Abstract
Ubiquitin-mediated proteolysis has emerged as a key mechanism of regulation in eukaryotic cells. During cell division, a multi-subunit ubiquitin ligase termed the anaphase promoting complex (APC) targets critical regulatory proteins such as securin and mitotic cyclins, and thereby triggers chromosome separation and exit from mitosis. Previous studies in the yeast Saccharomyces cerevisiae identified the conserved WD40 proteins Cdc20 and Hct1 (Cdh1) as substrate-specific activators of the APC, but their precise mechanism of action has remained unclear. This study provides evidence that Hct1 functions as a substrate receptor that recognizes target proteins and recruits them to the APC for ubiquitylation and subsequent proteolysis. By co-immunoprecipitation, we found that Hct1 interacted with the mitotic cyclins Clb2 and Clb3 and the polo-related kinase Cdc5, whereas Cdc20 interacted with the securin Pds1. Failure to interact with Hct1 resulted in stabilization of Clb2. Analysis of Hct1 derivatives identified the C-box, a motif required for APC association of Hct1 and conserved among Cdc20-related proteins. We propose that proteins of the Cdc20 family are substrate recognition subunits of the ubiquitin ligase APC.
Keywords: anaphase promoting complex/C-box/cell cycle/protein degradation/Saccharomyces cerevisiae
Introduction
Passage through the eukaryotic cell division cycle requires ubiquitin-mediated proteolysis (King et al., 1996). In this pathway, ubiquitin ligases (E3) select protein substrates and cooperate with ubiquitin-activating (E1) and -conjugating (E2) enzymes to assemble polyubiquitin chains on substrates, thereby targeting them for degradation by the 26S proteasome (Hochstrasser, 1996; Varshavsky, 1997; Hershko and Ciechanover, 1998). A multi-subunit ubiquitin ligase termed anaphase promoting complex (APC) or cyclosome serves essential functions during mitosis and G1 (Townsley and Ruderman, 1998; Morgan, 1999; Zachariae and Nasmyth, 1999). The APC mediates degradation of securin, an inhibitor of chromosome separation (Nasmyth, 1999). Moreover, the APC triggers proteolysis of mitotic cyclins whose associated kinase activities interfere with the completion of nuclear division, cytokinesis and the resetting of replication origins. Further substrates of the APC include the spindle-associated protein Ase1 and the polo-related kinase Cdc5 in budding yeast, and the replication inhibitor geminin in vertebrate cells (Juang et al., 1997; Charles et al., 1998; McGarry and Kirschner, 1998; Shirayama et al., 1998). Substrates of the APC typically contain a destruction box, a short motif originally identified in mitotic cyclins (Glotzer et al., 1991). Being required and, in certain cases, sufficient for APC-mediated ubiquitylation, the destruction box is thought to function as a recognition sequence for the APC.
The APC is a large particle consisting of at least 10 conserved subunits (Peters, 1999; Zachariae and Nasmyth, 1999). These include the putative assembly factor Cdc26; the DOC domain protein Apc10 (Grossberger et al., 1999); the tetratricopeptide repeat (TPR) proteins Cdc16, Cdc23 and Cdc27; the cullin-domain protein Apc2; and the RING-H2 finger protein Apc11. TPRs are thought to mediate protein–protein interaction (Lamb et al., 1995). Cullin and RING-H2 proteins also occur in other ubiquitin ligases and may form part of the catalytic centre (Tyers and Willems, 1999). In fact, recent studies show that the RING-H2 protein Apc11 is sufficient to support the synthesis of polyubiquitin chains in the presence of E1 and E2 enzymes (Gmachl et al., 2000; Leverson et al., 2000). In general, however, relatively little is known about the function of individual APC subunits.
The activity of the APC is regulated during the cell cycle, being low in S, G2 and early M phase, rising in metaphase and persisting until the end of G1 (Amon et al., 1994; King et al., 1995; Sudakin et al., 1995; Brandeis and Hunt, 1996). APC activity requires association of the core particle with a member of a conserved subfamily of WD40 proteins called Cdc20 and Hct1 (or Cdh1) in budding yeast or fizzy and fizzy-related in Drosophila (Schwab et al., 1997; Sigrist and Lehner, 1997; Visintin et al., 1997; Fang et al., 1998; Kramer et al., 1998; Zachariae et al., 1998). Several lines of evidence indicate that Cdc20 and Hct1 are stage-specific APC regulators with Cdc20 activating the APC in mitosis and Hct1 sustaining APC activity during G1. In yeast, Cdc20 is essential and rate limiting for proteolysis of the securin Pds1 at the onset of anaphase, but dispensable for proteolysis of the mitotic cyclin Clb2 in G1 (Schwab et al., 1997; Visintin et al., 1997; Shirayama et al., 1998). Cdc20 is an unstable protein whose level peaks in mitosis and whose binding to the APC is stimulated by cyclin-dependent kinases (Cdks) through phosphorylation of APC core subunits (Shirayama et al., 1998; Kramer et al., 2000; Rudner and Murray, 2000). The activity of APC–Cdc20 is restrained by the spindle checkpoint, which monitors the proper attachment of chromosomes to the spindle (Elledge, 1998; Amon, 1999). Hct1, on the other hand, is dispensable for Pds1 proteolysis in mitosis, but necessary and rate limiting for proteolysis of Clb2. Unlike Cdc20, Hct1 is inhibited by Cdk1. Hct1 is present throughout the division cycle, but phosphorylation by Cdk1 prevents its association with the APC (Zachariae et al., 1998; Jaspersen et al., 1999). Thus, Hct1 can activate the APC only in late mitosis and G1 when Cdk activity is low. Consistent with its role as a G1-specific activator, Hct1 orthologues in Drosophila and Xenopus are not expressed during the embryonic division cycles, but only later during development when cell cycles include a G1 phase (Sigrist and Lehner, 1997; Lorca et al., 1998). Furthermore, the Schizosaccharomyces pombe orthologue of Hct1 promotes degradation of mitotic cyclins in G1 (Blanco et al., 2000; Yamaguchi et al., 2000).
In addition to being stage-specific activators, evidence from budding yeast suggests that Cdc20 and Hct1 also determine the substrate specificity of the APC. Overexpression studies indicated that Cdc20 and Hct1 preferentially trigger proteolysis of Pds1 and Clb2, respectively (Schwab et al., 1997; Visintin et al., 1997). In fact, Pds1 and the S-phase cyclin Clb5 disappear shortly before anaphase and their proteolysis requires Cdc20, whereas Clb2 and Cdc5 disappear later in mitosis and their proteolysis largely depends on Hct1 (Charles et al., 1998; Shirayama et al., 1998, 1999). Similar to B-type cyclins in other organisms, the mitotic cyclin Clb3 is targeted by both Cdc20 and Hct1, and even in the case of Clb2, a small fraction of the protein appears to be targeted by Cdc20 (Zachariae et al., 1998; Alexandru et al., 1999; Bäumer et al., 2000; Yeong et al., 2000). Thus, Cdc20 and Hct1 may determine the efficiency with which the APC selects distinct substrates.
We have been interested in defining the mechanism by which Cdc20 and Hct1 activate the APC. Here we report that Hct1 binds to a subset of APC substrates including Clb2. A derivative of Clb2, which failed to bind to Hct1, also failed to be destroyed during G1, indicating that the observed interaction is relevant for APC-mediated proteolysis. Unexpectedly, the destruction box of Clb2 was neither necessary nor sufficient for interaction with Hct1. We identified a short motif of Hct1 required for its association with the APC. We named this motif the C-box because it is conserved among members of the Cdc20 family. APC association, however, was not required for Hct1 to interact with Clb2, suggesting a direct role of Hct1 in substrate recognition.
Results
Hct1 interacts with Clb2
Yeast Hct1 is thought to function as a substrate-specific activator of the ubiquitin ligase APC. Overproduction of Hct1 in growing cells triggered APC-dependent proteolysis of Clb2 and Clb3, but hardly affected Pds1 and Clb5 (Schwab et al., 1997; Visintin et al., 1997; Zachariae et al., 1998). Moreover, a deletion of HCT1 stabilized Clb2, but Pds1 and Clb5 were still destroyed (Schwab et al., 1997; Visintin et al., 1997; Shirayama et al., 1999). In late mitosis and G1 when Cdk activity is low, Hct1 was unphosphorylated and associated with the APC (Zachariae et al., 1998; Jaspersen et al., 1999). However, the precise mechanism by which Hct1 activates the APC in a substrate-specific fashion has remained unclear. One obvious possibility would be that Hct1 binds specific substrates and recruits them to the APC. To test this hypothesis, we performed co-immunoprecipitation assays to analyse protein interaction in vivo. In a first experiment, haemagglutinin (HA) epitope-tagged HCT1 was expressed from the galactose-inducible GAL1 promoter in strains carrying Myc epitope-tagged CLB2. Strains lacking epitope tags served as controls. This experiment was performed in cdc23-1 mutants defective in APC activity to avoid Hct1-induced Clb2 degradation. Whole-cell extracts were prepared and proteins were analysed by western blotting either directly or following precipitation with HA- or Myc-specific antibodies. As seen in Figure 1A, precipitates of HA3-Hct1 contained Clb2-Myc12 and Cdk1. Likewise, precipitates of Clb2-Myc12 contained HA3-Hct1 and Cdk1. Co-precipitated HA3-Hct1 migrated heterogeneously, indicating that Clb2-associated Hct1 is phosphorylated. Co-precipitation was specific because control precipitates produced no signal. These data show that Hct1 can bind Clb2-Cdk1 in vivo.
