Abstract
TAFIIs are conserved components of the TFIID, TFTC and SAGA-related mRNA transcription complexes. In yeast (y), yTAFII17 is required broadly for transcription, but various other TAFIIs appear to have more specialized functions. It is important to determine how TAFIIs contribute to transcription in metazoans, which have larger and more diverse genomes. We have examined TAFII functions in early Caenorhabditis elegans embryos, which can survive without transcription for several cell generations. We show that taf-10 (yTAFII17) and taf-11 (yTAFII25) are required for a significant fraction of transcription, but apparently are not needed for expression of multiple developmental and other metazoan-specific genes. In contrast, taf-5 (yTAFII48; human TAFII130) seems to be required for essentially all early embryonic mRNA transcription. We conclude that TAF-10 and TAF-11 have modular functions in metazoans, and can be bypassed at many metazoan-specific genes. The broad involvement of TAF-5 in mRNA transcription in vivo suggests a requirement for either TFIID or a TFTC-like complex.
Keywords: C.elegans/gene regulation/TAFIIs/TFIID/transcription
Introduction
Eukaryotic mRNA transcription requires assembly of a multiprotein pre-initiation complex (PIC) at promoters. This machinery includes RNA polymerase (Pol II), general transcription factors (GTFs) required for Pol II activity (TFIIA, B, D, E, F and H) and a mediator-related complex (Hampsey, 1998; Lemon and Tjian, 2000; Malik and Roeder, 2000). Some PIC components are essential for transcription, but in yeast others may act as modu lar interfaces through which gene groups can be regulated coordinately (Holstege et al., 1998; Green, 2000; Lee et al., 2000). In metazoans, additional PIC components and transcription cofactors have evolved that are not present in yeast (Lemon and Tjian, 2000). Most metazoan genes do not appear to correspond directly to yeast genes, even though many encode conserved domains (Chervitz et al., 1998; Rubin et al., 2000; Rubin, 2001). Given these differences, it is important to determine how conserved PIC components contribute to transcription in metazoans.
The GTF TFIID, which recognizes the transcription start site, is remarkably conserved from yeast to humans (Burley and Roeder, 1996; Albright and Tjian, 2000; Green, 2000). TFIID consists of the TATA-binding protein (TBP), along with ∼12 polypeptides known as the TAFIIs (TBP-associated factors). Some TAFIIs interact with core promoter sequences, and various individual TAFIIs can bind a diverse array of upstream transactivators. In addition, human (h) TAFII250 and its orthologs have enzymatic activities that include a conserved histone acetyl transferase (HAT) (Albright and Tjian, 2000; Green, 2000; Matangkasombut et al., 2000; Pham and Sauer, 2000). The TAFIIs may thus provide a functional link between proximal and distal promoter regions, and activities that promote transcription. Consistent with this idea, a TFIID structure reveals surfaces that could mediate extensive core promoter and protein–protein contacts (Andel et al., 1999; Brand et al., 1999a). Some TAFIIs are also present in the human TBP-free TAFII-containing complex (TFTC), and in the related complexes PCAF and STAGA (human) and SPT–ADA–GCN5 (SAGA) (yeast) (Martinez et al., 1998; Ogryzko et al., 1998; Wieczorek et al., 1998; Brand et al., 1999b; Sterner and Berger, 2000). We refer to these as TFTC-related complexes. They lack TBP, and contain either the GCN5 or PCAF HAT instead of an hTAFII250 ortholog. In addition to TAFIIs, TFTC-related complexes contain subunits that are related to TFIID-specific TAFIIs, suggesting possible functional overlap. Supporting this idea, TFTC is structurally similar to TFIID, and can mediate transcription initiation in vitro (Wieczorek et al., 1998; Brand et al., 1999a).
Analysis of conditional yeast mutants indicates that expression of most genes depends upon either the TFIID or SAGA HAT, and that many yeast genes may be regulated through the action of either complex (Lee et al., 2000). Individual yeast TAFIIs are each necessary for cell viability, but the extent to which they are required for Pol II transcription in vivo remains controversial (Albright and Tjian, 2000; Green, 2000; Kuras et al., 2000; Li et al., 2000). Some yeast TAFIIs are broadly required, but others appear to have more specific functions that derive from interactions with core promoters, and possibly with other proteins.
It appears likely that individual metazoan TAFIIs function analogously to yeast TAFIIs in regulating genes that are conserved in all eukaryotes. It is an open question, however, to what extent they are important at genes that do not have yeast counterparts, which we refer to as metazoan-specific genes. Analysis of metazoan TAFII function in vivo has been hampered by cell lethality, and by the complexity of terminal developmental phenotypes (Zhou et al., 1998; Pham et al., 1999; Wassarman et al., 2000). To circumvent the problem of cell lethality, we are studying metazoan TAFIIs in the Caenorhabditis elegans embryo. Caenorhabditis elegans embryonic mRNA transcription appears to begin at the 4-cell stage, but in its absence maternally produced mRNAs maintain viability until around the 100-cell stage (Powell-Coffman et al., 1996; Seydoux and Dunn, 1997). In this context, we can investigate the functions of otherwise essential transcription factors in living cells.
We have used RNA-mediated interference (RNAi) to investigate the functions of three C.elegans TAFIIs: TAF-5, TAF-10 and TAF-11 (Figure 1; Table I). TAF-10 is of considerable interest because it is orthologous to yeast (y)TAFII17, which is very broadly essential for Saccharomyces cerevisiae transcription (Apone et al., 1998; Michel et al., 1998; Moqtaderi et al., 1998; Lee et al., 2000) (Figure 1B). TAF-11 corresponds to yTAFII25, which has been proposed to be either universally (Sanders et al., 1999) or narrowly (Lee et al., 2000) required (Figure 1C). TAF-5 corresponds to yTAFII48, the requirements for which are unknown (Sanders and Weil, 2000). TAF-5 is particularly interesting because it corresponds to hTAFII130, which contains metazoan-specific motifs that are targeted directly by numerous activators (Figure 1A) (Saluja et al., 1998; Rojo-Niersbach et al., 1999). In addition to being present in TFIID, TAF-10 and TAF-11 orthologs are found in all TFTC-related complexes, and a TAF-5 ortholog is present in TFTC (see Brand et al., 1999b). We show that C.elegans taf-10 and taf-11 are required for a significant proportion of embryonic transcription but, strikingly, are not limiting for activation of multiple developmental and other metazoan-specific genes. In contrast, taf-5 appears to be essential for virtually all early transcription. Our findings suggest that TAF-10 and TAF-11 form part of a functional module that is not required for activation of many metazoan-specific genes, and that TAF-5 may have a more fundamental mechanistic role.
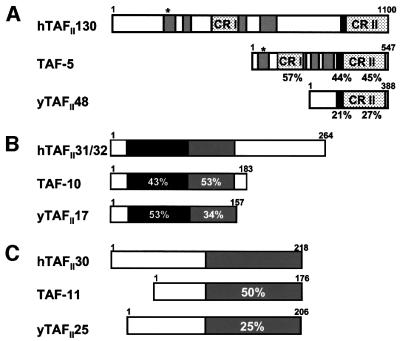
Fig. 1. Similarities between C elegans TAFIIs and their human and yeast counterparts. (A) C.elegans TAF-5 is compared with hTAFII130 and yTAFII48. TAF-5 includes conserved regions (CR) I and II (speckled boxes), the predicted histone fold (black box) (Gangloff et al., 2000) and glutamine-rich regions (gray boxes) present in hTAFII130. The metazoan-specific conserved elements are important for activator binding by hTAFII130 (Saluja et al., 1998; Rojo-Niersbach et al., 1999), particularly a small motif indicated by an asterisk. In (A), (B) and (C), the percentage similarity to the corresponding human TAFII is indicated below or within the relevant motifs. (B) TAF-10 is related to hTAFII31/32 and yTAFII17 within the histone fold (dark gray) (Burley and Roeder, 1996) and an adjacent conserved region (light gray). (C) TAF-11, hTAFII30 and yTAFII25 are most similar within the histone fold region (in gray) (Gangloff et al., 2001b).
