Abstract
Proteins of Gram-negative bacteria destined to the extracellular milieu must cross the two cellular membranes and then fold at the appropriate time and place. The synthesis of a precursor may be a strategy to maintain secretion competence while preventing aggregation or premature folding (especially for large proteins). The secretion of 230 kDa filamentous haemagglutinin (FHA) of Bordetella pertussis requires the synthesis and the maturation of a 367 kDa precursor that undergoes the proteolytic removal of its ∼130 kDa C-terminal intramolecular chaperone domain. We have identified a specific protease, SphB1, responsible for the timely maturation of the precursor FhaB, which allows for extracellular release of FHA. SphB1 is a large exported protein with a subtilisin-like domain and a C-terminal domain typical of bacterial autotransporters. SphB1 is the first described subtilisin-like protein that serves as a specialized maturation protease in a secretion pathway of Gram-negative bacteria. This is reminiscent of pro-protein convertases of eukaryotic cells.
Keywords: autotransporter protein/filamentous haemagglutinin/precursor maturation/protein secretion/subtilisin-like protein
Introduction
The addressing of proteins to their specific compartment is a fundamental cellular process that involves complex molecular signals and machineries. In Gram-negative bacteria, proteins destined for the cell surface or the extracellular milieu have to be transported through two distinct membranes (the cytoplasmic and outer membranes) separated by the periplasm. Therefore, these organisms have developed sophisticated protein translocation systems. Schematically, these protein secretion pathways can be divided into Sec-dependent and Sec-independent pathways. The Sec-independent secretory proteins have no cleavable N-terminal signal peptides and are transported across the envelope in one step (Fath and Kolter, 1993; Plano et al., 2001). Conversely, the secretory proteins synthesized with N-terminal signal peptides cross the cytoplasmic membrane via the ubiquitous Sec machinery and have their signal peptide proteolytically removed by a signal peptidase in the periplasm (Pugsley, 1993). They are then translocated across the outer membrane by one of several transport systems. These include the autotransporter systems independent of specific accessory proteins (Henderson et al., 1998), the two-partner secretion (TPS) pathway involving one accessory protein (Jacob-Dubuisson et al., 2001), the general secretory pathway (Sandkvist, 2001) and some of the type IV secretion systems (Christie, 2001), which make use of multi-component machineries.
The TPS pathway appears to be dedicated to the secretion of very large proteins (>100 kDa) in various bacterial genera. Following Sec-dependent export across the cytoplasmic membrane (Grass and St Geme III, 2000), these secretory proteins cross the outer membrane via specific transporters of the TpsB family (for component B of the TPS pathway, component A being the secreted protein). The secretion of filamentous haemagglutinin (FHA) by Bordetella pertussis represents a model system for the TPS pathway. FHA is the major adhesin of B.pertussis produced and secreted in large amounts despite its size of ∼220 kDa (Locht et al., 1993). Its secretion depends on an outer membrane protein called FhaC (Jacob-Dubuisson et al., 2001). As the prototype of the TpsB family, FhaC forms channels and folds into a transmembrane β-barrel, which probably serves as an FHA-specific secretion pore in the outer membrane (Jacob-Dubuisson et al., 1999; Guédin et al., 2000).
FHA derives from an even larger, 367 kDa precursor called FhaB (Domenighini et al., 1990; Renauld-Mongénie et al., 1996). fhaB expression is positively regulated by the two-component system, BvgAS, which controls the virulence regulon in B.pertussis (Akerley and Miller, 1996). The secreted mature form of FHA corresponds approximately to the N-terminal two-thirds of FhaB. The function of the C-terminal one-third of the precursor is not fully understood, but it has been proposed to act as an intramolecular chaperone preventing premature folding of the secreted protein (Renauld-Mongénie et al., 1996).
The role of this proteolytic maturation, the nature of the protease and the cellular compartment in which precursor cleavage occurs are unknown. Here we report the identification of a protease, called SphB1, which is specifically involved in the maturation of FhaB. SphB1 is a member of the autotransporter superfamily of Gram-negative bacteria (Henderson and Nataro, 2001).
Results
Identification of a protease involved in the extracellular release of FHA
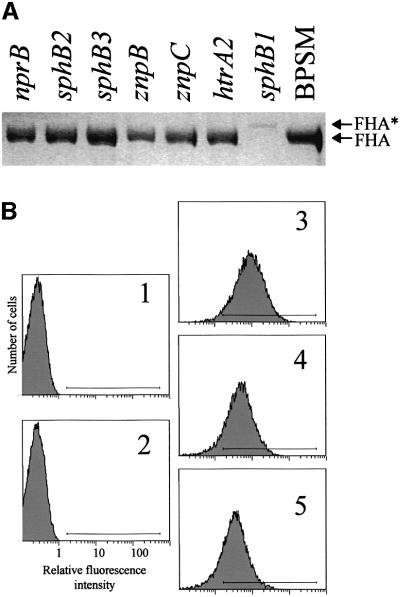
Since FHA derives from the precursor FhaB, we reasoned that this maturation might involve a specific protease that is yet to be identified. Scanning of the B.pertussis genomic sequences (www.sanger.ac.uk/Projects/B-pertussis) was therefore performed to identify open reading frames potentially coding for proteases. Genes encoding homologues of housekeeping proteases such as aminopeptidases, oligopeptidases, Lon, ClpP, HslUV, FtsH and signal peptidases were found but not analysed further. In addition, several genes encoding other putative proteases were identified: two degP/htrA homologues; a tsp homologue; three metalloprotease genes nprB, znpB and znpC; and three serine protease genes sphB1, sphB2 and sphB3. We attempted to generate null alleles for each of these nine genes by the targeted insertion of an integrational plasmid (Antoine et al., 2000). Mutants were obtained for htrA2, nprB, znpB, znpC, sphB1, sphB2 and sphB3 but not for htrA1 and tsp. The culture supernatants of the seven B.pertussis mutants were analysed by SDS–PAGE. The sphB1 mutant called BPLC1 (sphB1′–′lacZ) was deficient in FHA secretion (Figure 1A), suggesting that the product of sphB1 is involved in the extracellular release of FHA. However, a slightly larger protein present in low amounts in the culture supernatants of BPLC1 (FHA* in Figure 1A) was found to be an FHA derivative by immunoblot analyses (not shown). FHA secretion was not compromised in any of the other six mutants (Figure 1A). The secretion of pertussis toxin, haemolysin/adenylate cyclase and pertactin was not affected in any of the strains tested (data not shown), attesting at least some degree of specificity of the sphB1 mutation on the extracellular release of FHA. It should be noted that sphB1 is the only one of the seven genes to be positively regulated by BgvAS (Antoine et al., 2000).
