Abstract
The KIF1 subfamily members are monomeric and contain a number of amino acid inserts in surface loops. A particularly striking insertion of several lysine/arginine residues occurs in L12 and is called the K-loop. Two recent studies have employed both kinetic and single-molecule methods to investigate KIF1 motor properties and have produced very different conclusions about how these motors generate motility. Here we show that a hitherto unstudied member of this group, KIF1D, is not chemically processive and drives fast motility despite demonstrating a slow ATPase. The K-loop of KIF1D was analysed by deletion and insertion mutagenesis coupled with characterization by steady state and transient kinetics. Together, the results indicate that the K-loop not only increases the affinity of the motor for the MT, but crucially also inhibits its subsequent isomerization from weak to strong binding, with coupled ADP release. By stabilizing the weak binding, the K-loop establishes a pool of motors primed to undergo their power stroke.
Keywords: KIF1 family/kinesin/L12 loop/processivity/weak to strong transition
Introduction
Conventional kinesin or KHC was isolated in 1985 as a microtubule (MT)-based motor protein (Brady, 1985; Vale et al., 1985). Since then kinesin-related proteins have been isolated and are referred to as kinesins or KIFs. All are characterized by a conserved motor domain that uses ATP for directional transport of cargoes along MTs. The kinesin superfamily has been further subdivided into closely related subfamilies, on the basis of sequence similarities both inside and outside the motor domain (for reviews see Endow, 1991; Hirokawa, 1997; Hirokawa et al., 1998).
Most kinesin superfamily members are homodimeric. An exception is the KIF1/unc104 family, whose members are devoid of any substantial coiled-coil region, which together with biophysical data indicates that these proteins are naturally occurring monomeric motors. In vivo the monomeric motor KIF1A and its Caenorhabditis elegans homologue unc104 have been shown to transport synaptic vesicles in neurons (Hall and Hedgecock, 1991; Okada et al., 1995). In mouse knock-out experiments KIF1A’s absence abrogates synaptic vesicle precursor transport and consequently leads to neuronal cell death (Yonekawa et al., 1998). These KIF1A-deficient mice show motor and sensory disturbances, which result in death 24 h after birth, and hence KIF1A transport is critical for the maintenance of neurons. KIF1B has also been shown to transport mitochondria in vitro (Nangaku et al., 1994). KIF1B and KIF1A produce motility in multiple-motor assays of 0.66 and 1.5 µm/s, respectively (Nangaku et al., 1994; Okada et al., 1995). Conventional kinesin, which is a dimeric motor, is both mechanically and chemically processive, which means it can take in excess of 100 mechanical steps along the MT without detachment (Hackney, 1994; Coppin et al., 1996). Stepping is believed to occur by a ‘hand over hand’ mechanism where one head is always attached to the MT (Cross, 1999). At each step, one ATP molecule is hydrolysed and 8 nm of progress made, which may allow single kinesin molecules to transport cargoes over long distances (Hua et al., 1997; Schnitzer and Block, 1997; Coy et al., 1999). Recently, it has been shown that a 15 amino acid sequence called the neck-linker (which is C-terminal to the catalytic core and directly precedes the α-helical coiled-coil sequence) can undergo a nucleotide-dependent conformational change (Rice et al., 1999). The neck-linker can either be docked on to the catalytic core or in a ‘free’ undocked state. ATP binding to the bound head of kinesin may produce the conformational change required to drive the other free head to the next MT binding site (Cross, 2000; Vale and Milligan, 2000; Schief and Howard, 2001).
This single molecule ‘hand over hand’ mechanism cannot operate in KIF1 motility as the molecule contains only one head. This raises some important questions. Can a single kinesin head be processive in its own right by adopting an alternative mechanism or do single-headed kinesins have to work in teams in order to transport cargoes?
Recently Okada and Hirokawa (1999) observed that a modified KIF1A motor can undergo episodes of directional movement along MTs in single-molecule assays and that it hydrolyses almost 700 ATP molecules per MT interaction, with an ATPase rate of 110 s–1. This motor produced smooth 1.5 µm/s transport in multiple motor assays but the overall velocity of transport in the single-molecule assays was 0.14 µm/s. A biased-diffusion model was proposed to explain this single-molecule monomeric motor processivity (Okada and Hirokawa, 2000). In this model, single-headed KIF1A molecules are able to diffuse in both directions along the MT in a weakly bound state (ADP in the active site) but the overall motion is directional because ATP induces a directional-biased transition to the strongly bound state. Interestingly, the addition of the L12 loop of KIF1A to monomeric conventional kinesin also resulted in a processive motor with similar characteristics in single-motor assays.
A study of unc104 (a C.elegans KIF1A homologue) showed that this motor has a very low ATPase but generated smooth 1.5 µm/s transport in multiple-motor assays (Pierce et al., 1999). In contrast to the results of Okada and Hirokawa, this motor produces no motility in single-molecule assays.
A distinct feature of the KIF1 family is the addition of a number of amino acid motifs in the loop regions. A particularly striking example is the addition of several arginine/lysine residues in the L12 loop, which has been called the K-loop. Okada and Hirokawa have presented evidence that the K-loop maintains the motor–MT interaction in the ADP weakly bound state in the KIF1A motor (Kikkawa et al., 2000; Okada and Hirokawa, 2000). Interestingly, the L12 loop of conventional kinesin adopts a different conformational state when different nucleotides are in the active site, and is at the MT–motor interface (Alonso et al., 1998). In addition, the importance of the L12 loop in MT binding is illustrated by a triple mutation of residues Y274, R278 and K281 to alanine in conventional kinesin’s L12 loop, which results in a motor that binds MTs very weakly but has normal basal ATP turnover and binding rates (Woehlke et al., 1997; Shimizu et al., 2000).
We have recently isolated a hitherto unstudied member of the KIF1 family called KIF1D (Rogers et al., 1997, 1998). Here we characterize the properties of this KIF1 family member using steady state and transient kinetics. We show that this kinesin is neither chemically nor mechanically processive, and also demonstrate for the first time that the isomerization to the strongly bound nucleotide-free state is controlled by the K-loop, whose function appears to be to significantly increase the lifespan of the weakly bound state.
Results
KIF1D kinetics and motility
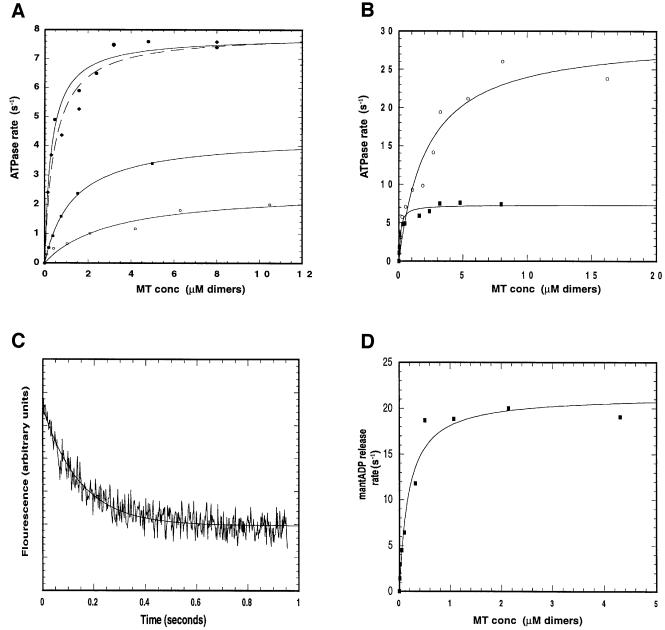
The purified KIF1D motor has a basal ATPase rate of 0.04 s–1, which is activated ∼200-fold to a maximum of 7.9 s–1 by addition of MTs. The degree of activation is sensitive to salt concentration >25 mM NaCl as shown in Figure 1A. As expected the MT concentration required for half-maximal ATPase activation (K(0.5)MT) increases from 0.08 µM at no salt to 3.1 µM at 100 mM NaCl, indicating a strong electrostatic interaction. At 200 mM salt no activation of KIF1D ATPase was evident under these conditions. The single-headed rat kinesin motor (i.e. rkin340GFP construct) has a maximum rate of 28.5 s–1 and a K(0.5)MT of 1.85 in the low ionic strength buffer [25 mM PIPES, 1 mM MgCl2, 1 mM dithiothreitol (DTT) pH 6.9]. Thus, rkin340GFP has both a higher kcat and K0.5(MT) than the naturally occurring single-headed KIF1D motor (Figure 1B), consistent with KIF1D having sequences that modify its motor domain in such a way as to specify stronger MT binding. The MT-activated ATPase rates of the single- and double-headed kinesin motors (rkin340GFP and rkin430GFP) are consistent with published results for similar constructs (Hackney, 1995; Lockhart et al., 1995b; Case et al., 2000).
