Abstract
We have investigated the causal factors behind the age-related oxidation of proteins during arrest of cell proliferation. A proteomic approach demonstrated that protein oxidation in non-proliferating cells is observed primarily for proteins being produced in a number of aberrant isoforms. Also, these cells exhibited a reduced translational fidelity as demonstrated by both proteomic analysis and genetic measurements of nonsense suppression. Mutants harboring hyperaccurate ribosomes exhibited a drastically attenuated protein oxidation during growth arrest. In contrast, oxidation was augmented in mutants with error-prone ribosomes. Oxidation increased concomitantly with a reduced rate of translation, indicating that the production of aberrant, and oxidized proteins, is not the result of titration of the co-translational folding machinery. The age-related accumulation of the chaperones, DnaK and GroEL, was drastically attenuated in the hyperaccurate rpsL mutant, demonstrating that the reduced translational fidelity in growth-arrested cells may also be a primary cause for the induction of the heat shock regulon. The data point to an alternative way of approaching the causal factors involved in protein oxidation in eukaryotic G0 cells.
Keywords: aging/chaperones/protein carbonylation/translation accuracy
Introduction
It has been proposed that aging results from random deleterious events, and oxidative damage has been suggested to be one major contributor to the stochastic degeneration of organisms and their cells. Denham Harman was perhaps the first to suggest that free radicals produced during aerobic respiration cause cumulative oxidative damage, resulting in mandatory aging and death (Harman, 1956). This theory gained in credibility with the identification of superoxide dismutase, which provided compelling evidence for in vivo generation of superoxide anions (McCord and Fridovich, 1969). The hypothesis was supported later by experimental data demonstrating that steady-state levels of oxidation-damaged macromolecules, including DNA, protein and lipids, increase with age in all species examined thus far, and that oxidation-modified proteins lose their catalytic activity and structural integrity. Perhaps the strongest support for the theory comes from experiments in which the lifespan of fruitflies was prolonged by overproducing antioxidants (Orr and Sohal, 1994; Parkes et al., 1998), and the identification of gerontogenes (genes whose alteration causes life extension) in Caenorhabditis elegans further supports the notion of a strong correlation between mandatory aging and oxidative damage (Larsen, 1993; Johnson et al., 2000).
The lifespan of unicellular organisms, such as Escherichia coli (Dukan and Nyström, 1998) and Saccharomyces cerevisiae (Longo et al., 1996) undergoing conditional aging (elicited by starvation-induced arrest of proliferation), appears to be limited similarly by the cell’s ability to combat reactive oxygen species (ROS). A large number of E.coli mutants that are specifically hypersensitive to various oxidative agents exhibit a shorter lifespan during reproductive arrest (Benov and Fridovich, 1995; Eisenstark et al., 1996), while the lifespan of growth-arrested wild-type E.coli cells can be increased >100% by omitting oxygen during stasis (Dukan and Nyström, 1999). Like aging of higher eukaryotes, the conditional aging process of growth-arrested E.coli cells is accompanied by increased oxidation modifications, including protein carbonylation and disulfide bond formation (Dukan and Nyström, 1998). Similarly to aged flies (Yan et al., 1998), this age-related oxidation in E.coli targets enzymes of the Krebs cycle and other proteins, including the universal stress protein (Nyström and Neidhardt, 1992, 1994, 1996). Chaperones, translation elongation factors and histone-like proteins were found to be additional targets for oxidation (Dukan and Nyström, 1998, 1999).
The key task of pinpointing the causal factors behind the increased oxidation of macromolecules during aging has proved difficult. Some attempts have been made to correlate oxidation in aging cells with a reduced activity (or abundance) of the antioxidant defense and repair systems. However, these attempts have generated conflicting results. For example, catalases have been demonstrated to either increase or decrease with age depending on the tissues analyzed (Sohal et al., 1995), and in other studies it has been demonstrated that some antioxidant defense proteins may increase while others decrease with age in the same tissues (Ji et al., 1990). In the prokaryotic model system E.coli, the situation is paradoxical rather than conflicting since in a reproductively arrested population of E.coli cells the levels of oxidative defense proteins increase markedly and the population becomes increasingly resistant to external oxidative stresses (Matin, 1991; Hengge-Aronis, 1993). However, the levels of oxidation-damaged proteins in such an E.coli population increase (Dukan and Nyström, 1998, 1999).
Attempts to explain this paradox have been inspired by the ‘rate of living’ hypothesis (e.g. Dukan and Nyström, 1999). This hypothesis states that there is a close correlation between species-specific metabolic rate and lifespan, such that species with lower metabolic rates consume less O2 per gram of tissue and have a longer maximum lifespan potential. Moreover, a factor called the ‘life energy potential’, defined as the product of the specific metabolic rate and the maximum lifespan potential, has been argued to be more or less constant for all species. Therefore, it is argued that the rate of energy consumption is a priori responsible for senescence (reviewed in Sohal, 1976). With the demonstration that respiring mitochondria generate ROS, an updated version of the hypothesis states that a faster rate of respira tion hastens aging by the greater generation of oxygen radicals. The rate of living hypothesis has, in this form, essentially merged with the free radical hypothesis of aging (Beckman and Ames, 1998). When applied to non-proliferating cells, the hypothesis could explain increased oxidation of proteins in such cells simply by the fact that they have an ongoing respiratory activity but little or no ability to replace damaged proteins. Thus, the prediction of the hypothesis is that the higher the rate of respiration, the greater the oxidation damage and the shorter the lifespan.
In this work, we approached this possibility by correlating protein carbonyl levels with total metabolic activity, oxygen consumption and carbon dioxide production in non-proliferating cells arrested by starvation for different nutrients. We found no strict correlation between respiratory activity and protein oxidation. In fact, the condition generating the lowest increase in oxidation in non-proliferating cells (phosphate starvation) exhibited the highest rate of oxygen consumption and cell viability. Instead, using a proteomic/immunochemical approach to analyze protein oxidation, we report that the increased oxidation of proteins in growth-arrested cells is linked intimately to the production of aberrant protein isoforms. Moreover, oxidation is drastically attenuated in mutants with hyperaccurate ribosomes, whereas oxidation is enhanced in mutants with error-prone ribosomes. Our observations have prompted us to hypothesize that oxidation of proteins in non-proliferating cells may occur in the absence of an increased oxidative stress, elevated levels of ROS or a diminished defense system, but may instead be due to an increased concentration of substrates available for oxidative attack. The data also provide a plausible molecular explanation for how the cell keeps the ribosomes free from aberrant components and feedback errors in translation processivity since oxidized proteins are more susceptible to proteolytic degradation (e.g. Starke et al., 1987; Dukan et al., 2000). We discuss how this new information may help to explain the contradicting literature on the role and functionality of oxidative stress defense proteins in age-related oxidation.