Fig. 1. FHA secretion and cell surface exposure in B.pertussis parental and protease mutant strains. (A) Identical volumes (20 µl) of non-concentrated supernatants from each culture at late logarithmic phase of growth were analysed by SDS–PAGE. The gel was stained with Coomassie Blue. BPSM is the parental strain, and each derived strain is denoted by its genotype relative to BPSM. FHA and FHA* represent the positions of mature FHA and a slightly larger derivative, respectively. (B) BPGR4 (fhaB null, panel 1), BPEC (fhaC null, panel 2), BPSM (parental strain, panel 3), BPLC5 (sphB1 null, panel 4) and BPLC7 (S412→A sphB1 mutant, panel 5) were incubated with the 12.6F8 anti-FHA monoclonal antibody followed by an anti-mouse FITC conjugate. The fluorescent cells were detected by flow cytometry, with 20 000 events counted for each sample. The fluorescence threshold (left end of the horizontal bar) was set such that 99% of non-labelled cells had intensities of autofluorescence below the threshold value. A representative experiment is shown, with percentages of fluorescent cells of 0.14% for BPGR4, 0.13% for BPEC, 87.8% for BPSM, 77.3% for BPLC5 and 70.9% for BPLC7.
To minimize the chance of a polar effect, we created an unmarked sphB1 null mutant by allelic exchange. The resulting strain, called BPLC5, was found to have the same phenotype as BPLC1 regarding FHA secretion (not shown). In addition, we created a strain in which the predicted catalytic Ser of SphB1 was replaced by Ala (Ser412→Ala, with residue #1 being the putative initiator Met). A gene fragment carrying this mutation was introduced by allelic exchange into BPLC5, thereby generating a complete chromosomal copy of sphB1 with a single point mutation. The resulting strain, called BPLC7, produced SphB1 (see below) but exhibited the same defect as BPLC5 (sphB1 null) for FHA secretion (not shown).
To determine whether FHA is nevertheless translocated to the cell surface in the sphB1 mutants, surface immunolabelling of the bacteria was performed using monoclonal antibodies directed against FHA and followed by flow cytometry analyses. The parental and sphB1 mutant strains were labelled, in contrast to BPGR4 (fhaB null) or BPEC (fhaC null), as shown in Figure 1B for the 12.6F8 antibody. Similar results were obtained using the other antibodies (not shown). Thus, translocation of FHA to the cell surface does not require SphB1.
In the culture supernatants of BPLC1 (sphB1′–′lacZ) and BPLC5 (sphB1 null), a slightly larger FHA derivative was present in low amounts (FHA* in Figure 1A). To determine whether it results from inefficient processing of the FHA precursor at a cleavage site distinct from that used in the parental strain, FHA and FHA* were purified from culture supernatants of BPSM (parental) and BPLC5 (sphB1 null), respectively, and analysed by mass spectrometry. The molecular mass determined for FHA was 232 760 ± 558 Da (mean ± SD) and that of FHA* was 246 954 ± 386 Da, confirming that FhaB was not processed at the same site in the parental and mutant strains. The mass measured for FHA localizes the cleavage site of FhaB in the sequence Pro–Leu–Phe–Glu–Thr–Arg–Ile– Lys–Phe–Ile–Asp, which corresponds to residues 2292– 2302 of FhaB after the removal of its signal peptide.
Role of SphB1 in the processing of FhaB
The most likely reason for the FHA secretion defect of the sphB1 mutant strains is that the proteolytic maturation of FhaB into FHA is compromised in the absence of SphB1. Therefore, we attempted to detect FhaB by immunoblot analyses of cellular extracts of the parental and mutant strains with anti-FHA antibodies. Cell-associated FHA was present in BPSM (parental strain), together with a major species of lower mass (FHAd) and a smear of smaller protein bands, most likely resulting from proteolytic degradation (Figure 2, lane 1). These molecular species were absent from the fhaB null strain BPGR4 (Figure 2, lane 2) and present in very low amounts in the fhaC null strain BPEC (Figure 2, lane 3), in agreement with previous observations that cell-associated forms of FHA are heavily proteolysed (Jacob-Dubuisson et al., 1997). In BPLC5 (sphB1 null) and BPLC7 (Ser412→Ala sphB1 mutant) extracts, a form of high molecular mass that probably corresponds to FhaB was detected (Figure 2, lanes 4 and 5). In addition, an immunoreactive species slightly larger than FHA and probably corresponding to FHA* appeared to accumulate, as well as FHAd. As with BPSM, a smear of smaller proteins was also found. These patterns suggest an extensive proteolytic degradation of cell-associated FhaB, probably initiated at preferential cleavage sites. That FhaB was absent from the parental extracts indicates that its processing is affected by the absence of an active SphB1.
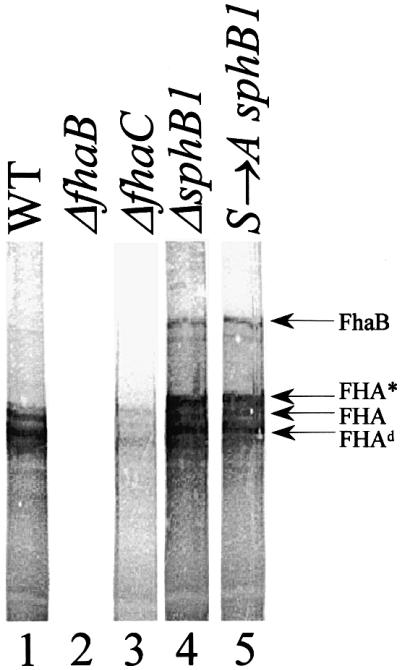
Fig. 2. Detection of cell-associated FhaB. Cellular lysates of BPSM [wild type (WT), lane 1], BPGR4 (ΔfhaB, lane 2), BPEC (ΔfhaC, lane 3), BPLC5 (ΔsphB1, lane 4) and BPLC7 (S→A sphB1, lane 5) were subjected to SDS–PAGE and immunoblotting with a mix of the anti-FHA F1, F4 and F5 antibodies. FhaB indicates the position of the FHA precursor and FHA and FHA* are as in Figure 1. Fhad represents a major degradation product of FhaB.
To corroborate this notion, we performed an in vivo pulse–chase experiment to follow the maturation of FhaB into FHA over time. In BPSM, FhaB was the major species at the beginning of the chase and was gradually chased into FHA to disappear totally after 60 min (Figure 3A). Both proteins were absent from fhaB null BPGR4 (Figure 3B). In contrast, in BPEC (fhaC null), BPLC5 (sphB1 null) and BPLC7 (Ser412→Ala sphB1 mutant), no processing of FhaB was detected over 60 min (Figure 3C–E). Thus, the kinetics of FhaB processing is affected in the sphB1 mutants, demonstrating that SphB1 is involved in the proteolytic maturation of FhaB. That this maturation also depends on FhaC, which transports FHA across the outer membrane, further indicates that it is a late step in the secretion pathway probably occurring at the cell surface.
Fig. 3. Kinetics of FhaB maturation. Pulse labelling was performed with BPSM (WT, A), BPGR4 (ΔfhaB, B), BPEC (ΔfhaC, C), BPLC5 (ΔsphB1, D) and BPLC7 (S→A sphB1, E). The cells were pulse-labelled for 20 min and chased for the indicated times. The proteins were precipitated and analysed by SDS–PAGE and autoradiography.
