Abstract
Chromatin is composed of nucleosomes, the universally repeating protein–DNA complex in eukaryotic cells. The crystal structure of the nucleosome core particle from Saccharomyces cerevisiae reveals that the structure and function of this fundamental complex is conserved between single-cell organisms and metazoans. Our results show that yeast nucleosomes are likely to be subtly destabilized as compared with nucleosomes from higher eukaryotes, consistent with the idea that much of the yeast genome remains constitutively open during much of its life cycle. Importantly, minor sequence variations lead to dramatic changes in the way in which nucleosomes pack against each other within the crystal lattice. This has important implications for our understanding of the formation of higher order chromatin structure and its modulation by post-translational modifications. Finally, the yeast nucleosome core particle provides a structural context by which to interpret genetic data obtained from yeast. Coordinates have been deposited with the Protein Data Bank under accession number 1ID3.
Keywords: chromatin/crystal structure/histone/nucleosome core particle/Saccharomyces cerevisiae
Introduction
The packaging of DNA in the eukaryotic nucleus is achieved by a hierarchical scheme of folding and compaction into protein–DNA assemblies, collectively called chromatin (Widom, 1998). At the first level of organization, 1.65 tight superhelical turns of 147 bp of DNA are wrapped around a histone octamer to form the nucleosome core. In higher eukaryotes, the addition of linker histone H1 to linker DNA forms the nucleosome, the basic repeating unit of chromatin. Hundreds of thousands of nucleosomes are compacted further into multiple higher organizational levels. Because of DNA packaging into chromatin, the structure and accessibility of nucleosomal DNA deviate dramatically from those of linear, ‘naked’ DNA, as seen in the high-resolution X-ray structure of the nucleosome core particle comprised of recombinant Xenopus laevis histone proteins and a palindromic 146 bp DNA fragment derived from human α-satellite DNA (Luger et al., 1997a, 2000). Depending on the structural context, chromatin can both promote and impede transcription, replication, recombination and DNA repair. Thus, chromatin plays a central role in the regulation of these vital processes. Eukaryotic cells have developed elaborate mechanisms to modulate the inherently dynamic chromatin structures in a regulated manner (Workman and Kingston, 1998).
The histone octamer consists of two copies each of the four histone proteins H2A, H2B, H3 and H4. Two histone pairs, composed either of H2A and H2B, or H3 and H4, form tight dimers that each organize 30 bp of DNA (Luger and Richmond, 1998a). Two H3–H4 dimers form a tetramer that binds the central 60 bp of the nucleo somal DNA. By structurally similar interactions, one H2A–H2B dimer is tethered to one half of the histone (H3–H4)2 tetramer. The H2A–H2B dimer organizes 30 bp towards either end of the DNA. The penultimate 10 bp of nucleosomal DNA are bound by a region of H3 that does not form an integral part of the (H3–H4)2 tetramer, and most probably is not able to bind DNA in the absence of the H2A–H2B dimer (Luger et al., 1997a; Luger and Richmond, 1998a; see also Figure 1B–E). The massive distortion of the DNA is brought about mainly by the tight interaction between the structured regions of the histone proteins and the minor groove of the DNA at 14 independent DNA-binding locations, using either L1L2 loops or α1α1 DNA-binding motifs (Luger and Richmond, 1998a). At the molecular level, the interaction of the histone octamer with the DNA is formed mainly by tight hydrogen bonds between the main chain amide and the phosphate oxygen of the DNA, assisted by electrostatic interaction with basic side chains. In the context of a mononucleosome, the flexible histone tails do not contribute to the stability of the complex (Luger et al., 1997b). Other nucleosome structures containing chicken histone proteins (Harp et al., 2000) or the histone variant H2A.Z (Suto et al., 2000) have confirmed this basic design.
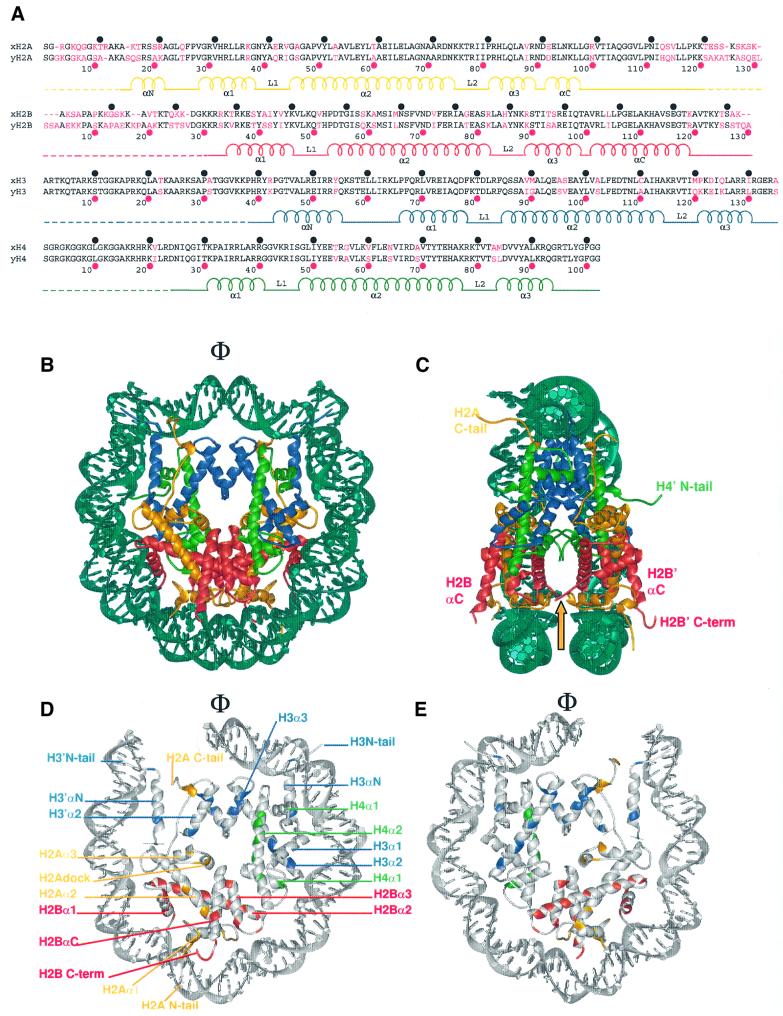
Fig. 1. Secondary and tertiary structure of the yeast nucleosome core particle. (A) Sequence alignment of X.laevis (top line) and S.cerevisiae histone proteins (bottom line). Amino acid differences are colored in magenta. Intervals of 10 amino acids for X.laevis (black circles) and S.cerevisiae (magenta circles) are indicated. The α-helices and loops located within the structured regions are labeled, and the flexible histone tails are indicated by dashed lines. (B) The crystal structure of the yeast nucleosome core particle, viewed down the superhelical axis. Histone chains are colored yellow for H2A, red for H2B, blue for H3 and green for H4. The DNA is shown in turquoise. α-helices and the location of the N- and C-terminal tails are shown. The position of the molecular dyad axis is indicated (Φ). (C) Side view of the yeast nucleosome core particle, obtained by rotation of 90° around the axis of non-crystallographic symmetry, with part of the DNA removed for clarity. The arrow denotes the location of the L1 loop. (D and E) Amino acid differences in the yeast octamer [as shown in (A)] are colored according to the histone coloring scheme in (B). The conserved amino acids and DNA are shown in gray. Only 73 bp of the DNA and associated proteins are shown. (D) The solvent-exposed surface view of one half of the nucleosome is shown, while (E) shows the same half of the nucleosome viewed from the interior surface between the two gyres of the DNA supercoil.
