO5 promoter region. In conclusion, the rate of PHO5 activation is strongly delayed in a Δgcn5 background, and this kinetic effect is already seen at the level of chromatin opening at the promoter, prior to actual transcription of the gene.
Fig. 3. The rate of chromatin remodeling at the PHO5 promoter is strongly decreased in the absence of Gcn5. Nuclei isolated from cells of a wt strain (YS18, lanes 1, 2, 5, 6, 9 and 10) and a Δgcn5 strain (YS5319, lanes 3, 4, 7, 8, 11 and 12) 3 h after transfer to phosphate-free medium were treated for 30 min at 37°C with 50 U (odd numbered lanes) or 200 U (even numbered lanes) of the restriction enzyme indicated. In order to monitor the extent of cleavage, DNA was isolated, cleaved with HaeIII, analyzed on a 1.5% agarose gel, blotted and hybridized with a probe (Almer et al., 1986). The positions of the restriction sites with respect to the nucleosomal organization of the repressed PHO5 promoter are shown schematically beneath. In all cases, the appearance of the lower band represents accessibility of the corresponding site. The nucleosomes that are perturbed during normal activation are shown as open circles, the Pho4 binding sites as squares and the TATA box as T.
Components transducing the phosphate starvation signal are not the target of Gcn5
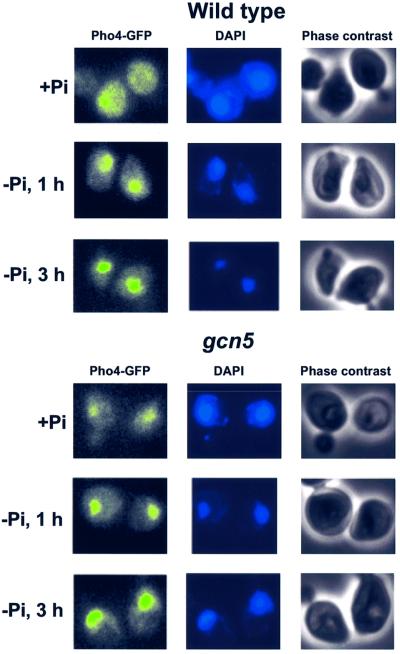
An early step of PHO5 induction by phosphate starvation is the activation of the cyclin/CDK inhibitor Pho81. It acts to block the Pho80/Pho85 cyclin/CDK pair which leads to the inhibition of Pho4 phosphorylation. Non-phosphorylated Pho4 accumulates in the nucleus and is capable of binding its cognate recognition sites in association with Pho2. Based on the results above, we could not rule out the possibility that the absence of Gcn5 simply affected one of the steps in the signal transduction pathway leading to the activation of Pho4 and thereby (indirectly) affected chromatin remodeling and induction of PHO5. If this were the case we would expect the nuclear accumulation of Pho4 to occur more slowly in a Δgcn5 strain. To address this possibility, we made use of a green fluorescent protein (GFP) derivative of Pho4 (O’Neill et al., 1996). The fluoromicroscopic images in Figure 4 show that there was no difference between a wt and a gcn5 strain in the rate of Pho4 accumulation in the nucleus after switching cells to phosphate starvation conditions (compare panels ‘–Pi’ for wt and Δgcn5). Thus, the kinetic delay occurs after the accumulation of non-phosphorylated, active Pho4 in the nucleus, and therefore directly at the subsequent nucleosome remodeling step of the PHO5 promoter.
Fig. 4. The kinetics of Pho4 accumulation in the nucleus following phosphate starvation is not affected by Gcn5. A wt and a Δgcn5 strain expressing a Pho4–GFP fusion protein were grown in high phosphate medium (+Pi) and at time zero transferred to phosphate-free medium (–Pi). Pho4 localization was monitored at the times indicated by fluorescence microscopy (left panel). 4′,6-diamidine-2-phenylindole (DAPI)-stained cells are shown in the center panel, and cells visualized by phase contrast microscopy on the right.