Characterization of SphB1
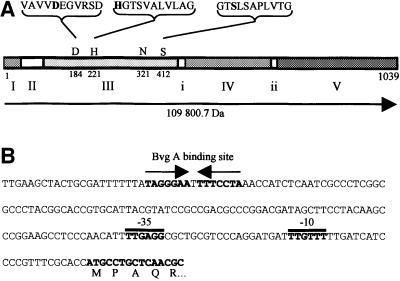
SphB1 presents 24% identity with the Serratia marcescens SSP-h1 serine protease (Ohnishi et al., 1997; Antoine et al., 2000). Other homologues include the S.marcescens PrtS (formerly SSP) (Miyazaki et al., 1989), Ssa1 of Pasteurella haemolytica (Lo et al., 1991) and the Pseudomonas fluorescens SspA and SspB (Kawai et al., 1999). The B.pertussis sphB1 sequence released on the Sanger Center web site was confirmed by resequencing a 3600 bp region encompassing sphB1. This sequence has been submitted to the DDBJ/EMBL/GenBank database under accession No. AJ318229 (BPE318229). The 3117 bp open reading frame of sphB1 is separated by >300 and 400 bp from the most likely upstream and downstream genes encoding a phospho-enolpyruvate carboxylase and an acyl-CoA dehydrogenase, respectively, suggesting that it is most likely in a monocistronic operon. A possible initiation codon was found that initiates a protein of 1039 residues ending with a Tyr. Potential promoter regions and a BvgA binding site were found close to this Met codon (Figure 4), in agreement with the observation that sphB1 is regulated by the global virulence control system BvgAS (Antoine et al., 2000). The presumed initiator Met is followed by a positively charged segment and a hydrophobic segment that are both typical of signal peptides, although no signal peptide cleavage site was readily predicted (www.cbs.dtu.dk/services/SignalP). Alternat ively, SphB1 might be a lipoprotein, as its putative signal peptide presents a Leu–Ala–Ala–Cys motif matching the consensus sequence of bacterial lipoproteins, in which Cys is the site of lipid modification (Sankaran and Wu, 1994). Immediately after the putative signal peptide (I in Figure 4), SphB1 contains a stretch of 14 Gly followed by a 60-residue-long Pro-rich region (II in Figure 4). This region is followed by a 350-residue long domain (III in Figure 4) with a putative Asp–His–Ser catalytic triad characteristic of subtilisin-like proteins according to the Prosite database (www.expasy.ch). The putative protease domain is followed by a 200-residue-long region conserved among SphB1 homologues (domain IV in Figure 4) and then by a predicted β-sheet-rich domain homologous to the C-terminal β-barrel domain of autotransporters (domain V in Figure 4). Thus, SphB1 has the main features of autotransporters with a ‘passenger’ domain and a C-terminal outer membrane ‘transporter’ domain (Henderson et al., 1998).
Fig. 4. Schematic representation of SphB1 and the promoter region of its gene. (A) The protein is represented in a linear form based on the amino acid sequence derived from the sphB1 gene. The precursor has a calculated mass of 109 800.7 Da (horizontal arrow). The areas numbered I–V denote the signal sequence region (I), the proline-rich region (II), the subtilisin-like domain (III), a 200-residue domain of unknown function (IV) and the β-barrel domain (V). According to the autotransporter nomenclature, the passenger domain would consist of regions II–IV and the transporter domain of region V. The catalytic Asp184–His221–Ser412 triad is shown in bold letters with the sequences surrounding each of the three catalytic residues. These sequences match the consensus sequences defined by ProSite for 10/11, 10/11 and 11/11 residues, respectively. Asn321 is the conserved oxyanion hole residue (Siezen and Leunissen, 1997). Probable linker regions between the domains, as suggested by multiple sequence alignments of SphB1 and 16 homologues (using the Clustal program available in the LaserGene package), are denoted by i and ii. (B) Nucleotide sequence of the 5′ portion of the gene. Putative promoter regions (denoted –10 and –35) and a potential BvgA binding site are indicated. The predicted start of the protein is shown with the first five aminoacyl residues.
Cellular localization of SphB1
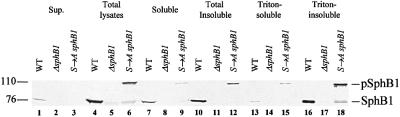
To detect SphB1 in B.pertussis, an antiserum was raised against a SphB1 peptide predicted to be antigenic. This peptide sequence corresponds to residues 600–614 of the protein. Fractionation experiments were performed on BPSM (parental strain), BPLC5 (sphB1 null) and BPLC7 (Ser412→Ala sphB1 mutant). Culture supernatants, as well as soluble and insoluble fractions following cell lysis, were analysed (Figure 5). The insoluble fractions were further fractionated by differential solubilization into Triton X-100-soluble and -insoluble fractions. SphB1 migrated as ∼75 and 105 kDa proteins in the BPSM (parental strain) and BPLC7 (Ser412→Ala sphB1 mutant) fractions, respectively (Figure 5), raising the possibility that SphB1 is responsible for its own maturation, like several autotransporter proteases (Pohlner et al., 1987; Henderson et al., 1998). As expected, SphB1 was not detected in any of the BPLC5 (sphB1 null) fractions.
Fig. 5. Distribution of SphB1 in B.pertussis cellular fractions. Proteins of BPSM (WT), BPLC5 (ΔsphB1) and BPLC7 (S→A sphB1) were separated between supernatants (Sup., lanes 1–3) and cell lysates (lanes 4–6). These lysates were further fractionated between soluble and insoluble fractions by ultracentrifugation (lanes 7–9 and 10–12, respectively). The insoluble fractions were then extracted with Triton X-100 to separate the Triton-soluble and -insoluble fractions (lanes 13–15 and 16–18, respectively). Twenty microlitres of 10-fold concentrated culture supernatants were loaded in lanes 1–3. The proteins loaded in lanes 4–12 were extracted from 0.4 OD600 equivalents of bacterial cells, while the proteins loaded in lanes 13–18 were extracted from 1.2 OD600 equivalents of bacterial cells. Samples were analysed by immunoblotting using the anti-SphB1 antiserum. pSphB1 and SphB1 denote the precursor and mature forms of the protein, respectively. The masses in kDa of the molecular markers are shown at the left.
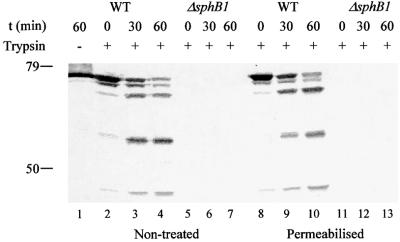
Mature SphB1 was released partly into the extracellular milieu, but was also found in the Triton X-100-insoluble fractions that usually contain outer membrane proteins, while the precursor form was essentially found in the Triton X-100-insoluble fractions. To determine whether cell-associated SphB1 was exposed at the cell surface, immunolabelling of BPSM (parental strain), BPLC5 (sphB1 null) and BPLC7 (Ser412→Ala sphB1 mutant) was performed using the anti-SphB1 antiserum followed by flow cytometry analyses. No surface labelling was observed for any of the three strains (not shown), suggesting that the protein might not be present at the surface. Alternatively, the peptide recognized by the antiserum might not be accessible in the folded protein. In a further attempt to detect SphB1 at the bacterial cell surface, BPSM (parental strain) and BPLC5 (sphB1 null) were subjected to trypsin digestion of surface proteins with or without prior permeabilization of the outer membrane. The proteins were analysed by immunoblotting with the anti-SphB1 antiserum. Mature SphB1 from intact BPSM cells was digested into smaller species over time using trypsin, and the same proteolytic fragments were present in the permeabilized cells (Figure 6). This suggests that SphB1 exposes several sites at the cell surface that are cleaved by externally added trypsin. This localization is compatible with the notion that SphB1 processes the precursor of FHA as it reaches the cell surface, after its translocation across the outer membrane via FhaC.
