Abstract
The heavy chain of dynein forms a globular motor domain that tightly couples the ATP-cleavage region and the microtubule-binding site to transform chemical energy into motion along the cytoskeleton. Here we show that, in the fungus Ustilago maydis, two genes, dyn1 and dyn2, encode the dynein heavy chain. The putative ATPase region is provided by dyn1, while dyn2 includes the predicted microtubule-binding site. Both genes are located on different chromosomes, are transcribed into independent mRNAs and are translated into separate polypeptides. Both Dyn1 and Dyn2 co-immunoprecipitated and co-localized within growing cells, and Dyn1–Dyn2 fusion proteins partially rescued mutant phenotypes, suggesting that both polypeptides interact to form a complex. In cell extracts the Dyn1–Dyn2 complex dissociated, and microtubule affinity purification indicated that Dyn1 or associated polypeptides bind microtubules independently of Dyn2. Both Dyn1 and Dyn2 were essential for cell survival, and conditional mutants revealed a common role in nuclear migration, cell morphogenesis and microtubule organization, indicating that the Dyn1–Dyn2 complex serves multiple cellular functions.
Keywords: AAA proteins/microtubules/nuclear migration/structure/Ustilago maydis
Introduction
Cytoplasmic dynein is a large multi-subunit motor complex that is involved in numerous essential processes within eukaryotic cells, including organelle transport, organization of the cytoskeleton and mitosis (summarized in Holzbaur and Vallee, 1994; Sharp et al., 2000). The central component of the dynein motor complex is the dynein heavy chain (DHC), a protein of 470–540 kDa, which utilizes ATP to move its cargo along microtubules (MTs). The N-terminal portion of this molecule contains a region for dimerization of two heavy chains and interacts with associated polypeptides (Habura et al., 1999; Tynan et al., 2000), while the C-terminal part of the DHC forms a globular motor domain of 380 kDa (Samso et al., 1998). The motor domain carries four P-loops (P1–4, see Figure 1; Gibbons et al., 1991; Ogawa, 1991), which bind several ATP molecules, although the ATPase activity is restricted to P1 (Gibbons et al., 1987). It is thought that during the ATPase cycle a conformational change of the motor domain is induced, which is transmitted to an exposed MT-binding region (Gee et al., 1997; Koonce, 1997), resulting in mechanical work along MTs.
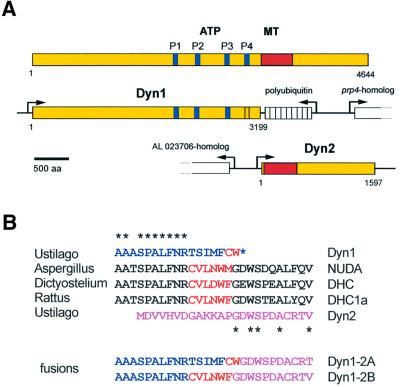
Fig. 1. Domain structure and organization of cytoplasmic dynein from U.maydis. (A) DHC from rat contains four P-loops (P1–4) that are also found in the predicted sequence of Dyn1, although the fourth P-loop differs from the consensus. Dyn1 lacks the putative MT-binding site (shown in red), which is encoded by a second gene, dyn2. (B) The transition from the C-terminus of Dyn1 (blue) to the N-terminus of Dyn2 (purple) lies within a highly conserved sequence region. Together, Dyn1 and Dyn2 contain most of the conserved amino acids (asterisks). However, typical cysteine and tryptophan residues are missing, or are misplaced at the C-terminus of Dyn1 (marked in red). The two fusion proteins generated in this study are either a direct fusion of Dyn1 and Dyn2, which eliminates the N-terminal 13 amino acids of Dyn2 (Dyn1-2A), or contain the missing amino acid stretch typical for cytoplasmic DHCs (Dyn1-2B).
Recently, ultrastructural data have shown that the dynein head consists of a ring of globular domains (Samso et al., 1998), which is likely to be assembled through six AAA modules within the motor domain (Neuwald et al., 1999). Therefore, dynein is considered to be a member of the AAA family of ATPases (King, 2000; Vale, 2000), among which are NSF, proteases and proteins involved in DNA replication and recombination (Patel and Latterich, 1998). The detailed mechanism by which this motor moves along MTs is not known. However, evidence exists that the DHC undergoes conformational changes during its duty cycle (Burgess, 1995), and it has been suggested that this is due to a combined action of the tightly coupled AAA modules (Vale, 2000) finally resulting in directed motion along MTs.
Here we set out to investigate cytoplasmic dynein from the pathogenic heterobasidiomycete Ustilago maydis, the causative agent of corn smut. This dimorphic fungus is amenable to molecular studies (Banuett, 1995) and was used to investigate the role of the MT cytoskeleton in polar growth and pathogenic development (Wedlich-Söldner et al., 2000; Steinberg et al., 2001). In order to study the role of dynein in these processes we performed a PCR approach to identify the heavy chain. Much to our surprise we found that the DHC is split within the fourth AAA module in such a way that the expected ATP-hydrolysis site is most likely provided by one gene, dyn1, while the putative MT-binding region is encoded by another gene, dyn2.
Results
The heavy chain of cytoplasmic dynein of U.maydis is encoded by two genes
As a result of our PCR approach we identified two genes, dyn1 and dyn2, which together encode the DHC from U.maydis. Compared with the DHC from rat, dyn1 provides the N-terminal 3199 amino acids that contain the assumed ATPase region, while dyn2 encodes for the C-terminal 1597 amino acids, including the MT-binding region (Figure 1A). The predicted amino acid sequence of Dyn1 shares 53.5% identity with the N-terminal part of NUDA (DHC from Aspergillus nidulans), and it has 50.9 and 45.3% identity to the DHCs from rat and Dictyostelium discoideum, respectively. Dyn1 contains the highly conserved N-terminal dimerization region (amino acids 667–903; Habura et al., 1999) and three of the four predicted P-loops (GXXXGKS/T) that are typical for DHCs (Gibbons et al., 1991; Ogawa, 1991; Figure 1A). The fourth potential P-loop (GASGSGRT) differs from this motif. Evidence exists to suggest that only the first P-loop is directly involved in ATP cleavage (Gibbons et al., 1987), but the other P-loops might also bind ATP (Mocz and Gibbons, 1996). It remains to be clarified whether the alteration in P4 is of any physiological relevance and whether this is a consequence of the unusual organization of U.maydis dynein.
Dyn2 shows high sequence conservation with the C-terminal part of DHC from rat (47.0% identity) and D.discoideum (43.8% identity), while the similarity to NUDA is slightly reduced (39.9% identity). According to the program COILS (Lupas et al., 1991), dyn2 encodes two putative coiled-coil regions (amino acids 82–175; amino acids 296–391) that include a globular region of ∼120 amino acids. Such regions are characteristic for DHC sequences and were recently defined as the putative MT-binding site of these motors (Gee et al., 1997; Koonce, 1997), suggesting that dyn2 codes for the part of the Ustilago dynein that interacts with MTs.
The DHC from U.maydis is separated within a conserved region of 27 amino acids that is typical for cytoplasmic dyneins (Figure 1B), and which is located within the fourth AAA module (Neuwald et al., 1999). The central part of this peptide is characterized by conserved cysteine and tryptophan residues that are separated by three amino acids (Figure 1B). This peptide is not present in the Ustilago DHC, but a cysteine and a tryptophan residue are found in the C-terminus of Dyn1 (Figure 1B).