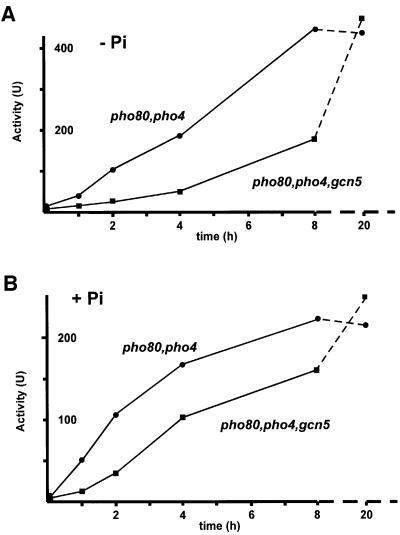
To confirm that the kinetic effect of Gcn5 on chromatin remodeling has nothing to do with signal transduction in the PHO system, the rate of PHO5 activation was examined in strains disrupted for PHO80. This way, any effect of Gcn5 on the process of Pho80/Pho85-mediated modification of Pho4 can be eliminated. However, a pho80 strain produces acid phosphatase constitutively, as Pho4 is uncoupled from the regulatory system that controls its activity, thus precluding kinetic measurements. To get around this problem, we used a PHO4 gene that was under the control of the GAL10 promoter (Jayaraman et al., 1994). With this hybrid construct, activation of PHO5 can be induced through addition of galactose to the medium. We introduced such a Pho4 expression plasmid into a pho80 strain and also into a pho80/gcn5 strain, both growing in the presence of phosphate in raffinose medium. Induction of PHO5 was then initiated by transferring the cells to phosphate-free medium containing galactose. As shown in Figure 5A, there was again a clear effect of Gcn5 on the rate of phosphatase production. In its presence, induction proceeded much more rapidly than in its absence.
Fig. 5. The effect of Gcn5 on the kinetics of PHO5 induction is maintained in pho80 cells. Expression of PHO5 in a pho80, pho4 strain (YS33, circles) and a pho80, pho4, gcn5 strain (YS53389, squares) carrying a plasmid with a PHO4 gene controlled by the GAL10 promoter (pKV701-PHO4) (Jayaraman et al., 1994), was followed by measuring acid phosphatase activity upon galactose induction in phosphate-free (A) or phosphate-containing media (B). The scale of the time axes is different for the first 8 h (solid lines) and the remaining 12 h (broken lines).
To rule out the possibility that the Gcn5 effect was specific for phosphate starvation conditions, the galactose induction experiment was repeated in high phosphate conditions. As shown in Figure 5B, there was a delay in activation when Gcn5 was absent that was indistinguishable from that observed in phosphate-free media. These results, taken together with the data on Pho4 translocation into the nucleus (Figure 4), effectively rule out the possibility that any of the pathway(s) transmitting the phosphate starvation signal to Pho4 is a primary target of Gcn5 and that the delay in PHO5 activation has its origin at that level. In addition, since Pho4 was expressed under the control of the GAL promoter, these experiments eliminate the possibility that the effect of Gcn5 on PHO5 induction is via the PHO4 promoter.
The kinetic effect of Gcn5 on PHO5 promoter activation is maintained even if the promoter is under the control of Gal4
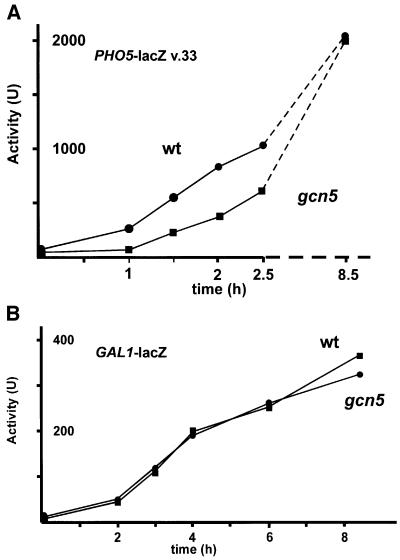
The kinetic effect of Gcn5 on PHO5 promoter activation could be an intrinsic property of the way Pho4 interacts with its upstream activator sequence (UAS) elements. Alternatively, it could be a property of the chromatin at the promoter that is remodeled in the course of activation. In order to differentiate between these two alternatives, we made use of a promoter derivative in which the two Pho4 binding, UASp1 and UASp2, were replaced by Gal4 binding sites (Ertinger, 1998). We have shown that this promoter derivative responds in exactly the same way to galactose induction as the wt PHO5 promoter responds to phosphate starvation. The same four nucleosomes are remodeled, and PHO5 is powerfully activated (Ertinger, 1998). This promoter derivative was fused to a lacZ reporter and activation was measured through β-galactosidase measurements in a wt and a gcn5 strain. As seen in Figure 6A, a virtually identical delay in the activation of this promoter in the absence of Gcn5 was observed, confirming that the delay is a property of the chromatin structure of the PHO5 promoter rather than due to the way Pho4 interacts with it.