Fig. 6. Trypsin digestion of SphB1 at the cell surface. BPSM (WT, lanes 1–4 and 8–10) and BPLC5 (ΔsphB1, lanes 5–7 and 11–13) cells were treated with trypsin with (lanes 8–13) or without (lanes 2–7) a prior Tris–EDTA treatment to permeabilize the outer membrane. In lane 1, BPSM cells were mock treated for 1 h. At the indicated times, aliquots were withdrawn and trypsin was inactivated with AEBSF. The proteins were precipitated and analysed by immunoblotting with the anti-SphB1 antiserum. The masses in kDa of the molecular markers are indicated at the left.
Self-processing of SphB1
SphB1 itself appears to derive from a larger precursor by the proteolytic cleavage of an ∼35 kDa fragment. Therefore, we sought to determine the region of the protein where the cleavage occurs. Triton X-100-insoluble proteins of BPSM (parent), BPLC5 (sphB1 null) and BPLC7 (Ser412→Ala sphB1 mutant) were separated by preparative SDS–PAGE, and the gel was stained with colloidal Coomassie Blue G-250. Protein bands most likely corresponding to the mature and precursor forms of SphB1 were present at the expected positions in the BPSM (parent) and BPLC7 (Ser412→Ala sphB1 mutant) extracts, respectively (Figure 7), and absent from BPLC5 (sphB1 null). These protein bands were excised and analysed by mass fingerprinting following tryptic digestion. Fifteen matching peptides were found for the mature form of SphB1 from BPSM (parent). The first N-terminal peptide was T142DFVTPTGGPFFAK155, and the peptide found closest to the C-terminus of the protein was L644DYR PQAVYLTMR656. Most of these peptides were also present in the tryptic digests of the precursor form of SphB1 from BPLC7. In addition, 10 tryptic peptides matching SphB1 between residues 718 and 995 were found for the precursor only. Therefore, the maturation of SphB1 involves the removal of its C-terminal β-domain by autoproteolysis, and the cleavage site of SphB1 is probably located between residues 656 and 718, which includes a putative linker region joining the C-terminal β-barrel domain to the rest of the protein (ii in Figure 4). Following maturation, the passenger domain appears to be partly released and partly associated with the outer membrane (Figure 5).
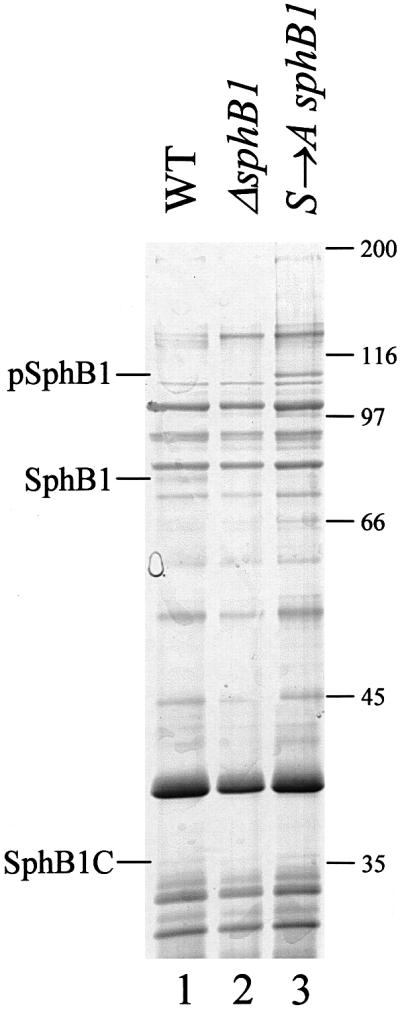
Fig. 7. Detection of SphB1 in Triton-insoluble membrane fractions. Triton X-100-insoluble proteins of BPSM (WT, lane 1), BPLC5 (ΔsphB1, lane 2) and BPLC7 (S →A sphB1, lane 3) were separated by SDS–PAGE, and the gel was stained with colloidal Coomassie Blue G-250. Protein bands corresponding to the mature and precursor forms of SphB1 and to its C-terminal domain, denoted SphB1, pSphB1 and SphB1C, respectively, were analysed by mass fingerprinting. The masses in kDa of the molecular markers are indicated at the right.
No protein band that might correspond to the C-terminal domain of SphB1 after maturation was readily apparent in the 30–40 kDa mass range. Nevertheless, we excised several bands from the gel corresponding to 30–40 kDa proteins of BPSM (parent) and BPLC7 (Ser412→Ala sphB1 mutant), and analysed each of them by mass fingerprinting. A protein corresponding to the C-terminus of SphB1 (SphB1C in Figure 7) was found in the BPSM extract, as eight tryptic peptides matched SphB1 between residues 796 and 995. No peptide matching SphB1 was detected for the protein band with an apparently similar mass in the BPLC7 extract. Thus, the C-terminal domain of SphB1 is found in the outer membrane fraction after the self-maturation of the protein. Incidentally, these analyses also identified the C-terminal domains of two B.pertussis autotransporters, Tcf and BrkA (Fernandez and Weiss, 1994; Finn and Stevens, 1995), in both BPSM and BPLC7, which indicates that the proteolytic maturation of these proteins from their respective precursors occurs independently from SphB1.
Discussion
Post-translational modification in the late compartments of the secretory pathway is essential for the maturation of many secreted proteins and peptides in both eukaryotic and prokaryotic cells. Limited endoproteolysis of precursors by specialized proteases is a widespread process within secretory pathways. In eukaryotes, the maturation of hormones and many other secreted proteins is performed by pro-protein convertases that belong to the superfamily of subtilisin-like proteases (Nakayama, 1997; Bergeron et al., 2000). In bacteria also, numerous secreted proteins and peptides derive from larger pro-proteins. In this work, we have shown that the subtilisin-related SphB1 is responsible for the maturation of the precursor of a large secreted protein of B.pertussis. These findings extend the notion of specialized subtilisin-related pro-protein convertases to a secretion pathway of Gram-negative bacteria.
The secretion of large extracellular proteins requires them to be kept in a secretion-competent conformation during synthesis, to avoid aggregation, and to fold at the appropriate time and place. The Gram-negative specific TPS pathway appears to have evolved for the secretion of a protein class, including FHA, which comprises some of the largest proteins of the prokaryotic world (Jacob-Dubuisson et al., 2001; Locht et al., 2001). FHA derives from a 367 kDa precursor, FhaB. The efficient extracellular release of mature FHA depends on the presence of the ∼130 kDa C-terminal region of FhaB, which is proteolytically removed in the course of secretion (Renauld-Mongénie et al., 1996). This suggested that this domain acts as an intramolecular chaperone to assist FHA secretion by preventing premature folding (Renauld-Mongénie et al., 1996). In agreement with this notion, it has been suggested that FHA may cross the cytoplasmic and outer membranes in a kinetically coupled fashion and hence may pass unfolded through the periplasm (Guédin et al., 1998). The timely processing of the FHA precursor by SphB1 then allows for the extracellular release of the mature protein. The synthesis and processing of a precursor with an intramolecular chaperone domain may well represent a widespread strategy for the secretion of large proteins.
