Abstract
The crystal structure of elongation factor 1α from the archaeon Sulfolobus solfataricus in complex with GDP (SsEF-1α⋅GDP) at 1.8 Å resolution is reported. As already known for the eubacterial elongation factor Tu, the SsEF-1α⋅GDP structure consists of three different structural domains. Surprisingly, the analysis of the GDP-binding site reveals that the nucleotide– protein interactions are not mediated by Mg2+. Furthermore, the residues that usually co-ordinate Mg2+ through water molecules in the GTP-binding proteins, though conserved in SsEF-1α, are located quite far from the binding site. [3H]GDP binding experiments confirm that Mg2+ has only a marginal effect on the nucleotide exchange reaction of SsEF-1α, although essential to GTPase activity elicited by SsEF-1α. Finally, structural comparisons of SsEF- 1α⋅GDP with yeast EF-1α in complex with the nucleotide exchange factor EF-1β shows that a dramatic rearrangement of the overall structure of EF-1α occurs during the nucleotide exchange.
Keywords: archaeal EF-1α/crystal structure/magnesium ion/nucleotide exchange/Sulfolobus solfataricus
Introduction
Among all biological processes, protein synthesis is one of the most attractive; one of the central steps in protein biosynthesis is the elongation cycle (Nyborg and Liljas, 1998; Al-Karadaghi et al., 2000). In this process, a pivotal role is played by the elongation factors (EFs) Tu in eubacteria (Sprinzl, 1994) and EF-1α in eukarya and archaea (Klink, 1985; Merrick and Nyborg, 2000) that carry the aminoacyl-tRNA to the ribosome. In performing their physiological functions, these proteins, switching from an inactive GDP-bound to an active GTP-bound form, interact with a variety of cellular components (Abel and Jurnak, 1996). The activation is catalysed by the nucleotide exchange factor EF-Ts in eubacteria and EF-1β in eukarya and archaea (Klink, 1985). In the GTP-bound form, the elongation factors EF-Tu and EF-1α bind the aminoacyl-tRNA. This ternary complex interacts with the ribosome, which activates the site for GTP hydrolysis located on the elongation factor (Masullo et al., 1994; Krab and Parmeggiani, 1998). Finally, the low affinity of the GDP-bound elongation factor for the ribosome favours its release.
In the last few years, a number of three-dimensional structures of EF-Tu have shed light on the structural details of the elongation cycle in eubacteria. Indeed, the crystal structure of EF-Tu in complex with GDP (Kjeldgaard and Nyborg, 1992; Abel et al., 1996; Polekhina et al., 1996; Song et al., 1999; Andersen et al., 2000b), with the GTP analogue GDPNP (Berchtold et al., 1993; Kjeldgaard et al., 1993), with the aminoacyl-tRNA (Nissen et al., 1995, 1999) and with EF-Ts (Kawashima et al., 1996; Wang et al., 1997) has been reported. In contrast, very little is known about the structure of the proteins involved in the elongation cycle of eukarya and archaea. The three-dimensional structures of the nucleotide exchange factor EF-1β from human (Perez et al., 1999) and the archaeon Methanobacterium thermoautotrophicum (Kozlov et al., 2000) and the complex of EF-1α with the catalytic C-terminus of EF-1β from Saccharomyces cerevisiae (ScEF-1α⋅EF-1β) (Andersen et al., 2000a) have been determined recently.
Here we report the high resolution crystal structure of EF-1α from the archaeon Sulfolobus solfataricus (Ss) in complex with GDP (SsEF-1α⋅GDP). In the fulfilment of its biochemical functions, SsEF-1α interacts with GDP, GTP (Masullo et al., 1991), aminoacyl-tRNA (Raimo et al., 2000) and other proteins (Raimo et al., 1996) and elicits an intrinsic GTPase activity (Masullo et al., 1994); in addition, SsEF-1α is endowed with a great resistance against chemical and physical agents (Masullo et al., 1993). Interestingly, the amino acid sequences of archaeal EF-1α are significantly more similar to those of eukaryal (49–57% identical residues) rather than to the eubacterial elongation factors (33–41% identical residues) (Amils et al., 1993). Furthermore, biochemical studies have indeed shown that, regarding the affinity for GTP and GDP, SsEF-1α resembles eukaryotic EF-1α more than eubacterial EF-Tu (Masullo et al., 1991). Therefore, the comparison of the SsΕF-1α⋅GDP structure with that of ScEF-1α in its complex with EF-1β provides reliable information about the guanosine nucleotide exchange mechanism in EF-1α.
Results and discussion
Overall structure and domain organization of SsEF-1α⋅GDP
The crystal structure of SsEF-1α⋅GDP has been solved by molecular replacement (see Materials and methods) and refined to an Rcryst of 0.220 (Rfree 0.269) by using diffraction data extending up to 1.8 Å resolution. The refinement statistics are given in Table I. The asymmetric unit contains two independent copies of the complex, related by non-crystallographic symmetry, and 479 water sites. In both molecules, the first three and the last five residues could not be modelled. Furthermore, the electron density shows that residues 66–76, belonging to the switch I region (residues 32–76), are disordered. The copy in the asymmetric unit with higher B-factors (hereafter referred to as molecule B) displays other regions with weak density, which correspond to residues 33–37 and 126–130. Therefore, the structure of molecule A will be described here since the quality of its electron density is better than that of molecule B.
