Abstract
The LCCL domain is a recently discovered, conserved protein module named after its presence in Limulus factor C, cochlear protein Coch-5b2 and late gestation lung protein Lgl1. The LCCL domain plays a key role in the autosomal dominant human deafness disorder DFNA9. Here we report the nuclear magnetic resonance (NMR) structure of the LCCL domain from human Coch-5b2, where dominant mutations leading to DFNA9 deafness disorder have been identified. The fold is novel. Four of the five known DFNA9 mutations are shown to involve at least partially solvent-exposed residues. Except for the Trp91Arg mutant, expression of these four LCCL mutants resulted in misfolded proteins. These results suggest that Trp91 participates in the interaction with a binding partner. The unexpected sensitivity of the fold with respect to mutations of solvent-accessible residues might be attributed to interference with the folding pathway of this disulfide-containing domain.
Keywords: deafness disorder/DFNA9/LCCL domain/NMR structure
Introduction
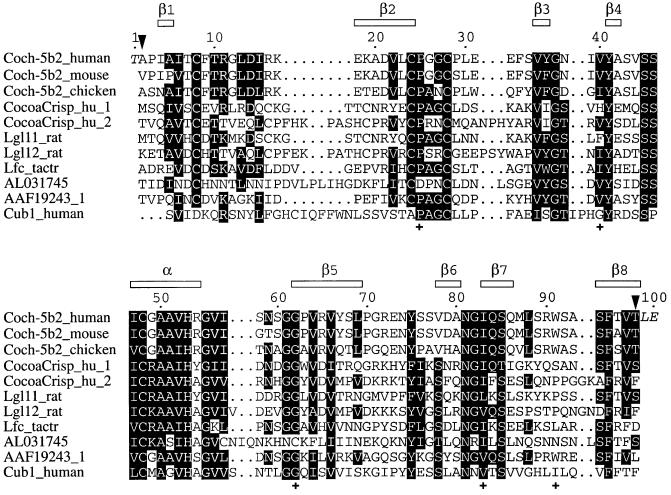
The LCCL domain was identified as an autonomous folding domain first by amino acid sequence comparisons and subsequently by circular dichroism (CD) of an overexpressed construct (Trexler et al., 2000). It is a module named after its presence in Limulus factor C, cochlear protein Coch-5b2, and late gestation lung protein Lgl1, but it has also been found in several other proteins (Figure 1). The LCCL domain plays a central role in the autosomal dominant human deafness disorder DFNA9, a non-syndromic sensorineural deafness associated with vestibular dysfunction, resulting in progressive hearing loss. Molecular analysis of cases of DFNA9 have identified several unrelated kindreds with five different mutations in the human COCH gene (Robertson et al., 1997, 1998; de Kok et al., 1999; Fransen et al., 1999; Kamarinos et al., 2001). All these mutations are in the LCCL domain of Coch-5b2.
Fig. 1. Multiple alignment of the amino acid sequences of LCCL domains. The top sequence is the construct used in the present study. The residues of the construct are numbered from 1 to 100. This numbering is used throughout this article and differs by 26 from the residue numbering by Trexler et al. (2000). The construct thus comprises residues 28–124 of human Coch-5b2 (positions indicated by arrows) and contains three additional residues at the termini from the expression system used (shown in italics). The locations of the α-helix and β-strands in human Coch-5b2 are shown at the top. The following sequences are from mouse Coch-5b2 (Coch-5b2_mouse, residues 30–126), chicken Coch-5b2 (Coch-5b2_chicken, residues 24–120), the two LCCL domains of the human CocoaCrisp protein (CocoaCrisp_hu_1, residues 289–387, and CocoaCrisp_hu_2, residues 390–497), the two LCCL domains of rat late gestation lung protein Lgl1 (Lgl11_rat, residues 224–322, and Lgl12_rat, residues 325–433), Limulus factor C (Lfc_tactr, residues 325–424), a predicted protein of P. falciparum (DDBJ/EMBL/GenBank accession No. AL031745), the predicted human Coch-5b2-related protein (DDBJ/EMBL/GenBank accession No. AAF19243, residues 40–136) and the predicted human Cub-1 protein (Cub1_human, residues 116–213). To highlight conserved features of the LCCL domain, similar residues present in more than half of the LCCL modules are shaded. Crosses mark the locations of residues, where mutations in the human Coch-5b2 protein correlate with the deafness disorder DFNA9.