Many groundbreaking studies that address the complex interplay between chromatin structure and transcription regulation in the living cell stem from yeast genetics (see Hartzog and Winston, 1997; reviewed by Gregory, 2001). These studies were made possible by the obvious suitability of yeast for genetic studies, and by the fact that the Saccharomyces cerevisiae genome contains only two genes for each of the four core histone proteins. Many of the characteristics of chromatin in higher organisms are seen in yeast. For example, S.cerevisiae contains histone variants, such as the histone H2A variant H2A.Z (HTZ1) (Jackson and Gorovsky, 2000; Santisteban et al., 2000) and the centromere-specific H3 variant CenpA (cse4) (Glowczewski et al., 2000). Saccharomyces cerevisiae also uses targeted ATP-dependent chromatin remodeling factors (Aalfs and Kingston, 2000) and reversible modification of histone tails, such as acetylation, de-acetylation (Vogelauer et al., 2000), methylation (Strahl et al., 1999), phosphorylation (Hsu et al., 2000) and ubiquitylation (Robzyk et al., 2000), in order to regulate the level of DNA accessibility in a chromatin context.
However, fundamental differences between the yeast genome and that of higher organisms suggest that chromatin might be organized in a different manner in yeast. Yeast is a unicellular organism whose entire genome is only ∼0.5% the size of that of humans. Much of the yeast genome is constitutively open for transcription, as opposed to the small percentage of actively transcribed genes at any given time in the cells of higher eukaryotes. Yeast nucleosomes are very closely spaced, with a repeat length of 162 ± 6 bp (Horz and Zachau, 1980), resulting in a linker length of only 15–20 bp. In contrast, the repeat length in metazoans ranges from 175 to 240 bp, with an average of ∼190 bp. No linker histones have been found to be associated with yeast chromatin in a stoichiometric manner. Recently, the presence of a gene (HHO) encoding a protein with linker histone homology (Hho1p) has been identified, and it has been suggested that this gene product may function as a linker histone. However, deletion of this gene has neither growth nor mating defects, suggesting that this protein does not play an important role in chromatin organization in yeast (Patterton et al., 1998).
Histone proteins are highly conserved among eukaryotic organisms, with yeast histones being among the most divergent from mammalian histones (Baxevanis and Landsman, 1998). The differences are distributed throughout the length of the amino acid sequence, with many of the more divergent sequence changes clustered in the flexible histone tails. The distinct sequence divergence between the histone proteins of S.cerevisiae and metazoans may well reflect the different requirements for DNA compaction between unicellular and multicellular organisms. Some earlier reports suggest that yeast mononucleosomes are indeed destabilized towards salt-dependent and thermal unfolding, suggesting a less constrained structure (Lee et al., 1982; Pineiro et al., 1991). In addition, yeast nucleosomes appear to be more permissive to thermal untwisting of the DNA (Morse et al., 1987).
Here we present the crystal structure of the S.cerevisiae nucleosome core particle at 3.1 Å resolution. Analysis of the structure shows that subtle sequence variations may affect the overall stability of the core particle in this organism, as well as lead to differences in the interaction between neighboring nucleosomes. This may be a reflection of differences in nucleosome organization in vivo. In addition, the structure is now available to analyze in a structural context the numerous genetic studies obtained from yeast. In particular, our data suggest a straightforward mechanism by which the recently described ubiquitylation of a specific residue in H2B (Robzyk et al., 2000) may affect chromatin compaction in vivo.
Results
Structure determination
Crystals of the nucleosome core particle from S.cerevisiae (Sce-NCP) were obtained under conditions similar to those previously published for the crystal structure of the nucleosome core particle containing X.laevis histone proteins (Xla-NCP; Luger et al., 1997a). We have found previously that crystallization conditions and diffraction quality are highly dependent on the DNA fragment being used. Despite the fact that the same palindromic DNA sequence derived from human α-satellite repeats was used to reconstitute yeast nucleosomes, Sce-NCP crystals behaved rather differently. They took much longer to grow, exhibited a different morphology, were extremely fragile and diffracted to a lower resolution at a synchrotron source (3.1 Å as opposed to a routinely obtained 2.2 Å resolution for Xla-NCP; Luger et al., 2000). Furthermore, although the original space group was maintained, the longest crystallographic axis deviated significantly in length (Table I).
Table I. Summary of the crystallographic analysis.
| Data collection statistics | |
| Space group | P212121 |
| Unit cell parameters (Å) | a = 104.9, b = 110.4, c = 192.6 |
| Resolution (Å) | 50–3.1 |
| Reflections (total/unique) | 517 812 (39 551) |
| Completeness (%) | 99.5 (99.9)a |
|
Rsymb |
0.056 (0.299)c |
| Refinement statisticsc | |
| Resolution (Å) | 40–3.1 |
| Reflections | 38 264 |
| Rcryst/Rfreed | 0.223/0.292 |
| R.m.s.ds | |
| bonds (Å) | 0.0069 |
| angles (°) | 1.14 |
| Average B-factors (Å2) | |
| protein | 62.05 |
| DNA | 132.93 |
| solvent | 56.7 |
aValue in parentheses is for the highest resolution shell: 3.15–3.1 Å.
bRsym = Σ|Ih – <Ih>|/ΣIh, where <Ih> is the mean of measurements for a single hkl.
cAtomic model: 757 amino acids (H2A, 15–124; H2A′, 12–119; H2B, 30–122; H2B′, 29–124; H3, 38–134; H3′, 38–134; H4, 24–102; H4′, 18–102), 60 water molecules and 17 manganese ions (a total of 12 129 atoms). The remainder of the histone tails was too disordered to be included in the final model.
dRcryst = Σ|Fobs – Fcalc|/ΣFobs.
Phase information was obtained using the previously published nucleosome core particle structure from X.laevis (Protein Data Bank entry 1AOI) as a search model. Data collection and refinement statistics are given in Table I. Sequence differences between S.cerevisiae and X.laevis were clearly visible in the original 2Fo – Fc electron density map. As in Xla-NCP, the histone fold regions, as well as the extensions that are responsible for the main protein–protein and protein–DNA interactions in the yeast nucleosome core particle, are highly ordered. However, the histone tails quickly become disordered as they extend past the DNA superhelix. Slightly less of the flexible histone tails is visible in the Sce-NCP structure as compared with Xla-NCP (Table I).