Table I. Crystallographic data.
| Crystal data | |
| Space group | P21 |
| Unit cell dimensions | |
| a (Å) | 62.11 |
| b (Å) | 113.72 |
| c (Å) | 80.32 |
| β (°) |
90.20 |
| Data collection | |
| Resolution limits (Å) | 20.0–1.8 |
| No. of observations | 762 914 |
| No. of unique reflections | 101 656 |
| Completenessa (%) | 98.6 (95.5) |
|
Rsyma,b |
0.034 (0.172) |
| Refinement | |
| Resolution limits (Å) | 20.0–1.8 |
| No. of reflections (Fo >0) | 99 196 |
| Rcrystc | 0.220 |
| Rfreed | 0.269 |
| No. of protein atoms | 6467 |
| No. of nucleotide atoms | 56 |
| No. of water molecules | 479 |
| R.m.s. deviation from ideal values | |
| bond lengths (Å) | 0.009 |
| bond angles (°) | 1.80 |
| dihedral angles (°) | 24.2 |
aValues in parentheses correspond to the highest resolution shell (1.86–1.80 Å).
bRsym = Σ|I(hi) – <I(h)>|/Σ<I(hi)>, where I(hi) is the scaled intensity of the ith symmetry-related observation of reflection h and <I(h)> is the mean value.
cRcryst = Σ|Fo(h) – Fc(h)|/ΣFo(h), where Fo(h) and Fc(h) are the observed and calculated structure factor amplitudes for reflection h.
dRfree was calculated against 8% of the complete data set excluded from refinement.
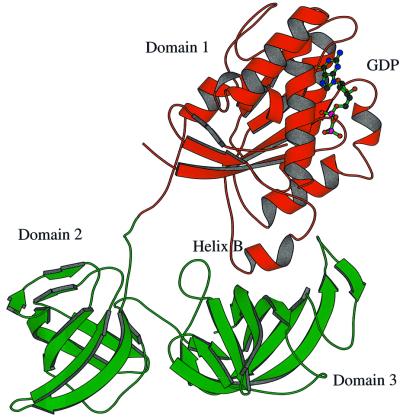
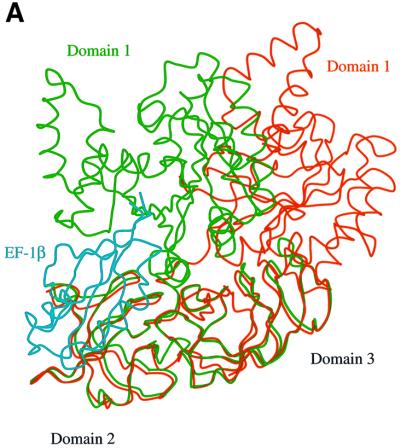
The structure of the complex exhibits a triangular shape with a peculiar large hole, typical of EF-Tu⋅GDP complexes (Kjeldgaard and Nyborg, 1992; Abel et al., 1996; Polekhina et al., 1996; Song et al., 1999; Andersen et al., 2000b), located on one side of the molecule. Three distinct structural domains can be identified (Figure 1), as found in eubacterial EF-Tu. The secondary structure elements, herein denoted according to the notation of Andersen et al. (2000a), of SsEF-1α are similar to those identified for ScEF-1α in the ScEF-1α⋅EF-1β complex. Domain 1, which carries the nucleotide-binding site, presents an α/β structure. It is worth noting that the orientation of helices B and C and those belonging to the switch I region is rather different in SsEF-1α and ScEF-1α. A long peptide (residues 223–232) joins domain 1 to domain 2, which has a β-barrel structure. Interestingly, this connecting peptide adopts a rather rigid structure, despite the absence of stabilizing interactions with the rest of the protein. The intrinsic rigidity of this peptide is probably due to the presence of several proline residues (Pro225, Pro226, Pro228 and Pro232) in its sequence. The restricted conformational freedom of the peptide may be important to select specific relative orientations between domains 1 and 2. Domain 3, which also has a β-barrel structure, interacts with domain 1 through a large interface (Figure 1). Indeed, a network of strong hydrogen bonds is observed between the residues of the switch II region (residues 90–109) and the residues of domain 3. In particular, the nitrogen and carbonyl oxygen of Gly93 form hydrogen bonds with the main chain and side chain of Arg368, respectively. Furthermore, the nitrogen of Val98 interacts with the oxygen of Pro367. Finally, the oxygen of Ile102 forms hydrogen bonds with the Nε and Nη2 atoms of the Arg409 side chain. These interactions are also found in molecule B, although the three domains have slightly different relative orientations. It is worth mentioning that Arg368 and Arg409, whose side chains strongly contribute to the stabilization of the domain 1–domain 3 interface, are conserved residues among almost all the known sequences of EF-1α. This type of interaction is specific for EF-1αs since the loop 360–375 is missing in all EF-Tu sequences. A different interface between domain 1 and 3 has indeed been reported for eubacterial and mitochondrial EF-Tu (Song et al., 1999; Andersen et al., 2000b). These results are in line with the biochemical properties of a chimeric elongation factor constituted by domain 1 of SsEF-1α and domains 2 and 3 of Escherichia coli EF-Tu (Arcari et al., 1999). In fact, it is likely that the impaired domain 1–domain 3 interface in this chimeric elongation factor makes the enzyme unable to support polyphenylalanine synthesis. In addition, a large interface between domains 1 and 3 of SsEF-1α could be relevant to the heat stability of the enzyme. Indeed, biochemical studies on truncated forms of SsEF-1α (Masullo et al., 1997) have shown that domain 3 is much more involved than domain 2 in the stability of the elongation factor.
Fig. 1. Ribbon representation of SsEF-1α⋅GDP. Residues 66–76 are missing as they are disordered in the crystal structure. The GDP is shown in ball-and-stick representation.