Results
Protein oxidation in proliferation-arrested cells is dependent on the missing nutrient
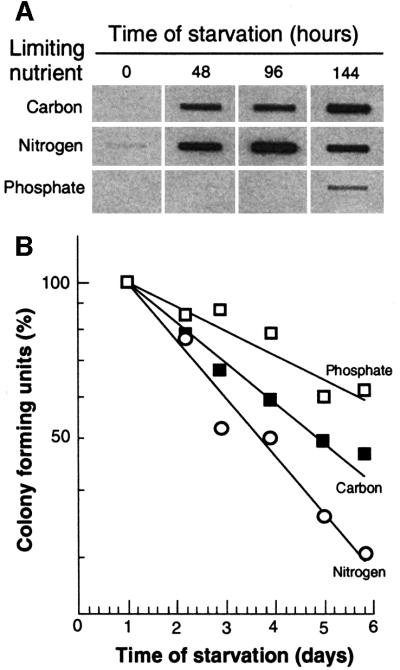
Previous experiments have demonstrated that the levels of oxidized proteins in growth-arrested E.coli cells, like somatic cells of eukaryotic organisms (e.g. Berlett and Stadtman, 1997), increase with the chronological age of the cells (Dukan and Nyström, 1998, 1999). In order to determine whether this oxidation results from growth arrest per se, we subjected the cells to different starvation conditions (see Materials and methods) and assayed the degree of protein carbonylation. Crude protein extracts were obtained during exponential growth and at different times after growth and cell division had ceased due to either carbon, nitrogen or phosphate depletion. A representative dot-blot autoradiogram from these experiments is shown in Figure 1A. As seen in this figure, protein oxidation is dependent on the starvation condition. Most significantly, carbonylation levels in the phosphate-starved cell population were exceedingly low and increased only slightly after prolonged incubation in the starvation regimen (Figure 1A). The highest levels of protein carbonyls were found in nitrogen-starved cells (Figure 1A). Samples were also taken at intervals to determine cell viability (reproductive potential), and we found that the lifespan (culture half-life) of phosphate-starved cells was longer than that of carbon-starved cells, which, in turn, exhibited a somewhat longer lifespan than that of nitrogen-starved cells (Figure 1B; see also Siegele et al., 1993).
Fig. 1. (A) Protein oxidation in non-proliferating cells arrested by starvation for different nutrients. The autoradiogram shows carbonyl contents in wild-type E.coli cells during starvation for carbon, nitrogen or phosphate (as indicated). Equal amounts of protein (1 µg) were loaded in each slot and analysis was repeated three times to confirm reproducibility. Time zero denotes the sample obtained from exponentially growing cells. (B) Viability, measured as colony-forming units, during starvation for different nutrients. The highest colony count obtained immediately after the nutrients were depleted was assigned a value of 100%.
Protein oxidation in non-proliferating cells is not related to the rate of respiration
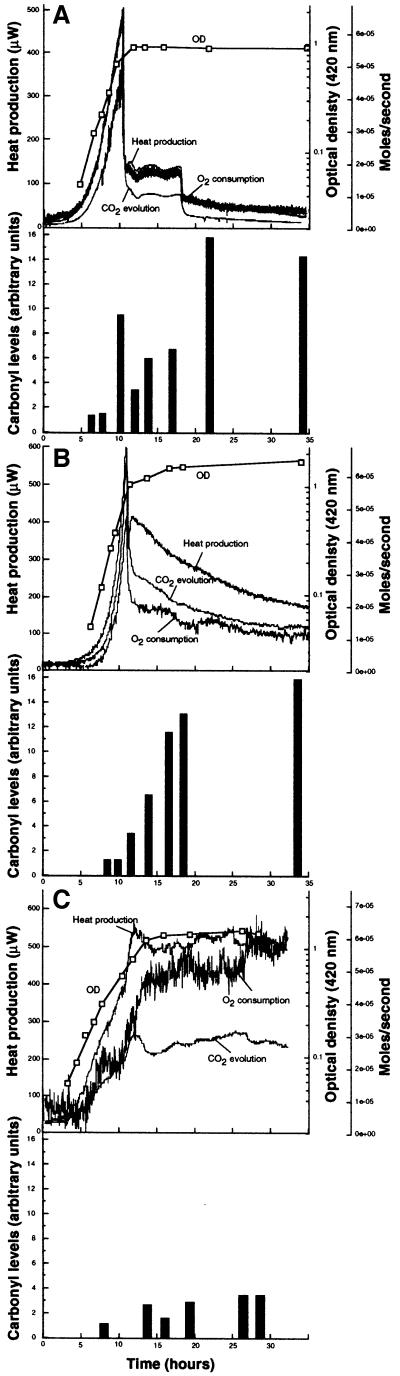
By using the concept of the ‘rate of living’ hypothesis, the gradual increase in protein oxidation during stasis can be explained by an imbalance between macromolecular synthesis and endogenous catabolism such that continued respiration in the absence of self-replacement will gradually result in elevated levels of oxidized macromolecules. We asked whether this hypothesis could explain the differences in oxidation described in the previous section (i.e. if the conditions generating a high oxidation were correlated to a high metabolic activity). We used microcalorimetry combined with on-line gas monitoring to register the physiology of the cells subjected to different starvation conditions. Cells were cultivated in a bioreactor under controlled conditions (stirring, temperature, pH and aeration), and heat dissipation, CO2 production and O2 consumption were measured throughout the experiments. The results are summarized in Figure 2. As seen in this figure, the physiological response of the cells is highly dependent on the starvation condition. When glucose is exhausted from the culture, the metabolic activity falls precipitously to a new level at which the cells use residual acetate excreted into the medium (Holms, 1986; Figure 2A). There is no growth during the period of acetate utilization. Subsequent to acetate depletion, the metabolic activities drop again to very low levels throughout the carbon starvation period studied (Figure 2A). Protein carbonylation increased immediately glucose was exhausted, and the respiratory activity dropped (Figure 2A). During the acetate utilization period, carbonyl levels initially were reduced but subsequently increased again (Figure 2A). Nitrogen starvation also caused an immediate, but less dramatic, decrease in metabolic activities (Figure 2B). Subsequent to this immediate drop, a more gradual reduction in heat production and respiration was observed (Figure 2B). Also, this gradual reduction in metabolic activities occurred concomitantly with a gradual increase in protein carbonyls (Figure 2B). In contrast to carbon- and nitrogen-starved cells, phosphate-starved cells maintained a very high metabolic activity throughout the experiment (Figure 2C; see also Gérard et al., 1999). However, only a modest increase in the levels of oxidized proteins was observed in these cells (Figure 2C). Thus, there is no direct relationship between respiratory activity and protein oxidation in non-proliferating cells of E.coli. In fact, the condition (phosphate starvation) generating the highest levels of respiratory activity was the least effective in causing protein oxidation. Moreover, protein oxidation during carbon starvation occurs as the respiratory activity falls precipitously to very low levels. We wanted to elucidate this apparently paradoxical oxidation further in carbon-starved cells and obtained useful hints as to the causation by using diagnostic proteomic analysis of the pattern of protein carbonylation (see below).
Fig. 2. Correlation between metabolic activity and protein oxidation. Protein carbonyl levels and metabolic activity were analyzed during growth and starvation for carbon (A), nitrogen (B) or phosphate (C). The upper panel in each graph shows growth (measured as OD at 420 nm), heat production, carbon dioxide production and oxygen consumption. The lower panels show protein carbonyl levels quantified using the NIH 1.62 software. All carbonyl values were related to that obtained during exponential growth, which was assigned a value of 1.0.
