Abstract
We recently reported that spliceosomes alter messenger ribonucleoprotein particle (mRNP) composition by depositing several proteins 20–24 nucleotides upstream of mRNA exon–exon junctions. When assembled in vitro, this so-called ‘exon–exon junction complex’ (EJC) contains at least five proteins: SRm160, DEK, RNPS1, Y14 and REF. To better investigate its functional attributes, we now describe a method for generating spliced mRNAs both in vitro and in vivo that either do or do not carry the EJC. Analysis of these mRNAs in Xenopus laevis oocytes revealed that this complex is the species responsible for enhancing nucleocytoplasmic export of spliced mRNAs. It does so by providing a strong binding site for the mRNA export factors REF and TAP/p15. Moreover, by serving as an anchoring point for the factors Upf2 and Upf3, the EJC provides a direct link between splicing and nonsense-mediated mRNA decay. Finally, we show that the composition of the EJC is dynamic in vivo and is subject to significant evolution upon mRNA export to the cytoplasm.
Keywords: exon–exon junction complex/mRNA export/nonsense-mediated decay/pre-mRNA splicing
Introduction
In higher eukaryotes, the vast majority of nascent RNA transcripts contain several introns (Lander et al., 2001). These intervening sequences are excised by the splicing machinery in the nucleus to generate mRNAs that are then exported to the cytoplasm. In addition to removing the introns, splicing specifically alters the protein composition of the product messenger ribonucleoprotein particle (mRNP) in the vicinity of exon–exon junctions (Le Hir et al., 2000a,b). This alteration consists of a stable protein complex deposited 20–24 nucleotides (nt) upstream of mRNA exon–exon junctions, independent of the sequence there. Immunoprecipitation experiments revealed that this so-called ‘exon–exon junction complex’ (EJC) contains at least five proteins: SRm160, DEK, RNPS1, REF and Y14 (Le Hir et al., 2000b). SRm160, DEK and RNPS1 are all either directly or indirectly associated with pre-mRNA splicing (Blencowe et al., 1998; Mayeda et al., 1999; McGarvey et al., 2000). REF is an RNA binding protein required for mRNA export (Stutz et al., 2000; Zhou et al., 2000; Rodrigues et al., 2001). REF interacts with the TAP/p15 heterodimer, which in turn makes specific contacts with components of the nuclear pore complex (Katahira et al., 1999; Bachi et al., 2000). Y14 is also an RNA binding protein that associates with TAP in vivo (Kataoka et al., 2000).
Numerous studies have documented metabolic effects of splicing on downstream mRNA metabolism. An early report was the observation that exon–exon junctions allow for distinction between premature and normal termination codons by the machinery involved in nonsense-mediated mRNA decay (NMD; Cheng et al., 1994). That is, if an in-frame termination codon is located some distance upstream of at least one exon–exon junction, it is generally recognized as premature and the mRNA is targeted for degradation (Li and Wilkinson, 1998; Nagy and Maquat, 1998; Hentze and Kulozik, 1999; Maquat, 2000). More recently, other studies have reported that pre-mRNA splicing can enhance both mRNA translation (Matsumoto et al., 1998) and mRNA export efficiency (Luo and Reed, 1999). Most of the latter experiments were conducted by comparing mRNAs produced by splicing with mRNAs directly transcribed from cDNA.
Although our previous study clearly demonstrated that the pre-mRNA splicing machinery deposits multiple proteins upstream of mRNA exon–exon junctions (Le Hir et al., 2000b), it did not specifically address whether this complex is the species responsible for the documented effects of splicing on downstream mRNA metabolism. Here, we describe a means to generate spliced mRNAs both in vitro and in vivo that either do or do not carry this complex. This has enabled us to specifically address the functional attributes of the EJC apart from any other possible effects that pre-mRNA splicing may have on mRNP composition or mRNA metabolism. These studies reveal that by providing a binding platform for the export factors REF and TAP/p15, the EJC is the species responsible for enhanced export efficiency of spliced mRNAs. Furthermore, we show that the composition of the complex is subject to dynamic changes as the mRNA travels from the nucleus to the cytoplasm. In addition to dissociation of export factors upon transport, the EJC successively serves as an anchoring point in the nuclear and cytoplasmic compartment, respectively, for the Upf3 and Upf2 factors required for NMD.
Results
Generation of spliced mRNAs that do not carry the EJC
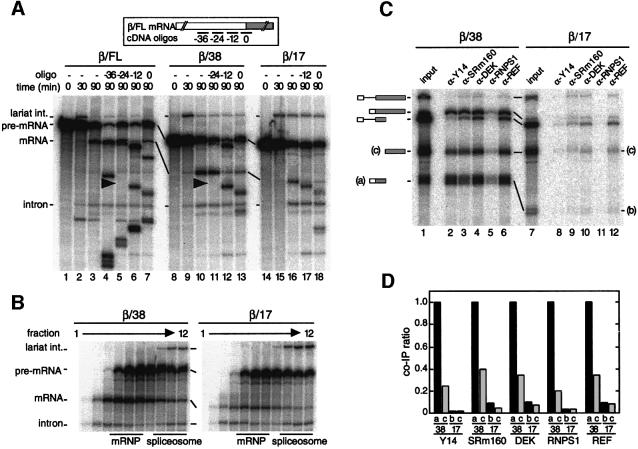
To investigate the function(s) of the EJC apart from any other possible effects of pre-mRNA splicing on mRNA metabolism, we wanted to generate mRNAs that had been produced by splicing but did not carry the complex. It was previously reported that pre-mRNAs having a 5′ exon as short as 12 nt can support both steps of splicing in vitro (Duchene et al., 1988; Chanfreau et al., 1999). Since the EJC is deposited on spliced mRNAs >20 nt upstream of the 3′ end of the 5′ exon, we hypothesized that spliced mRNAs with 5′ exons <20 nt might not carry the complex. To test this, we synthesized body-labeled β-globin pre-mRNAs that contained either a 38 or 17 nt exon 1 (named β/38 and β/17, respectively). When incubated in HeLa cell nuclear extract, β/38 pre-mRNA spliced with similar efficiency to our standard full-length β-globin pre-mRNA (β/FL; 130 nt exon 1, Figure 1A). Splicing of β/17 pre-mRNA was somewhat less efficient, although spliced β/17 mRNA was readily apparent. Glycerol gradient fractionation revealed that both β/38 and β/17 mRNAs were released from spliceosomes with equal efficiencies (Figure 1B).
Fig. 1. The EJC is not deposited on spliced β/17 mRNA. (A) Labeled β/FL (lanes 1–7), β/38 (lanes 8–13) or β/17 pre-mRNA (lanes 14–18) was incubated under splicing conditions in HeLa cell nuclear extract for the times indicated. Aliquots from 90 min reactions were further incubated with the indicated cDNA oligos (short bars underneath the mRNA schematic at top; named according to the center position of the oligo relative to the exon–exon junction, which was defined as 0). Splicing substrates, intermediates and products are indicated to the left. (B) Glycerol gradient fractionation of 90 min splicing reactions containing β/38 (left) or β/17 pre-mRNA (right). Bars indicate positions of mRNP and spliceosome.(C) Co-immunoprecipitation of β/38 (lanes 1–6) and β/17 (lanes 7–12) RNA species after splicing (2 h), separation of mRNPs from spliceosomes by glycerol gradient fractionation. 5′ (a or b) and 3′ (c) mRNA fragments resulted from subsequent RNase H cleavage with a single cDNA oligo centered 49 nt downstream of the exon–exon junction. Reactions were then subjected to immunoprecipitation with the antibodies indicated. Lanes 1 and 7 correspond to 1/15th of input RNA. (D) Co-immunoprecipitation ratios of fragments a, b and c from β/38 and β/17 mRNAs. Ratios shown were determined by dividing the absolute co-immunoprecipitation efficiency for each RNA with the indicated antibody by the absolute co-immunoprecipitation efficiency of β/38 fragment ‘a’ with that same antibody. In all experiments, RNAs were separated by 10% denaturing PAGE.