Table I. Caenorhabditis elegans TAFIIs.
| Human | Drosophila | Saccharomyces cerevisiae | C.elegans clone | Predicted mol. wt | C.elegans name |
|---|---|---|---|---|---|
| 250 | 250/230 | 130/145 | W04A8.7 | 204.0 | taf-1 |
| 150 | 150 | 150 | Y37E11B.4 | 137.0 | taf-2 |
| 80/70 | 60/62 | 60 | W09B6.2 | 80.9 | taf-3.1 |
| 80/70 | 60/62 | 60 | Y37E11AL.8 | 91.8 | taf-3.2 |
| 100 | 80/58 | 90 | F30F8.8 | 72.1 | taf-4 |
| 130 | 110 | 48 | R119.6 | 60.4 | taf-5 |
| 18 | 19 | C14A4.10 | 47.0 | taf-6 | |
| 28 | 30β | 40 | F48D6.1 | 37.8 | taf-7.1 |
| 28 | 30β | 40 | K10D3.3 | 37.6 | taf-7.2 |
| 28 | 30β | 40 | F43D9.5 | 24.5 | taf-7.3 |
| 55 | 67 | F54F7.1 | 29.2 | taf-8.1 | |
| 55 | 67 | Y111B2A.16 | 26.4 | taf-8.2 | |
| 20 | 30α | 68/61 | Y56A4.3 | 28.0 | taf-9 |
| 31/32 | 40/42 | 17 | T12D8.7 | 20.6 | taf-10 |
| 30 | 24 | 25 | K03B4.3 | 19.6 | taf-11 |
Caenorhabditis elegans homologs of TAFIIs were identified by searching WORMpep or genomic databases (Sanger Centre) with human, Drosophila or S.cerevisiae sequences. The open reading frame, predicted molecular weight and gene name are listed for the C.elegans homologs. Caenorhabditis elegans TAFII genes have been identified and described independently by Aoyagi and Wassarman (2000).
Results
Caenorhabditis elegans TAFIIs
By searching C.elegans databases, we have identified at least one well-conserved homolog for each TAFII in human TFIID (Table I). We have named these C.elegans TAFIIs in order of their predicted molecular weights. Each contains conserved sequence motifs found in their metazoan and yeast orthologs (not shown), including the histone fold domains through which multiple TAFIIs form heterodimers (Burley and Roeder, 1996; Gangloff et al., 2000, 2001a,b). TAF-5, TAF-10 and TAF-11 each contain a characteristic histone fold (Figure 1), through which they are each predicted to pair with a different TAFII (Burley and Roeder, 1996; Gangloff et al., 2000, 2001b). These similarities predict that TFIID structure and function have been conserved in C.elegans. We have also searched for C.elegans orthologs of other TFTC-related complex components. Caenorhabditis elegans encodes a GCN5/PCAF-related HAT (Y.Shi, unpublished) and a TRA1/TRRAP homolog (not shown) but, by our search criteria (Materials and methods), we did not identify orthologs of ADA/SPT proteins (ADA1, ADA2, ADA3, SPT3, SPT7, SPT8 and SPT20), which are not essential for yeast viability (Sterner and Berger, 2000). In C.elegans, ADA/SPT functions may be fulfilled by more distantly related proteins, or a streamlined version of TFTC may be formed by TAFIIs, the GCN5/PCAF-related HAT and TRA1/TRRAP.
tafII(RNAi) embryos arrest development early, without differentiation
Caenorhabditis elegans embryonic development is orchestrated initially by maternally derived proteins and mRNAs, which establish early cell asymmetries and patterns of new embryonic transcription (Newman-Smith and Rothman, 1998). To determine whether TAF-5, TAF-10 and TAF-11 proteins are present in the early embryo, we examined their expression by staining with peptide-derived antibodies. Under staining conditions that were optimized for early embryos (Figure 2 and data not shown), TAF-5 was apparent in nuclei from the 2-cell stage through early morphogenesis, and TAF-10 and TAF-11 were readily detectable in nuclei from the 4-cell stage through late gastrulation. TAF-5, TAF-10 and TAF-11 were similarly detectable in adult germline and oocyte nuclei (not shown), suggesting that they are maternally expressed.
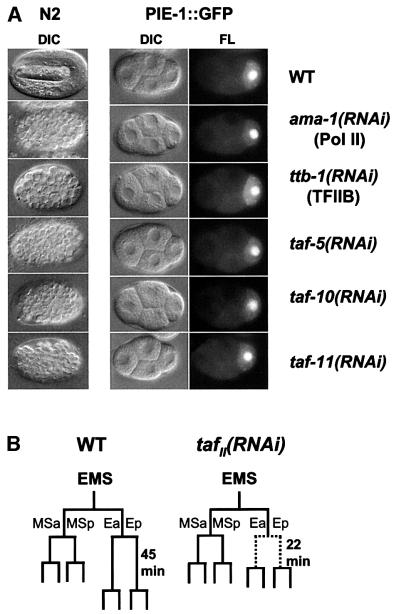
Fig. 2. Expression of TAF-5, TAF-10 and TAF-11 in wild-type and RNAi embryos. Representative wild-type or TAFII RNAi embryos (indicated in rows) were stained with antibodies to TAF-5, TAF-10 or TAF-11, or with DAPI to visualize DNA, as indicated above the columns. RNAi embryos were collected 24 h after injection of hermaphrodite mothers, when a uniformly affected population was being produced (see Materials and methods).
To investigate taf-5, taf-10 and taf-11 functions in the early embryo, we inhibited their expression by RNAi (Fire et al., 1998). As a benchmark for phenotypes caused by pleiotropic transcription defects, we compared tafII(RNAi) embryos with ama-1(RNAi) and ttb-1(RNAi) embryos. ama-1 encodes the Pol II large subunit (Powell-Coffman et al., 1996) and ttb-1 encodes TFIIB, a Pol II GTF required for transcription initiation (Lemon and Tjian, 2000). We determined whether maternal gene expression was generally intact in these RNAi embryos by monitoring early cell division patterns and timing, and by performing parallel RNAi experiments in a transgenic strain that expresses a fusion of the maternally derived germline protein PIE-1 to green fluorescent protein (GFP). This PIE-1::GFP protein recapitulates the complex patterns of PIE-1 expression and localization, which depend upon >20 other maternal genes (Tenenhaus et al., 1998; Reese et al., 2000b).
All ama-1, ttb-1, taf-5, taf-10 and taf-11(RNAi) embryos arrested development at 90–100 cells and lacked signs of differentiation (Figure 3A), as reported previously for ama-1 (Powell-Coffman et al., 1996). At every stage prior to terminal arrest, maternal PIE-1::GFP expression and localization patterns appeared normal in these RNAi embryos (Figure 3A and data not shown). Their early cell division timing and cleavage planes were also generally normal, except for the cell cycle period of the two E cell daughters (E2 cells), which form the endoderm (Figure 3B and data not shown). For gastrulation to occur, the E2 cells must divide after 45 min instead of the 22 min characteristic of their cousins, the two MS2 cells. This cell cycle lengthening requires new mRNA transcription and endodermal specification (Powell-Coffman et al., 1996; Zhu et al., 1998). In ama-1(RNAi), ttb-1(RNAi) and each set of tafII(RNAi) embryos, the E2 cells divided immediately after the MS2 cells (Figure 3B). Our findings suggest that in tafII(RNAi) embryos, maternal mRNA stores appear generally to be intact, but new mRNA transcription may be severely impaired.
