Abstract
Important morphogenetic transitions in fungi are triggered by starvation-induced changes in the expression of structural surface proteins. Here, we report that nutrient deprivation causes a strong and reversible up-regulation of TbSP1, a surface-associated, Ca2+-dependent phospholipase from the mycorrhizal fungus Tuber borchii. TbSP1 is the first phospholipase A2 to be described in fungi and identifies a novel class of phospholipid-hydrolyzing enzymes. The TbSP1 phospholipase, which is synthesized initially as a pre-protein, is processed efficiently and secreted during the mycelial phase. The mature protein, however, also localizes to the inner cell wall layer, close to the plasma membrane, in both free-living and symbiosis-engaged hyphae. It thus appears that a dual localization phospholipase A2 is involved in the adaptation of a symbiotic fungus to conditions of persistent nutritional limitation. Moreover, the fact that TbSP1-related sequences are present in Streptomyces and Neurospora, and not in wholly sequenced non-filamentous microorganisms, points to a general role for TbSP1 phospholipases A2 in the organization of multicellular filamentous structures in bacteria and fungi.
Keywords: cell wall/mycorrhizal fungi/phospholipase A2/secretion/starvation
Introduction
More than 5000 species of ascomycete and basidiomycete fungi form symbiotic associations, called ectomycorrhizas, with the fine roots of most shrubs and trees found in temperate and boreal forests (Molina et al., 1992). By promoting nutrient exchange between the two partners, mycorrhizas exert a positive influence on plant survival under a variety of unfavorable environmental conditions (Read, 1999). Ectomycorrhizal fungi, however, are not obligate symbionts, and some of them, including the ascomycete truffle Tuber borchii studied in the present work, can grow in pure mycelial culture by exploiting their limited saprotrophic capabilities. The mycorrhizal transition is triggered by as yet largely unidentified, environmental and plant-generated signals that are sensed by the free-living mycelium and transduced into a multifaceted morphogenetic program (Martin and Tagu, 1999; Buscot et al., 2000). Ectomycorrhiza formation is generally considered as a very soft colonization process, not causing any irreversible damage, nor any drastic defense response in the host plant. However, various cytological and biochemical evidence clearly points to the existence, at least in early infection stages, of some basic functional similarities between mycorrhiza formation and host invasion by pathogenic fungi (Hebe et al., 1999; Martin and Tagu, 1999).
As revealed by the fungal cell wall alterations that take place during symbiosis development (Bonfante et al., 1998), a central aspect of mycorrhiza formation is an extensive remodeling of the surface and aggregation state of the hyphae. Fungal molecular components that are thought to be involved in such events (albeit not exclusively associated with mycorrhiza development) have been identified recently by both large-scale (Voiblet et al., 2001) and conventional gene search approaches. These include a protein involved in vesicular transport and autophagocytosis (Kim et al., 1999), a transporter for monosaccharide movement from the plant to the mycobiont (Nehls et al., 1998), hydrophobins (Tagu et al., 1996) and the adhesin-like, symbiosis-regulated acidic polypeptides (SRAPs) of Pisolithus tinctorius (Laurent et al., 1999). The latter two are structural proteins that have been found extracellularly as well as surface associated in both free-living and symbiosis-engaged hyphae. The mRNAs for both proteins are up-regulated concomitantly with symbiosis formation, but the signals triggering such a response are still largely unknown. As revealed by the results obtained in some multicellular non-mycorrhizal fungi, many important morphogenetic transitions, such as aerial hyphae (Wosten et al., 1999) and conidiophore (Lauter et al., 1992) formation, and the development of invading appressoria in phytopathogenic fungi (Talbot et al., 1993), similarly are accompanied by the overexpression of secretable surface proteins. Many of these responses, e.g. hydrophobin mRNA up-regulation in the rice pathogen Magnaporthe grisea (Talbot et al., 1993) and in the cellulolytic filamentous fungus Trichoderma reesei (Nakari-Setala et al., 1997), can be mimicked in vitro by growth under conditions of nutrient deprivation. This link between nutritional status and surface protein expression has been interpreted as a sort of adaptive response, in which starvation for essential nutrients is perceived by specific receptors and transduced into morphogenetic changes aimed at inducing either a metabolically quiescent state (e.g. conidia) or a state of improved nutrient acquisition (e.g. invasive hyphal structures) (Madhani and Fink, 1998; Lengeler et al., 2000). It thus appears that nutritional factors, especially the conditions of inorganic nitrogen shortage often found in the root microenvironment of ectosymbiosis-susceptible plants (Keeney, 1980; Read, 1991), may also trigger adaptive changes in surface protein expression (as well as morphogenetic transitions) in mycorrhizal fungi.
Here, we report on the isolation of a novel phospholipase, named TbSP1, that is strongly and reversibly up-regulated by nutrient deprivation and accumulates in the inner cell wall layer of free-living mycelia, fruitbodies and mycorrhizas of the symbiotic fungus T.borchii. TbSP1, which is also released extracellularly and contains the adhesin-like motif RGD, is reminiscent of similarly localized phospholipid-hydrolyzing enzymes involved in host invasion by pathogenic fungi (Nespoulous et al., 1999; Ghannoum, 2000; Cox et al., 2001). However, at variance with such enzymes, all of which are (lyso)phospholipases B unable to discriminate between the sn-1 and the sn-2 ester bond of glycerophospholipids, TbSP1 is absolutely specific for the middle (sn-2) position and can thus participate in both membrane remodeling and cell signaling events. TbSP1 is the first phospholipase A2 (PLA2) thus far described in fungi and identifies a new family of phospholipases that are only present in filamentous microbes. To the best of our knowledge, the nutrient-regulated expression of a phospholipid-hydrolyzing enzyme has never been reported before.
Results
Isolation of the cDNA for an abundant low molecular weight protein extracellularly released by suspension cultured T.borchii mycelia
We initially analyzed by SDS–PAGE the growth medium of mycelia cultured for different lengths of time under suspension culture conditions. As shown in Figure 1A (lanes 1 and 2), many soluble polypeptides could be detected even at relatively early stages of in vitro culture. However, given the extremely slow growth rate of T.borchii mycelia and the correspondingly long lag phase (∼15 days under standard culture conditions; Saltarelli et al., 1998), it is possible that many of these polypeptides are the remnants of surface proteins shed from the starting inoculum rather than the products of de novo synthesis. Indeed, the overall polypeptide pattern changed strikingly after ∼2 weeks of culture. We thus focused on 3- to 5-week-old mycelial cultures and, particularly, on a highly represented polypeptide with an apparent molecular mass of 23 kDa (p23) that showed maximal accumulation around day 28 (Figure 1A, lane 4). This polypeptide was gel purified, subjected to N-terminal sequencing and the resulting sequence (p23/20; Figure 1A) was employed for the design of degenerate oligonucleotides that were utilized as primers for a PCR amplification programmed with a cDNA library prepared from 30-day-old, liquid medium-grown mycelia (SLM-30). A single DNA fragment of 54 bp, coding for a conceptual translation product perfectly matching the inner 17 amino acids of the p23/20 peptide, was obtained from such amplification and used as a probe to screen the SLM-30 library. A phage plaque harboring a cDNA insert of 899 bp was thus identified. This cDNA, designated TbSP1 (submitted to the DDBJ/EMBL/GenBank databases under accession No. AF162269), was utilized as a probe for DNA and RNA gel blot analyses. As revealed by the DNA gel blot data reported in Figure 1B, a single-band pattern was produced by hybridization of the TbSP1 probe to genomic DNA digested with three different restriction enzymes, thus indicating that TbSP1 is encoded by a single copy gene in the T.borchii genome. As shown in Figure 1C, an mRNA of ∼900 nucleotides was identified by the TbSp1 probe in total RNA extracted from T.borchii mycelia. Such a transcript is present in free-living mycelia grown in either liquid (SLM) or solid (SSM) synthetic medium, but accumulates at higher levels (∼4-fold) in the latter condition.
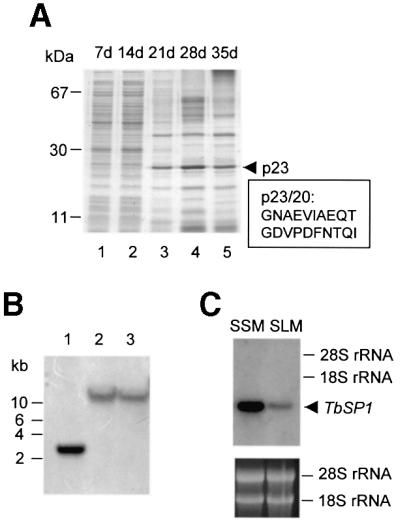
Fig. 1. TbSP1 identification. (A) Equal amounts of SLM-released protein were subjected to SDS–PAGE and stained with Coomassie Blue R-250. Days of in vitro culture (d) and the migration positions of molecular mass markers are indicated at the top and at the left, respectively. The migration position (p23) and the N-terminal sequence (p23/20) of the TbSP1 polypeptide are shown on the right. (B) Genomic DNA digested with EcoRI (lane 1), BamHI (lane 2) or HindIII (lane 3) was probed with the TbSP1 cDNA. The migration positions of DNA size markers are indicated on the left. (C) Balanced amounts of total RNA extracted from synthetic solid (SSM) or liquid (SLM) mycelial cultures were gel fractionated and probed with the 32P-labeled TbSP1 cDNA. The migration positions and the amounts of the 28S and 18S rRNAs, utilized as internal references, are shown on the right and in the lower panel, respectively.
