Abstract
Yeast endosomes, like those in animal cells, invaginate their membranes to form internal vesicles. The resulting multivesicular bodies fuse with the vacuole, the lysosome equivalent, delivering the internal vesicles for degradation. We have partially purified internal vesicles and analysed their content. Besides the known component carboxypeptidase S (Cps1p), we identified a polyphosphatase (Phm5p), a presumptive haem oxygenase (Ylr205p/Hmx1p) and a protein of unknown function (Yjl151p/Sna3p). All are membrane proteins, and appear to be cargo molecules rather than part of the vesicle-forming machinery. We show that both Phm5p and Cps1p are ubiquitylated, and that in a doa4 mutant, which has reduced levels of free ubiquitin, Cps1p, Phm5p and Hmx1p are mis-sorted to the vacuolar membrane. Mutation of Lys 6 in the cytoplasmic tail of Phm5p disrupts its sorting, but sorting is restored, even in doa4 cells, by the biosynthetic addition of a single ubiquitin chain. In contrast, Sna3p enters internal vesicles in a ubiquitin-independent manner. Thus, ubiquitin acts as a signal for the partitioning of some, but not all, membrane proteins into invaginating endosomal vesicles.
Keywords: endosome/multivesicular body/ubiquitin/vacuole/yeast
Introduction
The endosomes of both yeast and mammalian cells undergo a process of membrane segregation that is crucial to their function. Regions of the limiting endosomal membrane invaginate to form internal vesicles and the resulting structure is known as a multivesicular body. Membrane proteins that enter the internal vesicles are sequestered from the cytoplasm, thus terminating any interactions they may have with signal transduction pathways. In yeast, the internalization process occurs mainly in late endosomes and seems to be irreversible; mature multivesicular bodies fuse with the vacuole, releasing the internal vesicles into the vacuolar interior where they are destroyed by hydrolytic enzymes (Odorizzi et al., 1998; Lemmon and Traub, 2000). Proteins that do not enter the internal vesicles are either removed by conventional vesicular transport prior to fusion with the vacuole, or become constituents of the delimiting vacuolar membrane.
The sorting process that delivers some membrane proteins to the internal membranes for destruction whilst others remain outside is not well understood. Most of the proteins known to reach the vacuole interior are plasma membrane proteins which undergo regulated turnover, such as the pheromone receptors and some permeases (Berkower et al., 1994; Volland et al., 1994; Egner et al., 1995; Lai et al., 1995; Hicke and Riezman, 1996; Medintz et al., 1996; Beck et al., 1999; Springael et al., 1999), but one hydrolytic enzyme, carboxypeptidase S (Cps1p), is also known to follow this route (Odorizzi et al., 1998). Once inside the vacuole, Cps1p is processed to release a soluble form of the active enzyme. We have recently shown that sorting of membrane proteins into the internal vesicles can be influenced by the nature of their transmembrane domains, the insertion of a potentially charged residue being sufficient to cause the internalization and destruction of Pep12p, a SNARE protein that normally remains on the endosomal surface (Reggiori et al., 2000).
To gain further insight into the sorting process, we have partially purified the vesicles that accumulate inside the vacuoles of a hydrolase-deficient yeast strain, identified proteins that are in them, and studied the targeting of these components. We detected three new proteins that follow the same pathway as Cps1p: a polyphosphatase (Phm5p), a protein with homology to haem oxygenases (Hmx1p) and a possible proton transporter (Sna3p). All are membrane proteins and appear to represent cargo molecules rather than components of the sorting machinery itself.
In animal cells, the fate of internalized epidermal growth factor (EGF) receptors has been shown to be controlled by ubiquitylation, which prevents their recycling to the surface and instead promotes their degradation in lysosomes (Levkowitz et al., 1998). Similarly, transport of the interleukin-2 receptor from early to late endosomes is ubiquitin dependent (Rocca et al., 2001). Turnover of plasma membrane proteins in yeast is also known to be regulated by ubiquitylation. Conjugation of ubiquitin, a 76 residue polypeptide, to the cytosolic regions of a variety of proteins stimulates their endocytosis (Hicke, 2001). Ubiquitin has also been implicated in the regulated sorting of tryptophan permease and of general amino acid permease from the trans-Golgi network to endosomes (Beck et al., 1999; Helliwell et al., 2001), and recent reports have suggested a possible involvement of ubiquitylation in vacuolar degradation of the a-factor transporter Ste6p and other membrane proteins (Amerik et al., 2000; Losko et al., 2001).
We show here that ubiquitylation of Phm5p is both necessary and sufficient for its incorporation into the internal vesicles of multivesicular bodies. Ubiquitin is also involved in the targeting of Cps1p and Hmx1p to these vesicles. However, Sna3p follows the same pathway in a ubiquitin-independent manner. Thus, ubiquitylation marks some, but not all, membrane proteins for sorting to the vacuolar interior.
Results
Multivesicular body purification
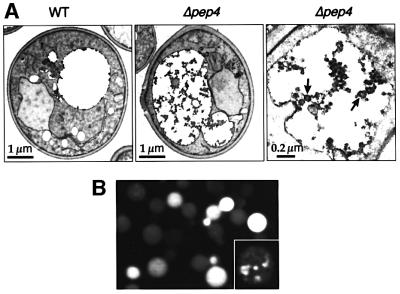
It has been shown that vesicles derived from multivesicular bodies accumulate inside the vacuole in Δvma4 cells. Vma4p is a subunit of the vacuolar H+-ATPase and in its absence vacuoles are not properly acidified, leading to a reduction of vacuolar hydrolase activity and a consequent stabilization of the internal vesicles (Morano and Klionsky, 1994; Odorizzi et al., 1998; Wurmser and Emr, 1998). Δpep4 cells also have a reduced vacuolar protease activity and similarly accumulate 40- to 50-nm vesicles inside the vacuole (Figure 1A; Baba et al., 1997). Some of these structures are derived from the cytoplasm to vacuole targeting (Cvt) pathway (Baba et al., 1997), a pathway akin to autophagy in which the cytosolic protein aminopeptidase I (API) is wrapped in membranes and delivered directly to the vacuole (Kim and Klionsky, 2000), but the remaining vesicles should be derived from multivesicular endosomes. In agreement with this, transmembrane proteins internalized into multivesicular bodies are not degraded in this Δpep4 strain (Reggiori et al., 2000).
Fig. 1. Vesicles accumulate inside protease-deficient vacuoles. (A) EM sections of wild-type (WT) and Δpep4 yeast. Vacuoles appear as large white areas in these permanganate-fixed cells, which are empty in the wild-type cells but contain aggregates with vesicular profiles in the mutant (arrows). (B) Isolated vacuoles purified from a Δpep4 strain expressing a GFP-tagged mutant form of Pep12p show internal fluorescence. The inset shows an enlarged single confocal scan of a vacuole, revealing punctate structures within it.
To mark endosome-derived vesicles, the Δpep4 strain was transformed with a green fluorescent protein (GFP)-tagged version of Pep12p with an aspartate in the third position of the transmembrane domain (TMD) (GFP–PEP3D). This construct is transported to late endosomes, enters internal vesicles, and is delivered to the vacuolar lumen (Reggiori et al., 2000). Figure 1B shows vacuoles purified from this strain (Haas, 1995), which on visual inspection could be seen to contain numerous rapidly moving internal fluorescent structures. Their motion created a homogeneous blur in confocal images obtained with multiple laser scans, but single-scan pictures revealed structures within this (Figure 1B, inset).