Fig. 3. Terminal and early cell division phenotypes of ama-1 (RNA pol II), ttb-1 (TFIIB), taf-5, taf-10 and taf-11 RNAi embryos. (A) TAFII RNAi embryo phenotypes. RNAi embryos produced by N2 (wild-type) or pie-1::gfp mothers were examined by differential interference (DIC) or fluorescence (FL) microscopy. Typical examples of wild-type (WT) or RNAi embryos are shown, as indicated to the right of each row. The left column compares terminally arrested RNAi embryos with a wild-type embryo that is about to hatch. ama-1(RNAi), ttb-1(RNAi), taf-5(RNAi), taf-10(RNAi) and taf-11(RNAi) embryos each arrested with 90–100 cells (n = 5).The right two columns show 4-cell pie-1::gfp WT and RNAi embryos. In these RNAi embryos, each aspect of PIE-1::GFP germline and subcellular localization was indistinguishable from wild type, including the presence of PIE-1 in germline RNA– protein P granules (Reese et al., 2000b). Embryos measure ∼50 µm. (B) Shortened E2 cell cycle in tafII(RNAi) embryos. Lineage analysis of each set of tafII(RNAi) embryos (n ≥5) revealed that their early cell division planes and times were normal, except that their E2 cells (Ea and Ep) divided prematurely. Only the EMS cell lineage is shown.
Nuclear antibody staining for TAF-5, TAF-10 or TAF-11 was eliminated in each respective set of RNAi embryos (Figure 2), indicating a penetrant loss of function. In yeast, loss of some TAFIIs destabilizes other TFIID components (Apone et al., 1998; Michel et al., 1998; Moqtaderi et al., 1998; Chen and Manley, 2000). To investigate whether this might have occurred, we stained tafII(RNAi) embryos with antibodies against each TAFII that we analyzed (Figure 2). TAF-10 and TAF-11 were both present at normal levels in taf-5(RNAi) embryos. Interference with either taf-10 or taf-11 did not affect TAF-5 expression, but caused loss of both TAF-10 and TAF-11. Because we have not detected evidence of maternal gene expression defects in these tafII(RNAi) embryos, we conclude that TAF-10 and TAF-11 proteins may each depend upon each other for stability.
Inhibition of Pol II CTD phosphorylation in tafII(RNAi) embryos
To investigate overall transcription levels in tafII(RNAi) embryos, we analyzed phosphorylation of the Pol II large subunit C-terminal domain (CTD). The CTD consensus repeat (YSPTSPS; 42 copies in C.elegans) is phosphorylated on actively transcribing Pol II (Hirose and Manley, 2000). CTD phosphorylation is important for promoter clearance, elongation and integration of transcription with mRNA processing. At the promoter, yeast Pol II is phosphorylated on Ser5 of the CTD repeat by the TFIIH kinase (Komarnitsky et al., 2000; Schroeder et al., 2000). As Pol II moves away from the start site, the distribution of CTD phosphorylation shifts to Ser2, but the kinase responsible has not been identified (Komarnitsky et al., 2000). In C.elegans embryos, CTD phosphorylation patterns are tightly correlated with transcriptional activity (Seydoux and Dunn, 1997; Tenenhaus et al., 1998). In somatic nuclei, antibody staining first detects Ser5 phosphorylation as a bright punctate pattern at the 4-cell stage, when transcription begins (Seydoux and Dunn, 1997) (Figure 4, columns 2 and 3). In the transcriptionally silent early germline precursor nucleus, this staining is confined to two distinct foci. Ser2 phosphorylation is first detectable at the 4-cell stage, and is absent in the early embryonic germline (Seydoux and Dunn, 1997) (Figure 4, column 5).
Fig. 4. Broader requirement for taf-5 than taf-10 or taf-11 for Pol II CTD phosphorylation. Wild-type or RNAi embryos were stained with Pol II CTD antibodies (see text) prior to terminal developmental arrest. Representative embryos of comparable stages are presented in rows, as indicated. Columns 1, 4 and 6 show nuclei stained by DAPI. α-P-Ser5 staining is shown in column 2, and an expansion of the nucleus marked by the red arrow is shown in column 3. Column 5 shows embryos stained with the α-P-Ser2 antibody. Column 7 shows staining with α-UnP CTD; identical results were obtained with an antibody against a different Pol II region (POL 3/3; not shown). α-PSer5 and α-PSer2 recognize Pol II isoforms associated with transcription initiation and elongation, respectively (Komarnitsky et al., 2000; Schroeder et al., 2000), and the α-UnP CTD antibody provides a Pol II expression control. In columns 2 and 5, germline nuclei that are in the focal plane shown are marked with an asterisk. In α-P-Ser5-stained germline nuclei, note the lack of nucleoplasmic staining and the presence of two discrete dots (Seydoux and Dunn, 1997). Some germline nuclei stained with α-PSer2 have perinuclear background deriving from cross-reactivity of the secondary antibodies with P granules. The relative differences in α-PSer5 and α-PSer2 staining intensities were comparable when embryos were photographed at multiple different exposure times.
We stained embryos with the P-CTD antiserum to detect phospho-Ser5 (Schroeder et al., 2000), the H5 antibody to detect phospho-Ser2 (Bregman et al., 1995; Patturajan et al., 1998) and the 8WG16 antibody to detect the unphosphorylated CTD (Patturajan et al., 1998). To avoid confusion, we refer to these antibodies as α-PSer5, α-PSer2 and α-UnP CTD, respectively. In contrast to ama-1(RNAi) embryos, in which staining with α-UnP CTD was abolished, in ttb-1(RNAi) and tafII(RNAi) embryos Pol II levels were not significantly affected (Figure 4, column 7). α-PSer5 staining of wild-type embryos (Figure 4, columns 2 and 3) recapitulated the pattern obtained previously with the phospho-Ser5 antibody H14 (see above) (Seydoux and Dunn, 1997). In contrast, nuclear α-PSer5 staining was reduced to background levels in ama-1(RNAi) and ttb-1(RNAi) embryos. In taf-5(RNAi) embryos, diffuse nucleoplasmic α-PSer5 staining was reduced similarly, but each somatic nucleus contained two distinct foci like those in the transcriptionally silent germline cell (Figure 4, columns 2 and 3). In parallel to staining with α-PSer5, α-PSer2 reactivity was comparably severely reduced in ama-1(RNAi), ttb-1 (RNAi) and taf-5(RNAi) embryos (Figure 4, column 5). These staining reductions were representative of these RNAi embryos from the 4-cell stage until arrest (not shown), suggesting that in ama-1(RNAi), ttb-1(RNAi) and taf-5(RNAi) embryos overall transcription levels are extremely low at each embryonic stage.
In contrast, CTD phosphorylation was less severely affected in taf-10(RNAi), taf-11(RNAi) and taf-10(RNAi); taf-11(RNAi) embryos. In the somatic nuclei, at all embryonic stages, two α-PSer5 foci were accompanied by nucleoplasmic staining that was decreased, but more prominent than in taf-5(RNAi) embryos (Figure 4, columns 2 and 3; not shown). In taf-10(RNAi), taf-11(RNAi) and taf-10(RNAi); taf-11(RNAi) embryos, α-PSer2 staining levels were similarly not eliminated (Figure 4, column 5). The comparable reduction in CTD phosphorylation accompanying simultaneous interference with taf-10 and taf-11 is consistent with the interdependence of TAF-10 and TAF-11 protein levels (Figure 2). These findings suggest that some transcription can occur independently of taf-10 and taf-11.
taf-10 and taf-11 are not rate limiting for many metazoan-specific promoters
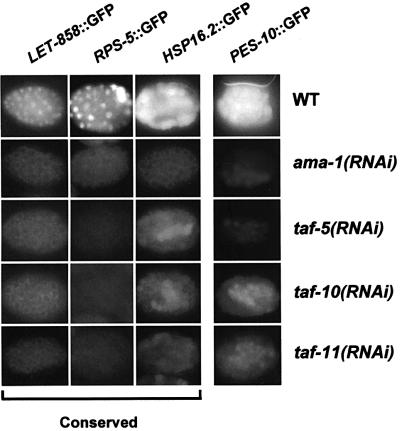
To investigate how these TAFIIs are involved in expression of individual genes, we performed RNAi experiments in C.elegans that carry transgenic reporters that are transcribed in the early embryo. These reporters include intact regulatory regions along with GFP-fused coding regions, and are expressed in patterns that parallel the corresponding endogenous genes. We examined expression of three genes that are common to yeast and metazoans: let-858, rps-5, and hsp16.2, a heat shock gene. In yeast, rps-5 transcription is highly dependent upon TAFIIs (Li et al., 2000). Interference with taf-5, taf-10 or taf-11 abolished LET-858::GFP and RPS-5::GFP expression, and significantly decreased expression of HSP-16.2::GFP in response to heat shock (Figure 5). Interference with each of these tafIIs comparably impaired expression of these reporters (Figure 5), suggesting that the less severe decrease in CTD phosphorylation associated with interference with taf-10 or taf-11 (Figure 4) reflects a difference in function, not RNAi penetrance.