Fig. 1. Kinetic properties of KIF1D. (A) Effect of salt concentration on KIF1D MT-activated ATPase. Assays in (A)–(C) were carried out in 25 mM PIPES, 1 mM MgCl2, 1 mM DTT pH 6.9 with the addition of NaCl to the specified concentration at 22°C. The motor concentration was 0.1 µM. Each data set was fitted to a rectangular hyperbola to give a maximum ATPase rate (Kcat) and the MT concentration required for half-maximal activation [K(0.5)MT]. Open circles, 100 mM NaCl; squares, 50 mM NaCl; diamonds, 25 mM NaCl; filled circles, no NaCl. (B) Comparison of the MT-activated ATPase of KIF1D and single-headed kinesin (rkin340GFP). Open circles, rkin340GFP; filled squares, KIF1D. (C) Representative stop-flow trace of KIF1D. 0.5 µM KIF1D was labelled with 2 µM mantATP and rapidly mixed with MT (20 µM). Concentrations are after mixing in the stop-flow chamber. Solid line is the best fit to a single exponential. (D) MT-activated mantADP release from the KIF1D motor. Each point is the average of three traces. Solid line shows the best fit to a rectangular hyperbola and gives the maximum mantADP release rate K(mantADP) of 21.35 s–1 and the MT concentration required for half-maximal activation K(0.5mantADP) of 0.18 µM.
During the initial part of this study we attempted to express numerous different truncations of untagged and N- and C-terminally tagged KIF1D constructs. However, all of these suffered from either very low expression levels, insolubility or degradation in the bacteria. The KIF1D construct that we developed allowed us to conduct a complete kinetic analysis.
MT-activated mantADP release
MantADP was used to measure the rate constant for transient release of ADP in KIF1D’s enzymatic cycle. The experiment mixes the KIF1D–mantADP complex with MT and chase nucleotide. At saturating MT concentration the MT-activated mantADP release rate is a measure of the ADP release step. For several other kinesins the release of ADP has been shown previously to be the rate-limiting step in the kinesin’s enzymatic cycle (Lockhart et al., 1995b; Jiang et al., 1997; Ma and Taylor, 1997). However, Gilbert et al. (1995) reported a far higher mantADP release rate of 306 s–1. The maximum rate of mantADP release for KIF1D was 21.4 s–1 with a K0.5(MT) of 0.18 µM (Figure 1C and D). This value suggests that the mantADP release step is the rate-limiting transition in KIF1D’s enzymatic cycle in the sense of being the slowest chemical transition. Note, however, that as we argue below, a preceding physical transition (a conformational change) could limit the rate of this chemical step. In addition, we found that premixing the MT with mantADP-loaded motor and the chasing with ATP resulted in no fluorescence signal and therefore all the mantADP is released in a single step.
KIF1D motility
In multiple-motor assays we found that the 373-amino-acid KIF1D motor did not produce reproducible motility. We believe this is due to the motor binding to the glass surface in an unproductive manner. We therefore used a KIF1D679 construct, which terminates at amino acid 679 of KIF1D [this corresponds to the equivalent position of amino acid 693 of unc104, as used by Pierce et al. (1999)]. This construct produced a fast motility in multiple motor assays (2.01 µm/s; see Figure 2). This is consistent with previous work that shows that both KIF1A and unc104 are fast motors in multiple-motor assays (Nangaku et al., 1994; Okada et al., 1995; Pierce et al., 1999). The ATPase rate of KIF1D is insufficient to support mechanical processivity at this speed; if the motor hydrolyses approximately eight ATP molecules per second, and has an 8 nm step distance (the distance between adjacent motor binding sites on the MT), then the maximum velocity would be 64 nm/s for a tightly coupled processive motor, which is clearly much slower than the observed rate.
Fig. 2. Histogram of MT motility of KIF1D in multiple-motor assays. Assays were carried out in 25 mM PIPES, 25 mM NaCl, 1 mM MgCl2, 1 mM DTT, 1 mM ATP, 0.1 mg/ml casein pH 6.9. Mean velocity is 2.01 ± 0.21 µM/s. Bin size, 0.1 µm/s.
Deletion of the K-loop increases both the ATPase and transient mantADP release rate of KIF1D
Sequence comparison of the KIF1 subfamily with other members of the kinesin family reveals that there are amino acid inserts in the L1, L2 and L12 loop regions. To investigate the role of this K-loop motif we deleted the lysine/arginine residues in KIF1D’s L12 loop. We also made constructs that add the KIF1D K-loop to single- and double-headed rkin (see Figure 3).
Fig. 3. Schematic representation of KIF1D and kinesin motor constructs. The wild-type L12 loop regions are shown in grey and the mutant L12 loop outlined in black. The numbers above each construct show the position of the first and last amino acids of the motor sequence. The L12 loop region is expanded to show the amino acid sequence of wild-type KIF1D and rat kinesin (rkin) together with the KIF1D-K and rkin+K mutants. GST, glutathione S-transferase; GFP, green fluorescence protein; His, His6 affinity tag.
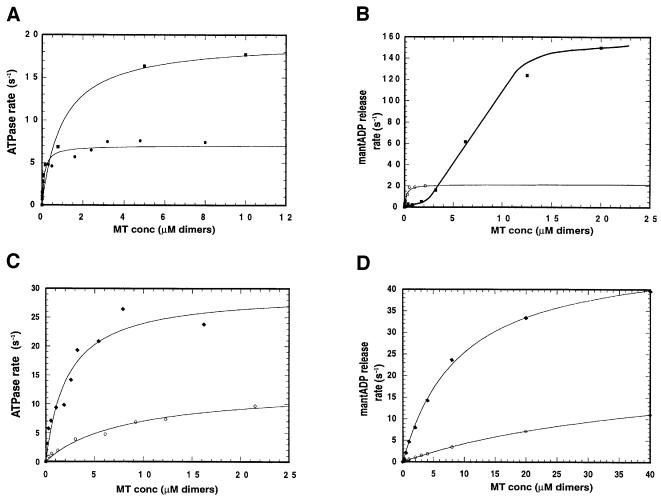
Deletion of the lysines from KIF1D had little effect on the basal ATPase rate but surprisingly increased the MT-activated ATPase by ∼2-fold to 19.3 s–1, bringing the total degree of MT activation to ∼1000-fold (Figure 4A). K0.5(MT) was increased to 0.99 µM, consistent with a decreased electrostatic attraction between the motor and MT. Furthermore, there was a dramatic effect on the MT-induced release of mantADP (Figure 4B). At low concentration of MT the ADP release rate is low, due to the low encounter rate, but at higher MT concentration the release rate is increased 10-fold to ∼298 s–1. We suggest below that deletion of the K-loop dramatically accelerates an ordinarily rate-limiting conformational change that immediately precedes ADP release.
Fig. 4. Kinetic properties of the K-loop mutants. (A) Steady state ATPase rates of KIF1D and KIF1D-K motors. The MT-activated ATPase assays were performed as in Figure 1 in the low salt buffer. Circles, KIF1D; squares, KIF1D-K. (B) MT-activated mantADP release of KIF1D and KIF1D-K. Each point is the average of at least three traces. Experiments were performed as in Figure 1D. Open circles, KIF1D; squares, KIF1D-K. (C and D) MT-activated ATPase and MT-activated mantADP release of rkin340GFP and rkin340+KGFP motors, respectively. Circles, rkin340+KGFP; diamonds, rkin340GFP.
