Abstract
A C-terminally modified ubiquitin (Ub) derivative, ubiquitin vinyl sulfone (UbVS), was synthesized as an active site-directed probe that irreversibly modifies a subset of Ub C-terminal hydrolases (UCHs) and Ub-specific processing proteases (UBPs). Specificity of UbVS for deubiquitylating enzymes (DUBs) is demonstrated not only by inhibition of [125I]UbVS labeling with N-ethylmaleimide and Ub aldehyde, but also by genetic analysis. [125I]UbVS modifies six of the 17 known and putative yeast deubiquitylating enzymes (Yuh1p, Ubp1p, Ubp2p, Ubp6p, Ubp12p and Ubp15p), as revealed by analysis of corresponding mutant strains. In mammalian cells, greater numbers of polypeptides are labeled, most of which are likely to be DUBs. Using [125I]UbVS as a probe, we report the association of an additional DUB with the mammalian 26S proteasome. In addition to the 37 kDa enzyme reported to be part of the 19S cap, we identified USP14, a mammalian homolog of yeast Ubp6p, as being bound to the proteasome. Remarkably, labeling of 26S-associated USP14 with [125I]UbVS is increased when proteasome function is impaired, suggesting functional coupling between the activities of USP14 and the proteasome.
Keywords: active site-directed probe/deubiquitylating enzyme/proteasome/ubiquitin/vinyl sulfone
Introduction
Covalent modification of proteins by ubiquitin (Ub), a small 76 amino acid protein, is important in the control of many cellular processes (Hershko and Ciechanover, 1998). Ubiquitylation primarily serves as a targeting signal, and proteins carrying the most common type of poly-Ub chain are targeted for destruction by the ubiquitin–proteasome pathway, responsible for the majority of cytosolic proteolysis (Ciechanover and Schwartz, 1998). Ub is attached to proteins through an isopeptide linkage, involving the C-terminal carboxylate of Ub and the ε-NH2 of a lysine side chain or, less commonly, the N-terminus of the substrate protein (Hodgins et al., 1996; Ciechanover et al., 1999). The enzyme cascade involved in Ub conjugation and poly-Ub chain formation comprises at least three distinct sets of enzymatic activities including the Ub-activating enzyme E1, Ub-conjugating enzymes (E2) and E3 ligases (reviewed in Hershko and Ciechanover, 1998). In addition, other non-enzymatic factors may also regulate poly-Ub chain length, such as E4 in a positive (Koegl et al., 1999), or Rad 23 and hPLIC in a negative manner (Kleijnen et al., 2000; Ortolan et al., 2000). Recent studies suggest that ubiquitin-dependent events are also controlled at the level of deubiquitylation (Wilkinson, 1997). Removal of Ub is carried out by deubiquitylating enzymes (DUBs), a large family of proteases that can release poly-Ub chains from proteins to be degraded by the 26S proteasome, recycle monomeric Ub, liberate Ub from the Ub fusion protein precursors, reverse regulatory ubiquitylation and edit inappropriately ubiquitylated proteins (reviewed in Chung and Baek, 1999).
DUBs can be subdivided into Ub C-terminal hydrolases (UCHs) and Ub-specific processing proteases (UBPs). In vitro, UBPs hydrolyze isopeptide bonds between Ub and folded protein domains, such as additional Ub moieties or target proteins. Thus, UBPs exhibit broad substrate specificity (Wilkinson, 1997). UCHs generally cleave bonds between Ub and an unfolded polypeptide or Ub and small substituents (Pickart and Rose, 1985; Wilkinson et al., 1986; Wilkinson, 1997). Deletion studies in yeast suggest that the substrate specificities of UCHs and UBPs overlap (Baker et al., 1992; Amerik et al., 2000). Both UBPs and UCHs can associate with the 26S proteasome and are involved in the regulation of Ub-dependent proteolysis (Voges et al., 1999). A UCH enzyme within the 19S regulatory complex has been reported in mammals (UCH37) (Lam et al., 1997a), Drosophila (p37a) (Holzl et al., 2000), and Schizosaccharomyces pombe (Uch2p) (Li et al., 2000) and is thought to edit Ub chains on protein substrates before they are degraded (Lam et al., 1997a,b). Doa4 in yeast and Ap-UCH in Aplysia are associated more transiently with the proteolytic complex and may cleave Ub from remnants of degraded proteins (Papa and Hochstrasser, 1993; Hegde et al., 1997; Papa et al., 1999) In higher organisms, UCHs are expressed in a tissue-specific manner and are linked to a variety of cellular functions and diseases, such as Parkinson’s disease (UCH-L1) (Wilkinson et al., 1992), the function of BRCA1 (BAP-1) (Jensen et al., 1998) and long-term nerve potentiation in Aplysia (Ap-UCH) (Hegde et al., 1997). UBPs also play a role in determination of cell fate (fat facets) (Huang et al., 1995), transcriptional silencing (Ubp3) (Moazed and Johnson, 1996), response to cytokines (DUB1 and 2) (Zhu et al., 1996) and oncogenic transformation (tre-2) (Papa and Hochstrasser, 1993).
The yeast genome encodes 17 DUBs whereas the number of UBPs and UCHs in mammals is likely to be far greater (Chung and Baek, 1999). Enzymatic activity has been demonstrated for many DUBs using model substrates (Amerik et al., 2000), whereas for others the assignment was made based on homology with known family members (Wilkinson, 1997). The multiplicity of DUBs raises the possibility of distinct functions and/or protein substrates for each DUB. All known UBPs and UCHs are thiol proteases with specificity for Ub. We reasoned that the introduction of a suitable electrophile at the C-terminus of Ub should allow irreversible trapping of UBPs and UCHs as covalent adducts. By labeling such probes with 125I, it should be possible to visualize active DUBs directly.
Vinyl sulfones are versatile functional groups ideally suited to inhibit thiol proteases (Palmer et al., 1995). Their reactivity is not necessarily limited to thiol proteases, as proteasomes, N-terminal threonine hydrolases, are inhibited efficiently by peptide vinyl sulfones (Bogyo et al., 1997). Using the well-established method of trypsin-catalyzed transpeptidation to modify Ub (Wilkinson et al., 1986), we synthesized the C-terminally modified vinyl sulfone derivative of Ub, hereafter referred to as UbVS. We demonstrate specificity of labeling and show that [125I]UbVS allows the visualization of active UBPs/UCHs in crude cell extracts. In addition to the previously character ized 37 kDa enzyme (Lam et al., 1997a), we observe a second DUB associated with the mammalian proteasome. We identify this enzyme as USP14, a mammalian homolog of yeast Ubp6. USP14 associates with the 26S complex and its activity, unlike that of p37, is affected by proteasome inhibition both in vitro and in living cells.
Results
Synthesis and characterization of UbVS
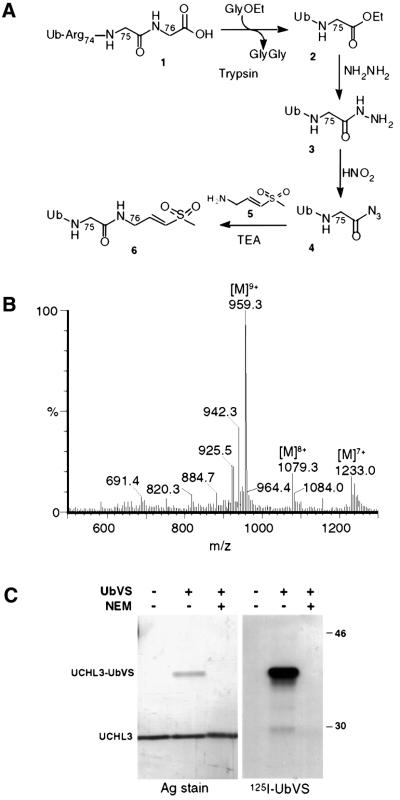
The strategy for the synthesis of UbVS is outlined in Figure 1A. Ubiquitin 1 was digested with trypsin in the presence of 2.5 M glycine ethyl ester to obtain Ub75-ethyl ester 2 in ∼40% yield (Wilkinson et al., 1986). Trypsin was inactivated by addition of soybean trypsin inhibitor and phenylmethylsulfonyl fluoride (PMSF). Ub75-ethyl ester was purified by Sephadex G-50 gel filtration, followed by CM Sepharose cation-exchange chromatography. Treat ment of 2 with hydrazine converted the ethylester moiety into Ub75-hydrazine 3, and subsequent exposure of 3 to dilute nitrous acid yielded Ub75-azide 4 (Wilkinson et al., 1990). Both reactions were near quantitative, and little residual Ub-ethylester and hydrazine were detected. UbVS 6 was obtained by coupling 4 with an excess of NH2-glycyl-vinyl sulfone 5 (Bogyo et al., 1998). Purification by cation-exchange chromatography allowed separation of UbVS from the remaining precursors and side products generated by the Michael reaction of additional glycine vinyl sulfone molecules with UbVS. The molecular weight of the isolated UbVS was in agreement with the predicted mass, as assessed by liquid chromatography/electrospray mass spectrometry (LC/ES-MS) (Figure 1B). The overall yield was 3%.