Fig. 6. Effect of Gcn5 on induction of galactose-inducible promoters. (A) Activation of pPHO5-lacZ variant 33 [both Pho4 sites in the PHO5 promoter replaced by Gal4 binding sites (Ertinger, 1998)] was followed in a wt (YS18, squares) and a gcn5 strain (YS5189, circles) by measuring β-galactosidase activity. At time zero, galactose was added to the medium. The scale of the time axis is different for the first 2.5 h (solid lines) and the remaining 6 h (broken lines). (B) Galactose-mediated activation of the p416-GAL1-lacZ plasmid containing the GAL1 promoter was measured in the same two strains.
Absence of Gcn5 does not delay activation of the GAL1 promoter
The results presented so far raise the possibility that a delay in the kinetics of activation is a general property of inducible promoters in the absence of Gcn5. The GAL1 promoter is an obvious choice when it comes to testing other promoters, since it is one of the best studied promoters in yeast. At the same time, comparing the native GAL1 promoter with the galactose-inducible PHO5 promoter variant described in the previous section (which did show a delay) might resolve the question of whether the delay is specific to cis features of the PHO5 promoter or features of transactivation.
As shown in Figure 6B, a GAL1–lacZ construct (p416-GAL1-lacZ) was activated at the same rate regardless of whether Gcn5 was present or not. Therefore, the delay we observe for chromatin remodeling and activation of the PHO5 promoter can not be due to a general effect of Gcn5 on activation of regulated genes, but rather must be due to features of the PHO5 promoter.
The absence of histone tails can also lead to a delay in PHO5 activation
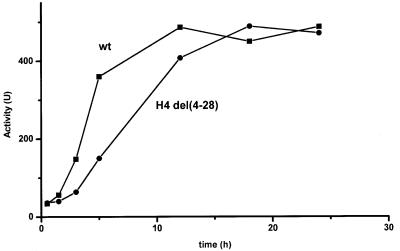
Although it appears plausible to interpret the effect of Gcn5 in accelerating chromatin remodeling at the PHO5 promoter as due to histone acetylation at the promoter, it is still conceivable that other targets of Gcn5 are responsible for that. Efforts to demonstrate targeted acetylation of the PHO5 promoter in the course of normal activation, i.e. in a wild-type strain (and its absence in a gcn5 background) have so far not been successful (see Discussion). As a complementary approach we therefore initiated experiments using yeast strains lacking histone termini which are the targets of HAT activity. Strains lacking the H3 termini grew very poorly under our conditions of activation and therefore made reliable measurements of the kinetics of induction quite difficult. We therefore assayed a strain lacking the H4 termini which did not give this complication. The results of these experiments are shown in Figure 7. It is striking that the absence of the histone H4 N-termini generated the same kind of delay in PHO5 induction as the absence of Gcn5. This result demonstrates that histone tails do have an effect on the kinetics of PHO5 activation and strongly suggest that Gcn5 exerts its effects on the rate of PHO5 activation via histone tail acetylation.
Fig. 7. Effect of the histone H4 N-termini on the kinetics of PHO5 induction. The time course of PHO5 induction in strains PKY899 (HHF2; squares) and PKY813 [hhf2(del 4–28); circles] was followed by measuring acid phosphatase activity at the indicated time points after transferring cells from phosphate-containing media to phosphate-free media.
Discussion
A novel role for Gcn5 in yeast
Gcn5 was originally discovered as a coactivator required by the Gcn4 activator and the HAP activation complex in order to promote normal levels of transcriptional activation (Georgakopoulos and Thireos, 1992), and later found to be required also for full activity of other transcriptional activators (Marcus et al., 1994). Thus, Gcn5 was concluded to be a new member of a class of transcriptional regulators that collaborate with specific DNA binding activators to promote high levels of transcription. The discovery that Gcn5 has intrinsic HAT activity provided a crucial link between histone acetylation and transcription. Interestingly however, genome-wide analyses demonstrated that just 5% of yeast genes are dependent on GCN5 for normal expression (Lee et al., 2000). It is important to realize that under the conditions used in the yeast DNA microarray studies, the majority of the regulated genes are in the repressed state. This is succinctly demonstrated for PHO5, the expression of which is 10-fold lower in the absence of Gcn5 in standard high phosphate media, that is, under repressing conditions (Gregory et al., 1998). However, we have shown that the steady-state level of PHO5 expression under activating conditions reaches about the same level in a gcn5 strain as in a wt strain. This is consistent with the finding of only a handful of regulated promoters such as HO (Pollard and Peterson, 1997; Cosma et al., 1999; Krebs et al., 1999), HIS3 (Chong et al., 1996) and PHO8 (Gregory et al., 1999) that require Gcn5 to reach full activation.