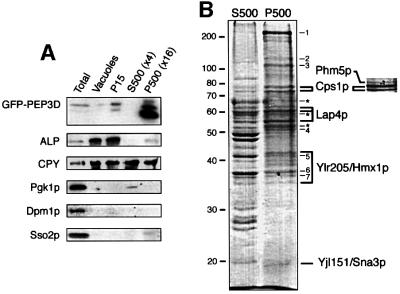
To identify proteins in these internal vesicles, we developed a procedure to enrich for them. Vacuoles were purified, collected by sedimentation, then osmotically lysed and the outer vacuole membranes pelleted at 15 000 g (P15). Soluble vacuolar proteases and internal vesicles were partially released into the supernatant, which was then centrifuged at 500 000 g to separate the small vesicles (P500) from soluble proteins (S500). GFP– PEP3D was strongly enriched in the P500 pellet, whereas the soluble vacuolar protease carboxypeptidase Y (CPY) and the vacuole membrane protein alkaline phosphatase (ALP) were not (Figure 2A). Other membranes such as the endoplasmic reticulum (ER), marked by Dpm1p, and the plasma membrane, marked by Sso2p, were largely removed during initial purification of the vacuoles, as was the cytosolic enzyme Pgk1p (Figure 2A).
Fig. 2. Isolation of internal vesicles from vacuoles. (A) The fractionation procedure described in the text and Materials and methods was carried out on pep4 cells expressing GFP–PEP3D. Immunoblotting was used to follow the distribution of proteins in total cell extract (total), purified vacuoles, the P15 pellet, the S500 supernatant and the P500 pellet. The P500 and S500 samples were overloaded as indicated; the total extract was substantially underloaded since only a few percent of the vacuoles were recovered in the initial purification step. Intact GFP–PEP3D is under-represented in the total vacuole fraction due to proteolysis during sample preparation (data not shown). Markers are alkaline phosphatase (ALP, vacuolar membrane marker), carboxypeptidase Y (CPY, vacuole lumen), phosphoglycerate kinase (Pgk1p, cytosolic), dolicholphosphomannose synthase (Dpm1p, ER) and the syntaxin Sso2p (plasma membrane). (B) Coomassie Blue-stained gel of the S500 and P500 fractions from a scaled-up preparation. Molecular weight markers are indicated on the left (kDa). Proteins in the P500 sample were identified by tryptic digestion and mass spectrometry. These were often present in multiple bands, presumably due to proteolysis, and these are indicated by the linked lines. Asterisks mark bands containing the GFP–PEP3D marker, which also contaminated all three of the Lap4p bands. Numbered bands were identified as follows: 1, Fas1p (fatty acyl CoA synthase); 2, Ena2p (plasma membrane Na+-ATPase); 3, Pma1p (plasma membrane H+-ATPase); 4, a mixture of Tef2p (translation elongation factor) and Eno2p (enolase); 5, Adh1p (alcohol dehydrogenase); 6, Tdh3p; 7, Tdh2p (glyceraldehyde-3-P dehydrogenases). Phm5p was not identified from this gel, but the relevant part of a silver-stained gel from which it was obtained is shown on the right.
Identification of proteins in internal vesicles
The purification procedure was scaled up, and the proteins in the various fractions separated by SDS–PAGE (Figure 2B). Bands enriched in the P500 fraction were identified by mass spectrometry of their tryptic peptides. In addition to the Pep12 chimera used as a marker, five other proteins were identified that might be in internal vesicles: Cps1p, Phm5p, Lap4p, Ylr205p and Yjl151p (Figure 3). Phm5p was found in only one preparation, perhaps because its synthesis depends on growth conditions (see below), but the others were each found at least twice. A number of other proteins from the P500 fraction were also identified, and found to be abundant cytosolic or plasma membrane proteins (see legend to Figure 2 for details). It is not clear whether these are simply contaminants, or whether some are partially present within the vacuole, but we did not investigate them further.
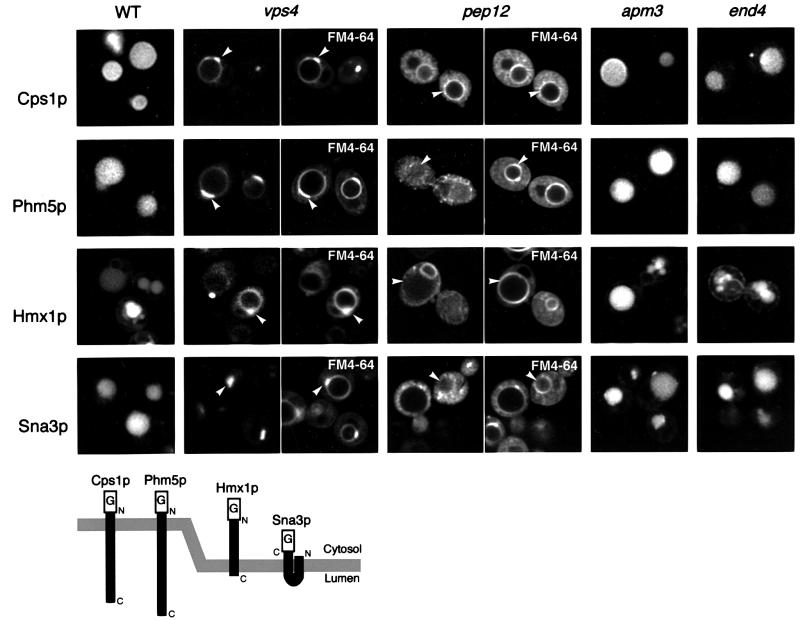
Fig. 3. Delivery of proteins to the vacuole interior occurs via endosomes. GFP-tagged versions of the four proteins identified in internal vesicles were expressed in the indicated deletion mutant strains. The vps4 and pep12 cells were double labelled with the dye FM4-64 to reveal vacuoles and endosomal compartments. The diagram at the bottom indicates the position of the GFP tag (G) and the topology of the proteins in the membrane, as predicted by the TMHMM program (http://www.cbs.dtu.dk/services).
Two of the enriched proteins were expected components of internal vesicles. Cps1p, a single transmembrane protein with a short cytoplasmic N-terminus and a large luminal C-terminus, was previously known to reach the vacuole in the vesicles delivered from endosomes (Odorizzi et al., 1998). Aminopeptidase I (Lap4p) is a soluble protease transported from the cytosol to the vacuole via the Cvt pathway and, as mentioned above, accumulates in internal vacuolar vesicles in a Δpep4 strain (Baba et al., 1997).
The other three proteins are all predicted to be integral membrane proteins. Phm5p is a polyphosphate endophosphatase. Yeast cells accumulate large amounts of polyphosphate chains in the vacuole which are used as a reservoir for phosphate (Urech et al., 1978). Phm5p is required to mobilize phosphate from these chains and its transcription is induced when extracellular phosphate levels fall (Ogawa et al., 2000). Phm5p has a single TMD and is predicted to have a topology similar to that of Cps1p.
Ylr205p is a C-tail anchored protein with a high similarity to members of the family of haem oxygenases; on the basis of this similarity, we propose the name HMX1 for the corresponding gene. These oxygenases cleave the haem ring to form biliverdin and carbon monoxide, and are found in both eukaryotes and photosynthetic bacteria. Despite extensive studies of these proteins in animal cells, no precise subcellular localization has been determined. However, human monocytes phagocytose the malarial pigment haemozoin, a polymer of haem rings, and this compound seems to be degraded by haem oxygenase in the lysosome (Schwarzer et al., 1999). The catalytic domain of Hmx1p is predicted to be exposed initially to the cytosol, but would end up in the vacuolar lumen; where it performs its function is unclear.
Yjl151p is a very small protein with two TMDs. It has three homologues in yeast and several in fungi, Caenorhabditis elegans and plants. One of the yeast homologues, Pmp3p/Sna1p, has been shown to be localized to the plasma membrane and to be involved in salt and charge homeostasis. It has been hypothesized to form a proton channel that limits the polarization of the plasma membrane, thus reducing the uptake of sodium ions (Navarre and Goffeau, 2000). Because of its similarity to SNA1, we propose the name SNA3 for YJL151C, and for the other two homologues SNA2 (YDR525W-A) and SNA4 (YDL123W), the numerical order reflecting the intracellular locations of the gene products (see below).