To determine whether either spliced mRNA carried the EJC, we first performed RNase H analysis as previously described (Le Hir et al., 2000b). For this analysis, β/FL, β/38 and β/17 pre-mRNAs were incubated in HeLa nuclear extract for 90 min to generate spliced mRNAs. Aliquots of each reaction were then incubated with individual cDNA oligos complementary to exon 1 or the splice junction to activate cleavage by endogenous RNase H (Figure 1A, top). Both β/FL- and β/38-spliced mRNAs exhibited protection to RNase H digestion in the region centered 24 nt upstream of the exon–exon junction (between positions –30 and –18). In contrast, β/17-spliced mRNA exhibited no protection either within exon 1 or at the exon–exon junction. These results suggest that the EJC is deposited on β/38- but not β/17-spliced mRNA.
We next examined the ability of antibodies against known EJC components to co-immunoprecipitate the two spliced mRNAs (Le Hir et al., 2000b). After generation by splicing, β/38 and β/17 mRNPs were separated from spliceosomes by glycerol gradient fractionation as shown in Figure 1B. The mRNP-containing fractions were then subjected to RNase H digestion with a cDNA oligo centered at +49 in exon 2 to divide each mRNA into two fragments: one containing exon 1 and the exon–exon junction (bands a and b) and another comprising the remainder of exon 2 (band c). These reactions were then incubated with different antibodies and the co-immunoprecipitation efficiencies of RNA fragment monitored by denaturing PAGE (Figure 1C). Relative to input, the immunoprecipitation efficiencies varied somewhat from one antibody to another, but all were readily quantifiable as previously observed (Le Hir et al., 2000b). For each antibody, the co-immunoprecipitation efficiencies of fragments b and c relative to the co-immunoprecipitation efficiency of fragment a are summarized in Figure 1D. This method of signal normalization facilitates comparison of relative co-immunoprecipitation efficiencies of individual fragments both in and between different immunoprecipitation samples, and is therefore used throughout the remainder of the paper.
Antibodies against Y14, SRm160, DEK, RNPS1 and REF all precipitated the β/38-spliced mRNA and the fragment from β/38 mRNA containing exon 1 and the exon–exon junction (fragment a). However, no antibody significantly precipitated either full-length spliced β/17 mRNA or the equivalent β/17 fragment (fragment b). Exactly parallel results were obtained with spliced AdML mRNAs containing either a 33 or 19 nt exon 1 (data not shown), demonstrating that this effect is not specific to β-globin mRNAs. Unexpectedly, despite the mRNA fragment derived from exon 2 (fragment c) being identical for both the β/38 and β/17 constructs, this fragment exhibited differential co-immunoprecipitation efficiencies dependent on the length of exon 1. Although fragment c was precipitated much less efficiently than fragment a when both derived from β/38 mRNA, β/38 fragment c was nonetheless precipitated 4- to 15-fold more efficiently with all antibodies than was fragment c from β/17 mRNA. Thus, when the EJC is present, it may associate weakly with other species bound to the remainder of mRNA. Alternatively, the higher co-immunoprecipitation efficiency of fragment c from β/38 mRNA could simply reflect that the 5′ and 3′ mRNA fragments do not completely dissociate after RNase H treatment, possibly because of secondary structure or interacting heterogeneous nuclear ribonucleoproteins (hnRNPs). In any case, the spliceosome apparently does not deposit any of the EJC components elsewhere on mRNAs whose 5′ exons are not long enough to accept the complex.
Taken together, the above results confirm that deposition of the EJC is spatially restricted to the region located 19–33 nt upstream of exon–exon junctions in spliced mRNAs. Deposition of this complex is not required either for splicing or for efficient release of product mRNP from the spliceosome. Finally, this complex may promote interactions between relatively distant regions within the mRNA.
The EJC is required for efficient export of short spliced mRNAs
Since the EJC contains at least one known RNA export factor, REF, we previously proposed that the EJC is the species that facilitates export of spliced mRNAs (Le Hir et al., 2000b). Having found a way to generate spliced mRNAs that either did or did not carry the complex, it became possible to test this hypothesis directly. To do so, body-labeled β/FL, β/38 and β/17 pre-mRNAs were separately injected into Xenopus laevis oocyte nuclei along with a mixture of labeled control RNAs, and the export of all RNAs was monitored over time (Figure 2A). Control RNAs consisted of U6Δss, U1ΔSm, U5ΔSm RNAs and/or the human initiator methionyl-tRNA, depending on the experiment. U6Δss RNA is not exported from the nucleus and lacks protein binding sites required for nuclear import (Vankan et al., 1992). It therefore serves as a control for nuclear injection and nuclear envelope integrity. Nuclear export of U1 and U5 snRNAs and of the initiator methionyl-tRNA is mediated by CRM1 and exportin-t, respectively, and is distinct from the export pathway of mRNAs (Mattaj and Englmeier, 1998). These latter RNAs served as controls for nuclear export.
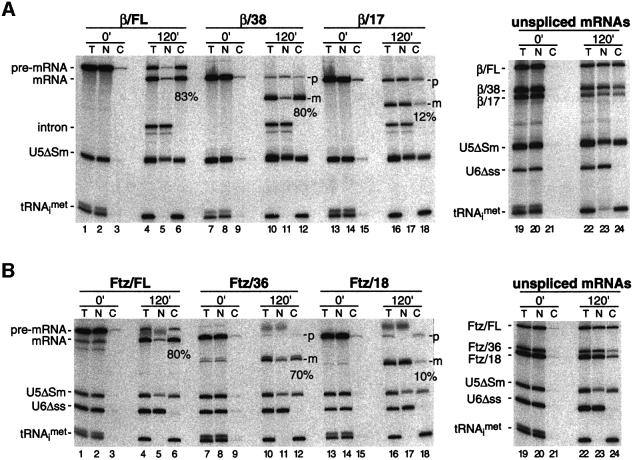
Fig. 2. The EJC promotes efficient export of spliced mRNAs. (A) Xenopus oocyte nuclei were injected with control RNAs plus the indicated pre-mRNA (lanes 1–18) or unspliced control mRNAs (lanes 19–24). RNA samples from total oocyte (T), nuclear (N) and cytoplasmic (C) fractions were collected immediately (0 min) or 120 min after injection and analyzed by 10% denaturing PAGE. One oocyte equivalent of RNA, from a pool of 10 oocytes, was loaded per lane in all panels. Splicing substrates, products and control RNAs are identified to the left of each gel. (B) Same as (A), with the corresponding Ftz-derived RNAs. Percentages indicate the proportion of each spliced mRNA in the cytoplasm 120 min after injection.
Immediately after injection, all RNAs were nuclear (Figure 2A, lanes 2, 8 and 14). After 2 h, 100% of tRNA and ∼65% of U5ΔSm RNA had been exported to the cytoplasm. During the same period, more than half of all three pre-mRNAs had been converted to spliced mRNA (lanes 4, 10 and 16) and a portion of that mRNA was exported to the cytoplasm. However, whereas ∼80% of spliced β/FL and β/38 mRNAs was cytoplasmic after 2 h, only 10–15% of spliced β/17 mRNA had been exported by this time (efficiency range observed in several independent experiments). Thus, while removal of the first 92 nt of β-globin exon 1 (the difference between β/FL and β/38 mRNAs) did not significantly affect export efficiency, removal of 21 nt more (the difference between β/38 and β/17 mRNAs) had a very pronounced effect. To verify that this was not simply due to the 21 nt size difference between β/38 and β/17 mRNAs, we performed similar export assays with the corresponding unspliced control mRNAs. When not generated by splicing, β/38 and β/17 mRNAs were exported with similar efficiencies, ∼30–50% cytoplasmic after 2 h (lane 24), but less efficiently than β/FL, as previously reported (Braun et al., 2001; Rodrigues et al., 2001). These results clearly indicate that the EJC is the species that provides the strong export advantage to spliced β/FL and β/38 mRNAs over β/17 mRNA.