Fig. 1. Synthesis and characterization of UbVS. (A) C-terminal modification of ubiquitin 1 to generate ubiquitin vinyl sulfone 6. A 25 mg aliquot of 1 was converted to ubiquitin75-ethyl ester 2 by treatment with 2.5 mg of trypsin in the presence of 2.5 M glycine ethyl ester (GlyOEt) and 20% PEG 20 000. Ubiquitin75-ethyl ester was treated with hydrazine monohydrate and HCl to generate ubiquitin75-hydrazine 3 which was dialyzed against water and converted to ubiquitin75-azide 4 by treatment with 0.5 M nitrous acid for 1 min at –5°C. Ubiquitin75-azide immediately reacted with the TFA salt of glycine vinyl sulfone 5 in the presence of TEA, generating 6. (B) Purified UbVS is resolved on an analytical C4 column using a 0.1% formic acid/acetonitrile buffer system. Eluate was analyzed by on-line ES-MS. The indicated multicharged species correspond to a mol. wt of 8624.9 Da, in agreement with the predicted molecular weight of UbVS (8625 Da). (C) Recombinant, purified UCH-L3 was incubated with a substochiometric amount of [125I]UbVS in PBS for 45 min at 37°C with or without pre-treatment with 2 mM N-ethylmaleimide. Protein conjugates were resolved by 12.5% SDS–PAGE under reducing conditions, and visualized by silver stain (left panel) and autoradiography (right panel).
UbVS specifically labels the ubiquitin C-terminal hydrolase UCH-L3
The UCH-L3 enzyme was produced in recombinant form in Escherichia coli and purified to apparent homogeneity (Larsen et al., 1996). UbVS was radioiodinated with Na[125I] and added in substoichiometric amounts to UCH-L3. The appearance of an additional polypeptide of a molecular mass consistent with a covalent UCH-L3–UbVS adduct was observed on silver-stained gels (Figure 1C, left panel). The reaction was blocked completely by inclusion of the alkylating agent N-ethylmaleimide, consistent with the requirement for the active site cysteine of UCH-L3 in the reaction. Autoradiography of the same samples confirmed that the UCH-L3–[125I]UbVS adduct was the single predominant radiolabeled species (Figure 1C, right panel). We conclude that UbVS efficiently labels the Ub C-terminal hydrolase UCH-L3 through a Michael reaction of the active site thiol of UCH-L3 with the vinyl sulfone moiety. The resulting thioether is stable under the conditions routinely used for reducing SDS–PAGE.
Labeling of extracts from yeast with deletions of known DUB genes
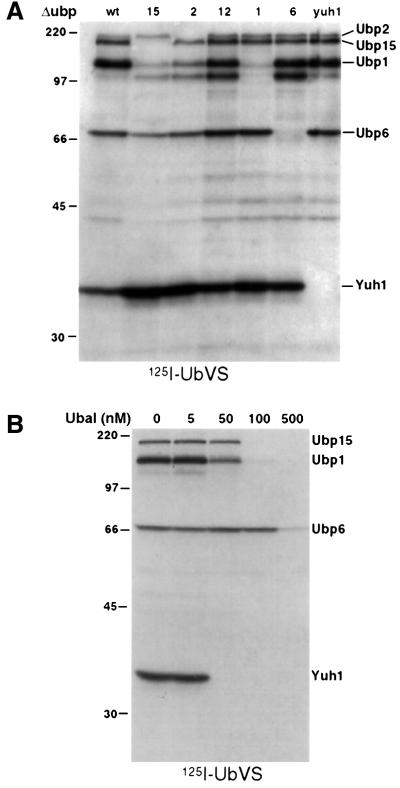
The yeast genome specifies 16 UBPs and a UCH, Yuh1 (Chung and Baek, 1999). We obtained yeast strains deficient in the individual genes encoding these enzymes. Cytosolic extracts were prepared from each strain and exposed to [125I]UbVS. We observed at least five clearly labeled polypeptides, all of which could be assigned to Yuh1p and UBPs 1, 2, 6 and 15, based on the analysis of mutant strains (Figure 2A; Table I). On longer exposures, labeling of Ubp12p was also detected. These results provide a genetic corroboration of the specificity of labeling by UbVS.
Fig. 2. [125I]UbVS specifically labels a subset of yeast DUBs. (A) A 100 µg aliquot of post-nuclear lysates from DUB deletion strains was incubated with 1 × 106 c.p.m. of [125I]UbVS for 45 min at 37°C. Reactions were quenched with sample buffer, resolved by 10% SDS–PAGE and analyzed by autoradiography. The identity of the bands was assigned based on the absence of a band corresponding to the molecular weight of a DUB deleted in that strain. (B) Lysates from wild-type yeast were pre-incubated with increasing concentrations of Ubal competitor for 30 min at room temperature prior to addition of [125I]UbVS as described in (A).
Table I. Deubiquitylating enzymes of S.cerevisiae.
| Gene | Mol. wt (kDa) | UbVS labeling | Characterization |
|---|---|---|---|
| Ubp1 | 93 | ++ | known Ubp |
| Ubp2 | 146 | ++ | known Ubp |
| Ubp3 | 110 | – | known Ubp |
| Ubp4/Doa4 | 105 | – | known Ubp |
| Ubp5 | 92 | – | known Ubp |
| Ubp6 | 57 | ++ | known Ubp |
| Ubp7 | 123 | – | putative |
| Ubp8 | 54 | – | putative |
| Ubp9 | 86 | – | putative |
| Ubp10/Dot4 | 88 | – | known Ubp |
| Ubp11 | 83 | – | known Ubp |
| Ubp12 | 143 | + | putative |
| Ubp13 | 77 | ND | putative |
| Ubp14 | 91 | – | known Ubp |
| Ubp15 | 143 | ++ | known Ubp |
| Ubp16 | 57 | ND | putative |
| Yuh1 | 26 | ++ | known Uch |
Labeling with [125I]UbVS was determined as described in Figure 3; ++ indicates strong labeling, + weak labeling, – no labeling; ND = not determined. The molecular weights of DUBs are based on the predicted sizes listed in the YPD database (http://www.proteome.com/databases/index.html).
Specificity of labeling was ascertained further by inclusion of ubiquitin aldehyde (Ubal), a known inhibitor of DUBs (Hershko and Rose, 1987). Labeling of DUBs by [125I]UbVS was competed by pre-treatment with Ubal in a concentration-dependent manner (Figure 2B). Ubp6p appeared less susceptible to inhibition than the other UBPs. Importantly, UbVS labels only five prominent polypeptides in wild-type yeast, even though 17 DUBs are encoded by the genome (Table I).