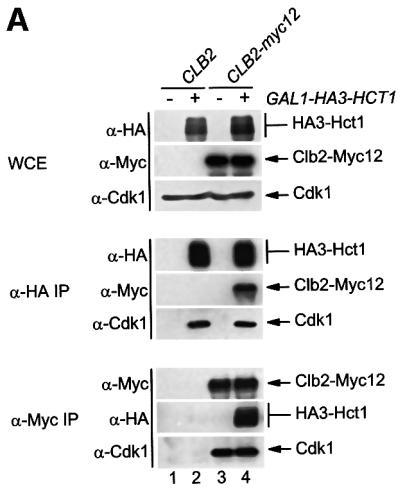
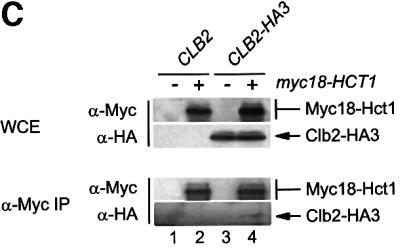
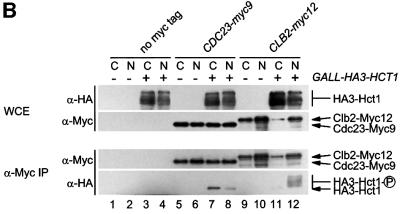
Fig. 1. Interaction of Hct1 with Clb2 and the APC. Whole-cell extracts (WCE) and proteins immunoprecipitated with antibodies to HA (α-HA IP) or Myc (α-Myc IP) were analysed by immunoblotting with HA- (α-HA), Myc- (α-Μyc) or Cdk1- (α-Cdk1) specific antibodies. (A) Hct1 binds Clb2–Cdk1 complexes. cdc23-1 strains carrying CLB2 or CLB2-myc12 and expressing untagged (–) or HA3-tagged (+) HCT1 from the GAL1 promoter were grown to exponential phase at 25°C and treated with galactose for 2 h. Lane 1, W989; lane 2, W987; lane 3, W1067; lane 4, W1066. (B) Phospho-Hct1 binds Clb2 but not the APC. Wild-type (no myc tag), CDC23-myc9 and CLB2-myc12 strains lacking (–) or containing (+) GALL-HA3-HCT1 were grown to exponential phase (C) or arrested with nocodazole for 3 h (N) and then treated with galactose for 2 h. Lanes 1 and 2, K699, lanes 3 and 4, W1158, lanes 5 and 6, W1641, lanes 7 and 8, W1644, lanes 9 and 10, W1079, lanes 11 and 12, W1156. (C) Hct1 binds Clb2 at normal protein levels. Wild-type strains carrying CLB2 or CLB2-HA3 and untagged (–) or Myc18-tagged (+) HCT1 were arrested with nocodazole for 3 h. Lane 1, K699; lane 2, W2541; lane 3, W9317; lane 4, W2542.
To substantiate this result and to rule out effects of the cdc23-1 mutation, a similar experiment was carried out in wild-type cells which expressed HA3-HCT1 from the relatively weak GALL promoter (Mumberg et al., 1994). Strains carrying Myc epitope-tagged CDC23 or CLB2 were used so that the interaction of Hct1 with the APC and Clb2 could be compared. Strains lacking epitope tags served as controls. Protein extracts were prepared from growing and nocodazole-arrested cells. Cdc23-Myc9 and Clb2-Myc12 were immunoprecipitated and analysed for the presence of HA3-Hct1. Epitope-tagged proteins in extracts and precipitates were detected by western blotting (Figure 1B). Consistent with previous work which had shown that phosphorylation of Hct1 inhibits its binding to the APC (Zachariae et al., 1998), a single fast-migrating species of HA3-Hct1 was detected in precipitates of Cdc23-Myc9. The signal was weaker in nocodazole-arrested cells where more Hct1 is phosphorylated due to high Cdk1 activity. In contrast, precipitates of Clb2-Myc12 contained all species of HA3-Hct1 present in the cell extract, indicating that both unphosphorylated and phosphorylated forms of Hct1 bind Clb2. In this case, co-precipitation was more efficient in nocodazole-arrested cells where Clb2 levels are high. These results confirm that Hct1 can bind Clb2. Unlike the association of Hct1 with the APC, its interaction with Clb2 was not affected by phosphorylation, suggesting that Hct1 can recognize Clb2 without binding to the APC.
Hct1 is a protein of low abundance. To ask whether interaction with Clb2 is detectable when Hct1 is present at its normal cellular level, we analysed a strain in which the chromosomal copies of HCT1 and CLB2 carried sequences for epitope tags. Clb2-HA3 was detectable in precipitates of Myc18-Hct1 (Figure 1C). The signal was quite weak but specific, since it was absent from precipitates of control strains. Thus, Hct1 interacts with Clb2 under physiological conditions.
Hct1 and Cdc20 interact with specific APC substrates
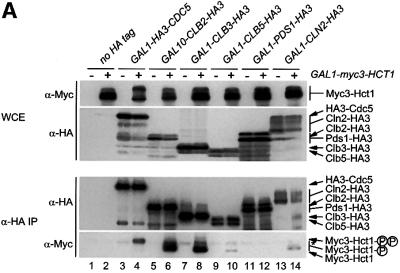
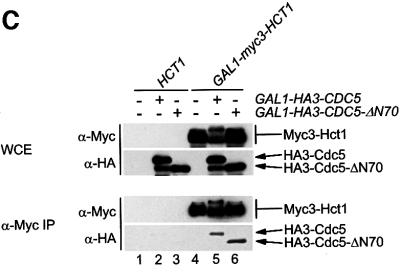
In a next step, we examined the ability of Hct1 to bind different substrates of the APC. Besides Clb2, we tested the mitotic cyclin Clb3 and the polo-related kinase Cdc5, whose instability in G1 is Hct1 dependent (Charles et al., 1998; Zachariae et al., 1998). We also tested the S-phase cyclin Clb5 and the securin Pds1, whose proteolysis in mitosis depends on Cdc20 (Visintin et al., 1997; Shirayama et al., 1999). We were also interested in comparing binding with different cyclins, since Hct1 is phosphorylated by various Cln– and Clb–Cdk1 kinases (Zachariae et al., 1998). We therefore included the G1-phase cyclin Cln2 in this assay. Cln2 is not a substrate of the APC, but its proteolysis is mediated by a distinct ubiquitin ligase termed Skp1–Cullin–F-box protein (SCF) (Patton et al., 1998; Peters, 1998). To examine protein interaction, myc3-HCT1 was co-expressed with HA-tagged versions of CDC5, CLB2, CLB3, CLB5, PDS1 or CLN2. Protein extracts were prepared and HA-tagged proteins were immunoprecipitated and analysed for the presence of Myc3-Hct1 (Figure 2A). Relatively high levels of Myc3-Hct1 co-precipitated with Clb2-HA3 and Clb3-HA3, and somewhat lower levels with HA3-Cdc5. Only trace amounts of Myc3-Hct1 were found in precipitates of Clb5-HA3 and Cln2-HA3, and none was detectable in precipitates of Pds1-HA3 even though high levels of Pds1-HA3 were present in cell extracts and precipitates. Thus, Hct1 can bind Clb2, Clb3 and Cdc5, but not Pds1, thus supporting a role of Hct1 in the recruitment of specific APC substrates. Moreover, Hct1 distinguished between different cyclins, arguing against the view that the Hct1–cyclin interaction simply reflects phosphorylation of Hct1 by cyclin–Cdk1 kinases.