The structures of nucleosome core particles from S.cerevisiae and X.laevis are very similar
Amino acid sequence alignments between S.cerevisiae and X.laevis histones show that H2A and H2B are more divergent (72 and 67% identity) than H3 and H4 (84 and 92% identity) (Figure 1A). Although changes are more numerous in the flexible histone tails (especially in those of H2A and H2B), the folded regions of all four histone proteins are also divergent in their amino acid sequence. Despite this sequence divergence, the overall structure of the nucleosome core particle from S.cerevisiae (Figure 1B and C) is very similar to that of the previously reported structure of the nucleosome core particle from X.laevis (Luger et al., 1997a). This shows that the function of the histone octamer is identical at the level of histone–DNA interaction, despite the differences in sequence and chromatin organization between yeast and higher eukaryotes. The Cα atoms of the histone octamers of Sce-NCP and Xla-NCP superimpose with an overall root mean square deviation (r.m.s.d.) of 1.14 Å, and the phosphates of the two DNA strands align with an r.m.s.d. of 1.15 Å. The two structures (including side chains and DNA bases) superimpose with an r.m.s.d. of 1.57 Å. The most striking deviations in the Cα trace of the two structures are found in the C-terminal tail of the second copy of histone H2A (H2A′) as well as in the N-terminal tail of histone H4′ (Table II, compare rows ‘All’ with rows ‘Δtail’). These tails take completely different paths in the two structures as a result of variations in their structural environment (see below).
Table II. Structural alignment of Sce-NCP and Xla-NCP: r.m.s.ds between the S.cerevisiae and X.laevis structuresa,b.
| H2A | H2B | H3 | H4 | DNA | |
|---|---|---|---|---|---|
| Chain 1 | |||||
| allc | 1.06 (0.51) | 1.23 (0.76) | 1.44 (0.47) | 0.83 (0.47) | 1.23 (1.15) |
| Δtaild | 0.89 (0.46) | 1.20 (0.71) | 1.43 (0.47) | 0.81 (0.43) | |
| Chain 2 | |||||
| allc | 1.85 (1.06) | 1.20 (0.80) | 1.07 (0.59) | 3.53 (2.77) | 1.11 (1.14) |
| Δtaild | 1.08 (0.56) | 1.18 (0.79) | 1.07 (0.59) | 1.04 (0.58) |
aValues given in angstroms.
bValues given include all side chains (values in parentheses are for the Cα backbone or phosphate atoms only).
cAll: includes all residues of the entire model.
dΔtail: includes only the structured regions of the model, as defined by Luger et al. (1997a).
The regions of the histone proteins that are responsible for protein–protein and protein–DNA interaction are structurally much more conserved than the histone tails. Analysis of the r.m.s.d. for each chain, based on the same alignment (Table II, row ‘Δtail’), and a distance plot for each Cα atom (not shown), shows that both copies of histone H2B are the most structurally divergent. This is expected from the relatively high degree of sequence divergence between X.laevis and S.cerevisiae for this particular histone (Figure 1A). However, the L1 loops and C-terminal helix of H2B, where the sequence is very similar (Figure 1A), also exhibit noticeable structural deviations (∼2.4 Å r.m.s.d.) compared with the corresponding regions in Xla-NCP. In Sce-NCP, both H2B L1 loops are involved in crystal contacts, whereas they are not involved in any internucleosome contacts in Xla-NCP (see below). These structural differences in L1 loop conformation lead to local deviations (∼2.5 Å r.m.s.d.) in the nearby DNA phosphates.
The overall architecture of the histone octamer, as well as all of the residues that are involved in direct protein– DNA interactions, is unchanged between S.cerevisiae and X.laevis (Figure 1D and E). Thus, the general mechanism by which the histone octamer distorts linear DNA into a tight superhelix is maintained between yeast and higher eukaryotes. The path of the DNA around the histone octamer is conserved between the two structures, with only local deviations between the positions of the phosphates in regions of high B-factors. This is indicated by the relatively low r.m.s.d. values for the superposition of the DNA in the Xla-NCP and Sce-NCP structures (Table II).
Sequence differences cause changes in molecular surface and in histone–histone interaction
Sequence differences in the histone fold regions and extensions (Figure 1A) are not limited to amino acid residues on the surface of the nucleosome core particle, but equally affect residues that are buried deep within the histone octamer core. This is demonstrated in Figure 1D and E, which shows the location of amino acid differences on one half of the yeast nucleosome core particle, viewed from the outside (Figure 1D), and from between the two symmetry-related halves (Figure 1E). Sequence differences that change the molecular surface of the yeast histone octamer (Figure 1D) could reveal how inter-particle interactions are altered in yeast in vivo. Evidence for this is seen in the dramatic differences in the packing of yeast nucleosomes within the crystal lattice (see below). In addition, interactions with other cellular factors may be affected by surface variations. In contrast, amino acid differences that are buried within the histone octamer (Figure 2E) may locally affect the stability of histone– histone interactions, thus contributing to an overall change in nucleosome stability.
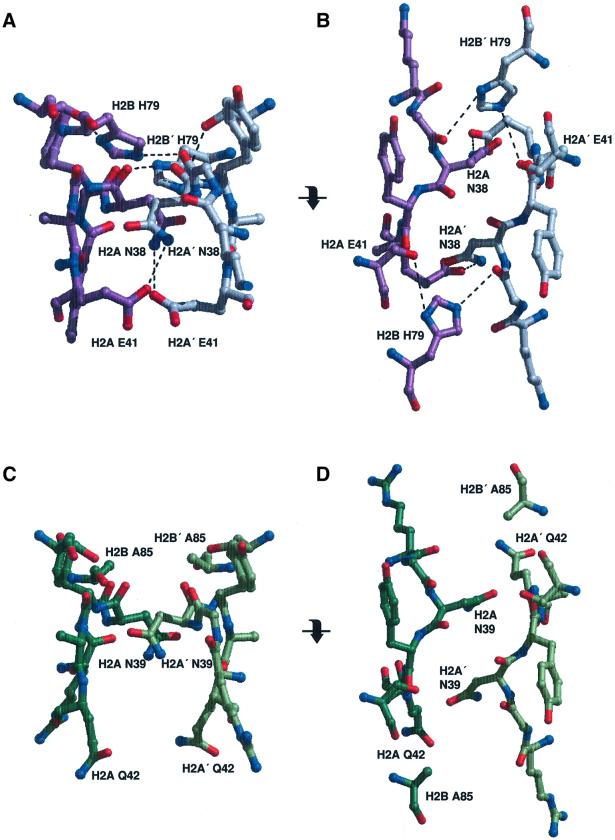
Fig. 2. Interactions between two H2A–H2B dimers within a nucleosome. (A) The side chain interactions of the H2A and H2A′ L1 loops in Xla-NCP, in the same orientation as shown in Figure 1C. Xla-H2A and H2A′ are colored in purple and gray, respectively. Dashed lines indicate hydrogen bonds. (B) The same region is shown in a view obtained by a 90° rotation around the horizontal axis (as indicated). (C and D) The equivalent region in Sce-NCP, viewed in the same orientation as in (A) and (B), respectively. Note the total absence of intermolecular hydrogen bonds.