Labeling of DUBs in mammalian cell extracts
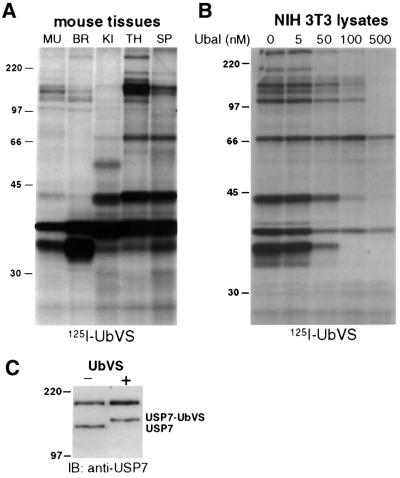
Given the conserved reaction mechanism used by eukaryotic DUBs, we expected that [125I]UbVS would label a diverse set of DUBs in mammalian cells as well. Indeed, when extracts prepared from different mouse tissues were labeled with [125I]UbVS (Figure 3A), we noted the appearance of multiple polypeptides with significant differences in labeling patterns for different tissues, the most striking of which was seen for brain (Figure 3A, lane 2). The intensely labeled polypeptide detected at a mol. wt of 30 kDa most probably corresponds to the UCH-L1 enzyme, known to be expressed abundantly in brain (Wilkinson et al., 1989). In all, we detected at least 12 and, upon longer exposures of autoradiograms, a substantially larger number of labeled species. Again, as seen with yeast, all labeling is abolished by inclusion of Ubal (Figure 3B), establishing the specificity of labeling. While confirming the known complexity of the DUB family, this observation demonstrates that active DUBs can be detected conveniently in a crude extract by simple incubation with [125I]UbVS, followed by SDS–PAGE and autoradiography.
Fig. 3. [125I]UbVS labels mammalian DUBs. (A) Single cell suspensions were prepared from tissues of a male B6 mouse: muscle (MU), brain (BR), kidney (KI), thymus (TH) and spleen (SP). A 50 µg aliquot of lysate was treated with 1 × 106 c.p.m. [125I]UbVS as described in Figure 2A and resolved by 10% SDS–PAGE. (B) A 20 µg aliquot of post-nuclear lysates from NIH 3T3 cells was pre-treated with increasing concentrations of Ubal as competitor for 30 min at 37°C followed by labeling with [125I]UbVS. (C) A 50 µg aliquot of lysates from EL-4 cells was treated with 2 µM UbVS at 37°C for 1 h, reactions were resolved by 10% SDS–PAGE and immunoblotted with the r201 rabbit antiserum against USP7.
To demonstrate modification of a known UBP by UbVS, we incubated EL-4 cell lysates with UbVS, followed by immunoblotting with antiserum specific for USP7 (HAUSP) (Everett et al., 1997). We observed a shift in one of the two immunoreactive species consistent with the formation of a USP7–UbVS conjugate (Figure 3C). The upper polypeptide cross-reacting with anti-USP7 serum has been observed previously (Everett et al., 1997) and is not reactive with UbVS. Thus, we demonstrate that UbVS labels not only mammalian UCHs, but also UBPs, as shown here for USP7.
Association of DUBs with higher molecular weight complexes
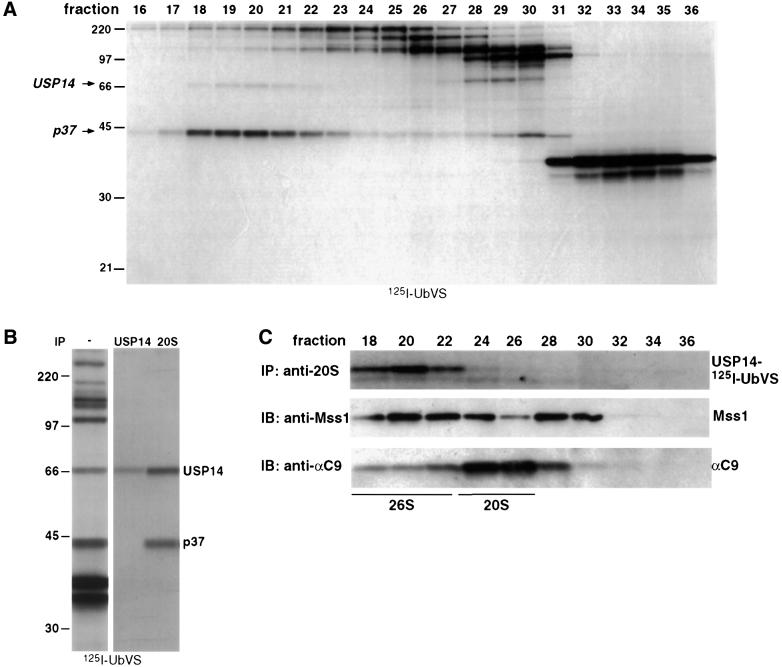
Several reports have described the association of DUBs with the proteasome (Hegde et al., 1997; Papa et al., 1999; Voges et al., 1999; Verma et al., 2000); a DUB of 37 kDa (referred to here as p37) is thought to be a subunit of the 19S cap (Lam et al., 1997b; Holzl et al., 2000). Since [125I]UbVS labeling allows us to monitor the activity of multiple DUBs at the same time, it is possible to examine directly the association of DUBs with higher molecular weight complexes such as the 26S proteasome. Labeling of fractions obtained by gel filtration on a Superose 6 column with [125I]UbVS shows that most DUBs elute at positions consistent with their observed molecular mass as determined by SDS–PAGE (Figure 4A). However, two polypeptides (mol. wts 45 and 66 kDa, which includes 8.5 kDa for UbVS) appear to be part of a larger complex (Figure 4A, fractions 17–23). The 45 kDa polypeptide is identical in mass to the ubiquitin isonitrile-modified p37 DUB present in the 19S cap (Lam et al., 1997b), and is likely to be the mammalian homolog of a UCH found in the 19S caps of Drosophila and S.pombe proteasomes (Holzl et al., 2000; Li et al., 2000). The identity of p37 in mammals remains to be established. The identity of the 58 kDa DUB was investigated further.
Fig. 4. USP14 associates with the 26S proteasome. (A) EL-4 cell lysates were fractionated on a Superose 6 FPLC column to isolate high molecular weight complexes, as described in Materials and methods. Fractions were labeled with 0.5 × 106 c.p.m. of [125I]UbVS and resolved by SDS–PAGE. (B) A 80 µg aliquot of EL-4 cell lysates was treated with 2 × 106 c.p.m. of [125I]UbVS and immunoprecipitated with anti-20S proteasome antiserum using proteasome IP buffer or denatured with 1% SDS and immunoprecipitated with anti-USP14 antiserum HM433 using NET buffer. (C) EL-4 lysates were fractionated on a Superose 6 column as described in (A). A 1 ml aliquot of each fraction was incubated with [125I]UbVS and immuno precipitated for the proteasome as in (B) (top panel). A 50 µl aliquot of each fraction was analyzed for the presence of proteasome subunits by immunoblot with the indicated antibodies against components of the 20S and 19S complexes (lower panels).
USP14 is associated with the 26S proteasome
We observe that p58 co-migrates on SDS–PAGE with yeast Ubp6p. Moreover the [125I]UbVS labeling of p58 and Ubp6p is competed similarly by Ubal (Figures 2B and 3B). This suggested that p58 may correspond to the mammalian homolog of Ubp6, USP14 (Yin et al., 2000). We raised polyclonal antibodies against synthetic peptides corresponding to USP14. In an immunoblot, these antisera detect a single polypeptide of a molecular weight consistent with that of USP14 (data not shown). The antiserum immunoprecipitated a [125I]UbVS-labeled polypeptide that co-migrates with the 66 kDa [125I]UbVS-labeled DUB observed in anti-20S immunoprecipitations (Figure 4B), suggesting that USP14 is associated with the proteasome. The observed difference in the USP14 to p37 ratio in whole lysates versus the immunoprecipitated samples is likely to be due to the fact that not all of the protein present in the lysate is proteasome associated and the differences in the affinity of p37/USP14 for the proteasome. Since the anti-USP14 antiserum recognizes only SDS-denatured protein, we were unable to co-immunoprecipitate the proteasome with USP14 directly with antibodies directed against USP14. To confirm the association of USP14 with the proteasome, we labeled Superose 6 fractions with [125I]UbVS and immunoprecipitated the 20S complex with antibodies against the 20S core. Parallel samples were immunoblotted for different proteasome subunits (Figure 4C). Indeed [125I]UbVS-modified USP14 is found in the fractions corresponding to 26S proteasome complex (fractions 18–22) and not in fractions containing the free 20S proteasomes (fractions 24–28). These results establish a physical association of USP14 with the 26S proteasome complex.