Protein carbonylation is associated with the production of multiple protein isoforms in carbon-starved cells
It has been demonstrated previously, using immunochemical proteomics, that stasis-induced carbonylation targets specific proteins (Dukan and Nyström, 1998, 1999). We used this assay for the detection of specific carbonylated proteins in carbon-starved cells during the peak in oxidation that occurs as an immediate response to glucose depletion (see Figure 2A). This analysis indicated that carbonylation was strongly associated with protein stuttering (Figure 3). The phenomenon called protein stuttering has been shown to be the result of erroneous incorporation of amino acids into proteins and can be detected on autoradiograms of two-dimensional gels as satellite spots with molecular weights similar to the authentic protein but separated from it in the isoelectric focusing dimension (O’Farrell, 1978; Parker et al., 1978). We subsequently analyzed the pattern of protein carbonylation in cells subjected to H2O2 exposure (Figure 3) and exponentially growing mutant cells (rpsD; Andersson et al., 1982) harboring intrinsically error-prone ribosomes (Figure 3). Interestingly, the pattern of protein oxidation in carbon-starved cells was much more similar to that observed in exponentially growing rpsD mutants than to that in the H2O2-stressed cells (Figure 3). Analysis of the pattern of carbonylation in streptomycin-treated cells and cells lacking the oxidative stress defense regulator OxyR gave similar results (not shown; Dukan and Nyström, 1999), i.e. the pattern of protein carbonylation in carbon-starved cells was mimicked by conditions reducing translation accuracy rather than increasing oxidative stress. These results, together with the fact that aberrant proteins have been reported to be more susceptible to oxidation than their native counterpart (Dukan et al., 2000), prompted us to hypothesize that protein carbonylation in non-proliferating cells could be the result of a reduced translational fidelity in these cells.
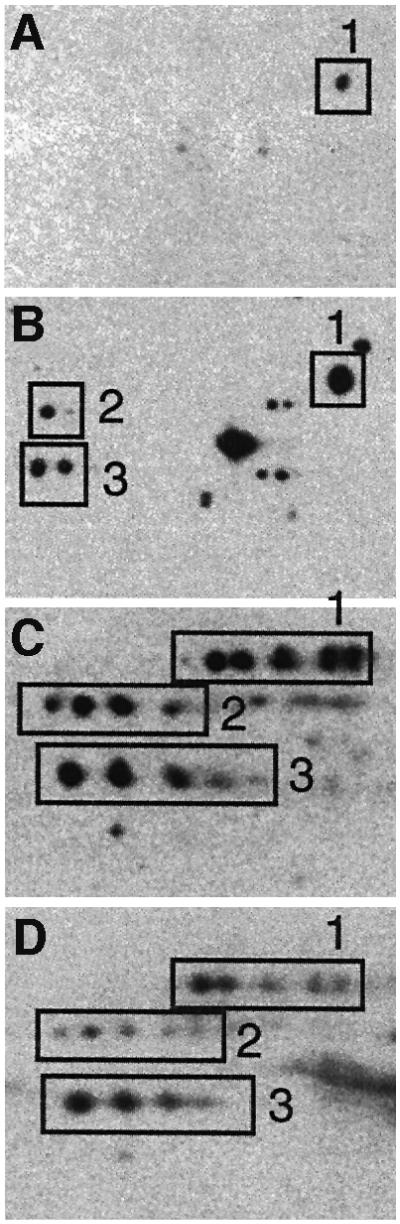
Fig. 3. Correlation between production of aberrant protein isoforms and carbonylation. The pattern of protein carbonylation was determined by two-dimensional western blot immunoassay. The films were obtained after carbonyl immunoassay of exponentially growing wild-type cells (A), exponentially growing wild-type cells exposed to 200 µM H2O2 (B), exponentially growing rpsD mutant (C) and 1 h carbon-starved wild-type cells (D). The autoradiogram depicts the area around the oxidation-sensitive enzyme glutamine synthetase (GlnA; boxed protein no. 1) since three proteins in this area exhibit extensive stuttering.
Translational errors increase in starved non-proliferating cells.
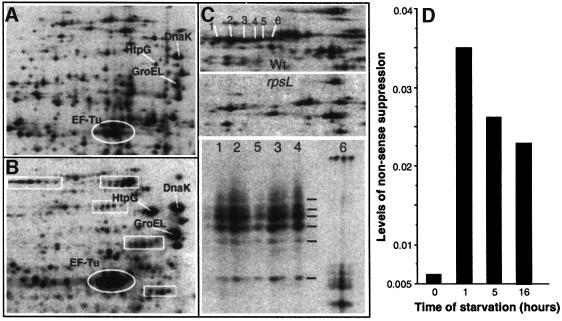
Ribosomal accuracy is known to be growth phase dependent. Both stop codon read-through and shifts in the translation reading frame (frameshifting) have been shown to increase in E.coli cultures upon entry into the stationary phase (Barak et al., 1996; Wenthzel et al., 1998). Using two-dimensional gel electrophoresis, we observed that the carbon starvation conditions used in this study triggered a significant increase in the production of protein isoforms (seen as protein stuttering on the two-dimensional autoradiograms; Figure 4A and B), accompanied by an elevated synthesis of heat shock proteins (Figure 4A and B). To confirm that the protein satellites were indeed protein isoforms rather than new, and different, proteins synthesized in response to starvation, we isolated the satellite spots from the two-dimensional gels and subjected them to V8 protease peptide mapping. We used the method of Cleveland et al. (1977), which allows the identity of a protein to be determined based on its peptide fingerprint produced by partial digestion with the Staphylococcus aureus V8 protease. As seen in Figure 4C, all satellite spots except one generated identical peptides, confirming that the satellites were indeed isoforms of the same protein. Other areas of intense stuttering generated similar results (not shown). It is theoretically possible that the satellites are produced by post-translational modifications (i.e. phosphorylation) rather than by translational errors. However, we found that stuttering was significantly reduced in a strain carrying the rpsL141 allele, which encodes a modified ribosomal S12 protein (Figure 4C). As described by Ruusala et al. (1984), mutant rpsL ribosomes have an increased proofreading activity that leads to an increased translational fidelity compared with the wild-type strain. The rpsL mutations have been demonstrated previously to reduce protein stuttering following asparagine starvation (Parker and Friesen, 1980). Taken together, the results suggest that translational errors increase in response to starvation and that the charged heterogeneity of proteins is a result of mistranslation. In addition to the occurrence of aberrant protein isoforms, we also observed an increased stop codon read-through as cells became starved for glucose (Figure 4D). This nonsense suppression was analyzed using a strain carrying a reporter lacI–lacZ fusion with a nonsense codon in the 5′ end of lacI such that β-galactosidase production is dependent on occasional read-through of this stop codon (see Materials and methods). As seen (Figure 4D), stop codon read-through increased immediately during starvation-induced growth arrest, followed by a gradual decrease.
Fig. 4. Mistranslation in starved, non-proliferating cells. Two-dimensional autoradiograms of (A) growing and (B) starving (1 h glucose starvation) cells obtained after [35S]methionine pulse labeling. The circles show elongation factor, EF-Tu, whereas the heat shock proteins DnaK, GroEL and HtpG are marked with white lines. The boxes mark areas of intense protein stuttering (production of multiple protein isoforms). (C) Protein stuttering in wild-type (upper panel) compared with the rpsL mutant (middle panel). The lower panel shows the V8 protease peptide fingerprints (see Materials and methods) of the protein spots indicated and numbered in the upper panel. (D) In vivo read-through of a nonsense codon in wild-type cells during carbon starvation. The assay for calculating the levels of nonsense suppression is described in Materials and methods. A value of 0.01 indicates that one out of 100 transcripts generates a full-length protein due to nonsense read-through.