To test the generality of the above results, we performed parallel experiments with truncated versions of Fushi tarazu (Ftz) pre-mRNA (Luo and Reed, 1999). When generated by splicing in oocytes, Ftz/FL, Ftz/36 and Ftz/18 mRNAs behaved exactly as had their respective β-globin counterparts (Figure 2B). That is, while spliced Ftz/FL and Ftz/36 mRNAs were efficiently exported (>70% export after 2 h, Figure 2B, lanes 4–6 and 10–12), spliced Ftz/18 mRNA was not (<10% export after 2 h, lanes 16–18). However, unspliced control Ftz/36 and Ftz/18 mRNAs were exported with comparable efficiencies (20–30%, lanes 22–24), but less so than full-length unspliced control Ftz mRNA, again paralleling the results with control β-globin mRNAs. Thus deposition of the EJC does impart a general export advantage on spliced mRNAs. Moreover, once the 5′ exon is long enough to accept the complex, there is no further advantage of increased mRNA length for more efficient export. In contrast, the export of unspliced mRNAs is highly length dependent, with longer mRNAs being more efficiently exported than short mRNAs (Rodrigues et al., 2001).
REF and TAP/p15 stimulate export of spliced mRNAs not carrying the EJC
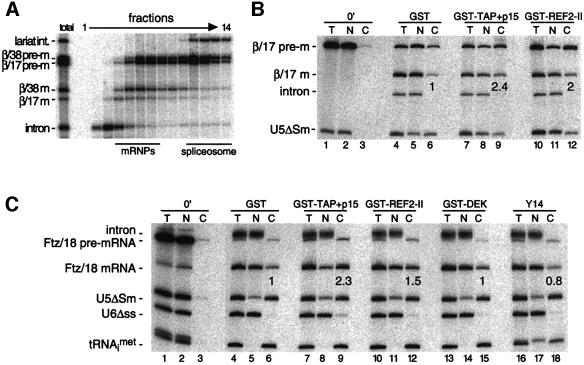
The most poorly exported of all mRNAs tested above were the β/17 and Ftz/18 mRNAs produced by splicing. In fact, greater proportions of these spliced mRNAs were retained in the nucleus than when the same species were injected as unspliced control mRNAs (Figure 2). This difference can potentially be attributed either to active retention, such as inefficient spliceosome release, or to lack of bound export factors. To assess the efficiency of spliceosome release in vivo, oocyte nuclei were co-injected with both β/38 and β/17 pre-mRNAs and then isolated nuclei were subjected to glycerol gradient fractionation 30 min later. As observed in vitro (Figure 1B), both spliced mRNAs were almost completely released from spliceosomes when generated in vivo (Figure 3A). Therefore, nuclear retention of spliced β/17 mRNA cannot be explained by defective spliceosome release.
Fig. 3. (A) Glycerol gradient fractionation of splicing substrates and products present in Xenopus oocyte nuclei 30 min after injection of β/38 and β/17 pre-mRNAs shows that both spliced mRNAs are released from spliceosomes in vivo. RNAs were extracted and analyzed as in Figure 1B. Bars indicate positions of mRNPs and spliceosomes. (B and C) Stimulation of spliced β/17 and Ftz/18 mRNA export by co-injection of recombinant proteins. Xenopus oocyte nuclei were injected with indicated control RNAs (at left) and either β/17 pre-mRNA (B) or Ftz/18 pre-mRNA (C). Where indicated, purified recombinant proteins (25 nl of 1 mg/ml stocks) were co-injected with RNAs (lanes ≥7) and co-injection of purified GST served as a control (lanes 4–6). RNA samples from total oocytes (T), nuclear (N) and cytoplasmic (C) fractions were collected immediately after injection (0 min; lanes 1–3) or 3 h after injection (lanes 4–18) and analyzed as in Figure 2. Export efficiencies of β/17 and Ftz/18 mRNAs in the presence of test proteins are indicated relative to their export efficiencies with GST alone.
Export of both spliced and unspliced mRNAs can be stimulated by co-injection of the recombinant export factors REF and TAP/p15 (Braun et al., 2001; Rodrigues et al., 2001). To test whether addition of these proteins can also rescue export of spliced β/17 and Ftz/18 mRNAs, we co-injected purified recombinant TAP/p15 heterodimers or REF2-II protein along with β/17 or Ftz/18 pre-mRNA into oocyte nuclei (Figure 3B and C). Both spliced mRNAs were exported almost twice as efficiently in the presence of excess TAP/p15 or REF2-II than in their absence (Figure 3B and C, lanes 7–12 versus 4–6). In contrast, co-injection of purified recombinant DEK or Y14, two other EJC components, did not stimulate mRNA export (Figure 3C, lanes 13–18). Thus, when supplemented as recombinant proteins, not all of the complex components are capable of stimulating mRNA export. The ability of TAP/p15 or REF2-II to partially rescue export of spliced β/17 and Ftz/18 mRNAs suggests that poor export of these RNAs in control oocytes is due to a general lack of bound export factors.
The EJC provides a binding platform for mRNA export factors
Since the mRNA export factor REF is an integral component of the EJC in vitro (Le Hir et al., 2000b) and is known to interact directly with TAP (Stutz et al., 2000), REF has been proposed to facilitate recruitment of the TAP/p15 heterodimer to cellular mRNPs (Stutz et al., 2000; Rodrigues et al., 2001). However, antibodies against TAP could not co-immunoprecipitate spliced mRNA produced in vitro (Le Hir et al., 2000b). This raised the possibility that TAP is recruited at an intermediate stage between splicing and export that is not replicated in HeLa cell nuclear extract.
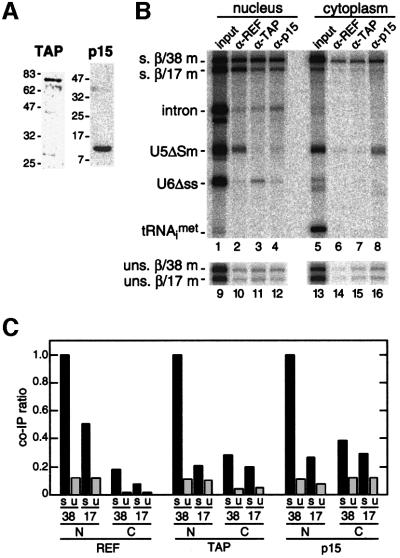
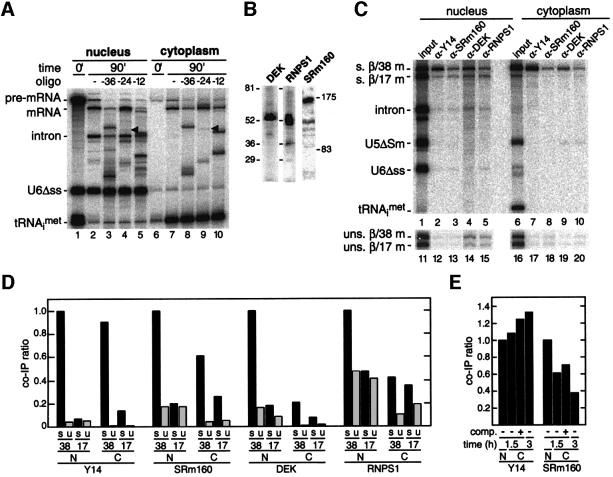
It was previously shown that an antiserum raised against the murine REF2-II recognizes a single band of the appropriate molecular weight in Xenopus oocyte lysates (Rodrigues et al., 2001). We now find by western blotting that antisera raised against human TAP and p15 also recognize single Xenopus proteins (Figure 4A). To determine whether these proteins are recruited to the EJC in vivo, we tested the ability of all three antisera to co-immunoprecipitate β/38 and β/17 mRNAs generated by splicing in oocyte nuclei (Figure 4B). Because the two mRNAs were sufficiently different in size that they could be distinguished by denaturing PAGE, co-injection of their respective pre-mRNAs allowed for direct comparison of the ability of each antiserum to co-immunoprecipitate a spliced mRNA that carries the EJC (β/38 mRNA) to one that does not (β/17 mRNA). Separate injection of unspliced control mRNAs allowed for comparison of identical mRNAs that were not produced by splicing. For each antibody, the co-immunoprecipitation efficiency of nuclear or cytoplasmic mRNA relative to that for nuclear spliced β/38 mRNA is indicated as a ratio in Figure 4C. Ratios shown represent averages from two to three independent experiments.