[125I]UbVS labeling of USP14 is increased upon proteasome inhibition
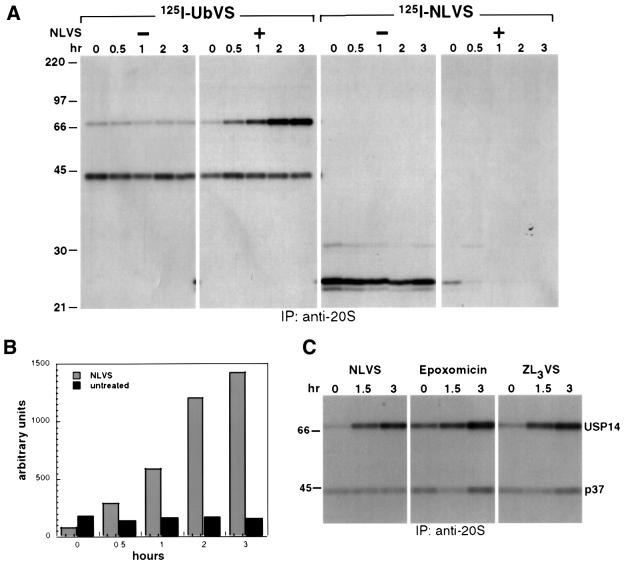
Complete Ub removal is thought to precede proteasomal proteolysis. Conversely, when proteasomal proteolysis is blocked, the resultant accumulation of Ub-conjugated substrates may elicit enhanced activity of DUBs. In other words, the activities of the proteasome and associated DUBs may be interdependent. Vinyl sulfones are mechanism-based inhibitors and, consequently, the increase in labeling of a given target enzyme is directly proportional to that enzyme’s ability to bind and hydrolyze the peptide bond at the C-terminus of Ub. [125I]UbVS thus provides a convenient tool to examine the enzymatic activity of DUBs in response to proteasome inhibition. We incubated intact EL-4 cells with 50 µM 4-hydroxy-3-iodo-2-nitro phenyl-leucinyl-leucinyl-leucine vinyl sulfone (NLVS) for different times followed by preparation of cell extracts. After incubation of the extracts with [125I]UbVS or [125I]NLVS, we immunoprecipitated the proteasome with an anti-20S serum (Figure 5A). We observed two [125I]UbVS-labeled polypeptides, a 45 kDa polypeptide (corresponding to p37) and a 66 kDa polypeptide (USP14). Whereas labeling of p37 does not change upon NLVS treatment (Figure 5A, second panel), the labeling of proteasome associated USP14 is increased up to 15-fold (Figure 5B) upon inclusion of NLVS (Figure 5A, second panel) in a time-dependent manner. The observed increase in [125I]UbVS labeling of USP14 is not unique to proteasome inhibition by NLVS, as treatment of cells with other proteasome inhibitors such as epoxomicin (Meng et al., 1999), ZL3VS and lactacystin (Fenteany et al., 1995) produces a similar effect (Figure 5C and data not shown). This increase may be due to recruitment of more USP14 to the proteasome complex or could be caused by increased activity of USP14 already associated with the proteasome.
Fig. 5. [125I]UbVS labeling of proteasome-associated USP14 is increased upon proteasome inhibition. (A) A total of 5 × 106 EL-4 cells were treated with 50 µM NLVS for the indicated times. Lysates were normalized for total protein and incubated with [125I]UbVS or [125I]NLVS for 1 h. Proteasomes were immunoprecipitated as described in Figure 4B. (B) Intensities of USP14 bands were quantified by densitometry. (C) EL-4 cells were incubated with 50 µM NLVS or ZL3VS or 4 µM epoxomicin for the indicated times. Proteasomes were immunoprecipitated as described in Figure 4B.
Activation of USP14 in response to proteasome inhibition
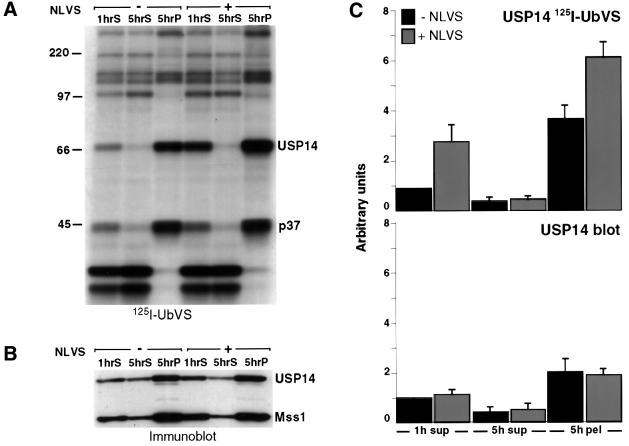
The observed increase in USP14 labeling is independent of protein synthesis, as inclusion of the protein synthesis inhibitor puromycin does not abolish increased USP14 labeling (data not shown). Is pre-existing free USP14 recruited to the proteasome in response to NLVS treatment? We incubated EL-4 cells with NLVS and carried out subcellular fractionation of cell extracts. The fractions were labeled with [125I]UbVS and analyzed by SDS– PAGE. Both USP14 and p37 are abundantly present in the 5 h pellet fraction, enriched for proteasomes and other large subcellular particles, while most other DUBs remain cytosolic or are distributed over both cytosol and the 5 h pellet (Figure 6A). Analysis of the same fractions by immunoblot demonstrates that USP14 indeed co-sediments with Mss1, an ATPase in the 19S cap of the proteasome (Figure 6B). As assessed by immunoblot, the protein levels of USP14 are comparable in cells treated or not with NLVS either before (Figure 6B) or after lysis (Figure 6C), while the [125I]UbVS labeling of USP14 in NLVS-treated 5 h pellets and 1 h supernatants is increased by 2- to 3-fold (average of four experiments; Figure 6C). The similar increase in USP14 labeling observed when EL-4 subcellular fractions are treated with NLVS after lysis (Figure 6C) confirms the lack of a requirement for ongoing protein synthesis. The 5 h pellet fraction contains only a small amount of soluble USP14. The recruitment of this material to the proteasome upon NLVS treatment would not account for the observed increase in labeling (Figure 6C). Combined, our observations are consistent with the notion that the increase in [125I]UbVS labeling of proteasome-associated USP14 is due to an increase in activity of USP14, and not due to recruitment of more USP14 to the proteasome complex. The increase in labeling intensity is less pronounced in the 5 h pellets than in anti-20S immunoprecipitations (Figures 5A and B, and 6A). This may be due to the presence of USP14 associated with other high molecular weight complexes in the 5 h pellets, which are unaffected by proteasome inhibitor or the technical differences between the two methods, i.e. immunoisolation versus subcellular fractionation, used to purify or enrich proteasomes. Immuno isolation does not result in quantitative recovery of proteasomes and may yield only a subset of the total proteasome population.
Fig. 6. Activity of USP14 is increased in response to proteasome inhibition. (A) Subcellular fractions from EL-4 cells previously treated with 50 µM NLVS for 5 h were incubated with [125I]UbVS, resolved by 10% SDS–PAGE and visualized by autoradiography. 1hS, 1 h supernatant; 5hS, 5 h supernatant; 5hP, 5 h pellet. (B) Parallel samples were resolved by 10% SDS–PAGE and immunoblotted with anti-USP14 and anti-Mss1 antisera. (C) Subcellular fractions prepared from EL-4 cell extracts were incubated with 50 µM NLVS for 3 h, resolved by 10% SDS–PAGE and visualized by autoradiography (upper panel), and by immunoblot using anti-USP14 (lower panel). The intensities of USP14 bands obtained from four independent experiments were quantified by densitometry and normalized to the amount of USP14 labeled by [125I]UbVS (upper panel) and USP14 protein (lower panel) observed in 1 h supernatant fractions.