Adding the K-loop to single- and double-headed rkin reduces the ATPase
If the removal of the K-loop from KIF1D increases its rate of mantADP release, adding the K-loop to single- and double-headed kinesin (rkin340GFP and rkin430GFP) should have the opposite effect. We made these constructs and found that the basal ATPase was largely unaffected in rkin340+KGFP, but the MT-activated ATPase rate of rkin340GFP was reduced from 28.54 s–1 in the wild type to 12.2 s–1 in rkin340+KGFP (i.e. when the K-loop was added) (Figure 4C). K0.5(MT) also increased from 1.85 to 8.2 µM. Consistent with the single-headed data the ATPase rate of the double-headed kinesin, rkin430GFP, was reduced from 30.37 to 12.3 s–1 in rkin430+KGFP. Hence, the addition of the K-loop also slows down the ATPase of double-headed kinesin. In this case, however, K(0.5)MT is almost halved (see Table I for summary of results). The single- and double-headed rkin steady state ATPase rates are similar to previously published values for similar constructs (Lockhart et al., 1995b; Case et al., 2000).
Table I. Summary of kinetic data and calculated chemical processivity index.
| MT-activated ATPase (s–1) | K0.5(MT) (µM) | Kbi(ATPase) (µM–1 s–1) | MT-activated mantADP release (s–1) | K0.5(MT) (µM) | Kbi(ADP) (µM–1 s–1) | Kbi(ratio) [Kbi(ATPase)/Kbi(ADP)] | |
|---|---|---|---|---|---|---|---|
| KIF1D | 7.9 | 0.08 | 98.7 | 21.3 | 0.18 | 106.1 | 0. 83 |
| KIF1D-K | 19.3 | 0.94 | 20.5 | ∼298 | ∼17.4 | 22.9 | 0.90 |
| rkin340GFP | 28.5 | 1.8 | 15.43 | 49.15 | 9.38 | 5.2 | 2.97 |
| rkin340+KGFP | 12.8 | 8.2 | 1.52 | 18.5 | 33 | 0.56 | 2.8 |
| rkin430GFP | 30.4 | 0.08 | 374.3 | 52.1 | 13.1 | 3.9 | 95 |
| rkin430+KGFP | 12.3 | 0.05 | 249 | 18.2 | 24.3 | 0.75 | 332 |
K0.5(MT) data have been corrected for mutual depletion.
The K-loop mutants of kinesin reduce the rate of MT-activated mantADP release
We next studied the MT-activated transient mantADP release rates for rkin340+KGFP and rkin430+KGFP. The maximum release rate for unmodified monomeric kinesin (i.e. rkin340GFP) was 49.15 s–1, with a K0.5(MT) of 9.38 µM (Figure 4B). With the mutant rkin340+KGFP the maximum rate of release was reduced to 18.5 s–1, with a K0.5(MT) of 33.2 µM (Figure 4D). For the double-headed kinesin constructs rkin430+KGFP the release rate is once again slowed from 52.1 s–1 in the rkin430GFP to 18.2 s–1 in rkin430+KGFP. A typical fluorescence trace of mantADP release is shown in Figure 5A. mantADP is released from the two heads of kinesin sequentially (Hackney, 1994; Ma and Taylor, 1997). We therefore used these double-headed constructs to test whether the rates of release of the two individual heads are affected separately. The rates of release of the mantADP from the two heads of double-headed rkin430GFP and rkin430+KGFP motors were investigated in two further experiments; first, the rate of release of mantADP from the first head binding was monitored by rapid mixing with MT (10 µM) without chase ATP nucleotide (a condition where the first head only binds the MT (Figure 5B), and secondly, preloaded motor (with mantATP) was premixed with MT (10 µM) so that just the first head binds (and releases its mantADP) but the second head is free with its mantADP in the active site. This complex was then rapidly mixed with ATP in the stopped flow, and the release of mantADP from the free head measured (Figure 5C). Results are summarized in Table II. The mantADP release rates from both heads were reduced when the K-loop was added (i.e. rkin430+KGFP) to the double-headed protein. The change in mantADP release of the second head is particularly large (7.6 versus 118.6). This rate is not affected by the MT concentration.
Fig. 5. rkin430+KGFP mantADP release kinetics and motility. (A and B) Representative fluorimeter traces of rkin430+KGFP. The motor was labelled with mantATP and rapidly mixed with: (A) MT (10 µM) with chase 1 mM ATP nucleotide; (B) MT (10 µM) alone. In (C) the motor was labelled with mantATP mixed with MTs to form a motor–MT complex. The release of the mantADP was monitored after rapid mixing with 1 mM ATP. The solid lines show the best fit to single exponentials. Assay conditions were as in Figure 3B. (D) Histogram of MT velocity supported by rkin430GFP (white bars) and rkin430+KGFP (grey bars). Bin size is 0.2 µM/s. The mean velocities of rkin430+KGFP and rkin430GFP are 139 ± 0.019 and 295 nm/s ± 0.032, respectively. Assays were carried out in BRB80 supplemented with 1 mM DTT, 0.5 mg/ml casein and 1 mM ATP.
Table II. Summary of mantADP release from rkin430GFP and rkin430+KGFP upon MT binding.
| Basal rate | mantADP release (1st and 2nd head) | mantADP release (2nd head) | mantADP release (1st head) | |
|---|---|---|---|---|
| rkin430GFP | 0.00149 | 28.9 (0.124) | 118.6 (0.076) | ND |
| rkin430+KGFP | 0.00143 | 6.78 (0.116) | 7.64 (0.066) | 5.00 (0.063) |
The release rates shown are for both the heads of double-headed kinesin. Experiments were performed as described in Materials and methods with 10 µM MTs and 1 µM motor in 25 mM PIPES, 1 mM MgCl2, 1 mM DTT pH 6.9 at 22°C.
The lysine-rich K-loop thus reduces the rate of MT-activated ADP release from both conventional kinesin and KIF1D motors. Since the lysines also increase the overall affinity for binding, the reduced rate of MT-activated release suggests strongly that a conformational change following binding is limiting the rate of ADP release (see Discussion).
The K-loop reduces the double-headed kinesin velocity of motility
The single-headed kinesin rkin340GFP and rkin340+KGFP proteins did not produce measurable motility so we analysed the motility of rkin430GFP and rkin430+KGFP. The double-headed kinesin wild-type rkin430GFP motor produced motility at 290 nm/s under present conditions. The K-loop mutant rkin430+KGFP velocity of transport was reduced to 130 nm/s (Figure 5D). The reduction in motility of the K-loop mutant is quantitatively similar to the reduction in its MT-activated ATPase activity. This is what would be expected for a tightly coupled processive motor and consistent with the mutant maintaining tight coupling.
Chemical processivity index of KIF1D and the K-loop mutants
The kinetic measurements allow us to calculate the chemical processivity index or Kbi(ratio) [KATPase/K0.5(MT) divided by KmantADP release/K0.5(MT)] of our motor constructs. This measurement was first introduced by Hackney (1995) and gives a measure of the number of ATP molecules that are hydrolysed each time the motor binds to the MT (results shown in Table I). The KIF1D motor hydrolyses 0.83 ATP molecules per MT encounter (which within experimental error shows that approximately one ATP is actually consumed), indicating therefore that the motor is not chemically processive. As hydrolysing multiple ATP molecules per MT binding event is a prerequisite for mechanical processivity we conclude that our KIF1D motor construct is also not mechanically processive. For rkin340GFP and rkin430GFP the Kbi(ratio) is 2.97 and 95, respectively, which is consistent with the previously published values of ∼4 and ∼61 for similar single- and double-headed constructs (Jiang et al., 1997). Hence, only double-headed kinesin is highly chemically processive. For the single-headed motors rkin340GFP and KIF1D, addition or removal of the K-loop did not have a significant effect on the Kbi(ratio), which indicates that the number of ATP molecules hydrolysed per MT encounter is not affected. In contrast, however, addition of the K-loop to double-headed kinesin did increase the Kbi(ratio) from 95 to 332, which indicates that the run lengths are increased in this mutant.