The central conclusion of the present work is that in addition to either affecting steady-state repressed or steady-state activated levels, Gcn5 can have a third type of effect: it can significantly increase the rate of activation rather than the final level of expression. Conversely, in its absence, the response to activating conditions can be strikingly delayed. This finding strongly suggests that the measurement of absolute expression levels, under basal or steady-state activating conditions, may miss an important role for acetylases in the dynamics of gene regulation.
Kinetics of PHO5 activation
From the time a cell senses lack of an adequate supply of phosphate to the increase in expression of the relevant genes, many steps are involved. The phosphate concentration is monitored in a way that is not fully understood but involves the high affinity phosphate transporter Pho84. The primary signal is transmitted through a signal transduction pathway involving the Pho80/Pho85 cyclin/CDK pair and a CDK inhibitor, Pho81, and culminates in the regulation of Pho4, the dedicated DNA binding transcription factor that then activates genes such as PHO5 and PHO8 (Lenburg and Oshea, 1996). The delay in PHO5 activation due to the absence of Gcn5 could therefore be due in principle to a delay at any of these steps. We were able to show, however, that activated Pho4 accumulates in the nucleus at the same rate in a wt and a gcn5 strain. Furthermore, bypassing the phosphate starvation signal transduction pathway by using pho80 cells that express PHO5 constitutively and putting expression of the Pho4 activator under control of the inducible GAL10 promoter (whose activation is not delayed in gcn5 cells) does not diminish the delay in PHO5 activation. As an ultimate control, we brought the PHO5 promoter completely under galactose control by replacing the Pho4 binding sites with Gal4 sites. Even in this promoter variant, which is now under the direct control of the Gal4 activator, activation is still delayed to a similar extent in the absence of Gcn5. These results rule out the possibility that the Gcn5 effects are at the level of phosphate signal transduction, Pho4 activation or expression, or that Pho4 is the relevant target of Gcn5.
We can also rule out the possibility that the coding sequence of the PHO5 gene plays a significant role, because activation and promoter chromatin remodeling of a PHO5–lacZ construct showed the same kinetic dependence on Gcn5 as the chromosomal PHO5 locus. This makes the structure of the promoter the most probable target of Gcn5. In this respect it is important to note that the PHO8 promoter, which is completely dependent on Gcn5, appears to offer maximal resistance to remodeling, since accessibility even in a fully active wild-type strain still seems significantly restricted through persisting nucleosomes. In contrast, PHO5 is completely accessible in the active state, whereas the GAL1-10 promoter does not undergo such extensive chromatin remodeling upon activation (Lohr, 1997). In conclusion, we think that the quality of the nucleosomal structure at the promoter and the impediment it presents to the act of transcription are responsible for the difference between the promoters we studied, and determine the extent to which they depend on Gcn5.
We have not been able to demonstrate hyperacetylation across the PHO5 promoter during activation in the presence of Gcn5 (data not shown). This was also not possible in the case of the PHO8 promoter, where hyperacetylation does take place (Reinke et al., 2001), presumably because rapid deacetylation occurs in the course of remodeling. Only by freezing chromatin in the repressed state by eliminating Snf2 could we demonstrate transient histone hyperacetylation across the PHO8 promoter. This strategy is not possible for PHO5 since chromatin remodeling at this promoter is not Snf2 dependent (Gaudreau et al., 1997). That it is indeed acetylation of the histone tails by which Gcn5 accelerates remodeling of the PHO5 promoter, and not of some pleiotropic non-histone protein, is made highly likely by the finding that strains that lack the histone H4 N-termini are similarly delayed in their activation of PHO5 as by the absence of Gcn5 (compare Figures 7 and 1).
Delay in activation is not a universal consequence of the absence of Gcn5
For the majority of genes the effect of Gcn5 on the rate of activation has not been explicitly addressed. It was therefore conceivable that the Gcn5-dependent delay we witnessed at PHO5 was a general property of regulated promoters. This is however not the case. Our results with the GAL1 promoter, the paradigm of a regulated promoter in yeast, show that activation here proceeds at the same rate regardless of whether Gcn5 is present or not. This result gains even more significance from the fact that activation of the PHO5 promoter put under control of the same galactose activation system remains delayed when Gcn5 is absent, and strongly suggests that a specific promoter architecture can determine the kinetic effect of the coactivator on gene activation.