The function of the LCCL domain seems to be either structural or antimicrobial. The domain is found in extracellular proteins in conjunction with other modular domains, including EGF-like, complement B-type, serine protease, von Willebrand type A, CAP, CUB, discoidin-lectin and C-type lectin modules (Trexler et al., 2000). In Coch-5b2, as in the predicted protein from the related gene AAF19243_1 (Figure 1), the LCCL domain is joined to two domains with extensive homology to the collagen-binding type A domains of von Willebrand factor (Robertson et al., 1997, 1998). von Willebrand type A domains are present in a variety of extracellular matrix components, e.g. cartilage matrix protein and collagens type VI, VII, XII and XIV, suggesting that Coch-5b2 plays a structural role in the architecture of the cochlea by binding to components of the extracellular matrix of this sensory organ (Robertson et al., 1997, 1998). von Willebrand type A domains are, however, also found in proteins involved in defense mechanisms. For example, the von Willebrand factor itself is involved in hemostasis, the complement proteases factor C2 and factor B participate in the complement system, and the integrins LFA-1, Mac-1, VLA-1, VLA-2, p150 and p95 are part of the immune system (Colombatti and Paolo, 1991; Colombatti et al., 1993). Therefore, a role in host defense presents an alternative possible function of Coch-5b2. According to this hypothesis, loss-of-function mutations in DFNA9 cause increased susceptibility to infection, which in turn result in progressive hearing loss (de Kok et al., 1999). A broader role of Coch-5b2 is further indicated by the fact that DFNA9 patients also develop cardiovascular disease in many cases (Bom et al., 1999).
There is further incidental evidence for involvement of LCCL domains in antibody-independent host defense. Limulus factor C is a trypsin-type serine protease that is activated by binding of lipopolysaccharides (LPSs) to the N-terminal extension of the zymogen (Nakamura et al., 1988a,b). The LCCL domain is one of eight modules constituting the N-terminal extension. Activation of factor C initiates the Limulus coagulation cascade, which is a host defense mechanism to protect the organism from bacterial infection.
The late gestation lung protein Lgl1 is expressed in fetal lungs coincident with the production of pulmonary surfactant (Kaplan et al., 1999). In analogy to the antimicrobial activity of the surfactant proteins SP-A and SP-D, Lgl1 may serve to protect the lung from pathogens. Antifungal activity has been demonstrated for plant PR-1 proteins, which share a CAP module with Lgl1 although they do not contain an LCCL domain (Niderman et al., 1995). The recently identified CocoaCrisp proteins of chicken (DDBJ/EMBL/GenBank accession No. AAK16497), mouse (DDBJ/EMBL/GenBank accession No. NP_113579) and human (DDBJ/EMBL/GenBank accession No. NP_113649) contain an LCCL domain (Figure 1) and have a domain organization similar to that of Lgl1. It is not clear whether an antimicrobial role could be attributed to these proteins. The DDBJ/EMBL/GenBank database associates CocoaCrisp with the process of septation in the developing chicken midbrain.
No biological data are available for other proteins containing an LCCL domain (Figure 1). In order to shed more light on the function of this domain and the molecular effect of the point mutations causing the deafness disorder DFNA9, we determined the three-dimensional (3D) structure of the LCCL domain of human Coch-5b2 by nuclear magnetic resonance (NMR) spectroscopy. In addition, mutant LCCL domains, corresponding to four of the five DFNA9 point mutations, were expressed and characterized by CD spectroscopy and differential scanning calorimetry (DSC). The structure presents an unusual, novel fold, which is affected by most of the point mutations. The mutation data indicate at least one interaction surface with a binding partner that is remote from the N- and C-terminal ends of the domain, in agreement with a cross-linking function of this module.