The DHC consists of two interacting polypeptides
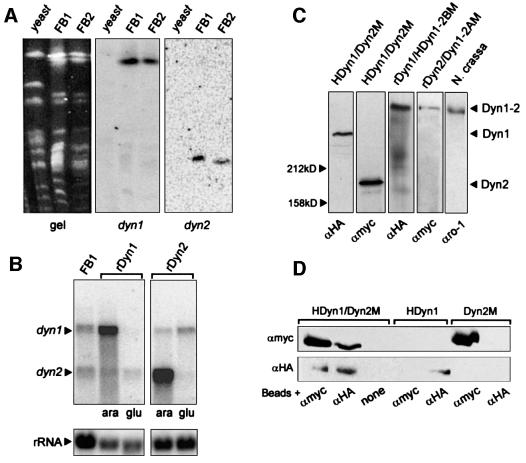
The cleavage of the motor domain within the presumed enzymatically active region raised the possibility that both dyn1 and dyn2 are pseudogenes. However, neither low-stringency Southern analysis of genomic DNA nor extensive PCR approaches revealed evidence for the existence of additional dynein sequences in the genome of U.maydis (not shown). Moreover, both genes are essential for cell survival (see below). Pulse field gel electrophoresis allowed us to map dyn1 and dyn2 to different chromosomes in the two wild-type strains FB1 and FB2 (Figure 2A). Both genes were transcribed into two separate mRNAs (Figure 2B). Finally, we detected two polypeptides of the expected size of 360 kDa (Dyn1) and 180 kDa (Dyn2) in extracts from strain FB2HDyn1/Dyn2M (Figure 2C), which contained dyn1 N-terminally fused to an HA epitope and dyn2 C-terminally fused to a Myc epitope, respectively. Insertion of these tags into wild-type loci of dyn1 and dyn2 did not alter cell shape, growth or nuclear migration (data not shown), indicating that tagged Dyn1 and Dyn2 can substitute for the endogenous dynein polypeptides. Comparative western analysis using extracts of FB1HDyn1 and FB1HDyn2 demonstrated that both polypeptides are expressed at comparable levels (HA-Dyn1:HA-Dyn2, 1:1.3), which is in agreement with the formation of a heterodimer of Dyn1 and Dyn2.
Fig. 2. Southern, northern and western analysis of dynein from U.maydis. (A) Chromosomes were separated on a pulse field gel. After blotting, the same filter was incubated with a dyn1- and a dyn2-specific probe. Both genes hybridized to distinct chromosomes in U.maydis strains FB1 and FB2, but no signal was obtained with genomic DNA of Saccharomyces cerevisiae (yeast). (B) Northern blot of total RNA from wild type (FB1) and the conditional mutants in dyn1 (rDyn1) and dyn2 (rDyn2) was incubated simultaneously with 32P-labelled probes against dyn1 and dyn2. In FB1 two distinct mRNAs corresponding to the two genes are detected. In the conditional mutants at permissive conditions (ara) dyn1 and dyn2 are strongly over expressed. After 5 h in glucose-containing medium (glu) the amount of either dyn1 (rDyn1) or dyn2 (rDyn2) decreased significantly. (C) In western blots using epitope antibodies (αHA, αMyc) and extracts of strain FB2HDyn1/Dyn2M both polypeptides appear at their expected size of ∼360 kDa (HA-Dyn1) and ∼180 kDa (Dyn2-Myc), respectively. Both were significantly smaller than the DHC from N.crassa that was detected in extracts of N.crassa by an antibody against N.crassa dynein (αro1, kindly provided by M.Plamann). Integration of epitope-tagged Dyn1–Dyn2 fusion constructs in FB1rDyn1 (rDyn1/HDyn1-2BM) and FB1rDyn2 (rDyn2/Dyn1-2AM) led to the expression of large fusion proteins. (D) Extracts of the double-tagged strain FB2HDyn1/Dyn2M were incubated with magnetic beads loaded with antibodies directed against the Myc epitope (αMyc), the HA epitope (αHA) or no antibody (none). Each antibody precipitated both HA-Dyn1 and Dyn2-Myc. Neither antibodies nor beads non-specifically precipitated tagged Dyn1 or Dyn2.
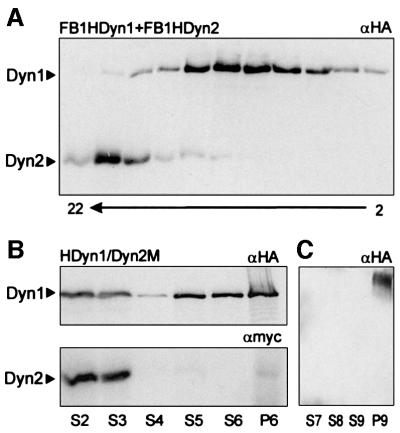
To obtain direct evidence for an interaction of Dyn1 and Dyn2 we performed co-immunoprecipitation experiments using extracts from strain FB2HDyn1/Dyn2M. Antibodies against the Myc epitope at the C-terminus of Dyn2 led to the precipitation of HA-Dyn1, but did not precipitate HA-Dyn1 from extracts of a strain expressing HA-Dyn1 only (FB1HDyn1; Figure 2D). In addition, antibodies against the HA tag fused to the N-terminus of Dyn1 isolated Dyn2-Myc from FB2HDyn1/Dyn2M, but did not precipitate Dyn2-Myc from extracts of FB2Dyn2M in the absence of HA-Dyn1 (Figure 2D). These experiments provide direct evidence that both Dyn1 and Dyn2 are part of a complex in vivo. We aimed to purify further the Dyn1–Dyn2 complex from extracts of FB2HDyn1/Dyn2M by sucrose density centrifugation. However, the complex dissociated during this procedure and Dyn1 and Dyn2 appeared at 21.6 ± 2.5 and 7.6 ± 1.8S, respectively (n = three experiments, using FB2HDyn1/Dyn2M or FB1HDyn1 and FB1HDyn2; Figure 3A).
Fig. 3. Dyn1 and Dyn2 behaviour in sucrose density gradients and MT affinity purification. (A) Extracts from FB1HDyn1 and FB1HDyn2 were loaded on 10–25% sucrose gradients and run for 13 h at 150 000 g. Fractions of 200 µl were collected and each second fraction was analysed by western analysis using αHA and αMyc antibodies. Note that HA-Dyn1 is completely separated from Dyn2-Myc, suggesting that the heavy chain complex dissociated during preparation. Fraction numbers are given below. (B) MT affinity experiments were performed using extracts of FB2HDyn1/Dyn2M. Both HA-Dyn1 and Dyn2-Myc can be detected in the same high speed supernatant (S2), but only a minor fraction binds to MTs and most protein remains in the following supernatant (S3). Washing the MT pellet released only small amounts of HA-Dyn1, while no Dyn2-Myc was found in S4. Surprisingly, treatment with 10 mM MgATP released Dyn1 (S5, S6), while only very small amounts of Dyn2 were present in the S5, S6 or final MT pellet (P6). (C) S5 and S6 were loaded on a 10–25% sucrose density gradient. After centrifugation the two peak fractions containing HA-Dyn1 were pooled and used for an additional round of MT binding. Again, HA-Dyn1 was able to bind MTs (P9) and no HA-Dyn1 remained in the supernatant (S7). However, HA-Dyn1 could not be released by MgATP (S8, S9).
To determine whether the complex has ATP-dependent MT-binding capacity we performed MT affinity purification experiments using extracts of FB2HDyn1/Dyn2M (Figure 3B). Both HA-Dyn1 and Dyn2-Myc could be detected in high-speed supernatants (S2). Under ATP depletion <2% of Dyn2-Myc was able to bind MTs and only traces were released by MgATP (S5; Figure 3B). By contrast, ∼10% of HA-Dyn1 bound to MTs, and of this up to 50% was released in the presence of MgATP (S5 and S6; Figure 3B). To confirm that Dyn1 binds independently of Dyn2 we pooled the ATP-release fractions S5 and S6, loaded them on sucrose density gradients and repeated the MT affinity purification experiments using the HA-Dyn1-containing fractions. MT sedimented all HA-Dyn1 from the peak fractions, but no release by MgATP could be detected (Figure 3C; unbound HA-Dyn1 in S7; ATP-release, S8, S9; MT pellet, P9). Interestingly, after partial purification of S2 in sucrose density gradients HA-Dyn1 behaved similarly, with more HA-Dyn1 binding to MTs, but no release in the presence of ATP. This suggests that MT binding of HA-Dyn1 is regulated by unknown factors that are lost during density centrifugation.