Why is chromatin remodeling at the PHO5 promoter delayed in the absence of Gcn5 yet the final level not affected?
Recent data we have obtained for the PHO8 promoter indicate that Gcn5-dependent acetylation of nucleosomes at this promoter provides a transient signal (Reinke et al., 2001). This signal does not in itself affect the stability of the nucleosomes, but rather seems to mark them for remodeling by Swi/Snf, which is critically required for generating an accessible chromatin structure at that promoter (Gregory et al., 1999). The central question then is how chromatin is remodeled at the PHO5 promoter. Swi/Snf might play a role, although not as conspicuous as at PHO8 since chromatin remodeling can be accomplished even in the absence of Snf2 (Gaudreau et al., 1997). We have found, however, that the kinetics of activation at PHO5 is significantly delayed also in the absence of Snf2 (our unpublished results), pointing to a subtler role of Swi/Snf at this promoter or to redundant activities involved. We have also found that stable Pho4 binding to the PHO5 promoter as detected by in vivo dimethylsulfate footprinting only occurs as chromatin remodeling nears completion (S.Barbaric and W.Hörz, unpublished results). However, Pho4 (and its association with the PHO5 promoter) is absolutely required for remodeling to occur. Immediately after its entry into the nucleus, Pho4 can only access UASp1 since nucleosome –2 prevents its binding to UASp2 (Venter et al., 1994). Binding to only UASp1 at the PHO5 promoter is not detectable in vivo and must therefore be weak and transient. However, the ability of the factor to bring chromatin remodeling complexes such as SAGA and Swi/Snf to the promoter would soon lead to the establishment of a more open chromatin structure and allow transient binding of Pho4 also at UASp2. Eventually, the continued generation of more accessible chromatin would permit stable DNA association of Pho4 with both UAS elements, allowing it to now recruit the basal transcription machinery to activate transcription. Indeed, the very long delay from nuclear entry of activated Pho4 (∼0.5 h; see Figure 4) to actual full activation of transcription (2–3 h; see Figure 2) might be due to such a mechanism. Chromatin structure at PHO5 would in this scenario be a barrier which, once overcome, no longer represented an obstacle to transcriptional activation.
This model could in turn explain why the absence of Gcn5 merely results in a delay in PHO5 chromatin remodeling and transcriptional activation; in the absence of Gcn5 recruitment, the affinity of chromatin at the PHO5 promoter for remodeling activities such as the Swi/Snf complex would be reduced. Remodeling would therefore be slowed down, and as a consequence, the persistent Pho4 DNA binding required for transcriptional activation would be established more slowly. Such a scenario would be consistent with our recent in vivo data for the PHO8 promoter which point to acetylation being a transient signal marking nucleosomes for remodeling (Reinke et al., 2001). It would also be consistent with the demonstration that hyperacetylated nucleosomes bind the Swi/Snf complex more tightly in vitro (Hassan et al., 2001).
A variation of this scheme might be found in the fact that Gcn5 also affects the global level of chromatin acetylation (Krebs et al., 1999; Kuo et al., 2000; Vogelauer et al., 2000). Global chromatin underacetylation (also affecting the PHO5 locus) would generate a chromatin environment that is harder to remodel by Swi/Snf or other remodeling complexes. Consequently, PHO5 activation in gcn5 cells would be delayed.
Concluding remarks
It is clear that the speed at which a cell responds to a stimulus by turning on a set of genes and turning off another is of great importance to the fate of that cell. However, this aspect of gene regulation is not generally appreciated. Instead, the amplitude of regulation or the absolute levels of expression typical of the fully active or the fully repressed state are generally seen as the hallmarks of a regulated gene. We have shown that the rate of activation of the PHO5 promoter is strongly dependent on the HAT Gcn5, whereas the final level of induction is not. The evidence points to the remodeling of the chromatin structure of the PHO5 promoter as being the rate-limiting step. It is likely that this role of chromatin remodeling may be a general phenomenon, and that one important contribution of enzymes such as Gcn5, Elp3, and Swi2/Snf2 is to increase the rate of chromatin remodeling and thereby the rate of gene induction, rather than to affect the final steady-state levels of expression.