The main signature of the molecular surface of Xla-NCP is a large acidic patch formed by seven amino acids from the structured regions of H2A and H2B (Luger et al., 1997a). Several basic regions create a favorable interaction interface for DNA surrounding this area. Both features are maintained in Sce-NCP, but there are subtle changes in the charge distribution and shape of the yeast histone octamer surface as compared with that of X.laevis. Additional positively charged amino acids are located on the surface, and thus render it slightly more basic (Supplementary figure in Supplementary data available at The EMBO Journal Online).
In all nucleosome core particle structures reported to date (Luger et al., 1997a; Suto et al., 2000; Harp et al., 2000), the two H2A–H2B dimers interact through a small but significant interface formed by the L1 loops of two adjacent H2A molecules, indicated by an arrow in Figure 1C. In Xla-NCP, two hydrogen bonds and two salt bridges stabilize this interface (Figure 2A and B). Xla-H2A Asn38 forms tight bonds with Xla-H2A′ Glu41 and Xla-H2B′ His79. Owing to non-crystallographic symmetry, these interactions are mirrored between Xla-H2A′ Asn38 and Xla-H2A/H2B (Figure 2A and B). Although the path of the main chain within the L1 loop is almost identical in the yeast structure, the corresponding residues are changed, resulting in a loss of all interactions at this interface (Figure 2C and D). In yeast, Glu41 is changed to a glutamine, and His79 is changed to an alanine (H2A Glu42 and H2B Ala85 in yeast, Figure 1A). In order to avoid steric clashes between the two amino groups of the asparagine and glutamine residues, the two glutamine residues point downward towards the DNA (Figure 2C). As a result, no stabilizing interactions exist between the two H2A–H2B dimers in this region, and the buried surface is reduced to 90 Å2 in Sce-NCP, compared with 150 Å2 in Xla-NCP. Together, these findings strongly suggest a significantly weaker interaction at this particular interface in the S.cerevisiae core particle as compared with that of X.laevis.
Most of the amino acid sequence differences are found in the N-terminal tails of H2A and H2B, and in the C-terminal tail of H2A. However, it is the N-terminal tail of H4′ that shows the largest deviations between the two structures (Table II). The involvement of this tail in transcription regulation and gene silencing has been particularly well documented, and its importance is demonstrated by the fact that the sequence of this region is almost completely conserved between S.cerevisiae and X laevis (Figure 1A). In Xla-NCP, the H4 N-terminal tail forms essential crystal contacts with the acidic patch on the surface of a neighboring nucleosome (Luger and Richmond, 1998b), and it has been speculated that these two regions may interact in a similar fashion in vivo to stabilize higher order structure. No such contact is made in Sce-NCP (see below), which allows the H4 tail to adopt an alternative conformation that is completely different from that observed in either chain in Xla-NCP. Instead of interacting with the acidic patch of a neighboring core particle, this tail is now poised to interact with the DNA of a neighboring nucleosome core particle within the crystal lattice (data not shown). However, since H4 amino acids 1–17 are too disordered to be included in the molecular model, interactions of these amino acids with the DNA phosphates are probably dislocalized. Similar differences in the path of the amino acid main chain are seen for the C-terminal tail of H2A. This confirms that histone tails can indeed assume different conformations that depend on the structural context.
Sce-NCP and Xla-NCP crystals are held together by different types of interactions
Perhaps the most striking difference between the S.cerevisiae and X.laevis structures is the way in which the nucleosome core particles pack within the crystal lattice. As has been pointed out previously (Finch et al., 1981), the packing of nucleosomes within a crystal most probably does not reflect the way in which particles are organized in condensed chromatin. The most obvious difference is that individual nucleosomes are located on long contiguous DNA in the cell, while they exist as individual, unconnected particles in the crystal. Furthermore, the presence of linker histones promotes the compaction of chromatin at least in higher eukaryotes. The packing of nucleosomes within a crystal represents the most obvious shape to pack wedge-shaped particles in an ordered manner (namely, in an alternating up-and-down packing; Finch et al., 1981), whereas this arrangement is not consistent with current models of the 30 nm fiber (for a recent review see Woodcock and Dimitrov, 2001). However, we believe that crystal packing may reflect some important aspects of internucleosome interaction in vivo for the following reasons. First, crystallization of nucleosome core particles is performed at relatively low ionic strength and at very high (micromolar) nucleosome concentrations. Secondly, crystallization is the result of the tight packaging of particles into a regular arrangement, and is driven by the search for an energetically favorable mode of interaction between neighboring particles. Necessarily, these same constraints also apply to chromatin condensation in vivo, either at the level of the 30 nm fiber or at higher levels of compaction.
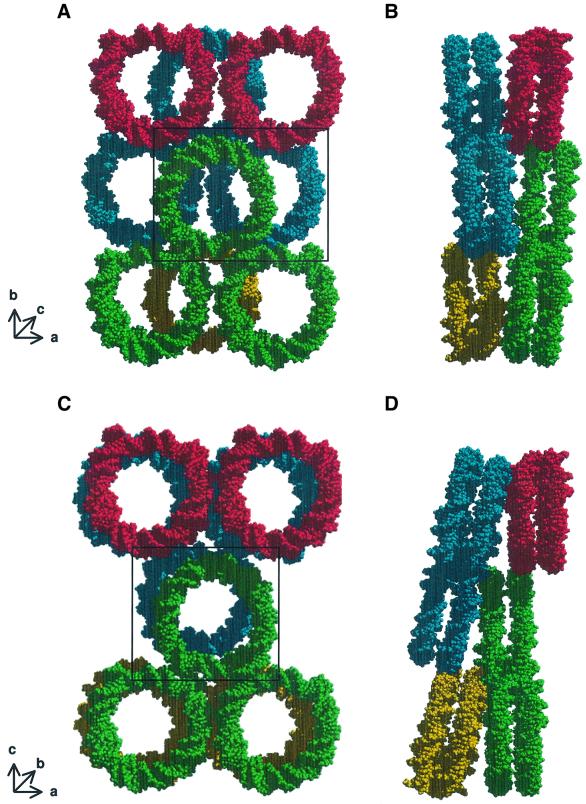
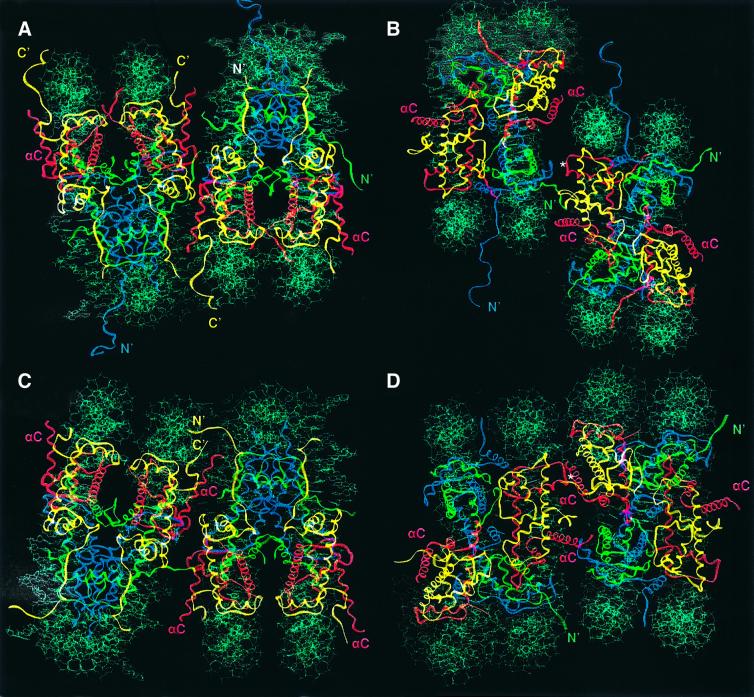
Three types of major interactions drive the formation of Xla-NCP crystals into staggered layers of nucleosome cores. First, the end-to-end stacking of nucleosomal DNA is the major driving force in forming one plane of the crystal lattice (green particles in Figure 3A). Secondly, the histone octamers contact one another through a series of interactions between the basic H4 tail of one nucleosome and the acidic patch region of a neighboring particle (Figure 4B; and Luger and Richmond, 1998b). These contacts are essential for crystallization, as has been shown by the mutation of Xla-H4 Lys20 to cysteine (K.Luger, unpublished results). Thirdly, an Mn2+-mediated crystal contact directly chelates the base of H3 α1 to the H2B L1 loop of the neighboring particle (denoted by an asterisk in Figure 4B). These areas of protein–protein interaction connect two consecutive layers of nucleosome core particles (Figure 3B). Additional protein–protein and DNA–DNA interactions further stabilize the crystal lattice. Importantly, each of the residues involved in the contacts mentioned above is conserved in yeast.