Dyn1 and Dyn2 co-localize
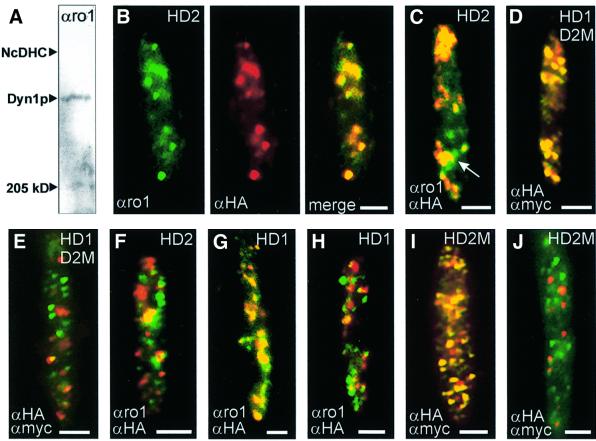
Antibodies raised against the first P-loop region of the DHC from Neurospora crassa (αro1; Kumar et al., 2000; kindly provided by M.Plamann) were specific for Dyn1, as confirmed by western blots (Figure 4A) and co-localization experiments with αHA antibodies in cells of strain FB1HDyn1 (Figure 4G). Therefore, αro1 and αHA antibodies were used to detect Dyn1 and HA-Dyn2 in strain FB2HDyn2, in which HA-Dyn2 was under the control of the native dyn2 promoter. Consistent with the formation of a Dyn1–Dyn2 complex, Dyn1 and Dyn2 co-localized in the cell (Figure 4B). This was most obvious in growing cells, where Dyn1 and Dyn2 accumulated at the growth region (Figure 4C). However, some single αro1 signals were also detected (arrow in Figure 4C). We confirmed the co-localization of Dyn1 and Dyn2 in strain FB2HDyn1/Dyn2M using αHA and αMyc antibodies (Figure 4D). Again, both HA-Dyn1 and Dyn2-Myc were expressed under their endogenous promoters. To our surprise, in one experiment Dyn1 and Dyn2 did not co-localize in 5–7% of cells of FB2HDyn1/Dyn2M (Figure 4E), and the same was detected in FB2HDyn2 using αro1 and αHA (Figure 4F). The percentage of cells showing this phenomenon varied between experiments, ranging from 100% co-localization in one experiment to no co-localization in another. In FB1HDyn1, where HA-Dyn1 is recognized by both αHA and αro1 antibodies (Figure 4G), co-localization was also missing in some cells (Figure 4H). This phenomenon was also seen for FB2HDyn2M when αHA and αMyc antibodies, both recognizing Dyn2, were used (Figure 4I and J). However, further studies are clearly needed to clarify whether the observed separations are of any cellular significance.
Fig. 4. Localization of Dyn1 and Dyn2 in haploid U.maydis cells. (A) Western blot using the αro1 antibody that was raised against the P-loop region of the DHC from N.crassa. The antibody clearly recognized a band at the expected size for Dyn1. However, the signal on blots was weak and could not be used for further western analysis. The position of the DHC from N.crassa (NcDHC) and myosin (250 kDa) is indicated. (B) Co-localization of Dyn1 and HA-Dyn2 in yeast-like cells of strain FB2HDyn2. The αro1 antibody gave a punctuate staining of Dyn1 (green). The same distribution was seen for HA-Dyn2 visualized by αHA antibodies (red). The overlay revealed a clear co-localization of Dyn1 and Dyn2 (yellow). (C) Overlay of signals for Dyn1 and HA-Dyn2, detected by αro1 and αHA, in a growing cell of FB2HDyn2. At the onset of budding, signals for both polypeptides co-localize (yellow) and the complex is concentrated at the growth region of the cell. Occasionally, single spots of Dyn1 were detected (green, arrow). (D and E) Detection of HA-Dyn1 and Dyn2-Myc by specific antibodies directed against both epitope tags in cultures of FB2HDyn1/Dyn2M. In agreement with the results described above, signals for HA-Dyn1 and Dyn2-Myc co-localize (D). However, some unbudded cells showed a clear separation of most αHA (green dots) and αMyc signals (red dots), suggesting that the HA-Dyn1–Dyn2-Myc complex disassembled in these cells (E). (F) Localization of Dyn1 and HA-Dyn2 by αro1 and αHA in FB2HDyn2. Again, some unbudded cells showed a separation of Dyn1 (green dots) and HA-Dyn2 (red dots). (G and H) Detection of HA-Dyn1 by αro1 and αHA antibodies in strain FB1HDyn1. Most signals co-localize (G), however in some unbudded cells almost no correspondence was observed (H). (I and J) Detection of HA-Dyn2-Myc by αHA and αMyc antibodies in FB2HDyn2M. As expected, both antibodies recognize the double-tagged Dyn2 (I; overlay results in yellow). However, not all signals co-localize and in densely grown cultures no correspondence was seen (J). All bars correspond to 2 µm. Antibodies are given in the lower left, strains are indicated in the upper right (HD1, FB1HDyn1; HD2, FB2HDyn2; HD1D2M, FB2HDyn1/Dyn2M; HD2M, FB2HDyn2M).
Ustilago maydis dynein functions in nuclear migration
To analyse the function of dyn1 and dyn2 we attempted to generate deletion mutants by gene replacement. These experiments failed, suggesting that both genes are essential. Therefore, we expressed both genes under the control of the crg promoter, which allowed the controlled down-regulation of both dyn1 and dyn2 by switching from arabinose- to glucose-containing medium (Bottin et al., 1996). The resulting strains FB1rDyn1 and FB1rDyn2 grew on solid media containing arabinose (CM-A), but were unable to form colonies when shifted to glucose (CM-G, not shown). In both mutant strains, overexpression of the dyn1 and dyn2 mRNA was observed under inducing conditions (Figure 2B). The high expression level did not affect cell growth (not shown). Shift of the conditional mutants to CM-G repressed the crg promoter and led to a drastic decrease of the dyn1 and dyn2 message (Figure 2B), which allowed detailed analysis of the mutant phenotype.
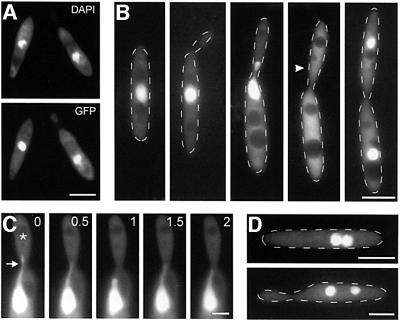
A role for cytoplasmic dynein in migration of fungal nuclei is well established (summarized in Steinberg, 2000). Therefore, we attempted to obtain direct evidence for a role of the DHC complex in nuclear migration. We fused green fluorescent protein (GFP) to the N-terminal nuclear localization signal of the GAL4 DNA-binding domain (C.Aichinger and R.Kahmann, unpublished; see Materials and methods) and expressed the construct in the conditional mutant strain FB1rDyn2, resulting in strain FB1rDyn2/nGFP. The fusion protein co-localized with the nuclear DNA that was stained with DAPI (Figure 5A), allowing in vivo observation of GFP-marked nuclei in the presence and absence of Dyn2. Under permissive conditions, nuclear distribution was normal (Figure 5B). Prior to mitosis the nucleus migrated into the bud, where mitosis occurs. The GFP staining was absent from mitotic nuclei (arrowhead in Figure 5B), which was confirmed by immunostaining of spindle MTs (not shown), suggesting that a fenestrated nuclear envelope (O’Donnell and McLaughlin, 1984) released the GFP fusion protein during mitosis. Finally, both nuclei are positioned within the mother and the daughter cell, respectively. Unexpectedly, nuclei stopped migration during microscopic observation. Only the formation of tubular extensions was seen at permissive conditions (arrow in Figure 5C). These nuclear tubules appeared in all budded cells analysed (n = 12) and in all cases were pulled into the growing bud, where most MT minus ends are supposed to be located (Steinberg et al., 2001). In agreement with a role of minus-end-directed dynein in this motion, tubule formation was not found after shift of the conditional mutant to CM-G (n = 11), and motionless nuclei accumulated in the mother cells under restrictive conditions (Figure 5D).