Protein oxidation is dictated by the accuracy of the ribosomes
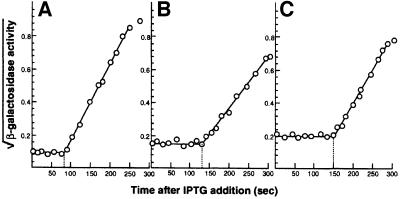
The results presented thus far suggest that there may be a close correlation between protein carbonylation and the degree of translational error in non-proliferating cells. To test this more directly, we assayed protein oxidation in a strain carrying the rpsL141 allele, which results in an increased translational fidelity compared with the wild-type strain. We determined the levels of protein carbonyls in this hyperaccurate mutant, alongside the parental strain and a strain carrying the rpsD allele, which renders the ribosomes error-prone, during exponential growth and entry into stationary phase (glucose starvation). As seen in Figure 5A, protein carbonylation was drastically attenuated in the rpsL mutant and enhanced in the rpsD strain. In addition, the attenuated production of carbonylated proteins in the rpsL mutant could be counteracted by the addition of streptomycin (Figure 5A). However, the reduced oxidation observed in the rpsL mutant apparently is not enough to allow an extended lifespan, since no difference in the survival of wild-type and rpsL mutant could be detected during the first days of stationary phase (not shown). When the kinetics of protein oxidation and metabolic activities were analyzed, we found that the respiratory activity and superoxide dismutase activity were very similar in the wild-type and the rpsL mutant (Figure 5B), indicating that the production of superoxide or its dismutation is not affected by the rpsL allele. We also confirmed that the rpsL mutant exhibits increased translational fidelity by assaying stop codon read-through (Figure 5C). The degree of carbonylation in all strains (wild-type, rpsL and rpsD) and conditions (carbon, nitrogen and phosphate starvation) analyzed was plotted as a function of nonsense suppression, and this exercise demonstrates a close correlation between the two phenomena (Figure 5D). It is known that translational fidelity is linked intimately to translation rates, and the rpsL mutations reduce the rate of peptide elongation whereas the rpsD mutations have the opposite effect (e.g. Kurland et al., 1996). Thus, it is possible that protein oxidation is the result of an increased rate of translation rather than an increased error frequency. For example, an increased elongation rate may overwhelm the co-translational folding machinery and result in increased levels of unfolded and oxidation-sensitive domains. To approach this possibility, we determined the peptide chain elongation rate, as measured by the synthesis time for β-galactosidase, during growth and starvation. As seen in Figure 6, we found that the peptide chain elongation time decreased rather than increased during growth phase transition. The elongation rate corresponds to ∼14 amino acids per second in exponentially growing cells and this rate decreases ∼2-fold during the time of starvation when protein oxidation increases.
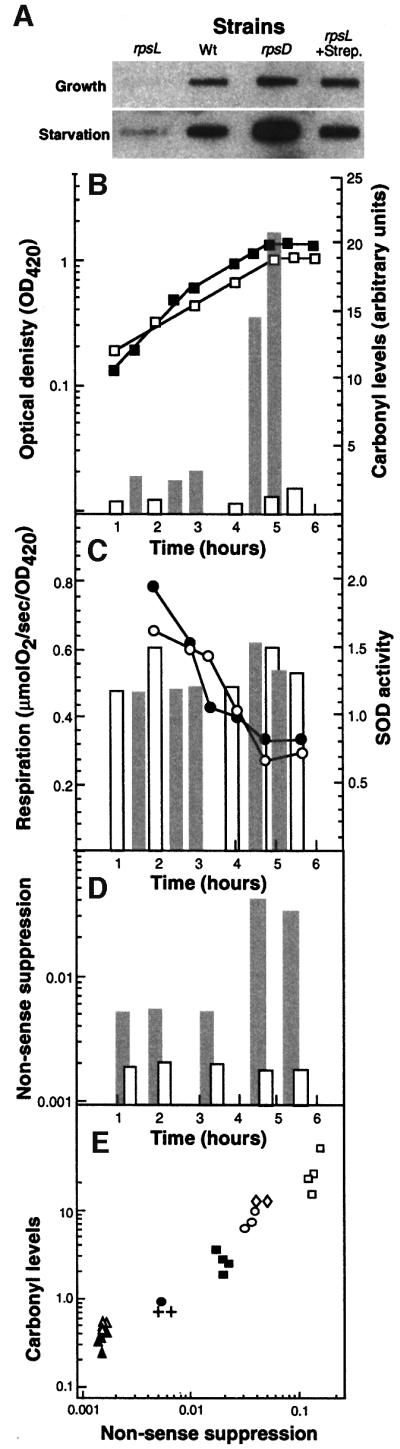
Fig. 5. Effects of translational errors on protein oxidation. (A) Levels of carbonylation during growth and starvation (1 h carbon starved) in wild-type, hyperaccurate (rpsL), with and without 1 µg/ml streptomycin, and sloppy (rpsD) mutants. (B) Carbonylation, respiration, superoxide dismutase activity and nonsense suppression in the rpsL mutant are compared with the wild-type strain. Protein carbonyl contents (bars) and growth (squares) in wild-type (gray bars, closed squares) and the rpsL mutant (open bars, open squares) during growth and entry into stationary phase (glucose starvation). Protein carbonyls were quantified from the autoradiograms using the NIH 1.62 software. (C) Oxygen consumption (circles) and SOD activity (bars) for wild-type (closed circles, gray bars) and rpsL mutant (open circles, open bars) were determined as described in Materials and methods. (D) The frequency of read-through of a nonsense codon in the wild-type (gray bars) and the rpsL mutant (open bars). (E) Correlation between nonsense suppression and protein carbonylation. The data shown are for wild-type (circles), rpsL mutant (triangles) and rpsD mutants (squares) obtained during growth (closed symbols) and carbon starvation (open symbols). Open diamonds and crosses represent values for nitrogen- and phosphate-starved wild-type cells, respectively. In the rpsL mutant, no increase in nonsense suppression was observed in stationary phase. All carbonylation values were compared with the level of the wild-type strain growing exponentially in glucose minimal M9 medium, which was assigned a value of 1.0.
Fig. 6. Determination of peptide chain elongation rates as measured by induction kinetics of β-galactosidase during exponential growth (A) and at 15 (B) and 60 min (C) of glucose starvation. The lac operon was induced by addition of IPTG and, at frequent intervals thereafter, samples were withdrawn and measured for β-galactosidase activity as described in Materials and methods.
Hyperaccurate ribosomes attenuate the production of heat shock chaperones in non-proliferating cells
The heat shock regulon is induced in starved, non-proliferating cells of E.coli (Matin, 1991) and this induction has been demonstrated to be due partly to oxidation-dependent production of aberrant proteins (Dukan and Nyström, 1998). Thus, overproduction of the superoxide dismutase, SodA, mitigates the induction of the heat shock regulon during entry of cells into stationary phase, whereas mutations in katE, katG, sodA and sodB cause a superinduction of the regulon (Dukan and Nyström, 1998). We entertained the idea that the reduced translation accuracy observed in growth-arrested cells could contribute to stasis induction of the heat shock response. We approached this notion by determining the levels of the heat shock chaperones GroEL and DnaK, using western blotting, in wild-type E.coli compared with the rpsL mutant harboring hyperaccurate ribosomes. The result of this analysis is depicted in Figure 7, demonstrating that the levels of the chaperones studied are significantly lower in the rpsL mutant during glucose starvation. We obtained the same results using a groE–lacZ transcriptional fusion, demonstrating that the increased accuracy of the ribosomes affected the transcription of the heat shock genes rather than the stability of the corresponding proteins (Figure 7). Thus, stasis-induced induction of the heat shock regulon can be mitigated by increasing either ribosome fidelity (this study) or superoxide dismutase activity (Dukan and Nyström, 1998).