The newly identified proteins are delivered to the vacuole lumen through late endosomes
We tagged Phm5p, Hmx1p, Sna3p and Cps1p with GFP and expressed them in a wild-type strain (Figure 3, WT). All constructs showed a clear staining of the vacuole lumen. Since each was tagged on the predicted cytosolic portion of the protein, the luminal fluorescence suggests that they entered internal vesicles, as expected from their fractionation behaviour. Part of the GFP–Hmx1p remained in the ER, as indicated by fluorescence of the nuclear envelope. This may reflect a normal dual location, but we cannot exclude the possibility that the presence of the GFP tag slows the exit of Hmx1p from the ER.
Formation of multivesicular bodies is an endosomal function and seems to occur primarily in late endosomes in yeast. The process is blocked in class E vacuolar protein sorting mutants such as vps4 (Odorizzi et al., 1998; Reggiori et al., 2000). In vps4 cells, abnormal enlarged prevacuolar compartments accumulate (Babst et al., 1997), which can be visualized using the endocytic tracer dye FM4-64 (Vida and Emr, 1995). As shown in Figure 3, all our chimeras were trapped in these structures in vps4 cells, indicating that they normally pass through late endosomes.
Because one vps4 allele is known to affect autophagy (Shirahama et al., 1997), and thus conceivably other processes such as the Cvt pathway, we also expressed the chimeras in a Δpep12 mutant. Removal of Pep12p does not affect the Cvt pathway (Abeliovich et al., 1999), but prevents fusion of vesicles with the late endosome (Becherer et al., 1996). In the Δpep12 strain, Cps1p and the three newly identified proteins accumulated in very small structures, probably vesicles, without reaching the vacuole lumen (Figure 3); this was less clear for Hmx1p, due to its tendency to accumulate in the ER. Some of the GFP–Cps1p did reach the vacuole in Δpep12 cells, but it remained on the outer membrane. This may reflect some transport by the direct route from the Golgi to the vacuole, mediated by the AP-3 coat protein, which bypasses late endosomes and hence the invagination process that forms internal vesicles (Cowles et al., 1997; Stepp et al., 1997). This route is blocked in a Δapm3 mutant, which lacks one of the AP-3 subunits, but as expected the apm3 mutation did not prevent any of the GFP chimeras from reaching the vacuole lumen when the endosomal pathway was intact (Figure 3).
These results confirm that Cps1p, Phm5p, Hmx1p and Sna3p all normally reach the vacuole from late endosomes, and that passage through endosomes is essential for their delivery to the vacuole interior. Their localization was also largely unaffected in the endocytosis-deficient mutant end4, indicating that they pass directly from the Golgi to endosomes rather than via the plasma membrane (Figure 3).
To confirm the predicted membrane orientation of the proteins, we analysed the GFP-tagged versions by immunoblotting extracts of wild-type and vps4 cells with anti-GFP antibodies. In wild-type cells, entry into the vacuole should eventually expose the GFP tag to vacuolar proteases, whereas in vps4 cells, which cannot form multivesicular bodies, it should remain exposed to the cytosol. Figure 4 shows that partial cleavage of all of the proteins occurred in wild-type cells to yield free GFP (band 1). In vps4 cells, this band was not observed with any chimera, confirming that the GFP was not exposed to proteases. However, both Cps1p and Phm5p showed a GFP product that was 4 kDa larger (band 2), corresponding to cleavage immediately beyond the transmembrane domain. Such cleavage, mediated by proteases within the prevacuolar compartment or vacuoles, is a known feature of Cps1p biogenesis (Spormann et al., 1992). Hmx1p and Sna3p did not show cleavage in vps4 cells, consistent with their minimal luminal domains. Thus, this analysis fully supports the predicted structures of the proteins.
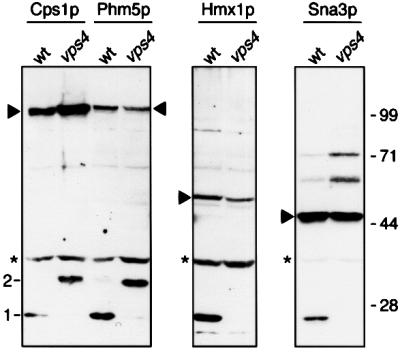
Fig. 4. Analysis of GFP chimeras. Western blots of total extracts from cells expressing the GFP-tagged versions of the indicated proteins were probed with anti-GFP antibodies. Triangles indicate full-length chimeras; asterisks mark a yeast protein that cross-reacts with the anti-GFP antibody. Bands at position 1 correspond to free GFP, those at position 2 contain GFP linked to the cytoplasmic tail and TMD of Cps1p or Phm5p. Molecular weight markers (kDa) are indicated on the right. The nature of the bands migrating more slowly than GFP–Sna3p is unknown; they may represent conformational isomers or protein complexes.
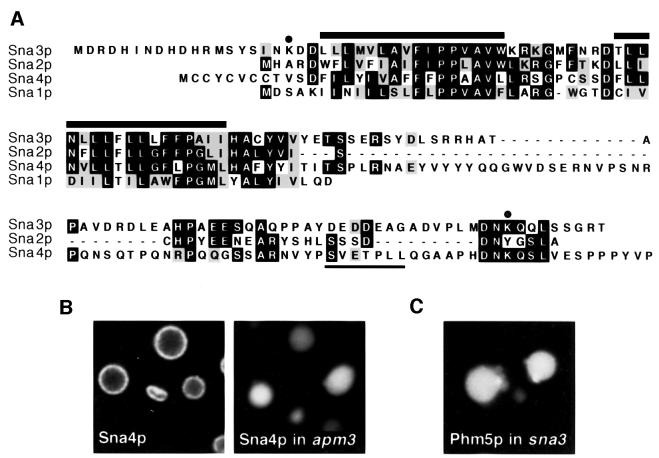
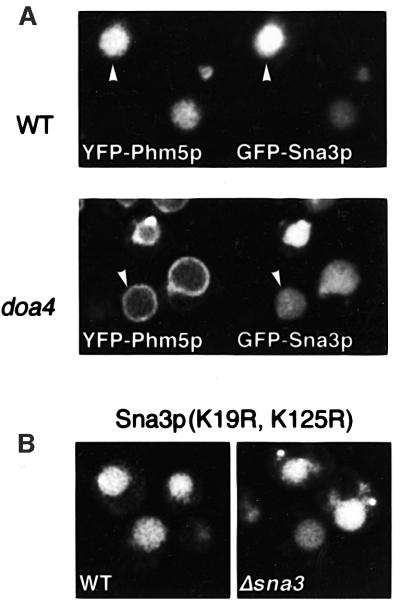
Since Sna3p is a member of a family of proteins in yeast, we investigated whether any other members of this family shared its location. Alignment of the four sequences shows that they are highly similar in the transmembrane domains, but diverge at the C-terminus (Figure 5A). GFP tagging confirmed the plasma membrane location of Sna1p and revealed a punctate pattern for Sna2p (not shown). However, Sna4p was on the outer vacuolar membrane (Figure 5B), raising the question of how it escapes the fate of its relative, Sna3p. One possibility is that it bypasses late endosomes by following the AP-3 pathway directly to vacuoles (Odorizzi et al., 1998; Reggiori et al., 2000), and indeed it contains an acidic di-leucine motif (Figure 5A), which is a likely AP-3 sorting signal (Darsow et al., 1998). We tested this hypothesis by expressing GFP–Sna4p in a Δapm3 mutant. In these cells, Sna4p is forced to pass through late endosomes to reach the vacuole, and like Sna3p is found in the lumen (Figure 5B). Hence, the ability to enter multivesicular bodies is not unique to Sna3p, but may be a common property of these proteins, perhaps mediated by their similar transmembrane segments.