Solution binding assays: the K-loop increases MT affinity in the ADP-bound state
The dissociation equilibrium constant for binding of motor to MTs was analysed by pelleting assays in the nucleotide-free, ADP and AMPPNP states. Several studies have shown that kinesin interacts with the MT track in a strongly and weakly (ADP) bound state. We performed these assays with KIF1D, KIF1D-K, rkin340GFP and rkin340+KGFP, and the results are shown in Table III. The KIF1D and KIF1D-K assays were performed in 25 mM PIPES, 50 mM NaCl, 1 mM MgCl2, 1 mM DTT pH 6.9, because in the lower strength buffer the binding was very tight and hence a measure of the dissociation constant was problematic. As expected, the AMPPNP and no nucleotide (apyrase-treated) states are strongly bound. This is true for all four motor constructs. The rkin340GFP motor is in a weakly bound state when ADP is present. This motor’s properties are therefore in good agreement with binding schemes described previously for kinesins (the ATP and nucleotide-free states being strongly MT bound and the ADP state weakly MT bound) (Shimizu et al., 1995; Crevel et al., 1996; Rosenfeld et al., 1996). Interestingly, however, the addition of the K-loop (in rkin340+KGFP) decreased the dissociation constant of the ADP state so that the dissociation constants are similar for each nucleotide state. The stoichiometry suggests that between 1.3 and 2.1 heads bind to each heterodimer of the MT and the higher values may represent over-binding of the motor construct particularly in the ADP state (see Crevel et al., 1999). For the wild-type KIF1D motor, where the K-loop is present naturally, the ADP state is also strongly MT bound and the dissociation constants are again similar in the AMPPNP, ADP and no nucleotide states. Deletion of the K-loop from KIF1D also significantly reduced MT binding strength when ADP is in the active site but the other binding states were largely unaffected. The stoichiometry of binding was different for KIF1D and KIF1D-K. For KIF1D and KIF1D-K the binding was between 2 and 3, and 1.2 and 1.4 heads per tubulin heterodimer, respectively. This suggests that the K-loop may induce excess binding to the MT at high motor concentration via the E hook of both α and β tubulin. These results are in good agreement with Okada and Hirokawa and show that the lysines help to maintain a more stable contact with the MT in the weakly bound state of the cycle. In this way the weakly bound state is stabilized in the naturally occurring single-headed KIF1 monomeric motors.
Table III. Estimated dissociation constants of kinesin constructs with and without the K-loop.
| ADP (µM) | AMPPNP (µM) | Apyrase (µM) | |
|---|---|---|---|
| KIF1D | 0.62 (1.9) | 0.75 (2.7) | 0.61 (2.9) |
| KIF1D-K | 2.60 (1.3) | 0.54 (1.2) | 0.32 (1.3) |
| rkin340GFP | 6.7 (1.9) | 0.19 (1.3) | 0.15 (1.7) |
| rkin340+KGFP | 0.35 (2.1) | 0.34 (1.1) | 0.30 (1.8) |
The stoichiometry of binding is shown in brackets as heads per tubulin heterodimer.
The K-loop does not have a significant effect on dissociation of the motor–MT complex
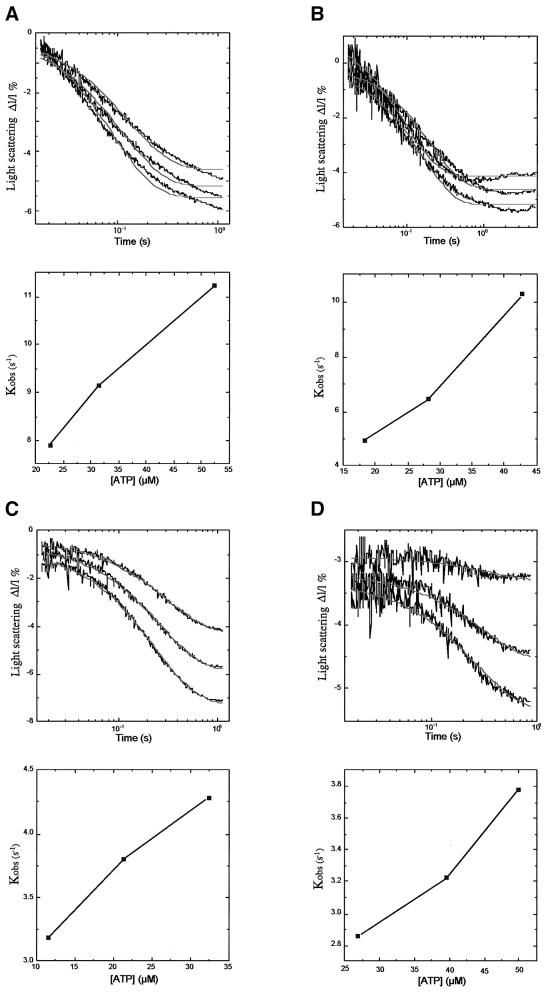
Next we tested whether the dissociation of the motor from the MT track is modulated by the K-loop by employing a flash photolysis apparatus. ATP was released from caged ATP and the dissociation of the preformed motor from the MT was monitored for the KIF1D, KIF1D-K, rKin340GFP and rkin340+KGFP motors (Figure 6A–D, respectively). The decrease in light scattering was fitted to a single exponential and the dissociation rate constants plotted against concentration of ATP released (lower panels). The single exponential fits for KIF1D and KIF1D-K were not optimal, which may suggest that an addition process is occurring in the sample. The fastest measured dissociation rate constants for the KIF1D and KIF1D-K motors were 11.4 s–1 at 52 µM ATP and 10.3 s–1 at 45 µM ATP, respectively. The removal of the K-loop from KIF1D therefore had little effect on the dissociation rate at the ATP concentrations used in this study. Signifcantly, however, these data show that the KIF1D motor dissociates from the MT at a faster rate than the overall ATPase rate. Hence the motor must be released from the MT for each ATP turnover cycle, confirming that this motor is not processive. The fastest measured rates of dissociation for rkin340GFP and rkin340+KGFP were 3.78 s–1 at 50 µM and 4.3 s–1 at 32.6 µM ATP, respectively. These data also support the conclusion that the K-loop has little effect on the process limiting the ATP-induced dissociation of the motor from the MT.
Fig. 6. Flash photolysis measurement of the decrease in light scattering of the motor–MT complex after release of ATP. (A) KIF1D; (B) KIF1D-K; (C) rkin340GFP; and (D) rkin340+KGFP. Upper traces show the light scattering versus time of the motor–MT complex (i.e. dissociation) after the release of three different concentrations of ATP by three consecutive flashes of laser light. The lower panels show the rate constants from single exponential fits of each trace plotted against the concentration of released ATP.
Discussion
In this study we have characterized the role of the positively charged residues of the L12 loop on the hitherto unstudied mechanochemistry of KIF1D.
The KIF1D motor domain is not chemically processive
Several lines of evidence indicate that KIF1D is not chemically processive, meaning that it turns over one ATP molecule per MT encounter, and then dissociates. First, KIF1D has a low maximum MT-activated ATPase (kcat). The non-processive kinesins Eg5, Ncd and unc104 have also been found to possess a low kcat when compared with conventional kinesin (Lockhart et al., 1995a; Lockhart and Cross, 1996; Pierce et al., 1999). This ATPase rate could not produce the very fast motility seen in multiple-motor assays if KIF1D was a tightly coupled processive motor. Secondly, our kinetic data, which allowed us to calculate the chemical processivity index, show that approximately one ATP molecule is hydrolysed per MT encounter. Thirdly, the dissociation rate of the motor–MT complex shows that the rate of release from the MT is faster than the overall ATPase rate, which again indicates that the motor dissociated from the MT during each ATP turnover cycle.