Fig. 5. Nuclear migration in FB1rDyn2 expressing GFP fused to a nuclear localization signal. (A) Co-localization of nuclear DNA, stained with DAPI, and GFP fused to the GAL4 nuclear localization signal. The fusion protein localized to the nuclei of haploid U.maydis cells. (B) Nuclear distribution during the cell cycle of U.maydis. During polar budding the nucleus remains stationary in the middle of the mother cell, until pre-mitotic migration into the bud occurs. During mitosis the GFP signal disappears while a short spindle is formed (Steinberg et al., 2001; position of spindle is marked by an arrowhead). Finally, the two nuclei show post-mitotic migration and are positioned in the middle of mother and daughter cell, respectively. (C) Motion of tubular extensions at the nucleus. In growing cells, tubular extensions are pulled towards the bud (asterisk). Previous studies have indicated that the minus ends of MTs are located in the neck region (Steinberg et al., 2001), suggesting that a minus-end-directed motor is responsible for this motility. (D) Cells of strain FB1rDyn2/nGFP after 10 h in CM-G. All motion is gone and nuclei accumulate in the mother cell, indicating that dynein participates in nuclear migration. Bars in (A), (B) and (D) correspond to 5 µm, and in (C) to 2 µm.
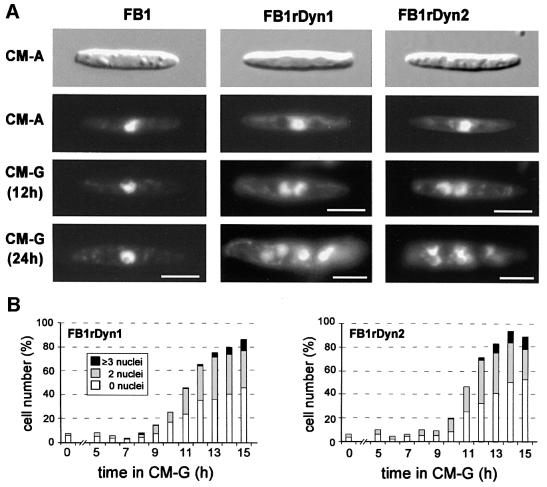
The nuclear migration phenotype was also quantitatively analysed in conditional mutant strains FB1rDyn1 and FB1rDyn2 stained with DAPI (Figure 6). While in CM-A and CM-G >95% of all wild-type FB1 cells contained a single nucleus that was positioned in the cell centre (Figure 6A), nuclei accumulated in FB1rDyn1 as well as FB1rDyn2 cells after a shift to restrictive conditions (Figure 6A). In three experiments (100 cells each) the nuclear phenotype of FB1rDyn1 and FB1rDyn2 was monitored for 15 h in CM-G (Figure 6B). Both mutant strains accumulated an increasing portion of aberrant cells with either no or more than one nucleus. The portion of defective cells rose to >90% after 25 h (not shown). In summary, depletion of Dyn1 and Dyn2 results in a severe nuclear phenotype, suggesting that both genes act together in promoting nuclear migration into the bud of U.maydis.
Fig. 6. Nuclear distribution phenotype of conditional mutants in dyn1 and dyn2. (A) Wild-type strain FB1 as well as conditional mutant strains FB1rDyn1 and FB1rDyn2 show normal cell shape when grown under permissive conditions (CM-A). All strains contain a single nucleus positioned in the cell centre. After shift and growth under restrictive conditions (CM-G) expression of dyn1 and dyn2 is repressed, resulting in accumulations of nuclei in the mother cell due to a nuclear migration defect. Bars correspond to 5 µm. (B) Quantification of the nuclear migration defect in FB1rDyn1 and FB1rDyn2 after growth under restrictive conditions. The number of aberrant cells containing no, two or three and more nuclei began to increase after 8–10 h growth in CM-G. The graph is based on three experiments with 100 cells at each time point.
Depletion of Dyn2 affects the MT cytoskeleton in haploid U.maydis cells
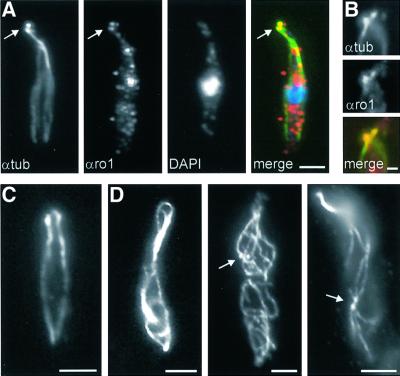
Besides the defect in nuclear migration, mutant strains showed growth defects in CM-G. Cells grew larger and often became spherical (Figure 7A, arrow), while they formed bud-like extensions at random sites (Figure 7A, arrowhead). This phenotype was very similar to that described for conditional α-tubulin mutants (Steinberg et al., 2001). We therefore analysed the MT cytoskeleton in conditional strains FB1rDyn1 and FB1rDyn2. Both mutants behaved almost identically and we have, therefore, restricted our experiments to FB1rDyn2. Staining of wild-type strain FB1 with anti-α-tubulin antibodies revealed 3–5 strands of MTs and pairs of globular tubulin structures (PTS, paired tubulin structures; Figure 8A). These structures are thought to be polar MT organizers that focus the minus ends of MTs towards the growing bud (Steinberg et al., 2001). Correspondingly, double staining of wild-type cells with the αro1 and anti-α-tubulin antibodies revealed in all cases a co-localization of Dyn1 and the PTS (n = 14; Figure 8A and B), indicating that the dynein complex localizes to these structures. FB1rDyn2 grown under permissive conditions showed normal MT organization (Figure 8C), while a shift to restrictive medium led to more and longer MTs (Figure 8D). After a shift to CM-G most cells were filled with MTs. Occasionally, cells were observed that contained pairs of globular tubulin structures randomly distributed within the cell (Figure 8D, arrows), suggesting that in these cases the PTS no longer located to the growing cell pole. These results indicate that the dynein complex participates in MT organization and regulates MT length.
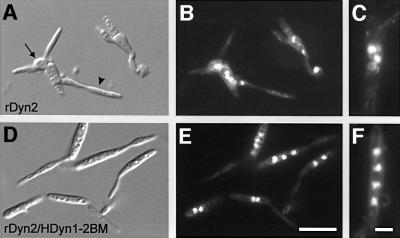
Fig. 7. Partial rescue of the morphological and nuclear phenotype of conditional Dyn2 by Dyn1–Dyn2 fusion proteins. (A) After 24 h in CM-G, cells of strain FB1rDyn2 change their morphology and tend to lose their cell polarity (arrow) while large bud-like extensions are formed (arrowhead). (B) Corresponding DAPI staining of DNA in FB1rDyn2. Note that numerous nuclei accumulate in the deformed mother cell. (C) In FB1rDyn2, nuclei tend to cluster within the cell, suggesting that no nuclear migration occurred. (D) Expression of the fusion protein HDyn1-2BM in strain FB1rDyn2/HDyn1-2BM leads to a partial rescue of morphological phenotype of FB1rDyn2, although cells grew slightly larger. (E and F) Corresponding DAPI staining of DNA revealed that most cells still contain numerous nuclei. However, nuclei are lined up in the presence of HDyn1-2BM and almost no clusters were observed (F). Bars in (A), (B), (D) and (E) correspond to 10 µm, and in (C) and (F) to 3 µm.