Materials and methods
Yeast strains and media
The YS series Saccharomyces cerevisiae strains used in this study are isogenic with strain YS18 (MATα, his3-11, his3-15, leu2-3, leu2-112, canR, ura3Δ5). YS5189 contains a disruption of the GCN5 gene, YS33 of the PHO4 and PHO80, and YS53389 of the PHO4, PHO80 and GCN5 genes. PKY899 carrying the plasmid pUK499 (HHF2) and the isogenic PKY813 carrying pK613 (hhf2 del(4–28) are described in Kayne et al. (1988). Yeast strains were grown in YPDA or in YNB medium supplemented with the required amino acids (high phosphate conditions) and PHO5 induction was initiated by transferring cells to phosphate-free synthetic medium (Svaren et al., 1995). For galactose induction of p416-GAL1-lacZ or pPHO5-lacZ variant 33, cells were pregrown in YPA or YNB media containing 2% raffinose, and induction was initiated by addition of 2% galactose to the medium. For simultaneous induction by galactose and by phosphate starvation, cells pregrown in YPA + 2% raffinose media were transferred to phosphate-free synthetic medium containing 2% raffinose and 2% galactose.
Plasmids
The plasmids used in this study were previously described: pKV701-PHO4 is a 2µ based vector containing the PHO4 gene under control of the GAL10 promoter (Jayaraman et al., 1994). pPHO5-lacZ variant 33 is a derivative of the pPHO5-lacZ reporter plasmid (Straka and Hörz, 1991) in which the two Pho4 binding sites, UASp1 and UASp2, were replaced by Gal4 binding sites (Ertinger, 1998). p416-GAL1-lacZ contains a GAL1 promoter–lacZ fusion in the pRS416 vector (Sikorski and Hieter, 1989).
Functional assays and chromatin analysis
The acid phosphatase (Haguenauer-Tsapis and Hinnen, 1984) and β-galactosidase assays (Straka and Hörz, 1991) were carried out as described before. All activity values shown are the average of at least three independent measurements (SD 10–20% for β-galactosidase and 5–10% for acid phosphatase). PHO5 mRNA was detected by northern blotting analysis (Sambrook et al., 1989) of total mRNA isolated by using Qiagen RNAeasy mini kits. Quantification of mRNA levels was performed using a Phosphoimager. The preparation of yeast nuclei (Almer and Hörz, 1986) and restriction nuclease digestion of isolated nuclei (Svaren et al., 1995) have been described. Restriction nuclease accessibility values were determined by phosphoimaging the Southern blots and calculating the ratio of the two fragments in each lane.
Microscopic analysis of Pho4–GFP localization
Cells were fixed in methanol and resuspended in acetone before mounting on polylysine coated slides. Nuclei were visualized in 0.3 mg/ml DAPI, 50% glycerol. Microscopy was performed on a Zeiss Axioplan microscope with 100× oil-immersion objective. Images were captured on a Hamamatsu C4742–95 digital camera using Openlab 2.0.4 software.
Acknowledgments
Acknowledgements
We thank E.O’Shea, C.Goding and M.Grunstein for kind gifts of plasmids and Philip Gregory for careful reading of the manuscript. This work was supported by grants from the Deutsche Forschungsgemeinschaft (SFB190) and Fonds der Chemischen Industrie to W.H., from PLIVA, Zagreb, to S.B., and by funds from the Imperial Cancer Research Fund to J.Q.S.
References
- Almer A. and Hörz,W. (1986) Nuclease hypersensitive regions with adjacent positioned nucleosomes mark the gene boundaries of the PHO5/PHO3 locus in yeast. EMBO J., 5, 2681–2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almer A., Rudolph,H., Hinnen,A. and Hörz,W. (1986) Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. EMBO J., 5, 2689–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbaric S., Fascher,K.D. and Hörz,W. (1992) Activation of the weakly regulated PHO8 promoter in S.cerevisiae: chromatin transition and binding sites for the positive regulator protein Pho4. Nucleic Acids Res., 20, 1031–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbaric S., Münsterkötter,M., Goding,C. and Hörz,W. (1998) Cooperative Pho2–Pho4 interactions at the PHO5 promoter are critical for binding of Pho4 to UASp1 and for efficient transactivation by Pho4 at UASp2. Mol. Cell. Biol., 18, 2629–2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C.E., Lechner,T., Howe,L. and Workman,J.L. (2000) The many HATs of transcription coactivators. Trends Biochem. Sci., 25, 15–19. [DOI] [PubMed] [Google Scholar]
- Chong J.P., Thommes,P. and Blow,J.J. (1996) The role of MCM/P1 proteins in the licensing of DNA replication. Trends Biochem. Sci., 21, 102–106. [PubMed] [Google Scholar]
- Cosma M.P., Tanaka,T. and Nasmyth,K. (1999) Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell, 97, 299–311. [DOI] [PubMed] [Google Scholar]
- Ertinger G. (1998) Rolle der Transkriptionsfaktoren bei der Öffnung der Chromatinstruktur am PHO5-Promoter in Saccharomyces cerevisiae. PhD thesis, Universität München, Germany.