Fig. 4. The EJC promotes recruitment of TAP/p15 heterodimers to spliced mRNAs. (A) Protein samples from Xenopus oocyte nuclei were analyzed by western blotting with anti-TAP and anti-p15 antibodies. (B) Control RNAs plus either β/38 and β/17 pre-mRNAs (upper panel, lanes 1–8) or unspliced β/38 and β/17 mRNAs (lower panel, lanes 9–16) were co-injected in Xenopus oocyte nuclei. Two hours after injection, RNA samples from nuclear and cytoplasmic fractions were subjected to immunoprecipitation with the antibodies indicated. Lanes 1, 5, 9 and 13 correspond to 1/15th of input RNA. RNAs were analyzed as in Figure 2. (C) Co-immunoprecipitation ratios of spliced (s) or unspliced (u) β/38 and β/17 mRNAs isolated from nuclear (N) or cytoplasmic (C) compartments with antibodies against the species indicated. For each antibody, ratios were calculated by dividing the absolute co-immunoprecipitation efficiency of the species indicated by the absolute co-immunoprecipitation efficiency of nuclear spliced β/38 mRNA with that antibody.
As expected from previous data (Le Hir et al., 2000b; Zhou et al., 2000; Kim et al., 2001), α-REF antisera precipitated the spliced β/38 mRNA in the nuclear fraction at least twice as efficiently as any other nuclear mRNA. Both the α-TAP and α-p15 antisera also precipitated spliced β/38 mRNA 4- to 5-fold more efficiently than spliced β/17 mRNA or the unspliced control mRNAs in the nuclear fraction. Thus, the EJC does provide a binding site for the TAP/p15 heterodimer in vivo.
We also tested whether antisera against REF, TAP and p15 were able to precipitate mRNAs that had been exported to the cytoplasm. Because REF and TAP are known nucleocytoplasmic shuttling proteins, it is possible that these proteins remain associated with mRNA after its export (Katahira et al., 1999; Bachi et al., 2000; Zhou et al., 2000; Rodrigues et al., 2001). However, none of the three antisera precipitated any of the cytoplasmic mRNAs as efficiently as nuclear spliced β/38 mRNA (Figure 4B and C). Therefore, REF, TAP and p15 most likely dissociate from the EJC soon after mRNA export.
Some EJC components do remain associated with spliced mRNA after export
Mechanistic studies of NMD have indicated that some feature marking exon–exon junctions must survive mRNA transport to the cytoplasm (Li and Wilkinson, 1998; Nagy and Maquat, 1998; Hentze and Kulozik, 1999; Maquat, 2000). To test whether the protection to RNase H digestion imparted by splicing in the nucleus (Le Hir et al., 2000b) survives nucleocytoplasmic export, we performed RNase H analysis of spliced β/FL mRNA in both cellular compartments (Figure 5A). When RNase H cleavage was activated 90 min after injection of β/FL pre-mRNA, exactly the same cleavage and protection pattern of spliced mRNA was observed in the cytoplasm as in the nucleus (Figure 5A). This suggests that one or more components of the EJC remain bound to mRNA at the same location after export to the cytoplasm. Consistent with our results, Y14 has been shown to be associated with several spliced mRNAs both before and after export (Kataoka et al., 2000; Kim et al., 2001).
Fig. 5. Y14 and SRm160 remain associated with spliced mRNAs after export. (A) Spliced β/FL mRNA exhibits a similar protection to RNase H digestion in both nucleus and cytoplasm. Following injection of β/FL pre-mRNA into oocyte nuclei, nuclear (lanes 1–5) and cytoplasmic (lanes 6–10) fractions were collected immediately (lanes 1 and 6) or after 90 min (lanes 2–5 and 7–10). Aliquots of 90 min samples were further incubated alone (lanes 2 and 7) or with the cDNA oligos indicated (lanes 3–5 and 8–10). RNAs were separated by 8% denaturing PAGE. (B) Protein samples from Xenopus oocyte nuclei were analyzed by western blotting with anti-DEK, anti-RNPS1 and anti-SRm160 antibodies. (C and D) Same as Figure 4B and C, respectively, except that experiments were performed with antisera raised against Y14, SRm160, RNPS1 and DEK. (E) Co-immunoprecipitation ratios of spliced β/38 mRNA isolated from nuclear (N) or cytoplasmic (C) fractions with antibodies against the species indicated.Co-immunoprecipitations were performed after 1.5 or 3 h incubation and in the absence (–) or presence (+) of competitor unspliced β/38 mRNA (comp.) in the cytoplasm. In (D) and (E), co-immunoprecipitation ratios were calculated as in Figure 4C.
The monoclonal antibody 4C4 recognizes both the human and the Xenopus Y14 proteins (Kataoka et al., 2000). Similarly, we find that antisera against human SRm160, DEK and RNPS1 all recognize single protein bands from homogenized Xenopus nuclei (Figure 5B), indicating that all of these epitopes are conserved between humans and amphibians. In the nucleus, antibodies against Y14, SRm160, DEK and RNPS1 all precipitated spliced β/38 mRNA more efficiently than spliced β/17 mRNA or the corresponding unspliced control mRNAs. The difference of co-immunoprecipitation efficiency between the two spliced mRNAs was 4- to 5-fold for Y14, SRm160 and DEK, and 2-fold for RNPS1 (Figure 5C). Thus, the EJC contains the same components when generated in vitro or in vivo.
When RNAs were precipitated from the cytoplasmic fraction, only the Y14 antibodies precipitated spliced β/38 mRNA as efficiently as from the nuclear fraction (Figure 5B and C). Antibodies against DEK and RNPS1 exhibited little specificity for any cytoplasmic RNA; in both cases the co-immunoprecipitation ratios of cytoplasmic spliced β/38 mRNA were comparable to the ratios for mRNAs not carrying the EJC. Thus, both DEK and RNPS1 apparently dissociate from the exon–exon junction just before or soon after export.
With antibodies against SRm160, spliced β/38 mRNA in the cytoplasmic fraction was precipitated with an efficiency ratio (∼0.6) intermediate to those of nuclear spliced β/38 mRNA (1.0) and mRNAs not carrying the EJC (≤0.25). This suggested that SRm160 may accompany spliced β/38 mRNA out of the nucleus, but once in the cytoplasm is not as stably bound as Y14. To test this, we asked whether the co-immunoprecipitation efficiency ratios with α-SRm160 and α-Y14 antibodies varied with time (Figure 5E). When immunoprecipitates were performed 1.5 or 3 h after pre-mRNA injection (as in Figure 5D), the co-immunoprecipitation ratio for cytoplasmic spliced β/38 mRNA with α-SRm160 antibodies decreased from ∼0.6 to ∼0.4 (Figure 5E). However, as expected for a stably bound protein, these co-immunoprecipitation ratios did not decrease with α-Y14 antibodies. In fact, the ratio actually increased slightly between 1.5 and 3 h for Y14, suggesting that the Y14 epitope may become somewhat more accessible over time.