Fig. 5. The Sna family of proteins in yeast. (A) Alignment of the Sna3p sequence with the other three, with identities and similarities indicated by black and grey backgrounds. Thick bars over the sequences indicate the TMD. Dots indicate cytoplasmic lysines in Sna3p; the acidic di-leucine motif in Sna4p is underlined. (B) GFP-tagged Sna4p is on the vacuolar membrane in wild-type cells, but inside the vacuoles in Δapm3 cells. (C) GFP–Phm5p shows its normal intra-vacuolar pattern in a Δsna3 mutant.
We also investigated whether Sna3p is itself required for the formation of multivesicular bodies, but found that delivery of Phm5p to the vacuole interior was normal in a Δsna3 strain (Figure 5C). We conclude that the three new proteins we have identified are cargo molecules that are sorted into the interior of multivesicular bodies. Although we cannot rule out a redundant role, we have no evidence that they are part of the machinery for the formation of these structures. Nevertheless, they provide tools to investigate the basis of this endosomal sorting step.
Ubiquitylation affects internalization in multivesicular bodies
A role for ubiquitin in endosomal sorting has been suggested recently (Amerik et al., 2000; Losko et al., 2001). These studies used a deletion strain lacking Doa4p, a ubiquitin-specific protease that removes ubiquitin from protein conjugates (Papa and Hochstrasser, 1993). Doa4p is important in the recycling of ubiquitin and a loss of its function results in a depletion of free ubiquitin, thus interfering with all ubiquitin-dependent processes (Swaminathan et al., 1999). When GFP-tagged versions of Cps1p, Phm5p and Hmx1p were expressed in the Δdoa4 strain, all three proteins were found predominantly on the vacuolar membrane, rather than inside the vacuole, indicating that their efficient internalization was dependent on ubiquitin (Figure 6).
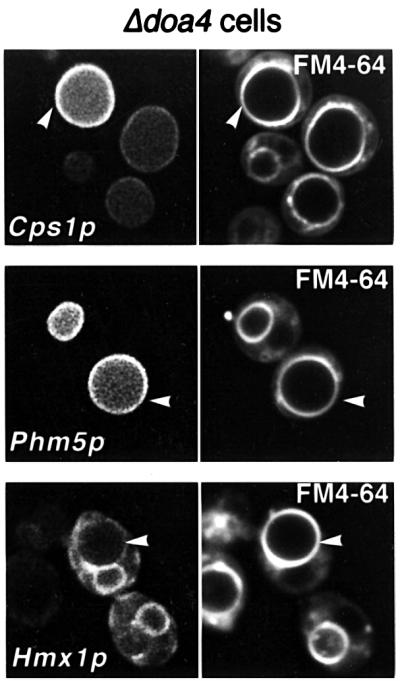
Fig. 6. Delivery to the vacuole interior is inhibited in Δdoa4 cells. GFP-tagged Cps1p, Phm5p and Hmx1p were expressed in Δdoa4 cells, which were also labelled with FM4-64. GFP fluorescence is more prominent on the vacuolar membrane than inside the vacuoles, in contrast to wild-type cells (see Figure 3). The brightest rings visible with GFP–Hmx1p correspond to the nuclear envelope.
Ubiquitin could have either a direct effect, marking proteins for internalization, or a more indirect effect on the formation of multivesicular bodies in general. As a first step to distinguish these possibilities, we investigated whether the proteins themselves were ubiquitylated. We focused on Cps1p and Phm5p, because they were delivered to the vacuole more efficiently than Hmx1p. These proteins were each tagged with protein A, with a tobacco etch virus (TEV) protease cleavage site at the junction to allow subsequent removal of the protein A. The constructs were introduced into a Δdoa4 strain that expressed myc-tagged ubiquitin from the CUP1 promoter, thus allowing ubiquitylated proteins to be detected by their acquisition of the myc epitope (Ellison and Hochstrasser, 1991). Expression of the tagged ubiquitin was induced by adding copper to the medium, the cells lysed, and the protein A chimeras isolated using an IgG–Sepharose column. They were released from the Sepharose beads by cleavage with TEV protease, and the presence of myc-ubiquitin tested by immunoblotting. As shown in Figure 7, both Cps1p and Phm5p yielded several myc-containing bands, their mobilities indicating that multiple ubiquitins could be added in each case. These modified bands were hard to see in total cell extracts, because it proved difficult to inhibit the numerous de-ubiquitylating enzymes during sample preparation.
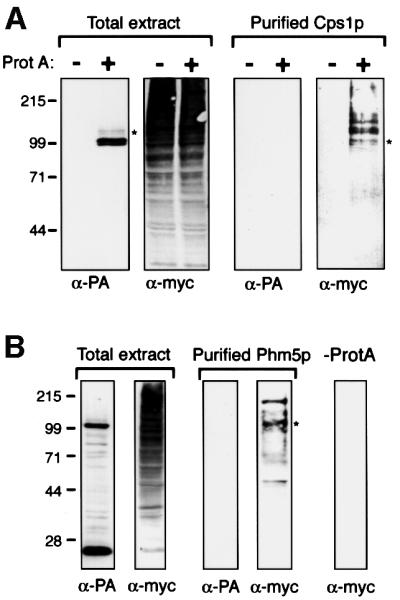
Fig. 7. Ubiquitylation of Cps1p and Phm5p. (A) Δdoa4 cells expressing myc-tagged ubiquitin and either protein A-tagged Cps1p, or only endogenous Cps1p, were processed in parallel. Total extracts, and protein purified by binding to IgG–Sepharose followed by cleavage with TEV protease to release Cps1p from the protein A tag, were immunoblotted with anti-protein A (PA) or anti-myc. Note that protein A is absent from the purified material. Cps1p migrates as a doublet, due to variable glycosylation (Spormann et al., 1992). The faint bands above the protein A chimera are likely to be ubiquitylated forms, but these are under-represented in total cell extracts due to de-ubiquitylation during sample preparation. (B) As (A), but cells expressed protein A-tagged Phm5p. The lower protein A-containing band results from processing close to the transmembrane domain (see Figure 4). In addition, some more random degradation was detectable, which accounts for the smaller myc-tagged proteins in the purified fraction. In both panels, the asterisks indicate the approximate mobility expected for a mono-ubiquitylated protein.
Ubiquitin chains are added to lysine residues, so if ubiquitylation is necessary for internalization of the proteins in late endosomes, mutation of the relevant lysines should prevent delivery to the vacuole interior. The cytoplasmic domain of Phm5p contains four lysines (Figure 8). Mutation of Lys6 to either glycine (not shown) or arginine caused the protein to accumulate on the vacuole membrane, just as it did in the Δdoa4 strain (Figure 8). This indicates that Lys6 is an important site of ubiquitylation. However, an effect of mutating this lysine was apparent in only ∼70% of the cells, whereas changing all four lysines to arginine gave vacuolar membrane fluorescence in all the cells (data not shown). Thus, one or more of the other, membrane-proximal lysines is also likely to be a substrate for ubiquitylation. Lysines present in GFP were evidently not able to substitute for those in the Phm5p tail.
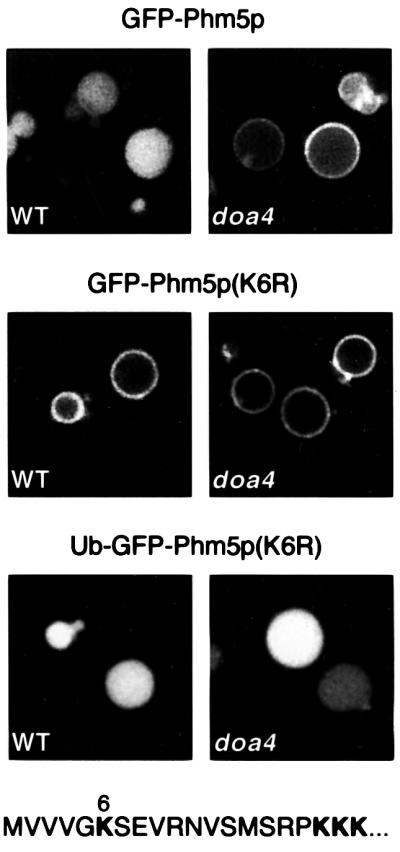
Fig. 8. Sorting of Phm5p is blocked by mutation of Lys6, but restored by the addition of ubiquitin. GFP–Phm5p, the K6R mutant, and the mutant with ubiquitin added to the N-terminus of the GFP were expressed in both wild-type cells (WT) and the Δdoa4 mutant. The sequence of the Phm5p cytoplasmic tail is shown at the bottom.