Fig. 3. Crystal packing of X.laevis and S.cerevisiae nucleosome core particles. (A) Xla-NCP crystal packing viewed approximately down the superhelical axis. Short arrows show the approximate location of the three crystallographic axes. Only the DNA is shown for clarity; the same colors denote nucleosomes that lie within the same plane. (B) The same arrangement of molecules is rotated by 90° around the crystallographic b-axis. (C and D) Sce-NCP crystal packing in the same views as (A) and (B), respectively. The discrepancy in the notation of the crystallographic b- and c-axes stems from the fact that the previous study used different programs to index the data (Luger et al., 1997a). Particles whose molecular interaction is shown in Figure 4 are boxed.
Fig. 4. Protein–protein interactions within the crystal lattice. (A) Crystal contacts between two neighboring nucleosomes in X.laevis in a view that places the superhelical axis in a horizontal orientation (as seen in Figure 1C). (B) The same Xla-NCP packing as shown in (A), but viewed down the molecular 2-fold axis. This view is achieved by a 90° rotation around the superhelical axis (horizontal). In both views, the nucleosome core particle to the left of the pair corresponds to the center green particle in Figure 3A, whereas the right-hand particle corresponds to the blue particle in Figure 3A (boxed in Figure 3A and C, respectively). Histones are colored as in Figure 1. The location of the H2A and H4 histone tails is indicated. The position of the Mn2+ ion that is crucially involved in forming crystal contacts is shown (*). (C and D) Two yeast nucleosome core particles shown in the same orientation as seen in (A) and (B), respectively. With respect to Figure 3C, the same two particles are depicted as for Xla-NCP.
The DNA end-to-end crystal contacts are essentially maintained in Sce-NCP, with only minor differences in stacking geometry. However, the protein–protein interactions between nucleosomes are completely different. The result is that nucleosomes in neighboring planes stack on top of each other with their superhelical axes almost superimposed (Figure 3C), whereas the arrangement in X.laevis is much more staggered (Figure 3B). Despite this fact, the total surface area that is actually buried upon crystal formation is very similar (∼4000 Å2) between the two structures. One consequence of this arrangement is that individual layers are more tilted with respect to each other in Sce-NCP (compare Figure 3B and D). The different contacts between the histone octamers in Sce-NCP pull the adjacent nucleosome upward, and thus cause the loss of the back-to-back crystal packing between nucleosomes within the same layer. This leads to the observed increase in the length of the crystallographic c-axis from 181 Å in Xla-NCP to 193 Å in Sce-NCP. The loss of this contact is evident when comparing the interaction of the central green particles with the neighboring red particles in Figure 3A and C. As a practical consequence of these changes in crystal packing, Sce-NCP crystals were extremely fragile and would often shatter at the slightest touch.
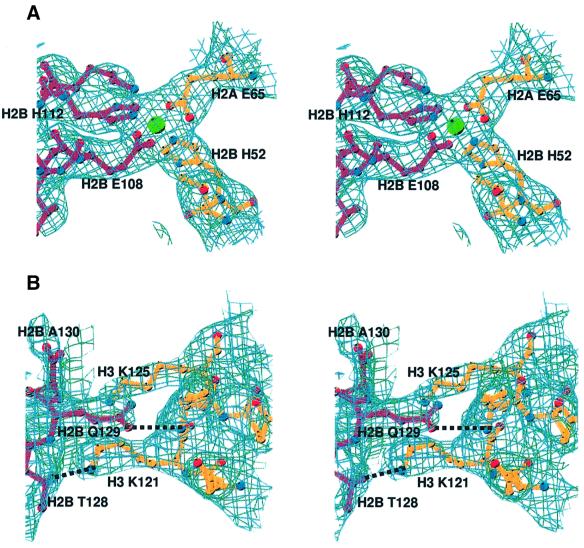
The dramatic changes in nucleosome–nucleosome interactions seen in yeast are a result of contacts involving the C-terminal helix (αC) of H2B. This extremely well ordered helix lies exposed on the surface of the histone octamer, and plays an important role in defining the surface of the nucleosome core particle (Figure 1C). In Xla-NCP, its C-terminal end is involved in some minor contacts with the DNA of a neighboring particle. In contrast, the entire helix is involved in forming essential crystal contacts in Sce-NCP (Figure 4C and D). Within one region, a manganese coordination site is formed by two histidine and two glutamate residues from two neighboring nucleosomes. Glu65 on α2 of H2A and His52 on the L1 loop of H2B chelate a manganese ion that is held in turn by H2B′ Glu108 and H2B′ His112 at the base of αC of the adjacent nucleosome (Figure 5A). If such a contact was made in vivo (and we emphasize that the involvement of these residues in in vivo nucleosome packing is not supported by any experimental evidence), zinc would probably replace manganese.
Fig. 5. Details of nucleosome–nucleosome interactions in yeast NCP. (A) Stereo view of a section of the |2Fo – Fc| electron density map, calculated at 3.1 Å and contoured at 1σ, showing an Mn2+-mediated crystal contact between H2A Glu65 and H2B His52 of one nucleosome (dark red) and H2B′ Glu108 and H2B′ His112 of the neighboring nucleosome (gold). (B) Stereo view of the hydrogen bonds between the H2B C-terminal end of H2B αC (dark red) and residues in histone H3 of a neighboring nucleosome core particle (gold), showing a section of the |2Fo – Fc| electron density map calculated at 3.1 Å and contoured at 8–1σ.