In SphB1, the subtilisin-like domain is joined to a presumably β-barrel C-terminal domain characteristic of autotransporters (Henderson and Nataro, 2001). Auto transporters make up a large family of proteins represented in all major branches of the Gram-negative phylogenetic tree, and most frequently found in pathogenic bacteria. As implied by their name, they are translocated to the cell surface by a distinctive mechanism involving a specific domain of the autotransporter proteins themselves. The unifying structure of autotransporters consists of an N-terminal signal peptide for export across the cytoplasmic membrane, a secreted mature protein called the passenger (or α) domain, and a dedicated C-terminal (or β) domain forming a β-barrel pore in the outer membrane through which the passenger domain passes to the cell surface. In contrast to the well-conserved structure and function of the β-domains, the passenger proteins diverge more widely, which reflects their disparate roles. The α-domains may function as adhesins, proteolytic toxins or IgA proteases (Henderson and Nataro, 2001). SphB1 is the first reported autotransporter whose passenger protein serves as a maturation factor for another protein secreted by the same organism. This represents a novel function for autotransporters.
The subfamily of autotransporter proteins to which SphB1 belongs includes recognized proteins such as the S.marcescens PrtS (formerly SSP) (Miyazaki et al., 1989) and the P.fluorescens SspA and SspB (Kawai et al., 1999), as well as putative members identified by scanning sequenced genomes. The secretion pathway and self-processing of PrtS have been thoroughly investigated (Miyazaki et al., 1989; Shikata et al., 1992, 1993), but no function has been assigned to this protein or any other of this subfamily so far (Henderson and Nataro, 2001). Interestingly, genes encoding SphB1 homologues are often found in bacterial species that also have genes of FHA homologues (or TpsA proteins), including Haemo philus ducreyi, Pseudomonas aeruginosa, Neisseria meningitidis and Xylella fastidiosa (Locht et al., 2001; our unpublished data). At least in the case of H.ducreyi, the 260 kDa extracellular FHA homologue LspA1 also derives from a much larger precursor (Ward et al., 1998). Therefore, it is possible that some SphB1 homologues are involved in the proteolytic maturation of such large secreted proteins in their respective organisms. Interest ingly, a region of FhaB that includes its maturation site and extends into the intramolecular chaperone region is conserved among TpsA proteins (Locht et al., 2001).
A role in the late steps of FHA secretion is in good agreement with the cellular localization of SphB1. Association of SphB1 with the outer face of the outer membrane makes it well suited to process FHA upon translocation across the outer membrane via FhaC. The timely proteolytic cleavage of the precursor FhaB and the consequent extracellular release of FHA may be important for the in vivo role of this major virulence protein. That sphB1 belongs to the Bvg virulence regulon-like fhaB argues in favour of this hypothesis. We have obtained preliminary evidence that the sphB1 mutation affects the colonization of the respiratory tract in a mouse model of infection (our unpublished results).
SphB1 does not seem to be involved in the maturation of other secreted proteins. Bordetella pertussis produces several virulence proteins including pertactin, the tracheal colonization factor and the serum resistance factor BrkA, which are proteolytically processed in the course of their secretion (Charles et al., 1989; Fernandez and Weiss, 1994; Finn and Stevens, 1995). Whether they self-process or rely on a protease or proteases in the cell envelope is unknown. Our immunoblot or mass fingerprinting analyses argue that SphB1 is not responsible for the proteolytic maturation of any of these three precursors. This suggests that SphB1 is probably not a general-purpose envelope protease, and that the maturation of FHA might be its major, if not unique, function.
Subtilisin-like proteases make up a superfamily composed of at least six subfamilies (Siezen and Leunissen, 1997). In most bacteria, archae and lower eukaryotes, subtilisin-like proteases are extracellular, rather unspecific enzymes required for defence or for growth on proteinaceous substrates. In contrast, the eukaryotic ‘kexins’ such as Kex2 of Saccharomyces cerevisiae and mammalian furin have developed into specialized subtilisin-like proteases involved in pro-protein processing in the late compartments of the secretory pathway (Nakayama, 1997; Siezen and Leunissen, 1997). Similarly, the subtilisin-like lantibiotic peptidases of Gram-positive bacteria process the precursors of secreted antibiotic peptides such as nisin after secretion (van der Meer et al., 1993; Siezen and Leunissen, 1997). SphB1 now extends the concept of specialized subtilisin-like proteases to a secretion pathway of Gram-negative bacteria. The addition of an autotransporter β-barrel module can be viewed as an adaptation to the presence of the Gram-negative outer membrane. Similarly, while most subtilisin-like proteases are synthesized with an inhibitor pro-peptide removed by self-processing after membrane translocation (Siezen and Leunissen, 1997), the autotransporter mode of secretion may ensure that the subtilisin-like domain of SphB1 acquires its active conformation late in secretion (Henderson et al., 1998). Eukaryotic pro-protein convertases are also structurally more complex than degradative enzymes, with the addition to the core catalytic domain of a so-called P domain important for activity (Gluschankof and Fuller, 1994). It is intriguing that SphB1 has an additional 200-residue region between its subtilisin-like and β-barrel domains, which appears to be conserved among its homologues. It will be interesting to investigate the function of this domain in the protein that may well be the prototype of a new family of maturation proteases.
Materials and methods
Construction of strains
The B.pertussis strains used in this study were derived from BPSM (Menozzi et al., 1994). BPGR4 (ΔfhaB), BPEC (ΔfhaC) and BPLC1 (sphB1′–′lacZ) have been described previously (Locht et al., 1992; Guédin et al., 1998; Antoine et al., 2000). BPLC5 and BPLC7 were obtained by homologous recombination (Stibitz, 1994). The recombinant plasmids were introduced into B.pertussis by conjugation with Escherichia coli SM10 (Simon et al., 1983). BPLC5 carries a 626 bp internal deletion between the unique BglII site of sphB1 and the XmaI site located 626 bp downstream of the BglII site. It was obtained as follows. Two ∼600 bp gene fragments flanking the region to be deleted were obtained by PCR on BPSM genomic DNA with the oligonucleotide pairs 5′-TATAGAGCTCACCCGCCCCTTCGCCACGCCCCGAACCT-3′ and 5′-TATAAGATCTTGATGCCGTGCCCGAGCAGGTC-3′, and 5′-AGTACGGCTGGGGCGTGCTCGACGTG-3′ and 5′-TATACTGCAGGGTAGTCCAGCCGGGCGTTGAGCAGCAG-3′. The PstI–XmaI fragment and the BglII–EcoRV fragment (with the EcoRV site derived from an intermediate construct in pCR2.1-TOPO; InVitrogen Life Technology, Groningen, The Netherlands) were successively introduced into the PstI–XmaI and BglII–NruI sites of pIC20H (Marsh et al., 1984), yielding pIC1-5. The 1.3-kb HindIII fragment of pIC1-5 was inserted into pJQ200 mp18 rpsL (Quandt and Hynes, 1993). This construct was used for allelic exchange in BPSM, yielding BPLC5. BPLC7 carries a complete chromosomal copy of sphB1 with a point mutation resulting in the substitution of Ala for Ser412 in SphB1. Site-directed mutagenesis was performed by the megaprimer method. The mutagenic primer 5′-AACAGGGCACGGCGCTATCC-3′ and the primer 5′-TATACCCGGGCGACGAAGTCGCCGAAGGCGAA-3′ were used to synthesize a 210 bp PCR fragment that then served as a primer for a second PCR with the primer 5′-TATAAGATCTTCAACAACAGTTTCGCCACCGA-3′. The mutated PCR fragment was inserted into XmaI-restricted and BglII-restricted pIC1-5, yielding pIC1-3-5. The 1.9 kb HindIII fragment of pIC1-3-5 was inserted into pJQ200 mp18 rpsL, and this construct was used for allelic exchange in BPLC5, yielding BPLC7. All PCR fragments were first cloned into pCR2.1 TOPO and checked by sequencing using the ABI 377 prism sequencer of Perkin Elmer (Courtaboeuf, France). For the sequencing of other portions of sphB1, several pairs of primers were used to generate ∼400 bp fragments by PCR. At least two independent clones were sequenced for each amplified region.