Features of the TbSP1 polypeptide
The full-length TbSP1 cDNA codes for a 211 amino acid polypeptide comprising a sequence identical to that of the previously determined p23/20 peptide between positions 32 and 51. This is preceded by an N-terminal stretch of 31 amino acids (underlined in Figure 2A) corresponding to the main hydrophobic region revealed by hydropathy analysis (not shown). The above observations, together with the presence of a Lys–Arg, KEX2-like protease cleavage site at the junction, suggest that the 31 N-terminal amino acids represent a secretion signal sequence, which is proteolytically processed to produce the mature TbSP1 protein found in the culture medium. The TbSP1 polypeptide is largely hydrophilic with a theoretical pI of 5.0, and the difference between the observed (23 kDa) and predicted (19 kDa) molecular mass of the mature protein is likely to be due to anomalous gel migration or post-translational modification (see below). Also, similarly to the adhesin-like SRAPs of P.tinctorius (Laurent et al., 1999), TbSp1 contains an RGD cell attachment motif (positions 60–62; boxed in Figure 2A). Apart from this single feature, however, there is no other similarity between TbSP1 and SRAPs.
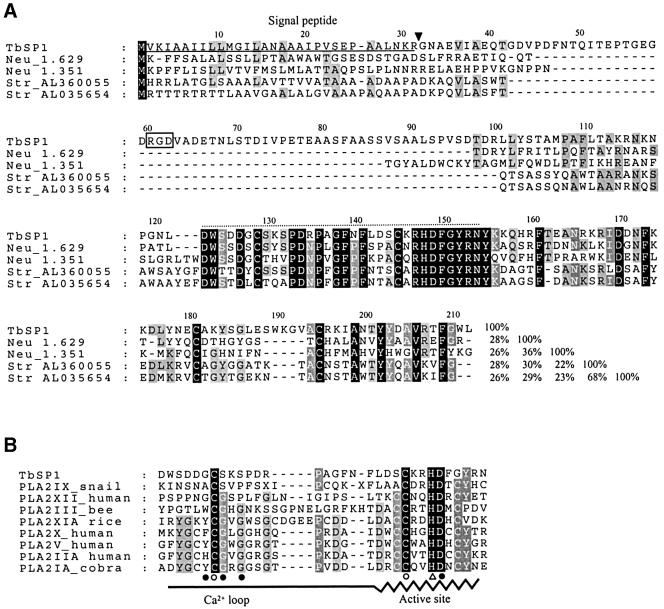
Fig. 2. Alignment of TbSP1 with related polypeptides and with the active site amino acid sequences of known phospholipases A2. (A) The TbSP1 polypeptide is aligned with the amino acid sequences of Neurospora crassa and Streptomyces coelicolor homologs. The predicted secretion signal peptide is underlined and the pre-protein cleavage site is marked with an arrowhead; the tripeptide RGD motif is boxed. The region utilized for the comparison with known PLA2s is indicated by an overlying dotted line. Amino acids that are identical in all or the majority of the sequences are indicated by black or gray shading, respectively. Percentage identity values are reported in the bottom right part of the alignment. (B) The most highly conserved portion of the TbSP1 polypeptide (positions 122–153) is aligned with the active site regions of various small secreted PLA2s. The sequences of archetype PLA2s (Gelb et al., 2000; Six and Dennis, 2000) were utilized for this comparison. The indicated structural elements refer to the X-ray structure of the cobra venom PLA2IA (Fremont et al., 1993): catalytic histidine (open triangle); Ca2+-binding residues (filled circles); disulfide-bonded cysteines (open circles).
Shown in Figure 2A is the alignment of TbSP1 with four homologous amino acid sequences identified by a search of the non-redundant protein database and of the unfinished whole genome sequence of Neurospora crassa. TbSP1 relatives, all corresponding to small hypothetical proteins (150–250 amino acids) with putative signal sequences at the N-terminus, were identified in Streptomyces coelicor (AL360055 and AL035654) and N.crassa (Neu_1.629 and Neu_1.351) and were used to query the protein database. Interestingly, a low sequence similarity with the conodipine-M PLA2 from the marine snail Conus magus (McIntosh et al., 1995) was revealed by the Streptomyces AL03564 sequence. Although only marginally significant (E = 7.1), this similarity mainly pertains to amino acid residues that are conserved in all TbSP1 relatives and, particularly, to a His–Asp pair within the conserved block evidenced in Figure 2A. An additional clue was provided by the identification in the patent section of DDBJ/EMBL/GenBank of an unpublished sequence from Streptomyces violaceoruber having the same nucleotide sequence as AL035654 and annotated as PLA2 (E08479, Patent: JP 1994327468-A; not included in the non-redundant set of the NCBI).
Small extracellular PLA2s are divided into eight groups that share little sequence similarity except for the active site region, i.e. a stretch of ∼30 amino acids containing a helix with the His–Asp catalytic dyad and a metal-binding loop for the activating calcium cation (Gelb et al., 2000; Six and Dennis, 2000). As shown in Figure 2B, the TbSP1 protein exhibits some of the distinctive features shared by the different PLA2 groups. Two other conserved amino acids, besides the HD dyad, are a pair of cysteine residues known to form an invariant disulfide bond within the active site region.
TbSP1 is a novel Ca2+-activated phospholipase A2
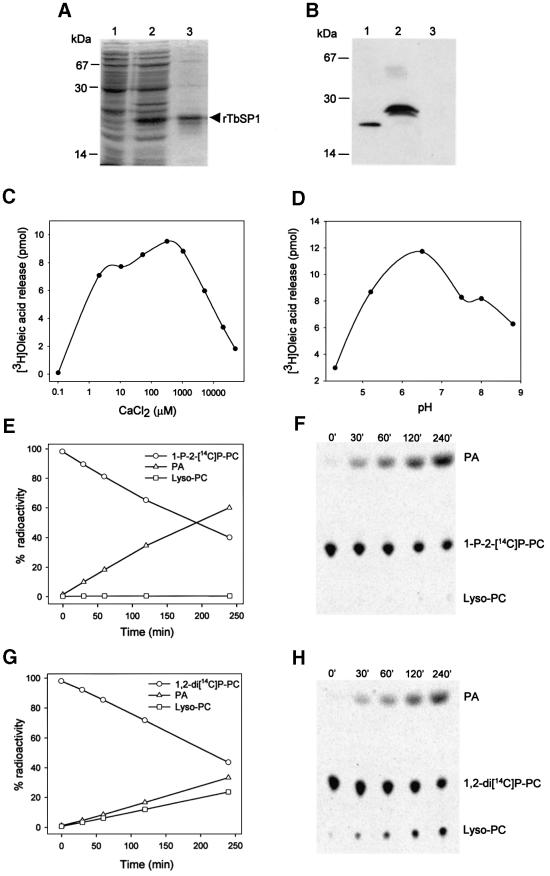
To verify the above prediction as to the function of TbSP1, the T.borchii polypeptide lacking the putative signal peptide was expressed in Escherichia coli as an N-terminal fusion with a metal-binding His6 tag. The resulting recombinant protein, initially identified with the use of a monoclonal anti-His tag antibody (not shown), was purified to near homogeneity (Figure 3A) and used for the production of polyclonal anti-TbSP1 antibodies. As shown by the immunoblot reported in Figure 3B, a single polypeptide with an apparent molecular mass of 23 kDa was recognized specifically by the anti-TbSP1 antibody in the growth medium of T.borchii suspension cultures (lane 1). The difference in size between natural (lane 1) and recombinant (lane 2) TbSP1 is all accounted for by the artificial 20 amino acid extension incorporated into the His-tagged TbSP1 protein. This indicates that the previously mentioned discrepancy between the predicted (19 kDa) and the estimated (23 kDa) molecular mass of TbSP1 is most probably due to an asymmetric protein conformation, and consequent anomalous gel migration, rather than to post-translational modification. Accord ingly, no glycoprotein–periodic acid–Schiff (PAS) staining was observed with natural TbSP1 (not shown).