The C-terminal region of H2B αC is also crucial in producing crystal interactions. Sequence alignments shows that this region is quite divergent between yeast and higher eukaryotes. Saccharomyces cerevisiae contains two additional amino acids along with other changes on the C-terminus of H2B (Figure 1A). H2B Thr128 and Gln129 form hydrogen bonds with Lys121 and Lys125 of the H3 chain of the neighboring nucleosome (Figure 5B). None of these residues are present in X.laevis (Figure 1A). We propose that the contacts located along H2B αC are responsible for the alternative crystal packing observed in yeast.
As a result of the change in the relative position of neighboring nucleosomes within the crystal lattice (best viewed by comparing Figure 4B and D), the yeast H4 N-terminal tail is no longer able to interact with the acidic patch of the neighboring nucleosome. As discussed above, this tail (whose first 17 amino acids are too disordered to be observed in the crystal structure) is now poised to interact with the DNA of a neighboring nucleosome core particle, although no direct interactions are observed. Disorder is most probably caused by the ability of this basic tail to interact with the DNA in multiple locations. As a consequence of alternative packing, neither acidic patch is involved in crystal contacts. Instead, both regions tightly coordinate a manganese ion, using the side chains of H2A Asp90, Glu92 and Glu61 (not shown). The altered arrangement of nucleosomes within the yeast crystal lattice also brings the N-terminal tail of H2A into close proximity with the C-terminal tail of H2A of a neighboring particle (Figure 4C). Although the electron density in this region is rather weak, these tails are positioned to cross over and make crystal contacts. One contact between Sce-H2A Lys120 and Sce-H2A Thr12 in the neighboring nucleosome is clearly observed in the crystal structure. Incidentally, the long H3 N-terminal tail from yet another nucleosome core particle is headed in the same direction (data not shown). These observations further support the role of histone tails in inter-particle contacts.
Discussion
The crystal structure of the yeast nucleosome core particle shows that the overall principles of DNA organization in the nucleosome are maintained between lower and higher eukaryotes. The path of the DNA as well as the overall conformation of the structured regions of the histone proteins are conserved, as is the location of many critical structural water molecules and divalent ions (K.Luger, in preparation). Sequence variations between yeast and higher eukaryotes are distributed throughout the histones, and are located both on the surface of the histone octamer and buried deep within the nucleosome structure. The latter could be collectively responsible for a subtle destabilization of the yeast nucleosome core particle. This finding is consistent with a more open chromatin structure in yeast. Importantly, all of the residues that are critically involved in the organization of the DNA are conserved between the two organisms.
Sequence variations are distributed throughout the nucleosome structure and may result in subtle destabilization of the yeast nucleosome
The most significant changes in protein–protein interactions within the yeast nucleosome core particle, compared with nucleosomes from higher organisms (Luger et al., 1997a; Harp et al., 2000; Suto et al., 2000), are located in the H2A L1 loops where the two H2A–H2B dimers interact. We found a complete absence of all stabilizing interactions (including hydrophobic contacts) in this area in yeast. This is in contrast to four strong hydrogen bonds and salt bridges located in this region in the structures of metazoan nucleosome core particles (Luger et al., 1997a; Harp et al., 2000). How much does this relatively small interface contribute to nucleosome stability? We are currently investigating this question using fluorescence energy transfer to compare the relative stability of nucleosomes containing X.laevis and S.cerevisiae histones. Considering that this region seemingly is involved in holding together the two gyres of the DNA superhelix (Figure 1C), even a subtle destabilization could have a relatively large effect on overall nucleosome stability during transcription, which involves depletion of histone dimers from the nucleosome. This is of particular interest in view of the finding that eukaryotic RNA polymerase II prefers nucleosome cores that are depleted in one H2A–H2B dimer (Baer and Rhodes, 1983). In addition, our findings are consistent with earlier studies presenting experimental evidence for a significant destabilization of yeast nucleosome core particles, either as mononucleosomes or in arrays (Lee et al., 1982; Morse et al., 1987; Pineiro et al., 1991). The same set of residues that we have shown to be prohibitive for the formation of a stable interface is also seen in the sequences of major H2A and H2B from Schizosaccharomyces pombe. Similarly prohibitive combinations of amino acids are found only in Euglena, Leishmania and Aspergillus nidulans, and in some genes from Psammechinus miliaris (Baxevanis et al., 1995). Generally, this region is rather divergent both among species and among histone variants (Baxevanis and Landsman, 1998). We have speculated earlier that the L1 loops could provide a mechanism to ensure that the gene product from one particular H2A gene is incorporated into a single nucleosome (Suto et al., 2000). The results presented here suggest a more general role for this region in modulating nucleosome stability.
Histone tails can assume various distinct conformations
The role of the histone N-terminal tails in the regulation of transcription, exerted mainly by reversible post-translational modification of conserved lysine and serine residues, is undisputed (for a recent review see Strahl and Allis, 2000). The N-terminal tails have also been shown to be involved in the condensation of the chromatin fiber (Carruthers and Hansen, 2000), and in the formation of silent chromatin regions by interaction with other factors (Grunstein, 1997b). The detrimental effects of N-terminal tail deletions in vivo suggest that they have many different, yet partially redundant, functions (Grunstein et al., 1995; Grunstein, 1997a; Hansen et al., 1998). We show for the first time that the H4 N-terminal tail can assume completely different conformations depending upon the structural context. We have evidence that this holds true for all histone tails (unpublished data). This structural heterogeneity might allow the histone tails to interact with a variety of different protein factors and to perform a large number of different functions (Hansen et al., 1998).
Sequence variations lead to changes in nucleosome–nucleosome interactions
It is reasonable to suggest that organisms such as S.cerevisiae, which maintain a relatively open chromatin structure and lack histone H1, organize nucleosomes into higher order structures that differ significantly from that found in higher eukaryotes. The folding of nucleosomal arrays into compact fibers involves, by definition, the close packing of nucleosome cores in an energetically favorable manner. Regardless of the source of histones, this process is not dependent on linker histone H1 in vitro, but has been shown to depend on the presence of the N-terminal tails at least in higher eukaryotes (Carruthers and Hansen, 2000).
To date, the architecture of the 30 nm chromatin fiber and its higher order assemblies is not known, nor have the forces that drive the formation of these assemblies been identified. The precise molecular mechanism by which nucleosomes pack together in the context of a chromatin fiber most probably is not reflected directly in crystal packing. However, it has been shown recently in the crystal structure of bacterial flagellin that protein crystals can indeed mimic reality (Samatey et al., 2001). Our results demonstrate that internucleosomal interactions can be completely altered by very minor changes in the amino acid sequence, such as the addition of two amino acids to the C-terminal end of H2B αC, while maintaining the contacts that are mediated by the quasi-helical end-to-end stacking of the DNA. This shows that there are indeed several ways in which nucleosomes may be packaged in an energetically favorable way, and that small changes in the histone surface are sufficient to provoke these changes. This finding has important implications for our understanding of the formation of higher order structure. The role of the flexible histone tails in the formation of higher order structure and crystal packing is well established (Moore and Ausio, 1997; Carruthers and Hansen, 2000), and it has been speculated that reversible modification may be responsible for different stages of chromatin compaction. Thus, it is tempting to speculate that changes imparted by the reversible modification in histone tails provide a means to switch between different modes of nucleosome–nucleosome interactions in vivo. Furthermore, methylation of lysine residues in the structured regions of the histones could have similar effects.