The nucleotide-binding site of SsEF-1α⋅GDP
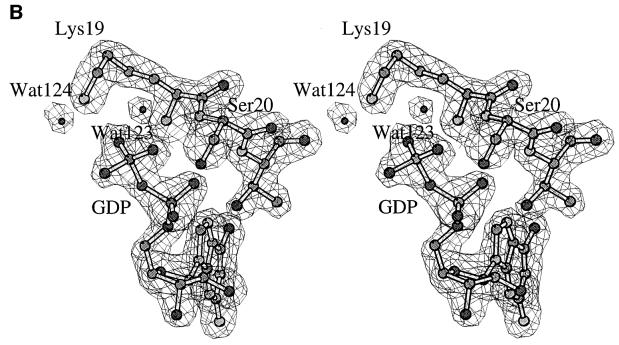
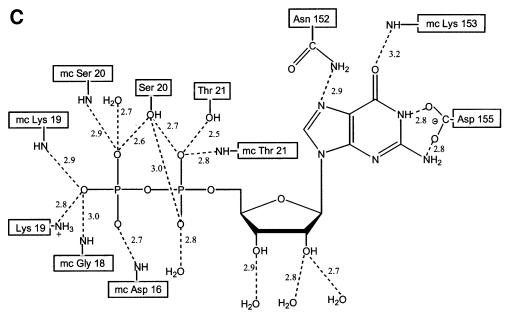
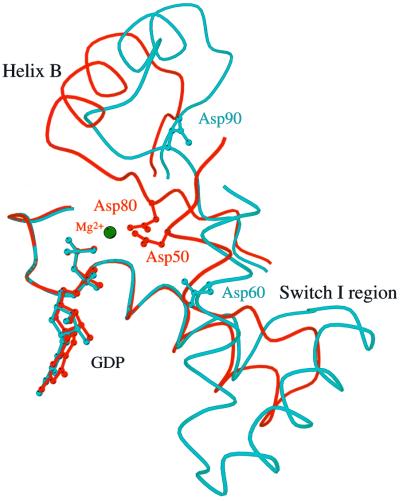
In SsEF-1α⋅GDP, the GDP-binding site is located in a cleft of domain 1. The well-defined electron density of the active site region of SsEF-1α⋅GDP is shown in Figure 2A and B. All of the interactions between SsEF-1α and GDP are reported schematically in Figure 2C. As in EF-Tu, the N1 and N2 atoms of the guanine base form strong hydrogen bonds with the side chain of Asp155 (Asp135 in the E.coli sequence). In addition, the purine base is anchored to the protein through an interaction between O6 and the main chain nitrogen of Lys153. Several main chain nitrogen atoms of the P-loop region 16–21 contribute to the binding of the oxygen atoms of the diphosphate moiety. The side chain of Lys19, belonging to the consensus sequence GKST of EF-1αs (Dever et al., 1987), contributes to the binding of the β-phosphate. The analysis of the electron density of the region surrounding the diphosphate moiety revealed an unexpected feature. In fact, despite the presence of 5 mM MgCl2 during the purification and in the crystallization medium as well, no electron density corresponding to Mg2+ was observed (Figure 2B). Close inspection of the omit electron density map of the nucleotide diphosphate moiety region revealed that only two peaks, assigned as water molecules, could be identified even at a low sigma level. Both peaks are located at hydrogen-bonding distance from hydrogen bond acceptors and donors (Figure 2B) and none of them exhibit the octahedral coordination typical of Mg2+. In particular, Wat123 forms a hydrogen bond with an oxygen atom of the β-phosphate (distance 2.7 Å). The magnesium ion has been identified in all the structures of EF-Tu in complex with GDP reported so far (Kjeldgaard and Nyborg, 1992; Abel et al., 1996; Polekhina et al., 1996; Song et al., 1999; Andersen et al., 2000b), and its absence from the active site has severe consequences on the local structure of the nucleotide-binding site. Indeed, residues such as Asp90 and Asp60, which in EF-Tu take part in the co-ordination of Mg2+ through water molecules, are located rather far from the active site (Figure 3). In the structure of SsEF-1α⋅GDP, the regions that include these residues are rather flexible, as indicated by their relatively high B-factors, since they interact weakly with the rest of the protein. Nevertheless, their electron density is well defined. As an example, the omit electron density map corresponding to the region of Asp90 is reported in Figure 4. It is worth mentioning that the observed conformations of switch I and II regions are not influenced by crystal packing as these residues are distant from any symmetry-related molecule.
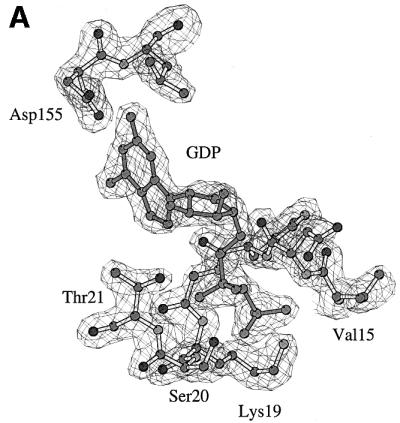
Fig. 2. The GDP-binding site. (A and B) Final omit Fo – Fc electron density maps contoured at 3.0σ superimposed on a stick model of the GDP site. (B) A differently oriented stereo view including water molecules. (C) Schematic representation of the protein–nucleotide interactions.
Fig. 3. Comparison of the nucleotide-binding site in SsEF-1α⋅GDP (cyan) and E.coli EF-Tu⋅GDP (red). For clarity, only the traces of residues 17–53 and 75–99 of SsEF-1α⋅GDP and 12–66 and 85–109 of E.coli EF-Tu⋅GDP are shown. The GDP molecule and the Mg2+ are also shown.