Fig. 3. Phospholipase A2 activity of purified rTbSP1. (A) SDS–PAGE of total lysates from uninduced (lane 1) or IPTG-induced (lane 2) E.coli cells and of purified mature rTbSP1 (lane 3). (B) Immunodetection of natural (lane 1) and recombinant (lane 2) TbSP1; a control reaction using rTbSP1-pre-saturated antibodies is shown in lane 3. The migration positions of molecular mass markers and of the recombinant TbSP1 protein (rTbSP1) are indicated. (C) Calcium dependence of rTbSP1-catalyzed [3H]oleic acid release from radiolabeled bacterial membranes. (D) pH dependence of TbSP1 phospholipase activity. (E) Time course of rTbSP1-catalyzed hydrolysis of 1-palmitoyl-2-[1-14C]palmitoyl, l-α-phosphatidylcholine (1-P-2-[14C]P-PC; open circles); radiolabeled hydrolysis products are indicated as follows: palmitic acid (PA; open triangles), 1-palmitoyl lysophosphatidylcholine (Lyso-PC; open squares). (F) TLC fractionation and phosphorimager visualization of radiolabeled hydrolysis products from a representative lipolytic assay conducted on 1-P-2-[14C]P-PC. The results of similar assays using double-labeled 1,2-di[1-14C]palmitoyl, l-α-phosphatidylcholine (1,2-di[14C]P-PC) as a substrate are shown in (G) and (H); see Materials and methods for details. Data reported in (C), (D), (E) and (G) are the average of at least three independent determinations which differed by no more than 15% of the mean.
Escherichia coli membranes labeled in vivo with [3H]oleic acid (a phospholipid precursor that is incorporated preferentially at the sn-2 position) were utilized initially as a substrate to test the predicted phospholipase activity of TbSP1. As shown in Figure 3C and D, purified recombinant TbSP1 (rTbSP1) catalyzed the calcium-dependent release of [3H]oleic acid from bacterial membranes (half-activating Ca2+ concentration <1 µM) with maximum activity at ∼pH 6.5. Under optimized assay conditions, 300 pmol of [3H]oleic acid/min/mg of protein were released by TbSP1 and a similar (albeit ∼20-fold lower) fatty acid release was catalyzed by a commercial PLA2 preparation from S.violaceoruber that was used as a control for these experiments (not shown). As revealed by the additional data reported in Figure 3E–H, the TbSP1 phospholipase is specific for the sn-2 ester bond position and has very little (if any) lysophospholipase activity. In fact, [1-14C]palmitic acid was the only product generated by the action of rTbSP1 on 1,2-dipalmitoyl-phosphatidyl choline carrying a 14C-labeled palmitoyl residue at the sn-2 position (Figure 3E and F). Instead, radioactively labeled palmitic acid and 1-palmitoyl lysophosphatidyl choline both accumulated in parallel lipolytic assays conducted on the double-labeled phospholipid 1,2-di[1-14C]palmitoyl-phosphatidyl choline, containing radiolabeled palmitoyl moieties at positions sn-1 and sn-2 (Figure 3G and H).
Localization of the TbSP1 phospholipase
To gain insight into TbSP1 localization, an immunoblot analysis was conducted on various fractions generated by aqueous washing and subcellular fractionation of mycelia grown on solid medium. As shown in Figure 4A, a single polypeptide having the same electrophoretic mobility as extracellularly released TbSP1 (SLM; lane 7) was present in mycelial washings (lanes 1 and 2), whereas no signal could be detected in any of the fractions generated upon disruption and subcellular fractionation of pre-washed mycelia (lanes 3–6). This indicates that a portion of TbSP1 is loosely bound to the cell surface from which it can be extracted by aqueous washing.
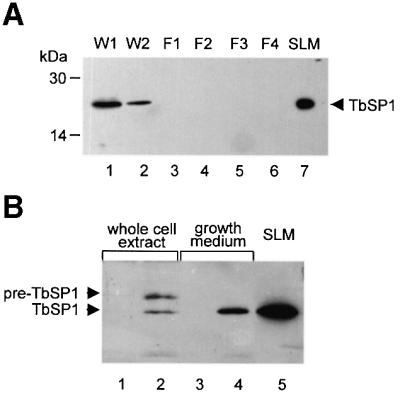
Fig. 4. Immunoblot analysis of TbSP1 localization in Tuber mycelia and in yeast. (A) Mycelial washings (W1 and W2) and particulate fractions (F1, F2 and F3) generated by a series of differential centrifugation steps carried out at increasing sedimentation velocities were probed with the anti-TbSP1 antibody; fraction F4 is the final cytosolic supernatant (see Materials and methods for details). The migration positions of molecular mass markers and of SLM-secreted TbSP1 are indicated. (B) Immunodetection of cell-associated (lane 2) and extracellularly secreted (lane 4) TbSP1 in yeast cells transformed with a ‘sense’ TbSP1 construct; the migration positions of the unprocessed pre-protein (pre-TbSP1) and of the mature polypeptide (TbSP1) are indicated on the left. The results of control experiments conducted on a yeast ‘antisense’ TbSP1 transformant are shown in lanes 1 and 3; TbSP1 secreted by SLM-grown Tuber mycelia is shown for comparison in lane 5.
To investigate TbSP1 localization further, the full-length TbSP1 protein, including the N-terminal hydrophobic peptide, was expressed in Saccharomyces cerevisiae under the control of a constitutive yeast promoter. As shown in Figure 4B, a single immunopositive band having the same gel mobility as the mature polypeptide released by T.borchii mycelia (SLM; lane 5) was detected specifically in the growth medium of yeast cells transformed with a ‘sense’ TbSP1 construct (lane 4). By comparison, a doublet of immunopositive polypeptides, with the sizes expected for the immature (pre-TbSP1) and the proteolytically processed (TbSP1) forms of the protein, was found in whole-cell extracts derived from the same ‘sense’ transformant (lane 2). These data, along with the initial identification of TbSP1 in the culture medium of T.borchii mycelia, thus point to TbSP1 as a dual localization protein that is both secreted extracellularly and surface associated.
Nutrient-regulated expression of TbSP1
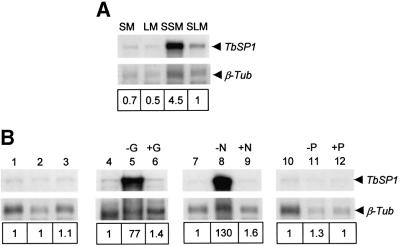
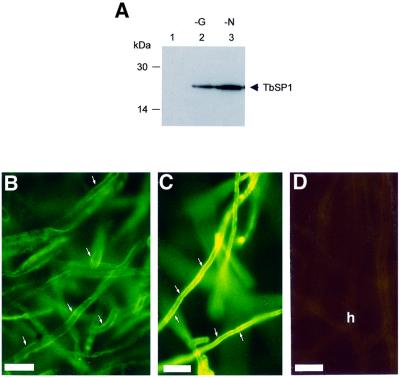
We next wished to find out whether TbSP1 expression is influenced by nutrient starvation. That this may indeed be the case initially was suggested by the results of RNase protection assays, reported in Figure 5A, showing that TbSP1 transcripts accumulate at higher levels upon culture of T.borchii mycelia in SSM or SLM than in the corresponding rich media (SM and LM). To examine this nutrient-dependent up-regulation in more detail, TbSP1 mRNA levels were determined in mycelia cultured for 21 days in SSM and then shifted for 10 days to incomplete culture media lacking a source of either carbon, nitrogen or phosphate. Data reported in Figure 5B show that the TbSP1 messenger was strikingly up-regulated by growth in either carbon- (∼80-fold; lane 5) or nitrogen- (∼130-fold; lane 8) deficient media, whereas basal TbSP1 levels, similar to those present in mock-shifted controls (lane 2), were present in mycelia grown for the same length of time in phosphate-deficient medium (lane 11). As further shown in Figure 5B (compare lanes 5 and 8 with lanes 6 and 9, respectively), this strong up-regulatory response is fully reversible, since basal levels of the TbSP1 transcript were recovered following an additional 5 day incubation in complete SSM. Furthermore, as revealed by the results of immunological analyses conducted on nutrient-deprived mycelia, starvation-induced up-regulation of the TbSP1 mRNA was accompanied by a parallel increase in the levels of the mature TbSP1 protein (Figure 6A), with a concomitantly increased accumulation of TbSP1 on the surface of nutrient-starved hyphae (cf. Figure 6B and C).
Fig. 5. Nutrient-regulated expression of the TbSP1 mRNA. (A) RNase protection analysis of TbSP1 mRNA levels in mycelia grown for 35 days on rich solid (SM) or liquid (LM) medium, or in the corresponding synthetic media (SSM and SLM). (B) RNase protection assays conducted on mycelia grown for 21 days on SSM (lanes 1, 4, 7 and 10) and then shifted for 10 days to the same medium (mock-shift control; lane 2) or to SSM lacking either glucose (–G, lane 5), ammonium (–N, lane 8) or phosphate (–P, lane 11). TbSP1 mRNA levels in parallel mycelial cultures starved for the above nutrients and returned for 5 days to complete SSM are shown in lanes 6, 9 and 12, respectively; the corresponding mock-shift control is shown in lane 3. A T.borchii β-tubulin (β-Tub) antisense riboprobe was included in all assays as an internal standard. The bands shown, which correspond to full-length protection products of the TbSP1 and β-Tub riboprobes were visualized by autoradiography and quantified by phosphorimaging. Relative transcript abundance values (reported below each lane) were calculated by dividing the volumes of the TbSP1 signals by the volumes of the corresponding β-Tub signals, followed by normalization with respect to TbSP1 abundance in SLM (A) or SSM (B; lane 1) cultured mycelia.