Discussion
Here we report the synthesis of UbVS, a covalent inhibitor of DUBs. We use genetic criteria as well as a comparison with a reversible inhibitor (Ubal) to establish specificity of the affinity probe. We used UbVS to detect DUBs in crude extracts and to uncover the presence of an additional DUB, USP14, in association with the 26S proteasome. Remarkably, the intensity of USP14 labeling with UbVS is inversely proportional to the activity of the proteasome, thus demonstrating physical and functional interaction among different components of the ubiquitin–proteasome pathway.
A key advantage of using [125I]UbVS as a probe for UBPs and UCHs is its mechanism-based, irreversible mode of labeling conferred by the vinyl sulfone moiety (Palmer et al., 1995). The thioether linkage results from attack of the DUB’s active site thiol on the vinyl sulfone of UbVS, a reactive Michael acceptor. This type of linkage is resistant to the reducing sample buffers used in SDS–PAGE, unlike the adduct formed with the Ub-isonitrile derivative described by Lam and co-workers (Lam et al., 1997b). It is therefore possible to use [125I]UbVS on crude lysates and directly visualize a subset of the active UBPs and UCHs, as proposed for other thiol proteases (Palmer, 1995). The intensity of labeling of a given enzyme by [125I]UbVS must correspond to its activity. However, comparisons of relative activities of different DUBs cannot be made without knowledge of their substrate specificity and affinity for UbVS. The use of UbVS obviates the need for purification of DUBs of interest to assess their activities.
Prior treatment of the recombinant UCH-L3 enzyme or crude extracts with an alkylating agent, N-ethylmaleimide, abolishes labeling (Figure 1 and data not shown). This is consistent with the requirement for an active site cysteine for the catalytic activity of DUBs (Wilkinson, 1997). Ubal, a known inhibitor of this class of enzymes, is an effective competitor of DUB [125I]UbVS labeling in cell extracts (Figures 2B and 3B). Both Ubal and UbVS bind UCH-L3 with a submicromolar binding constant (Dang et al., 1998; and data not shown).
The strongest argument for specific labeling by [125I]UbVS is based on a genetic approach. Deletion of DUB genes in budding yeast results in loss of [125I]UbVS labeling of polypeptides of the expected molecular weight, such that six of the 17 known and putative DUBs account for all bands labeled in crude yeast extracts, demonstrating the specificity of UbVS for this class of enzymes. The multiple bands seen for Ubp1p (Figure 2A) are likely to be due to proteolytic cleavage. Not all yeast DUBs are labeled by [125I]UbVS; this observation can be explained in several ways. First, the vinyl sulfone substitution may hinder interaction or not be reactive with a particular UBP’s active site and thus render that UBP refractory to labeling. Secondly, several of the UBPs may have higher affinity for poly-Ub chains than for monomeric Ub, as seen for isopeptidase T (Wilkinson et al., 1995). Finally, expression levels of some UBPs in logarithmically growing or stationary phase yeast (unpublished observations) may be too low for detection by this method. Inspection of the mRNA levels for the DUB family based on yeast genomic array analysis shows that three of the DUBs labeled (Ubp1, 2 and 6) have higher mRNA transcript levels than the majority of other DUBs, suggesting that overexpression may reveal other labelable DUBs (Holstege et al., 1998). Other studies have addressed the non-essential nature of yeast UBPs: deletions of single or multiple DUBs have been without severe phenotypes (Baker et al., 1992; Amerik et al., 2000; R.Casagrande and H.L.Ploegh, unpublished observations). These results suggest that functions of many UBPs may overlap or be regulatory and restricted to particular substrates or pathways, and that no single enzyme is obligatory for removal of Ub from substrates prior to proteasomal proteolysis. Synthesis of Ub derivatives with electrophiles other than vinyl sulfone could help identify additional active members of the DUB family.
Labeling of a variety of mammalian cell extracts, prepared from tissues or cultured cells, reveals a greater complexity of active DUBs (Figure 3), reflecting the larger number of mammalian UBPs and UCHs (Chung and Baek, 1999). One of the polypeptides labeled is similar in size to and has an affinity for Ubal comparable with yeast Ubp6p (Figures 2B and 3B). Using immunological criteria, we confirm that this polypeptide corresponds to the mammalian homolog of Ubp6, USP14 (Figure 4B). USP14 and Ubp6p have been studied in vitro, but their physiological function remains unclear. A deletion of Ubp6 in yeast is viable; reported phenotypes include sensitivity to canavinine and stabilization of the Ub-Pro-β-galactosidase (Wyndham et al., 1999). In vitro studies of recombinant USP14 show that the monomeric protein has low affinity for both Ub and non-hydrolyzable Ub dimers, consistent with the relatively poor competition of UbVS labeling by Ubal. USP14 is apparently unable to disassemble poly-Ub protein conjugates in vitro (Yin et al., 2000).
USP14 and its homologs possess a type II ubiquitin-like domain (Ubl) at the N-terminus (Schauber et al., 1998; Jentsch and Pyrowolakis, 2000). Ubp6 lacking a Ubl does not complement Ubp6 deletion phenotypes, but the Ubl is not necessary for in vitro processing of linear Ub fusions by Ubp6p (Wyndham et al., 1999). This suggests that the Ubl may be necessary for targeting USP14/Ubp6 to its interacting partner or substrate, but not for intrinsic catalytic activity. Proteins containing a type II Ubl, such as hPLIC1, Rad 23 and BAG-1, require this domain for interaction with the 26S proteasome (Schauber et al., 1998; Kleijnen et al., 2000; Luders et al., 2000). The 19S subunit, S5a, binds poly-Ub chains, and a direct interaction between S5a and the Ubl of hHR23 has been reported, suggesting that S5a may also bind other Ubl-containing proteins (Hiyama et al., 1999). The binding of Rad23 to S5a does not lead to its degradation. The role of the Ubl in USP14–proteasome association currently is under investigation. Preliminary data suggest that the Ubl does not target USP14 for degradation by the proteasome, as 35S-labeled USP14 is stable for long chase periods (6–24 h) (data not shown).
Transient association of several DUBs with the 26S proteasome has been observed, including Doa4 in yeast and Ap-UCH in Aplysia (Hegde et al., 1997; Papa et al., 1999). However, these enzymes are not detected in purified proteasome preparations. The only DUB considered to be a stable part of the 19S complex is a 37 kDa UCH found in pure proteasomes from human and Drosophila sources, and has been localized to the hinge region between the lid and the base of the 19S complex (Lam et al., 1997b; Holzl et al., 2000). A Ubal-insensitive deubiquitylating activity associated with the 26S complex was reported in an earlier study, but its identity was never established (Eytan et al., 1993). Recently, low amounts of Ubp6p were detected by mass spectrometry performed on affinity-purified yeast proteasomes (Verma et al., 2000; and data not shown). However, we do not detect USP14 in conventionally purified mammalian proteasomes, indicating that the USP14–proteasome association is less stable than that of p37 and that recovery of USP14 is highly dependent on the experimental method used.
Labeling of p37 with [125I]UbVS is unaffected by the presence of proteasome inhibitors (Figure 5A), whereas USP14 labeling is increased as much as 15-fold in a time-dependent manner (Figure 5B). This difference in response to proteasome inhibition indicates that USP14 functions differently from p37, an activity thought to edit the poly-Ub chains on proteasome substrates by cleaving from the distal end of the chain (Lam et al., 1997a,b). Proteasome-associated USP14 is likely to encounter poly-Ub-conjugated proteins as its substrates. Preliminary observations indicate that only the proteasome-bound form of USP14 can be labeled with [125I]UbVS (Figure 6 and data not shown), suggesting that the affinity of proteasome-bound USP14 for Ub may be different from that determined with the free recombinant protein (Yin et al., 2000). If so, USP14 is the first example of a DUB whose substrate specificity and activity are regulated by association with a binding partner.