Fig. 8. Dyn1 and MTs in cells of wild-type FB1 and conditional mutant strain FB1rDyn2. (A) Triple staining of MTs, Dyn1 and nuclear DNA in FB1. During growth the MT cytoskeleton becomes polarized and a PTS appears at the growth region (green, arrow; Steinberg et al., 2001). Dyn1, detected with αro1 antibodies (red), co-localizes with these spherical structures (yellow). The nucleus, stained with DAPI, is shown in blue. (B) Double staining of MTs and Dyn1. At higher magnification, antibodies against tubulin (αtub, green) and DHC (αro1, red) clearly co-localize (yellow). (C) The MT cytoskeleton in an unbudded cell of FB1rDyn2 under permissive conditions. (D) Alterations of MT organization in FB1rDyn2 after 13 h in CM-G. Cells contain more and longer MTs. Occasionally, pairs of spherical tubulin structures that most likely represent MTOCs lose their polar localization (arrows). Bars in (A), (C) and (D) correspond to 3 µm, and in (B) to 1 µm.
Fusions of Dyn1 and Dyn2 can partially rescue the mutant phenotypes
If the separation of dyn1 and dyn2 is required for dynein function in U.maydis, a complete DHC might not be expected to complement the phenotypes of conditional dyn1 or dyn2 mutants. To investigate this we fused both dynein genes and introduced the constructs ectopically into FB1 and mutant strains FB1rDyn1 and FB1rDyn2, respectively. First, we generated a fusion of dyn1 to dyn2, which conserved Dyn1 completely, but deleted the first 13 amino acids of Dyn2 to maintain spacing within this conserved region (Figure 1B). In addition, a Myc tag was fused to the C-terminus of the fusion construct (Dyn1-2AM; Figure 1B). A second fusion contained the conserved peptide CVLNWF at the transition site, as well as an HA tag and a Myc epitope at the N-terminus and the C-terminus, respectively (HDyn1-2BM; Figure 1B). Western blots of cell extracts of strains FB1rDyn1/HDyn1-2BM and FB1rDyn2/Dyn1-2AM revealed proteins of approximately the same size as the DHC from N.crassa (Figure 2C), which migrated at 22.4 ± 2.5S (n = three experiments). The double-tagged HDyn1-2BM fusion protein was recognized by both αHA and αMyc antibodies, demonstrating that the complete fusion protein was made (not shown). Both fusion proteins were able to enhance growth of FB1rDyn1 and FB1rDyn2 in CM-G, and partially rescued the morphological defect (Figure 7; only the effect of HDyn1-2BM is shown). In addition, instead of being clustered, nuclei were lined up within the cell (Figure 7C and F), although >80–90% of all cells still contained no or more than one nucleus after 25 h in CM-G. However, quantitative western analysis demonstrated that the fusion proteins, although under the control of the dyn1 promoter, reached up to ∼10% of wild-type HA-Dyn1 (not shown), suggesting that the protein is unstable or low in expression and, therefore, might not be able to supplement for Dyn1 or Dyn2.
Discussion
The heavy chain of cytoplasmic dynein consists of two polypeptides
We have shown that the heavy chain of cytoplasmic dynein from U.maydis is encoded by two separate genes that are transcribed into two mRNAs and translated into two polypeptides, each providing an essential part of the motor domain. Co-immunoprecipitation and co-localization experiments, as well as the partial rescue of the Dyn1–Dyn2 fusion proteins, indicate that both polypeptides interact to form a heavy chain complex in vivo. Such a separation into two distinct polypeptides is most surprising as all motors described so far show a tight coupling of the ATP hydrolysis and the cytoskeleton-binding site (Gee et al., 1997; Song and Endow, 1998; Knetsch et al., 1999). Recently, ultrastructural analysis of recombinant motor domains and the identification of AAA sequence motifs within the motor domain of cytoplasmic dynein (Neuwald et al., 1999) suggested that the globular domains of the dynein motor head represent these AAA modules (King, 2000; Vale, 2000). It is characteristic of AAA proteins that they undergo conformational changes by a combined action of their AAA modules. In the case of dynein, the tight coupling of the AAA modules is thought to transmit conformational changes from the ATP site through the large motor to the MT-binding stalk (Vale, 2000). The dynein head most likely evolved by gene duplication, thereby generating the sequence repeats (Gibbons, 1995). It has been argued that AAA modules 5 and 6 might have evolved separately, as they show the most divergence from the other AAA repeats (Asai and Koonce, 2001). In agreement with this model, Dyn2 encodes for these two AAA repeats and therefore the cleaved U.maydis DHC might represent just such an ancient evolutionary step. However, cleavage occurred within the fourth AAA module at the end of box VII, which is, in analogy to other AAA proteins, thought to be the transition between two domains within a single AAA module (Neuwald et al., 1999). Therefore, it is likely that both Dyn1 and Dyn2 provide a domain of the fourth AAA repeat, and this could argue for a more recent splitting of the heavy chain.
Instability of the Dyn1–Dyn2 complex
Since the complex dissociated almost completely during sucrose density centrifugation, the interaction of Dyn1 with Dyn2 appears to be rather weak. Dyn2 has an estimated S20,w of 7.6S and a molecular weight of ∼180 kDa. Compared with d-amino acid oxidase (S20,w, 7.6; mol. wt, 182 kDa; Smith, 1973), this suggests that outside the Dyn1–Dyn2 complex Dyn2 exists as a globular monomer. By contrast, the S20,w of HA-Dyn1 (21.6S) was found to be similar to the expected 20S estimated for purified cytoplasmic dyneins (Paschal et al., 1987; Koonce and McIntosh, 1990), suggesting that Dyn1 dimerizes and binds to accessory subunits. Correspondingly, we determined a higher S value for the fusion of Dyn1 and Dyn2 (22.4S). It remains to be seen whether associated polypeptides are responsible for the slightly higher S values of the Ustilago dynein complex compared with other cytoplasmic dyneins.
MT binding of the Dyn1-containing complex
Recently, the putative MT-binding region of cytoplasmic dyneins was identified (Gee et al., 1997; Koonce, 1997). The region consists of two coiled coils that are, according to structure predictions, most likely to be encoded by Dyn2. However, our MT affinity purification experiments suggest that Dyn2 alone is not able to bind MTs. This is surprising, as recombinant peptides containing the putative MT-binding region of dynein interact specifically with MTs (Gee et al., 1997; Koonce, 1997). Therefore, we speculate that Dyn2 MT-binding activity is under cellular control. Moreover, our experiments indicate that Dyn1 is able to bind MTs. This raises the possibility of a second MT binding site, which might be encoded by Dyn1. However, intensive analysis of the MT-binding capacity of cytoplasmic dyneins revealed no evidence for additional MT-binding regions in the heavy chain (Gee et al., 1997; Koonce, 1997; Koonce and Tikhonenko, 2000). Alternatively, associated polypeptides might mediate the unexpected MT-binding activity of the Dyn1-containing complex.
Both Dyn1 and Dyn2 function in nuclear migration
Dyn1, as well as Dyn2 mutants, displayed a drastic defect in nuclear migration, and such a role of dynein in nuclear migration is well established for filamentous fungi (Plamann et al., 1994; Xiang et al., 1994; Inoue et al., 1998) and S.cerevisiae (Eshel et al., 1993; Li et al., 1993). In general, dynein is not essential in fungi (Steinberg, 2000) and yeast DHC null-mutants grow at normal rates (Eshel et al., 1993; Li et al., 1993), suggesting that additional motors might substitute for the deletion of the DHC (Xiang et al., 1995; Cottingham and Hoyt, 1997). Contrarily, depletion of Dyn1 or Dyn2 in U.maydis was lethal. Compared with budding yeast, where deletion of dynein led to defects in nuclear distribution in only 4–19% of all mutant cells grown at 30°C (Li et al., 1993), depletion of either Dyn1 or Dyn2 in U.maydis resulted in >90% defective cells after extended time in restrictive medium. Therefore, functional dynein is crucial for nuclear motion in U.maydis, and this might account for the essential role of both Dyn1 and Dyn2.