Fig. 5. Comparable requirements for taf-5, taf-10 and taf-11 at conserved genes. GFP fluorescence was examined in wild-type or tafII(RNAi) embryos (in rows) that were produced by the reporter strains indicated above the columns. Each of these reporters was expressed in most embryonic cells. In a representative experiment, the RPS-5::GFP reporter, which is non-integrated, was expressed in 23/47 wild-type embryos but in none of >50 of each set of RNAi embryos. Embryos shown are otherwise representative of the entire population analyzed in each of multiple independent experiments, in which >40 embryos were scored per reporter strain. HSP16.2::GFP expression varied slightly within each set of embryos, but those depicted correspond to average levels of expression and to representative differences between WT and RNAi embryos. Genes that are conserved between yeast and metazoans are indicated at the bottom.
We next tested how interference with these TAFII genes affects expression of genes that are not present in yeast, but are widely expressed. pes-10 has been identified only in C.elegans, and is expressed at the onset of embryonic transcription (Seydoux and Fire, 1994). PES-10::GFP expression was severely affected by interference with ama-1 or taf-5 expression, and was decreased in taf-10 (RNAi) or taf-11(RNAi) embryos (Figure 5). cki-2 (CDK inhibitor) and sur-5 (MAP kinase pathway component) are broadly conserved among metazoans (Gu et al., 1998; Hong et al., 1998). cki-2 and sur-5 reporters required ama-1 and taf-5, but were unaffected by interference with either taf-10 or taf-11 expression (Table II).
Table II. Requirements for TAFIIs for metazoan-specific gene expression.
| SUR-5::GFP (MAP kinase pathway) | CKI-2::GFP (cell cycle) | MED-1::GFP (endoderm/mesoderm) | MED-2::GFP (endoderm/mesoderm) | PHA-4::GFP (digestive tract) | ELT-5::GFP (ectoderm) | |
|---|---|---|---|---|---|---|
| Wild type | + | + | + | + | + | + |
| ama-1(RNAi) | – | – | – | – | – | – |
| taf-5(RNAi) | – | – | – | – | – | – |
| taf-10(RNAi) | + | + | + | + | + | + |
| taf-11(RNAi) | + | + | + | + | + | + |
Reporter strains were scored as + when GFP was expressed at wild-type levels in all embryos. They were scored as – when GFP was undetectable, or was present at comparable trace levels in tafII and ama-1 RNAi embryos. In each case, >40 embryos were analyzed in multiple independent experiments. In three independent reporter strains, PHA-4::GFP was expressed at normal levels, but in fewer cells than wild type.
We also investigated the importance of these TAFIIs for activation of cell type-specific genes. The redundant GATA factor-encoding genes med-1 and med-2 specify mesendodermal lineages (Maduro et al., 2001), and are required for expression of the related gene end-1, which specifies the endoderm (Zhu et al., 1998). pha-4 is a forkhead-family gene that later specifies the pharynx and rectum (Horner et al., 1998; Kalb et al., 1998), and elt-5 encodes an epidermally expressed GATA factor (J.Rothman, unpublished). In taf-5(RNAi) embryos, GFP reporters corresponding to these genes were not expressed above the trace or undetectable levels characteristic of ama-1(RNAi) embryos (Figure 6; Table II). In contrast, med-1, med-2 and elt-5 reporters were expressed robustly in all taf-10(RNAi) and taf-11(RNAi) embryos (Table II). All taf-10(RNAi) and taf-11(RNAi) embryos also expressed PHA-4::GFP in many cells (Table II), a striking finding because pha-4 transcription requires upstream zygotic gene expression (Horner et al., 1998; Kalb et al., 1998). END-1::GFP normally appears in the E2 cells, then persists in their E4–E8 descendants (Figure 6 and data not shown). As predicted from their shortened E2 cell cycle (Figure 3B), in most taf-10(RNAi), taf-11(RNAi) and taf-10(RNAi); taf-11(RNAi) embryos, END-1::GFP initially appeared at normal levels in E4 cells, then was present in E8 cells (Figure 6). These reporter experiments confirm the general importance of taf-5, and suggest that taf-10 and taf-11 are required for a significant fraction of embryonic transcription, but not for expression of many metazoan-specific genes.
Fig. 6. taf-5, taf-10 and taf-11 are essential for gastrulation, but vary in importance for END-1::GFP expression. END-1::GFP expression was examined in RNAi embryos, as indicated to the right of each row. Differential interference (DIC) and fluorescent (FL) images of an E4 and an E8 stage embryo from each set are shown. In a representative experiment, END-1::GFP was not expressed in any ama-1(RNAi) or taf-5(RNAi) embryos (n >100), but at the E4 and E8 stages was expressed at normal levels in most taf-10(RNAi) (90%; n = 71) and taf-11(RNAi) (80%; n = 82) embryos. In parallel experiments, END-1::GFP was expressed in a similar proportion of taf-10(RNAi); taf-11(RNAi) embryos. Within these last RNAi embryo sets, only a small proportion (<5%) expressed END-1::GFP in E2 cells (not shown). Lineage analysis (n >5) revealed that all END-1::GFP-positive cells derived from the E cell. Asterisks mark the E4 cells in the ama-1(RNAi) and taf-5(RNAi) embryos. In each of these RNAi embryo sets, E2 descendants were mislocalized to the posterior edge of the embryo because their abnormally short cell cycle resulted in defective gastrulation.
Discussion
We have investigated how three TAFIIs contribute to gene expression in the developing early C.elegans embryo. We have found that TAF-10 and TAF-11 are required for a significant but not complete fraction of Pol II transcription, indicating that they have broad but modular functions. This conclusion is consistent with models suggested by some studies of the TAF-10 ortholog yTAFII17, which appears to be as broadly required for transcription as any TAFII studied in S.cerevisiae (Table III) (Apone et al., 1998; Michel et al., 1998; Moqtaderi et al., 1998). Our findings extend these yeast models, however, by indicating that many developmental and other metazoan-specific genes are regulated independently of TAF-10 and TAF-11. In contrast, TAF-5 appears to be generally essential for early embryonic transcription, suggesting that it has a broader role in transcription than has been demonstrated previously for other TAFIIs.
Table III. Requirements for yeast and C.elegans TAFIIs for transcription in vivo.
|
S.cerevisiae |
C.elegans embryo |
||
|---|---|---|---|
| TAFII | Transcriptional requirement | TAFII | Transcriptional requirement |
| TAFII48 | unknown | TAF-5 | essentially complete |
| TAFII17 | 59% of genesa; broad or essentially completeb | TAF-10, TAF-11 | significant fraction, not including many metazoan-specific genes |
| TAFII25 | 16% of genesa; essentially completeb | ||
Results of the present study are compared with in vivo analyses of S.cerevisiae TAFII17 and TAFII25 (see text).
aPercentages indicate the proportion of yeast genes that required the indicated TAFII for normal expression, as indicated by microarray analysis (Lee et al., 2000).
bIn separate studies, the approximate proportion of transcription that appeared to depend upon yTAFII17 (Michel et al., 1998; Moqtaderi et al., 1998) or yTAFII25 (Sanders et al., 1999).