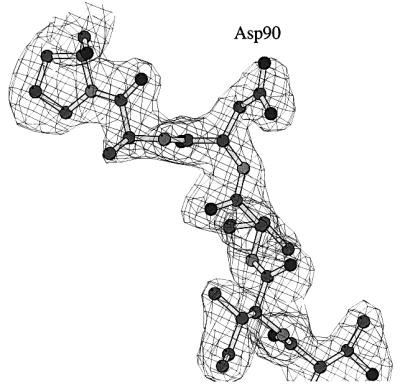
Fig. 4. Omit Fo – Fc electron density map of the Asp90 region contoured at 3.0σ.
Finally, in the current model of SsEF-1α⋅GDP, the Ser20 side chain is linked directly to the oxygens of both α- and β-phosphate (Figure 2). However, the quality of the electron density (Figure 2B) does not rule out the possibility that the Ser20 side chain may assume the conformation that is usually assumed by this residue in EF-Tu structures, with the Oγ atom interacting with Wat123 and the oxygen of the β-phosphate.
Magnesium dependence of the nucleotide exchange in SsEF-1α
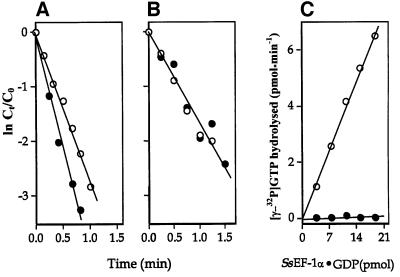
In order to investigate the role of Mg2+ in the binding of GDP or GTP to SsEF-1α, the interaction between the elongation factor and guanine nucleotides was studied in both the absence and presence of MgCl2. As reported in Figure 5, the absence of added Mg2+ and the addition of EDTA provoked only a marginal effect or no effect at all on the exchange reactions [3H]GDP/GDP (Figure 5A) and [3H]GDP/GTP (Figure 5B), respectively. In contrast, the intrinsic NaCl-stimulated GTPase of SsEF-1α (Masullo et al., 1994) showed an absolute requirement for Mg2+ (Figure 5C). The overall results indicated that Mg2+ is required in SsEF-1α for the hydrolytic GTP breakdown but not for the nucleotide exchange. On the other hand, it has been reported that the nucleotide binding to EF-Tu strongly depends on the presence of Mg2+ (Arai et al., 1974; Rutthard et al., 2001). In fact, in eubacterial elongation factors, the absence of Mg2+ reduced the affinity of EF-Tu for guanosine nucleotides by ∼2–3 orders of magnitude (Rutthard et al., 2001). This finding is also supported by mutagenesis analyses on the residues involved in the binding of Mg2+ in eubacterial EF-Tu (Zeidler et al., 1995; Krab and Parmeggiani, 1999a,b). The different role Mg2+ plays in EF-Tu and SsEF-1α is somehow surprising since all of the residues that participate in the Mg2+ co-ordination in EF-Tu are conserved in SsEF-1α as well as in all the other known EF-1α sequences. The differences in the nucleotide-binding properties of SsEF-1α and EF-Tu might be attributed to the different orientation of helix B (Figure 3), which in SsEF-1α⋅GDP interacts strongly with domain 3 (Figure 1). These interactions cause the Asp90 region to be located far from the nucleotide-binding site in the GDP complex.
Fig. 5. Nucleotide exchange and NaCl-dependent GTPase activity of SsEF-1α. [3H]GDP/GDP (A) and [3H]GDP/GTP (B) exchange reactions in the presence of either 10 mM MgCl2 (open circles) or 10 mM EDTA (filled circles). Co and Ct represent the initial concentration and the concentration at time t of SsEF-1α⋅[3H]GDP, respectively. (C) Determination of the NaCl-dependent GTPase activity of SsEF-1α⋅GDP was carried out in a reaction mixture containing either 10 mM MgCl2 (open circles) or 10 mM EDTA (filled circles).
Structural modifications of EF-1α in the nucleotide exchange process
The availability of the high resolution structures of ScEF-1α bound to the catalytic C-terminus of EF-1β (Andersen et al., 2000a) and SsEF-1α bound to GDP allows an understanding of the mechanism of nucleotide exchange in EF-1α. In fact, although ScEF-1α and SsEF-1α belong to different microorganisms, several indications suggest that the structural basis of the nucleotide exchange mechanism may be somewhat similar in S.solfataricus and S.cerevisiae. First, the sequence identity (52%) and the secondary structure similarity of EF-1αs in ScEF-1α⋅EF-1β and in SsEF-1α⋅GDP are very high. Secondly, although the amino acid sequence of the catalytic C-terminus of EF-1β from both yeast and S.solfataricus shares low homology (18%) (Raimo et al., 1996), most of the residues that play a major role in the formation and stabilization of the ScEF-1α⋅EF-1β complex are either conserved or replaced by similar amino acid residues in the SsEF-1β sequence. In particular, Phe163 and Asp183 (S.cerevisiae sequence numbering), which mediate the interactions between ScEF-1β and domain 2 of ScEF-1α (Andersen et al., 2000a), are conserved in archaeal EF-1β. Finally, the NMR structure of EF-1β from the archaeon M.thermoautotrophicum (Kozlov et al., 2000), which has 36% sequence identity with SsEF-1β, shows a close similarity to its human and yeast analogues despite the low sequence homology (<20%). It is worth noting that, among EF-1βs, residues interacting with domain 1 of EF-1α are not conserved. Altogether, these observations suggest that EF-1α and EF-1β probably associate in similar ways in eukarya and archaea, although the fine details of these interactions may be different in these organisms.