Results
Solution structure of the LCCL domain
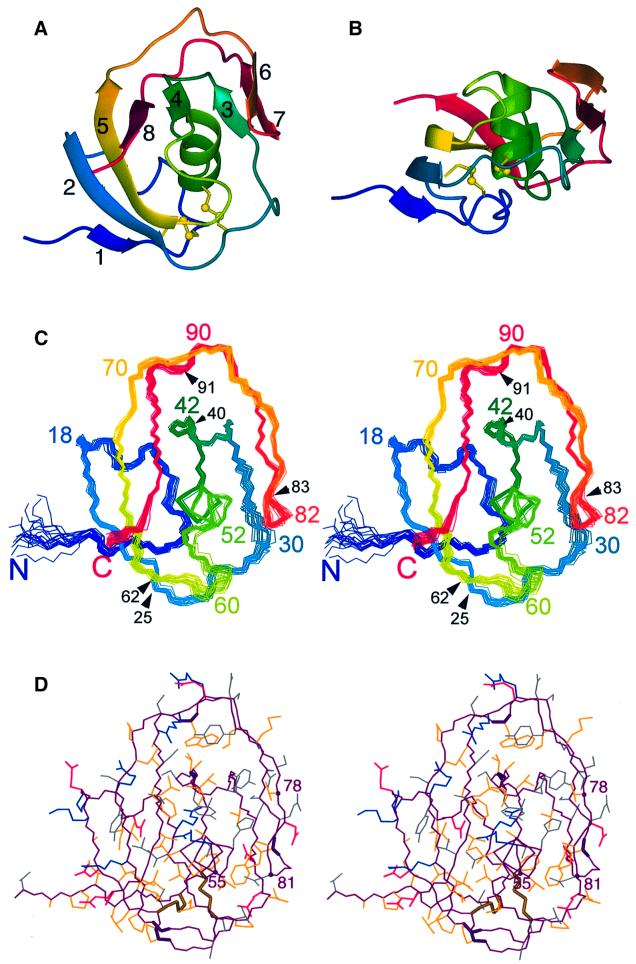
The fold of the LCCL domain comprises a centrally located α-helix wrapped by two β-sheets, reminiscent of a hot-dog (Figure 2). The helix is composed of residues 47–54, and the β-strands, as identified by the Kabsch– Sander algorithm (Kabsch and Sander, 1983), include residues 4–5 (β1), 19–24 (β2), 35–36 (β3), 41–42 (β4), 62–69 (β5), 78–80 (β6), 83–86 (β7) and 94–98 (β8). Strands β1, β2, β5, β8 and β4 form a five-stranded β-sheet that is antiparallel except for strand β1, which aligns with strand β2 in a parallel fashion. Similarly, the strands β3, β7 and β6 arrange into a mixed β-sheet, where the outer strands β3 and β6 align with the central strand β7 in a parallel and antiparallel way, respectively. Several of the β-strands are remarkably short, comprising only two or three residues, so that the secondary structure of most of the domain is irregular.
Fig. 2. Solution structure of the LCCL domain of human Coch-5b2. (A) Ribbon representation. The disulfide bonds are shown in a ball-and-stick representation. The β-strands are numbered as in Figure 1. (B) Ribbon representation as in (A), but in a different orientation. (C) Stereo view showing a superposition of the backbone atoms in the 20 conformers representing the NMR structure (Table I), in a similar orientation to that in (A). Approximately every tenth residue is identified by its sequence number. In addition, black arrows and sequence numbers identify the locations of the five known DFNA9 mutations. (D) Stereo view of the conformer closest to the mean structure of the 20 conformers shown in (C), using a heavy atom representation in the same orientation as in (C). The polypeptide backbone is drawn in purple. The following colors were used for the side chains: blue, Arg, Lys; red, Glu, Asp; yellow, Ala, Cys, Ile, Leu, Met, Phe, Pro, Trp, Val; gray, Asn, Gln, His, Ser, Thr, Tyr. Bold lines label the backbone of the residues mutated in the deafness disorder DFNA9 (Pro25, Val40, Gly62, Ile 83, Trp91) and the two disulfide bridges. Spheres and residue numbers highlight the Cα atoms of the fully conserved residues Gly55 and Asn81, and of residue 78, where all LCCL domains have an uncharged and solvent-exposed residue (Figures 1 and 3).
All peptide bonds are in trans conformation. Disulfide bonds are formed between Cys8 and Cys24, and Cys28 and Cys48. This disulfide pattern was predicted based on the simultaneous mutation of Cys8 and Cys24 in the LCCL domain of Cub1 (Figure 1; Trexler et al., 2000) and agrees with the analysis of tryptic peptides. While the digestion experiment could not rule out the alternative disulfide pattern linking Cys8–Cys28 and Cys24–Cys48, NMR structures computed with this disulfide pattern did not converge as well.
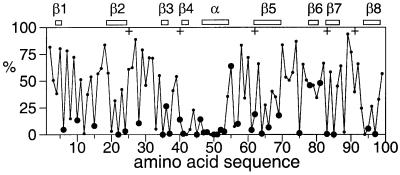
The side chains of Thr10, Tyr41, Ser43, Ser45, Ser46, Tyr75, Ser85 and Ser94 are buried (Figure 3), which was experimentally manifested by slow exchange of their hydroxyl protons with water, allowing the assignment of these resonances.