To rule out the possibility that some fraction of the co-immunoprecipitation of spliced β/38 mRNA from the cytoplasm by α-Y14 or α-SRm160 antibodies represented rebinding of these proteins to mRNA after export, we also performed competition experiments (Figure 5E). Ten minutes prior to nuclear pre-mRNA injection, 20 times more unlabeled, unspliced β/38 mRNA was injected directly into the cytoplasm. After another 1.5 h, cytoplasmic RNAs were precipitated as above. Because addition of competitor RNA did not decrease the co-immunoprecipitation ratio of spliced β/38 mRNA with either antibody, we conclude that the cytoplasmic co-immunoprecipitates do result from loading of the proteins in the nucleus. In fact, as was observed above with the longer incubation (3 h) for Y14, addition of competitor slightly increased the co-immunoprecipitation ratios for both Y14 and SRm160 at 1.5 h. This small effect might be attributed to the titration by the competitor of less stably bound species that affect co-immunoprecipitation of the spliced β/38 mRNA from the cytoplasm. In summary, both SRm160 and Y14 likely accompany the mRNA out of the nucleus, but only Y14 remains stably bound in the cytoplasm.
The EJC provides a binding platform for factors involved in NMD
To date, four human proteins involved in NMD have been identified: hUpf1, hUpf2, hUpf3a and hUpf3b (also known as hUpf3-X). Interestingly, these proteins have distinct cellular locations: hUpf3a and 3b are nucleocytoplasmic shuttling proteins, hUpf2 is perinuclear, and hUpf1 is cytoplasmic (Applequist et al., 1997; Lykke-Andersen et al., 2000; Serin et al., 2001). The proteins hUpf3a and 3b associate selectively with spliced mRNA in vivo (Lykke-Andersen et al., 2000) and all four proteins can be found complexed with one another (Lykke-Andersen et al., 2000; Serin et al., 2001). Therefore, it has been speculated that hUpf3 associates with spliced mRNAs in the nucleus, and after export, hUpf2 and hUpf1 join the hUpf3-containing mRNP in the cytoplasm (Lykke-Andersen et al., 2000). However, because antibodies against hUpf3a and hUpf3b do not co-immunoprecipitate spliced mRNA generated in vitro in HeLa cell nuclear extract (Lykke-Andersen et al., 2000), there was heretofore no direct evidence to support some aspects of this model.
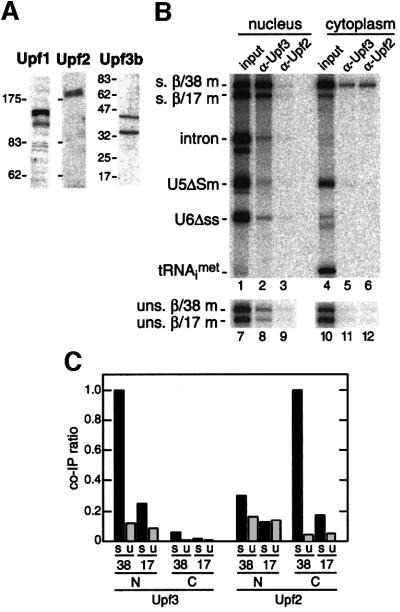
To test whether antibodies against hUpf1, hUpf2 and hUpf3b recognized homologous proteins in Xenopus, we performed western blots with homogenized oocyte lysates (Figure 6A). Antisera against hUpf1 and hUpf2 both recognized single Xenopus proteins of the appropriate molecular weight to be true orthologs. Antisera against hUpf3b revealed two major bands in oocytes (Figure 6A), with the apparent molecular weight of the upper (45 kDa) being comparable to the mass of the Saccharomyces cerevisiae and human Upf3 proteins (45 and 52 kDa, respectively).
Fig. 6. The EJC promotes efficient recruitment of hUpf3 and hUpf2 to spliced mRNAs. (A) Protein samples from total Xenopus oocyte lysates were analyzed by western blotting using anti- hUpf3b, hUpf2 and hUpf1 antibodies. (B and C) Same as Figure 4B and C except that experiments were performed with antisera raised against hUpf3b and hUpf2.
To determine whether any of the Xenopus Upf proteins selectively associated with the EJC in vivo, the same co-immunoprecipitation strategy as above was performed with all three antibodies (Figure 6B and C). No mRNAs, regardless of EJC or subcellular localization, were precipitated by the anti-Upf1 serum (data not shown). In contrast, antibodies against Upf2 and Upf3b both precipitated spliced β/38 mRNA with much greater efficiency than any other RNA. However, their selective precipitation of spliced β/38 mRNA occurred in different cellular compartments: Upf3b in the nucleus and Upf2 in the cytoplasm. Thus, when assayed in vivo, the EJC binds factors involved in NMD in both cellular compartments.
Discussion
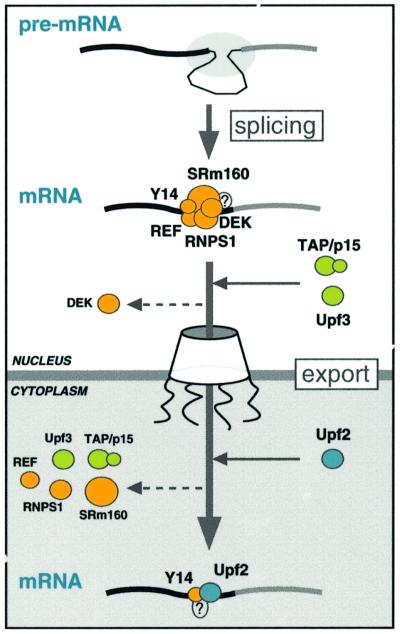
In this study, we showed that the EJC, deposited on mRNA as a consequence of splicing, is the entity that provides a selective advantage for the export of spliced mRNAs to the cytoplasm. This effect is most likely to be due to the ability of the complex to act as a strong binding site for the export factors REF and TAP/p15. We also show that the composition of the complex evolves upon mRNA export. Furthermore, two factors involved in NMD specifically associate with the complex in different cellular compartments. These observations provide direct functional links between the process of splicing and the downstream processes of mRNA export and NMD. A model of how the composition of the EJC evolves upon mRNA export is illustrated in Figure 7.
Fig. 7. A model for the evolution of the EJC upon mRNA export. Pre-mRNA splicing deposits the EJC upstream of the splice junction. At this stage, the complex contains at least five proteins, SRm160, DEK, RNPS1, REF and Y14. After splicing, the TAP/p15 heterodimer and Upf3 join the complex in the nucleus. DEK probably dissociates (dashed arrow) before mRNA export to the cytoplasm, whereas SRm160 and the nucleocytoplasmic shuttling proteins RNPS1, REF, TAP/p15 and Upf3 are all likely to leave after mRNA export. Y14 remains stably associated with spliced mRNA in both compartments, while Upf2 only joins the complex in the cytoplasm. Question marks symbolize potential unidentified components.
Deposition of the EJC is not required for splicing or mRNA release
We found that pre-mRNAs with 5′ exons <20 nt are spliced, but the product mRNPs lack the EJC. In contrast, pre-mRNAs with 5′ exons only 14 nt longer (33 nt total) do permit association of all five complex components with spliced mRNA (Figures 1, 4 and 5). These observations confirm our previous results showing that deposition of the EJC is spatially restricted to the region of mRNA located 20–24 nt upstream of the exon–exon junction (Le Hir et al., 2000b). Although pre-mRNAs with short 5′ exons (<20 nt) are not spliced quite as efficiently as those with longer 5′ exons (<33 nt), mRNAs with either length 5′ exon are efficiently released from the spliceosome (Figures 1B and 3A). Therefore, complex deposition is not necessary either for splicing or for mRNA release.
Because truncation of the 5′ exon had minimal effects on splicing and no detectable effect on mRNA release, it is not yet clear when or how the EJC is assembled during the splicing process. Another unanswered question is which and how many of the EJC components contact the mRNA directly. The latter question is of particular interest because the complex interacts very stably with mRNA independently of sequence (Le Hir et al., 2000b). Here we observed that two of the EJC components, SRm160 and Y14, accompany spliced mRNA to the cytoplasm (Figure 5C). Therefore, it is tempting to speculate that one or both of these proteins is the species making direct contact with mRNA. This hypothesis is in agreement with some preliminary results from photocrosslinking experiments using pre-mRNAs containing site-specific photoreactive groups in the 5′ exon (H.Le Hir and M.J.Moore, unpublished results). Analysis of the dynamic protein– RNA interactions with this region of the 5′ exon over the course of the entire splicing pathway is ongoing.