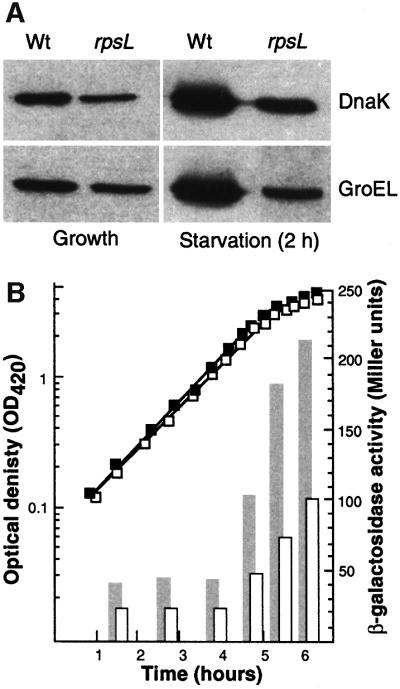
Fig. 7. Effects of translational accuracy on the induction of heat shock proteins during starvation. (A) Analysis of GroEL and DnaK levels in exponentially growing and starving (glucose starvation, 2 h) wild-type and rpsL mutant cells. (B) GroEL promoter activity (bars) during growth and starvation (OD; squares) in wild-type (closed squares, gray bars) and rpsL mutant (open squares, open bars).
Discussion
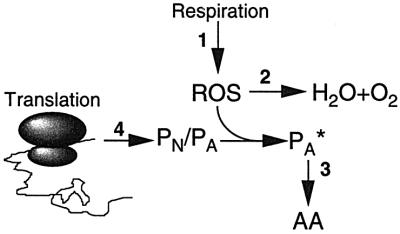
The causal factors behind the elevated oxidation of macromolecules in aging organisms and individual cells of both eukaryotic and prokaryotic origin is an unsolved problem of gerontology. Figure 8 depicts some activities and mechanisms that have been suggested to be involved in the oxidation of proteins during aging in non-proliferating somatic cells. One traditional idea holds that a continued respiration in somatic G0 cells will inevitably increase the levels of oxidized macromolecules because such cells have little ability to dilute any damage with de novo macromolecular synthesis (activity 1; Figure 8). This proposal is in line with the ‘rate of living’ hypothesis. In its simplest form, this model predicts that the higher the metabolic activity (i.e. respiration) in a non-growing system, the higher the protein oxidation and the shorter the lifespan. As described above, our data concerning non-proliferating E.coli cells do not support this notion, since the correlation between respiratory activity and protein carbonylation in growth-arrested cells was poor or non-existent in the set of starvation experiments performed. For example, phosphate-starved cells exhibited the highest rates of respiration during growth arrest, yet protein oxidation was only marginally increased. In addition, the culture half-life was longer in the phosphate-starved cultures despite the continued high metabolic activity in these non-proliferating cells. Again, this result is at odds with the rate of living hypothesis but not the free radical hypothesis of aging since phosphate-starved cells exhibited very low levels of oxidized proteins. We conclude that the rate of respiration in a non-growing aerobic system does not, per se, determine the degree of oxidative damage to the proteins of the system.
Fig. 8. Schematic representation of activities suggested to be involved in causing increased oxidation during aging of cells. Activity 1 is the respiratory activity generating reactive oxygen species (ROS), whereas activity 2 denotes the oxidation defense system, including the superoxide dismutases, catalases and peroxidases. Activity 3 denotes the proteolytic apparatus responsible for degrading oxidation-damaged proteins and peptides, and activity 4 the production of aberrant (PA) and native (PN) proteins by the translation process. PA* denotes aberrant and oxidized proteins and AA amino acids. See text for details.
Another possibility is that the efficiency of the oxidation defense system is diminished as cells grow older (activity 2, Figure 8). In E.coli, this does not appear to be the case, as non-proliferating and old cells in a stationary phase culture become more resistant to both hydrogen peroxide- and superoxide-generating agents (e.g. Matin, 1991). In addition, the notion that increased oxidation in aging cells is due to a diminished ability of the defense systems to counteract such oxidation begs the question of why such systems fail to function in old cells. The suggestion that this is due primarily to free radicals and oxidative damage is, of course, a circular argument, and attempts to correlate oxidation in aging eukaryotic cells with a reduced activity of the antioxidant defense and repair systems have generated conflicting results (see e.g. Beckman and Ames, 1998). However, an increase in the levels of oxidized proteins could be due also to a diminished proteolytic activity in old cells (activity 3; Figure 8). In E.coli, this is unlikely since the induction of both the heat shock regulon and the stringent response network in stationary phase cells would provide these cells with an enhanced proteolytic activity. It should be noted, however, that the regulators and the proteases degrading carbonylated proteins are not yet identified.
The work presented here suggests that there may be another mechanism behind protein oxidation in non-proliferating cells and that this oxidation may occur in the absence of an increased oxidative stress but may instead be due to an increased concentration of substrates available for oxidative attack. We argued that aberrant and misfolded proteins (PA, Figure 8) may be such substrates that are more susceptible to oxidation than their native (PN, Figure 8) counterparts. The concentration of PA increases during growth arrest due to increased translational errors (activity 4, Figure 8). Thus, the potential to oxidize proteins may be exceedingly high in the cell but the process of coupled translation and folding may have evolved to escape such oxidation, e.g. by rapidly hiding oxidation-sensitive domains in the peptides being produced. Any condition causing an increased production of aberrant, misfolded proteins may then also cause elevated levels of oxidized proteins. Based on the results presented here and by Dukan et al. (2000), we can conclude that at least two types of erroneous proteins are modified oxidatively by carbonylation; full-length aberrant isoforms and truncated polypeptides.
The increased oxidation observed during reduced translational fidelity could conceivably be an indirect effect related to the titration of proteases having a role in the degradation of both mistranslated and oxidation-modified proteins. In other words, if both mistranslated and oxidized proteins were degraded by the same proteolytic system, an elevated rate of production of erroneous proteins could sequester the proteases such that the stability of oxidation-modified proteins increases. However, Dukan et al. (2000) found no support for this notion as the half-life of carbonylated proteins was not affected by treatments that increase translation error rates. Moreover, the data presented here and by Dukan et al. (2000) provide an explanation for how E.coli cells may avoid an error catastrophe elicited by an increased production of aberrant proteins. Orgel (1963) suggested that a breakdown in fidelity of information transfer from DNA to protein may contribute to aging, and he presented a conceptual and mathematical account of how an error feedback loop in macromolecular synthesis may cause an irreversible and exponential increase in error levels, leading to an ‘error catastrophe’ (Orgel, 1963). The feedback loop in Orgel’s original model concerned ribosomes and translational accuracy such that errors in the sequences of proteins that themselves functioned in protein synthesis (e.g. ribosomal proteins, elongation factors) might lead to additional errors. Such a positive feedback loop was argued to lead towards an inexorable decay of translational accuracy and, as a result, cellular senescence. The hypothesis is thus based on the assumption that mistranslated proteins can escape degradation and be incorporated into functional (but less accurate) ribosomes. However, it is not clear why such aberrant proteins would cause a decreased accuracy of the ribosome rather than a reduced efficiency, and several shortcomings of the hypothesis have been pointed out (see for example Gallant et al., 1997). Moreover, the data demonstrating that aberrant isoforms are oxidatively modified by carbonylation (this study) and that carbonylated proteins are intrinsically unstable (Dukan et al., 2000) suggest that this provides the cell with a mechanism for avoiding feedback errors in cyclic cascade processes. We believe that the elevated mistranslation of stationary phase cells is the result of ribosomes being increasingly starved of charged tRNAs rather than being intrinsically error-prone.