Fig. 3. Solvent accessibility of the amino acid side chains in the LCCL domain of human Coch-5b2. The solvent accessibilities are expressed as a percentage of their accessibilities calculated for the situation where the respective amino acid residues are located in a hypothetical poly-Gly α-helix with a fully extended side chain (Sevilla-Sierra et al., 1994). The values were averaged over the 20 NMR conformers. Large filled circles identify highly conserved amino acid residues that are uncharged in all LCCL domains known to date (Figure 1), including His in the group of uncharged residues. The locations of the α-helix and β-strands are indicated at the top. Crosses identify the locations of DFNA9 mutations.
All charged amino acid side chains are solvent exposed. His53, which is strictly conserved in all LCCL domains (Figure 1), is almost completely buried in the protein interior and uncharged. The Nε2 nitrogen carries a proton that is hydrogen bonded to the carbonyl carbon of Val78. It exchanges sufficiently slowly with the water protons to yield an observable resonance in the 1H NMR spectrum at 11.35 p.p.m. The Nδ1 nitrogen of the side chain of His53 is unprotonated and hydrogen bonded to the hydroxyl proton of Tyr41, which was observed at 12.77 p.p.m. This hydrogen bond seems to be a conserved feature of LCCL domains, as both residues are completely conserved (Figure 1). The aromatic ring of Tyr41 is rigidly held in place, as shown by non-degenerate side chain resonances. Tyr75 was the only other aromatic residue for which the ring rotation was observed to be slow on the NMR time scale (ms), albeit only at low temperature (10°C).
Conservation of the LCCL fold
As expected for a conserved fold, all insertions are in segments of irregular secondary structure (Figure 1). The side chains of residues 57 and 62 are close in space, suggesting the presence of a disulfide bridge in the LCCL domain of the predicted Plasmodium falciparum protein (DDBJ/EMBL/GenBank accession No. AL031745; Figure 1), as predicted from sequence alignments (Trexler et al., 2000). In contrast, the β carbons of residues 16 and 19 are ∼10.5 Å apart, i.e. formation of the corresponding intra-domain disulfide bridge in the LCCL modules of the Lgl1 and CocoaCrisp proteins would have to be accompanied by a local structural difference. The formation of this disulfide bridge is probably facilitated by the 2–6 residue insert between strands β1 and β2, which will change the local conformation in these proteins anyway (Figure 1).
Figure 3 shows the solvent accessibility of the side chains, highlighting the highly conserved residues that are uncharged in all known LCCL domains. The solvent accessibility is very small for most of these residues (Figure 3), suggesting that they are crucial features of a conserved 3D fold. Only three of these residues (Gly55, Val78 and Asn81) show high solvent accessibility. The side chain of Asn81 forms a hydrogen bond to the backbone carbonyl of Val52 in nearly all NMR conformers, effectively providing a C-cap for the α-helix (Richardson and Richardson, 1988). A non-glycine residue at position 55 would interfere with this interaction, explaining the complete conservation of both residues. The conservation at position 78 is not as high and thus of lesser significance. Furthermore, residues in this position are hydrophilic in most LCCL domains (Figure 1).
Structural impact of DFNA9 mutations
Five genetic variants of the LCCL domain of human Coch-5b2 have been reported to cause the DFNA9 deafness disorder. These mutations are Pro25Ser, Val40Gly, Gly62Glu, Trp91Arg and Ile83Asn (Robertson et al., 1997, 1998; de Kok et al., 1999; Fransen et al., 1999; Kamarinos et al., 2001). Their locations are identified in Figure 2C and D. To assess the impact of these mutations on domain stability, the mutant variants Pro25Ser, Val40Gly, Gly62Glu and Trp91Arg were constructed and expressed. The mutants Pro25Ser, Val40Gly and Gly62Glu failed to refold properly as shown by the fact that most of the protein precipitated during the standard refolding protocol. Furthermore, CD spectroscopy and DSC studies on the soluble proteins revealed that these three mutants did not fold into 3D structures characteristic of the wild-type LCCL domain (data not shown). In contrast, the Trp91Arg mutant yielded data indistinguishable from those of the wild-type protein, including a denaturation temperature (63°C) that was identical within experimental error to that of the wild-type protein.
Among the five DFNA9 mutation sites, only the side chain of residue 83 is completely buried (Figure 3). Ile83 is located near the tip of the hairpin loop formed by the strands β6 and β7, and the contacts formed by its side chain with the α-helix are probably as important as the totally conserved Gly55–Asn81 interaction (Figure 2D). The point mutation Ile83Asn presents a non-conservative change of residue that is hydrophobic in all known LCCL domains (Figure 1). This mutation was not tested experimentally. It is most likely to be disruptive for the native fold.