The EJC stimulates mRNA export by providing a strong binding site for export factors
It was previously reported that when injected into Xenopus oocytes, unspliced AdML mRNA is much less efficiently exported to the cytoplasm than is the same mRNA generated by splicing (Luo and Reed, 1999; Zhou et al., 2000). However, in most other cases the difference in export efficiency between unspliced and spliced mRNAs is much less striking. In the Xenopus system, the export efficiencies of unspliced mRNAs are highly dependent on their lengths (Rodrigues et al., 2001). The unspliced AdML mRNA used in most transport studies is quite short (181 nt) and is therefore almost completely retained in the nucleus. Longer unspliced mRNAs, such as the full-length β-globin and Ftz mRNAs or the naturally intron-less DHFR mRNA (360, 343 and 815 nt, respectively), are much more efficiently exported (∼50% cytoplasmic 2 h after injection, Figure 2; Rodrigues et al., 2001). This length effect is most likely due to the ability of export factors such as REF and/or TAP/p15 to interact with mRNAs in a relatively weak and sequence-unspecific manner (Braun et al., 1999; Stutz et al., 2000). Therefore, the shorter an unspliced mRNA, the lower the probability that it will interact productively with the export machinery. Consistent with this idea, injection of additional REF and/or TAP/p15 into oocytes promotes the export of all inefficiently exported mRNAs tested to date (Figure 3; Braun et al., 2001; Rodrigues et al., 2001). Furthermore, titration of TAP with antibodies or with an RNA element that binds tightly to TAP (SRV1-CTE), impairs export of both spliced and intronless mRNAs (Pasquinelli et al., 1997; Saavedra et al., 1997; Gruter et al., 1998). This suggests that both types of mRNA are exported via the same pathway.
When produced by splicing, AdML, β-globin and Ftz mRNAs are all exported quite efficiently (80–90% exported within 2 h; Figure 2). Thus, spliced mRNAs are generally well exported regardless of length. By studying the behavior of spliced mRNAs that do or do not carry the EJC, we showed that efficient export of spliced mRNA can be entirely attributed to deposition of the complex as opposed to any other possible effect of splicing on mRNP composition. The EJC apparently provides an export advantage by serving as a strong binding site in vivo for the export factor TAP/p15 (Figure 4). The TAP/p15 heterodimer is required for mRNA export (Braun et al., 2001), and TAP is known to contact REF (Stutz et al., 2000; Rodrigues et al., 2001) as well as a component of the nuclear pore complex (Katahira et al., 1999; Bachi et al., 2000; reviewed by Conti and Izaurralde, 2001). However, specific association between TAP and spliced mRNPs had not previously been demonstrated. Our finding that antibodies against TAP and p15 selectively precipitate nuclear mRNAs carrying the EJC now provides direct evidence that this complex constitutes the functional link between pre-mRNA splicing and the nuclear export machinery.
The EJC provides a link between splicing and NMD
In addition to export factors, the EJC provides a specific binding site for the proteins Upf3b and Upf2 involved in NMD. Our data suggest that these two proteins join the EJC in different subcellular compartments: Upf3 in the nucleus and Upf2 in the cytoplasm (Figure 6). Given the fact that splicing and translation are both required to elicit NMD in mammals (Carter et al., 1996; Thermann et al., 1998; Zhang et al., 1998; Maquat and Li, 2001; Neu-Yilik et al., 2001), our results show how these two spatially distinct cellular events could be functionally linked (Figure 7). They also agree with the current sequential model of NMD (Culbertson, 1999; Lykke-Andersen et al., 2000). Briefly, splicing in the nucleus marks mRNAs by loading the EJC. Subsequently, Upf3, a known shuttling protein with predominantly nuclear localization (Lykke-Andersen et al., 2000; Serin et al., 2001), joins the complex in the nucleus and accompanies the mRNA through the nuclear pore. During or immediately after export, Upf2, which is predominantly perinuclear (Lykke-Andersen et al., 2000; Serin et al., 2001), associates with the spliced mRNP. It most likely does so by interacting with Upf3, since direct contact between these two proteins has been observed in yeast (He et al., 1997). At that stage, we cannot currently differentiate between two possibilities: (i) Upf3 remains associated with the mRNP but it is masked from immunoprecipitation by other species (e.g. Upf2) that join the complex in the cytoplasm; or (ii) Upf3 simply leaves the mRNP when Upf2 joins. Subsequently, ribosomes displace the EJCs located within the open reading frame (ORF) during the first round of translation. However, if the mRNA contains a premature termination codon located upstream of at least one EJC, Upf1 joins the mRNP via its interaction with the translation termination release factors eRF1 and eRF3 (Czaplinski et al., 1998). Finally, interactions between Upf1p and factors, perhaps Upf2p (He et al., 1997), bound to any downstream EJC trigger NMD. Our observation that α-Upf1 antibodies could not co-immunoprecipitate the β-globin mRNAs used in this study is consistent with the fact that these mRNAs do not contain optimal translation initiation signals or complete ORFs, and so most probably do not interact productively with the cytoplasmic translation machinery.
The EJC composition evolves upon mRNA export
Although the splicing-dependent protection to RNase H digestion observed upstream of mRNA exon–exon junctions is identical in the nucleus and cytoplasm (Figure 4A), the exact complement of proteins associated with this region evolves upon nucleocytoplasmic export (Figure 7). The two proteins that we found associated with spliced mRNA in both compartments were Y14 and SRm160 (Figure 5). In the case of Y14, our results agree with a recent report that Y14 accompanies spliced mRNA to the cytoplasm (Kim et al., 2001). However, our finding that α-SRm160 antibodies precipitate a significant fraction of spliced β/38 mRNA from the cytoplasm was less expected since there is no corroborating evidence at this time that SRm160 is a shuttling protein. Our data do indicate that SRm160 is not as tightly associated with the exon–exon junction in the cytoplasm as is Y14.
Six other complex components, DEK, RNPS1, REF, TAP, p15 and Upf3, were not found associated with spliced mRNAs in the cytoplasm (Figures 4, 5 and 6). Since DEK is exclusively a nuclear protein (Mayeda et al., 1999; McGarvey et al., 2000), it probably leaves the mRNP prior to export. In contrast, RNPS1, REF, TAP and Upf3p are known shuttling proteins (Katahira et al., 1999; Lykke-Andersen et al., 2000, 2001; Zhou et al., 2000; Rodrigues et al., 2001; Serin et al., 2001). Therefore, they either dissociate from the mRNP soon after export or are masked by the association of new proteins, such as Upf2p, in the cytoplasm. Future studies will explore the mechanisms behind the structural evolution of the EJC as it journeys from the nucleus to the cytoplasm.
Materials and methods
DNA constructs and in vitro splicing
Uniformly labeled splicing substrates containing a GpppG dinucleotide cap were synthesized by standard run-off transcription. DNA templates for β-globin (Reed and Maniatis, 1988) and Ftz (Luo and Reed, 1999) have been previously described. DNA templates for the corresponding intron-less control mRNAs were constructed by PCR. DNA templates for truncated pre-mRNAs and corresponding truncated mRNAs were also constructed by PCR using 5′ end primers, all containing the T7 promoter sequence followed by the sequence indicated: β/38, T7-GGGAACGTG GATG; β/17, T7-GGTGAGGCCCTGG; Ftz/36, T7-GGGATTGGT CGCACATC; Ftz/18, T7-GGGAGACTTTGGCATC. In vitro splicing reactions, analysis of splicing products and glycerol gradients were performed as previously described (Le Hir et al., 2000b).