Mitosis of U.maydis occurs within the bud. Therefore, nuclear migration in haploid U.maydis cells can be subdivided into a pre-mitotic and a post-mitotic step (Steinberg et al., 2001). The former process depends on spindle-pole-independent MTs, which are thought to be organized by polar MT organizing centres (PTS; Steinberg et al., 2001). Our in vivo observations of GFP-stained nuclei demonstrate that under permissive conditions forces are exerted on the nuclear envelope. These forces extend the nucleus towards the neck region, where, based on analysis of MT-dynamics in GFP-Tub1-expressing strain, the minus ends of the MTs are localized (Steinberg et al., 2001). In contrast, nuclei remain motionless and accumulate in the mother cell in the absence of Dyn2. Therefore, we conclude that dynein is most likely to be responsible for the pre-mitotic nuclear migration. However, in contrast with S.cerevisiae, where dynein in the cell cortex interacts with astral MTs (Shaw et al., 1997), this motion appears to be independent of astral MTs (Steinberg et al., 2001). The detailed mechanism by which dynein exerts force on the nucleus remains to be elucidated.
Dynein mutant cells show an altered MT cytoskeleton and defective morphology
Cytoplasmic dynein is a minus-end-directed motor, and in agreement with the assumed MT orientation dynein localizes to the PTS at growth regions. Interestingly, the polar localization of the PTS appears to depend on the presence of an intact DHC complex, as the PTS lose their polar localization and mother cells become rounded, while the formation of elongated buds is almost unimpaired. This phenotype is reminiscent of a conditional α-tubulin mutant of U.maydis (Steinberg et al., 2001), indicating that dynein participates in MT organization and, thereby, in polar growth of U.maydis. In addition, depletion of Dyn2 leads to more and longer MTs, which points to a function of dynein in the regulation of MT dynamics, a phenomenon already described for dynein from filamentous fungi (Willins et al., 1995). In summary, depletion of Ustilago dynein could affect MT-dependent growth processes in various ways and this might hold as an explanation for the observed morphological defects of our conditional mutants.
Conclusions
Our experiments suggest that the U.maydis DHC functions as a heterodimer of Dyn1 and Dyn2. This organization is highly unexpected and has so far not been described for any other motor. In addition, both dyn1 and dyn2 are essential genes, which is in contrast to the minor importance of dyneins in other fungi (Steinberg, 2000). Since expression of two separate and important polypeptides that form a heavy chain complex requires costly regulation, this should be a disadvantage in evolutionary terms. At present we can only speculate on the biological reason for this unexpected phenomenon. One possibility for the separation of the DHC might be that it allows novel combinatorial associations with polypeptides as yet unidentified. The unexpected MT-binding capacity and high S value of Dyn1 could reflect such a situation, and it will be a challenge for the future to elucidate whether this possibility is realized.
Materials and methods
Identification and cloning of dyn1 and dyn2
To identify dyn1, primers against the first P-loop (K13: AAGGCTTGG NACNGGAARACNGA and K15: GAATTCRTTRAASTCRTCRAARCA; both kindly provided by Dr E.Kube-Granderath) were used in PCR with genomic DNA from wild-type strains (35 cycles of 94°C, 45 s; 58°C, 45 s; 72°C, 45 s). One microlitre of this reaction was used in a second PCR containing 5% glycerol and 10% dimethylsulfoxide (DMSO) at an annealing temperature of 55°C. The dyn2 gene was identified using primers C7 (CTBTGYACNGARAAYGC) and C10 (ATVARVACDCGDCCDCC), both directed against the MT-binding region, in 30 cycles (94°C, 60 s; 42–58°C, 60 s; 72°C, 60 s). The amplified DNA fragments were used to obtain the complete genomic DNA from a cosmid library (Bölker et al., 1995). Both genes were subcloned according to standard procedures and sequenced on both strands.
Strains and plasmids
All strains had the genetic background of FB1 (a1b1) or FB2 (a2b2; Banuett and Herskowitz, 1989) and are summarized in Table I. To generate strains FB1rDyn1 and FB1rDyn2 a phleomycin resistance cassette (ble; Table I) followed by the 3.6 kb crg promoter (Bottin et al., 1996) was fused to the ATG start codon of dyn1 or dyn2, respectively. This construct was inserted into the respective gene locus by homologous recombination. To generate FB1rDyn2/nGFP the N-terminal nuclear localization signal of the GAL4 DNA-binding domain from the yeast two-hybrid vector pC-ACT.1 (ClonTech) was fused to the eGFP gene, resulting in plasmid pnGFP, and integrated ectopically into FB1rDyn2. FB1HDyn1 carries an insertion of a hygromycin resistance cassette (hph; Table I) at position –1600 of dyn1 and a triple HA tag at the start codon of dyn1. In FB2Dyn2M, dyn2 is fused to a triple Myc tag at the 3′ end followed by the nos terminator and the nourseothricin resistance cassette (nat, Table I). In FB2HDyn1/Dyn2M, both epitope tags were introduced in FB2 by homologous integration. FB1HDyn2 and FB2HDyn2 carry a hygromycin resistance cassette –1700 of dyn2 and a triple HA tag at the start codon of dyn2. Strain FB2HDyn2M contained an additional triple Myc tag at the 3′ end. In the fusion constructs pDyn1-2A and pDyn1-2B, a hygromycin resistance cassette is followed by 1600 bp of dyn1 promoter and the fused genes. A triple Myc tag and a following nos terminator were fused to the 3′ end of fusion genes and, in the case of pDyn1-2B, an additional triple HA tag was inserted at the start codon, resulting in plasmids pFUSA and pFUSB. Strains FB1Dyn1-2AM, FB1rDyn1/HDyn1-2BM, FB1rDyn2/HDyn1-2BM and FB1rDyn2/Dyn1-2AM were created by ectopic integration of pFUSA or pFUSB in FB1, FB1rDyn1 and FB1rDyn2, respectively. Cloning and transformation were carried out according to published protocols (Schulz et al., 1990) and integration was confirmed by Southern blotting. Strains were grown at 28°C in CM medium (Holliday, 1974) supplemented with either 1% arabinose (CM-A) or 1% glucose (CM-G), respectively. For medium shift experiments cells were usually grown overnight to OD600 <1 in CM-A, washed twice in water and transferred to CM-G.
Table I. Strains and plasmids used in this study.
| Name | Genotype |
|---|---|
| FB1a | a1b1 |
| FB2a | a2b2 |
| FB1rDyn1 | a1b1 Δdyn1(p)::[ble, crg(p)] |
| FB1rDyn2 | a1b1 Δdyn2(p)::[ble, crg(p)] |
| FB1rDyn2/nGFP | a1b1 Δdyn2(p)::[ble, crg(p)] pnGFP (EC) |
| FB1HDyn1 | a1b1 Δdyn1::[hph, 3HA-dyn1] |
| FB2Dyn2M | a2b2 Δdyn2::[dyn2-3myc, nat] |
| FB1HDyn2 | a1b1 Δdyn2::[hph, 3HA-dyn2] |
| FB2HDyn2 | a2b2 Δdyn2::[hph, 3HA-dyn2] |
| FB2HDyn2M | a2b2 Δdyn2::[hph, 3HA-dyn2-3myc, nat] |
| FB2HDyn1/Dyn2M | a2b2 Δdyn2::[dyn2-3myc, nat] Δdyn1::[hph, 3HA-dyn1] |
| FB1Dyn1-2AM | a1b1 pFUSA (EC) |
| FB1rDyn2/Dyn1-2AM | a1b1 Δdyn2(p)::[ble, crg(p)] pFUSA (EC) |
| FB1rDyn1/HDyn1-2BM | a1b1 Δdyn1(p)::[ble, crg(p)] pFUSB (EC) |
| FB1rDyn2/HDyn1-2BM | a1b1 Δdyn2(p)::[ble, crg(p)] pFUSB (EC) |
| pnGFP | gal4(s)::egfp, cbx |
| pFUSA | dyn1-dyn2-3myc, hph |
| pFUSB | 3HA-dyn1-CVLNWF-dyn2-3myc, hph |
aBanuett and Herskowitz (1989).
a, b, mating type loci; Δ, deletion; (p), promoter; ::, homologous replacement; -, gene fusion; hph, hygromycin resistance; ble, phleomycin resistance; nat, nourseothricin resistance; cbx, carboxin resistance; gal4(s), nuclear localization signal of the GAL4 DNA-binding domain from pC-ACT1 (Clontech); EC, ectopically integrated.