Discussion
Functional implications of the fold
The LCCL domain presents a structure with an unusual fold, where a centrally located helix is wrapped by extended polypeptide segments of mostly irregular secondary structure. The β-sheets are small and some of the β-strands are exceedingly short. The fold has not previously been observed as judged by a search of the Protein Data Bank (PDB) using the program DALI (Holm and Sander, 1993), where the highest Z-score was 1.9 for less than half of the residues aligned. Protein pairs with structural similarity yield DALI Z-scores >2.0.
In the full-length proteins, the LCCL domain is found in association with various other domains. The N- and C-terminal ends of the LCCL module are fairly close in space, which may result in contacts between the preceding and following modules in the polypeptide chain, in analogy to inter-module contacts observed in fibronectin (Pickford et al., 2001). Interestingly, all disulfide bridges, including any additional disulfide bridge formed in the LCCL domains of the Lgl1, CocoaCrisp and AL031745 proteins (Figure 1), are located near the face with the N- and C-terminal ends (Figure 2), suggesting that structural stability in this area is important.
Remarkably, no insertions or deletions occur in the long double-stranded loop between residues 70 and 93 in any of the LCCL domains (Figure 1), although the solvent exposure of this segment is quite high on average and its secondary structure is irregular except for the short strands β6 and β7 (Figures 2C and 3). This segment buries a few hydrophilic side chains, including two side-chain hydroxyls (Tyr75 and Ser85) and, to a large extent, the side chain of the totally conserved residue His53 (Figure 2D). The superficial attachment of this segment to the rest of the protein may infer a low tolerance towards insertions and deletions that destabilize its contacts with the core of the protein structure.
Deafness disorder mutations
Most of the mutations leading to the deafness disorder DFNA9 result in misfolding of the LCCL domain of Coch-5b2. This was experimentally confirmed for the mutants Pro25Ser, Val40Gly and Gly62Glu, and is likely in the case of the mutant Ile83Asn because of the very low solvent accessibility of this side chain in the wild-type protein and its high conservation between different LCCL domains.
In contrast, the Trp91Arg mutant is correctly folded and as stable as the wild-type protein. The side chain of Trp91 is largely exposed on the surface of the protein (Figures 2D and 3). This residue is conserved between all LCCL domains of Coch-5b2 and the predicted Coch-5b2-like protein AAF19243_1, but not between the LCCL domains in general (Figure 1). These features strongly suggest that this residue participates in the interaction with a binding partner.
Based on the locations of Pro25 and Gly62 on the surface of the LCCL domain, it was unexpected that mutations to Ser and Glu, respectively, should result in misfolded structures. Both residues are spatially close to each other and to the two disulfide bridges (Figure 2C and D). As all four cysteine sulfurs are close in space, incorrect pairing of the sulfurs could easily occur during oxidative formation of the disulfide bridges. The disruptive effect of these mutations could thus be explained as an effect on the protein folding pathway rather than as a destabilization of the native fold. Alternatively, the mutants yield correctly folded, stable structures in vivo, in which case the pathogenic effect of these mutations would indicate a second interaction site with a binding partner. As expected for an interaction site, no charged residues are present in the immediate vicinity (Figure 2D).
The mutants Val40Gly and Ile83Asn probably destabilize the association of the long double-stranded loop between residues 70 and 93 with the core of the protein. Alternatively, we note that the mutation of Ile83 to Asn creates the amino acid sequence …GNQSQ…, which represents an NxS motif for N-glycosylation. This new glycosylation site could also disrupt the normal function of the Coch-5b2 protein.
In conclusion, the NMR structure determination of the LCCL domain revealed a novel fold, where most of the residues mutated in the autosomal dominant human deafness disorder DFNA9 are located on the protein surface, yet interfere with proper folding of the domain. While mutations of tryptophan to charged residues are likely to disrupt the fold of most proteins, Trp91 is highly solvent exposed in the LCCL domain and the Trp91Arg mutant results in a protein with a fold and stability characteristic of the wild-type protein. The structure of the LCCL domain will now facilitate the search for binding partners. Preliminary tests for binding of LPS and carbohydrates (glucose, mannose, cellobiose, 2-N-acetyl-glucosamine) failed to show binding (data not shown), motivating the search for a protein binding partner.