Fig. 6. Enhanced accumulation of the TbSP1 protein in nutrient-starved mycelia. (A) Immunoblot analysis of TbSP1 levels in mycelia grown for a total of 31 days in SSM (control; lane 1) and in parallel mycelial cultures shifted for the last 10 days to SSM lacking glucose (–G, lane 2) or ammonium (–N, lane 3). Equal amounts of total protein were loaded onto each lane and probed with the anti-TbSP1 antibody; an immunopositive TbSP1 band was also detectable in control mycelia with a longer exposure time. Samples of the same control (B) and nitrogen-starved (C) mycelia used for the experiment reported in (A) were subjected to immunofluorescence analysis using an FITC-labeled secondary antibody (bars = 18 µm); a control section of hyphae (h) from which the primary anti-TbSP1 antibody was omitted is shown in (D) (bar = 9 µm).
TbSP1 accumulation in the inner cell wall layer of free-living and symbiosis-engaged hyphae
The surface localization of TbSP1 and its production by hyphal structures representative of different stages of the Tuber life cycle were investigated finally by transmission electron microscopy (TEM)-immunolabeling experiments. As shown in Figure 7, cell wall-associated gold granules were detected in free-living hyphae transversally sectioned close to the subapical (Figure 7A) or the fully differentiated (Figure 7B) area. An identical surface localization was revealed by TEM-immunolabeling experiments conducted on T.borchii fruitbodies, in which TbSP1-associated gold granules were found in the cell walls of both vegetative hyphae and asci as well as on sporal envelopes (data not shown).
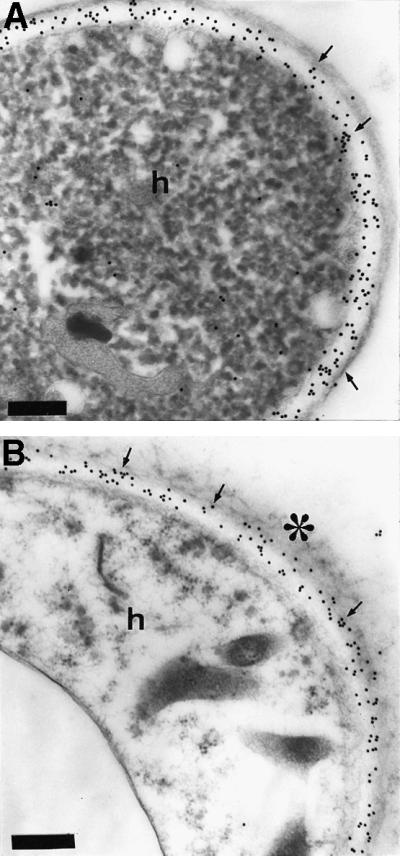
Fig. 7. Immunogold TbSP1 labeling of free-living hyphae. (A) Immunogold-TEM localization of TbSP1 in 30-day-old, SM-grown hyphae (h) transversally cut close to the subapical zone. (B) TbSP1 localization in the fully differentiated area of the same mycelium; the electron-dense material coating the external surface of the hyphae (h) is marked with an asterisk. Arrows point to TbSP1-associated gold granules accumulated in the electron-transparent, inner cell wall layer; bars = 0.3 µm.
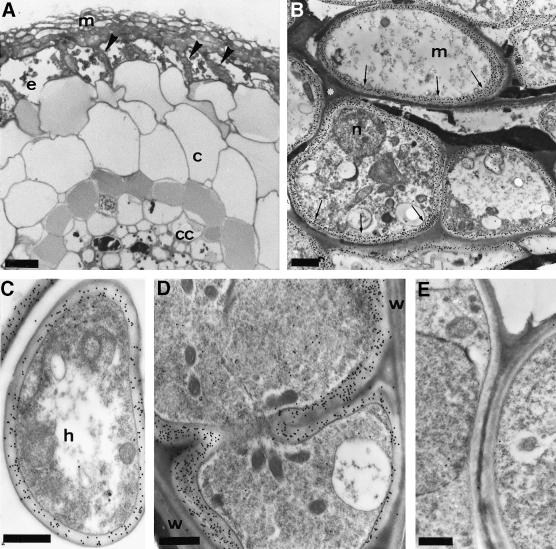
Tuber–hazelnut ectomycorrhizas with a fully developed fungal sheath (mantle) and with hyphae spread among root cells (Balestrini et al., 1996a) were analyzed next. As revealed by the data reported in Figure 8, TbSP1 also accumulates in symbiosis-engaged hyphae, where anti-TbSP1 antibodies produced a strong immunogold labeling along the cell walls of both root-surrounding (Figure 8B) and extramatrical (Figure 8C) hyphae as well as in the cell walls of hyphae penetrating between root cells to form the Hartig net (Figure 8D). More specifically, gold granules were found to be localized in transversal (septa) and longitudinal walls, whereas no labeling occurred in the electron-opaque layer separating mantle-forming hyphae. Similarly, no immunogold labeling could be detected in plant cell walls (Figure 8D), nor in control sections where the primary anti-TbSP1 antibody was omitted (Figure 8E).
Fig. 8. Immunogold localization of TbSP1 in ectomycorrhizas. (A) Light microscopy, cross-sectional view of a Tuber–hazelnut ectomycorrhiza. Arrowheads indicate the Hartig net penetrating between root cells. The epidermis (e), the cortex (c) and the central cylinder (cc) of the root as well the fungal sheath (mantle, m) are also indicated; bar = 15 µm. (B) Immunogold TEM analysis of mantle-forming hyphae (m) labeled with the anti-TbSP1 antibody. The immunonegative, electron-opaque material separating the hyphae is marked with an asterisk; n, nucleus. Arrows point to TbSP1-associated gold granules accumulated in the chitin-containing, inner cell wall layer (bar = 1.3 µm). Gold granules in the inner cell wall layers of a growing extramatrical hypha (h) and of hyphae progressing between root cells in the Hartig net region are shown in (C) and (D) (bars = 0.4 µm). Also apparent in (D) is the absence of immunopositive material in the host cell walls (w). A control section from which the primary anti-TbSP1 antibody was omitted is shown in (E) (bar = 0.5 µm).
Tuber cell walls are made up of structurally distinct microdomains and exhibit different morphologies and layerings depending on the age and life cycle stage of the fungus (Balestrini et al., 1996a). In young proliferating hyphae, a thin and continuous electron-dense layer delimits an inner, electron-transparent layer composed mainly of chitin (Figure 7A), whereas mature hyphae present an irregular external surface due to the presence of a loose electron-opaque material coating the outer layer (Figure 7B). Regardless of these differences, however, TbSP1 labeling was always found to be restricted to the chitin-containing inner cell wall layer, in close proximity to the underlying plasma membrane. This peculiar cell wall microdomain localization was observed in both free-living mycelia and fruitbodies (Figure 7 and data not shown) as well as in symbiosis-engaged hyphae (Figure 8).
Discussion
In fungi, nutrient starvation is a prime environmental cue that through the overexpression of surface proteins leads to changes in invasiveness, nutrient foraging or cell–cell interaction capacity as well as to important morphogenetic transitions (Madhani and Fink, 1998; Lengeler et al., 2000). As demonstrated by the results of this work, a striking up-regulation of a novel cell wall-associated and extracellularly secreted PLA2 is elicited by nutrient starvation in the mycorrhizal fungus T.borchii.
TbSP1 identifies a novel family of phospholipases A2
PLA2s are a broad class of enzymes defined by their ability to hydrolyze the sn-2 ester bond of glycerophospholipids. PLA2s utilizing a catalytic histidine residue typically are extracellular enzymes of small size with a high disulfide bond content and Ca2+ requirement, whereas those utilizing a catalytic serine are usually larger intracellular enzymes with no disulfide bonds and no Ca2+ requirement for catalysis (Six and Dennis, 2000). Biochemical properties and sequence similarities with the active site region of histidine PLA2s indicate that TbSP1 is a new small extracellular PLA2. Moreover, the high similarity of TbSP1 with paralogous coding sequences from Streptomyces and Neurospora implies the existence of a family of fungal/bacterial PLA2s. Since all the PLA2s documented so far are from metazoans and plants, it is not surprising that the TbSP1-PLA2 family displays some unique sequence features that distinguish it from all of the other PLA2 groups. The two major differences highlighted by the alignment reported in Figure 2B are: (i) the substitution of the first and the last cysteine in the catalytic site consensus sequence CCXXHDC with serine/alanine and glycine residues, respectively; and (ii) the lack of three partially conserved Ca2+-coordinating residues (the fourth being the aspartate adjacent to the catalytic histidine) within the Ca2+-binding loop that always precedes the HD segment, where the only shared feature is a cysteine residue involved in a universally conserved disulfide bond. More specifically, we find that two conserved glycines belonging to the consensus segment X1CG1XG2 and involved in metal binding are replaced by other small amino acids (S,T,V,A) in the TbSP1-PLA2 and in its relatives. Since the substitution of the G1 residue with a serine is known to cause a 10- to 20-fold reduction in catalytic activity (Bekkers et al., 1991), it is unlikely that the structural context of the Ca2+ coordination site of TbSP1 is the same as that of the other PLA2 groups. It is interesting to note, in this regard, the presence in the Ca2+-binding region of TbSP1-PLA2s of three conserved aspartate residues (Figure 2). Because aspartate is a metal-coordinating amino acid that recurs in many calcium-binding motifs (e.g. EF-hands and EGF-like domains), it is conceivable to hypothesize a role for these residues in metal coordination. Also, amino acid sequence differences in the Ca2+-binding region may explain the much lower half-activating calcium ion concentration of TbSP1 as compared with that of other histidine PLA2s. The sole exception is the conodipine-M PLA2, which also has a non-canonical metal-binding site and a micromolar Ca2+ requirement (McIntosh et al., 1995). Another remarkable difference between TbSP1 and the other PLA2s is the number of disulfide bonds, which ranges from five to eight in the various PLA2 groups as opposed to only two disulfides in the case of TbSP1. One of them is probably formed by residues C128 and C144 in the active site region, whereas the other involves residues C180 and C194, which are conserved in the alignment of TbSP1 homologs, but not in other PLA2 groups (not shown).