The observed increase in USP14 labeling is not due to up-regulation of de novo synthesis of USP14, as a similar increase is seen in cell-free lysates treated with NLVS and in whole cells in which translation is blocked by addition of puromycin (Figure 6 and data not shown). The increase in labeling could be due to the stabilization of the proteasome complex in the presence of inhibitor, resulting in binding of additional USP14 protein. A similar observation was made for the proteasome association of hPLIC1 and 2 (Kleijnen et al., 2000). However, the USP14 protein level in proteasome-enriched fractions is unaffected by NLVS treatment (Figure 6B), even though an increase in [125I]UbVS labeling of USP14 is observed, and the recruitment of the small amount of soluble USP14 protein present in the 5 h supernatant fraction would not explain this effect (Figure 6B and C).
We propose a functional coupling between the activities of the proteasome and that of proteasome-bound USP14. How can such coupling be achieved? One possible mechanism is the activation of USP14 by a conformational change propagated through the entire 26S complex upon proteasome inhibition. Evidence for the transmission of a conformational change upon engagement of HslU in the active site of HslV has been reported recently (Sousa et al., 2000), demonstrating the possibility of long-range conformational changes within large protein complexes such as the proteasome. On the other hand, USP14 activity may be regulated by the level of poly-ubiquitylated substrates, which accumulate upon proteasome inhibition if Ub conjugation continues under our experimental conditions. The DUB-specific affinity probe, UbVS, will be a useful tool to define further the role of USP14 in proteasome-mediated protein degradation.
Materials and methods
Cell lines and antibodies
EL-4 and NIH 3T3 mouse cell lines were maintained under standard cell culture conditions (Bogyo et al., 1998). The rabbit anti-human HAUSP peptide serum r201 was a generous gift of Dr Roger Everett (Everett et al., 1997) and the rabbit anti-20S proteasome and anti-αC9 antisera were generous gifts of Dr John Monaco (Brown et al., 1991). Rabbit antiserum against Mss1 was purchased from Affinity Research Products Ltd (Exeter, UK). Antiserum against mouse USP14 (HM433 and HM434) was raised in New Zealand White rabbits immunized with keyhole limpet hemocyanin (KLH) coupled to four synthetic peptides from the USP14 sequence (Y3SVTVKWGKEKFEGVELNT21C; CK239SLIDQYFGVEF ETTMK256; CK289LRLQEEITKQSPTLQRNAL308; and C358TPELQE KMVSFRSKFKDLED378) synthesized on an Advanced ChemTech 440 MOS synthesizer (Louisville, KY, USA).
Inhibitors
The proteasome inhibitors NLVS and ZL3VS were synthesized as described (Bogyo, 1997), and epoxomicin and ubiquitin aldehyde were purchased from Affinity Research Products Ltd (Exeter, UK).
Yeast strains, media and methods
Media were prepared as described (Sherman, 1991). All yeast cultures were grown in rich media (YPD) at 30°C. A wild-type strain MHY501 (Matα, his3-Δ200, leu2-3,112, ura-52, lys2-801, trp1-1) was used. Strains with deletions in UBP genes were otherwise genetically identical to MHY501 (MHY526, Δubp1::URA3; MHY648, Δubp2::TRP1; MHY821, Δubp6::HIS3; MHY887, Δubp12::HIS3; MHY989, Δubp15::HIS3; MHY525, Δyuh1::LEU2). All yeast strains were a generous gift of the Hochstrasser laboratory.
To prepare yeast lysates, 8 OD of exponentially growing yeast cells were harvested. Cells were resuspended in phosphate-buffered saline (PBS) containing 2 mM ATP, 1.5 mM dithiothreitol (DTT), 20 µM PMSF, 1 µM N-tosyl-l-phenylalanine chloromethyl ketone (TPCK), 1 µM leupeptin, 1 µM pepstatin and lysed by vortexing with glass beads. Lysates were centrifuged to remove cell debris and nuclei. An aliquot of 50–100 µg of lysate was used for labeling with [125I]UbVS.
Purification of UCH-L3
The E.coli expression vector for UCH-L3 was a kind gift of Dr Christopher Larsen. The protein was overexpressed in E.coli and purified as described (Larsen et al., 1996) except that Q Sepharose and Sephadex 75 FPLC columns were used. A 66 ng aliquot of UCH-L3 was used for labeling with [125I]UbVS.
Synthesis of ubiquitin vinyl sulfone (UbVS)
Ubiquitin75-ethyl ester (Ub75OEt) was synthesized and purified by gel filtration and cation-exchange chromatography as described (Wilkinson et al., 1986, 1990), except that a final purification step on a Pharmacia MonoS 1.6/5 column was included. Ub75OEt was converted to Ub75-hydrazine as described and used without further purification (Wilkinson et al., 1990). Trans-Boc-Gly-VS was synthesized as previously described (Bogyo et al., 1998) and deprotected prior to use by treatment with 50% trifluoroacetic acid in methylene chloride or treatment with p-toluene sulfonic acid. Ub75-hydrazine was converted to Ub75-azide by treatment with 0.5 M nitrous acid at –5°C for 1 min and immediately coupled with NH2-Gly-VS in the presence of triethylamine (Wilkinson et al., 1990). After a 5 min incubation at –5°C, the reaction was dialyzed against 50 mM sodium acetate pH 4.5 and purified on the Pharmacia SMART system MonoS 1.6/5 column to 95% purity. UbVS was identified by liquid chromatography/mass spectroscopy, using an LCZ electrospray mass spectrometer instrument (Micromass, UK) coupled with an HP1100 HPLC system (Hewlett Packard, USA). A C4 reverse phase HPLC column with a 0–80% gradient over 20 min and a 0.1% formic acid/acetonitrile buffer system was used.
Preparation of mammalian cell extracts and labeling with [125I]UbVS
A total of 5 × 108 EL-4 cells were harvested and washed three times with PBS. Cell pellets were lysed with glass beads in buffer HR (50 mM Tris pH 7.4, 5 mM MgCl2, 250 mM sucrose, 1 mM DTT, 2 mM ATP). Nuclei were removed by centrifugation, and 20 µg of lysate was used for labeling with [125I]UbVS. A 40 µg aliquot of UbVS was iodinated as described for Ub (Ciechanover et al., 1980), except Iodo-gen was used as a catalyst and 1 mg/ml hen egg lysozyme was added as carrier protein after quenching the reaction. A total of 0.5 × 106–1 × 106 c.p.m. of [125I]UbVS was incubated with cell extracts for 1 h at 37°C. Samples were resolved by reducing SDS–PAGE, and analyzed by autoradiography.
Immunoprecipitation
Anti-proteasome immunoprecipitations were carried out on 80 µg of EL-4 lysates, prepared as above, and previously labeled with 2 × 106 c.p.m. of [125I]UbVS or [125I]NLVS. Immunoprecipitation conditions described by Luders and co-workers were used (Luders et al., 2000). Briefly, samples were resuspended in proteasome IP buffer (25 mM Tris pH 7.5, 100 mM KCl, 0.5% Tween-20, 2 mM MgCl2, 1 mM ATP, 1 mM PMSF, 2 µg/ml aprotinin, 0.5 µg/ml leupeptin), and protease inhibitors were omitted after the pre-clearing step. Samples were pre-cleared twice with normal rabbit serum and immunoprecipitated with 3 µl of anti-20S proteasome serum; immune complexes were recovered with fixed Staphylococcus aureus. Pellets were washed three times with 1 ml of proteasome IP buffer before addition of SDS–PAGE sample buffer, followed by analysis by SDS– PAGE and autoradiography. Immunoprecipitations with anti-USP14 antibodies were carried out on lysates denatured with 1% SDS in PBS. SDS concentration was decreased to <0.01% with NP-40 lysis buffer (10 mM Tris pH 7.8, 0.5% NP-40, 150 mM NaCl, 5 mM MgCl2) for subsequent pre-clear and immunoprecipitation. StaphA immunoprecipitates were washed three times with NET buffer (0.5% NP-40, 50 mM Tris pH 7.4, 150 mM NaCl, 5 mM EDTA).