Materials and methods
Production of wild-type and mutant LCCL domains of human Coch-5b2
Expression, refolding and purification of the wild-type LCCL domain of Coch-5b2 were performed as described previously (Trexler et al., 2000). The protocol includes expression as a fusion protein with β-galactosidase, isolation and purification of inclusion bodies, and tryptic cleavage of the fusion protein to obtain the isolated LCCL domain. A 15N-labeled sample was prepared in the same way, except that M9 minimal medium was used. Mutant versions of the LCCL domain were generated in a two-step PCR according to Higuchi (1990). In the first reaction we produced two overlapping fragments containing the appropriate mutations. In this reaction we used the mutagenesis primers paired with the amplimers complementary to the 5′ or 3′ end of the LCCL domain. The sequences of the amplimers were: 5′-GCGGGATCCGTCGACCGCTCCCATTGCTATCACATC-3′ (sense) and 5′-GCGAAGCTTACTCGAGAGTTACTGTGAAAGAAGCAG-3′ (antisense). The sequences of the mutagenesis primers were as follows: 5′-GATGTCCTCTGCTCAGGGGGCTG-3′ (sense) and 5′-CAGCCCCCTGAGCAGAGGACACT-3′ (antisense) for the Pro25Ser mutation, 5′-ATGGGAACATAGGATATGCTTCTG-3′ (sense) and 5′-CAGAGGCATATCCTATGTTCCCAT-3′ (antisense) for the Val40Gly mutation, 5′-GCAACTCAGGGGAACCTGTACGAG-3′ (sense) and 5′-CTCGTACAGGTTCCCCTGAGTTGC-3′ (antisense) for the Gly62Glu mutation, and 5′-GTGAAAGAAGCAGACCGTCTAGAAAGC-3′ (antisense) for the Trp91Arg mutation. The template was the M13mp18/LCCL clone described previously (Trexler et al., 2000). In the second reaction we linked the two overlapping cDNA fragments with the amplimers.
The PCR products were cloned into M13mp18 for sequence verification by sequencing on both strands. The inserts from the mutant clones were cut with HincII and HindIII restriction enzymes, and ligated in PvuII–HindIII-digested pmed23 expression vector. Expression, refolding and purification of the mutant LCCL proteins were performed using the same protocol as for the wild-type protein (Trexler et al., 2000).
In the case of the Trp91Arg mutant fusion protein, its β-galactosidase moiety was removed by limited elastase digestion: β-galLCCLW117R fusion protein (1 mg/ml) was dissolved in 0.1 M ammonium bicarbonate pH 8.0 buffer and was incubated with 0.001 mg/ml elastase at 25°C for 30 min. Reaction was arrested with 2 mM phenylmethylsulfonyl fluoride and the digest was applied to a Sephadex G-50 column. Porcine pancreatic elastase and phenylmethylsulfonyl fluoride were commercial products (Serva, Heidelberg, Germany).
The concentration of DEL β-galLCCLW117R protein was determined using the extinction coefficient 5240 M–1 cm–1. The extinction coefficients were determined according to a described method (Mach et al., 1992). The composition of protein samples was analysed by SDS–PAGE using 6–16 and 11–22% linear polyacrylamide gradient slab gels under both reducing and non-reducing conditions (Laemmli, 1970).
CD spectroscopy
Far-UV (190–250 nm) CD spectra of recombinant DEL β-galLCCL and DEL β-galLCCLW117R were measured using a JASCO J-720 spectropolarimeter thermostatted with a Neslab RT-100 water bath. The CD spectropolarimeter and the optical cells were calibrated with recrystallized d-10-camphorsulfonic acid. The measurements were carried out with protein solutions of ∼0.1–0.2 mg/ml. All spectra were measured at 25°C, averaging over five scans recorded with an 8 s time constant and a scan rate of 10 nm/min.
Differential scanning calorimetry
Calorimetric measurements were carried out on a VP-DSC Micro Calorimeter at a heating rate of 1°C/min and a solution concentration of 0.1–0.2 mg/ml. Experiments were conducted at pH 8.0 in 20 mM Tris–HCl buffer. Buffer base lines were obtained under the same conditions and subtracted from sample tracings. The VP-DSC MicroCalorimeter was calibrated according to the instructions of the manufacturer. DSC data analysis was performed with the Microcal Origin Version 5.0 program.