Expression of recombinant proteins in Escherichia coli
Plasmids allowing the expression of glutathione S-transferase (GST) fusions of full-length human TAP (pGEXCS-TAP) and murine REF2-II (pGEXCS-REF2-II) in Escherichia coli have been previously described (Braun et al., 1999; Stutz et al., 2000). Human p15-1 cDNA was cloned within the SphI–BamHI sites of vector pQE70zz, kindly provided by D.Görlich (ZMBH, Heidelberg). This vector is a derivative of pQE70 (Qiagen) and has two immunoglobulin binding domains of protein A from Staphylococcus aureus (zz-tag) inserted within the EcoRI and BglII restriction sites. Proteins expressed using pQE70zz vector have a zz-tag at their N-termini and a His6 tag at their C-termini. For rabbit immunizations, p15-1 was expressed in E.coli with an N-terminal His6 tag. To this end, p15-1 cDNA was cloned within the BamHI–EcoRI sites of vector pRSETC (Invitrogen). Full-length human Y14 cDNA was amplified by PCR using human testis Marathon-Ready™ cDNA library (Clontech) as a template and primers introducing unique restriction sites. A cDNA encoding human DEK protein was kindly provided by B.Blencowe (Toronto). Plasmids allowing expression of human Y14 and DEK GST fusions in E.coli were generated by inserting the corresponding cDNAs into the BamHI and NotI sites of pGEX4T-1 (Y14) (Pharmacia) or the NcoI and BamHI sites present in pGEXCS (DEK). Proteins were purified on glutathione–agarose beads (Sigma) or Ni+–NTA Sepharose fast flow (Qiagen) as described by Gruter et al. (1998). The GST moiety of purified GST–Y14 was cleaved with thrombin (Pharmacia). For oocyte injections recombinant proteins were dialyzed against 1.5× phosphate-buffered saline supplemented with 10% glycerol.
RNase H digestions
RNase H digestions of radiolabeled in vitro spliced mRNAs were performed as previously described (Le Hir et al., 2000b). For RNase H cleavage of RNAs isolated from Xenopus oocytes, nuclear and cytoplasmic fractions corresponding to 10 oocytes were collected 90 min after injection and homogenized in 35 µl of RNase H buffer (100 mM Tris–HCl pH 7.5, 50 mM MgCl2, 250 mM KCl and 0.5 mM dithiothreitol) containing 5 µM corresponding cDNA oligo and incubated for 15 min at 30°C. RNase H cleavage was stopped by addition of 200 µl of SDS–proteinase K buffer (300 mM NaCl, 50 mM Tris–HCl pH 7.5, 5 mM EDTA, 1.5% SDS and 2 mg/ml proteinase K). RNAs were extracted, precipitated and separated by 10% denaturing PAGE.
Xenopus laevis oocyte microinjections
The DNA templates for in vitro synthesis of U1ΔSm, U5ΔSm, U6Δss snRNAs and human initiator methionyl-tRNA have been described (Jarmolowski et al., 1994). Synthesis of pre-mRNAs, mRNAs, U1ΔSm and U5ΔSm was primed with the m7GpppG cap dinucleotide. Oocyte injections and analysis of microinjected RNA by denaturing gel electrophoresis and autoradiography were performed as previously described (Jarmolowski et al., 1994) and quantified with a FluorImager (Fuji FLA-2000). All injections were 25 nl of RNA mixture (∼0.1 µM each RNA) per nucleus and, when indicated, contained 1 mg/ml recombinant proteins. For glycerol gradient fractionation, 20 oocyte nuclei were homogenized in 20 µl of TNE buffer (10 mM Tris–HCl pH 7.6, 50 mM NaCl and 5 mM EDTA). For competition experiments, 25 nl of unlabeled, unspliced β/38 mRNA (2 µM) were injected into the cytoplasm of each oocyte 10 min prior to nuclear injection of labeled RNAs.
Antibodies, immunoblotting and immunoprecipitations
The anti-TAP and anti-REF (KJ70) sera have been described previously (Braun et al., 1999; Rodrigues et al., 2001). Rabbit antibodies were raised against recombinant His6–p15 protein expressed in E.coli using vector pRSETC-p15-1. The following antibodies were kindly provided by the person indicated: anti-SRm160 (mAb B1C8, B.J.Blencowe); anti-DEK (G.Grosveld); anti-RNPS1 (A.Mayeda); anti-Y14 (mAb-4C4, G.Dreyfuss); anti-hUpf1, anti-hUpf2 and anti-hUpf3b (J.Lykke-Andersen/J.Steitz). Western blotting was performed as previously described (Rodrigues et al., 2001). For co-immunoprecipitation experiments, monoclonal antibodies B1C8 and 4C4 were bound to protein A–Sepharose (PAS) beads (Pharmacia) via rabbit anti-mouse IgG + IgM (Pierce). All other antibodies were bound directly to PAS beads. Co-immunoprecipitates were performed as previously described (Le Hir et al., 2000b). For RNAs isolated from Xenopus oocytes, isolated nuclear and cytoplasmic fractions were homogenized in 100 µl of TNE buffer after dissection. Nuclear fractions were added directly to PAS-bound antibodies, whereas cytoplasmic fractions were first clarified by centrifugation (5 min at 3800 g). About 10–20 injected oocytes were used per co-immunoprecipitation. Co-immunoprecipitation efficiencies were calculated by dividing the intensity of a given precipitated fragment (measured by PhosphorImaging; Molecular Dynamics) by the intensity of the same fragment present in the input reaction, taking into account that only 1/15th of each input reaction was loaded on every gel.
Acknowledgments
Acknowledgements
We are grateful to the following persons for providing antibodies: Ben Blencowe, Gideon Dreyfuss, Gerard Grosveld, Jens Lykke-Andersen and Akila Mayeda. We thank Kwang Hee Hong for technical assistance, members of the Moore laboratory for helpful discussions, and Torben Heick-Jensen, Melissa Jurica, Ajit Nott and Michael Rosbash for critical comment on the manuscript. M.J.M. is an HHMI assistant investigator.