In addition to the attenuated oxidation of proteins in the rpsL mutant, the induction of heat shock genes was mitigated markedly, indicating that increased translational errors in non-proliferating cells may be part of the mechanism behind the induction of the RpoH regulon. There are two conditions that have now been shown to counteract the induction of heat shock genes during stasis: (i) a reduction in superoxide levels by elevating the levels of SodA and Mn2+ (Dukan and Nyström, 1998); and (ii) an increased translational fidelity (this study). Both the ROS-dependent and translation-dependent mechanisms of inducing the heat shock regulon in stationary phase cells can be explained by the DnaJ/DnaK/GrpE titration model (Bukau, 1993; Gamer et al., 1996). In the case of ROS, these reactive molecules could oxidize native proteins to the extent that they lose their structural integrity and become recognized as substrates for DnaJ, DnaK and GrpE. In addition, DnaK, being an intrinsically oxidation-sensitive protein (Dukan and Nyström, 1998; Tamarit et al., 1998), may, under oxidative conditions, become inactive and incapable of participating in the pathway directing σ32 to the proteolysis apparatus. This would provide a more direct link between oxidative stress and the regulation of the heat shock regulon. Translational errors, on the other hand, are known to generate aberrant unfolded proteins, titrating out the DnaJ, DnaK and GrpE chaperones and allowing σ32 to become sufficiently stable to direct RNA polymerase to heat shock promoters. One important question is whether translational errors and oxidation participate in parallel pathways to produce aberrant proteins, during stasis as described above or work in concert to induce the heat shock regulon during stasis. For example, it is possible that carbonylation tagging of aberrant proteins may augment the signal inducing the heat shock regulon. Mechanistically one could, for example, envisage that a carbonylation-tagged substrate has even higher affinity for the heat shock chaperones (perhaps by exposing more hydrophobic residues due to structural alterations occurring by the addition of carbonyl groups). If so, the attenuated induction of heat shock genes during overproduction of SodA (Dukan and Nyström, 1998) may be the result of a diminished ability of these cells to tag aberrant proteins by means of ROS-dependent carbonylation. Studies are under way to address these questions.
In summary, we propose that oxidation of proteins can occur in the absence of an increased oxidative stress due to an increased concentration of substrates available for oxidative attack. We suggest that aberrant and misfolded proteins may be such substrates and that they accumulate in non-proliferating E.coli due to a reduced translational fidelity. This mechanism of oxidation may also deserve some attention in elucidating the cause of oxidation during aging of higher eukaryotes.
Materials and methods
Chemicals and reagents
Protein assay reagents were from Pierce Inc. All chemicals used for radiolabeling of proteins were from Amersham Corp. X-Omat AR-5 film was purchased from Eastman Kodak Co. The ampholines (Resolyte 4–8) used for two-dimensional electrophoresis were from BDH.
Bacterial strains, media and growth conditions
The E.coli K12 strains used in this study are listed in Table I. Strain ÅF1 was obtained by P1-mediated transduction (Miller, 1972) of the rpsL141 allele into MG1655-Δlac using streptomycin resistance as a marker. ÅF2 is a derivative of ÅF1 that was lysogenized with λpF13-PgroE::lacZ. This vector contains a transcriptional fusion of the groE promoter region to the reporter lacZ gene (Yano et al., 1987). Cultures were grown in Ehrlenmeyer flasks using a rotary shaker at 37°C or, when indicated, in a 1.5 l bioreactor (Chemap) under controlled conditions. The bioreactor temperature was kept at 37°C and the pH adjusted between 7.0 and 7.4. Central palettes fixed on a magnetic stirrer operated the stirring at a fixed rate of 800 r.p.m. The normal growth medium used was M9 (Miller, 1972) with 0.4% glucose (standard medium). When required, thiamine (20 mM) and arginine (0.4 mM) were added. The carbon starvation medium contained 0.05% glucose, the nitrogen starvation medium, 1.87 mM NH4Cl, and the phosphate starvation medium, 0.036 mM KH2PO4 and 0.083 mM N2HPO4. The phosphate starvation medium was buffered with MOPS.
Table I. Escherichia coli strains.
| Designation | Sex, extrachromosomal markers | Chromosomal markers | Origin |
|---|---|---|---|
| MG1655 | F– | wild type | D.Touati |
| MG1655-Δlac | F–Δlac | ||
| Wt/Δ14 | F′proAB lacIZYA | ara argE Δ (lac proAB)gyrA thi | L.A.Isaksson |
| Wt/U4 | F′proAB lacI(UGA)ZYA | ara argE Δ (lac proAB)gyrA thi | L.A.Isaksson |
| S12/Δ14 | F′proAB lacIZYA | as above, rpsL141 | L.A.Isaksson |
| S12/U4 | F′proAB lacI(UGA)ZYA | as above, rpsL141 | L.A.Isaksson |
| S4/Δ14 | F′proAB lacIZYA | as above, rpsD12 | L.A.Isaksson |
| S4/U4 | F′proAB lacI(UGA)ZYA | as above, rpsD12 | L.A.Isaksson |
| ÅF1 | F– | MG1655Δlac rpsL141 | this work |
| ÅF2 | F– | MG1655Δlac rpsL141groELp::lacZ | this work |
General methods
Crude cell extracts were obtained using a 20 K French Pressure Cell (Spectronic Instruments). Culture samples were processed to produce extracts for resolution on two-dimensional polyacrylamide gels by the methods of O’Farrell (1975) with modifications (VanBogelen et al., 1990). Isoelectric focusing (IEF) was performed as described in VanBogelen et al. (1990). Gel electrophoresis was carried out using 11.5% SDS–polyacrylamide gels and the gels were stained regularly with Coomassie Blue. Immunoblotting was performed according to standard procedures, using mouse monoclonal antibodies specific for the relevant protein as primary antibody. For detection, we used the ECL-plus blotting kit (Amersham) using alkaline phosphatase-conjugated anti-mouse IgG as secondary antibody (Sigma). Blots were then exposed to films (Kodak X-Omat). Anti-GroEL and DnaK monoclonal antibodies were from Sigma.
V8 protease peptide mapping
Protein spots were excised directly from two-dimensional gels and re-hydrated as described (Cleveland et al., 1977). Partial digestion of protein spots, after being mashed thoroughly in the gel, was performed using the V8 protease at a concentration of 100 µg/µg of polypeptide. The buffers and gel conditions subsequently used for separating the generated peptides were the same as described in Cleveland et al. (1977).
Determination of nonsense suppression in vivo
Nonsense suppression was determined as described in Andersson et al. (1982) by measuring a stop codon read-through in a lacI–lacZ fusion. The frequency of nonsense suppression was calculated as the levels of β-galactosidase produced by the lacI–lacZ allele, carrying a nonsense codon in lacI, divided by the activity produced by the wild-type allele (transcribed from the same promoter) under the same conditions. Thus, a value of 0.01 indicates that one out of 100 transcripts generates a full-length protein due to nonsense read-through. Both alleles were carried on the F′ factor in the wild-type strain, and the rpsL and rpsD mutants. Measurements of β-galactosidase activity were performed as described (Miller, 1972).