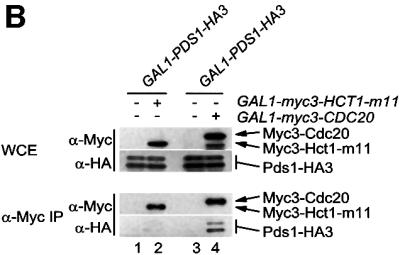
Fig. 2. Binding of Hct1 and Cdc20 to specific APC substrates. (A) Hct1 binds specific APC substrates. cdc23-1 strains (no HA tag) and cdc23-1 strains carrying GAL1-HA3-CDC5, GAL10-CLB2-HA3, GAL1-CLB3-HA3, GAL1-CLB5-HA3, GAL1-PDS1-HA3 or GAL1-CLN2-HA3 and lacking (–) or containing (+) GAL1-myc3-HCT1 were grown to exponential phase at 25°C and galactose was added for 2 h. Epitope-tagged proteins in whole-cell extracts and in immunoprecipitates prepared with HA antibodies cross-linked to protein A-agarose (α-HA IP) were detected on western blots with HA- (α-HA) or Myc- (α-Myc) specific antibodies. The asterisk in the bottom panel marks a cross-linked antibody species arising from incomplete coupling of antibodies to protein A–agarose. Hct1 coprecipitated with Cdc5 was hyperphosphorylated, indicating that Cdc5 kinase may phosphorylate Hct1. The significance of this hyperphosphorylation is unclear. Lane 1, W1500; lane 2, W1502; lane 3, W1619; lane 4, W1621; lane 5, W1504; lane 6, W1506; lane 7, W1615; lane 8, W1617; lane 9, W1508; lane 10, W1510; lane 11, W1623; lane 12, W1625; lane 13, W1653; lane 14, W1655. (B) Cdc20 but not Hct1-m11 binds Pds1. cdc23-1 strains carrying GAL1-PDS1-HA3 and lacking (–) or containing (+) GAL1-myc3-HCT1-m11 as well as cdc16-1 strains carrying GAL1-PDS1-HA3 and lacking (–) or containing (+) GAL1-myc3-CDC20 were grown to exponential phase at 25°C and treated with galactose for 2 h. Whole-cell extracts and proteins immunoprecipitated with antibodies to Myc (α-Myc IP) were analysed by immunoblotting with HA- (α-HA) or Myc- (α-Myc) specific antibodies. Lane 1, W1623; lane 2, W1741; lane 3, W2538; lane 4, W1588.
The failure of Hct1 to interact with Pds1 prompted us to perform a similar interaction assay with Cdc20 (Figure 2B). To this end, we immunoprecipitated Myc3-Cdc20 from extracts of cells expressing PDS1-HA3. While a control precipitate produced no signal, Pds-HA3 was easily detectable in the Myc3-Cdc20 precipitate, indicating that Cdc20 can bind Pds1. A recent study suggested that during G1, when Cdc20 levels are low, Hct1 is responsible for Pds1 proteolysis (Rudner et al., 2000). We therefore repeated the interaction assay with Hct1-m11, a phosphosite mutant mimicking the unphosphorylated, G1-specific form of Hct1 (Zachariae et al., 1998). However, we could not detect co-precipitation of Pds1-HA3 with Myc3-Hct1-m11 (Figure 2B). Thus, Cdc20 and Hct1 differ in their ability to recognize Pds1.
Hct1 does not recognize the destruction box
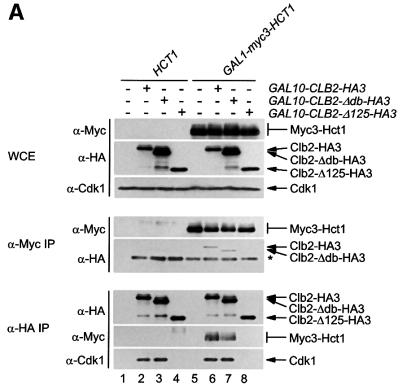
Ubiquitylation of a substrate protein by the APC depends on a short motif in the substrate termed the destruction box (Glotzer et al., 1991). We wanted to know whether the destruction box serves as a recognition signal for Hct1. To address this question, we analysed the interaction of Hct1 with a derivative of Clb2 lacking the destruction box motif (Clb2-Δdb). Unlike Clb2, Clb2-Δdb is stable during exit from mitosis, it is not ubiquitylated by the APC and resistant to Hct1-induced proteolysis (Amon et al., 1994; Zachariae and Nasmyth, 1996; Schwab et al., 1997). We also looked at a Clb2 derivative lacking amino acids 263–387 (Clb2-Δ125). This deletion removes sequences conserved among cyclins and needed for interaction with Cdk1, but leaves the N-terminal region with the destruction box intact. HA3-tagged versions of Clb2, Clb2-Δdb and Clb2-Δ125 were co-expressed with Myc3-tagged Hct1, and protein interaction was again determined by immunoprecipitation and western analysis. As seen in Figure 3A, Clb2-HA3 and Clb2-Δdb-HA3, but not Clb2-Δ125-HA3, co-precipitated with Myc3-Hct1. Likewise, Myc3-Hct1 and Cdk1 co-precipitated with Clb2-HA3 and Clb2-Δdb-HA3, but not with Clb2-Δ125-HA3. Thus, the central portion of Clb2 containing sequences for Cdk1 association is needed for interaction with Hct1, but the destruction box of Clb2 is neither required nor sufficient for interaction with Hct1.
Fig. 3. Binding of Hct1 to derivatives of Clb2 and Cdc5 lacking a destruction box. Whole-cell extracts and proteins immunoprecipitated with antibodies to HA (α-HA IP) or Myc (α-Myc IP) were examined by western analysis using HA- (α-HA), Myc- (α-Myc) or Cdk1- (α-Cdk1) specific antibodies. (A) Hct1 binds Clb2 lacking the destruction box. cdc23-1 strains containing HCT1 or GAL1-myc3-HCT1 and lacking (–) or carrying (+) the indicated GAL10-CLB2-HA3 construct were grown to exponential phase at 25°C and treated with galactose for 1 h. The asterisk marks the IgG heavy chain of the Myc antibody. Lane 1, W1500; lane 2, W1504; lane 3, W1684; lane 4, W1744; lane 5, W1502; lane 6, W1506; lane 7, W1686; lane 8, W1746. (B) Non-phosphorylatable Hct1 binds Clb2 lacking the destruction box. cdc23-1 strains carrying HCT1 or GAL1-myc3-HCT1-m11 and lacking (–) or containing (+) the indicated GAL10-CLB2-HA3 construct were treated as described in (A). Lane 1, W1500; lane 2, W1504; lane 3, W1684; lane 4, W1739; lane 5, W1740; lane 6, W1889. (C) Hct1 binds a truncated version of Cdc5 lacking destruction boxes. cdc23-1 strains carrying HCT1 or GAL1-myc3-HCT1 and lacking (–) or containing (+) the indicated GAL1-CDC5-HA3 construct were treated as described in (A) except that galactose was added for 2 h. Lane 1, W1500; lane 2, W1619; lane 3, W1930; lane 4, W1502; lane 5, W1621; lane 6, W1931.
Precipitates of Clb2 and Clb2-Δdb contained similar amounts of Hct1 (Figure 3A). As indicated by the heterogeneous mobility, co-precipitated Hct1 was phosphorylated. We wondered whether the destruction box of Clb2 may be important for binding specifically to unphosphorylated Hct1—the form that associates with the APC (Zachariae et al., 1998). We therefore analysed the interaction of Clb2 and Clb2-Δdb with Hct1-m11, a derivative from which all serine and threonine residues of potential Cdk1 phosphorylation sites have been removed (Zachariae et al., 1998). Myc3-tagged Hct1-m11 was co-expressed with HA3-tagged Clb2 and Clb2-Δdb. Tagged proteins were precipitated from cell extracts and analysed by western blotting. Similar amounts of Clb2-HA3 and Clb2-Δdb-HA3 co-precipitated with Myc3-Hct1-m11, and in the reverse experiment similar amounts of Myc3-Hct1-m11 co-precipitated with Clb2-HA3 and Clb2-Δdb-HA3 (Figure 3B). These results show that the destruction box of Clb2 is not required for the interaction of unphosphorylated Hct1 with Clb2.
The polo-related kinase Cdc5 is a substrate of the APC–Hct1 pathway and carries two destruction boxes in the N-terminal region. Unlike Cdc5, a truncated version lacking these sequences, Cdc5-ΔN70, is not ubiquitylated in vitro and stable in G1 cells (Shirayama et al., 1998). We asked whether the destruction boxes of Cdc5 are necessary for the observed interaction with Hct1 (Figure 2A). To this end, Myc3-tagged Hct1 was co-expressed with HA3-tagged Cdc5 or Cdc5-ΔN70. Protein interaction was again examined by immunoprecipitation of tagged proteins from cell extracts and subsequent western analysis. As seen in Figure 3C, similar levels of HA3-Cdc5 and HA3-Cdc5-ΔN70 co-precipitated with Myc3-Hct1. The same was true for the phosphosite mutant Myc3-Hct1-m11 (not shown). Thus, the destruction box-containing region of Cdc5 is not required for the interaction of Cdc5 with Hct1. Together, these results suggest that the destruction box functions in a step after the initial recognition of the substrate by Hct1.