Similarly restricted requirements for taf-10 and taf-11 in vivo
The reduced but significant levels of Pol II CTD Ser2 and Ser5 phosphorylation in taf-10(RNAi) and taf-11(RNAi) embryos (Figure 4 and data not shown) predicted a substantial but incomplete transcriptional defect. Accordingly, these RNAi embryos expressed multiple reporter transgenes at normal levels (Figure 6; Table II). Various lines of evidence indicate that this restricted transcriptional requirement for taf-10 and taf-11 reflects their biological functions, rather than incomplete RNAi penetrance. These RNAi effects were highly reproducible, appeared with consistent timing after injection, were accompanied by lack of TAF-10 and TAF-11 antibody staining (Figure 2) and were not enhanced by simultaneous interference with taf-10 and taf-11 expression (Figures 4 and 6). Finally, expression of the conserved genes let-858, rps-5 and hsp-16.2 was decreased as severely in these RNAi embryos as in taf-5(RNAi) embryos (Figure 5; Table II). Although our experiments do not eliminate the possibility that trace levels of TAF-10 and TAF-11 remain in these RNAi embryos, they suggest that these TAFIIs are not required for transcription of a significant proportion of C.elegans embryonic genes.
In yeast, taf-10 and taf-11 orthologs have been inactivated conditionally by mutation or expression shut-off (Table III). Our findings are consistent with evidence that yTAFII17 is broadly required for transcription (Apone et al., 1998; Moqtaderi et al., 1998; Lee et al., 2000), but argue against the model that yTAFII17 and yTAFII25 are generally essential (Michel et al., 1998; Sanders et al., 1999). Caenorhabditis elegans TAF-10 and TAF-11 levels are mutually dependent (Figure 2). This finding suggests that the discrepancy between evidence that yTAFII25 is required very broadly (Sanders et al., 1999) or to transcribe 16% of yeast genes (Lee et al., 2000) might involve effects on stability of other TAFIIs. While some yeast heat shock genes are yTAFII17 independent (Apone et al., 1998; Moqtaderi et al., 1998), hsp-16.2 was partially dependent upon taf-10 and taf-11 (Figure 5), making it of interest to investigate the TAFII dependence of other C.elegans heat shock genes.
A significant aspect of our findings is that taf-10 and taf-11 generally did not appear to be required for expression of the metazoan-specific genes that we tested, including multiple developmental genes (Figures 5 and 6; Table II). The single exception was pes-10 (Figure 5), which was partially dependent upon these TAFIIs. In contrast, taf-10 and taf-11 were essential for expression of each gene we analyzed that is conserved between metazoans and fungi (Figure 5). In these TAFII RNAi embryos, proliferation ceased at ∼100 cells (Figure 2), presumably as maternal proteins that are critical for cell division or function became limiting, suggesting that taf-10 and taf-11 are needed to express genes that are required for these functions. We conclude that taf-10- and taf-11-dependent genes are likely to include many conserved genes involved in cell division or viability, but a much smaller proportion of the specialized genes that have evolved in metazoans. Consistent with our results, murine embryonic carcinoma cells from which TAFII30 (TAF-11) was depleted conditionally could be induced by retinoic acid to leave the cell cycle and differentiate into endodermal cells, but otherwise underwent apoptosis (Metzger et al., 1999). Conditional depletion of a chicken TAF-10 ortholog from a cell line also triggered apoptosis, in conjunction with an apparently modest reduction in mRNA transcription (Chen and Manley, 2000).
Our data suggest that TAF-10 and TAF-11 are part of a functional module within TFIID- and TFTC-related complexes that can be bypassed during transcription of many, or possibly most, metazoan-specific genes. We predict that at TAF-10- and TAF-11-independent genes, regulatory mechanisms have evolved that depend upon other PIC components, some of which are unique to metazoans. For example, the metazoan-specific co-activator HAT p300/CBP is targeted by many signal- and tissue-specific activators (Goodman and Smolik, 2000). In sharp contrast to taf-10 and taf-11, the C.elegans p300/CBP ortholog cbp-1 prevents inappropriate cell proliferation, and is required for multiple differentiation pathways (Shi and Mello, 1998). The mediator complex also contains metazoan-specific components (Malik and Roeder, 2000), one of which has been implicated in C.elegans developmental gene expression (Zhang and Emmons, 2000).
TAF-5 has properties of an essential transcriptional regulator
In contrast to taf-10 and taf-11, taf-5 is generally required for early embryonic transcription (Table III). In taf-5(RNAi) embryos, at each stage somatic cells were indistinguishable from the transcriptionally silent germline precursor in their Pol II CTD Ser2 and Ser5 phosphorylation patterns (Figure 4 and data not shown). taf-5 was also comparable with ama-1 in its importance for reporter gene expression (Figures 5 and 6; Table II). These genes included med-1, med-2 and pes-10, which are expressed approximately when embryonic transcription starts (Seydoux and Fire, 1994; Maduro et al., 2001). TAF-5 appears to be more generally required for early embryonic transcription than even TBP because, in C.elegans and various other metazoans, a considerable proportion of embryonic transcription involves the TBP-related protein TLF (Dantonel et al., 2000; Kaltenbach et al., 2000; Veenstra et al., 2000; Muller et al., 2001). Previous studies implicated the Drosophila TAF-5 ortholog in transcription mediated by the activators Twist and Dorsal (Zhou et al., 1998; Pham et al., 1999), and indicated that an hTAFII130 isoform found in B cells (hTAFII105) is required for some transcription driven by the Dorsal-related factor NF-κB (Yamit-Hezi et al., 2000). Our results suggest that a TAF-5 ortholog or isoform may be generally essential for metazoan mRNA transcription. Interactions between hTAFII130 and polyglutamines encoded by CAG repeat expansions have been implicated in neurodegenerative diseases and neuronal apoptosis (Shimohata et al., 2000). It has been proposed that sequestering of hTAFII130 inhibits CREB-driven transcription, but the broader effect predicted by our results could cause apoptosis analogously to lack of taf-10 or taf-11 orthologs (see above).
In taf-5(RNAi) embryos, α-PSer5 staining appeared in somatic nuclei when transcription normally would begin, but was confined primarily to two discrete foci, as is characteristic of the transcriptionally silent germline cells in early C.elegans and Drosophila embryos (Figure 4 and data not shown) (Seydoux and Dunn, 1997). These foci of α-PSer5 staining may be analogous to structures that appear in mammalian cells when Pol II activity is inhibited (Bregman et al., 1995), and could represent recycling or storage particles (Komarnitsky et al., 2000). These foci did not appear in ttb-1(RNAi) embryos (Figure 4), suggesting that TFIIB may be required at an earlier transcription step than TAF-5. Consistent with this idea, during activation of a mammalian gene in vivo, TFIIB and the Pol II holoenzyme (including the TFIIH kinase) were recruited to the promoter before TFIID (Agalioti et al., 2000). Levels of α-PSer2 staining correlated with nucleoplasmic α-PSer5 staining and reporter expression defects in the respective tafII(RNAi) embryos (Figures 4, 5 and 6; Table II), supporting the model that CTD Ser2 phosphorylation levels reflect early embryonic transcription activity (Seydoux and Dunn, 1997; Tenenhaus et al., 1998). CTD Ser2 can be phosphorylated in vitro by the CDK9 kinase of the elongation factor pTEFb (Zhou et al., 2000), but it has not been determined which kinase is primarily responsible for CTD Ser2 phosphorylation in vivo. Our findings predict that CTD Ser2 phosphorylation is likely to be broadly important for mRNA transcription.