These results are in stark contrast to those of Okada and Hirokawa (1999), who found that mouse KIF1A hydrolysed ATP at 110 s–1, which is one of the fastest ATPase rates published for any kinesin. Our results are in agreement with Pierce et al. (1999), who concluded that the unc104 motor is not processive. We conclude that KIF1D motors must work in teams where individual members have a short duty cycle. Each individual motor would spend a large portion of its cycle not contributing to forward motion but as other members of the team are generating force, motility is maintained (see model below). In this case there is no direct relationship between the ATPase rate and velocity of motility. The concentration of MT required for half-maximal ATPase activation of KIF1D [K0.5(MT)] is significantly lower than that for engineered single-headed kinesin (i.e. rkin340GFP; see Figure 1B). Hence, the single-headed KIF1D motor has a higher affinity for MT through its enzymatic cycle than rkin340GFP. K0.5(MT) of KIF1D is also strongly affected by salt concentration. In this respect our results are consistent with the findings of Okada and Hirokawa (2000), and show that specific amino acid sequences in the KIF1 family are involved in increasing the MT affinity via an ionic interaction. However, our conclusion that KIF1D is not processive (and those obtained with unc104) is somewhat different to the single molecule and kinetic data presented by Okada and Hirokawa on KIF1A. These authors provide elegant and convincing evidence that KIF1A is both chemically and mechanically processive via a biased-diffusional mechanism. Essentially this model is based around the K-loop maintaining association with the MT in the weakly bound state via an electrostatic interaction with the MT’s charged C-terminal tail (called the E hook). The exact reason for these discrepancies is not clear but there are a number of possible explanations, some of which have been discussed elsewhere (Bloom, 2001). The most likely reason is the difference in the constructs used for the studies. The construct used by Okada and Hirokawa was a fusion protein between the first 320 amino acids of murine KIF1A and 23 amino acids (amino acids 323–344) of murine kinesin (Okada and Hirokawa, 1999). This is particularly important as the 23 amino acids encompass conventional kinesin’s neck-linker and the start of the neck-coil region. The neck-linker has been shown to undergo a large nucleotide-dependent conformational change that results in it docking and undocking the motor core (Rice et al., 1999), and is critical for processivity and motility (Case et al., 2000; Tomishige and Vale, 2000). The neck-coil contains a highly positively charged motif (the KIF1A constructs correspond to Drosphila kinesin 351, which is equivelent to mouse amino acid 344, which includes the first three lysines of the charged motif). As the neck-coil is joined directly to the neck-linker, which undergoes a large conformational change, its position relative to the motor core and tubulin molecule undergoes a substantial change during the enzymatic cycle. It is not clear whether such a conformational change occurs in the native KIF1 family neck-linker, but the charged neck-coil region is certainly absent. As both the KIF1D and unc104 constructs do not contain the conventional kinesin neck-linker it is possible that this region as well as the K-loop is involved in KIF1A single-headed processivity. Significantly, the neck-coil has also been shown to bind the E hook of MTs and, furthermore, Tomishige and Vale (2000) have also observed biased-diffusional behaviour of double-headed kinesin that has had its neck-linker immobilized by an engineered disulfide crosslink. In addition, the mutants produced by Okada and Hirokawa (2000), where the KIF1A’s L12 loop is substituted for the single-headed kinesin L12 loop, also demonstrated biased-diffusional motility. Since the biased diffusion observed is not unique to KIF1A fusion it is possible that Okada and Hirokawa by addition of the conventional kinesin sequence have artificially created a situation that exaggerates biased-diffusional behaviour and in this way have identified an important aspect of KIF1 and possibly other kinesin’s force-generating mechanism. Due to the rather flexible nature of the E hook both the KIF1A and crosslinked double-headed kinesin mechanisms would probably produce random behaviour, which fits well with the observed erratic nature of the biased diffusion motion. Also consistent with the E hook interaction with the neck-coil and its importance in processivity is the finding that increasing or decreasing its positive charge increases or decreases kinesin run lengths, respectively.
The production of untagged or modified KIF1D protein is extremely problematic and hence we used a glutathione S-transferase (GST)-tagged version of the motor in these studies as this allowed us to perform a complete analysis. The difficulty in producing unmodified KIF1A has also been reported in Okada and Hirokawa (1999). The GST construct used to express the KIF1D protein does tend to dimerize. We believe, however, that this does not invalidate the kinetic data presented here, as the KIF1D preparation behaves kinetically as a monomer, indicating no communication between the heads in the antiparallel dimerization (as shown by mantADP release). Further more, several studies have used GST-tagged proteins to study kinesin and show that the GST tag does not invalidate the kinetic data (Lockhart and Cross, 1994, 1996; Endow and Waligora, 1998; Crevel et al., 1999; Endow and Higuchi, 2000). We believe that the conclusion that KIF1D behaves as a non-processive monomer is robust, simply because it is difficult to imagine how antiparallel dimerization of a processive monomer would abolish processivity.
Despite these arguments, however, it remains possible that some of KIF1D’s properties are affected by the GST tag and future work will be directed to producing full-length unmodified KIF1D.
The K-loop reduces the transition to the strongly MT-bound state and thus stabilizes the K-loop state
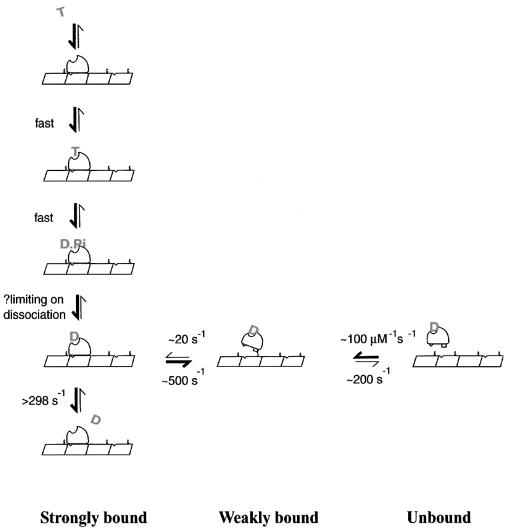
Our data indicate a crucial role for the K-loop in determining mechanism. Even though the K-loop of KIF1D is less positively charged than in other KIF1s, it nonetheless considerably increases the affinity of the motor for the MT, as assayed both by deletion of the loop from KIF1D and insertion of the loop into rkin. The increase in affinity coupled with the decrease in the rate of transition to strong binding indicates that the K-loop stabilizes an attached state that is distinct from, and precedes, the strong binding state. By stabilizing this ‘K-loop state’, the K-loop slows down the transition into strong binding, and thereby slows down MT-activated ADP release. The processivity index, however, is largely unaffected and indicates that one ATP molecule is still hydrolysed for each MT encounter. Existing model mechanisms for kinesin family motors have a weak to strong binding transition, which is coupled to ADP release. MT binding biases the motor conformation in such a way that ADP is released, and the motor–MT complex drops into its lowest energy ground state (apo or rigor state). Subsequent ATP binding, hydrolysis and Pi release drive the motor out of its ground state and progressively back into its weakly binding/detaching K.ADP conformation. Figure 7 shows a kinetic scheme incorporating KIF1D’s features. In the scheme, the K.ADP state is assigned as the dissociating state, based on the present finding for KIF1D, and on previous findings for other kinesins that K.ADP is the weakest binding state (Cross et al., 2000). There is no information about dissociation from other states of KIF1D, e.g. the K.ADP.Pi state, but as we show, a consistent scheme can be built on the simplest assumption that dissociation is significant from the K.ADP state. Rebinding will occur dominantly from K.ADP, because the K.ADP state overwhelmingly dominates in solution (Hackney, 1994). In this scheme, binding to and release from the MTs occur via a non-stereospecific (weak) binding state that is stabilized by the K-loop.