To confirm that the lysine requirement was solely for ubiquitylation, we added ubiquitin to the Lys6 mutant, by incorporating its sequence at the N-terminus of the GFP tag. The ubiquitin sequence was modified at the junction with GFP to prevent its removal (Gly76 to Val; see Weissman, 2001). In addition, lysines 29, 48 and 63, which are the known sites for the addition of further ubiquitins in yeast (Arnason and Ellison, 1994), were replaced with arginines. These modifications ensure that a single ubiquitin is present on the construct. As shown in Figure 8, the addition of ubiquitin was sufficient to restore delivery of the Phm5p K6R mutant to the vacuole interior, in all cells. Furthermore, delivery was restored even in Δdoa4 cells, indicating that the sorting defect in these cells is due to the lack of ubiquitylation of Phm5p, rather than to any general effect on the sorting machinery.
From these results we conclude that Phm5p is sorted into internal vesicles in late endosomes by a mechanism that requires it to be ubiquitylated. The same is likely to be true for Cps1p and Hmx1p, since both are affected in a doa4 strain, and we have shown that Cps1p is also ubiquitylated. Although both Cps1p and Phm5p can receive several ubiquitins, at least in the doa4 strain, a single ubiquitin is a sufficient signal for this sorting process.
Sna3p is internalized into multivesicular bodies without being ubiquitylated
In contrast to the other proteins studied, we found that Sna3p reached the inside of the vacuole even in Δdoa4 cells. One possibility was that overexpression of this protein might induce vigorous invagination of endosomal membranes and make internalization signal independent. To check this, we co-expressed GFP–Sna3p with yellow fluorescent protein (YFP)–Phm5p. In wild-type cells, both were present inside vacuoles; in Δdoa4 cells, Sna3p was inside while Phm5p was on the outer membrane of the very same vacuoles (Figure 9A). Thus, Sna3p is specifically internalized in a manner that appears independent of ubiquitylation.
Fig. 9. Ubiquitylation of Sna3p is not required for it to enter the vacuole. (A) Double labelling with YFP–Phm5p and GFP–Sna3p in wild-type (WT) and Δdoa4 cells. (B) GFP–Sna3p with both cytoplasmic lysines changed to arginines expressed in wild-type cells and in a Δsna3 mutant.
To confirm this, we changed the two lysine residues (19 and 125) in the predicted cytoplasmic portions of Sna3p to arginines, thus making ubiquitylation of these residues impossible. As shown in Figure 9B, this had no effect on the sorting of the protein. The same result was obtained in a Δsna3 strain, eliminating the possibility that sorting of the mutant Sna3p was dependent on oligomerization with wild-type protein. An analogous observation was that Sna4p truncated at position 108, which lacks lysines altogether, also reached internal vesicles (data not shown). We conclude that Sna3p (as well as Sna4p) has features that allow its selective inclusion in invaginating vesicles, and that this process does not require ubiquitylation of either Sna3p itself or of any other component that is sensitive to the effects of the doa4 mutation.
Discussion
Components of multivesicular bodies
Our analysis of the vesicles that accumulate within the vacuole of a protease-deficient yeast strain identified four proteins that are specifically enriched: Cps1p, Phm5p, Hmx1p and Sna3p. Three of these are enzymes and none is an obvious candidate for components of the machinery that forms such vesicles, or receptors that might serve to sort proteins into them. Although the difficulties of purification mean that negative results must be interpreted with caution, we found no proteins present in significantly higher molar amounts than these cargo molecules. We therefore consider it likely that the vesicles invaginate without stoichiometric coat components that would end up inside them. Nor are there major transmembrane proteins that could be considered structural. Sorting of proteins into internal vesicles does not, therefore, seem to involve their sequestration either by a vesicle-associated transmembrane receptor or by a coat. In agreement with this, we have seen little evidence for saturation of the sorting mechanism by overexpression of the proteins (Reggiori et al., 2000 and our unpublished observations).
Ubiquitylation as a sorting signal in late endosomes
Our experiments indicate that Phm5p enters multivesicular bodies because it is ubiquitylated. We show that lysine residues in its cytoplasmic tail are crucial for efficient internalization, that the protein is ubiquitylated, and that the absence of a cytoplasmic lysine can be compensated for by the addition of a single, non-removable, ubiquitin to the N-terminus of the construct. In a doa4 mutant in which free ubiquitin levels are limiting (Swaminathan et al., 1999), internalization is also blocked. We have also shown that internalization of Cps1p and Hmx1p is affected in doa4 cells, and that Cps1p is ubiquitylated. Parallel studies by Emr and colleagues have also documented ubiquitin-dependent sorting of Cps1p (Katzmann et al., 2001).
The Phm5p data rule out two alternative interpretations of the doa4 effect. Thus, it cannot be that doa4 cells fail to carry out an essential ubiquitylation of some component of the internalization machinery, rather than the cargo proteins themselves, because sorting of Phm5p can be restored in these cells by addition of ubiquitin specifically to Phm5p. It remains possible that other ubiquitylation events are required for endosome function, but if so, they can still occur in doa4 cells. This is not implausible, since doa4 cells maintain a low level of ubiquitylation, which is essential for viability. Secondly, complete removal of ubiquitin from the substrates by the Doa4p ubiquitin-specific protease is not essential for their internalization. Such removal does normally occur (Dupré and Haguenauer-Tsapis, 2001), but a non-removable ubiquitin moiety does not prevent invagination of Phm5p.
It is striking that addition of a single ubiquitin to Phm5p is sufficient. In this respect, sorting in endosomes resembles ubiquitin-dependent endocytosis of cell surface receptors and permeases (Terrell et al., 1998; Nakatsu et al., 2000; Hicke, 2001), rather than proteasome-dependent proteolysis, which requires a chain of at least four ubiquitins (Weissman, 2001). Vacuolar proteins can, however, clearly receive multiple ubiquitins, at least when their removal is inhibited by the doa4 mutation. It may be that extension of ubiquitin chains improves the efficiency of sorting, as has been shown for endocytosis of uracil permease (Galan and Haguenauer-Tsapis, 1997).
The vacuolar proteins do not reach the cell surface in an end4 mutant, and thus their modification must occur entirely within the cell, either at the late endosome or at an earlier point such as the Golgi. It seems that ubiquitin is used as a signal at multiple points, to induce transport to endosomes from either the plasma membrane or the Golgi, and to induce invagination in endosomes and hence delivery to the vacuole. Indeed, the ubiquitin that marks a plasma membrane protein for endocytosis would, if retained, also ensure its entry into multivesicular bodies and its eventual demise in the vacuole. Diversion from this fate requires specific removal of the ubiquitin, which may therefore be an important regulatory step in protein recycling.
How ubiquitin is recognized in endosomes, and the tagged proteins directed to internal vesicles, remains to be seen. Interestingly, Vps23p, which is required for multivesicular body formation, has a domain with homology to ubiquitylating enzymes, yet lacks the Cys residue to which ubiquitin is conjugated in these enzymes (Babst et al., 2000; Bishop and Woodman, 2001). This domain could, therefore, have a recognition, rather than enzymatic, function (Katzmann et al., 2001).