TAF-5 orthologs have been identified in both TFIID and TFTC, each of which can mediate transcription initiation in vitro (Wieczorek et al., 1998; Brand et al., 1999b). Based upon this biochemical framework, our findings suggest that in the early C.elegans embryo, essentially all Pol II transcription may require at least one of these two complexes. Like hTAFII130, TAF-5 could be essential for the structural integrity or assembly of TFIID and TFTC (Furukawa and Tanese, 2000), even though TAF-10 and TAF-11 remained present in taf-5(RNAi) embryos (Figure 2). Alternatively, TAF-5 orthologs might fulfill a critical function of these complexes. In yeast, transcription of most genes appears to depend on either TFIID or SAGA, or both complexes (Lee et al., 2000). Yeast SAGA lacks the TAF-5 ortholog (yTAFII48; Figure 1A), but contains a related histone fold protein (ADA-1) (Sterner and Berger, 2000), for which we have not identified a C.elegans ortholog. A critical structural or mechanistic role for TAF-5 predicts that most yeast transcription may require either yTAFII48 or ADA-1, each of which pairs with the same histone fold partner (yTAFII61/68) (Gangloff et al., 2000; Reese et al., 2000a). It will be of interest to determine whether interactions with activators are important for TAF-5 functions, because these yeast proteins lack metazoan-specific elements that bind activators (Figure 1A). Elucidating how TAF-5 orthologs function in vivo will provide important insights into how the TFIID- and TFTC-related complexes contribute to transcription.
Materials and methods
Worm strains and maintenance
Caenorhabditis elegans were maintained at 20°C according to standard protocols (Brenner, 1974). N2 was the wild-type strain. We used the following GFP reporter strains: end-1::gfp and elt-5::gfp (J.Rothman, unpublished), med-1::gfp and med-2::gfp (Maduro et al., 2001), pha-4 ::gfp (Horner et al., 1998), sur-5::gfp (Gu et al., 1998), cki-2::gfp (J.Rothman, unpublished), pie-1::gfp (Reese et al., 2000b), pes-10::gfp (G.Seydoux, unpublished), hsp-16.2::gfp (C.Link, unpublished), let-858::gfp ((Kelly et al., 1997) and rps-5::gfp (A.Fire, unpublished). These reporters differed somewhat in expression intensities, but these differences did not correlate with whether they corresponded to metazoan-specific or conserved genes.
Bioinformatics
The C.elegans homologs of TAFIIs or other transcriptional regulators were identified by searching WORMpep or genomic databases (Sanger Centre) with human, Drosophila or yeast sequences. Searches were performed using full-length and partial sequences, including predicted conserved domains. A C.elegans gene was considered orthologous to a human, Drosophila or yeast gene only if it re-identified that gene as its closest relative in a search of GenBank redundant databases. Alignments were produced by Megalign (DNAStar). taf-5, taf-10 and taf-11 open reading frames are R119.6, T12D8.7 and K03B4.3, respectively.
Immunostaining and fluorescence analysis
Rabbit antisera were raised against N-terminal peptides of TAF-5 (CKIAGERSTPGVSTPEPAPPQ), TAF-10 (CDTGEKDTETTASDTD GHSKE) and TAF-11 (CMNDPEQYEPSSSTESVL) (Cocalico), then affinity purified (Pierce). Staining by each antibody was competed by autologous but not heterologous peptides (not shown). Other antibodies used included P-CTD (α-PSer5) (Schroeder et al., 2000), H5 (α-PSer2) (Babco), 8WG16 (α-UnP CTD) (Babco) and POL 3/3 (Bellier et al., 1997). For staining, hermaphrodites were cut on polylysine-treated slides. α-PSer5, α-PSer-2 and anti-TAF-11 staining was performed as in Seydoux and Dunn (1997). Anti-TAF-5 or 3/3 staining was performed by fixation in paraformaldehyde then methanol, with incubations and washes in PBT [1× phosphate-buffered saline (PBS), 1% Triton X-100, 1% bovine serum albumin (BSA)]. Anti-TAF-10 was incubated in 100 mM Tris pH 7.5, 150 mM NaCl, 5% BSA after fixation in paraformaldehyde, then dimethylformamide. Secondary antibodies used were fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit and anti-mouse IgM, and Cy3-conjugated goat anti-mouse IgG (Jackson). To analyze GFP expression, embryos were transferred to 2% agarose pads. For heat shock, hsp16.2::gfp embryos were heated to 37°C for 1 min in 10 µl of PBS. Fluorescence was examined after a 20 min recovery. Embryos without GFP expression were re-evaluated 1–2 h later. Images were captured using a Zeiss AxioSKOP2 microscope and AxioCam digital camera, and GFP or antibody staining intensities were compared over a range of exposure times. Pixel intensities were standardized using Adobe Photoshop 5.0.
RNAi analysis
cDNAs corresponding to taf-5 (yk326f12), taf-10 (yk163f12), taf-11 (yk331g8), ama-1 (yk84f7) and ttb-1 (yk117e3) were obtained from Yuji Kohara (NIG, Japan). Each covered >90% of the predicted coding region. In vitro synthesized double-stranded (ds) RNA (Ribomax; Promega) was injected at 0.6–1.0 µg/µl into young adults (2–8 fertilized embryos). Uniform populations of terminally arrested embryos appeared 18–22 h later, and evidence of maternal gene expression defects (rounded embryos, equal cell division planes) did not appear until 48 h. For GFP analysis or immunostaining, embryos were collected from dissected hermaphrodites 24 h after injection. Embryos were generally obtained from worm pools, but for END-1::GFP, progeny of individual worms were scored. Because most analyses were performed before terminal arrest, RNAi effectiveness was confirmed by monitoring sibling embryos that were allowed to develop. Simultaneous taf-10 and taf-11 RNAi was performed with a 1:1 mixture of dsRNAs. In parallel, a 1:1 dilution of each individual dsRNA with either TE or an unrelated dsRNA (glp-1) resulted in appropriate terminal arrest, END-1::GFP expression and CTD epitope staining levels (not shown).
Acknowledgments
Acknowledgements
We acknowledge Morris Maduro, Geraldine Seydoux, Susan Mango, Chris Link, Min Han and Andy Fire for generously providing strains, Yuji Kohara for cDNA clones, and Olivier Bensaude and David Bentley for the POL 3/3 and P-CTD antibodies, respectively. For helpful discussions or reading the manuscript we are indebted to Rosa Navarro, other Blackwell and Shi lab members, Grace Gill, Geraldine Seydoux, Susan Mango, Laszlo Tora, Linda Kaltenbach, Steve Buratowski, Matt Wallenfang, Wanyuan Ao, Craig Hunter, Steve Hahn and Bruce Bowerman. This work was supported in part by grants from the NIH to A.K.W. (F32 DK0946), J.H.R. (HD37487) and Y.S. (R01 GM58012), and from the March of Dimes to J.H.R. (FY2000-659).