Fig. 7. A two-step binding model for KIF1D–MT interaction. The motor binds the MT in two steps; first, the weakly bound or diffusional state followed by the strongly bound state. Critically the rate of isomerization into the strongly bound state is slow and therefore increases the length of time the motor spends in the M–ADP weakly bound state. The rate constant for isomerization from weakly bound to strongly bound M–ADP is estimated based on the MT-activated mantADP release rate of KIF1D; we argue that the isomerization is rate limiting in this assay. The release of mantADP from the strongly bound motor is assumed to be ∼298 s–1 from MT-activated mantADP release rates from KIF1D-K. Other steps are assumed to be fast, except phosphate release, which may set a limit on dissociation.
This model introduces an important new aspect to KIF1D’s kinetic cycle; the binding and release from the MT are two-step processes, as postulated by Geeves et al. (1984) for binding and release of myosin from actin. The new scheme is only obviously necessary when the motor lingers in the weak binding state, as in the present case. Thus, upon binding the MT the motor is held in a weakly bound or diffusional K-loop state. The motor then undergoes an isomerization into the strongly bound state, which then allows the subsequent rapid chemical step in which ADP is released from the motor. This step is measured in the mantADP release experiments. The isomerization into the strongly bound state is slow when the K-loop is present and is the step that is effectively measured in the mantADP release experiments. Removing the K-loop from KIF1D eliminates this inhibition and the second fast chemical step of ADP release becomes limiting.
We thus propose that increasing the K-loop–E hook interaction reduces the rate of the conformational change to strong binding that is required to activate ADP release. This proposal is supported by the double-headed kinesin results, where introduction of the K-loop reduces the velocity of motility and slows the ATPase rate but increases the chemical processivity by increasing MT affinity. This result is similar to that of Tomishige and Vale (2000), who found that increasing the positive charge of the neck-coil increased kinesin’s run lengths.
Through this mechanism we conclude that the K-loop has two important roles in motor kinetics; first, to increase recruitment of the motor for the MT and secondly, to slow down the transition from the strongly to weakly bound states, establishing a pool of motors primed to undergo their power stroke.
Functional implications: model of single-headed kinesin transport
What is the biological role of the K-loop? The slowing of the transition to the strongly bound state may have significant implications for the mechanism of force generation and is an important part of our model (see Figure 8). If the KIF1 single-headed kinesin has to work in teams them it may be advantageous to maintain the motor in a conformation that is still associated with the MT but in a weak or diffusional state. This may be important for generating smooth movement if single-headed kinesin were to work in small teams. If some members of the team are in the strongly bound state, it will be favourable for the other members of the team to be in a weakly bound or diffusional state (as demonstrated by Okada and Hirokawa, 2000), as they would otherwise act as a drag on the strongly bound motors. A particularly attractive aspect of this model is that it would reduce unproductive diffusional behaviour seen in single molecule assays of KIF1A and could explain the difference in velocity seen in single- and multiple-motor measurements. As the cargo is held in close proximity to the MT by the K-loop interaction of the weakly bound motors, further members of the team can be recruited quickly to perform the next power stroke required to generate continued forward motion. Hence, if forward motion stops, transport of the cargo would pause but the cargo would not dissociate from the MT, which could allow the time for other motors to go into a strongly bound state and generate force to continue cargo transport. Hence, this mechanism may represent an adaptation to working in a small group of motors and the strength of the K-loop–E hook interaction would be expected to define the minimum team competent to drive sustained motion of the cargo along the track.
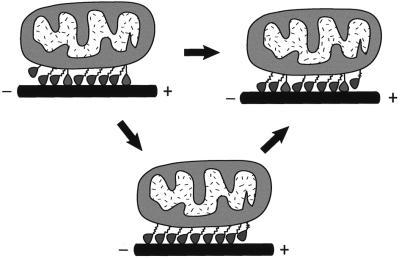
Fig. 8. Model of KIF1 cargo transport. We propose that the monomeric motors work in small teams (or arrays) on the cargo that is transported (e.g. a mitochondria). A large proportion of the motors in the team are in the weakly bound state (tilted, slack tether), with the K-loop tethering the cargo to the MT and inhibiting the transition to the strongly bound state. They do not exert significant drag on forward motion. Once strongly bound the motors undergo a power stroke to produce forward motion after which they revert to the weakly bound K-loop-tethered diffusional state. To maintain further forward motion, other weakly bound motors are recruited to undergo their strong interaction and consequent power stroke. If traction is lost the weakly bound motor maintains close association with the MT until other heads are recruited, thus maintaining cargo transport over longer distances.
Materials and methods
Clones
All molecular biology procedures were carried out using standard methods with the manufacturer’s recommended enzyme buffers. All primers for constructing expression plasmids were purchased from MWG-Biotech (Germany). The KIF1D–GST clone was constructed by PCR amplification of the KIF1D cDNA using pfu polymerase (Stratagene Europe, The Netherlands) between nucleotides 1 and 1119 (i.e. encoding amino acids 1–373) of the open reading frame. The forward and reverse primers were P1, 5′-GGATCCGGTGCCTCCGTGAAAGTG-3′ and P2, 5′-GTCGACTTAGTGTAGGTGTAGGTGTAGCCGGGCCACCTCCTCCTGCAG-3′, respectively. The forward primers introduced a BamHI site (shown in bold) before nucleotide 1, and the reverse primer introduced six codons for histidine after nucleotide 1119 followed by a stop codon (shown in italics) and a SalI restriction site. The PCR product was cloned directly into pGEMT vector (Promega, Southampton, UK) and sequenced (Oswel custom sequencing, Southampton, UK) to confirm the absence of unwanted nucleotide changes. The insert encoding for KIF1D was excised using BamHI and SalI restriction enzymes (New England Biolabs, Hitchin, UK) and ligated into pGEX2T vector digested with BamHI–XhoI restriction enzymes. A further similar construct that terminates at amino acid 679 of KIF1D was made as above with the reverse primer of 5′-GTCGACTTAGTGTAGGTGTAGGTGTAGGCGCTTGTCAGAGTCTTCGTTGTGCTTCTG-3′. Unless otherwise stated all experiments were performed with the KIF1D373 construct.
KIF1D-K was constructed by PCR mutagenesis (Higuchi, 1989) using the original KIF1D cDNA as a template and the forward and reverse primers P3, 5′-TCGGACTTCATCCCTTACAGAGACTCT-3′ and P4, 5′-TGATTGCAAATCTGCCAGGGCTGA-3′ in combination with the P1 and P2 primers. The final PCR product was again cloned into pGEMT for sequencing and subcloned into pGEX2T as above.
We also created a number of other KIF1D constructs: N-termninally His6-tagged as well as C-terminally His6- and GST-tagged KIF1D357. In addition, several alternative length deletions were made: KIF1D 360, 376 and 375 for the C-terminally tagged versions. However, none of these constructs when expressed in Escherichia coli yielded sufficient protein to allow a complete kinetic analysis. We also attempted to remove the GST tag from KIF1D using the thrombin site that is present in the pGEX2T vector, but despite using numerous different cleavage conditions were unable to retrieve the intact motor in this way due to internal cleavage.
rkin340GFP and rkin430GFP were generated by PCR using forward primer P5, 5′-CAAGACTGCATATGGCGGATCCAGCCGAATGCAGC-3′, which introduces an NdeI restriction site at nucleotide 1 of rkin coding sequence. The reverse primers for rkin340GFP and rkin430GFP were P6, 5′-AGTATCGTCGACGGTACCCTTGTCATCCAGCTGTCGGTACAG-3′ and P7, 5′-AGTATCGTCGACGGTACCCTCTTTCTCTTTTTCATATTTCTTC-3′, respectively, and introduce a KpnI restriction site after nucleotides 1020 and 1290. The PCR product was purified using Qiex resin (Qiagen, Crawley, UK), digested with NdeI/KpnI restriction enzymes and ligated into U653GTF vector (a kind gift from Prof. R.Vale) digested with NdeI/KpnI restriction enzymes. When translated the constructs code for rkin 340 or 430 followed by glycine and threonine residues and the S65T GFP protein (Case et al., 2000).
rkin430+KGFP were generated using PCR mutagenesis employing the original rkin cDNA with primers P5 and P7 and the additional forward and reverse primers P8, 5′-CAAAAAAGAAAAAGAAGAAGACACATGTGCCATACCGGGAC-3′ and P9, 5′-CATGTGTCTTCTTTTTCTTTTTTGTCCCTTCTGCCAAGGCAG-3′. The PCR products were cloned into U653GTF as described above. rkin340+KGFP was generated by PCR amplification of the rkin430+KGFP plasmid with the P5 and P6 primers and subcloned as described. All constructs were sequenced to confirm the absence of unwanted nucleotide changes. A schematic diagram of the construct is shown in Figure 3.