Pulse field gel electrophoresis
Chromosomes were isolated essentially as described (Kinscherf and Leong, 1988). In brief, ∼1 × 107 cells of strains FB1 and FB2 were incubated in 5 mg/ml novozyme pH 5.8 for wall digestion. Protoplasts were embedded in 1% agarose and incubated in 2% SDS, 1 mg/ml proteinase K and 0.5 M EDTA pH 8. After several washing steps the blocks were used in pulse field gel electrophoresis using the Chef-Drive II system from Bio-Rad following the manufacturer’s instructions. Gels were incubated in 0.4 M NaOH/1.5 M NaCl for 15 min and blotted with 0.4 M NaOH for 48 h. Hybridization with probes of dyn1 and dyn2 was carried out according to standard protocols.
Northern, Southern and western analysis
RNA and genomic DNA was isolated as described (Schulz et al., 1990). Northern and Southern blotting were carried out according to standard protocols. Protein extracts were obtained by disruption of N2-frozen Ustilago cells grown to OD600 <0.6, in a mixer mill MM 200 (RETSCH), and thawing in PME [100 mM PIPES pH 6.9, 5 mM MgSO4, 1 mM EDTA, 5 mM EGTA supplemented with 1 mM Pefabloc (Roth) and Complete protease inhibitor (Roche)]. Proteins were separated in 5% polyacrylamide gels and transferred to nitrocellulose membranes for 1 h at 400 mA in a wet blot chamber. Antibody detection followed standard procedures. Quantitative western analysis was performed using digital images and ImageProPlus (MediaCybernetics).
Sucrose gradients and immunoprecipitation
Protein extracts of FB1HDyn1, FB1HDyn2, FB2HDyn1/Dyn2M, FB1Dyn1-2AM, FB1rDyn1/HDyn1-2BM and FB1rDyn2/HDyn1-2BM were prepared as described above and loaded on linear 10–25% sucrose gradients (4.2 ml) in PME and centrifuged at 150 000 g in a Sorvall TH660 rotor for 13 h. Fractions of 200 µl were analysed by gel electrophoresis and western blotting. Sedimentation coefficients were estimated from three experiments from calibration curves, which were based on linear regression of the results of three sucrose density gradients using thyroglobulin, catalase, aldolase and bovine serum albumin (BSA; SIGMA). For immunoprecipitation, cells were disrupted in 50 mM Tris–HCl pH 8.0, 150 mM NaCl, 15 mM MgCl2, 1% NP-40, complete protease inhibitor mix. DYNABEADS Pan Mouse IgG (DYNAL) were blocked in phosphate-buffered saline (PBS) containing 0.1% BSA and incubated for 1 h with 80 µg/ml antibodies against the Myc and HA epitopes (Sigma). After washing with PBS, 0.1% BSA the beads were exposed to the cell extracts. The magnetic beads were harvested and attached proteins were analysed using SDS–PAGE.
MT affinity experiments
MT precipitation was modified from Koonce and McIntosh (1990). Briefly, cell extracts of FB2HDyn1/Dyn2M at OD600 <0.6 were prepared in PME and 0.9 M glycerol. All steps were performed at 4°C. A 150 000 g supernatant (S2) was supplemented with 3 U/ml apyrase (Sigma), 2 mM AMPPNP (Sigma), 10 µM taxol (Sigma) and 0.8 mg/ml polymerized tubulin (kindly provided by M.Schliwa). The mixture was incubated for 1 h at 4°C, and MTs were sedimented at 40 000 g for 30 min (S3). The pellet was resuspended in PME, 0.9 M glycerol, 0.5 mM AMPPNP, 10 µM taxol and centrifuged through a 20% sucrose cushion at 50 000 g for 30 min (S4). Dynein was released by incubation of the MTs for 15 min on ice in PME, 0.9 M glycerol, 10 mM MgATP, 10 µM taxol. MTs were sedimented at 50 000 g for 30 min, resulting in S5. The release was repeated to obtain S6 and P6.
Immunofluorescence and microscopy
Immunostaining was performed using epitope-tag antibodies (Sigma) or αro1 antibodies as primary antibodies and Cy2- and Cy3-conjugated secondary antibodies (Dianova) on cells from growing cultures (OD600 <1). All procedures followed published protocols (Steinberg et al., 2001). Light microscopy was performed using a Zeiss Axiophot and standard filter sets. Digital images were taken by a cooled CCD camera (4742-95; Hamamatsu) and processed using ImageProPlus (Media Cybernetics). DAPI staining was carried out after formaldehyde fixation and washes in PBS with 1 µg/ml DAPI in PBS for 15 min at 60°C.
Accession numbers
DDBJ/EMBL/GenBank accession Nos for dyn1 and dyn2 are AF403739 and AF403740, respectively.
Acknowledgments
Acknowledgements
We thank Michael Bölker for supporting the initial phase of the project and Jörg Kämper for help with chromosome isolation. We are grateful to Mike Plamann for the αro1 antibody, Eckhardt Granderath for PCR primers and Manfred Schliwa for purified tubulin. Michael Artmeier and Karl-Heinz Braun are acknowledged for technical assistance. We also wish to thank the anonymous referees for their constructive criticism. This work was supported by the Deutsche Forschungsgemeinschaft through SFB 413 and by the Max-Planck-Society.
References
- Asai D.J and Koonce,M.P. (2001) The dynein heavy chain: structure, mechanics and evolution. Trends Cell Biol., 11, 196–202. [DOI] [PubMed] [Google Scholar]
- Banuett F. (1995) Genetics of Ustilago maydis, a fungal pathogen that induces tumors in maize. Annu. Rev. Genet., 29, 179–208. [DOI] [PubMed] [Google Scholar]
- Banuett F. and Herskowitz,I. (1989) Different a alleles of Ustilago maydis are necessary for maintenance of filamentous growth but not for meiosis. Proc. Natl Acad. Sci. USA, 86, 5878–5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bölker M., Bohnert,H.U., Braun,K.H., Görl,J. and Kahmann,R. (1995) Tagging pathogenicity genes in Ustilago maydis by restriction-enzyme mediated integration (REMI). Mol. Gen. Genet., 248, 547–552. [DOI] [PubMed] [Google Scholar]
- Bottin A., Kämper,J. and Kahmann,R. (1996) Isolation of a carbon source-regulated gene from Ustilago maydis. Mol. Gen. Genet., 253, 342–352. [DOI] [PubMed] [Google Scholar]
- Burgess S.A. (1995) Rigor and relaxed outer dynein arms in replicas of cryofixed motile flagella. J. Mol. Biol., 250, 52–63. [DOI] [PubMed] [Google Scholar]
- Cottingham F.R. and Hoyt,M.A. (1997) Mitotic spindle positioning in Saccharomyces cerevisiae is accomplished by antagonistically acting MT motor proteins. J. Cell Biol., 138, 1041–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshel D., Urrestarazu,L.A., Vissers,S., Jauniaux,J.C., van Vliet-Reedijk, J.C., Planta,R.J. and Gibbons,I.R. (1993) Cytoplasmic dynein is required for normal nuclear segregation in yeast. Proc. Natl Acad. Sci. USA, 90, 11172–11176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gee M.A., Heuser,J.E. and Vallee,R.B. (1997) An extended microtubule-binding structure within the dynein motor domain. Nature, 390, 636–639. [DOI] [PubMed] [Google Scholar]
- Gibbons I.R. (1995) Dynein family of motor proteins: present status and future questions. Cell Motil. Cytoskeleton, 32, 136–144. [DOI] [PubMed] [Google Scholar]
- Gibbons I.R., Lee-Eiford,A., Mocz,G., Phillipson,C.A., Tang,W.J. and Gibbons,B.H. (1987) Photosensitized cleavage of dynein heavy chains. Cleavage at the ‘V1 site’ by irradiation at 365 nm in the presence of ATP and vanadate. J. Biol. Chem., 262, 2780–2786. [PubMed] [Google Scholar]
- Gibbons I.R., Gibbons,B.H., Mocz,G. and Asai,D.J. (1991) Multiple nucleotide-binding sites in the sequence of dynein β heavy chain. Nature, 352, 640–643. [DOI] [PubMed] [Google Scholar]
- Habura A., Tikhonenko,I., Chisholm,R.L. and Koonce,M.P. (1999) Interaction mapping of a dynein heavy chain—identification of dimerization and intermediate-chain binding domains. J. Biol. Chem., 274, 15447–15453. [DOI] [PubMed] [Google Scholar]
- Holliday R. (1974) Ustilago maydis. In King,R.C. (ed.), Handbook of Genetics. Plenum Press, New York, NY.