References
- Agalioti T., Lomvardas,S., Parekh,B., Yie,J., Maniatis,T. and Thanos,D. (2000) Ordered recruitment of chromatin modifying and general transcription factors to the IFN-β promoter. Cell, 103, 667–678. [DOI] [PubMed] [Google Scholar]
- Albright S.R. and Tjian,R. (2000) TAFs revisited: more data reveal new twists and confirm old ideas. Gene, 242, 1–13. [DOI] [PubMed] [Google Scholar]
- Andel F., Ladurner,A.G., Inouye,C., Tjian,R. and Nogales,E. (1999) Three-dimensional structure of the human TFIID–IIA–IIB complex. Science, 286, 2153–2156. [DOI] [PubMed] [Google Scholar]
- Aoyagi N. and Wassarman,D.A. (2000) Genes encoding Drosophila melanogaster RNA polymerase II general transcription factors: diversity in TFIIA and TFIID components contributes to gene-specific transcriptional regulation. J. Cell Biol., 150, F45–F50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apone L.M., Virbasius,C.A., Holstege,F.C., Wang,J., Young,R.A. and Green,M.R. (1998) Broad, but not universal, transcriptional requirement for yTAFII17, a histone H3-like TAFII present in TFIID and SAGA. Mol. Cell, 2, 653–661. [DOI] [PubMed] [Google Scholar]
- Bellier S., Dubois,M.F., Nishida,E., Almouzni,G. and Bensaude,O. (1997) Phosphorylation of the RNA polymerase II largest subunit during Xenopus laevis oocyte maturation. Mol. Cell. Biol., 17, 1434–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand M., Leurent,C., Mallouh,V., Tora,L. and Schultz,P. (1999a) Three-dimensional structures of the TAFII-containing complexes TFIID and TFTC. Science, 286, 2151–2153. [DOI] [PubMed] [Google Scholar]
- Brand M., Yamamoto,K., Staub,A. and Tora,L. (1999b) Identification of TATA-binding protein-free TAFII-containing complex subunits suggests a role in nucleosome acetylation and signal transduction. J. Biol. Chem., 274, 18285–18289. [DOI] [PubMed] [Google Scholar]
- Bregman D.B.X., Du,L., van der Zee,S. and Warren,S.L. (1995) Transcription-dependent redistribution of the large subunit of RNA polymerase II to discrete nuclear domains. J. Cell Biol., 129, 287–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. (1974) The genetics of Caenorhabditis elegans. Genetics, 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burley S.K. and Roeder,R.G. (1996) Biochemistry and structural biology of transcription factor IID (TFIID). Annu. Rev. Biochem., 65, 769–799. [DOI] [PubMed] [Google Scholar]
- Chen Z. and Manley,J.L. (2000) Robust mRNA transcription in chicken DT40 cells depleted of TAF(II)31 suggests both functional degeneracy and evolutionary divergence. Mol. Cell. Biol., 20, 5064–5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chervitz S.A. et al. (1998) Comparison of the complete protein sets of worm and yeast: orthology and divergence. Science, 282, 2022–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantonel J.C., Quintin,S., Lakatos,L., Labouesse,M. and Tora,L. (2000) TBP-like factor is required for embryonic RNA polymerase II transcription in C.elegans. Mol. Cell, 6, 715–722. [DOI] [PubMed] [Google Scholar]
- Fire A., Xu,S., Montgomery,M.K., Kostas,S.A., Driver,S.E. and Mello,C.C. (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature, 391, 806–811. [DOI] [PubMed] [Google Scholar]
- Furukawa T. and Tanese,N. (2000) Assembly of partial TFIID complexes in mammalian cells reveals distinct activities associated with individual TATA box-binding protein-associated factors. J. Biol. Chem., 275, 29847–29856. [DOI] [PubMed] [Google Scholar]
- Gangloff Y.G., Werten,S., Romier,C., Carre,L., Poch,O., Moras,D. and Davidson,I. (2000) The human TFIID components TAF(II)135 and TAF(II)20 and the yeast SAGA components ADA1 and TAF(II)68 heterodimerize to form histone-like pairs. Mol. Cell. Biol., 20, 340–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff Y., Romier,C., Thuault,S., Werten,S. and Davidson,I. (2001a) The histone fold is a key structural motif of transcription factor TFIID. Trends Biochem. Sci., 26, 250–257. [DOI] [PubMed] [Google Scholar]
- Gangloff Y.G., Sanders,S.L., Romier,C., Kirschner,D., Weil,P.A., Tora,L. and Davidson,I. (2001b) Histone folds mediate selective heterodimerization of yeast TAF(II)25 with TFIID components yTAF(II)47 and yTAF(II)65 and with SAGA component ySPT7. Mol. Cell. Biol., 21, 1841–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman R.H. and Smolik,S. (2000) CBP/p300 in cell growth, transformation and development. Genes Dev., 14, 1553–1577. [PubMed] [Google Scholar]
- Green M.R. (2000) TBP-associated factors (TAFIIs): multiple, selective transcriptional mediators in common complexes. Trends Biochem. Sci., 25, 59–63. [DOI] [PubMed] [Google Scholar]
- Gu T., Orita,S. and Han,M. (1998) Caenorhabditis elegans SUR-5, a novel but conserved protein, negatively regulates LET-60 Ras activity during vulval induction. Mol. Cell. Biol., 18, 4556–4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampsey M. (1998) Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol. Mol. Biol. Rev., 62, 465–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose Y. and Manley,J.L. (2000) RNA polymerase II and the integration of nuclear events. Genes Dev., 14, 1415–1429. [PubMed] [Google Scholar]
- Holstege F.C., Jennings,E.G., Wyrick,J.J., Lee,T.I., Hengartner,C.J., Green,M.R., Golub,T.R., Lander,E.S. and Young,R.A. (1998) Dissecting the regulatory circuitry of a eukaryotic genome. Cell, 95, 717–728. [DOI] [PubMed] [Google Scholar]
- Hong Y., Roy,R. and Ambros,V. (1998) Developmental regulation of a cyclin-dependent kinase inhibitor controls postembryonic cell cycle progression in Caenorhabditis elegans. Development, 125, 3585–3597. [DOI] [PubMed] [Google Scholar]
- Horner M.A., Quintin,S., Domeier,M.E., Kimble,J., Labouesse,M. and Mango,S.E. (1998) pha-4, an HNF-3 homolog, specifies pharyngeal organ identity in Caenorhabditis elegans. Genes Dev., 12, 1947–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalb J.M., Lau,K.K., Goszczynski,B., Fukushige,T., Moons,D., Okkema, P.G. and McGhee,J.D. (1998) pha-4 is Ce-fkh-1, a fork head/HNF-3α, β, γ homolog that functions in organogenesis of the C.elegans pharynx. Development, 125, 2171–2180. [DOI] [PubMed] [Google Scholar]
- Kaltenbach L., Horner,M.A., Rothman,J.H. and Mango,S.E. (2000) The TBP-like factor CeTLF is required to activate RNA polymerase II transcription during C.elegans embryogenesis. Mol. Cell, 6, 705–713. [DOI] [PubMed] [Google Scholar]
- Kelly W.G., Xu,S., Montgomery,M.K. and Fire,A. (1997) Distinct requirements for somatic and germline expression of a generally expressed Caenorhabditis elegans gene. Genetics, 146, 227–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarnitsky P., Cho,E.J. and Buratowski,S. (2000) Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev., 14, 2452–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuras L., Kosa,P., Mencia,M. and Struhl,K. (2000) TAF-containing and TAF-independent forms of transcriptionally active TBP in vivo. Science, 288, 1244–1248. [DOI] [PubMed] [Google Scholar]
- Lee T.I., Causton,H.C., Holstege,F.C., Shen,W.C., Hannett,N., Jennings, E.G., Winston,F., Green,M.R. and Young,R.A. (2000) Redundant roles for the TFIID and SAGA complexes in global transcription. Nature, 405, 701–704. [DOI] [PubMed] [Google Scholar]
- Lemon B. and Tjian,R. (2000) Orchestrated response: a symphony of transcription factors for gene control. Genes Dev., 14, 2551–2569. [DOI] [PubMed] [Google Scholar]
- Li X.Y., Bhaumik,S.R. and Green,M.R. (2000) Distinct classes of yeast promoters revealed by differential TAF recruitment. Science, 288, 1242–1244. [DOI] [PubMed] [Google Scholar]