Expression of constructs
Expression was performed in freshly transformed BL21 (DE3) cells carrying the plasmids. Cells were grown overnight and seeded at a 1:100 dilution into fresh 2× YT media supplemented with 100 µg/ml ampicillin. The cells were then grown at 37°C with vigorous shaking until the absorbance at 595 nm reached 0.8. The cells were then cooled to 20°C and expression induced by addition of 0.25 mM isopropyl-β-d-thio galactopyranoside. The cells were harvested by centrifugation after a further 6 h incubation, flash frozen in liquid nitrogen and stored at –70°C.
Protein purification
KIF1D and KIF1D-K bacterial lysates were generated essentially as described previously (Crevel et al., 1999). Briefly, bacterial cell pellets were resuspended at a ratio of 1 g to 3 ml of buffer A (50 mM sodium phosphate, 100 mM NaCl, 5 mM MgCl2, 10 µM ATP, 5 mM mercaptoethanol pH 8) supplemented with protease inhibitor cocktail (Boehringer Mannheim). The sample was homogenized and incubated on ice with 0.1 mg/ml lysozyme for 10 min. Deoxyribonuclease I (40 µg/ml) and Triton X-100 (0.1%) were added and the sample incubated for a further 20 min. The lysate was clarified by centrifugation (27 000 g for 50 min). The soluble supernatant fraction was mixed with 5 ml of pre-equilibrated GST–Sepharose (Pharmacia Biotech, St Albans, UK). This was mixed in a 200 ml bottle for 20 min on a rolling platform at 4°C. The column matrix was packed into a column and washed with 50 ml of 50 mM sodium phosphate, 300 mM NaCl, 5 mM MgCl2, 5 mM mercaptoethanol, 1 mM ATP pH 6.9. The column was further washed with 50 ml of buffer A and eluted with 25 mM glutathione in buffer A. The peak fractions were pooled, diluted 1:1 in buffer A and loaded on to a 5 ml column of Talon metal affinity resin (Clontech, Basingstoke, UK). The column was washed extensively and eluted with buffer A supplemented with 100 mM imidazole. Pooled peak fractions were concentrated on a HiTrapSP column eluted with a step increase in NaCl and buffer immediately exchanged into 25 mM PIPES, 100 mM NaCl, 1 mM MgCl2, 1 mM DTT, 10 µM ATP pH 6.9 using a Macrosep 50K spin column (Filtron, UK).
rkin340GFP, rkin430GFP, rkin340+KGFP and rkin430+KGFP cell pellets were all lysed as above in buffer B (50 mM sodium phosphate, 100 mM NaCl, 1 mM MgCl2, 10 µM ATP, 5 mM mercaptoethanol pH 7.4). The soluble fraction was then loaded on to a pre-equilibrated 5 ml Talon affinity column and washed extensively with buffer B and eluted in buffer B supplemented with 100 mM imidazole. The peak fractions were pooled, diluted 1:1 with buffer B and loaded on to a 1 ml HiTrap Q column. The column was washed with 20 ml of 50 mM PIPES, 50 mM NaCl, 5 mM MgCl2, 1 mM DTT pH 6.9. The proteins were then eluted by increasing salt steps of 50 mM NaCl increments. All protein concentrations were determined using absorbance measurements at 280 nm with the appropriate extinction coefficient. Proteins were made up to 30% glycerol and flash frozen and stored in liquid nitrogen.
Kinetic assays and other methods
MTs were purified from fresh porcine brain and assembled just prior to use as described previously (Crevel et al., 1999). ATPase activity, transient kinetics using a stopped-flow apparatus, pelleting assays and motility assays were all measured using standard previously described methods (Crevel et al., 1999). For fluorescence traces with mantADP release we found no energy transfer to GFP in rkin340GFP and rkin430GFP constructs.
Flash photolysis to assay MT–motor dissociation
The dissociation rate constants of motor–MT complex were measured by assessing the decrease in light scattering using a custom-built apparatus described in Weiss et al. (2000). MT–motor complex was formed with 0.5 µM motor and 0.5 µM MT in 25 mM PIPES, 2 mM MgCl2, 1 mM DTT pH 6.9 with 0.1 U of apyrase to remove any residual ATP/ADP in the solution. Caged ATP was then added to 1 mM and the sample loaded into the apparatus. ATP was released by flashing with 355 nm laser light. Light scattering was monitored at 90° using a 100 W halogen lamp. Dissociation rates were measured at three different ATP concentrations by varying the intensity of the laser flash. Rate constants were calculated by using single exponential fits to the decrease in light scattering.
References
- Alonso M.C., van Damme,J., Vandekerckhove,J. and Cross,R.A. (1998) Proteolytic mapping of kinesin/ncd-microtubule interface: nucleotide-dependent conformational changes in the loops L8 and L12. EMBO J., 17, 945–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom G.S. (2001) The unc104/KIF1 family of kinesins. Curr. Opin. Cell Biol., 13, 36–40. [DOI] [PubMed] [Google Scholar]
- Brady S.T. (1985) A novel brain ATPase with properties expected for the fast axonal transport motor. Nature, 317, 73–75. [DOI] [PubMed] [Google Scholar]
- Case R.B., Rice,S., Hart,C.L., Ly,B. and Vale,R.D. (2000) Role of the kinesin neck linker and catalytic core in microtubule-based motility. Curr. Biol., 10, 157–160. [DOI] [PubMed] [Google Scholar]
- Coppin C.M., Finer,J.T., Spudich,J.A. and Vale,R.D. (1996) Detection of sub-8-nm movements of kinesin by high-resolution optical-trap microscopy. Proc. Natl Acad. Sci. USA, 93, 1913–1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coy D.L., Wagenbach,M. and Howard,J. (1999) Kinesin takes one 8-nm step for each ATP that it hydrolyzes. J. Biol. Chem., 274, 3667–3671. [DOI] [PubMed] [Google Scholar]
- Crevel I.M., Lockhart,A. and Cross,R.A. (1996) Weak and strong states of kinesin and ncd. J. Mol. Biol., 257, 66–76. [DOI] [PubMed] [Google Scholar]
- Crevel I., Carter,N., Schliwa,M. and Cross,R. (1999) Coupled chemical and mechanical reaction steps in a processive Neurospora kinesin. EMBO J., 18, 5863–5872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross R.A. (1999) Walking talking heads. Curr. Biol., 9, R854–R856. [DOI] [PubMed] [Google Scholar]
- Cross R.A. (2000) Molecular motors: kinesin’s dynamically dockable neck. Curr. Biol., 10, R124–R126. [DOI] [PubMed] [Google Scholar]
- Cross R.A., Crevel,I., Carter,N.J., Alonso,M.C., Hirose,K. and Amos,L.A. (2000) The conformational cycle of kinesin. Phil. Trans. R. Soc. Lond. B, 355, 459–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endow S.A. (1991) The emerging kinesin family of microtubule motor proteins. Trends Biochem. Sci., 16, 221–225. [DOI] [PubMed] [Google Scholar]
- Endow S.A. and Higuchi H. (2000) A mutant of the motor protein kinesin that moves in both directions on microtubules. Nature, 406, 913–916. [DOI] [PubMed] [Google Scholar]
- Endow S.A. and Waligora K.W. (1998) Determinants of kinesin motor polarity. Science, 281, 1200–1202. [DOI] [PubMed] [Google Scholar]
- Geeves M.A., Goody,R.S. and Gutfreund,H. (1984) Kinetics of acto–S1 interaction as a guide to a model for the crossbridge cycle. J. Muscle Res. Cell Motil., 5, 351–361. [DOI] [PubMed] [Google Scholar]
- Gilbert S.P., Webb,M.R., Brune,M. and Johnson,K.A. (1995) Pathway of processive ATP hydrolysis by kinesin. Nature, 373, 671–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackney D.D. (1994) Evidence for alternating head catalysis by kinesin during microtubule-stimulated ATP hydrolysis. Proc. Natl Acad. Sci. USA, 91, 6865–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackney D.D. (1995) Highly processive microtubule-stimulated ATP hydrolysis by dimeric kinesin head domains. Nature, 377, 448–450. [DOI] [PubMed] [Google Scholar]
- Hall D.H. and Hedgecock,E.M. (1991) Kinesin-related gene unc-104 is required for axonal transport of synaptic vesicles in C. elegans. Cell, 65, 837–847. [DOI] [PubMed] [Google Scholar]
- Higuchi R. (1989) Using PCR to engineer DNA. In Erlich,H.A. (ed.), PCR Technology. Stockton Press, London, UK, pp. 61–70.