A further remaining question is how the proteins are initially recognized for ubiquitylation. Sequence features of their cytoplasmic tails might provide a signal, although there is no obvious motif common to the various proteins that enter internal vesicles. Alternatively, some other feature such as the TMD might be important (Reggiori et al., 2000), and lysine residues recognized more for their position than their context. Such distinctions will be easier to make when the enzymes responsible for the modification are known, and this is currently under investigation.
A second sorting mechanism
A surprising finding was that Sna3p is sorted by a mechanism that requires neither doa4-sensitive ubiquitylation nor cytoplasmic lysine residues. It is possible that Sna3p has some other signal that is specifically recognized by the invagination machinery. However, we favour the idea that the internal membrane differs in its composition and physical properties from the outer endosomal membrane (Kobayashi et al., 1998), and that Sna3p preferentially partitions into this domain due to its own physical properties. This fits with the observation that the related Sna4p is also invaginated when forced to pass through late endosomes, even though it normally uses the AP-3 pathway to the vacuolar membrane and has no need for a specific invagination signal. Other proteins may have similar properties: since completion of this work we have identified one more protein in internal vesicles (corresponding to the band above number 6 in Figure 2B) as Cos4p, a member of a large family of closely related tetraspanning membrane proteins, and this protein also enters multivesicular bodies even in doa4 cells.
If direct partitioning into the invaginating membrane is possible, it follows that proteins that are selectively retained on the outer endosomal membrane, such as the vacuolar H+-ATPase, must have properties that exclude them from this domain. Indeed, we have noticed that Phm5p is not completely absent from the vacuole interior even when its ubiquitylation is inhibited, perhaps because it lacks such properties.
Degradation inside the vacuole is a common fate for proteins that are endocytosed or mislocalized to the endosomal system, and for many foreign or abnormal proteins. Indeed, it is so common that it might be considered a ‘default’ pathway, with specific signals being required to avoid it. It is now clear, however, that sorting of membrane proteins into the vacuole is a specific process that in many cases requires the selective tagging of the proteins with ubiquitin. This provides many opportunities for regulation, just as the ubiquitin-dependent proteasomal degradation pathway does for cytosolic and nuclear proteins.
Materials and methods
Yeast strains and growth
Yeast strains are described in Table I. All were derived from SEY6210, except the doa4 strain SD20. The DOA4 gene was also disrupted in SEY6210 and the distributions of GFP-labelled proteins checked, with identical results to those shown for SD20. Strains were grown in yeast extract–peptone–dextrose (YEPD) or synthetic dextrose (SD) medium as appropriate.
Table I. Yeast strains used in this work.
| Strain | Genotype | Origin |
|---|---|---|
| SEY6210 | Matα ura3-52 leu2-3,-112 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 | S.Emr |
| JHY024 | Matα ura3-52 leu2-3,-112 his3-Δ 200 trp1-Δ901 lys2-801 suc2-Δ9 pep4::HIS3 | Holthuis et al. (1998) |
| SEY4-1 | Matα ura3-52 leu2-3,-112 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 vps4-1 | Babst et al. (1997) |
| FRY034 | Matα ura3-52 leu2-3,-112 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 end4::HIS5spL | this work |
| FRY024 | Matα ura3-52 leu2-3,-112 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 apm3::HIS5spL | Reggiori et al. (2000) |
| JHY005 | Matα ura3-52 leu2-3,-112 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 pep12::HIS3L | Holthuis et al. (1998) |
| SD20 | Matα ade2-1 ura3-1 leu2-3,-112 his3-11 trp1-1 can1-100 doa4::HIS3 | Dupré and Haguenauer-Tsapis (2001) |
| FRY050 | Matα ura3-52 leu2-3,-112 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 yjl151c::HIS5spL | this work |
For YJL151c (SNA3) and END4 gene disruptions, the entire coding region was replaced with the HIS5 gene of Schizosaccharomyces pombe and the TRP1 gene, respectively, flanked by coliphage loxP sites using PCR primers containing ∼50 bases of identity to the regions flanking the open reading frame.
Purification of internal vacuolar vesicles and protein analysis
Vacuoles were purified as described (Haas, 1995) from a Δpep4 strain expressing a GFP-tagged PEP3D construct [Pep12p with an aspartate in the third position of the TMD (Reggiori et al., 2000)]. Vacuoles from each gradient (600–800 µl) were collected in a microcentrifuge tube, diluted with 700 µl of ice-cold iso-osmotic buffer (10 mM PIPES–KOH pH 6.8, 200 mM sorbitol) and centrifuged at 15 000 g for 15 min (4°C). Supernatant was discarded and the pellet resuspended in 20–40 µl of iso-osmotic buffer containing 15% Ficoll. The protein concentration was determined by Bradford assay (Bio-Rad, Munich, Gemany) and adjusted to 1 µg/µl by diluting the samples with the same buffer. Aliquots were then frozen in liquid nitrogen.
Vacuoles (600–800 µg) were thawed on ice and 40 µl aliquots distributed in microcentrifuge tubes. Each aliquot was diluted by pipetting in 460 µl of ice-cold hypo-osmotic buffer [20 mM PIPES pH 6.8, 20 mM EDTA containing 2 mM phenylmethylsulfonyl fluoride (PMSF), 2 µg/ml pepstatin A and Cømplete™ EDTA-free protease inhibitor cocktail (Roche, Mannheim, Germany)] and then centrifuged at 15 000 g for 15 min (4°C). Supernatants were pooled and centrifuged again at 15 000 g for 15 min (4°C). The final supernatant was transferred to a 3 ml polycarbonate tube (Beckman, Palo Alto, CA) and centrifuged at 500 000 g for 30 min (4°C) in a TLA 100.3 Beckman rotor. The supernatant (S500) was removed, 3 ml of hypo-osmotic buffer gently added and the tube inverted 3–4 times before re-centrifugation at 500 000 g for 30 min (4°C). The final pellet (P500) was resuspended by sonication in 50 µl of sample buffer containing 2 mM PMSF and Cømplete™ EDTA-free protease inhibitor cocktail; 30 µl were loaded on a preparative gel for mass spectrometry. Tryptic digestion of protein bands and analysis by mass spectrometry were performed as described previously (Siniossoglou et al., 2000). The remaining 20 µl were diluted with 80 µl of sample buffer and 10 µl aliquots were used for western blot analysis. Incubations were carried out with monoclonal antibodies against ALP, Dpm1p and Pgk1 (Molecular Probes, Eugene, OR), and with rabbit polyclonal antibodies against GFP (a gift from Dr Derek McCusker, MRC Cambridge), CPY and Sso2p (Holthuis et al., 1998).
Plasmids
CPS1 was PCR generated and cloned as an EcoRI–BamHI fragment into pRS416 vector (Sikorski and Hieter, 1989) behind sequences expressing the mut2 GFP variant (Cormack et al., 1996) and the TPI1 promoter as described by Wooding and Pelham (1998). The 5′ primer introduced five glycines in front of the starting methionine. Ylr205c (HMX1) was also generated by PCR and cloned as a StuI–BamHI fragment into the expression plasmid described above, cut with EcoRI, blunted with Klenow, and finally digested with BamHI to eliminate CPS1. PHM5 and PHM5 point mutations were generated by PCR and cloned as an AscI–BamHI fragment in the same expression plasmid in which an AscI site was introduced at the 3′ end of GFP in place of an EcoRI site. YFP versions of the same vectors were made by replacing the GFP coding sequences with a PCR-generated HindIII–AscI fragment encoding YFP.