The crucial involvement of the C-terminal helix of yeast H2B in internucleosomal contacts is of particular interest in light of the recent discovery that H2B Lys123 is ubiquitylated in vivo by Rad6 in yeast. Ubiquitylation is predicted to disrupt these contacts and lead to local or global effects on chromatin folding. Consistent with this idea, mutation of Lys123 in yeast leads to defects in mitioic cell growth and meiosis (Robzyk et al., 2000). Additional in vivo studies also support the notion that ubiquitylation of the H2B C-terminus has an effect on chromatin accessibility. Ubiquitylated H2B has been found to have a role in activated transcription that partially overlaps with that of the nucleosome remodeling factors Swi/Snf and SAGA (M.A.Osley, personal communication). This suggests that internucleosomal contacts that are mediated by H2B αC may indeed be involved in forming inhibitory chromatin structures in vivo in yeast, and that the degree of chromatin compaction may be regulated by Rad6-dependent ubiquitylation.
In summary, our findings confirm that the overall structure of the nucleosome core particle is conserved between yeast and higher eukaryotes. However, localized sequence variations, particularly in the L1 loop of H2A, may serve to fine-tune nucleosome stability, providing a structural basis for earlier studies comparing the stability of yeast nucleosomes with those of higher eukaryotes. Importantly, we have shown that there are different ways to form stable inter-particle contacts. We have verified that the major driving force in the formation of such contacts are interactions made between the protein moiety of nucleosomes. We further show that packing is altered in response to subtle sequence variations. This has important implications for our understanding of how the reversible modification of histone tails and structured regions may be employed to alter higher order chromatin structure.
Materials and methods
Expression, purification and reconstitution of yeast NCP
Yeast histone expression plasmids (pet28a) were a kind gift from Drs Ali Hamiche and Xuetong Shen. Proteins were overexpressed in BL21 (DE3) CodonPlus RIL– plysS (Stratagene) and purified using previously published protocols (Luger et al., 1999). The histone proteins were refolded to a histone octamer, and reconstituted into nucleosome core particles using a 146 bp palindromic DNA fragment derived from human α-satellite regions (Luger et al., 1997a). Milligram amounts of yeast nucleosome core particles were subjected to heat shifting, followed by subsequent purification using preparative gel electrophoresis (Luger et al., 1999).
Crystallographic procedures
Crystals of Sce-NCP were obtained by vapor diffusion at a protein concentration of 4 mg/ml with 70 mM KCl, 76 mM MnCl2 and 10 mM potassium cacodylate pH 6.0 in the drop equilibrated against 35 mM KCl, 38 mM MnCl2 and 5 mM potassium cacodylate pH 6.0. Macro-seeding of sitting drops increased crystal size. For data collection, the crystals were transferred into cryo-protectant and were flash frozen in liquid propane at –130°C before transferring to the cryo-stream at –180°C, as previously described (Luger et al., 1997a). Data were collected at beamline 5.0.2 at the Advanced Light Source in Berkeley. Data from two crystals were processed using Denzo and Scalepack (Otwinowski and Minor, 1997).
Molecular replacement, with Protein Data Bank entry 1AOI as the search model, was used to obtain initial phases. Refinement was performed using CNS (Brünger et al., 1997), and model building into the |2Fo – Fc| and |Fo – Fc| electron density maps was done in program O (Jones et al., 1991). The entire model was checked using SA-OMIT maps during the early stages of model building to eliminate model bias. PROCHECK analysis (Laskowski et al., 1993) of the final model shows that the model has good overall geometry, with no residues falling in the disallowed regions of the Ramachandran map. Figures were prepared using MOLSCRIPT (Kraulis, 1991), BOBSCRIPT (Esnouf, 1999), MIDAS (Ferrin et al., 1988) and O (Jones et al., 1991).
Protein Data Bank coordinates
Coordinates have been deposited with the Protein Data Bank under accession number 1ID3.
Supplementary data
Supplementary data for this paper are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We would like to thank Drs A.Hamiche (LBME-IBCG-CNRS, Toulouse, France), Xuetong Shen and Carl Wu (Laboratory of Molecular Cell Biology, NCI, NIH) for yeast histone expression plasmids, and Drs T.Earnest and Gerry McDermott at the Advanced Light Source in Berkeley for support. We are particularly grateful to Dr M.A.Osley for critical reading of the manuscript and for the kind permission to cite unpublished data. This work was supported in part by the NIH (GM61909), by a Searle Scholar Award to K.L., by the Cancer League of Colorado and by grant RG0059/2000-M from the Human Frontiers Science Program.
References
- Aalfs J.D. and Kingston,R.E. (2000) What does ‘chromatin remodeling’ mean? Trends Biochem. Sci., 25, 548–555. [DOI] [PubMed] [Google Scholar]
- Baer B.W. and Rhodes,D. (1983) Eukaryotic RNA polymerase II binds to nucleosome cores from transcribed genes. Nature, 301, 482–488. [DOI] [PubMed] [Google Scholar]
- Baxevanis A.D. and Landsman,D. (1998) Histone Sequence Database: new histone fold family members. Nucleic Acids Res., 26, 372–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxevanis A.D., Arents,G., Moudrianakis,E.N. and Landsman,D. (1995) A variety of DNA-binding and multimeric proteins contain the histone fold motif. Nucleic Acids Res., 23, 2685–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger A.T., Adams,P.D. and Rice,L.M. (1997) New applications of simulated annealing in X-ray crystallography and solution NMR. Structure, 5, 325–336. [DOI] [PubMed] [Google Scholar]
- Carruthers L.M. and Hansen,J.C. (2000) The core histone N termini function independently of linker histones during chromatin condensation. J. Biol. Chem., 275, 37285–37290. [DOI] [PubMed] [Google Scholar]
- Esnouf R.M. (1999) Further additions to MOLSCRIPT version 1.4, including reading and contouring of electron density maps. Acta Crystallogr. D, 55, 938–940. [DOI] [PubMed] [Google Scholar]
- Ferrin T.E., Huang,C.C., Jarvis,L.E. and Langridge,R. (1988) The MIDAS display system. J. Mol. Graphics, 6, 13–27. [Google Scholar]