It thus seems that TbSP1 and its homologs constitute a new family of small secreted PLA2s that is defined by organism source, active site sequence similarity and cysteine residue conservation. Following the current PLA2 classification (Six and Dennis, 2000) and the recent identification of group XII PLA2s (Gelb et al., 2000), we propose to name it fungal/bacterial group XIII PLA2. A remarkable feature of this family is its peculiar distribution among fungi and bacteria. In fact, even though the complete genomes of many bacteria and of the yeast S.cerevisiae are available, this family appears to be represented only in filamentous fungi and in the Actino mycetales (Streptomyces spp.), bacteria that resemble fungi in their branching filamentous structure. It is likely that other group XIII PLA2s will soon be identified, since present members of this group have been found in organisms that have only partial genome coverage, and because neither of the two Neurospora homologs appears to be orthologously related to TbSP1 (see the identity values reported in Figure 2A). Furthermore, the most conserved portion of the TbSP1-PLA2 sequence was found as part of a much larger polypeptide (704 amino acids) of unknown function encoded by the Streptomyces clavuligerus Rhsa gene (Alexander and Jensen, 1998), thus suggesting that fungal/bacterial group XIII domains can also be used as catalytic modules in multidomain proteins.
Dual localization of the TbSP1 phospholipase
In keeping with the notion that filamentous fungi are highly efficient secretory machines (Wessels, 1999), no immature TbSP1 pre-protein was found in T.borchii mycelia, not even under conditions of persistent starvation that caused a massive increase in TbSP1 accumulation. Intracellularly entrapped, immature pre-TbSP1 was only detected as a consequence of heterologous expression in yeast cells, which nevertheless properly recognized the TbSP1 secretion signal and released sizeable amounts of the processed Tuber protein into the growth medium. The partitioning of TbSP1 between the cell surface and the extracellular space is reminiscent of the dual localization of other fungal proteins, such as various hydrophobins and adhesins (Martin et al., 1999; McCabe and Van Alfen, 1999; Wessels, 1999) as well as hydrolytic enzymes, including phospholipases B (Chaffin et al., 1998). Some of these proteins are associated only transiently with the cell wall during translocation to the extracellular environment. In other cases, such as that of the WI-1 adhesin of Blastomyces dermatitidis, final cell wall destination is achieved by initial release into the extracellular space, followed by reabsorption and stable cell wall association through interaction with exposed chitin fibrils (Brandhorst and Klein, 2000). Similarly to the WI-1 adhesin, TbSP1 is a largely hydrophilic protein that localizes specifically to a chitin-rich cell wall region from which it can be extracted simply by aqueous washing. At variance with the Blastomyces situation, however, chitin is present mainly in the inner layer of the Tuber cell wall, thus making any surface localization mechanism relying on extracellular release and chitin-mediated reattachment quite unlikely. A more viable alternative, supported by the extremely uniform inner cell wall layer localization of TbSP1 and by the fairly large amounts of TbSP1 extracellularly released by yeast cells, which are known to have a relatively chitin-poor inner layer (Smits et al., 1999), is that TbSP1 binds to chitin fibers as it is secreted, and that shedding from, or saturation of, such chitin-binding sites results in the release of the protein into the extracellular space. Also consistent with its partially extracellular localization is the presence in TbSP1 of the tripeptide RGD, a motif mediating the interaction of extracellular proteins with surface-associated integrin-like receptors that have been described in both fungi and plants (Hostetter, 2000). This motif is located in an extra segment of ∼40 amino acids (Figure 2A) that is predicted to be composed mainly of loops and distinguishes TbSP1 from other group XIII PLA2s. Interestingly, among the few RGD-containing proteins thus far identified in fungi, there are the SRAPS of P.tinctorius, which accumulate on the surface of both free-living and symbiosis-engaged hyphae and may be involved in ectomycorrhiza formation (Laurent et al., 1999). Also noteworthy, in this regard, is that the only precedent for an RGD-containing phospholipase is a sea snake venom (Hydrophis cyanocinctus) PLA2, in which the RGD motif is thought to mediate interaction with platelet surface receptors, thereby inhibiting platelet aggregation (Ali et al., 1999).
Starvation-induced up-regulation of TbSP1 and its possible significance
The remaining part of our work was aimed at verifying the existence in T.borchii of control mechanisms modulating the expression of TbSP1 in response to nutrient availability. We found that a strong and reversible up-regulation of the TbSP1 mRNA does indeed take place in response to carbon or nitrogen deprivation and results in a massive surface accumulation of the TbSP1 phospholipase. Nitrogen starvation was the most effective stimulus. Instead, no appreciable effect was elicited by phosphate deprivation, thus arguing that TbSP1 up-regulation genuinely reflects adaptation to the shortage of specific nutrients, rather than an indirect and non-specific consequence of a generalized growth perturbation. The paradigm of this kind of response is the induction of pseudohyphal growth in nitrogen-starved S.cerevisiae cells (Madhani and Fink, 1998). Similar nutrient-responsive transduction pathways, however, have been delineated in a variety of other unicellular and multicellular fungi (Lengeler et al., 2000), including the plant pathogen M.grisea, where nitrogen starvation up-regulates the expression of the MPG1 hydrophobin mRNA and of other mRNAs coding for as yet uncharacterized surface proteins involved in appressorium formation and disease symptom outbreak (Talbot et al., 1993, 1997). Given this high degree of functional conservation, it is likely that similar regulatory networks also operate in underground symbiotic fungi such as Tuber, which live in environments where the nitrogen supply is often limiting and the poorly mobile ammonium ion predominates as an inorganic nitrogen source (Keeney, 1980; Read, 1991). Other questions raised by this finding regard the generality of occurrence of TbSP1-like enzymes in other organisms and the possible adaptive significance of starvation-induced phospholipase up-regulation. The identification of TbSP1 homologs in distantly related, but all filamentous microorganisms, argues in favor of a rather widespread occurrence and clearly points to the existence of some specific functional link between group XIII PLA2s and the formation of mycelia through hyphal branching and extension. Based on the constitutive expression of TbSP1 in nutrient-sufficient free-living mycelia as well as in mycorrhizal hyphae, on its close plasma membrane proximity and on available data for related enzymes from other organisms (Six and Dennis, 2000), it is conceivable that TbSP1, in its basal function, may be involved in cell surface remodeling, either directly via phospholipid hydrolysis, or through the inhibition of enzymes [e.g. (1,3)-β-glucan synthase] involved in the synthesis of cell wall polysaccharides (Ko et al., 1994). Two additional possibilities can be envisaged, however. The first rests on the ability of TbSP1 to be released extracellularly and on the well-documented action of fungal pathogen phospholipases B as virulence factors promoting membrane disruption and cell invasion in both animal and plant hosts with the concomitant liberation of fatty acids (Nespoulous et al., 1999; Ghannoum, 2000; Cox et al., 2001). In the context of starvation-induced TbSP1 accumulation, one can thus imagine that free fatty acids, either endogenously generated or derived from closely associated soil microorganisms, may be oxidized and utilized by the fungus as gluconeogenic substrates through the glyoxylate shunt pathway. In support of this view, there is the recently discovered up-regulation of various mRNAs encoding glyoxylate cycle enzymes in nutritionally stressed, macrophage-phagocytosed Candida albicans and S.cerevisiae cells (Lorenz and Fink, 2001), as well as our observation that TbSP1 and the mRNA for the glyoxylate cycle enzyme isocitrate lyase (I.Lacourt and P.Bonfante, unpublished) both accumulate in mature T.borchii fruitbodies, where sugar metabolism is known to be depressed (Saltarelli et al., 1998). The other possibility bears upon the symbiotic capacity of Tuber and on the accumulation of TbSP1 in mycorrhizal hyphae, where the TbSP1 phospholipase may be involved in membrane remodeling or signaling events during early stages of plant colonization. Both actions are consistent with TbSP1-catalyzed phospholipid hydrolysis at the sn-2 position (often containing long chain unsaturated acyl groups) with the concomitant release of potentially bioactive 1-acyl lysophospholipid and unsaturated free fatty acid. Particularly interesting, in this regard, is the recently documented ability of the pathogenic yeasts Cryptococcus neoformans and C.albicans to convert exogenously supplied as well as endogenously produced arachidonic acid into bioactive prostaglandins, which strongly enhance both cell viability and filamentation capacity (Noverr et al., 2001).