All B.pertussis strains were grown on Bordet–Gengou agar (BG) or in liquid medium as described by Locht et al. (1992). The culture media were supplemented with 100 µg/ml streptomycin, 10 µg/ml gentamycin or 30 µg/ml nalidixic acid where appropriate.
Protein preparations and analyses
To harvest proteins from culture supernatants, B.pertussis strains were grown in liquid medium and the cells were collected by centrifugation in the late logarithmic phase of growth. The culture supernatants were analysed by SDS–PAGE. Proteins were detected by staining the gels with Coomassie Brilliant Blue R250, or the proteins were transferred onto nitrocellulose membranes for immunoblot analyses. Pertussis toxin and pertactin were detected with the monoclonal antibodies 1B7 and BPE3, respectively (Sato et al., 1984; Leininger et al., 1991). FHA was detected using a mixture of three monoclonal antibodies, F1, F4 and F5, the corresponding hybridomas of which were a gift of H.Sato (Japan). F4 and F5 have been determined by enzyme-linked immunosorbent assays to bind an 80 kDa N-terminal fragment of FHA, while F1 recognizes full-length FHA only (our unpublished data). A rat antiserum against the LHVDDIRPGYVGGDG peptide of SphB1 was purchased from Eurogentec (Seraing, Belgium) and used to detect SphB1 by immunoblot analyses. The blots were developed using alkaline phosphatase-conjugated secondary antibodies followed by the addition of 5-bromo-4-chloro-3-indolyl-phosphate and nitroblue tetrazolium. The secretion of adenylate cyclase/haemolysin was assessed by monitoring the haemolysis haloes surrounding the colonies grown on BG agar plates.
For fractionation experiments, bacteria were grown in liquid medium to an OD600 of 2, and harvested by centrifugation. Cells were washed and resuspended into 20 mM HEPES pH 7.4, 10 µg/ml DNase and 0.5 µg/ml 4-(2-aminoethyl)-benzene-sulfonyl hydrochloride (AEBSF) (Roche, Meylan, France). Cells were broken by two passages in a French pressure cell at 1000 psi. The lysates were centrifuged for 10 min at 6000 g to pellet large debris (total lysates). The clarified lysates were ultracentrifuged for 1 h at 100 000 g in a Beckman TL100 rotor 100.3 to pellet membranes. The soluble fractions were collected for analyses. The membrane pellets were washed and resuspended in 20 mM HEPES pH 7.4 (total insoluble fractions). Triton X-100 was then added to a final concentration of 2%. After 30 min incubation at room temperature, the samples were ultracentrifuged as above. The supernatants were collected (Triton-soluble), and the pellets were resuspended in 20 mM HEPES pH 7.4 (Triton-insoluble).
For trypsin digestions, B.pertussis cells were harvested by centrifugation in the logarithmic phase of growth (OD600 = 2). Cells were suspended in 10 mM Tris–HCl pH 8, 75 mM NaCl (intact cells), or into 30 mM Tris–HCl pH 8, 20% sucrose, 10 mM EDTA (permeabilized cells). Trypsin was then added to the cell suspensions to a final concentration of 5 mg/ml, and the digestions were performed on ice for 0, 30 or 60 min. Trypsin was inactivated by the addition of AEBSF to a final concentration of 0.5 mg/ml. The proteins were precipitated by the addition of ice-cold trichloroacetic acid (TCA) to a final concentration of 6%, and the precipitates were collected by centrifugation. SphB1 and its proteolytic fragments were detected by immunoblot analyses using the anti-peptide rat antiserum.
Pulse labelling of B.pertussis
Pulse labelling was performed as described by Lambert-Buisine et al. (1998) except that the samples were analysed using 4–15% acrylamide precast gradient gels (Bio-Rad, Ivry-sur Seine, France).
Flow cytometry
Bordetella pertussis cells freshly grown on BG agar plates were harvested by scraping the plates and resuspending the cells in phosphate-buffered saline containing 5% glycerol (PBSG). The samples were diluted in PBSG to an OD600 of 1, and incubated for 1 h with anti-FHA monoclonal antibodies. Preliminary experiments were performed to determine the optimal dilution of each antibody for the cytometry experiments. The 12.1F9, 12.2B1, 55.4G9, 12.6F8 and 55.6C4 antibodies have been characterized previously (Leininger et al., 1997); F4 and F5 have been described above. Bacteria were washed three times in PBSG and incubated at room temperature for 1 h with fluorescein isothiocyanate (FITC)-conjugated anti-mouse IgG (Dako, Trappes Cedex, France) diluted 300-fold in PBSG. After three washes, bacteria-associated fluorescence was quantified by flow cytometry using an EPICS Elite cytometer (Coulter, Fullerton, USA).
Mass spectrometry
FHA was purified from BPSM and BPLC5 culture supernatants as described by Menozzi et al. (1991), desalted using a C18 ZipTip column (Millipore, St Quentin-en Yvelines, France) and prepared for mass spectrometry. Briefy, the protein (∼1 pmol) was mixed with freshly dissolved sinapinic acid [10 mg/ml in 50% acetonitrile, 0.1% trifluoroacetic acid (TFA)], spotted onto the sample plate and allowed to dry. The samples were analysed by matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry using a Voyager-DE-STR (Applied Biosystems, Palo Alto, CA). Mass spectra were obtained in the delayed extraction, positive ion, linear mode with an accelerating voltage of 20 kV. The parameters were set as follows: grid, % 92; delay, 750 ns; and low mass gate, 1000 atomic mass units (a.m.u.). The instrument was calibrated externally with carbonic anhydrase and bovine serum albumin. As the mass of FHA is much higher than those of the proteins used in the calibration, its double, triple and quadruple ions were used for mass determination.