Interaction with Hct1 is required for Clb2 instability
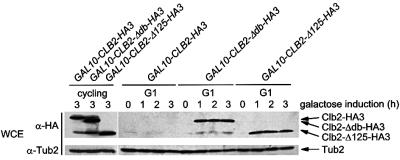
The idea that Hct1 recognizes specific protein substrates and delivers them to the APC predicts that these substrates must bind to Hct1 to be destroyed by this proteolysis pathway. The failure of Clb2-Δ125 to bind to Hct1 (Figure 3A) allowed us to test this prediction. To analyse the stability of Clb2-Δ125, HA3-tagged versions of Clb2, Clb2-Δdb and Clb2-Δ125 were expressed in cdc28-4 cells, which arrest in G1 after a shift to the restrictive temperature. Under these conditions, the APC–Hct1 pathway is active and interferes with accumulation of Clb2 (Amon et al., 1994; Schwab et al., 1997). When expressed in cdc28-4 cells growing at the permissive temperature, Clb2-HA3, Clb2-Δdb-HA3 and Clb2-Δ125-HA3 were present at similar levels (Figure 4). However, when expressed in G1-arrested cells, Clb2-HA3 was not detectable, whereas Clb2-Δdb-HA3 and Clb2-Δ125-HA3 accumulated to comparable levels (Figure 4), indicating that Clb2-Δ125-HA3 is stabilized. Moreover, Clb2-Δdb-HA3 and Clb2-Δ125-HA3 were unaffected by overexpression of HCT1, while Clb2-HA3 rapidly disappeared under these conditions (not shown); this is consistent with previous observations (Schwab et al., 1997). These results indicate that the binding of Clb2 to Hct1 is necessary for the stage-specific proteolysis of Clb2, and thus further support a role of Hct1 in substrate recruitment.
Fig. 4. Stability of Clb2 derivatives in G1-arrested cells. CLB2-HA3 (W1883), CLB2-Δdb-HA3 (W1884) and CLB2-Δ125-HA3 (W1885) were expressed from the GAL10 promoter in cycling or G1-arrested (G1) cdc28-4 bar1-Δ1 cells. Cultures grown at 25°C in raffinose medium were split. One part was further incubated at 25°C (cycling) and induced with galactose for 3 h. The other part was treated with α-factor for 3.5 h and, following arrest in G1, cultures were shifted to 37°C to inactivate Cdk1 (Cdc28) kinase and then induced with galactose. Samples were taken at the indicated times and the protein levels were determined by immunoblotting with HA-specific antibodies (α-HA). Blots were reprobed with an antiserum to β-tubulin (α-Tub2) as a loading control.
The C-box of Hct1 is required for binding to the APC
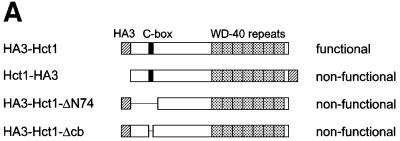
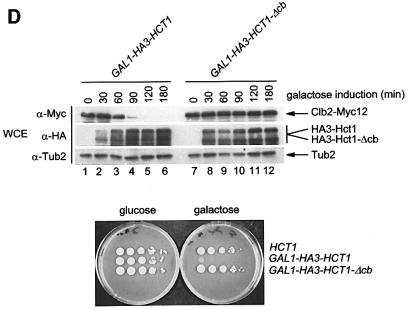
Hct1 consists of a highly conserved C-terminal domain containing seven WD40 repeats and a largely non-conserved N-terminal region (Figure 5A). In a search for sequences required for Hct1 function, we found that Hct1 is inactivated either by the addition of a HA3 epitope tag to the C-terminus (Hct1-HA3) or by a deletion of 74 amino acids from the N-terminus (HA3-Hct1-ΔN74). Interestingly, this N-terminal portion contains a short motif of seven amino acids, which is also present in Cdc20 as well as in Cdc20- and Hct1-related proteins from other organisms (Figure 5B). We named this motif the C-box (conserved in Cdc20 family proteins). To define its significance, we constructed derivatives of Cdc20 and Hct1 lacking the C-box motif and analysed their functional properties. Unlike wild-type CDC20, CDC20-Δcb did not complement the temperature sensitivity of a cdc20-3 mutant strain (Figure 5C), indicating that CDC20-Δcb is non-functional in vivo. Similarly, HA3-HCT1-Δcb failed to trigger Clb2 degradation when overexpressed, even though the protein accumulated to levels comparable to wild-type HA3-Hct1 (Figure 5D). Moreover, overexpression of HA3-HCT1-Δcb did not affect cell proliferation, whereas wild-type HA3-HCT1 inhibited colony formation under these conditions (Figure 5D). Thus, the C-box is essential for the cellular functions of Cdc20 and Hct1.
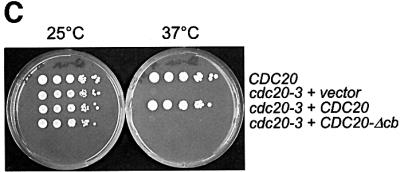
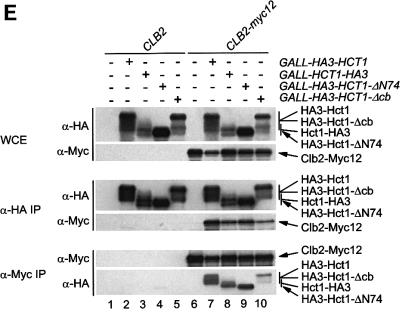
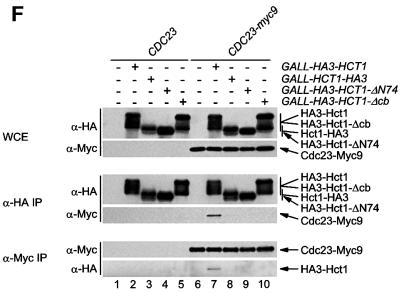
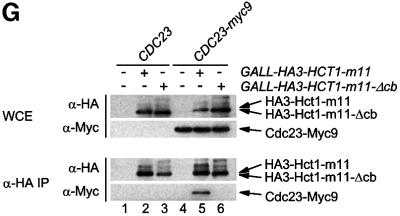
Fig. 5. Sequences of Hct1 required for APC binding. (A) Schematic representation of Hct1 derivatives. Positions of the HA3 epitope, the C-box motif and the seven WD40 repeats are shown. Deleted segments are indicated by a line. Non-functional denotes failure of the overproduced Hct1 derivative to induce Clb2 proteolysis and cell-cycle arrest. (B) The C-box, a conserved motif in proteins of the Cdc20 family. A sequence alignment of the C-box-containing regions of Hct1- and Cdc20-related proteins is shown with residues identical in at least 11 members in bold. A consensus sequence for the C-box is deduced with X indicating any amino acid. Accession numbers and positions of the first amino acids are given. S.c., Saccharomyces cerevisiae; S.p., Schizosaccharomyces pombe; D.m., Drosophila melanogaster; X.l., Xenopus laevis; M.m., Mus musculus; H.s., Homo sapiens. (C) The C-box of Cdc20 is required for function. We compared a wild-type CDC20 strain (W933) and a temperature sensitive cdc20-3 mutant strain (W1117) transformed with vector alone (pRS316) or with plasmids expressing either CDC20 (pWS923) or CDC20-Δcb (pWS924) from the natural CDC20 promoter. Cells were spotted in serial 10-fold dilutions on plates with glucose-supplemented synthetic medium lacking uracil. Plates were incubated at 25°C for 4 days or at 37°C for 3 days. (D) The C-box of Hct1 is required for function. Upper part: Strains carrying HA3-HCT1 (W2539) or HA3-HCT1-Δcb (W2540) under control of the GAL1 promoter were grown in raffinose medium and treated with nocodazole for 3.5 h. Samples were taken at the indicated times after addition of galactose and protein levels were followed by western analysis with Myc- (α-Myc) and HA- (α-HA) specific antibodies. Blots were reprobed with an antiserum to β-tubulin (α-Tub2) as a loading control. Lower part: We compared a wild-type HCT1 strain (K699) and strains expressing either HA3-HCT1 (W2539) or HA3-HCT1-Δcb (W2540) from the GAL1 promoter. Cells were spotted in serial 10-fold dilutions on XY-plates containing 2% glucose or 2% galactose. Plates were incubated at 25°C for 3 days. (E–G) Interaction of Hct1 derivatives with Clb2 and the APC. Whole-cell extracts and proteins immunoprecipitated with HA- (α-HA IP) or Myc- (α-Myc IP) specific antibodies were analysed by western blotting with the indicated antibodies. (E) Wild-type strains (CLB2) and CLB2-myc12 strains lacking (–) or containing (+) the indicated GALL-HA3-HCT1 construct were arrested with nocodazole for 3 h and then galactose was added for 2 h. Lane 1, K699; lane 2, W1158; lane 3, W1758; lane 4, W1759; lane 5, W1841; lane 6, W1079; lane 7, W1156; lane 8, W1760; lane 9, W1761; lane 10, W1842. (F) Wild-type strains (CDC23) and CDC23-myc9 strains lacking (–) or containing (+) the indicated GALL-HA3-HCT1 construct were grown to log phase and treated for 2 h with galactose. Lane 1, K699; lane 2, W1158; lane 3, W1758; lane 4, W1759; lane 5, W1841; lane 6, W1641; lane 7, W1644; lane 8, W1762; lane 9, W1763; lane 10, W1843. (G) Wild-type strains (CDC23) and CDC23-myc9 strains lacking (–) or containing (+) either the GALL-HA3-HCT1-m11 or GALL-HA3-HCT1-m11-Δcb construct were treated as described in (F). Lane 1, K699; lane 2, W2545; lane 3, W2546; lane 4, W1641; lane 5, W2547; lane 6, W2548.