Despite the many limitations presently imposed by the lack of genetic transformation in the Tuber system and by the difficulties of in vitro mycorrhization, future studies taking advantage of the molecular reagents reported here will allow better definition of the involvement of TbSP1 and related phospholipases in mycelium formation and in the adaptation to nutrient-limited growth conditions.
Materials and methods
Biological materials
Tuber borchii Vittad. mycelia (isolate ATCC 95640) were grown in the dark at 23°C. The modified Melin–Norkrans synthetic medium (referred to as SLM) or potato–dextrose broth (LM) were employed as liquid nutrient solutions, while synthetic dextrose–agar (SSM) or potato dextrose–agar (SM) (Marx, 1969) were utilized for solid medium cultures, which usually were conducted on semipermeable cellophane membranes (Bio-Rad). For nutrient deprivation experiments, mycelia were first cultured for 21 days on complete SSM, with one transfer to fresh SSM after 14 days, and subsequently shifted for 10 days to the same medium (mock-shift control), or to culture media in which either glucose was omitted (carbon starvation), (NH4)2HPO4 was replaced by K2HPO4 (nitrogen starvation), or (NH4)2HPO4 and KH2PO4 were replaced by the corresponding chloride salts (phosphate starvation). Hazelnut (Corylus avellana) seedlings grown in sterilized natural truffle soil (20–25°C; 70–90% humidity) were utilized for the production of mycorrhizas, which were obtained in 6–8 months after inoculation with a sporal suspension from T.borchii ascomata collected from natural truffle grounds in Piedmont, Italy (Balestrini et al., 1996a).
Gel electrophoretic and microsequence analysis
Balanced amounts of mycelium-conditioned SLM (filtered on 0.8 µm membranes and concentrated ∼1000-fold by ultrafiltration) were fractionated by gel electrophoresis on 12% polyacrylamide–SDS gels (Laemmli, 1970). Following electrophoresis and Coomassie Blue R-250 staining, the gel was electroblotted onto Immobilon-PSQ (Millipore) and the membrane area containing the 23 kDa TbSP1 protein was cut out and subjected to N-terminal amino acid sequence analysis using a blot cartridge device and a Procise 494A protein sequencer (Applied Biosystems).
cDNA isolation and sequence analysis
Two degenerate oligonucleotides (5′-AYGCIGARGTIATIGCIGARCARAC-3′ and 5′-TYTGIGTRTTRAARTCIGGNACRTC-3′) deduced from the sequence of the back-translated p23/20 peptide were used as PCR primers to amplify the corresponding region of the TbSP1 cDNA from an SLM-30, T.borchii mycelium library constructed in the phage vector Uni-Zap XR (a gift of B.Lazzari and A.Viotti, Istituto Biosintesi Vegetali, CNR, Milano). The supernatant of a phage suspension (∼107 phage particles) was used as a template for such reactions. The resulting DNA fragment (54 bp) was gel eluted, cloned into the pGEM-T Easy vector (Promega) and utilized as a probe for plaque hybridization analysis of the SLM-30 library (Sambrook and Russell, 2001). Phage DNA extracted from hybridization-positive plaques was then employed as a template for an additional PCR amplification, using a sequence-specific upstream primer (5′-GCGGAGCAGACCGGTGACGT-3′) and a universal (M13-40) downstream primer. The product of this reaction (800 bp) was cloned into the pGEM-T Easy vector, sequence verified and used for a second hybridization screening conducted on the positive plaques obtained from the previous hybridization. A full-length cDNA of 899 bp was thus identified, cloned into the pGEM-T Easy vector (pGEM-TbSP1) and sequenced on both strands. Protein sequence motifs were searched with the Motif program (GenomeNet WWW server). Sequence similarity searches were conducted with BLAST using the non-redundant and the patent databases at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov) and the Neurospora database at MIPS (http://mips.gsf.de/proj/neurospora/). Predicted sequences of N.crassa polypeptides were inferred from the BLAST search and refined using Genewise (Birney and Durbin, 2000). TbSP1 homologs from Neurospora were designated with the number of the corresponding contig. Sequence alignments were performed with ClustalW (Thompson et al., 1994) and manually edited with GeneDoc (http://www.psc.edu/biomed/genedoc/). Secondary structure predictions were made with the PredictProtein server (http://www.embl-heidelberg.de/predictprotein/predictprotein.html).
DNA and RNA analyses
Genomic DNA samples for gel blot analysis (4 µg each) were digested with EcoRI, BamHI and HindIII, followed by electrophoresis on a 0.8% agarose gel. A random priming labeling kit (Amersham Pharmacia Biotech) was used for 32P labeling of the TbSP1 hybridization probe; blotting onto Hybond-N (Amersham Pharmacia Biotech), pre-hybridization, hybridization and washing were conducted according to the manufacturer’s instructions. Total RNA for RNase protection and RNA gel blot analyses was isolated and quantified as described previously (Balestrini et al., 2000). A 32P-labeled antisense riboprobe (430 nucleotides) for RNase protection assays was prepared by in vitro SP6 RNA polymerase transcription of KpnI-digested pGEM-TbSP1. Saturating amounts of a 259 nucleotide, T.borchii β-tubulin riboprobe (E.Soragni and S.Ottonello, unpublished) were added to all reactions and used as an internal standard. Hybridization (5 µg of total RNA per assay), RNase A/T1 digestion and fractionation on denaturing polyacrylamide gels were carried out as described (Balestrini et al., 2000). Protected fragments were visualized by autoradiography and quantified with a Personal Imager FX using the Multi-Analyst/PC software (Bio-Rad). Total RNA and the 32P-labeled cDNA probe described above were also employed for RNA gel blot analysis: RNA samples (20 µg each) were heat denatured, electrophoresed on formaldehyde–agarose gels and transferred to Hybond-N membranes, which were processed and washed under high stringency conditions following the manufacturer’s instructions. The amount of RNA loaded onto each lane was quantified by UV absorbance and by densitometric analysis of ethidium bromide-stained rRNA bands.
Heterologous expression and purification of the TbSP1 protein
The mature TbSP1 coding region was PCR amplified (25 cycles) using pGEM-TbSP1 as a template (20 ng), a high fidelity DNA polymerase (VentR; New England Biolabs), a sequence-specific NdeI-tailed upstream primer (5′-ACCATATGGGAAACGCTGAGGTTATTGC-3′) and the M13 forward universal primer. The restriction fragment obtained from NdeI–XhoI digestion of the resulting PCR product was ligated into the pET-28b expression vector as an in-frame fusion with a vector-encoded His6 tag sequence (pET-TbSP1) and electroporated into BL21(DE3) cells (Novagen). Protein expression was induced by adding 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and allowed to proceed for 4 h at 30°C. After cell lysis, mature rTbSP1 bearing the N-terminal His6 tag was bound to a metal affinity resin (Talon; Clontech) and purified following the manufacturer’s instructions. Protein concentration was determined with the Coomassie dye method (Bio-Rad) or by UV absorbance using an estimated A280 molar extinction coefficient of 26 270. The composition and purity of protein fractions were assessed by gel electrophoresis on 12% polyacrylamide–SDS gels (Laemmli, 1970). Dithio-bis(2-nitrobenzoic acid) (1 mM final concentration) was utilized for sulfhydryl group determination. Monoclonal anti-His tag antibodies (Amersham Pharmacia Biotech) were utilized for the initial identification of rTbSP1. Rabbit polyclonal anti-TbSP1 antibodies, raised in two independent immunization cycles, were affinity purified on rTbSP1-derivatized Sepharose-4B (Harlow and Lane, 1988).
For yeast expression assays, the full-length pre-TbSP1 cDNA was excised from the pGEM-TbSP1 plasmid by NotI digestion and subcloned into the yeast expression vector pFL61. ‘Sense’ or ‘antisense’ pFL61-TbSP1 constructs, identified by restriction analysis, were transformed into chemically competent W303 S.cerevisiae cells. Yeast cells were grown in SD medium lacking uracil and, after centrifugation at 1500 g for 10 min, the cell pellet and the growth medium (filtered on 0.8 µm membranes and concentrated ∼200-fold by ultrafiltration) were subjected to SDS–PAGE and immunoblot analysis (see below).