- Maduro M.F., Meneghini,M.D., Bowerman,B., Broitman-Maduro,G. and Rothman,J.H. (2001) Restriction of mesendoderm to a single blastomere by the combined action of SKN-1 and a GSK-3β homolog is mediated by MED-1 and -2 in C.elegans. Mol. Cell, 7, 475–485. [DOI] [PubMed] [Google Scholar]
- Malik S. and Roeder,R.G. (2000) Transcriptional regulation through Mediator-like coactivators in yeast and metazoan cells. Trends Biochem. Sci., 25, 277–283. [DOI] [PubMed] [Google Scholar]
- Martinez E., Kundu,T.K., Fu,J. and Roeder,R.G. (1998) A human SPT3–TAFII31–GCN5–l acetylase complex distinct from transcription factor IID. J. Biol. Chem., 273, 23781–23785. [DOI] [PubMed] [Google Scholar]
- Matangkasombut O., Buratowski,R.M., Swilling,N.W. and Buratowski, S. (2000) Bromodomain factor 1 corresponds to a missing piece of yeast TFIID. Genes Dev., 14, 951–962. [PMC free article] [PubMed] [Google Scholar]
- Metzger D., Scheer,E., Soldatov,A. and Tora,L. (1999) Mammalian TAF(II)30 is required for cell cycle progression and specific cellular differentiation programmes. EMBO J., 18, 4823–4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel B., Komarnitsky,P. and Buratowski,S. (1998) Histone-like TAFs are essential for transcription in vivo. Mol. Cell, 2, 663–673. [DOI] [PubMed] [Google Scholar]
- Moqtaderi Z., Keaveney,M. and Struhl,K. (1998) The histone H3-like TAF is broadly required for transcription in yeast. Mol. Cell, 2, 675–682. [DOI] [PubMed] [Google Scholar]
- Muller F., Lakatos,L., Dantonel,J., Strahle,U. and Tora,L. (2001) TBP is not universally required for zygotic RNA polymerase II transcription in zebrafish. Curr. Biol., 11, 282–287. [DOI] [PubMed] [Google Scholar]
- Newman-Smith E.D. and Rothman,J.H. (1998) The maternal-to-zygotic transition in embryonic patterning of Caenorhabditis elegans. Curr. Opin. Genet. Dev., 8, 472–480. [DOI] [PubMed] [Google Scholar]
- Ogryzko V.V., Kotani,T., Zhang,X., Schlitz,R.L., Howard,T., Yang,X.J., Howard,B.H., Qin,J. and Nakatani,Y. (1998) Histone-like TAFs within the PCAF histone acetylase complex. Cell, 94, 35–44. [DOI] [PubMed] [Google Scholar]
- Patturajan M., Schulte,R.J., Sefton,B.M., Berezney,R., Vincent,M., Bensaude,O., Warren,S.L. and Corden,J.L. (1998) Growth-related changes in phosphorylation of yeast RNA polymerase II. J. Biol. Chem., 273, 4689–4694. [DOI] [PubMed] [Google Scholar]
- Pham A.D. and Sauer,F. (2000) Ubiquitin-activating/conjugating activity of TAFII250, a mediator of activation of gene expression in Drosophila. Science, 289, 2357–2360. [DOI] [PubMed] [Google Scholar]
- Pham A.D., Muller,S. and Sauer,F. (1999) Mesoderm-determining transcription in Drosophila is alleviated by mutations in TAF(II)60 and TAF(II)110. Mech. Dev., 84, 3–16. [DOI] [PubMed] [Google Scholar]
- Powell-Coffman J.A., Knight,J. and Wood,W.B. (1996) Onset of C.elegans gastrulation is blocked by inhibition of embryonic transcription with an RNA polymerase antisense RNA. Dev. Biol., 178, 472–483. [DOI] [PubMed] [Google Scholar]
- Reese J.C., Zhang,Z. and Kurpad,H. (2000a) Identification of a yeast transcription factor IID subunit, TSG2/TAF48. J. Biol. Chem., 275, 17391–17398. [DOI] [PubMed] [Google Scholar]
- Reese K.J., Dunn,M.A., Waddle,J.A. and Seydoux,G. (2000b) Asymmetric segregation of PIE-1 in C.elegans is mediated by two complementary mechanisms that act through separate PIE-1 protein domains. Mol. Cell, 6, 445–455. [DOI] [PubMed] [Google Scholar]
- Rojo-Niersbach E., Furukawa,T. and Tanese,N. (1999) Genetic dissection of hTAF(II)130 defines a hydrophobic surface required for interaction with glutamine-rich activators. J. Biol. Chem., 274, 33778–33784. [DOI] [PubMed] [Google Scholar]
- Rubin G.M. (2001) The draft sequences. Comparing species. Nature, 409, 820–821. [DOI] [PubMed] [Google Scholar]
- Rubin G.M. et al. (2000) Comparative genomics of the eukaryotes. Science, 287, 2204–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saluja D., Vassallo,M.F. and Tanese,N. (1998) Distinct subdomains of human TAFII130 are required for interactions with glutamine-rich transcriptional activators. Mol. Cell. Biol., 18, 5734–5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders S.L., Klebanow,E.R. and Weil,P.A. (1999) TAF25p, a non-histone-like subunit of TFIID and SAGA complexes, is essential for total mRNA gene transcription in vivo. J. Biol. Chem., 274, 18847–18850. [DOI] [PubMed] [Google Scholar]
- Sanders S.L. and Weil,P.A. (2000) Identification of two novel TAF subunits of the yeast Saccharomyces cerevisiae TFIID complex. J. Biol. Chem., 275, 13895–13900. [DOI] [PubMed] [Google Scholar]
- Schroeder S.C., Schwer,B., Shuman,S. and Bentley,D. (2000) Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev., 14, 2435–2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydoux G. and Dunn,M.A. (1997) Transcriptionally repressed germ cells lack a subpopulation of phosphorylated RNA polymerase II in early embryos of Caenorhabditis elegans and Drosophila melanogaster. Development, 124, 2191–2201. [DOI] [PubMed] [Google Scholar]
- Seydoux G. and Fire,A. (1994) Soma–germline asymmetry in the distributions of embryonic RNAs in Caenorhabditis elegans. Development, 120, 2823–2834. [DOI] [PubMed] [Google Scholar]
- Shi Y. and Mello,C. (1998) A CBP/p300 homolog specifies multiple differentiation pathways in Caenorhabditis elegans. Genes Dev., 12, 943–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimohata T. et al. (2000) Expanded polyglutamine stretches interact with TAFII130, interfering with CREB-dependent transcription. Nature Genet., 26, 29–36. [DOI] [PubMed] [Google Scholar]
- Sterner D.E. and Berger,S.L. (2000) Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev., 64, 435–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenenhaus C., Schubert,C. and Seydoux,G. (1998) Genetic requirements for inhibition of gene expression and PIE-1 localization in the embryonic germ lineage of Caenorhabditis elegans. Dev. Biol., 200, 212–224. [DOI] [PubMed] [Google Scholar]
- Veenstra G.J., Weeks,D.L. and Wolffe,A.P. (2000) Distinct roles for TBP and TBP-like factor in early embryonic gene transcription in Xenopus. Science, 290, 2312–2315. [DOI] [PubMed] [Google Scholar]
- Wassarman D.A., Aoyagi,N., Pile,L.A. and Schlag,E.M. (2000) TAF250 is required for multiple developmental events in Drosophila. Proc. Natl Acad. Sci. USA, 97, 1154–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieczorek E., Brand,M., Jacq,X. and Tora,L. (1998) Function of TAF(II)-containing complex without TBP in transcription by RNA polymerase II. Nature, 393, 187–191. [DOI] [PubMed] [Google Scholar]
- Yamit-Hezi A., Nir,S., Wolstein,O. and Dikstein,R. (2000) Interaction of TAFII105 with selected p65/RelA dimers is associated with activation of a subset of NF-κB genes. J. Biol. Chem., 275, 18180–18187. [DOI] [PubMed] [Google Scholar]
- Zhang H. and Emmons,S.W. (2000) A C.elegans mediator protein confers regulatory selectivity on lineage-specific expression of a transcription factor gene. Genes Dev., 14, 2161–2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Zwicker,J., Szymanski,P., Levine,M. and Tjian,R. (1998) TAFII mutations disrupt Dorsal activation in the Drosophila embryo. Proc. Natl Acad. Sci. USA, 95, 13483–13488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M., Halanski,M.A., Radonovich,M.F., Kashanchi,F., Peng,J., Price,D.H. and Brady,J.N. (2000) Tat modifies the activity of CDK9 to phosphorylate serine 5 of the RNA polymerase II carboxyl-terminal domain during human immunodeficiency virus type 1 transcription. Mol. Cell. Biol., 20, 5077–5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Fukushige,T., McGhee,J.D. and Rothman,J.H. (1998) Reprogramming of early embryonic blastomeres into endodermal progenitors by a Caenorhabditis elegans GATA factor. Genes Dev., 12, 3809–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]