- Finch J.T., Brown,R.S., Richmond,T.J.R., Rushton,B., Lutter,L.C. and Klug,A. (1981) X-ray diffraction study of a new crystal form of the nucleosome core showing higher resolution. J. Mol. Biol., 145, 757–769. [DOI] [PubMed] [Google Scholar]
- Glowczewski L., Yang,P., Kalashnikova,T., Santisteban,M.S. and Smith,M.M. (2000) Histone–histone interactions and centromere function. Mol. Cell. Biol., 20, 5700–5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory P.D. (2001) Transcription and chromatin converge: lessons from yeast genetics. Curr. Opin. Genet. Dev., 11, 142–147. [DOI] [PubMed] [Google Scholar]
- Grunstein M. (1997a) Histone acetylation in chromatin structure and transcription. Nature, 389, 349–352. [DOI] [PubMed] [Google Scholar]
- Grunstein M. (1997b) Molecular model for telomeric heterochromatin in yeast. Curr. Opin. Cell Biol., 9, 383–387. [DOI] [PubMed] [Google Scholar]
- Grunstein M., Hecht,A., Fisher Adams,G., Wan,J., Mann,R.K., Strahl Bolsinger,S., Laroche,T. and Gasser,S. (1995) The regulation of euchromatin and heterochromatin by histones in yeast. J. Cell Sci. Suppl., 19, 29–36. [DOI] [PubMed] [Google Scholar]
- Hansen J.C., Tse,C. and Wolffe,A.P. (1998) Structure and function of the core histone N-termini: more than meets the eye. Biochemistry, 37, 17637–17641. [DOI] [PubMed] [Google Scholar]
- Harp J.M., Hanson,B.L., Timm,D.E. and Bunick,G.J. (2000) Asymmetries in the nucleosome core particle at 2.5 Å resolution. Acta Crystallogr. D, 56, 1513–1534. [DOI] [PubMed] [Google Scholar]
- Hartzog G.A. and Winston,F. (1997) Nucleosomes and transcription: recent lessons from genetics. Curr. Opin. Genet. Dev., 7, 192–198. [DOI] [PubMed] [Google Scholar]
- Horz W. and Zachau,H.G. (1980) Deoxyribonuclease II as a probe for chromatin structure. I. Location of cleavage sites. J. Mol. Biol., 144, 305–327. [DOI] [PubMed] [Google Scholar]
- Hsu J.Y. et al. (2000) Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell, 102, 279–291. [DOI] [PubMed] [Google Scholar]
- Jackson J.D. and Gorovsky,M.A. (2000) Histone H2A.Z has a conserved function that is distinct from that of the major H2A sequence variants. Nucleic Acids Res., 28, 3811–3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.Y., Cowan,S.W. and Kjelgaard,M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kraulis P. (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Laskowski R., MacArthur,M., Moss,D. and Thornton,J. (1993) PROCHECK: a program to evaluate stereochemical quality of protein structures. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Lee K.P., Baxter,H.J., Guillemette,J.G., Lawford,H.G. and Lewis,P.N. (1982) Structural studies on yeast nucleosomes. Can. J. Biochem., 60, 379–388. [DOI] [PubMed] [Google Scholar]
- Luger K. and Richmond,T.J. (1998a) DNA binding within the nucleosome core. Curr. Opin. Struct. Biol., 8, 33–40. [DOI] [PubMed] [Google Scholar]
- Luger K. and Richmond,T.J. (1998b) The histone tails of the nucleosome. Curr. Opin. Genet. Dev., 8, 140–146. [DOI] [PubMed] [Google Scholar]
- Luger K., Maeder,A.W., Richmond,R.K., Sargent,D.F. and Richmond,T.J. (1997a) X-ray structure of the nucleosome core particle at 2.8 Å resolution. Nature, 389, 251–259. [DOI] [PubMed] [Google Scholar]
- Luger K., Rechsteiner,T.J., Flaus,A.J., Waye,M.M. and Richmond,T.J. (1997b) Characterization of nucleosome core particles containing histone proteins made in bacteria. J. Mol. Biol., 272, 301–311. [DOI] [PubMed] [Google Scholar]
- Luger K., Rechsteiner,T.J. and Richmond,T.J. (1999) Preparation of nucleosome core particle from recombinant histones. Methods Enzymol., 304, 3–19. [DOI] [PubMed] [Google Scholar]
- Luger K., Maeder,A.W., Sargent,D.F. and Richmond,T.J. (2000) The atomic structure of the nucleosome core particle. J. Biomol. Struct. Dynam., 11, 185–188. [DOI] [PubMed] [Google Scholar]
- Moore S.C. and Ausio,J. (1997) Major role of the histones H3–H4 in the folding of the chromatin fiber. Biochem. Biophys. Res. Commun., 230, 136–139. [DOI] [PubMed] [Google Scholar]
- Morse R.H., Pederson,D.S., Dean,A. and Simpson,R.T. (1987) Yeast nucleosomes allow thermal untwisting of DNA. Nucleic Acids Res., 15, 10311–10330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Patterton H.G., Landel,C.C., Landsman,D., Peterson,C.L. and Simpson,R.T. (1998) The biochemical and phenotypic characterization of hho1p, the putative linker histone H1 of Saccharomyces cerevisiae. J. Biol. Chem., 273, 7268–7276. [DOI] [PubMed] [Google Scholar]
- Pineiro M., Puerta,C. and Palacian,E. (1991) Yeast nucleosomal particles: structural and transcriptional properties. Biochemistry, 30, 5805–5810. [DOI] [PubMed] [Google Scholar]
- Robzyk K., Recht,J. and Osley,M.A. (2000) Rad6-dependent ubiquitination of histone H2B in yeast. Science, 287, 501–504. [DOI] [PubMed] [Google Scholar]
- Samatey F.A., Imada,K., Nagashima,S., Vonderviszt,F., Kumasaka,T., Yamamoto,M. and Namba,K. (2001) Structure of the bacterial flagellar protofilament and implications for a switch for supercoiling. Nature, 410, 331–337. [DOI] [PubMed] [Google Scholar]
- Santisteban M.S., Kalashnikova,T. and Smith,M.M. (2000) Histone H2A.Z regulates transcription and is partially redundant with nucleosome remodeling complexes. Cell, 103, 411–422. [DOI] [PubMed] [Google Scholar]
- Strahl B.D. and Allis,C.D. (2000) The language of covalent histone modifications. Nature, 403, 41–45. [DOI] [PubMed] [Google Scholar]
- Strahl B.D., Ohba,R., Cook,R.G. and Allis,C.D. (1999) Methylation of histone H3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in Tetrahymena. Proc. Natl Acad. Sci. USA, 96, 14967–14972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suto R.K., Clarkson,M.J., Tremethick,D.J. and Luger,K. (2000) Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nature Struct. Biol., 7, 1121–1124. [DOI] [PubMed] [Google Scholar]
- Vogelauer M., Wu,J., Suka,N. and Grunstein,M. (2000) Global histone acetylation and deacetylation in yeast. Nature, 408, 495–498. [DOI] [PubMed] [Google Scholar]
- Widom J. (1998) Structure, dynamics and function of chromatin in vitro. Annu. Rev. Biophys. Biomol. Struct., 27, 285–327. [DOI] [PubMed] [Google Scholar]
- Woodcock C.L. and Dimitrov,S. (2001) Higher-order structure of chromatin and chromosomes. Curr. Opin. Genet. Dev., 11, 130–135. [DOI] [PubMed] [Google Scholar]
- Workman J.L. and Kingston,R.E. (1998) Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu. Rev. Biochem., 67, 545–579. [DOI] [PubMed] [Google Scholar]