Determination of peptide chain elongation rate
The elongation rate was estimated by measuring the time it takes to make the first β-galactosidase molecule after induction with isopropyl-β-d-thiogalactopyranoside (IPTG; 1 mM) of the lac operon. The initial parabolic portion of the induction curve was linearized by plotting the square root of enzyme levels versus time; the time it takes for the first β-galactosidase molecule to be made can be extrapolated from such a plot (Schleif et al., 1973).
Microcalorimetry and measurement of oxygen consumption
The heat production rate (dQ/dt) was measured with a heat conduction-type multichannel microcalorimeter (Bioactivity Monitor LKB 2277; Thermometric AB, Järfälla, Sweden) equipped with flow-through cells as described in Larsson et al. (1991). The microcalorimeter was operated at 30.0°C at a measuring range of 900 µW. The cultures were pumped in at a rate of 75 ml/h from the growth bioreactor by a peristaltic pump as previously described (Ölz et al., 1993). Calibrations were performed as described by Ölz et al. (1993). Measurements of oxygen consumption were performed using a Cyclobios oxygraph (A.Paar KG, Graz, Austria; Haller et al., 1994). Samples (2.2 ml) were taken out directly from the culture flasks and analyzed immediately.
Gas analysis
Sterile air was pumped continuously into the bioreactor at a rate of 750 ml/min such that oxygen would never limit growth. The airflow out of the bioreactor was analyzed with an acoustic gas analyzer (carbon dioxide and oxygen monitor, type 1308; Brüel and Kjaær, Nærum, Denmark) as described by Christensen et al. (1995).
Carbonylation assays
Detection of carbonylated proteins was performed using the chemical and immunological reagents of the OxyBlot™ Oxidized Protein Detection Kit (Intergen, USA). The carbonyl groups in the protein side chains were derivatized, to 2,4-dinitrophenylhydrazone (DNPH) by reaction with 2,4-dinitrophenylhydrazine. The chemiluminescence blotting substrate (POD) was obtained from Pharmacia/Amersham and used according to instructions provided by the manufacturer. Immobilon-P polyvinylidene difluoride (PVDF) membrane was from Millipore Corp. As described (Dukan and Nyström, 1998), crude protein extracts were obtained during growth and at different times in stationary phase, the extracts reacted with the carbonyl reagent, DNPH, dot-blotted onto PVDF membranes and oxidatively modified proteins were detected with anti-DNP antibodies. In general, 1 and 10 µg of protein were loaded into the slot-blot apparatus and onto two-dimensional gels, respectively.
Superoxide dismutase activity
Superoxide dismutase activity was assayed using the xantine oxidase/cytochrome c method (Imlay and Fridovich, 1991). One unit of superoxide dismutase is defined as that amount of enzyme that inhibits the rate of cytochrome c reduction by 50% at 25°C.
Acknowledgments
Acknowledgements
We thank Professor Leif Isaksson, University of Stockholm, for providing strains and information necessary for this work, and Professor Glenn Björk, University of Umeå, for valuable discussions. This work was sponsored by grants from the Swedish Natural Science Research Council and the Foundation for Strategic Research, Sweden.
References
- Andersson D.I., Bohman,K., Isaksson,L.A. and Kurland,C.G. (1982) Translation rates and misreading characteristics of rpsD mutants in Escherichia coli. Mol. Gen. Genet., 187, 467–472. [DOI] [PubMed] [Google Scholar]
- Barak Z., Gallant,J., Lindsley,D., Kwieciszewki,B. and Heidel,D. (1996) Enhanced ribosome frameshifting in stationary phase cells. J. Mol. Biol., 263, 140–148. [DOI] [PubMed] [Google Scholar]
- Beckman K.B. and Ames,B.N. (1998) The free radical theory of aging matures. Physiol. Rev., 78, 547–581. [DOI] [PubMed] [Google Scholar]
- Benov L. and Fridovich,I. (1995) A superoxide dismutase mimic protects sodA sodB Escherichia coli against aerobic heating and stationary-phase death. Arch. Biochem. Biophys., 322, 291–294. [DOI] [PubMed] [Google Scholar]
- Berlett B.S. and Stadtman,E.R. (1997) Protein oxidation in aging, disease and oxidative stress. J. Biol. Chem., 272, 20313–20316. [DOI] [PubMed] [Google Scholar]
- Bukau B. (1993) Regulation of the Escherichia coli heat-shock response. Mol. Microbiol., 9, 671–680. [DOI] [PubMed] [Google Scholar]
- Christensen L.H., Schulze,U., Nielsen,J. and Villadsen,J. (1995) Acoustic off-gas analyser for bioreactors: precision, accuracy and dynamics of detection. Chem. Eng. Sci., 50, 2601–2610. [Google Scholar]
- Cleveland D.W., Fischer,S.G., Kirschner,M.W. and Laemmli,U.K. (1977) Peptide mapping by limited proteolysis in sodium dodecyl sulfate and analysis by gel electrophoresis. J. Biol. Chem., 84, 655–667. [PubMed] [Google Scholar]
- Dukan S. and Nystrom,T. (1998) Bacterial senescence: stasis results in increased and differential oxidation of cytoplasmic proteins leading to developmental induction of the heat shock regulon. Genes Dev., 12, 3431–3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dukan S. and Nystrom,T. (1999) Oxidative stress defense and deterioration of growth-arrested Escherichia coli cells. J. Biol. Chem., 274, 26027–26032. [DOI] [PubMed] [Google Scholar]
- Dukan S., Farewell,A., Ballesteros,M., Taddei,F., Radman,M. and Nystrom,T. (2000) Protein oxidation in response to increased transcriptional or translational errors. Proc. Natl Acad. Sci. USA, 97, 5746–5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenstark A., Calcutt,M.J., Becker-Hapak,M. and Ivanova,A. (1996) Role of Escherichia coli rpoS and associated genes in defense against oxidative damage. Free Radic. Biol. Med., 21, 975–993. [DOI] [PubMed] [Google Scholar]
- Gallant J., Kurland,C., Parker,J., Holliday,R. and Rosenberger,R. (1997) The error catastrophe theory of aging: point counterpoint. Exp. Gerontol., 32, 333–346. [DOI] [PubMed] [Google Scholar]
- Gamer J., Multhaup,G., Tomoyasu,T., McCarty,J.S., Rudiger,S., Schonfeld,H.J., Schirra,C., Bujard,H. and Bukau,B. (1996) A cycle of binding and release of the DnaK, DnaJ and GrpE chaperones regulates activity of the Escherichia coli heat shock transcription factor σ32. EMBO J., 15, 607–617. [PMC free article] [PubMed] [Google Scholar]
- Gérard F., Dri,A.M. and Moreau,P.L. (1999) Role of Escherichia coli RpoS, LexA and H-NS global regulators in metabolism and survival under aerobic, phosphate-starvation conditions. Microbiology, 145, 1547–1562. [DOI] [PubMed] [Google Scholar]
- Haller T., Ortner,M. and Gnaiger,E. (1994) A respirometer for investigating oxidative cell metabolism: toward optimization of respiratory studies. Anal. Biochem., 218, 338–342. [DOI] [PubMed] [Google Scholar]
- Harman D. (1956) Aging: a theory based on free radical and radiation chemistry. J. Gerontol., 2, 298–300. [DOI] [PubMed] [Google Scholar]
- Hengge-Aronis R. (1993) Survival of hunger and stress: the role of rpoS in early stationary phase gene regulation in E.coli. Cell, 72, 165–168. [DOI] [PubMed] [Google Scholar]
- Holms W.H. (1986) The central metabolic pathways of Escherichia coli: relationship between flux and control at a branch point, efficiency of conversion to biomass and excretion of acetate. Curr. Top. Cell Regul., 28, 69–105. [DOI] [PubMed] [Google Scholar]
- Imlay J.A. and Fridovich,I. (1991) Assay of metabolic superoxide production in Escherichia coli. J. Biol. Chem., 266, 6957–6965. [PubMed] [Google Scholar]
- Ji L.L., Dillon,D. and Wu,E. (1990) Alteration of antioxidant enzymes with aging in rat skeletal muscle and liver. Am. J. Physiol., 258, R918–R923. [DOI] [PubMed] [Google Scholar]
- Johnson T.E., Cypser,J., de Castro,E., de Castro,S., Henderson,S., Murakami,S., Rikke,B., Tedesco,P. and Link,C. (2000) Gerontogenes mediate health and longevity in nematodes through increasing resistance to environmental toxins and stressors. Exp. Gerontol., 35, 687–694. [DOI] [PubMed] [Google Scholar]
- Kurland C.G., Hughes,D. and Ehrenberg,M. (1996) Limitations in translational accuracy. In Neidhardt,F.C. (ed.), Escherichia coli and Salmonella: Cellular and Molecular Biology. ASM Press, Washington, DC, pp. 979–1004.