The comparison of the EF-1α structures in SsEF-1α⋅GDP and ScEF-1α⋅EF-1β reveals that the structure of the single domains in the two elongation factors is very similar. In fact, when Cα atoms of domains 2 and 3 of SsEF-1α⋅GDP and ScEF-1α⋅EF-1β are superimposed, the r.m.s. deviation is as low as 1.0 Å; when domains 1 are superimposed, the r.m.s. deviation is 1.9 Å. The larger deviations observed for domain 1 can be attributed to the different orientation of the helices of the switch I and II regions. Despite these similarities, the relative orientation of the nucleotide-binding domain 1 with respect to domains 2 and 3 is completely different. In particular, when domains 2 and 3 of the two EF-1α structures are superimposed, the position of domain 1 is rotated by 73° (Figure 6A). The distance of some equivalent atoms of domain 1 is >50 Å. The structure of SsEF-1α⋅GDP is much closer to the structure of EF-Tu in complex with GDP. Indeed, an analogous comparative analysis carried out on SsEF-1α⋅GDP and E.coli EF-Tu⋅GDP shows that domain 1 is rotated by 27°. It is worth noting that deviations up to 19° have been observed among EF-Tus isolated from different species (Andersen et al., 2000b).
Fig. 6. Comparison of SsEF-1α⋅GDP and ScEF-1α⋅EF-1β. (A) Superimposition of domains 2 and 3 of SsEF-1α⋅GDP (red) and ScEF-1α⋅EF-1β (green). The EF-1β is coloured in cyan. (B) Superimposition of the nucleotide-binding site in SsEF-1α⋅GDP and ScEF-1α⋅EF-1β (cyan).
These findings indicate that in the process of nucleotide exchange, EF-1α undergoes a dramatic domain rearrangement, in contrast to the eubacterial EF-Tu system where the overall domain rearrangement which occurs in the nucleotide exchange is rather limited (Kawashima et al., 1996; Wang et al., 1997). On the other hand, the domain rearrangement observed in EF-1α during the nucleotide exchange is comparable with that observed in the switch between the GTP and GDP forms of eubacterial EF-Tu (Berchtold et al., 1993; Kjeldgaard et al., 1993).
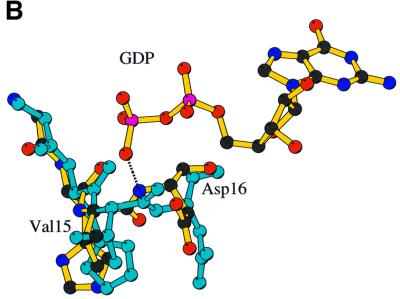
As a consequence of the domain rearrangement, the network of hydrogen bonds at the domain 1–domain 3 interface of SsEF-1α⋅GDP is disrupted in ScEF-1α⋅EF-1β, with helix B sliding on the surface of domain 3. The shift of helix B produces structural alterations that propagate to the P-loop region. Indeed, a comparison of the phosphate-binding site of SsEF-1α⋅GDP and ScEF-1α⋅EF-1β reveals a flip of the peptide bond between Val15 and Asp16 (Figure 6B). This flip disrupts the favourable hydro gen bonding interaction between the nitrogen atom of Asp16 and one of the β-phosphate oxygens of GDP. Simultaneously, it moves the carbonyl group of Val15 to a position that overlaps with the β-phosphate moiety of GDP, thus favouring nucleotide release. A similar flip occurs in the nucleotide exchange of EF-Tu (Kawashima et al., 1996; Wang et al., 1997). It is interesting to note that despite a completely different association between the elongation factor and the exchange factor in EF-Tu and EF-1α, similar structural alterations occur in the P-loop region.
Conclusions
The structure of the SsEF-1α⋅GDP complex reveals novel features of the nucleotide binding and the exchange mechanisms of EF-1α. In particular, we demonstrate that the magnesium ion is not required in the nucleotide-binding process. However, Mg2+ is essential for the GTPase activity. It is worth mentioning that somewhat similar data have been reported recently for the small GTPase Arl3 (Hillig et al., 2000). The ability of SsEF-1α to bind GDP productively, without the assistance of a magnesium ion, is a feature already found in other GTPases involved in the protein synthesis cycle, such as elongation factor G (Czworkowski et al., 1994) and the initiation factor IF2/eIF5B (Roll-Mecak et al., 2000).
The finding that Mg2+ is absent from the nucleotide-binding site of SsEF-1α⋅GDP and that this ion has a marginal effect on nucleotide exchange has important implications for the nucleotide exchange mechanism of EF-1αs. Indeed, an important step in the nucleotide exchange process of EF-Tu is the disruption of the magnesium co-ordination site (Kawashima et al., 1996; Wang et al., 1997). In contrast, it is likely that the first step of the exchange process in archaeal EF-1α is the release of the phosphate moiety from the binding site.
This finding is not in line with the recent hypothesis put forward by Andersen et al. (2001) on the essential role played by both magnesium and Lys205 of ScEF-1β in the yeast nucleotide exchange process. Considering that Lys205 is not a conserved residue among archaeal EF-1βs, the magnesium may play a different role in eukaryal yeast and archaeal elongation factor nucleotide exchange.
The similarity of the structural organization of the GDP complexes of EF-Tu and EF-1α and, conversely, the different association between elongation and exchange factors in eubacteria and archaea/eukarya provide further support for the proposal that nucleotide exchange factors may have appeared independently of GTPases (Cherfils and Chardin, 1999).