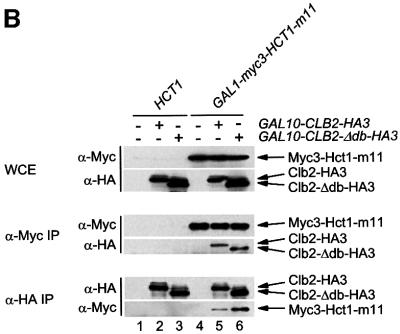
We next asked whether alterations that inactivate Hct1 may affect the binding of Hct1 to the substrate Clb2 or the APC, or both. To this end, a functional version of HCT1 (HA3-HCT1) and the three non-functional derivatives (HCT1-HA3, HA3-HCT1-ΔN74 and HA3-HCT-Δcb) were expressed from the GALL promoter in wild-type cells carrying either CLB2-myc12 or CDC23-myc9; interaction of tagged proteins was investigated by immunoprecipitation and western analysis (Figure 5E and F). Similar amounts of Clb2-Myc12 co-precipitated with HA3-Hct1 and the non-functional Hct1 derivatives. Consistent with this, wild-type and mutant versions of HA3-Hct1 were detected in precipitates of Clb2-Myc12 (Figure 5E). In contrast, Cdc23-Myc9 co-precipitated with HA3-Hct1, but not with the non-functional Hct1 derivatives. Likewise, only wild-type HA3-Hct1 was detected in precipitates of Cdc23-Myc9 (Figure 5F). Thus, alterations of Hct1 at the N- or C-terminus are compatible with the binding of Hct1 to Clb2, but interfere with binding to the APC, suggesting that different domains of Hct1 are responsible for substrate recognition and APC association. Furthermore, these results show that Hct1 requires the newly identified C-box motif to associate with the APC.
Previous studies established that phosphorylated Hct1 fails to interact with the APC (Zachariae et al., 1998; Jaspersen et al., 1999). The C-box deletion derivative of Hct1 appears to be hyperphosphorylated, since the protein accumulated predominately in a slow migrating form (Figure 5E and F). This raises the possibility that the C-box has a role in dephosphorylation of Hct1, and that loss of the C-box interferes with APC association only indirectly through stabilizing the phosphorylation of Hct1. To address this idea, we deleted the C-box from the phosphosite mutant Hct1-m11, which lacks Cdk1 phosphorylation sites and interacts with the APC constitutively (Zachariae et al., 1998). Unlike HA3-Hct1-m11, the C-box deletion derivative (HA3-Hct1-m11-Δcb) did not co-precipitate with Cdc23-Myc9 (Figure 5G), indicating that the C-box of Hct1 is required for APC binding in the absence of inhibitory phosphorylation as well. This result is consistent with the C-box of Hct1 serving as an interaction motif necessary for APC association.
Discussion
The ubiquitin ligase APC performs essential functions in the eukaryotic cell division cycle by triggering the stage-specific degradation of key regulatory proteins such as securin and mitotic cyclins (Zachariae and Nasmyth, 1999). It has therefore been of considerable interest to define the mechanism by which the APC selects its substrates. Previous studies in budding yeast identified the related WD40 repeat proteins Cdc20 and Hct1 as cell cycle-regulated and substrate-specific activators of the APC (Schwab et al., 1997; Visintin et al., 1997; Shirayama et al., 1998; Zachariae et al., 1998). This work provides evidence that Hct1 interacts with specific APC substrates.
Our data suggest that Hct1 is a substrate receptor of the APC, which recognizes target proteins and recruits them to the complex for ubiquitylation and subsequent proteolysis. This model is supported by the following results. First, by immunoprecipitation of epitope-tagged proteins from cell extracts we found that Hct1 interacts with the mitotic cyclins Clb2 and Clb3 and the polo-related kinase Cdc5, but not or only very weakly with the securin Pds1 and the S-phase cyclin Clb5 (Figures 1 and 2). Thus, the observed interaction pattern reflects the previously determined requirement of Hct1 for proteolysis of specific APC substrates. This pattern also argues against the view that the interaction of Hct1 with cyclins is only due to phosphorylation of Hct1 by Clb–Cdk1 kinases, since Clb5–Cdk1 was the most efficient Hct1 kinase in vitro (Zachariae et al., 1998) and the interaction of Clb5 with Hct1 was almost undetectable (Figure 2). Secondly, a deletion derivative of Clb2 unable to interact with Hct1 was protected against G1-specific proteolysis in vivo (Figure 4), supporting the idea that binding to Hct1 is necessary for degradation. Thirdly, association with the APC was not required for Hct1 to recognize substrates, since mutant derivatives of Hct1 unable to bind to the APC still interacted with Clb2 (Figure 5) and with Clb3 (not shown). This indicates that Hct1 carries binding sites for proteolytic substrates which can be separated from the sequences required for APC binding. Consistent with the notion that substrate recognition and APC association are independent, we also observed that phosphorylated forms of Hct1, which do not bind to the APC, interact with Clb2 (Figure 1). Thus, Hct1 may recognize and bind to proteolytic substrates before associating with the APC in late mitosis and G1. This could provide a mechanism for the rapid and efficient recruitment of substrates.
Its similarity in structure and function suggested that Cdc20 would also serve as a substrate receptor of the APC. Its specificity, however, should differ from that of Hct1 to provide efficient recognition of proteins to be destroyed at the onset of anaphase, such as the securin Pds1. This view was confirmed by the finding that, unlike Hct1, Cdc20 can bind Pds1 (Figure 2). Together, these data suggest that proteins of the Cdc20 family are substrate recognition subunits of the ubiquitin ligase APC.
Substrate recognition
The list of Hct1 substrates includes diverse proteins such as cyclins, Cdc5 kinase, the spindle protein Ase1 and the septin-associated kinase Hsl1 (Zachariae and Nasmyth, 1999; Burton and Solomon, 2000). This raises the question how a single protein can recognize a number of different targets. One possibility would be that Hct1 substrates, even though largely dissimilar, share short recognition sequences. Indeed, APC substrates contain short motifs needed for ubiquitylation and subsequent proteolysis, either a canonical destruction box or a more recently identified element termed KEN-box (Glotzer et al., 1991; Pfleger and Kirschner, 2000). However, Hct1 still interacted with derivatives of Clb2 and Cdc5 lacking destruction boxes (Figure 3), and similar results were reported for Hsl1 (Burton and Solomon, 2000). Moreover, Clb2-Δ125, whose destruction box-containing region remains intact, failed to bind to Hct1 (Figure 3). It is, therefore, unlikely that Hct1 uses the destruction box to recognize substrates. Another possibility would be that Hct1 associates with distinct sequences in individual substrates. The C-terminal portion of Hct1 consists of seven WD40 repeats predicted to fold into a seven-bladed β-sheet propeller (Smith et al., 1999). This structure has the potential to provide a number of specific contact sites. In fact, the yeast G-protein β-subunit Ste4, a prototype WD40 protein, not only interacts with the Gα and Gγ subunits, but also with the scaffold protein Ste5, the kinase Ste20 and others (Leberer et al., 1997). Thus, the architecture of the WD40 protein Hct1 may allow multiple specific interactions enabling Hct1 to recognize unrelated substrates.
The C-box of Hct1 is required for APC association
Within the largely non-conserved N-terminal part of Hct1, we identified a short sequence motif whose removal destroyed the ability of Hct1 to trigger Clb2 proteolysis (Figure 5D). Interestingly, a closely related motif of seven amino acids is located in the N-terminal portions of yeast Cdc20 as well as Cdc20- and Hct1-related proteins from other organisms (Figure 5B). We named this motif C-box, to indicate its conservation among proteins of the Cdc20 family. Binding studies showed that a Hct1 derivative lacking the C-box failed to associate with the APC, but still interacted with Clb2 and Clb3 (Figure 5E and F and our unpublished data). Thus, the C-box of Hct1 is specifically required for APC association. Given its conservation, the C-box may also mediate the APC association of other Cdc20 family members. Consistent with this view, the C-box of Cdc20 turned out to be essential for its cellular function (Figure 5C).
In addition to the C-box, integrity of the C-terminus of Hct1 was required for APC association (Figure 5F). Sequence comparison showed that all Cdc20 family proteins carry the amino acids isoleucine–arginine (IR-motif) at their C-termini. Thus, the invariant IR-motif is possibly part of the APC interaction domain. Consistent with the idea that the C-box cooperates with one or more other conserved motifs, database searches failed to detect a close match to the C-box consensus in proteins outside this subclass of WD40 proteins.
Identification of the C-box as a necessary domain for APC interaction may also explain how phosphorylation of Hct1 controls its binding to the APC. Most cyclin–Cdk1 phosphorylation sites are located in the N-terminal portion of Hct1 and their phosphorylation may render the C-box inaccessible.