Phospholipase activity assays
Phospholipase activity was measured by quantifying [3H]oleic acid release from in vivo labeled E.coli membranes (Franson et al., 1974; Ancian et al., 1995). Enzyme assays were conducted routinely at 30°C for 30 min in 100 µl reaction mixtures containing 140 mM NaCl, 0.5 mM CaCl2, 0.1% (w/v) bovine serum albumin (BSA), 20 mM HEPES–KOH pH 6.5, 2 × 105 d.p.m. of membrane-incorporated [9,10(n)-3H]oleic acid (7.5 Ci/mmol; Amersham Pharmacia Biotech) and 1 µg of purified rTbSP1. Streptomyces violaceoruber PLA2 (10 µg; Sigma) was used as a control. Assays conducted in the absence of Ca2+ (plus 2 mM EDTA) or in the presence of increasing concentrations of CaCl2 (0.001–50 mM) were used to determine the calcium dependence of the TbSP1 phospholipase. The pH optimum for activity was determined using 20 mM sodium acetate buffer for pH values ranging from 4.3 to 6.5, and 20 mM HEPES–KOH from pH 7.0 to 8.8. To determine positional specificity, experiments were conducted under the above-described reaction conditions using unlabeled dipalmitoyl, l-α-phosphatidylcholine (Sigma, 40 µM) plus either 1-palmitoyl-2-[1-14C]palmitoyl, l-α-phosphatidyl choline (2 × 105 d.p.m.) or 1,2-di[1-14C]palmitoyl, l-α-phosphatidyl choline (4 × 105 d.p.m.) (Amersham Pharmacia Biotech) as substrates. Reaction mixtures, containing 50 µg of rTbSP1 and 1.5 mM Triton X-100 in a final volume of 150 µl, were extracted as described (McIntosh et al., 1995). After TLC fractionation (Merkel et al., 1999), reaction products were visualized and quantified by phosphorimaging. Non-linear regression analysis of the data was performed with SigmaPlot (Jandel).
Mycelium extraction, fractionation and immunoblot analysis
After two washings with 5 vols of SLM (W1 and W2), mycelia were frozen and ground in liquid nitrogen, resuspended in 10 vols of a solution containing 1 mM EDTA, 2 mM phenylmethylsulfonyl fluoride (PMSF), 20 mM Na-phosphate buffer pH 7.5, and subjected to a series of differential centrifugation steps (30 min each) carried out at 1500 g (fraction F1), 12 000 g (fraction F2) and 100 000 g (fraction F3, plus the final cytosolic supernatant, F4). Following electrophoresis, balanced amounts of each fraction were electrotransferred to Hybond-ECL (Amersham Pharmacia Biotech) and analyzed with standard procedures using horseradish peroxidase-conjugated anti-rabbit immunoglobulin antibodies and enhanced chemiluminescence reagents (Pierce) according to the manufacturer’s instructions. Parallel reactions with no primary antibody or with rTbSP1-saturated primary antibodies were used as specificity controls.
Immunofluorescence and immunogold microscopic analyses
For immunofluorescence experiments, hyphae obtained from nutrient-sufficient or starved mycelia were incubated overnight at 4°C in the absence (specificity control) or presence (test sample) of the anti-TbSP1 antibody. Hyphae were then washed three times with phosphate buffer, saturated with 1% BSA in phosphate buffer and further incubated in the dark (2 h) with a fluorescein isothiocyanate (FITC)-conjugated, goat anti-rabbit IgG antibody (Sigma). Samples were then washed as before, mounted and visualized at 494 nm with a confocal scanning microscope (Optiphot-2 View Scan DVC-250, Nikon). Mycelia, fruitbodies and mycorrhizas were prepared for TEM analysis by overnight fixation at 4°C in 2.5% (v/v) glutaraldehyde dissolved in 10 mM sodium phosphate buffer pH 7.2. After washing with the same buffer, followed by osmium tetroxide (1% w/v) post-fixation, ethanol dehydration and ethanol/LR White (Sigma) infiltration, samples were embedded in LR White resin. Immunogold labeling was performed on thin sections (Balestrini et al., 1996b) and visualized with a Philips CM10 transmission electron microscope. Samples from which the primary antibody was omitted were used as labeling specificity controls. Semi-thin sections (1 µm) stained with 1% toluidine blue were used for light microscopy analysis.
Acknowledgments
Acknowledgements
We thank Hans Heid (DKFZ, Heidelberg) for microsequence analysis, Angelo Viotti and Barbara Lazzari (Istituto Biosintesi Vegetali, CNR, Milano) for the gift of the Tuber cDNA library, Alessandra Zambonelli (Dipartimento Protezione e Valorizzazione Agroalimentare, Università di Bologna) for Tuber mycelia, Michèle Minet (CNRS, Gif-sur-Yvette) for the pFL61 plasmid, Stefania Petrucco and Barbara Montanini for helpful discussions, and Gian Luigi Rossi for encouragement and support. This work was supported by the National Research Council of Italy, Progetto Strategico ‘Biotecnologia dei funghi eduli ectomicorrizici’ by Regione Emilia-Romagna and by a grant from the Ministry of University and Scientific and Technological Research to P.B.
References
- Alexander D.C. and Jensen,S.E. (1998) Investigation of the Streptomyces clavuligerus cephamycin C gene cluster and its regulation by the CcaR protein. J. Bacteriol., 180, 4068–4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali S.A., Alam,J.M., Stoeva,S., Schutz,J., Abbasi,A., Zaidi,Z.H. and Voelter,W. (1999) Sea snake Hydrophis cyanocinctus venom. I. Purification, characterization and N-terminal sequence of two phospholipases A2. Toxicon, 37, 1505–20. [DOI] [PubMed] [Google Scholar]
- Ancian P., Lambeau,G. and Lazdunski,M. (1995) Multifunctional activity of the extracellular domain of the M-type (180 kDa) membrane receptor for secretory phospholipases A2. Biochemistry, 34, 13146–13151. [DOI] [PubMed] [Google Scholar]
- Balestrini R., Hahn,M.G. and Bonfante,P. (1996a) Location of cell-wall components in ectomycorrhizae of Corylus avellana and Tuber magnatum. Protoplasma, 191, 55–69. [Google Scholar]
- Balestrini R., Hahn,M.G., Faccio,A., Mendgen,K. and Bonfante,P. (1996b) Differential localization of carbohydrate epitopes in plant cell walls in the presence and absence of arbuscular mycorrhizal fungi. Plant Physiol., 111, 203–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balestrini R., Mainieri,D., Soragni,E., Garnero,L., Rollino,S., Viotti,A., Ottonello,S. and Bonfante,P. (2000) Differential expression of chitin synthase III and IV mRNAs in ascomata of Tuber borchii Vittad. Fungal Genet. Biol., 31, 219–232. [DOI] [PubMed] [Google Scholar]
- Bekkers A.C., Franken,P.A., Toxopeus,E., Verheij,H.M. and de Haas,G.H. (1991) The importance of glycine-30 for enzymatic activity of phospholipase A2. Biochim. Biophys. Acta, 1076, 374–378. [DOI] [PubMed] [Google Scholar]
- Birney E. and Durbin,R. (2000) Using GeneWise in the Drosophila annotation experiment. Genome Res., 10, 547–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonfante P., Balestrini,R., Martino,E., Perotto,S., Plassard,C. and Mousain,D. (1998) Morphological analysis of early contacts between pine roots and two ectomycorrhizal Suillus strains. Mycorrhiza, 8, 1–10. [Google Scholar]
- Brandhorst T. and Klein,B. (2000) Cell wall biogenesis of Blastomyces dermatitidis. Evidence for a novel mechanism of cell surface localization of a virulence-associated adhesin via extracellular release and reassociation with cell wall chitin. J. Biol. Chem., 275, 7925–7934. [DOI] [PubMed] [Google Scholar]
- Buscot F., Munch,J.C., Charcosset,J.Y., Gardes,M., Nehls,U. and Hampp,R. (2000) Recent advances in exploring physiology and biodiversity of ectomycorrhizas highlight the functioning of these symbioses in ecosystems. FEMS Microbiol. Rev., 24, 601–614. [DOI] [PubMed] [Google Scholar]
- Chaffin W.L., Lopez-Ribot,J.L., Casanova,M., Gozalbo,D. and Martinez,J.P. (1998) Cell wall and secreted proteins of Candida albicans: identification, function and expression. Microbiol. Mol. Biol. Rev., 62, 130–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox G.M. et al. (2001) Extracellular phospholipase activity is a virulence factor for Cryptococcus neoformans. Mol. Microbiol., 39, 166–175. [DOI] [PubMed] [Google Scholar]
- Franson R., Patriarca,P. and Elsbach,P. (1974) Phospholipid metabolism by phagocytic cells. Phospholipases A2 associated with rabbit polymorphonuclear leukocyte granules. J. Lipid Res., 15, 380–388. [PubMed] [Google Scholar]