Finally, the comparison of SsEF-1α⋅GDP and ScEF-1α⋅EF-1β provides a new example of dramatic structural re-organization of enzymes occurring in biological processes.
Materials and methods
Crystallization and data collection
SsEF-1α⋅GDP was prepared and crystallized as previously reported. The preliminary crystallographic investigations, based on a limited number of diffraction images, suggested an orthorhombic symmetry (Zagari et al., 1994). Subsequent data collections (Table I) demonstrated that the crystals were monoclinic (space group P21) with an orthorhombic pseudo-symmetry. The two molecules of the asymmetric unit are related by a 21 axis in such a way as to generate a pseudo P21212 space group. Larger deviations from orthorhombic pseudo-symmetry are observed when crystals are frozen. Diffraction data were collected at ELETTRA synchrotron to 1.8 Å resolution, on a frozen crystal at 100 K. Data were processed using the HKL package (Otwinowski and Minor, 1997). A summary of the data processing statistics is given in Table I.
Structure determination and refinement
The structure of the complex was solved initially by molecular replacement by using the structure of Thermus aquaticus EF-Tu⋅GDP (PDB code 1TUI) as a starting model (Polekhina et al., 1996). The structure determination was particularly difficult because of (i) the low homology of the starting model with EF-1α; (ii) the presence of two molecules in the asymmetric unit; and (iii) the great flexibility of the elongation factors in complex with GDP. Preliminary attempts to locate either domain 1 or domains 2 and 3 together by using the standard molecular replacement procedure did not provide correct solutions to the problem. On the other hand, clear signals were obtained for domains 2 and 3 of both molecules in the asymmetric unit using the direct rotation function implemented in the program X-PLOR (Brünger, 1992). The AMoRe program (Navaza, 1994) was then used successfully to find the translation of domains 2 and 3. The location of domain 1 turned out to be even more difficult. More than 100 different orientations were tested before the correct translation for domain 1 could be identified. An indication of the correctness of this solution was that the two molecules in the asymmetric unit were oriented in such a way as to generate an orthorhombic pseudo-symmetry. Nevertheless, the quality of the electron density did not allow an easy trace of the protein structure in several regions of the protein. In the meantime, the co-ordinates of ScEF-1α⋅EF-1β became available (Andersen et al., 2000a). Using single domains of this protein as starting models, the molecular replacement was repeated and the previous solution was confirmed. In this case, the electron density was much clearer and the missing parts of the model were built by using the automated Arp procedure (Perrakis et al., 1999). The model was refined using the program X-PLOR (Brünger, 1992) to an Rcryst of 0.220. The final model presents a good stereochemistry (Table I) and no residue falling in the disallowed region of the Ramachandran plot. The figures were drawn using the programs O (Jones et al., 1991), MOLSCRIPT (Kraulis, 1991) and BOBSCRIPT (Esnouf, 1999).
Nucleotide exchange and GTPase activity of SsEF-1α
The nucleotide exchange activity of SsEF-1α was measured at 60°C as already reported (Raimo et al., 1996) in a reaction mixture containing 1 µM SsEF-1α⋅[3H]GDP (specific activity 520 c.p.m./pmol) in 350 µl of 20 mM Tris–HCl buffer pH 7.8, containing 50 mM KCl. The effect of Mg2+ was evaluated in the presence of either 10 mM MgCl2 or 10 mM EDTA. The exchange reaction was started by the addition of 1 mM final concentration of either GDP or GTP.
The GTPase activity of SsEF-1α was measured in a reaction mixture containing 0.075–0.375 µM SsEF-1α⋅GDP in 50 µl of 20 mM Tris–HCl buffer pH 7.8, containing 3.6 M NaCl, 50 µM [γ-32P]GTP (specific activity 120 c.p.m./pmol) in the presence of either 10 mM MgCl2 or 10 mM EDTA. The reaction was carried out for 30 min at 60°C and the amount of 32Pi released was determined as reported (Masullo et al., 1994).
Accession numbers
The structure factors and the co-ordinates of SsEF-1α⋅GDP have been deposited in the Protein Data Bank (code 1JNY).
Acknowledgments
Acknowledgements
We thank A.Podjarny for his help in the early stage of this work, J.Nyborg and G.A.Andersen for kindly providing the ScEF-1α⋅EF-1β co-ordinates prior to deposition and for useful discussions, and the CNR staff at Elettra, Trieste (Italy) for help in the use of the facility. This work was supported by the European Community Biotechnology Programme, contract no. BIO4-CT97-2188, CNR, MURST (Rome, PRIN 1997) and CNR Agenzia 2000.
References
- Abel K. and Jurnak,F. (1996) A complex profile of protein elongation: translating chemical energy into molecular movement. Structure, 4, 229–238. [DOI] [PubMed] [Google Scholar]
- Abel K., Yoder,M.D., Hilgenfeld,R. and Jurnak,F. (1996) An α to β conformational switch in EF-Tu. Structure, 4, 1153–1159. [DOI] [PubMed] [Google Scholar]
- Al-Karadaghi S., Kristensen,O. and Liljas,A. (2000) A decade of progress in understanding the structural basis of protein synthesis. Prog. Biophys. Mol. Biol., 73, 167–193. [DOI] [PubMed] [Google Scholar]
- Amils R., Cammarano,P. and Londei,P. (1993) Translation in archaea. In Kates,M., Kushner,D.J. and Matheson,A.T. (eds), The Biochemistry of Archaea. Elsevier, Amsterdam, The Netherlands, pp. 393–438.