For mass fingerprinting analyses, protein bands were excised from a gel stained with colloidal Coomassie Blue G-250 (Neuhoff et al., 1988). Trypsin digestions were performed as follows. Small pieces of gel were covered with 100 µl of 25 mM NH4HCO3/50% acetonitrile in a 0.65 ml siliconized tube, the suspension was vortexed for 10 min and the supernatant was discarded. These steps were repeated twice. The gel pieces were dried completely in a Speed Vac, and a solution of 20 ng/µl trypsin in 25 mM NH4HCO3 was added at 3 vols per volume of dried gel. The suspension was vortexed for 10 min and incubated for 30 min at 4°C. NH4HCO3 (25 mM) was added such as to cover the gel pieces, and the samples were incubated at 37°C overnight. An organic extraction of the peptides from the gel pieces was performed using 50 µl of 50% acetonitrile/1% TFA. The suspension was vortexed for 10 min, spun briefly and sonicated for 5 min. The extracted peptides were pooled and the organic extraction steps were repeated twice. The volume of the extracted digests was reduced to 10 µl in a Speed Vac. The extracted peptides were desalted using a C18 ZipTip column. They were then mixed with freshly dissolved α-cyano-4-hydroxycinnaminic acid (5 mg/ml in 50% acetonitrile, 0.1% TFA), spotted on the plate and allowed to dry. Mass spectra were obtained in the delayed extraction, positive ion, reflection mode, with an accelerating voltage of 20 kV. The parameters were set as follows: grid, % 61; delay, 90 ns; and low mass gate, 500 a.m.u. to obtain the best resolution in the mass range (typically >10 000). The laser energy required to desorb and ionize the samples was kept at the lowest value compatible with a signal/noise ratio >50. The instrument was calibrated externally using the MH+ monoisotopic ions from several peptides (angiotensin, 1296.685; ACTH clip 1–17, 2093.087; ACTH clip 18–39, 2465.199).
Identification of proteins by mass fingerprinting
No annotation of the B.pertussis genome is available yet. Therefore, in order to generate a B.pertussis protein database, all contigs were artificially concatenated, and the resulting DNA sequence was translated in the six possible reading frames. All resulting polypeptide sequences were retained as proteins independently of their length, codon usage and coding probability. This crude protein database was digested by trypsin in silicon for use with the GPMaw program.
Supplementary data
Supplementary data to this paper are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We wish to thank Drs H.Sato and E.Leninger for the gift of hybridomas and antibodies. The help of E.Willery and B.Quatannens for the sequencing and the flow cytometry experiments, respectively, is gratefully acknowledged. We are also grateful to Jean Dubuisson and Alain Baulard for their critical reading of the manuscript. L.C. is supported by the Ministère de l’Education Nationale, de la Recherche et de la Technologie and F.J.-D. is a Researcher of the CNRS. This work was supported in part by INSERM, the Institut Pasteur de Lille and the Région Nord-Pas-de Calais.
References
- Akerley B.J. and Miller,J.F. (1996) Understanding signal transduction during bacterial infection. Trends Microbiol., 4, 141–146. [DOI] [PubMed] [Google Scholar]
- Antoine R., Alonzo,S., Raze,D., Coutte,L., Lesjean,S., Willery,E., Locht,C. and Jacob-Dubuisson,F. (2000) New virulence-activated and virulence-repressed genes identified by systematic gene inactivation and generation of transcriptional fusions in Bordetella pertussis. J. Bacteriol., 182, 5902–5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeron F., Leduc,R. and Day,R. (2000) Subtilase-like pro-protein convertases: from molecular specificity to therapeutic applications. J. Mol. Endocrinol., 24, 1–22. [DOI] [PubMed] [Google Scholar]
- Charles I.G., Dougan,G., Pickard,D., Chatfield,S., Smith,M., Novotny,P., Morrissey,P. and Fairweather,N.F. (1989) Molecular cloning and characterization of protective outer membrane protein P69 from Bordetella pertussis. Proc. Natl Acad. Sci. USA, 86, 3554–3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie P.J. (2001) Type IV secretion: intercellular transfer of macromolecules by systems ancestrally related to conjugation machines. Mol. Microbiol., 40, 294–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domenighini M., Relman,D., Capiau,C., Falkow,S., Prugnola,A., Scarlato,V. and Rappuoli,R. (1990) Genetic characterization of Bordetella pertussis filamentous hemagglutinin: a protein processed from an unusually large precursor. Mol. Microbiol., 4, 787–800. [DOI] [PubMed] [Google Scholar]
- Fath M.J. and Kolter,R. (1993) ABC transporters: bacterial exporters. Microbiol. Rev., 57, 995–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez R.C. and Weiss,A.A. (1994) Cloning and sequencing of a Bordetella pertussis serum resistance locus. Infect. Immun., 62, 4727–4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn T.M. and Stevens,L.A. (1995) Tracheal colonization factor: a Bordetella pertussis secreted virulence determinant. Mol. Microbiol., 16, 625–634. [DOI] [PubMed] [Google Scholar]
- Gluschankof P. and Fuller,R.S. (1994) A C-terminal domain conserved in precursor processing proteases is required for intramolecular N-terminal maturation of pro-Kex2 protease. EMBO J., 13, 2280–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grass S. and St Geme III,J.W. (2000) Maturation and secretion of the non-typable Haemophilus influenzae HMW1 adhesin: roles of the N-terminal and C-terminal domains. Mol. Microbiol., 36, 55–67. [DOI] [PubMed] [Google Scholar]
- Guédin S., Willery,E., Locht,C. and Jacob-Dubuisson,F. (1998) Evidence that a folded conformation is not compatible with FhaC-mediated secretion of the Bordetella pertussis filamentous hemagglutinin. Mol. Microbiol., 29, 763–774. [DOI] [PubMed] [Google Scholar]
- Guédin S., Willery,E., Tommassen,J., Fort,E., Drobecq,H., Locht,C. and Jacob-Dubuisson,F. (2000) Novel topological features for FhaC, the outer membrane transporter involved in the secretion of Bordetella pertussis filamentous hemagglutinin. J. Biol. Chem., 274, 37731–37735. [DOI] [PubMed] [Google Scholar]
- Henderson I.R. and Nataro,J.P. (2001) Virulence functions of autotransporter proteins. Infect. Immun., 69, 1231–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson I.R., Navarro-Garcia,F. and Nataro,J.P. (1998) The great escape: structure and function of the autotransporter proteins. Trends Microbiol., 6, 370–378. [DOI] [PubMed] [Google Scholar]
- Jacob-Dubuisson F., Buisine,C., Willery,E., Renauld-Mongénie,G. and Locht,C. (1997) Lack of functional complementation between Bordetella pertussis filamentous hemagglutinin and Proteus mirabilis HpmA hemolysin secretion machineries. J. Bacteriol., 179, 775–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob-Dubuisson F., El-Hamel,C., Saint,N., Guédin,S., Willery,E., Molle,G. and Locht,C. (1999) Channel formation by FhaC, the outer membrane protein involved in the secretion of the Bordetella pertussis filamentous hemagglutinin. J. Biol. Chem., 274, 37731–37735. [DOI] [PubMed] [Google Scholar]