- Fascher K.D., Schmitz,J. and Hörz,W. (1993) Structural and functional requirements for the chromatin transition at the PHO5 promoter in Saccharomyces cerevisiae upon PHO5 activation. J. Mol. Biol., 231, 658–667. [DOI] [PubMed] [Google Scholar]
- Gaudreau L., Schmid,A., Blaschke,D., Ptashne,M. and Hörz,W. (1997) RNA polymerase II holoenzyme recruitment is sufficient to remodel chromatin at the yeast PHO5 promoter. Cell, 89, 55–62. [DOI] [PubMed] [Google Scholar]
- Georgakopoulos T. and Thireos,G. (1992) Two distinct yeast transcriptional activators require the function of the GCN5 protein to promote normal levels of transcription. EMBO J., 11, 4145–4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory P.D., Schmid,A., Zavari,M., Lui,L., Berger,S.L. and Hörz,W. (1998) Absence of Gcn5 HAT activity defines a novel state in the opening of chromatin at the PHO5 promoter in yeast. Mol. Cell, 1, 495–505. [DOI] [PubMed] [Google Scholar]
- Gregory P.D., Schmid,A., Zavari,M., Münsterkötter,M. and Hörz,W. (1999) Chromatin remodelling at the PHO8 promoter requires SWI-SNF and SAGA at a step subsequent to activator binding. EMBO J., 18, 6407–6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haguenauer-Tsapis R. and Hinnen,A. (1984) A deletion that includes the signal peptidase cleavage site impairs processing, glycosylation, and secretion of cell surface yeast acid phosphatase. Mol. Cell. Biol., 4, 2668–2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan A.H., Neely,K.E. and Workman,J.L. (2001) Histone acetyltransferase complexes stabilize Swi/Snf binding to promoter nucleosomes. Cell, 104, 817–827. [DOI] [PubMed] [Google Scholar]
- Jayaraman P.S., Hirst,K. and Goding,C.R. (1994) The activation domain of a basic helix–loop–helix protein is masked by repressor interaction with domains distinct from that required for transcription regulation. EMBO J., 13, 2192–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaffman A., Herskowitz,I., Tjian,R. and O’Shea,E.K. (1994) Phosphorylation of the transcription factor Pho4 by a cyclin–CDK complex, Pho80–Pho85. Science, 263, 1153–1156. [DOI] [PubMed] [Google Scholar]
- Kaneko Y., Hayashi,N., Toh-e,A., Banno,I. and Oshima,Y. (1987) Structural characteristics of the PHO8 gene encoding repressible alkaline phosphatase in Saccharomyces cerevisiae. Gene, 58, 137–148. [DOI] [PubMed] [Google Scholar]
- Kayne P.S., Kim,U.J., Han,M., Mullen,J.R., Yoshizaki,F. and Grunstein,M. (1988) Extremely conserved histone H4 N-terminus is dispensable for growth but essential for repressing the silent mating loci in yeast. Cell, 55, 27–39. [DOI] [PubMed] [Google Scholar]
- Komeili A. and O’Shea,E.K. (1999) Roles of phosphorylation sites in regulating activity of the transcription factor Pho4. Science, 284, 977–980. [DOI] [PubMed] [Google Scholar]
- Krebs J.E., Kuo,M.H., Allis,C.D. and Peterson,C.L. (1999) Cell cycle-regulated histone acetylation required for expression of the yeast HO gene. Genes Dev., 13, 1412–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo M.H., vom Baur,E., Struhl,K. and Allis,C.D. (2000) Gcn4 activator targets Gcn5 histone acetyltransferase to specific promoters independently of transcription. Mol. Cell, 6, 1309–1320. [DOI] [PubMed] [Google Scholar]
- Lee T.I., Causton,H.C., Holstege,F.C., Shen,W.C., Hannett,N., Jennings,E.G., Winston,F., Green,M.R. and Young,R.A. (2000) Redundant roles for the TFIID and SAGA complexes in global transcription. Nature, 405, 701–704. [DOI] [PubMed] [Google Scholar]
- Lenburg M.E. and Oshea,E.K. (1996) Signaling phosphate starvation. Trends Biochem. Sci., 21, 383–387. [PubMed] [Google Scholar]