- Andersen G.R., Pedersen,L., Valente,L., Chatterjee,I., Kinzy,T.G., Kjeldgaard,M. and Nyborg,J. (2000a) Structural basis for nucleotide exchange and competition with tRNA in the yeast elongation factor complex eEF1A:eEF1Bα. Mol. Cell, 6, 1261–1266. [DOI] [PubMed] [Google Scholar]
- Andersen G.R., Thirup,S., Spremulli,L.L. and Nyborg,J. (2000b) High resolution crystal structure of bovine mitochondrial EF-Tu in complex with GDP. J. Mol. Biol., 297, 421–436. [DOI] [PubMed] [Google Scholar]
- Andersen G.R., Valente,L., Pedersen,L., Kinzy,T.G. and Nyborg,J. (2001) Crystal structures of nucleotide exchange intermediates in the eEF1A–eEF1Bα complex. Nature Struct. Biol., 8, 531–534. [DOI] [PubMed] [Google Scholar]
- Arai K., Kawakita,M. and Kaziro,Y. (1974) Studies on the polypeptide elongation factors from E.coli. V. Properties of various complexes containing EF-Tu and EF-Ts. J. Biochem., 76, 293–306. [DOI] [PubMed] [Google Scholar]
- Arcari P., Masullo,M., Arcucci,A., Ianniciello,G., de Paola,B. and Bocchini,V. (1999) A chimeric elongation factor containing the putative guanine nucleotide binding domain of archaeal EF-1α and the M and C domains of eubacterial EF-Tu. Biochemistry, 38, 12288–12295. [DOI] [PubMed] [Google Scholar]
- Berchtold H., Reshetnikova,L., Reiser,C.O., Schirmer,N.K., Sprinzl,M. and Hilgenfeld,R. (1993) Crystal structure of active elongation factor Tu reveals major domain rearrangements. Nature, 365, 126–132. [DOI] [PubMed] [Google Scholar]
- Brünger A. (1992) X-PLOR v3.1. User’s Guide. A System for X-ray Crystallography and NMR. Yale University Press, New Haven, CT.
- Cherfils J. and Chardin,P. (1999) GEFs: structural basis for their activation of small GTP-binding proteins. Trends Biochem. Sci., 24, 306–311. [DOI] [PubMed] [Google Scholar]
- Czworkowski J., Wang,J., Steitz,T.A. and Moore,P.B. (1994) The crystal structure of elongation factor G complexed with GDP, at 2.7 Å resolution. EMBO J., 13, 3661–3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever T.E., Glynias,M.J. and Merrick,W.C. (1987) GTP-binding domain: three consensus sequence elements with distinct spacing. Proc. Natl Acad. Sci. USA, 84, 1814–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnouf R.M. (1999) Further additions to MolScript version 1.4, including reading and conturing of electron-density maps. Acta Crystallogr. D, 55, 938–940. [DOI] [PubMed] [Google Scholar]
- Hillig R.C., Hanzal-Bayer,M., Linari,M., Becker,J., Wittinghofer,A. and Renault,L. (2000) Structural and biochemical properties show ARL3-GDP as a distinct GTP binding protein. Structure Fold. Des., 8, 1239–1245. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kawashima T., Berthet-Colominas,C., Wulff,M., Cusack,S. and Leberman,R. (1996) The structure of the Escherichia coli EF-Tu⋅EF-Ts complex at 2.5 Å resolution. Nature, 379, 511–518. [DOI] [PubMed] [Google Scholar]
- Kjeldgaard M. and Nyborg,J. (1992) Refined structure of elongation factor EF-Tu from Escherichia coli. J. Mol. Biol., 223, 721–742. [DOI] [PubMed] [Google Scholar]
- Kjeldgaard M., Nissen,P., Thirup,S. and Nyborg,J. (1993) The crystal structure of elongation factor EF-Tu from Thermus aquaticus in the GTP conformation. Structure, 1, 35–50. [DOI] [PubMed] [Google Scholar]
- Klink F. (1985) Elongation factors. In Woese,C.R. and Wolfe,R. (eds), The Bacteria. Academic Press, London, UK, pp. 379–410.
- Kozlov G., Ekiel,I., Beglova,N., Yee,A., Dharamsi,A., Engel,A., Siddiqui,N., Nong,A. and Gehring,K. (2000) Rapid fold and structure determination of the archaeal translation elongation factor 1β from Methanobacterium thermoautotrophicum. J. Biomol. NMR, 17, 187–194. [DOI] [PubMed] [Google Scholar]
- Krab I.M. and Parmeggiani,A. (1998) EF-Tu, a GTPase odyssey. Biochim. Biophys. Acta, 1443, 1–22. [DOI] [PubMed] [Google Scholar]
- Krab I.M. and Parmeggiani,A. (1999a) Functional–structural analysis of threonine a residue coordinating the nucleotide-bound magnesium in elongation factor Tu. J. Biol. Chem., 274, 11132–11138. [DOI] [PubMed] [Google Scholar]
- Krab I.M. and Parmeggiani,A. (1999b) Mutagenesis of three residues, isoleucine-60, threonine-61 and aspartic acid-80, implicated in the GTPase activity of Escherichia coli elongation factor Tu. Biochemistry, 38, 13035–13041. [DOI] [PubMed] [Google Scholar]
- Kraulis P.J. (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Masullo M., Raimo,G., Parente,A., Gambacorta,A., De Rosa,M. and Bocchini,V. (1991) Properties of the elongation factor 1α in the thermoacidophilic archaebacterium Sulfolobus solfataricus. Eur. J. Biochem., 199, 529–537. [DOI] [PubMed] [Google Scholar]
- Masullo M., Raimo,G. and Bocchini,V. (1993) Resistance of archaebacterial aEF-1α⋅GDP against denaturation by heat and urea. Biochim. Biophys. Acta, 1162, 35–39. [DOI] [PubMed] [Google Scholar]
- Masullo M., De Vendittis,E. and Bocchini,V. (1994) Archaebacterial elongation factor 1α carries the catalytic site for GTP hydrolysis. J. Biol. Chem., 269, 20376–20379. [PubMed] [Google Scholar]
- Masullo M., Ianniciello,G., Arcari,P. and Bocchini,V. (1997) Properties of truncated forms of the elongation factor 1α from the archaeon Sulfolobus solfataricus. Eur. J. Biochem., 243, 468–473. [DOI] [PubMed] [Google Scholar]
- Merrick W.C. and Nyborg,J. (2000) The protein biosynthesis elongation cycle. In Sonenberg,N., Hershey,J.W.B. and Mathews,M.B. (eds), Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 89–126.