Yjl151c (SNA3), Yjl151c point mutations, SNA1/PMP3, Ydr525w-A (SNA2) and Ydl123w (SNA4) were all generated by PCR and cloned as HindIII–AgeI fragments in the pTL321 vector (a gift from T.Levine and S.Munro), creating a C-terminal GFP-tagged Yjl151p under the control of a TPI1 promoter. A plasmid encoding ubiquitin (Ub) with the point mutations K29R, K48R, K63R and G76V fused to the N-terminus of GFP was kindly provided by E.Hettema. This Ub–GFP fusion was swapped to the GFP–PHM5KR construct as a HindIII–MscI fragment. To construct a NOP1 promoter-driven protein A fusion with a TEV cleavage site after the protein A, CPS1 was cloned by PCR as an NdeI–BamHI fragment into the pNOPATA/NPL3 vector (Senger et al., 1998). PHM5 was cloned as an AscI–BamHI fragment in the same expression plasmid in which an AscI site was introduced at the 3′ end of the protein A coding sequence. In all cases, changes were confirmed by sequencing. All enzymes for manipulation of DNA were from New England Biolabs (Beverly, MA).
Electron microscopy and imaging of live cells
For electron microscopy, WT and Δpep4 cells were grown in YEPD medium to early log phase. Permanganate fixation, dehydratation and embedding in Spurr’s resin (Agar Scientific, Stansted, UK) were carried out as described by Kaiser and Schekman (1990). Sections were stained with 5% uranyl acetate for 10 min at 60°C followed by 5 min in Reynold’s lead citrate at room temperature. Sections were observed using a Philips CM10 transmission electron microscope.
Living cells were prepared for GFP fluorescence and stained with FM4-64 (Molecular Probes) as described previously (Reggiori et al., 2000). Double-label GFP/YFP analysis was performed as described (Siniossoglou et al., 2000). Purified vacuoles were resuspended in iso-osmotic buffer containing 15% Ficoll and trapped on slides with the coverslip prior to fluorescence imaging.
Affinity purification of protein A fusion proteins for ubiquitylation analysis
Δdoa4 strains expressing protein A fusions were transformed with a plasmid carrying a myc-tagged ubiquitin under the control of a CUP1 promoter (Ellison and Hochstrasser, 1991). These strains were grown in selective medium containing 0.1 mM CuSO4, and 3–4 g of cells harvested by centrifugation. Spheroplast lysis, purification on IgG–Sepharose and cleavage with TEV protease (Life Technologies, Paisley, UK) were performed as described (Siniossoglou et al., 2000), except that 1 mM PMSF (Sigma Chemical, St Louis, MO) and 10 mM N-ethylmaleimide (Sigma) were included in the lysis buffer. The final supernatants containing the released proteins and the successive bead washes were pooled and successively concentrated using Nanosep columns (MWCO 10 kDa; Pall-Gelman Laboratory, Ann Arbor, MI).
Total extracts and samples were loaded on SDS–PAGE gels and after western blotting were probed either with anti-protein A polyclonal serum (Dako, Glostrup, Denmark) or with anti-myc polyclonal serum (Santa Cruz Biotechnology, Santa Cruz, CA).
Acknowledgments
Acknowledgements
We thank Ewald Hettema in particular for the Ub–GFP plasmid, and also Mike Black, Mike Lewis and Symeon Siniossoglou for plasmids, reagents and helpful discussions; Douglas Kershaw for help with electron microscopy; and Sew Y.Peak-Chew and Farida Begum for mass spectrometry. We also thank Rosine Haguenauer-Tsapis for plasmids and strains. F.R. was supported by a postdoctoral fellowship from the Swiss National Science Foundation and an EMBO long-term fellowship.
References
- Abeliovich H., Darsow,T. and Emr,S.D. (1999) Cytoplasm to vacuole trafficking of aminopeptidase I requires a t-SNARE–Sec1p complex composed of Tlg2p and Vps45p. EMBO J., 18, 6005–6016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amerik A.Y., Nowak,J., Swaminathan,S. and Hochstrasser,M. (2000) The Doa4 deubiquitinating enzyme is functionally linked to the vacuolar protein-sorting and endocytic pathways. Mol. Biol. Cell, 11, 3365–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnason T. and Ellison,M.J. (1994) Stress resistance in Saccharomyces cerevisiae is strongly correlated with assembly of a novel type of multiubiquitin chain. Mol. Cell. Biol., 14, 7876–7883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba M., Osumi,M., Scott,S.V., Klionsky,D.J. and Ohsumi,Y. (1997) Two distinct pathways for targeting proteins from the cytoplasm to the vacuole/lysosome. J. Cell Biol., 139, 1687–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M., Sato,T.K., Banta,L.M. and Emr,S.D. (1997) Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J., 16, 1820–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M., Odorizzi,G., Estepa,E.J. and Emr,S.D. (2000) Mammalian tumor susceptibility gene 101 (TSG101) and the yeast homologue, Vps23p, both function in late endosomal trafficking. Traffic, 1, 248–258. [DOI] [PubMed] [Google Scholar]
- Becherer K.A., Rieder,S.E., Emr,S.D. and Jones,E.W. (1996) Novel syntaxin homologue, Pep12p, required for the sorting of lumenal hydrolases to the lysosome-like vacuole in yeast. Mol. Biol. Cell, 7, 579–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck T., Schmidt,A. and Hall,M.N. (1999) Starvation induces vacuolar targeting and degradation of the tryptophan permease in yeast. J. Cell Biol., 146, 1227–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkower C., Loayza,D. and Michaelis,S. (1994) Metabolic instability and constitutive endocytosis of STE6, the a-factor transporter of Saccharomyces cerevisiae. Mol. Biol. Cell, 5, 1185–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop N. and Woodman,P. (2001) TSG101/mammalian VPS23 and mammalian VPS28 interact directly and are recruited to VPS4-induced endosomes. J. Biol. Chem., 276, 11735–11742. [DOI] [PubMed] [Google Scholar]
- Cormack B.P., Valdivia,R.H. and Falkow,S. (1996) FACS-optimized mutants of the green fluorescent protein (GFP). Gene, 173, 33–38. [DOI] [PubMed] [Google Scholar]
- Cowles C.R., Odorizzi,G., Payne,G.S. and Emr,S.D. (1997) The AP-3 adaptor complex is essential for cargo-selective transport to the yeast vacuole. Cell, 91, 109–118. [DOI] [PubMed] [Google Scholar]
- Darsow T., Burd,C.G. and Emr,S.D. (1998) Acidic di-leucine motif essential for AP-3-dependent sorting and restriction of the functional specificity of the Vam3p vacuolar t-SNARE. J. Cell Biol., 142, 913–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupré S. and Haguenauer-Tsapis,R. (2001) Deubiquitination step in the endocytic pathway of yeast plasma membrane proteins: crucial role of doa4p ubiquitin isopeptidase. Mol. Cell. Biol., 21, 4482–4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egner R., Mahe,Y., Pandjaitan,R. and Kuchler,K. (1995) Endocytosis and vacuolar degradation of the plasma membrane-localized Pdr5 ATP-binding cassette multidrug transporter in Saccharomyces cerevisiae. Mol. Cell. Biol., 15, 5879–5887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison M.J. and Hochstrasser,M. (1991) Epitope-tagged ubiquitin. A new probe for analyzing ubiquitin function. J. Biol. Chem., 266, 21150–21157. [PubMed] [Google Scholar]
- Galan J. and Haguenauer-Tsapis,R. (1997) Ubiquitin lys63 is involved in ubiquitination of a yeast plasma membrane protein. EMBO J., 16, 5847–5854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas A. (1995) A quantitative assay to measure homotypic vacuole fusion in vitro. Methods Cell Sci., 17, 283–294. [Google Scholar]
- Helliwell S.B., Losko,S. and Kaiser,C.A. (2001) Components of a ubiquitin ligase complex specify polyubiquitination and intracellular trafficking of the general amino acid permease. J. Cell Biol., 153, 649–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicke L. (2001) Protein regulation by monoubiquitin. Nature Rev. Mol. Cell Biol., 2, 195–201. [DOI] [PubMed] [Google Scholar]