Common features of the SCF and the APC
The ubiquitin ligases SCF and APC control major cell-cycle events in eukaryotes. Both are multi-subunit complexes and, although their subunits are largely distinct, both complexes contain a cullin-related and a RING-H2 finger protein (Peters, 1998). Furthermore, the SCF uses multiple recognition subunits to target specific substrates. These recognition subunits contain a so-called F-box required for association with the core complex and a protein interaction domain such as the WD40 repeats (Patton et al., 1998). An analogous situation appears to apply to the APC. Previous work identified Cdc20 and Hct1 as substrate-specific activators of the APC (Zachariae and Nasmyth, 1999). This study provides evidence that Hct1 interacts with specific substrates of the APC and carries a short conserved motif, the C-box, required for binding to the APC. Most recently, Ama1 has been described as a third member of this protein family in budding yeast (Cooper et al., 2000). Ama1 was found to regulate APC functions specifically during meiosis. Thus, the SCF and the APC belong to a class of ubiquitin ligases that contain related subunits and use particular recognition factors as substrate-specific receptors.
Materials and methods
Yeast methods
Standard protocols were followed for transformation, mating, sporulation and tetrad dissection of yeast cells (Adams et al., 1997). Yeast strains used in this study are listed in Table I. Strains are isogenic derivatives of W303-1a (K699) (MATa ade2-1 can1-100 his3-11, -15 leu2-3, -112 ssd1-d trp1-1 ura3), except for cdc16-1 and cdc23-1 mutant strains, which were made congenic to W303 by backcrossing at least five times.
Table I. Yeast strains.
| Name | Relevant genotype |
|---|---|
| W987 | MATa cdc23-1 ura3::pGAL1-HA3-HCT1-URA3 |
| W989 | MATa cdc23-1 ura3::pGAL1-HCT1-URA3 |
| W1066 | MATa cdc23-1 clb2::CLB2-myc12-HIS3 ura3::pGAL1-HA3-HCT1-URA3 |
| W1067 | MATa cdc23-1 clb2::CLB2-myc12-HIS3 ura3::pGAL1-HCT1-URA3 |
| W1079 | MATa clb2::CLB2-myc12-HIS3 |
| W1156 | MATa clb2::CLB2-myc12-HIS3 ura3::pGALL-HA3-HCT1-URA3 |
| W1158 | MATa ura3::pGALL-HA3-HCT1-URA3 |
| W1641 | MATa cdc23::CDC23-myc9-LEU2 |
| W1644 | MATa cdc23::CDC23-myc9-LEU2 ura3::pGALL-HA3-HCT1-URA3 |
| W2541 | MATa hct1::myc18-HCT1-LEU2 |
| W2542 | MATa CLB2-HA3 hct1::myc18-HCT1-LEU2 |
| W9317 | MATa CLB2-HA3 |
| W1500 | MATa cdc23-1 |
| W1502 | MATa cdc23-1 trp1::pGAL1-myc3-HCT1-TRP1 |
| W1504 | MATa cdc23-1 ura3::pGAL10-CLB2-HA3-URA3 |
| W1506 | MATa cdc23-1 trp1::pGAL1-myc3-HCT1-TRP1 ura3::pGAL10-CLB2-HA3-URA3 |
| W1508 | MATa cdc23-1 ura3::pGAL1-CLB5-HA3-URA3 |
| W1510 | MATa cdc23-1 trp1::pGAL1-myc3-HCT1-TRP1 ura3::pGAL1-CLB5-HA3-URA3 |
| W1615 | MATa cdc23-1 ura3::pGAL1-CLB3-HA3-URA3 |
| W1617 | MATa cdc23-1 trp1::pGAL1-myc3-HCT1-TRP1 ura3::pGAL1-CLB3-HA3-URA3 |
| W1619 | MATa cdc23-1 ura3::pGAL1-HA3-CDC5-URA3 |
| W1621 | MATa cdc23-1 trp1::pGAL1-myc3-HCT1-TRP1 ura3::pGAL1-HA3-CDC5-URA3 |
| W1623 | MATa cdc23-1 ura3::pGAL1-PDS1-HA3-URA3 |
| W1625 | MATa cdc23-1 trp1::pGAL1-myc3-HCT1-TRP1 ura3::pGAL1-PDS1-HA3-URA3 |
| W1653 | MATa cdc23-1 leu2::pGAL1-CLN2-HA3-LEU2 |
| W1655 | MATa cdc23-1 trp1::pGAL1-myc3-HCT1-TRP1 leu2::pGAL1-CLN2-HA3-LEU2 |
| W1588 | MATa cdc16-1 CDC20::pGAL1-myc3-CDC20-URA3 ura3::pGAL1-PDS1-HA3-URA3 |
| W1741 | MATa cdc23-1 trp1::pGAL1-myc3-HCT1-m11-TRP1 ura3::pGAL1-PDS1-HA3-URA3 |
| W2538 | MATα cdc16-1 ura3::pGAL1-PDS1-HA3-URA3 |
| W1684 | MATa cdc23-1 ura3::pGAL10-CLB2-Δdb-HA3-URA3 |
| W1686 | MATa cdc23-1 trp1::pGAL1-myc3-HCT1-TRP1 ura3::pGAL10-CLB2-Δdb-HA3-URA3 |
| W1744 | MATa cdc23-1 ura3::pGAL10-CLB2-Δ125-HA3-URA3 |
| W1746 | MATa cdc23-1 trp1::pGAL1-myc3-HCT1-TRP1 ura3::pGAL10-CLB2-Δ125-HA3-URA3 |
| W1739 | MATa cdc23-1 trp1::pGAL1-myc3-HCT1-m11-TRP1 |
| W1740 | MATa cdc23-1 trp1::pGAL1-myc3-HCT1-m11-TRP1 ura3::pGAL10-CLB2-HA3-URA3 |
| W1889 | MATa cdc23-1 trp1::pGAL1-myc3-HCT1-m11-TRP1 ura3::pGAL10-CLB2-Δdb-HA3-URA3 |
| W1930 | MATa cdc23-1 ura3::pGAL1-HA3-CDC5-ΔN70-URA3 |
| W1931 | MATa cdc23-1 trp1::pGAL1-myc3-HCT1-TRP1 ura3::pGAL1-HA3-CDC5-ΔN70-URA3 |
| W1883 | MATa cdc28-4 bar1-Δ1::HIS3 ura3::pGAL10-CLB2-HA3-URA3 |
| W1884 | MATa cdc28-4 bar1-Δ1::HIS3 ura3::pGAL10-CLB2-Δdb-HA3-URA3 |
| W1885 | MATa cdc28-4 bar1-Δ1::HIS3 ura3::pGAL10-CLB2-Δ125-HA3-URA3 |
| W933 | MATa URA3 |
| W1117 | MATa cdc20-3 |
| W2539 | MATa clb2::CLB2-myc12-HIS3 ura3::pGAL1-HA3-HCT1-URA3 |
| W2540 | MATa clb2::CLB2-myc12-HIS3 ura3::pGAL1-HA3-HCT1-Δcb-URA3 |
| W1758 | MATa ura3::pGALL-HCT1-HA3-URA3 |
| W1759 | MATa ura3::pGALL-HA3-HCT1-ΔN74-URA3 |
| W1760 | MATa clb2::CLB2-myc12-HIS3 ura3::pGALL-HCT1-HA3-URA3 |
| W1761 | MATa clb2::CLB2-myc12-HIS3 ura3::pGALL-HA3-HCT1-ΔN74-URA3 |
| W1841 | MATa ura3::pGALL-HA3-HCT1-Δcb-URA3 |
| W1842 | MATa clb2::CLB2-myc12-HIS3 ura3::pGALL-HA3-HCT1-Δcb-URA3 |
| W1762 | MATa cdc23::CDC23-myc9-LEU2 ura3::pGALL-HCT1-HA3-URA3 |
| W1763 | MATa cdc23::CDC23-myc9-LEU2 ura3::pGALL-HA3-HCT1-ΔN74-URA3 |
| W1843 | MATa cdc23::CDC23-myc9-LEU2 ura3::pGALL-HA3-HCT1-Δcb-URA3 |
| W2545 | MATa ura3::pGALL-HA3-HCT1-m11-URA3 |
| W2546 | MATa ura3::pGALL-HA3-HCT1-m11-Δcb-URA3 |
| W2547 | MATa cdc23::CDC23-myc9-LEU2 ura3::pGALL-HA3-HCT1-m11-URA3 |
| W2548 | MATa cdc23::CDC23-myc9-LEU2 ura3::pGALL-HA3-HCT1-m11-Δcb-URA3 |
Cells were grown at 25°C in YEP complex medium containing adenine (100 mg/l), tryptophan (200 mg/l) and KH2PO4 (10 mM), and supplemented with raffinose (2%). Expression from the the GAL1, GAL10 or GALL promoter was induced by addition of galactose (2%) to exponentially growing cells or cells arrested either with α-factor (50 ng/ml) or nocodazole (15 µg/ml).