- Navaza J. (1994) AMoRe an automated package for molecular replacement. Acta Crystallogr. A, 50, 157–163. [Google Scholar]
- Nissen P., Kjeldgaard,M., Thirup,S., Polekhina,G., Reshetnikova,L., Clark,B.F. and Nyborg,J. (1995) Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu and a GTP analog. Science, 270, 1464–1472. [DOI] [PubMed] [Google Scholar]
- Nissen P., Thirup,S., Kjeldgaard,M. and Nyborg,J. (1999) The crystal structure of Cys–tRNACys–EF-Tu–GDPNP reveals general and specific features in the ternary complex and in tRNA. Structure Fold. Des., 7, 143–156. [DOI] [PubMed] [Google Scholar]
- Nyborg J. and Liljas,A. (1998) Protein biosynthesis: structural studies of the elongation cycle. FEBS Lett., 430, 95–99. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Perez J.M., Siegal,G., Kriek,J., K,H.r., Dijk,J., Canters,G.W. and Moller,W. (1999) The solution structure of the guanine nucleotide exchange domain of human elongation factor 1β reveals a striking resemblance to that of EF-Ts from Escherichia coli. Structure Fold. Des., 7, 217–226. [DOI] [PubMed] [Google Scholar]
- Perrakis A., Morris,R. and Lamzin,V.S. (1999) Automated protein model building combined with iterative structure refinement. Nature Struct. Biol., 6, 458–463. [DOI] [PubMed] [Google Scholar]
- Polekhina G., Thirup,S., Kjeldgaard,M., Nissen,P., Lippmann,C. and Nyborg,J. (1996) Helix unwinding in the effector region of elongation factor EF-Tu–GDP. Structure, 4, 1141–1151. [DOI] [PubMed] [Google Scholar]
- Raimo G., Masullo,M., Savino,G., Scarano,G., Ianniciello,G., Parente,A. and Bocchini,V. (1996) Archaeal elongation factor 1β is a dimer. Primary structure, molecular and biochemical properties. Biochim. Biophys. Acta, 1293, 106–112. [DOI] [PubMed] [Google Scholar]
- Raimo G., Masullo,M., Lombardo,B. and Bocchini,V. (2000) The archaeal elongation factor 1α bound to GTP forms a ternary complex with eubacterial and eukaryal aminoacyl-tRNA. Eur. J. Biochem., 267, 6012–6018. [DOI] [PubMed] [Google Scholar]
- Roll-Mecak A., Cao,C., Dever,T.E. and Burley,S.K. (2000) X-Ray structures of the universal translation initiation factor IF2/eIF5B: conformational changes on GDP and GTP binding. Cell, 103, 781–792. [DOI] [PubMed] [Google Scholar]
- Rutthard H., Banerjee,A. and Makinen,M.W. (2001) Mg2+ is not catalytically required in the intrinsic and kirromycin-stimulated GTPase action of Thermus thermophilus EF-Tu. J. Biol. Chem., 276, 18728–18733. [DOI] [PubMed] [Google Scholar]
- Song H., Parsons,M.R., Rowsell,S., Leonard,G. and Phillips,S.E. (1999) Crystal structure of intact elongation factor EF-Tu from Escherichia coli in GDP conformation at 2.05 Å resolution. J. Mol. Biol., 285, 1245–1256. [DOI] [PubMed] [Google Scholar]
- Sprinzl M. (1994) Elongation factor Tu: a regulatory GTPase with an integrated effector. Trends Biochem. Sci., 19, 245–250. [DOI] [PubMed] [Google Scholar]
- Wang Y., Jiang,Y., Meyering-Voss,M., Sprinzl,M. and Sigler,P.B. (1997) Crystal structure of the EF-Tu⋅EF-Ts complex from Thermus thermophilus. Nature Struct. Biol., 4, 650–656. [DOI] [PubMed] [Google Scholar]
- Zagari A., Sica,F., Scarano,G., Vitagliano,L. and Bocchini,V. (1994) Crystallization of a hyperthermophilic archaeal elongation factor 1α. J. Mol. Biol., 242, 175–177. [DOI] [PubMed] [Google Scholar]
- Zeidler W., Egle,C., Ribeiro,S., Wagner,A., Katunin,V., Kreutzer,R., Rodnina,M., Wintermeyer,W. and Sprinzl,M. (1995) Site-directed mutagenesis of Thermus thermophilus elongation factor Tu. Replacement of His85, Asp81 and Arg300. Eur. J. Biochem., 229, 596–604. [DOI] [PubMed] [Google Scholar]