- Lohr D. (1997) Nucleosome transactions on the promoter of the yeast GAL and PHO genes. J. Biol. Chem., 272, 26795–26798. [DOI] [PubMed] [Google Scholar]
- Marcus G.A., Silverman,N., Berger,S.L., Horiuchi,J. and Guarente,L. (1994) Functional similarity and physical association between GCN5 and ADA2: putative transcriptional adaptors. EMBO J., 13, 4807–4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münsterkötter M., Barbaric,S. and Hörz,W. (2000) Transcriptional regulation of the yeast PHO8 promoter in comparison to the coregulated PHO5 promoter. J. Biol. Chem., 275, 22678–22685. [DOI] [PubMed] [Google Scholar]
- Ng H.H. and Bird,A. (2000) Histone deacetylases: silencers for hire. Trends Biochem. Sci., 25, 121–126. [DOI] [PubMed] [Google Scholar]
- O’Neill E.M., Kaffman,A., Jolly,E.R. and O’Shea,E.K. (1996) Regulation of Pho4 nuclear localization by the Pho80-Pho85 cyclin-CDK complex. Science, 271, 209–212. [DOI] [PubMed] [Google Scholar]
- Otero G., Fellows,J., Li,Y., de Bizemont,T., Dirac,A.M., Gustafsson,C.M., Erdjument-Bromage,H., Tempst,P. and Svejstrup,J.Q. (1999) Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol. Cell, 3, 109–118. [DOI] [PubMed] [Google Scholar]
- Peterson C.L. and Workman,J.L. (2000) Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr. Opin. Genet. Dev., 10, 187–192. [DOI] [PubMed] [Google Scholar]
- Pollard K.J. and Peterson,C.L. (1997) Role for ADA/GCN5 products in antagonizing chromatin-mediated transcriptional repression. Mol. Cell. Biol., 17, 6212–6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard K.J. and Peterson,C.L. (1998) Chromatin remodeling: a marriage between two families? BioEssays, 20, 771–780. [DOI] [PubMed] [Google Scholar]
- Reinke H., Gregory,P.D. and Hörz,W. (2001) A transient histone hyperacetylation signal marks nucleosomes for remodeling at the PHO8 promoter. Mol. Cell, 7, 529–538. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner D.E. and Berger,S.L. (2000) Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev., 64, 435–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straka C. and Hörz,W. (1991) A functional role for nucleosomes in the repression of a yeast promoter. EMBO J., 10, 361–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svaren J. and Hörz,W. (1997) Transcription factors vs nucleosomes: regulation of the PHO5 promoter in yeast. Trends Biochem. Sci., 22, 93–97. [DOI] [PubMed] [Google Scholar]
- Svaren J., Venter,U. and Hörz,W. (1995) In vivo analysis of nucleosome structure and transcription factor binding in Saccharomyces cerevisiae. Methods Mol. Genet., 6, 153–167. [Google Scholar]
- Venter U., Svaren,J., Schmitz,J., Schmid,A. and Hörz,W. (1994) A nucleosome precludes binding of the transcription factor Pho4 in vivo to a critical target site in the PHO5 promoter. EMBO J., 13, 4848–4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignali M., Hassan,A.H., Neely,K.E. and Workman,J.L. (2000) ATP-dependent chromatin-remodeling complexes. Mol. Cell. Biol., 20, 1899–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelauer M., Wu,J., Suka,N. and Grunstein,M. (2000) Global histone acetylation and deacetylation in yeast. Nature, 408, 495–498. [DOI] [PubMed] [Google Scholar]
- Wang L., Liu,L. and Berger,S.L. (1998) Critical residues for histone acetylation by Gcn5, functioning in Ada and SAGA complexes, are also required for transcriptional function in vivo. Genes Dev., 12, 640–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittschieben B.O. et al. (1999) A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Mol. Cell, 4, 123–128. [DOI] [PubMed] [Google Scholar]