NMR spectroscopy and collection of structure restraints
NMR spectra were recorded using an ∼1 mM solution of the LCCL domain at pH 4.9 and 28°C at 600 and 800 MHz 1H NMR frequency on Bruker DMX 600 and Varian Unity INOVA 800 NMR spectrometers. Virtually complete sequential resonance assignments were obtained using NOESY and TOCSY spectra recorded in 90% H2O/10% D2O, and NOESY and DQF-COSY spectra recorded in D2O solution following several cycles of lyophilization and redissolving in D2O to remove the exchangeable protons. The 15N-HSQC spectrum was assigned using a 3D TOCSY-15N-HSQC spectrum recorded with a 0.3 mM solution of 15N-labeled LCCL domain. Distance restraints for the structure calculation were collected from a NOESY spectrum recorded in 90% H2O/10% D2O solution at 800 MHz with a mixing time of 40 ms. Dihedral angle restraints were derived from the 3JHNHα and 3JHαHβ coupling constants manifested in line-splittings observed for the NOESY and DQF-COSY cross-peaks. Stereospecific assignments were obtained for 101 pairs of diastereotopic methylene protons, side-chain amide protons and methyl groups. Residual dipolar 15N-1H couplings of the amide protons were measured from 15N-HSQC spectra with α/β-half-filter (Andersson et al., 1998) recorded of 15N-labeled LCCL domain in the presence and absence of 0.3% Pf1 phages (Hansen et al., 2000).
Structure calculations
The cross-peaks in the NOESY spectra were assigned and integrated using the program XEASY (Bartels et al., 1995). The NMR structure was calculated using the program DYANA (version 1.5) and the associated routines CALIBA, HABAS and GLOMSA (Güntert et al., 1997). Fifty random conformers were annealed in 30 000 steps using torsion-angle dynamics. The twenty conformers with the lowest residual restraint violations were energy minimized using the program OPAL (version 2.6) with standard parameters (Luginbühl et al., 1996).
Table I shows an overview of the restraints used and structural statistics. The Ramachandran plot was analyzed using PROCHECK-NMR (version 3.4) (Laskowski et al., 1996). Ile39 was the only residue consistently found in a forbidden region. It is located in a highly solvent-exposed loop.
Table I. Structural statistics for the NMR conformers.
| Parameter | Value |
|---|---|
| Number of assigned NOE cross peaks | 2002 |
| Number of non-redundant NOE upper-distance limits | 1187 |
| Number of scalar coupling constantsa | 199 |
| Number of residual 1HN-15N dipolar couplings | 69 |
| Number of dihedral-angle restraints | 270 |
| Intra-protein AMBER energy (kcal/mol) | –3870 ± 51 |
| Sum of residual NOE-restraint violations (Å) | 3.7 ± 0.3 |
| Maximum dihedral-angle restraint violations (°) | 2.4 ± 0.2 |
| R.m.s.d. to the mean for N, Cα and C′ (Å)b | 0.43 ± 0.06 |
| R.m.s.d. to the mean for all heavy atoms (Å)b | 0.74 ± 0.08 |
| Ramachandran plot appearancec | |
| most favored regions (%) | 75.7 |
| additionally allowed regions (%) | 22.6 |
| generously allowed regions (%) | 0.5 |
| disallowed regions (%) | 1.2 |
aEighty 3J(HN,Hα), 119 3J(Hα,Hβ).
bFor residues 3–99.
cFrom PROCHECK-NMR (Laskowski et al., 1996).
Secondary structure elements and root mean square deviation (r.m.s.d.) values were calculated using the program MOLMOL (version 2.6) (Koradi et al., 1996). MOLMOL was also used to generate Figure 2.
Coordinates
The coordinates of the twenty energy-refined DYANA conformers of the LCCL domain were deposited in the PDB with the accession code 1JB1. The NMR chemical shifts were deposited at the BioMagResBank (BMRB) under accession code 5047.
Acknowledgments
Acknowledgements
This work was supported by the Swedish Natural Science Research Council and by Grant AKP 2000-8 3,3 of the Research Fund of the Hungarian Academy of Sciences.