- Holzbaur E.L.F. and Vallee,R.B. (1994) Dyneins—molecular structure and cellular function. Annu. Rev. Cell Biol., 10, 339–372. [DOI] [PubMed] [Google Scholar]
- Inoue S., Turgeon,B.G., Yoder,O.C. and Aist,J.R. (1998) Role of fungal dynein in hyphal growth, microtubule organization, spindle pole body motility and nuclear migration. J. Cell Sci., 111, 1555–1566. [DOI] [PubMed] [Google Scholar]
- King S.M. (2000) AAA domains and organization of the dynein motor unit. J. Cell Sci., 113, 2521–2526. [DOI] [PubMed] [Google Scholar]
- Kinscherf T.G. and Leong,S.A. (1988) Molecular analysis of the karyotype of Ustilago maydis. Chromosoma, 96, 427–433. [DOI] [PubMed] [Google Scholar]
- Knetsch M.L., Uyeda,T.Q. and Manstein,J.D. (1999) Disturbed communication between actin- and nucleotide-binding sites in a myosin II with truncated 50/20-kDa junction. J. Biol. Chem., 274, 20133–20138. [DOI] [PubMed] [Google Scholar]
- Koonce M.P. (1997) Identification of a microtubule-binding domain in a cytoplasmic dynein heavy chain. J. Biol. Chem., 272, 19714–19718. [DOI] [PubMed] [Google Scholar]
- Koonce M.P. and McIntosh,J.R. (1990) Identification and immunolocalization of cytoplasmic dynein in Dictyostelium. Cell Motil. Cytoskeleton, 15, 51–62. [DOI] [PubMed] [Google Scholar]
- Koonce M.P. and Tikhonenko,I. (2000) Functional elements within the dynein microtubule-binding domain. Mol. Biol. Cell, 11, 523–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Lee,I.H. and Plamann,M. (2000) Cytoplasmic dynein ATPase activity is regulated by dynactin-dependent phosphorylation. J. Biol. Chem., 275, 31798–31804. [DOI] [PubMed] [Google Scholar]
- Li Y.Y., Yeh,E., Hays,T. and Bloom,K. (1993) Disruption of mitotic spindle orientation in a yeast dynein mutant. Proc. Natl Acad. Sci. USA, 90, 10096–10100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A., Van Dyke,M. and Stock,J. (1991) Predicting coiled coils from protein sequences. Science, 252, 1162–1164. [DOI] [PubMed] [Google Scholar]
- Mocz G. and Gibbons,I.R. (1996) Phase partition analysis of nucleotide binding to axonemal dynein. Biochemistry, 35, 9204–9211. [DOI] [PubMed] [Google Scholar]
- Neuwald A.F., Aravind,L., Spouge,J.L. and Koonin,E.V. (1999) AAA+: a class of chaperone-like ATPases associated with the assembly, operation and disassembly of protein complexes. Genome Res., 9, 27–43. [PubMed] [Google Scholar]
- O’Donnell K.L. and McLaughlin,D.J. (1984) Postmeiotic mitosis, basidiospore development and septation in Ustilago maydis. Mycologia, 76, 486–502. [Google Scholar]
- Ogawa K. (1991) Four ATP-binding sites in the midregion of the β heavy chain of dynein. Nature, 352, 643–645. [DOI] [PubMed] [Google Scholar]
- Paschal B.M., Shpetner,H.S. and Vallee,R.B. (1987) MAP 1C is a microtubule-activated ATPase which translocates microtubules in vitro and has dynein-like properties. J. Cell Biol., 105, 1273–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S. and Latterich,M. (1998) The AAA team: related ATPases with diverse functions. Trends Cell Biol., 8, 65–71. [PubMed] [Google Scholar]
- Plamann M., Minke,P.F., Tinsley,J.H. and Bruno,K.S. (1994) Cytoplasmic dynein and actin-related protein Arp1 are required for normal nuclear distribution in filamentous fungi. J. Cell Biol., 127, 139–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samso M., Radermacher,M., Frank,J. and Koonce,M.P. (1998) Structural characterization of a dynein motor domain. J. Mol. Biol., 276, 927–937. [DOI] [PubMed] [Google Scholar]
- Schulz B., Banuett,F., Dahl,M., Schlesinger,R., Schaefer,W., Martin,T., Herskowitz,I. and Kahmann,R. (1990) The B alleles of Ustilago maydis whose combinations program pathogenic development code for polypeptides containing a homeodomain-related motif. Cell, 60, 295–306. [DOI] [PubMed] [Google Scholar]
- Sharp D.J., Rogers,G.C. and Scholey,J.M. (2000) Roles of motor proteins in building microtubule-based structures: a basic principle of cellular design. Biochim. Biophys. Acta, 1496, 128–141. [DOI] [PubMed] [Google Scholar]
- Shaw S.L., Yeh,E., Maddox,P., Salmon,E.D. and Bloom K. (1997) Astral microtubule dynamics in yeast: a microtubule-based searching mechanism for spindle orientation and nuclear migration into the bud. J. Cell Biol., 139, 985–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. (1973) Molecular weights of proteins and some other materials including sedimentation, diffusion and frictional coefficients and partial specific volume. In Sobert,H.A. (ed.), Handbook of Biochemistry. CRC Press, Cleveland, OH, pp. C3–C35.
- Song H. and Endow,S.A. (1998) Decoupling of nucleotide and microtubule binding sites in a kinesin mutant. Nature, 396, 587–590. [DOI] [PubMed] [Google Scholar]
- Steinberg G. (2000) The cellular role of molecular motors in fungi. Trends Microbiol., 8, 162–168. [DOI] [PubMed] [Google Scholar]
- Steinberg G., Wedlich-Söldner,R., Brill,M. and Schulz,I. (2001) Microtubules in the fungal pathogen Ustilago maydis are highly dynamic and determine cell polarity. J. Cell Sci., 114, 609–622. [DOI] [PubMed] [Google Scholar]
- Tynan S.H., Gee,M.A. and Vallee,R.B. (2000) Distinct but overlapping sites within the cytoplasmic dynein heavy chain for dimerization and for intermediate chain and light intermediate chain binding. J. Biol. Chem., 275, 32769–32774. [DOI] [PubMed] [Google Scholar]
- Vale R.D. (2000) AAA proteins: Lords of the ring. J. Cell Biol., 150, F13–F19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedlich-Söldner R., Bölker,M., Kahmann,R. and Steinberg,G. (2000) A putative endosomal t-SNARE links exo- and endocytosis in the phytopathogenic fungus Ustilago maydis. EMBO J., 19, 1974–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willins D.A., Xiang,X. and Morris,N.R. (1995) An α-tubulin mutation suppresses nuclear migration mutations in Aspergillus nidulans. Genetics, 141, 1287–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang X., Beckwith,S.M. and Morris,N.R. (1994) Cytoplasmic dynein is involved in nuclear migration in Aspergillus nidulans. Proc. Natl Acad. Sci. USA, 91, 2100–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang X., Roghi,C. and Morris,N.R. (1995) Characterization and localization of cytoplasmic dynein heavy chain of Aspergillus nidulans. Proc. Natl Acad. Sci. USA, 92, 9890–9894. [DOI] [PMC free article] [PubMed] [Google Scholar]