- Hicke L. and Riezman,H. (1996) Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell, 84, 277–287. [DOI] [PubMed] [Google Scholar]
- Holthuis J.C., Nichols,B.J., Dhruvakumar,S. and Pelham,H.R. (1998) Two syntaxin homologues in the TGN/endosomal system of yeast. EMBO J., 17, 113–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser C.A. and Schekman,R. (1990) Distinct sets of SEC genes govern transport vesicle formation and fusion early in the secretory pathway. Cell, 61, 723–733. [DOI] [PubMed] [Google Scholar]
- Katzmann D.J., Babst,M. and Emr,S.D. (2001) Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell, 106, 145–155. [DOI] [PubMed] [Google Scholar]
- Kim J. and Klionsky,D.J. (2000) Autophagy, cytoplasm-to-vacuole targeting pathway, and pexophagy in yeast and mammalian cells. Annu. Rev. Biochem., 69, 303–342. [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Stang,E., Fang,K.S., de Moerloose,P., Parton,R.G. and Gruenberg,J. (1998) A lipid associated with the antiphospholipid syndrome regulates endosome structure and function. Nature, 392, 193–197. [DOI] [PubMed] [Google Scholar]
- Lai K., Bolognese,C.P., Swift,S. and McGraw,P. (1995) Regulation of inositol transport in Saccharomyces cerevisiae involves inositol-induced changes in permease stability and endocytic degradation in the vacuole. J. Biol. Chem., 270, 2525–2534. [DOI] [PubMed] [Google Scholar]
- Lemmon S.K. and Traub,L.M. (2000) Sorting in the endosomal system in yeast and animal cells. Curr. Opin. Cell Biol., 12, 457–466. [DOI] [PubMed] [Google Scholar]
- Levkowitz G., Waterman,H., Zamir,E., Kam,Z., Oved,S., Langdon,W.Y., Beguinot,L., Geiger,B. and Yarden,Y. (1998) c-Cbl/Sli-1 regulates endocytic sorting and ubiquitination of the epidermal growth factor receptor. Genes Dev., 12, 3663–3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losko S., Kopp,F., Kranz,A. and Kolling,R. (2001) Uptake of the ATP-binding cassette (ABC) transporter Ste6 into the yeast vacuole is blocked in the doa4 mutant. Mol. Biol. Cell, 12, 1047–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medintz I., Jiang,H., Han,E.K., Cui,W. and Michels,C.A. (1996) Characterization of the glucose-induced inactivation of maltose permease in Saccharomyces cerevisiae. J. Bacteriol., 178, 2245–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morano K.A. and Klionsky,D.J. (1994) Differential effects of compartment deacidification on the targeting of membrane and soluble proteins to the vacuole in yeast. J. Cell Sci., 107, 2813–2824. [DOI] [PubMed] [Google Scholar]
- Nakatsu F., Sakuma,M., Matsuo,Y., Arase,H., Yamasaki,S., Nakamura,N., Saito,T. and Ohno,H. (2000) A di-leucine signal in the ubiquitin moiety. Possible involvement in ubiquitination-mediated endocytosis. J. Biol. Chem., 275, 26213–26219. [DOI] [PubMed] [Google Scholar]
- Navarre C. and Goffeau,A. (2000) Membrane hyperpolarization and salt sensitivity induced by deletion of PMP3, a highly conserved small protein of yeast plasma membrane. EMBO J., 19, 2515–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odorizzi G., Babst,M. and Emr,S.D. (1998) Fab1p PtdIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell, 95, 847–858. [DOI] [PubMed] [Google Scholar]
- Ogawa N., DeRisi,J. and Brown,P.O. (2000) New components of a system for phosphate accumulation and polyphosphate metabolism in Saccharomyces cerevisiae revealed by genomic expression analysis. Mol. Biol. Cell, 11, 4309–4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa F.R. and Hochstrasser,M. (1993) The yeast DOA4 gene encodes a deubiquitinating enzyme related to a product of the human tre-2 oncogene. Nature, 366, 313–319. [DOI] [PubMed] [Google Scholar]
- Reggiori F., Black,M.W. and Pelham,H.R. (2000) Polar transmembrane domains target proteins to the interior of the yeast vacuole. Mol. Biol. Cell, 11, 3737–3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocca A., Lamaze,C., Subtil,A. and Dautry-Varsat,A. (2001) Involvement of the ubiquitin/proteasome system in sorting of the interleukin 2 receptor β chain to late endocytic compartments. Mol. Biol. Cell, 12, 1293–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzer E., De Matteis,F., Giribaldi,G., Ulliers,D., Valente,E. and Arese,P. (1999) Hemozoin stability and dormant induction of heme oxygenase in hemozoin-fed human monocytes. Mol. Biochem. Parasitol., 100, 61–72. [DOI] [PubMed] [Google Scholar]
- Senger B., Simos,G., Bischoff,F.R., Podtelejnikov,A., Mann,M. and Hurt,E. (1998) Mtr10p functions as a nuclear import receptor for the mRNA-binding protein Npl3p. EMBO J., 17, 2196–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirahama K., Noda,T. and Ohsumi,Y. (1997) Mutational analysis of Csc1/Vps4p: involvement of endosome in regulation of autophagy in yeast. Cell Struct. Funct., 22, 501–509. [DOI] [PubMed] [Google Scholar]
- Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S., Peak-Chew,S.Y. and Pelham,H.R. (2000) Ric1p and Rgp1p form a complex that catalyses nucleotide exchange on Ypt6p. EMBO J., 19, 4885–4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spormann D.O., Heim,J. and Wolf,D.H. (1992) Biogenesis of the yeast vacuole (lysosome). The precursor forms of the soluble hydrolase carboxypeptidase yscS are associated with the vacuolar membrane. J. Biol. Chem., 267, 8021–8029. [PubMed] [Google Scholar]
- Springael J.Y., Galan,J.M., Haguenauer-Tsapis,R. and Andre,B. (1999) NH4+-induced down-regulation of the Saccharomyces cerevisiae Gap1p permease involves its ubiquitination with lysine-63-linked chains. J. Cell Sci., 112, 1375–1383. [DOI] [PubMed] [Google Scholar]
- Stepp J.D., Huang,K. and Lemmon,S.K. (1997) The yeast adaptor protein complex, AP-3, is essential for the efficient delivery of alkaline phosphatase by the alternate pathway to the vacuole. J. Cell Biol., 139, 1761–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan S., Amerik,A.Y. and Hochstrasser,M. (1999) The Doa4 deubiquitinating enzyme is required for ubiquitin homeostasis in yeast. Mol. Biol. Cell, 10, 2583–2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrell J., Shih,S., Dunn,R. and Hicke,L. (1998) A function for monoubiquitination in the internalization of a G protein-coupled receptor. Mol. Cell, 1, 193–202. [DOI] [PubMed] [Google Scholar]
- Urech K., Durr,M., Boller,T., Wiemken,A. and Schwencke,J. (1978) Localization of polyphosphate in vacuoles of Saccharomyces cerevisiae. Arch. Microbiol., 116, 275–278. [DOI] [PubMed] [Google Scholar]
- Vida T.A. and Emr,S.D. (1995) A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol., 128, 779–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volland C., Urban-Grimal,D., Geraud,G. and Haguenauer-Tsapis,R. (1994) Endocytosis and degradation of the yeast uracil permease under adverse conditions. J. Biol. Chem., 269, 9833–9841. [PubMed] [Google Scholar]
- Weissman A.M. (2001) Themes and variations on ubiquitylation. Nature Rev. Mol. Cell Biol., 2, 169–178. [DOI] [PubMed] [Google Scholar]
- Wooding S. and Pelham,H.R. (1998) The dynamics of Golgi protein traffic visualized in living yeast cells. Mol. Biol. Cell, 9, 2667–2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurmser A.E. and Emr,S.D. (1998) Phosphoinositide signaling and turnover: PtdIns(3)P, a regulator of membrane traffic, is transported to the vacuole and degraded by a process that requires lumenal vacuolar hydrolase activities. EMBO J., 17, 4930–4942. [DOI] [PMC free article] [PubMed] [Google Scholar]