Immunoblotting
Immunoblotting was carried out according to published protocols (Bonifacino, 2000). Briefly, samples were resolved by SDS–PAGE and blotted onto PVDF membranes, blocked with 5% milk 0.1% Tween in PBS and incubated with primary antibody at 1:1000 dilution, unless otherwise indicated. Detection was by chemiluminesence, using horseradish peroxidase-coupled goat-anti-rabbit as secondary antibody. For the anti-USP7 immunoblot, 20 µg of EL-4 cell lysate were pre-incubated for 1 h with or without 2 µM UbVS, resolved by SDS–PAGE and immunoblotted for USP7 (HAUSP) with r201 antiserum as described (Everett et al., 1997).
Subcellular fractionations
Proteasome fractions were generated as described (Wang et al., 2000). Briefly, EL-4 cells were lysed in HR buffer and lysates were centrifuged for 1 h at 100 000 g to pellet membranes; the supernatants were spun for an additional 5 h at 100 000 g to produce the proteasome enriched fraction.
Superose 6 column
EL-4 lysates were centrifuged for 1 h at 100 000 g and fractionated in HR buffer on a preparative Pharmacia Superose 6 column using the AKTA FPLC system (Pharmacia, Sweden).
Acknowledgments
Acknowledgements
We would like to thank Roger Everett for the gift of HAUSP antibodies, Mark Hochstrasser for the gift of yeast DUB deletion strains, Chris Larsen for the UCH-L3 expression vector, and Brian Hekking for the synthesis of USP14 peptides. A.B. was a recipient of an NSF pre-doctoral fellowship, B.M.K. was supported by a Human Frontiers Science Program Organization long-term fellowship and currently is supported by a Roche Research Foundation fellowship. H.S.O. was supported by the Netherlands Organization for Scientific Research (NWO). K.D.W. is supported by NIH GM30308. This work was supported in part by a grant from Hoechst Marion Roussel.
References
- Amerik A.Y., Li,S.J. and Hochstrasser,M. (2000) Analysis of the deubiquitinating enzymes of the yeast Saccharomyces cerevisiae. Biol. Chem., 381, 981–992. [DOI] [PubMed] [Google Scholar]
- Baker R.T., Tobias,J.W. and Varshavsky,A. (1992) Ubiquitin-specific proteases of Saccharomyces cerevisiae. Cloning of UBP2 and UBP3 and functional analysis of the UBP gene family. J. Biol. Chem., 267, 23364–23375. [PubMed] [Google Scholar]
- Bogyo M.S. (1997) Peptide vinyl sulfones: inhibitors and active site directed probes for the study of proteasome function in vivo. PhD thesis, Massachusetts Institute of Technology.
- Bogyo M., McMaster,J.S., Gaczynska,M., Tortorella,D., Goldberg,A.L. and Ploegh,H. (1997) Covalent modification of the active site threonine of proteasomal β subunits and the Escherichia coli homolog HslV by a new class of inhibitors. Proc. Natl Acad. Sci. USA, 94, 6629–6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogyo M., Shin,S., McMaster,J.S. and Ploegh,H.L. (1998) Substrate binding and sequence preference of the proteasome revealed by active-site-directed affinity probes. Chem. Biol., 5, 307–320. [DOI] [PubMed] [Google Scholar]
- Bonifacino J.S. (2000) Electrophoresis and immunoblotting. In Bonifacino,J.S., Dasso,M., Harford,J.B., Lippincott-Schwartz,J. and Yamada,K.M. (eds), Current Protocols in Cell Biology. Vol. 1. John Wiley & Sons, New York, NY, pp. 6.2.1–6.2.16.
- Brown M.G., Driscoll,J. and Monaco,J.J. (1991) Structural and serological similarity of the MHC-linked LMP and proteasome (multicatalytic proteinase) complex. Nature, 353, 355–357. [DOI] [PubMed] [Google Scholar]
- Chung C.H. and Baek,S.H. (1999) Deubiquitinating enzymes: their diversity and emerging roles. Biochem. Biophys. Res. Commun., 266, 633–640. [DOI] [PubMed] [Google Scholar]
- Ciechanover A. and Schwartz,A.L. (1998) The ubiquitin–proteasome pathway: the complexity and myriad functions of protein death. Proc. Natl Acad. Sci. USA, 95, 2727–2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A., Heller,H., Elias,S., Haas,A.L. and Hershko,A. (1980) ATP-dependent conjugation of reticulocyte proteins with the peptide required for protein degradation. Proc. Natl Acad. Sci. USA, 77, 1365–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A., Breitschopf,K., Hatoum,O.A. and Bengal,E. (1999) Degradation of MyoD by the ubiquitin pathway: regulation by specific DNA-binding and identification of a novel site for ubiquitination. Mol. Biol. Rep., 26, 59–64. [DOI] [PubMed] [Google Scholar]
- Dang L.C., Melandri,F.D. and Stein,R.L. (1998) Kinetic and mechanistic studies on the hydrolysis of ubiquitin C-terminal 7-amido-4-methylcoumarin by deubiquitinating enzymes. Biochemistry, 37, 1868–1879. [DOI] [PubMed] [Google Scholar]
- Everett R.D., Meredith,M., Orr,A., Cross,A., Kathoria,M. and Parkinson,J. (1997) A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein [corrected and republished in EMBO J., 1997, 16, 1519–1530]. EMBO J., 16, 566–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eytan E., Armon,T., Heller,H., Beck,S. and Hershko,A. (1993) Ubiquitin C-terminal hydrolase activity associated with the 26S protease complex. J. Biol. Chem., 268, 4668–4674. [PubMed] [Google Scholar]
- Fenteany G., Standaert,R.F., Lane,W.S., Choi,S., Corey,E.J. and Schreiber,S.L. (1995) Inhibition of proteasome activities and subunit-specific amino-terminal threonine modification by lacta cystin. Science, 268, 726–731. [DOI] [PubMed] [Google Scholar]
- Hegde A.N., Inokuchi,K., Pei,W., Casadio,A., Ghirardi,M., Chain,D.G., Martin,K.C., Kandel,E.R. and Schwartz,J.H. (1997) Ubiquitin C-terminal hydrolase is an immediate-early gene essential for long-term facilitation in Aplysia. Cell, 89, 115–126. [DOI] [PubMed] [Google Scholar]
- Hershko A. and Ciechanover,A. (1998) The ubiquitin system. Annu. Rev. Biochem., 67, 425–479. [DOI] [PubMed] [Google Scholar]
- Hershko A. and Rose,I.A. (1987) Ubiquitin-aldehyde: a general inhibitor of ubiquitin-recycling processes. Proc. Natl Acad. Sci. USA, 84, 1829–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiyama H., Yokoi,M., Masutani,C., Sugasawa,K., Maekawa,T., Tanaka,K., Hoeijmakers,J.H. and Hanaoka,F. (1999) Interaction of hHR23 with S5a. The ubiquitin-like domain of hHR23 mediates interaction with S5a subunit of 26S proteasome. J. Biol. Chem., 274, 28019–28025. [DOI] [PubMed] [Google Scholar]
- Hodgins R., Gwozd,C., Arnason,T., Cummings,M. and Ellison,M.J. (1996) The tail of ubiquitin-conjugating enzyme redirects multi-ubiquitin chain synthesis from the lysine 48-linked configuration to a novel nonlysine-linked form. J. Biol. Chem., 271, 28766–28771. [DOI] [PubMed] [Google Scholar]
- Holstege F.C., Jennings,E.G., Wyrick,J.J., Lee,T.I., Hengartner,C.J., Green,M.R., Golub,T.R., Lander,E.S. and Young,R.A. (1998) Dissecting the regulatory circuitry of a eukaryotic genome. Cell, 95, 717–728. [DOI] [PubMed] [Google Scholar]