DNA constructs and genetic manipulations
The GAL1-HCT1 (pWS203) and myc18-HCT1 (pWZV109) constructs have been described (Schwab et al., 1997; Zachariae et al., 1998). A triple HA1 epitope or a triple myc epitope sequence was PCR amplified and fused to the 5′ end of HCT1 to construct the GAL1-HA3-HCT1 plasmid (pWS210) or GAL1-myc3-HCT1 plasmid (pWS434). In GAL1-myc3-HCT1-m11 constructs (pWS526), the open reading frame of wild-type HCT1 was replaced by the HCT1-m11 coding region, in which codons for serines and threonines of potential Cdk1 phosphorylation sites were changed to alanine codons (Zachariae et al., 1998). HA3-HCT1 was fused to the GALL promoter (Mumberg et al., 1994) in pWS383. In GALL-HA3-HCT1-m11 (pWS384), wild-type HCT1 was replaced by HCT1-m11. In the GALL-HCT1-HA3 plasmid (pWS534), a triple HA1 epitope sequence was added to the 3′ end of HCT1 fused to the GALL promoter. To construct GALL-HA3-HCT1-ΔN74 (pWS535), a HCT1 fragment lacking the first 74 codons was PCR amplified and used to replace the wild-type HCT1 coding region of pWS383. To construct GAL1-HA3-HCT1-Δcb (pWS920), GALL-HA3-HCT1-Δcb (pWS634) and GALL-HA3-HCT1-m11-Δcb (pWS922), codons 55–59 of HCT1 were deleted by site-directed mutagenesis (Stratagene). All plasmids carrying HCT1 were cut with EcoRV and integrated at the URA3 or TRP1 locus.
To express epitope-tagged CDC20, the myc3 sequence was fused to the 5′ end of the PCR-amplified CDC20 coding sequence and the construct was placed under the control of the GAL1 promoter (GAL1-myc3-CDC20: pWS255). The plasmid was linearized with EcoRI and integrated at the CDC20 locus. To delete the C-box of CDC20, codons 144–148 were replaced by a serine codon using directed mutagenesis. pWS923 and pWS924 are ARSH4 CEN6 URA3 plasmids that contain CDC20 or CDC20-Δcb under the control of the natural CDC20 promoter.
The CLB2-myc12 and CDC23-myc9 constructs have been described (Zachariae and Nasmyth, 1996; Zachariae et al., 1996). HA-tagged cyclin genes are fused at their 3′ ends with the triple HA1 epitope sequence CLB2-HA3 and CLB5-HA3 (Seufert et al., 1995), and CLN2-HA3 (Tyers et al., 1993). To construct GAL10-CLB2-HA3 (pWS942), GAL1-CLB3-HA3 (pWS295), GAL1-CLB5-HA3 (pWS253) and GAL1-PDS1-HA3 (pWS254; PDS1-HA3: Cohen-Fix et al., 1996) plasmids, the open reading frames were PCR amplified and fused to the GAL1 or GAL10 promoter. GAL10-CLB2-Δdb-HA3 (pWS293) was cloned by replacing the wild-type CLB2 coding region of pWS942 with a CLB2 allele lacking codons 17–33 (Schwab et al., 1997). To construct GAL10-CLB2-Δ125 (pWS516), wild-type CLB2 of pWS942 was replaced by a mutated version of CLB2 lacking codons 263–388. This deletion was introduced by removal of a 377 bp EcoRV fragment followed by an insertion of 2 bp with site-directed mutagenesis to restore the correct reading frame. GAL1-HA3-CDC5 (pWS448) and GAL1-HA3-CDC5-ΔN70 (pWS668) plasmids were constructed by addition of a triple HA1 epitope sequence to the 5′ end of PCR-amplified CDC5 and CDC5 without the first 70 codons (Shirayama et al., 1998) and fusion of these genes to the GAL1 promoter. For integration at the URA3 locus, plasmids were linearized with the following restriction enzymes: pWS942, pWS293, pWS295: StuI; pWS516: ApaI; pWS253, pWS254: BsmI; pWS448, pWS668: PstI.
Protein analysis
For preparation of crude extracts, cells were harvested by centrifugation and washed once with ice-cold lysis buffer (150 mM NaCl, 50 mM Tris–HCl pH 7.5, 50 mM NaF, 5 mM EDTA, 0.1% IGEPAL CA-630) containing 60 mM β-glycerophosphate. An equal volume of glass beads was added and cells were broken by shaking in a mixer mill (Retsch) for 5 min at 4°C. Glass beads and cell debris were removed by centrifugation for 10 min followed by centrifugation for another 15 min in a microfuge at 4°C.
For analysis of whole-cell extracts, 2× Lämmli sample buffer was added to the supernatant and the mix was boiled for 5 min. For immunoprecipitations, extracts containing ∼3 mg of total protein were mixed with either α-HA- (12CA5) or α-Myc- (9E10) specific antibodies. After 2 h incubation at 4°C, 50 µl of protein A–agarose were added and the mixtures were incubated for another 2 h at 4°C. Beads were collected by centrifugation and washed three times with lysis buffer. After the last washing step, the beads were resuspended in Lämmli sample buffer and boiled for 10 min.
Western analysis was performed essentially as described by Schwab et al. (1997). Mouse monoclonal antibodies 12CA5 and 9E10 were used for detection of HA- and Myc-tagged proteins, respectively. Cdk1 and Tub2 were probed with specific rabbit antisera.
Acknowledgments
Acknowledgements
We thank W.Zachariae and K.Nasmyth for providing strains and DNA constructs, P.Scheurich for 9E10 antibodies and C.Mann for a Cdk1 antiserum. This work was supported by the Deutsche Forschungsgemeinschaft and Fonds der chemischen Industrie.
References
- Adams A., Gottschling,D.E., Kaiser,C.A. and Stearns,T. (1997) Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Alexandru G., Zachariae,W., Schleiffer,A. and Nasmyth,K. (1999) Sister chromatid separation and chromosome re-duplication are regulated by different mechanisms in response to spindle damage. EMBO J., 18, 2707–2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amon A. (1999) The spindle checkpoint. Curr. Opin. Genet. Dev., 9, 69–75. [DOI] [PubMed] [Google Scholar]
- Amon A., Irniger,S. and Nasmyth,K. (1994) Closing the cell cycle circle in yeast: G2 cyclin proteolysis initiated at mitosis persists until the activation of G1 cyclins in the next cycle. Cell, 77, 1037–1050. [DOI] [PubMed] [Google Scholar]
- Bäumer M., Braus,G.H. and Irniger,S. (2000) Two different modes of cyclin Clb2 proteolysis during mitosis in Saccharomyces cerevisiae. FEBS Lett., 468, 142–148. [DOI] [PubMed] [Google Scholar]
- Blanco M.A., Sánchez-Diaz,A., de Prada,J.M. and Moreno,S. (2000) APCste9/srw1 promotes degradation of mitotic cyclins in G1 and is inhibited by cdc2 phosphorylation. EMBO J., 19, 3945–3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandeis M. and Hunt,T. (1996) The proteolysis of mitotic cyclins in mammalian cells persists from the end of mitosis until the onset of S phase. EMBO J., 15, 5280–5289. [PMC free article] [PubMed] [Google Scholar]
- Burton J.L. and Solomon,M.J. (2000) Hsl1p, a Swe1p inhibitor, is degraded via the anaphase-promoting complex. Mol. Cell. Biol., 20, 4614–4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles J.F., Jaspersen,S.L., Tinker-Kulberg,R.L., Hwang,L., Szidon,A. and Morgan,D.O. (1998) The polo-related kinase Cdc5 activates and is destroyed by the mitotic cyclin destruction machinery in S.cerevisiae. Curr. Biol., 8, 497–507. [DOI] [PubMed] [Google Scholar]
- Cohen-Fix O., Peters,J.-M., Kirschner,M.W. and Koshland,D. (1996) Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev., 10, 3081–3093. [DOI] [PubMed] [Google Scholar]
- Cooper K.F., Mallory, M.J., Egeland,D.B., Jarnik,M. and Strich,R. (2000) Ama1p is a meiosis-specific regulator of the anaphase promoting complex/cyclososme in yeast. Proc. Natl Acad. Sci. USA, 97, 14548–14553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge S.J. (1998) Mitotic arrest: Mad2 prevents sleepy from waking up the APC. Science, 279, 999–1000. [DOI] [PubMed] [Google Scholar]
- Fang G., Yu,H. and Kirschner,M.W. (1998) Direct binding of CDC20 protein family members activates the anaphase-promoting complex in mitosis and G1. Mol. Cell, 2, 163–171. [DOI] [PubMed] [Google Scholar]
- Glotzer M., Murray,A.W. and Kirschner,M.W. (1991) Cyclin is degraded by the ubiquitin pathway. Nature, 349, 132–138. [DOI] [PubMed] [Google Scholar]