- Jacob-Dubuisson F., Locht,C. and Antoine,R. (2001) Two-partner secretion in Gram-negative bacteria: a thrifty, specific pathway for large virulence proteins. Mol. Microbiol., 40, 306–313. [DOI] [PubMed] [Google Scholar]
- Kawai E., Idei,A., Kumura,H., Shimazaki,K., Akatsuka,H. and Omori,K. (1999) The ABC-exporter genes involved in the lipase secretion are clustered with the genes for lipase, alkaline protease and serine protease homologues in Pseudomonas fluorescens no. 33. Biochim. Biophys. Acta, 1446, 377–382. [DOI] [PubMed] [Google Scholar]
- Lambert-Buisine C., Willery,E., Locht,C. and Jacob-Dubuisson,F. (1998) N-terminal characterization of the Bordetella pertussis filamentous hemagglutinin. Mol. Microbiol., 28, 1283–1293. [DOI] [PubMed] [Google Scholar]
- Leininger E., Roberts,M., Kenimer,J.G., Charles,I.G., Fairweather,N., Novotny,P. and Brennan,M.J. (1991) Pertactin, an Arg-Gly-Asp-containing Bordetella pertussis surface protein that promotes adherence of mammalian cells. Proc. Natl Acad. Sci. USA, 88, 345–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leininger E., Bowen,S., Renauld-Mongenie,G., Rouse,J.H., Menozzi,F.D., Locht,C., Heron,I. and Brennan,M.J. (1997) Immunodominant domains present on the Bordetella pertussis vaccine component filamentous hemagglutinin. J. Infect. Dis., 175, 1423–1431. [DOI] [PubMed] [Google Scholar]
- Lo R.Y.C., Strathdee,C.A., Shewen,P.E. and Cooney,B.J. (1991) Molecular studies of Ssa1, a serotype-specific antigen of Pasteurella haemolytica. Infect. Immun., 59, 3398–3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locht C., Geoffroy,M.-C. and Renauld,G. (1992) Common accessory genes for the Bordetella pertussis filamentous hemagglutinin and fimbriae share sequence similarities with the papC and papD gene families. EMBO J., 11, 3175–3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locht C., Bertin,P., Menozzi,F.D. and Renauld,G. (1993) The filamentous hemagglutinin, a multifaceted adhesion produced by virulent Bordetella spp. Mol. Microbiol., 9, 653–660. [DOI] [PubMed] [Google Scholar]
- Locht C., Antoine,R. and Jacob-Dubuisson,F. (2001) Bordetella pertussis, molecular pathogenesis under multiple aspects. Curr. Opin. Microbiol., 4, 82–89. [DOI] [PubMed] [Google Scholar]
- Marsh J.L., Erfle,M. and Wykes,E.J. (1984) The pIC plasmid and phage vectors with versatile cloning sites for recombinant selection by insertional inactivation. Gene, 32, 481–485. [DOI] [PubMed] [Google Scholar]
- Menozzi F.D., Gantiez,C. and Locht,C. (1991) Interaction of the Bordetella pertussis filamentous hemagglutinin with heparin. FEMS Microbiol. Lett., 78, 59–64. [DOI] [PubMed] [Google Scholar]
- Menozzi F.D., Mutombo,R., Renauld,G., Gantiez,C., Hannah,J.H., Leininger,E., Brennan,M.J. and Locht,C. (1994) Heparin-inhibitable lectin activity of the filamentous hemagglutinin adhesin of Bordetella pertussis. Infect. Immun., 62, 769–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki H., Yanagida,N., Horinouchi,S. and Beppu,T. (1989) Characterization of the precursor of Serratia marcescens serine protease and COOH-terminal processing of the precursor during its excretion through the outer membrane of Escherichia coli. J. Bacteriol., 171, 6566–6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama K. (1997) Furin: a mammalian subtilisin/Kex2p-like endoprotease involved in processing of a wide variety of precursor proteins. Biochem. J., 327, 625–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhoff V., Arold,N., Taube,D. and Ehrhardt,W. (1988) Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis, 9, 255–262. [DOI] [PubMed] [Google Scholar]
- Ohnishi Y., Beppu,T. and Horinouchi,S. (1997) Two genes encoding serine protease homologues in Serratia marcescens and characterization of their products in Escherichia coli.J. Biochem. (Tokyo), 121, 902–913. [DOI] [PubMed] [Google Scholar]
- Plano G.V., Day,J.B. and Ferraci,F. (2001) Type III export: new uses for an old pathway. Mol. Microbiol., 40, 284–293. [DOI] [PubMed] [Google Scholar]
- Pohlner J., Halter,R., Beyreuther,K. and Meyer,T.F. (1987) Gene structure and extracellular secretion of Neisseria gonorrheae IgA protease. Nature, 325, 458–462. [DOI] [PubMed] [Google Scholar]
- Pugsley A.P. (1993) The complete general secretory pathway in Gram-negative bacteria. Microbiol. Rev., 57, 50–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quandt J. and Hynes,M.F. (1993) Versatile suicide vectors which allow direct selection for gene replacement in Gram-negative bacteria. Gene, 127, 15–21. [DOI] [PubMed] [Google Scholar]
- Renauld-Mongénie G., Cornette,J., Mielcarek,N., Menozzi,F.D. and Locht,C. (1996) Distinct roles of the N-terminal and the C-terminal precursor domains in the biogenesis of the Bordetella pertussis filamentous hemagglutinin. J. Bacteriol., 178, 1053–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandkvist M. (2001) Biology of type II secretion. Mol. Microbiol., 40, 271–283. [DOI] [PubMed] [Google Scholar]
- Sankaran K. and Wu,H.C. (1994) Lipid modification of bacterial prolipoprotein. J. Biol. Chem., 31, 19701–19706. [PubMed] [Google Scholar]
- Sato H., Ito,A., Chiba,J. and Sato,Y. (1984) Monoclonal antibody against pertussis toxin: effect on toxin activity and pertussis infections. Infect. Immun., 46, 422–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikata S., Shimada,K., Kataoka,H., Horinouchi,S. and Beppu,T. (1992) Detection of large COOH-terminal domains processed from the precursor of Serratia marcescens serine protease in the outer membrane of Escherichia coli. J. Biochem. (Tokyo), 111, 627–632. [DOI] [PubMed] [Google Scholar]
- Shikata S., Shimada,K., Ohnishi,Y., Horinouchi,S. and Beppu,T. (1993) Characterization of secretory intermediates of Serratia marcescens serine protease produced during its extracellular secretion from Escherichia coli cells. J. Biochem. (Tokyo), 114, 723–731. [DOI] [PubMed] [Google Scholar]
- Siezen R.J. and Leunissen,J.A.M. (1997) Subtilases: the superfamily of subtilisin-like serine proteases. Protein Sci., 6, 501–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R., Priefer,U. and Pühler,A. (1983) A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Biotechnology, 1, 784–790. [Google Scholar]
- Stibitz S. (1994) Use of conditionally counterselectable suicide vectors for allelic exchange. Methods Enzymol., 235, 458–465. [DOI] [PubMed] [Google Scholar]
- van der Meer J.R., Polman,J., Beerthuyzen,M., Siezen,R.J., Kuipers,O.P. and De Vos,W.M. (1993) Characterization of the Lactococcus lactis Nisin A operon genes nisP, encoding a subtilisin-like serine protease involved in precursor processing and nisR, encoding a regulatory protein involved in nisin biosynthesis. J. Bacteriol., 175, 2578–2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward C.K., Lumbley,S.R., Latimer,J.L., Cope,L.D. and Hansen,E.J. (1998) Haemophilus ducreyi secretes a filamentous hemagglutinin-like protein. J. Bacteriol., 180, 6013–6022. [DOI] [PMC free article] [PubMed] [Google Scholar]