References
- Applequist S.E., Selg,M., Raman,C. and Jack,H.M. (1997) Cloning and characterization of HUPF1, a human homolog of the Saccharomyces cerevisiae nonsense mRNA-reducing UPF1 protein. Nucleic Acids Res., 25, 814–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachi A. et al. (2000) The C-terminal domain of TAP interacts with the nuclear pore complex and promotes export of specific CTE-bearing RNA substrates. RNA, 6, 136–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blencowe B.J., Issner,R., Nickerson,J.A. and Sharp,P.A. (1998) A coactivator of pre-mRNA splicing. Genes Dev., 12, 996–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun I.C., Rohrbach,E., Schmitt,C. and Izaurralde,E. (1999) TAP binds to the constitutive transport element (CTE) through a novel RNA-binding motif that is sufficient to promote CTE-dependent RNA export from the nucleus. EMBO J., 18, 1953–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun I.C., Herold,A., Rode,M., Conti,E. and Izaurralde,E. (2001) Overexpression of TAP/p15 heterodimers bypasses nuclear retention and stimulates nuclear mRNA export. J. Biol. Chem., 276, 20536–20543. [DOI] [PubMed] [Google Scholar]
- Carter M.S., Li,S. and Wilkinson,M.F. (1996) A splicing-dependent regulatory mechanism that detects translation signals. EMBO J., 15, 5965–5975. [PMC free article] [PubMed] [Google Scholar]
- Chanfreau G., Gouyette,C., Schwer,B. and Jacquier,A. (1999) Trans-complementation of the second step of pre-mRNA splicing by exogenous 5′ exons. RNA, 5, 876–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J., Belgrader,P., Zhou,X. and Maquat,L.E. (1994) Introns are cis effectors of the nonsense-codon-mediated reduction in nuclear mRNA abundance. Mol. Cell. Biol., 14, 6317–6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti E. and Izaurralde,E. (2001) Nucleocytoplasmic transport enters the atomic age. Curr. Opin. Cell Biol., 13, 310–319. [DOI] [PubMed] [Google Scholar]
- Culbertson M.R. (1999) RNA surveillance. Unforeseen consequences for gene expression, inherited genetic disorders and cancer. Trends Genet., 15, 74–80. [DOI] [PubMed] [Google Scholar]
- Czaplinski K., Ruiz-Echevarria,M.J., Paushkin,S.V., Han,X., Weng,Y., Perlick,H.A., Dietz,H.C., Ter-Avanesyan,M.D. and Peltz,S.W. (1998) The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev., 12, 1665–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchene M., Low,A., Schweizer,A. and Domdey,H. (1988) Molecular consequences of truncations of the first exon for in vitro splicing of yeast actin pre-mRNA. Nucleic Acids Res., 16, 7233–7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruter P., Tabernero,C., von Kobbe,C., Schmitt,C., Saavedra,C., Bachi,A., Wilm,M., Felber,B.K. and Izaurralde,E. (1998) TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol. Cell, 1, 649–659. [DOI] [PubMed] [Google Scholar]
- He F., Brown,A.H. and Jacobson,A. (1997) Upf1p, Nmd2p and Upf3p are interacting component of the yeast nonsense-mediated mRNA decay pathway. Mol. Cell. Biol., 17, 1580–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentze M.W. and Kulozik,A.E. (1999) A perfect message: RNA surveillance and nonsense-mediated decay. Cell, 96, 307–310. [DOI] [PubMed] [Google Scholar]
- Jarmolowski A., Boelens,W.C., Izaurralde,E. and Mattaj,I.W. (1994) Nuclear export of different classes of RNA is mediated by specific factors. J. Cell Biol., 124, 627–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katahira J., Strasser,K., Podtelejnikov,A., Mann,M., Jung,J.U. and Hurt,E. (1999) The Mex67p-mediated nuclear mRNA export pathway is conserved from yeast to human. EMBO J., 18, 2593–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka N., Yong,J., Kim,V.N., Velazquez,F., Perkinson,R.A., Wang,F. and Dreyfuss,G. (2000) Pre-mRNA splicing imprint mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol. Cell, 6, 673–682. [DOI] [PubMed] [Google Scholar]
- Kim V.N., Yong,J., Kataoka,N., Abel,L., Diem,M.D. and Dreyfuss,G. (2001) The Y14 protein communicates to the cytoplasm the position of exon–exon junctions. EMBO J., 20, 2062–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E.S. et al. (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- Le Hir H., Moore,M.J. and Maquat,L.E. (2000a) Pre-mRNA splicing alters mRNP composition: evidence for stable association of proteins at exon–exon junctions. Genes Dev., 14, 1098–1108. [PMC free article] [PubMed] [Google Scholar]
- Le Hir H., Izaurralde,E., Maquat,L.E. and Moore,M.J. (2000b) The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon–exon junctions. EMBO J., 19, 6860–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S. and Wilkinson,M.F. (1998) Nonsense surveillance in lymphocytes? Immunity, 8, 135–141. [DOI] [PubMed] [Google Scholar]
- Luo M.J. and Reed,R. (1999) Splicing is required for rapid and efficient mRNA export in metazoans. Proc. Natl Acad. Sci. USA, 96, 14937–14942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J., Shu,M.D. and Steitz,J.A. (2000) Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell, 103, 1121–1131. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J., Shu,M.D. and Steitz,J.A. (2001) The protein RNPS1 communicates the position of exon–exon junctions to the mRNA surveillance machinery. Science, in press. [DOI] [PubMed] [Google Scholar]
- Maquat L.E. (2000) Nonsense-mediated RNA decay in mammalian cells: a splicing-dependent means to down-regulate the levels of mRNAs that prematurely terminate translation. In Hershey,J.W.B., Mathews,M.B. and Sonenberg,N. (eds), Translational Control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 849–868.
- Maquat L.E. and Li,X. (2001) Mammalian heat shock p70 and histone H4 transcripts, which derive from naturally intronless genes, are immune to nonsense-mediated decay. RNA, 7, 445–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto K., Wassarman,K.M. and Wolffe,A.P. (1998) Nuclear history of a pre-mRNA determines the translational activity of cytoplasmic mRNA. EMBO J., 17, 2107–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaj I.W. and Englmeier,L. (1998) Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem., 67, 265–306. [DOI] [PubMed] [Google Scholar]
- Mayeda A., Badolato,J., Kobayashi,R., Zhang,M.Q., Gardiner,E.M. and Krainer,A.R. (1999) Purification and characterization of human RNPS1: a general activator of pre-mRNA splicing. EMBO J., 18, 4560–4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarvey T. et al. (2000) The acute myeloid leukemia-associated protein DEK forms a splicing-dependent interaction with exon–product complexes. J. Cell Biol., 150, 309–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy E. and Maquat,L.E. (1998) A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem. Sci., 23, 198–199. [DOI] [PubMed] [Google Scholar]
- Neu-Yilik G., Gehring,N.H., Thermann,R., Frede,U., Hentze,M.W. and Kulozik,A.E. (2001) Splicing and 3′ end formation in the definition of nonsense-mediated decay-competent human β-globin mRNPs. EMBO J., 20, 532–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli A.E., Ernst,R.K., Lund,E., Grimm,C., Zapp,M.L., Rekosh,D., Hammarskjold,M.-L. and Dahlberg,J.E. (1997) The constitutive transport element (CTE) of Mason–Pfizer monkey virus (MPMV) accesses an RNA export pathway utilized by cellular messenger RNAs. EMBO J., 16, 7500–7510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R. and Maniatis,T. (1988) The role of the mammalian branchpoint sequence in pre-mRNA splicing. Genes Dev., 2, 1268–1276. [DOI] [PubMed] [Google Scholar]
- Rodrigues J.P., Rode,M., Gatfield,D., Blencowe,B.J., Carmo-Fonseca,M. and Izaurralde,E. (2001) REF proteins mediate the export of spliced and unspliced mRNAs from the nucleus. Proc. Natl Acad. Sci. USA, 98, 1030–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saavedra C., Felber,B. and Izaurralde,E. (1997) The simian retrovirus-1 constitutive transport element, unlike the HIV-1 RRE, uses factors required for cellular mRNA export. Curr. Biol., 7, 619–628. [DOI] [PubMed] [Google Scholar]
- Serin G., Gersappe,A., Black,J.D., Aronoff,R. and Maquat,L.E. (2001) Identification and characterization of human orthologues to Saccharomyces cerevisiae Upf2 protein and Upf3 protein (Caenorhabditis elegans SMG-4). Mol. Cell. Biol., 21, 209–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutz F., Bachi,A., Doerks,T., Braun,I.C., Seraphin,B., Wilm,M., Bork,P. and Izaurralde,E. (2000) REF, an evolutionary conserved family of hnRNP-like proteins, interacts with TAP/Mex67p and participates in mRNA nuclear export. RNA, 6, 638–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thermann R., Neu-Yilik,G., Deters,A., Frede,U., Wehr,K., Hagemeier,C., Hentze,M.W. and Kulozik,A.E. (1998) Binary specification of nonsense codons by splicing and cytoplasmic translation. EMBO J., 17, 3484–3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vankan P., McGuigan,C. and Mattaj,I.W. (1992) Roles of U4 and U6 snRNAs in the assembly of splicing complexes. EMBO J., 11, 335–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Sun,X., Qian,Y. and Maquat,L.E. (1998) Intron function in the nonsense-mediated decay of β-globin mRNA: indications that pre-mRNA splicing in the nucleus can influence mRNA translation in the cytoplasm. RNA, 4, 801–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Luo,M.J., Straesser,K., Katahira,J., Hurt,E. and Reed,R. (2000) The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature, 407, 401–405. [DOI] [PubMed] [Google Scholar]