- Larsen P.L. (1993) Aging and resistance to oxidative damage in Caenorhabditis elegans. Proc. Natl Acad. Sci. USA, 90, 8905–8909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson C., Liden,G., Niklasson,C. and Gustaffson,L. (1991) Calorimetric control of fed-batch cultures of Saccharomyces cerevisae. Bioprocess Eng., 7, 151–155. [Google Scholar]
- Longo V.D., Gralla,E.B. and Valentine,J.S. (1996) Superoxide dismutase activity is essential for stationary phase survival in Saccharomyces cerevisiae. Mitochondrial production of toxic oxygen species in vivo. J. Biol. Chem., 271, 12275–12280. [DOI] [PubMed] [Google Scholar]
- Matin A. (1991) The molecular basis of carbon-starvation-induced general resistance in Escherichia coli. Mol. Microbiol., 5, 3–10. [DOI] [PubMed] [Google Scholar]
- McCord J.M. and Fridovich,I. (1969) Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein). J. Biol. Chem., 244, 6049–6055. [PubMed] [Google Scholar]
- Miller J.H. (1972) Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Nyström T. and Neidhardt,F.C. (1992) Cloning, mapping and nucleotide sequencing of a gene encoding a universal stress protein in Escherichia coli. Mol. Microbiol., 6, 3187–3198. [DOI] [PubMed] [Google Scholar]
- Nyström T. and Neidhardt,F.C. (1994) Expression and role of the universal stress protein, UspA, of Escherichia coli during growth arrest. Mol. Microbiol., 11, 537–544. [DOI] [PubMed] [Google Scholar]
- Nyström T. and Neidhardt,F.C. (1996) Effects of overproducing the universal stress protein, UspA, in Escherichia coli K-12. J. Bacteriol., 178, 927–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Farrell P.H. (1975) High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem., 250, 4007–4021. [PMC free article] [PubMed] [Google Scholar]
- O’Farrell P.H. (1978) The suppression of defective translation by ppGpp and its role in the stringent response. Cell, 14, 545–557. [DOI] [PubMed] [Google Scholar]
- Ölz R., Larsson,K., Adler,L. and Gustafsson,L. (1993) Energy flux and osmoregulation of Saccharomyces cerevisiae grown in chemostats under NaCl stress. J. Bacteriol., 175, 2205–2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orgel L.E. (1963) The maintenance of the accuracy of protein synthesis and its relevance to ageing. Proc. Natl Acad. Sci. USA, 49, 517–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr W.C. and Sohal,R.S. (1994) Extension of life-span by overexpression of superoxide dismutase and catalase in Drosophila melanogaster. Science, 263, 1128–1130. [DOI] [PubMed] [Google Scholar]
- Parker J. and Friesen,J.D. (1980) ‘Two out of three’ codon reading leading to mistranslation in vivo. Mol. Gen. Genet., 177, 439–446. [DOI] [PubMed] [Google Scholar]
- Parker J., Pollard,J.W., Friesen,J.D. and Stanners,C.P. (1978) Stuttering: high-level mistranslation in animal and bacterial cells. Proc. Natl Acad. Sci. USA, 75, 1091–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkes T.L., Elia,A.J., Dickinson,D., Hilliker,A.J., Phillips,J.P. and Boulianne,G.L. (1998) Extension of Drosophila lifespan by overexpression of human SOD1 in motorneurons. Nature Genet., 19, 171–174. [DOI] [PubMed] [Google Scholar]
- Ruusala T., Andersson,D., Ehrenberg,M. and Kurland,C.G. (1984) Hyper-accurate ribosomes inhibit growth. EMBO J., 3, 2575–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleif R., Hess,W., Finkelstein,S. and Ellis,D. (1973) Induction kinetics of the l-arabinose operon of Escherichia coli.J. Bacteriol., 115, 9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegele D.A., Almirón,M. and Kolter,R. (1993) Approaches to the study of survival and death in stationary-phase Escherichia coli. In Kjelleberg,S. (ed.), Starvation in Bacteria. Plenum Press, New York, NY, pp. 151–167.
- Sohal R.S. (1976) Aging changes in insect flight muscle. Gerontology, 22, 317–333. [DOI] [PubMed] [Google Scholar]
- Sohal R.S., Agarwal,S. and Sohal,B.H. (1995) Oxidative stress and aging in the Mongolian gerbil (Meriones unguiculatus). Mech. Ageing Dev., 81, 15–25. [DOI] [PubMed] [Google Scholar]
- Starke P.E., Oliver,C.N. and Stadtman,E.R. (1987) Modification of hepatic proteins in rats exposed to high oxygen concentration. FASEB J., 1, 36–39. [DOI] [PubMed] [Google Scholar]
- Tamarit J., Cabiscol,E. and Ros,J. (1998) Identification of the major oxidatively damaged proteins in Escherichia coli cells exposed to oxidative stress. J. Biol. Chem., 273, 3027–3032. [DOI] [PubMed] [Google Scholar]
- VanBogelen R.A., Hutton,M.E. and Neidhardt,F.C. (1990) Gene–protein database of Escherichia coli K-12: edition 3. Electrophoresis, 11, 1131–1166. [DOI] [PubMed] [Google Scholar]
- Wenthzel A.M., Stancek,M. and Isaksson,L.A. (1998) Growth phase dependent stop codon readthrough and shift of translation reading frame in Escherichia coli. FEBS Lett., 421, 237–242. [DOI] [PubMed] [Google Scholar]
- Yan L.J. and Sohal,R.S. (1998) Mitochondrial adenine nucleotide translocase is modified oxidatively during aging. Proc. Natl Acad. Sci. USA, 95, 12896–12901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano R., Imai,M. and Yura,T. (1987) The use of operon fusions in studies of the heat-shock response: effects of altered σ32 on heat-shock promoter function in Escherichia coli. Mol. Gen. Genet., 207, 24–28. [DOI] [PubMed] [Google Scholar]