- Holzl H. et al. (2000) The regulatory complex of Drosophila melanogaster 26S proteasomes. Subunit composition and localization of a deubiquitylating enzyme. J. Cell Biol., 150, 119–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Baker,R.T. and Fischer-Vize,J.A. (1995) Control of cell fate by a deubiquitinating enzyme encoded by the fat facets gene. Science, 270, 1828–1831. [DOI] [PubMed] [Google Scholar]
- Jensen D.E. et al. (1998) BAP1: a novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene, 16, 1097–1112. [DOI] [PubMed] [Google Scholar]
- Jentsch S. and Pyrowolakis,G. (2000) Ubiquitin and its kin: how close are the family ties? Trends Cell Biol., 10, 335–342. [DOI] [PubMed] [Google Scholar]
- Kleijnen M.F., Shih,A.H., Zhou,P., Kumar,S., Soccio,R.E., Kedersha,N.L., Gill,G. and Howley,P.M. (2000) The hPLIC proteins may provide a link between the ubiquitination machinery and the proteasome. Mol. Cell, 6, 409–419. [DOI] [PubMed] [Google Scholar]
- Koegl M., Hoppe,T., Schlenker,S., Ulrich,H.D., Mayer,T.U. and Jentsch,S. (1999) A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell, 96, 635–644. [DOI] [PubMed] [Google Scholar]
- Lam Y.A., DeMartino,G.N., Pickart,C.M. and Cohen,R.E. (1997a) Specificity of the ubiquitin isopeptidase in the PA700 regulatory complex of 26S proteasomes. J. Biol. Chem., 272, 28438–28446. [DOI] [PubMed] [Google Scholar]
- Lam Y.A., Xu,W., DeMartino,G.N. and Cohen,R.E. (1997b) Editing of ubiquitin conjugates by an isopeptidase in the 26S proteasome. Nature, 385, 737–740. [DOI] [PubMed] [Google Scholar]
- Larsen C.N., Price,J.S. and Wilkinson,K.D. (1996) Substrate binding and catalysis by ubiquitin C-terminal hydrolases: identification of two active site residues. Biochemistry, 35, 6735–6744. [DOI] [PubMed] [Google Scholar]
- Li T., Naqvi,N.I., Yang,H. and Teo,T.S. (2000) Identification of a 26S proteasome-associated UCH in fission yeast. Biochem. Biophys. Res. Commun., 272, 270–275. [DOI] [PubMed] [Google Scholar]
- Luders J., Demand,J. and Hohfeld,J. (2000) The ubiquitin-related BAG-1 provides a link between the molecular chaperones Hsc70/hsp70 and the proteasome. J. Biol. Chem., 275, 4613–4617. [DOI] [PubMed] [Google Scholar]
- Meng L., Mohan,R., Kwok,B.H., Elofsson,M., Sin,N. and Crews,C.M. (1999) Epoxomicin, a potent and selective proteasome inhibitor, exhibits in vivo antiinflammatory activity. Proc. Natl Acad. Sci. USA, 96, 10403–10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D. and Johnson,D. (1996) A deubiquitinating enzyme interacts with SIR4 and regulates silencing in S.cerevisiae. Cell, 86, 667–677. [DOI] [PubMed] [Google Scholar]
- Ortolan T.G., Tongaonkar,P., Lambertson,D., Chen,L., Schauber,C. and Madura,K. (2000) The DNA repair protein Rad23 is a negative regulator of multi-ubiquitin chain assembly. Nature Cell Biol., 2, 601–608. [DOI] [PubMed] [Google Scholar]
- Palmer J.T., Rasnick,D., Klaus,J.L. and Bromme,D. (1995) Vinyl sulfones as mechanism-based cysteine protease inhibitors. J. Med. Chem., 38, 3193–3196. [DOI] [PubMed] [Google Scholar]
- Papa F.R. and Hochstrasser,M. (1993) The yeast DOA4 gene encodes a deubiquitinating enzyme related to a product of the human tre-2 oncogene. Nature, 366, 313–319. [DOI] [PubMed] [Google Scholar]
- Papa F.R., Amerik,A.Y. and Hochstrasser,M. (1999) Interaction of the Doa4 deubiquitinating enzyme with the yeast 26S proteasome. Mol. Biol. Cell, 10, 741–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart C.M. and Rose,I.A. (1985) Ubiquitin carboxyl-terminal hydrolase acts on ubiquitin carboxyl-terminal amides. J. Biol. Chem., 260, 7903–7910. [PubMed] [Google Scholar]
- Schauber C., Chen,L., Tongaonkar,P., Vega,I., Lambertson,D., Potts,W. and Madura,K. (1998) Rad23 links DNA repair to the ubiquitin/proteasome pathway. Nature, 391, 715–718. [DOI] [PubMed] [Google Scholar]
- Sherman F. (1991) Getting started with yeast. Methods Enzymol., 194, 3–21. [DOI] [PubMed] [Google Scholar]
- Sousa M.C., Trame,C.B., Tsuruta,H., Wilbanks,S.M., Reddy,V.S. and McKay,D.B. (2000) Crystal and solution structures of an HslUV protease–chaperone complex. Cell, 103, 633–643. [DOI] [PubMed] [Google Scholar]
- Verma R., Chen,S., Feldman,R., Schieltz,D., Yates,J., Dohmen,J. and Deshaies,R.J. (2000) Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol. Biol. Cell, 11, 3425–3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voges D., Zwickl,P. and Baumeister,W. (1999) The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem., 68, 1015–1068. [DOI] [PubMed] [Google Scholar]
- Wang E.W., Kessler,B.M., Borodovsky,A., Cravatt,B.F., Bogyo,M., Ploegh,H.L. and Glas,R. (2000) Integration of the ubiquitin– proteasome pathway with a cytosolic oligopeptidase activity. Proc. Natl Acad. Sci. USA, 97, 9990–9995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson K.D. (1997) Regulation of ubiquitin-dependent processes by deubiquitinating enzymes. FASEB J., 11, 1245–1256. [DOI] [PubMed] [Google Scholar]
- Wilkinson K.D., Cox,M.J., Mayer,A.N. and Frey,T. (1986) Synthesis and characterization of ubiquitin ethyl ester, a new substrate for ubiquitin carboxyl-terminal hydrolase. Biochemistry, 25, 6644–6649. [DOI] [PubMed] [Google Scholar]
- Wilkinson K.D., Lee,K.M., Deshpande,S., Duerksen-Hughes,P., Boss,J.M. and Pohl,J. (1989) The neuron-specific protein PGP 9.5 is a ubiquitin carboxyl-terminal hydrolase. Science, 246, 670–673. [DOI] [PubMed] [Google Scholar]
- Wilkinson K.D., Smith,S.E., O’Connor,L., Sternberg,E., Taggart,J.J., Berges,D.A. and Butt,T. (1990) A specific inhibitor of the ubiquitin activating enzyme: synthesis and characterization of adenosyl-phospho-ubiquitinol, a nonhydrolyzable ubiquitin adenylate analogue. Biochemistry, 29, 7373–7380. [DOI] [PubMed] [Google Scholar]
- Wilkinson K.D., Deshpande,S. and Larsen,C.N. (1992) Comparisons of neuronal (PGP 9.5) and non-neuronal ubiquitin C-terminal hydrolases. Biochem. Soc. Trans, 20, 631–637. [DOI] [PubMed] [Google Scholar]
- Wilkinson K.D., Tashayev,V.L., O’Connor,L.B., Larsen,C.N., Kasperek,E. and Pickart,C.M. (1995) Metabolism of the polyubiquitin degradation signal: structure, mechanism and role of isopeptidase T. Biochemistry, 34, 14535–14546. [DOI] [PubMed] [Google Scholar]
- Wyndham A.M., Baker,R.T. and Chelvanayagam,G. (1999) The Ubp6 family of deubiquitinating enzymes contains a ubiquitin-like domain: SUb. Protein Sci., 8, 1268–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L., Krantz,B., Russell,N.S., Deshpande,S. and Wilkinson,K.D. (2000) Nonhydrolyzable diubiquitin analogues are inhibitors of ubiquitin conjugation and deconjugation. Biochemistry, 39, 10001–10010. [DOI] [PubMed] [Google Scholar]
- Zhu Y., Pless,M., Inhorn,R., Mathey-Prevot,B. and D’Andrea,A.D. (1996) The murine DUB-1 gene is specifically induced by the β subunit of interleukin-3 receptor. Mol. Cell. Biol., 16, 4808–4817. [DOI] [PMC free article] [PubMed] [Google Scholar]