- Hirokawa N. (1997) The mechanisms of fast and slow transport in neurons: identification and characterization of the new kinesin superfamily motors. Curr. Opin. Neurobiol., 7, 605–614. [DOI] [PubMed] [Google Scholar]
- Hirokawa N., Noda,Y. and Okada,Y. (1998) Kinesin and dynein superfamily proteins in organelle transport and cell division. Curr. Opin. Cell Biol., 10, 60–73. [DOI] [PubMed] [Google Scholar]
- Hua W., Young,E.C., Fleming,M.L. and Gelles,J. (1997) Coupling of kinesin steps to ATP hydrolysis. Nature, 388, 390–393. [DOI] [PubMed] [Google Scholar]
- Jiang W., Stock,M.F., Li,X. and Hackney,D.D. (1997) Influence of the kinesin neck domain on dimerization and ATPase kinetics. J. Biol. Chem., 272, 7626–7632. [DOI] [PubMed] [Google Scholar]
- Kikkawa M., Okada,Y. and Hirokawa,N. (2000) 15 Å resolution model of the monomeric kinesin motor, KIF1A. Cell, 100, 241–252. [DOI] [PubMed] [Google Scholar]
- Lockhart A. and Cross,R.A. (1994) Origins of reversed directionality in the ncd molecular motor. EMBO J., 13, 751–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart A. and Cross,R.A. (1996) Kinetics and motility of the Eg5 microtubule motor. Biochemistry, 35, 2365–2373. [DOI] [PubMed] [Google Scholar]
- Lockhart A., Crevel,I.M. and Cross,R.A. (1995a) Kinesin and ncd bind through a single head to microtubules and compete for a shared MT binding site. J. Mol. Biol., 249, 763–771. [DOI] [PubMed] [Google Scholar]
- Lockhart A., Cross,R.A. and McKillop,D.F. (1995b) ADP release is the rate-limiting step of the MT activated ATPase of non-claret disjunctional and kinesin. FEBS Lett., 368, 531–535. [DOI] [PubMed] [Google Scholar]
- Ma Y.Z. and Taylor,E.W. (1997) Interacting head mechanism of microtubule-kinesin ATPase. J. Biol. Chem., 272, 724–730. [DOI] [PubMed] [Google Scholar]
- Nangaku M., Sato-Yoshitake,R., Okada,Y., Noda,Y., Takemura,R., Yamazaki,H. and Hirokawa,N. (1994) KIF1B, a novel microtubule plus end-directed monomeric motor protein for transport of mitochondria. Cell, 79, 1209–1220. [DOI] [PubMed] [Google Scholar]
- Okada Y. and Hirokawa,N. (1999) A processive single-headed motor:kinesin superfamily protein KIF1A. Science, 283, 1152–1157. [DOI] [PubMed] [Google Scholar]
- Okada Y. and Hirokawa,N. (2000) Mechanism of the single-headed processivity: diffusional anchoring between the K-loop of kinesin and the C terminus of tubulin. Proc. Natl Acad. Sci. USA, 97, 640–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y., Yamazaki,H., Sekine-Aizawa,Y. and Hirokawa,N. (1995) The neuron-specific kinesin superfamily protein KIF1A is a unique monomeric motor for anterograde axonal transport of synaptic vesicle precursors. Cell, 81, 769–780. [DOI] [PubMed] [Google Scholar]
- Pierce D.W., Hom-Booher,N., Otsuka,A.J. and Vale,R.D. (1999) Single-molecule behavior of monomeric and heteromeric kinesins. Biochemistry, 38, 5412–5421. [DOI] [PubMed] [Google Scholar]
- Rice S. et al. (1999) A structural change in the kinesin motor protein that drives motility. Nature, 402, 778–784. [DOI] [PubMed] [Google Scholar]
- Rogers K.R., Griffin,M. and Brophy,P.J. (1997) The secretory epithelial cells of the choroid plexus employ a novel kinesin-related protein. Brain Res. Mol. Brain Res., 51, 161–169. [DOI] [PubMed] [Google Scholar]
- Rogers K.R., Griffin,M. and Brophy,P.J. (1998) The secretory epithelial cells of the choroid plexus employ a novel kinesin-related protein. Brain Res. Mol. Brain Res., 55, 355. [DOI] [PubMed] [Google Scholar]
- Rosenfeld S.S., Rener,B., Correia,J.J., Mayo,M.S. and Cheung,H.C. (1996) Equilibrium studies of kinesin–nucleotide intermediates. J. Biol. Chem., 271, 9473–9482. [DOI] [PubMed] [Google Scholar]
- Schief W.R. and Howard,J. (2001) Conformational changes during kinesin motility. Curr. Opin. Cell Biol., 13, 19–28. [DOI] [PubMed] [Google Scholar]
- Schnitzer M.J. and Block,S.M. (1997) Kinesin hydrolyses one ATP per 8-nm step. Nature, 388, 386–390. [DOI] [PubMed] [Google Scholar]
- Shimizu T., Sablin,E., Vale,R.D., Fletterick,R., Pechatnikova,E. and Taylor,E.W. (1995) Expression, purification, ATPase properties, and microtubule-binding properties of the ncd motor domain. Biochemistry, 34, 13259–13266. [DOI] [PubMed] [Google Scholar]
- Shimizu T., Thorn,K.S., Ruby,A. and Vale,R.D. (2000) ATPase kinetic characterization and single molecule behavior of mutant human kinesin motors defective in microtubule-based motility. Biochemistry, 39, 5265–5273. [DOI] [PubMed] [Google Scholar]
- Tomishige M. and Vale,R.D. (2000) Controlling kinesin by reversible disulfide cross-linking. Identifying the motility-producing conform ational change. J. Cell Biol., 151, 1081–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale R.D. and Milligan,R.A. (2000) The way things move: looking under the hood of molecular motor proteins. Science, 288, 88–95. [DOI] [PubMed] [Google Scholar]
- Vale R.D., Reese,T.S. and Sheetz,M.P. (1985) Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell, 42, 39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss S., Chizhov,I. and Geeves,M.A. (2000) A flash photolysis fluorescence/light scattering apparatus for use with sub microgram quantities of muscle protein. J. Muscle Res. Cell Motil., 21, 423–432. [DOI] [PubMed] [Google Scholar]
- Woehlke G., Ruby,A.K., Hart,C.L., Ly,B., Hom-Booher,N. and Vale,R.D. (1997) Microtubule interaction site of the kinesin motor. Cell, 90, 207–216. [DOI] [PubMed] [Google Scholar]
- Yonekawa Y., Harada,A., Okada,Y., Funakoshi,T., Kanai,Y., Takei,Y., Terada,S., Noda,T. and Hirokawa,N. (1998) Defect in synaptic vesicle precursor transport and neuronal cell death in KIF1A motor protein-deficient mice. J. Cell Biol., 141, 431–441. [DOI] [PMC free article] [PubMed] [Google Scholar]