- Gmachl M., Gieffers,C., Podtelejnikov,A.V., Mann,M. and Peters,J.-M. (2000) The RING-H2 finger protein APC11 and the E2 enzyme UBC4 are sufficient to ubiquitinate substrates of the anaphase-promoting complex. Proc. Natl Acad. Sci. USA, 97, 8973–8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossberger R., Gieffers,C., Zachariae,W., Podtelejnikov,A.V., Schleiffer,A., Nasmyth,K., Mann,M. and Peters,J.-M. (1999) Characterization of the DOC1/APC10 subunit of the yeast and the human anaphase-promoting complex. J. Biol. Chem., 274, 14500–14507. [DOI] [PubMed] [Google Scholar]
- Hershko A. and Ciechanover,A. (1998) The ubiquitin system. Annu. Rev. Biochem., 67, 425–479. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. (1996) Ubiquitin-dependent protein degradation. Annu. Rev. Genet., 30, 405–439. [DOI] [PubMed] [Google Scholar]
- Jaspersen S.L., Charles,J.F. and Morgan,D.O. (1999) Inhibitory phosphorylation of the APC regulator Hct1 is controlled by the kinase Cdc28 and the phosphatase Cdc14. Curr. Biol., 9, 227–236. [DOI] [PubMed] [Google Scholar]
- Juang Y.-L., Huang,J., Peters,J.-M., McLaughlin,M.E., Tai,C.-Y. and Pellman,D. (1997) APC-mediated proteolysis of Ase1 and the morphogenesis of the mitotic spindle. Science, 275, 1311–1314. [DOI] [PubMed] [Google Scholar]
- King R.W., Peters,J.-M., Tugendreich,S., Rolfe,M., Hieter,P. and Kirschner,M.W. (1995) A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell, 81, 279–288. [DOI] [PubMed] [Google Scholar]
- King R.W., Deshaies R.J., Peters.J.-M. and Kirschner,M.W. (1996) How proteolysis drives the cell cycle. Science, 274, 1652–1659. [DOI] [PubMed] [Google Scholar]
- Kramer E.R., Gieffers,C., Hölzl,G., Hengstschläger,M. and Peters,J.-M. (1998) Activation of the human anaphase-promoting complex by proteins of the CDC20/Fizzy family. Curr. Biol., 8, 1207–1210. [DOI] [PubMed] [Google Scholar]
- Kramer E.R., Scheuringer,N., Podtelejnikov,A.V., Mann,M. and Peters,J.-M. (2000) Mitotic regulation of the APC activator proteins CDC20 and CDH1. Mol. Biol. Cell, 11, 1555–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb J.R., Tugendreich,S. and Hieter,P. (1995) Tetratrico peptide repeat interactions: to TPR or not to TPR? Trends Biochem. Sci., 20, 257–259. [DOI] [PubMed] [Google Scholar]
- Leberer E., Thomas,D.Y. and Whiteway,M. (1997) Pheromone signalling and polarised morphogenesis in yeast. Curr. Opin. Genet. Dev., 7, 59–66. [DOI] [PubMed] [Google Scholar]
- Leverson J.D., Joazeiro,C.A.P., Page,A.M., Huang,H.-K., Hieter,P. and Hunter,T. (2000) The APC11 RING-H2 finger mediates E2-dependent ubiquitination. Mol. Biol. Cell, 11, 2315–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorca T., Castro,A., Martinez,A.-M., Vigneron,S., Morin,N., Sigrist,S., Lehner,C., Dorée,M. and Labbé,J.C. (1998) Fizzy is required for activation of the APC/cyclosome in Xenopus egg extracts. EMBO J., 17, 3565–3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarry T.J. and Kirschner,M.W. (1998) Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell, 93, 1043–1053. [DOI] [PubMed] [Google Scholar]
- Morgan D.O. (1999) Regulation of the APC and the exit from mitosis. Nature Cell Biol., 1, E47–E53. [DOI] [PubMed] [Google Scholar]
- Mumberg D., Müller,R. and Funk,M. (1994) Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res., 22, 5767–5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. (1999) Separating sister chromatids. Trends Biochem. Sci., 24, 98–104. [DOI] [PubMed] [Google Scholar]
- Patton E.E., Willems,A.R. and Tyers, M. (1998) Combinatorial control in ubiquitin-dependent proteolysis: don’t Skp the F-box hypothesis. Trends Genet., 14, 236–243. [DOI] [PubMed] [Google Scholar]
- Peters J.-M. (1998) SCF and APC: the Yin and Yang of cell cycle regulated proteolysis. Curr. Opin. Cell Biol., 10, 759–768. [DOI] [PubMed] [Google Scholar]
- Peters J.-M. (1999) Subunits and substrates of the anaphase-promoting complex. Exp. Cell Res., 248, 339–349. [DOI] [PubMed] [Google Scholar]
- Pfleger K. and Kirschner,M.W. (2000) The KEN-box: an APC recogntion signal distinct from the D box targeted by Cdh1. Genes Dev., 14, 655–665. [PMC free article] [PubMed] [Google Scholar]
- Rudner A.D. and Murray,A.W. (2000) Phosphorylation by Cdc28 activates the Cdc20-dependent activity of the anaphase-promoting complex. J. Cell Biol., 149, 1377–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner A.D., Hardwick,K.G. and Murray,A.W. (2000) Cdc28 activates exit from mitosis in budding yeast. J. Cell Biol., 149, 1361–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab M., Schulze Lutum,A. and Seufert,W. (1997) Yeast Hct1 is a regulator of Clb2 cyclin proteolysis. Cell, 90, 683–693. [DOI] [PubMed] [Google Scholar]
- Seufert W., Futcher,B. and Jentsch,S. (1995) Role of a ubiquitin-conjugating enzyme in degradation of S- and M-phase cyclins. Nature, 373, 78–81. [DOI] [PubMed] [Google Scholar]
- Shirayama M., Zachariae,W., Ciosk,R. and Nasmyth,K. (1998) The polo-like kinase Cdc5p and the WD-repeat protein Cdc20p/fizzy are regulators and substrates of the anaphase-promoting complex in Saccharomyces cerevisiae. EMBO J., 17, 1336–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M., Toth,A., Galova,M. and Nasmyth,K. (1999) APCCdc20 promotes exit from mitosis by destroying the anaphase inhibitor Pds1 and cyclin Clb5. Nature, 402, 203–207. [DOI] [PubMed] [Google Scholar]
- Sigrist S. and Lehner,C.F. (1997) Drosophila fizzy-related down-regulates mitotic cyclins and is required for cell proliferation arrest and entry into endocycles. Cell, 90, 671–681. [DOI] [PubMed] [Google Scholar]
- Smith T.F., Gaitatzes,C., Saxena,K. and Neer,E.J. (1999) The WD repeat: a common architecture for diverse functions. Trends Biochem. Sci., 24, 181–185. [DOI] [PubMed] [Google Scholar]
- Sudakin V., Ganoth,D., Dahan,A., Heller,H., Hershko,J., Luca,F.C., Ruderman,J.V. and Hershko,A. (1995) The cyclosome, a large complex containing cyclin-selective ubiquitin ligase activity, targets cyclins for destruction at the end of mitosis. Mol. Biol. Cell, 6, 185–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsley F.M. and Ruderman,J.V. (1998) Proteolytic ratchets that control progression through mitosis. Trends Cell Biol., 8, 238–244. [DOI] [PubMed] [Google Scholar]
- Tyers M. and Willems,A.R. (1999) One ring to rule a superfamily of E3 ubiquitin ligases. Science, 284, 601–604. [DOI] [PubMed] [Google Scholar]
- Tyers M., Tokiwa,G. and Futcher,B. (1993) Comparison of the S.cerevisiae G1 cyclins: Cln3 may be an upstream activator of Cln1, Cln2 and other cyclins. EMBO J., 12, 1955–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A. (1997) The ubiquitin system. Trends Biochem. Sci., 22, 383–387. [DOI] [PubMed] [Google Scholar]
- Visintin R., Prinz,S. and Amon,A. (1997) CDC20 and CDH1: a family of substrate-specific activators of APC-dependent proteolysis. Science, 278, 460–463. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S., Okayama,H. and Nurse,P. (2000) Fission yeast fizzy-related protein srw1 is a G1-specific promoter of mitotic cyclin B degradation. EMBO J., 19, 3968–3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeong F.M., Lim,H.H., Padmashree,C.G. and Surana,U. (2000) Exit from mitosis in budding yeast: biphasic inactivation of the Cdc28-Clb2 mitotic kinase and the role of Cdc20. Mol. Cell, 5, 501–511. [DOI] [PubMed] [Google Scholar]
- Zachariae W. and Nasmyth,K. (1996) TPR proteins required for anaphase progression mediate ubiquitination of mitotic B-type cyclins in yeast. Mol. Biol. Cell, 7, 791–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachariae W. and Nasmyth,K. (1999) Whose end is destruction: cell division and the anaphase-promoting complex. Genes Dev., 13, 2039–2058. [DOI] [PubMed] [Google Scholar]
- Zachariae W., Shin,T.H., Galova,M., Obermaier,B. and Nasmyth,K. (1996) Identification of subunits of the anaphase-promoting complex of Saccharomyces cerevisiae. Science, 274, 1201–1204. [DOI] [PubMed] [Google Scholar]
- Zachariae W., Schwab,M., Nasmyth,K. and Seufert,W. (1998) Control of cyclin ubiquitination by CDK-regulated binding of Hct1 to the anaphase promoting complex. Science, 282, 1721–1724. [DOI] [PubMed] [Google Scholar]