- Fremont D.H., Anderson,D.H., Wilson,I.A., Dennis,E.A. and Xuong,N.H. (1993) Crystal structure of phospholipase A2 from Indian cobra reveals a trimeric association. Proc. Natl Acad. Sci. USA, 90, 342–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelb M.H., Valentin,E., Ghomashchi,F., Lazdunski,M. and Lambeau,G. (2000) Cloning and recombinant expression of a structurally novel human secreted phospholipase A2. J. Biol. Chem., 275, 39823–39826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghannoum M.A. (2000) Potential role of phospholipases in virulence and fungal pathogenesis. Clin. Microbiol. Rev., 13, 122–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E. and Lane,D. (1988) Antibodies. A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Hebe G., Hager,A. and Salzer,P. (1999) Initial signalling processes induced by elicitors of ectomycorrhiza-forming fungi in spruce cells can also be triggered by G-protein-activating mastoparan and protein phosphatase-inhibiting cantharidin. Planta, 207, 418–425. [Google Scholar]
- Hostetter M.K. (2000) RGD-mediated adhesion in fungal pathogens of humans, plants and insects. Curr. Opin. Microbiol., 3, 344–348. [DOI] [PubMed] [Google Scholar]
- Keeney D.R. (1980) Prediction of soil nitrogen availability in forest ecosystems: a literature review. Forest Sci., 26, 159–171. [Google Scholar]
- Kim S.J., Bernreuther,D., Thumm,M. and Podila,G.K. (1999) LB-AUT7, a novel symbiosis-regulated gene from an ectomycorrhizal fungus, Laccaria bicolor, is functionally related to vesicular transport and autophagocytosis. J. Bacteriol., 181, 1963–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko Y.T., Frost,D.J., Ho,C.T., Ludescher,R.D. and Wasserman,B.P. (1994) Inhibition of yeast (1,3)-β-glucan synthase by phospholipase A2 and its reaction products. Biochim. Biophys. Acta, 1193, 31–40. [DOI] [PubMed] [Google Scholar]
- Laemmli U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Laurent P. et al. (1999) A novel class of ectomycorrhiza-regulated cell wall polypeptides in Pisolithus tinctorius. Mol. Plant Microbe Interact., 12, 862–871. [DOI] [PubMed] [Google Scholar]
- Lauter F.R., Russo,V.E. and Yanofsky,C. (1992) Developmental and light regulation of eas, the structural gene for the rodlet protein of Neurospora. Genes Dev., 6, 2373–2381. [DOI] [PubMed] [Google Scholar]
- Lengeler K.B., Davidson,R.C., D’Souza,C., Harashima,T., Shen,W.C., Wang,P., Pan,X., Waugh,M. and Heitman,J. (2000) Signal transduction cascades regulating fungal development and virulence. Microbiol. Mol. Biol. Rev., 64, 746–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz M.C. and Fink,G.R. (2001) The glyoxylate cycle is required for fungal virulence. Nature, 412, 83–86. [DOI] [PubMed] [Google Scholar]
- Madhani H.D. and Fink,G.R. (1998) The control of filamentous differentiation and virulence in fungi. Trends Cell Biol., 8, 348–353. [DOI] [PubMed] [Google Scholar]
- Martin F. and Tagu,D. (1999) Developmental biology of a plant–fungus symbiosis: the ectomycorrhiza. In Varma,A. and Hock,B. (eds), Mycorrhiza: Structure, Function, Molecular Biology and Biotechnology. Springer-Verlag, Berlin, Germany, pp. 51–73.
- Martin F., Laurent,P., de Carvalho,D., Voiblet,C., Balestrini,R., Bonfante,P. and Tagu,D. (1999) Cell wall proteins of the ectomycorrhizal basidiomycete Pisolithus tinctorius: identification, function and expression in symbiosis. Fungal Genet. Biol., 27, 161–174. [DOI] [PubMed] [Google Scholar]
- Marx D.H. (1969) The influence of ectopic mycorrhizal fungi on the resistance of pine roots to pathogenic infection. I. Antagonism of mycorrhizal fungi to root pathogenic fungi and soil bacteria. Phytopathology, 59, 153–163. [PubMed] [Google Scholar]
- McCabe P.M. and Van Alfen,N.K. (1999) Secretion of cryparin, a fungal hydrophobin. Appl. Environ. Microbiol., 65, 5431–5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh J.M., Ghomashchi,F., Gelb,M.H., Dooley,D.J., Stoehr,S.J., Giordani,A.B., Naisbitt,S.R. and Olivera,B.M. (1995) Conodipine-M, a novel phospholipase A2 isolated from the venom of the marine snail Conus magus. J. Biol. Chem., 270, 3518–3526. [DOI] [PubMed] [Google Scholar]
- Merkel O., Fido,M., Mayr,J.A., Pruger,H., Raab,F., Zandonella,G., Kohlwein,S.D. and Paltauf,F. (1999) Characterization and function in vivo of two novel phospholipases B/lysophospholipases from Saccharomyces cerevisiae. J. Biol. Chem., 274, 28121–28127. [DOI] [PubMed] [Google Scholar]
- Molina R., Massicotte,H. and Trappe,J.M. (1992) Specificity phenomena in mycorrhizal symbioses: community–ecological consequences and practical implications. In Allen,M.F. (ed.), Mycorrhizal Functioning, an Integrative Plant–Fungal Process. Chapman and Hall, New York, NY, pp. 357–423.
- Nakari-Setala T., Aro,N., Ilmen,M., Munoz,G., Kalkkinen,N. and Penttila,M. (1997) Differential expression of the vegetative and spore-bound hydrophobins of Trichoderma reesei—cloning and characterization of the hfb2 gene. Eur. J. Biochem., 248, 415–423. [DOI] [PubMed] [Google Scholar]
- Nehls U., Wiese,J., Guttenberger,M. and Hampp,R. (1998) Carbon allocation in ectomycorrhizas: identification and expression analysis of an Amanita muscaria monosaccharide transporter. Mol. Plant Microbe Interact., 11, 167–176. [DOI] [PubMed] [Google Scholar]
- Nespoulous C., Gaudemer,O., Huet,J.C. and Pernollet,J.C. (1999) Characterization of elicitin-like phospholipases isolated from Phytophthora capsici culture filtrate. FEBS Lett., 452, 400–406. [DOI] [PubMed] [Google Scholar]
- Noverr M.C., Phare,S.M., Toews,G.B., Coffey,M.J. and Huffnagle,G.B. (2001) Pathogenic yeasts Cryptococcus neoformans and Candida albicans produce immunomodulatory prostaglandins. Infect. Immun., 69, 2957–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read D.J. (1991) Mycorrhizas in ecosystems. Experientia, 47, 376–391. [Google Scholar]
- Read D.J. (1999) Mycorrhiza—the state of the srt. In Varma,A. and Hock,B. (eds), Mycorrhiza: Structure, Function, Molecular Biology and Biotecnology. Springer-Verlag, Berlin, Germany, pp. 3–34.
- Saltarelli R., Ceccaroli,P., Vallorani,L., Zambonelli,A., Citterio,B., Malatesta,M. and Stocchi,V. (1998) Biochemical and morphological modifications during the growth of Tuber borchii mycelium. Mycol. Res., 102, 403–409. [Google Scholar]
- Sambrook J. and Russell,D.W. (2001) Molecular Cloning. A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Six D.A. and Dennis,E.A. (2000) The expanding superfamily of phospholipase A(2) enzymes: classification and characterization. Biochim. Biophys. Acta, 1488, 1–19. [DOI] [PubMed] [Google Scholar]
- Smits G.J., Kapteyn,J.C., van den Ende,H. and Klis,F.M. (1999) Cell wall dynamics in yeast. Curr. Opin. Microbiol., 2, 348–352. [DOI] [PubMed] [Google Scholar]
- Tagu D., Nasse,B. and Martin,F. (1996) Cloning and characterization of hydrophobins-encoding cDNAs from the ectomycorrhizal basidomycete Pisolithus tinctorius. Gene, 168, 93–97. [DOI] [PubMed] [Google Scholar]
- Talbot N.J., Ebbole,D.J. and Hamer,J.E. (1993) Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea. Plant Cell, 5, 1575–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot N.J., McCafferty,H.R.K., Ma,M., Moore,K. and Hamer,J.E. (1997) Nitrogen starvation of the rice blast fungus Magnaporthe grisea may act as an environmental cue for disease symptom expression. Physiol. Mol. Plant Pathol., 50, 179–195. [Google Scholar]
- Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voiblet C., Duplessis,S., Encelot,N. and Martin,F. (2001) Identification of symbiosis-regulated genes in Eucalyptus globulus–Pisolithus tinctorius ectomycorrhiza by differential hybridization of arrayed cDNAs. Plant J., 25, 181–191. [DOI] [PubMed] [Google Scholar]
- Wessels J.G.H. (1999) Fungi in their own right. Fungal Genet. Biol., 27, 134–145. [DOI] [PubMed] [Google Scholar]
- Wosten H.A., van Wetter,M.A., Lugones,L.G., van der Mei,H.C., Busscher,H.J. and Wessels,J.G. (1999) How a fungus escapes the water to grow into the air. Curr. Biol., 9, 85–88. [DOI] [PubMed] [Google Scholar]