References
- Andersson P., Weigelt,J. and Otting,G. (1998) Spin-state selection filters for the measurement of heteronuclear one-bond coupling constants. J. Biomol. NMR, 12, 435–441. [DOI] [PubMed] [Google Scholar]
- Bartels C., Xia,T., Güntert,P., Billeter,M. and Wüthrich,K. (1995) The program XEASY for computer-supported NMR spectral analysis. J. Biomol. NMR, 5, 1–10. [DOI] [PubMed] [Google Scholar]
- Bom S.J., Kemperman,M.H., de Kok,Y.J., Huygen,P.L., Verhagen,W.I., Cremers,F.P. and Cremers,C.W. (1999) Progressive cochleo-vestibular impairment caused by a point mutation in the COCH gene at DFNA9. Laryngoscope, 109, 1525–1530. [DOI] [PubMed] [Google Scholar]
- Colombatti A. and Paolo,B. (1991) The superfamily of proteins with von Willebrand factor type A-like domains: one theme common to components of extracellular matrix, hemostasis, cellular adhesion, and defense mechanisms. Blood, 77, 2305–2315. [PubMed] [Google Scholar]
- Colombatti A., Bonaldo,P. and Doliana,R. (1993) Type A modules: interacting domains found in several non-fibrillar collagens and in other extracellular matrix proteins. Matrix, 13, 297–306. [DOI] [PubMed] [Google Scholar]
- de Kok Y.J.M. et al. (1999) A Pro51 Ser mutation in the COCH gene is associated with late onset autosomal dominant progressive sensorineural hearing loss with vestibular defects. Hum. Mol. Genet., 8, 361–366. [DOI] [PubMed] [Google Scholar]
- Fransen E. et al. (1999) High prevalence of symptoms of Menière’s disease in three families with a mutation in the COCH gene. Hum. Mol. Genet., 8, 1425–1429. [DOI] [PubMed] [Google Scholar]
- Güntert P., Mumenthaler,C. and Wüthrich,K. (1997) Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol., 273, 283–298. [DOI] [PubMed] [Google Scholar]
- Hansen M.R., Hanson,P. and Pardi,A. (2000) Filamentous bacteriophage as a versatile method for aligning RNA, DNA and proteins for measurement of NMR dipolar coupling information. Methods Enzymol., 317, 220–240. [DOI] [PubMed] [Google Scholar]
- Higuchi R. (1990) Recombinant PCR. In Innis,M.A., Gelfand,D.H., Sninsky,J.J. and White,T.J. (eds), PCR Protocols. Academic Press, New York, NY, pp. 177–183.
- Holm L. and Sander,C. (1993) Protein structure comparison by alignment of distance matrices. J. Mol. Biol., 233, 123–138. [DOI] [PubMed] [Google Scholar]
- Kabsch W. and Sander,C. (1983) Dictionary of protein secondary structures: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers, 22, 2577–2637. [DOI] [PubMed] [Google Scholar]
- Kamarinos M., McGill,J., Lynch,M. and Dahl H. (2001) Identification of a novel COCH mutation, I109N, highlights the similar clinical features observed in DFNA9 families. Hum. Mutat., 17, 351–356. [DOI] [PubMed] [Google Scholar]
- Kaplan F., Ladoux,P., Kassamali,F.Q., Gagnon,S., Post,M., Koehler,D., Deimling,J. and Sweezey,N.B. (1999) A novel developmentally regulated gene in lung mesenchyme: homology to a tumor-derived trypsin inhibitor. Am. J. Physiol., 276, L1027–L1036. [DOI] [PubMed] [Google Scholar]
- Koradi R., Billeter,M. and Wüthrich,K. (1996) MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph., 14, 51–55. [DOI] [PubMed] [Google Scholar]
- Laemmli U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., Rullmann,J.A.C., MacArthur,M.W., Kaptein,R. and Thornton,J.M. (1996) AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR, 8, 477–486. [DOI] [PubMed] [Google Scholar]
- Luginbühl P., Güntert,P., Billeter,M. and Wüthrich K. (1996) The new program OPAL for molecular dynamics simulations and energy refinements of biological macromolecules. J. Biomol. NMR, 8, 136–146. [DOI] [PubMed] [Google Scholar]
- Mach H., Middaugh,C.R. and Lewis,R.V. (1992) Statistical determin ation of the average values of the extinction coefficients of tryptophan and tyrosine in native proteins. Anal. Biochem., 200, 74–80. [DOI] [PubMed] [Google Scholar]
- Nakamura T., Tokunaga,F., Morita,T., Iwanaga,S., Kusumoto,S., Shiba,P., Kobayashi,T. and Inoue,K. (1988a) Intracellular serine-protease zymogen factor C, from horseshoe crab hemocytes: its activation by synthetic lipid A analogues and acidic phospholipids. Eur. J. Biochem., 176, 89–94. [DOI] [PubMed] [Google Scholar]
- Nakamura T., Tokunaga,F., Morita,T. and Iwanaga,S. (1988b) Inter action between lipopolysaccharide and intracellular serine protease zymogen, factor C, from horseshoe crab (Tachypleus trdentatus) hemocytes. J. Biochem. (Tokyo), 103, 370–374. [DOI] [PubMed] [Google Scholar]
- Niderman T., Genetet,I., Bruyere,T., Gees,R., Stintzi,A., Legrand,M., Fritig,B. and Mosinger,E. (1995) Pathogenesis-related PR-1 proteins are antifungal. Plant Physiol., 108, 17–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickford A.R., Smith,S.P., Staunton,D., Boyd,J. and Campbell,I.D. (2001) The hairpin structure of the 6F11F22F2 fragment from human fibronectin enhances gelatin binding. EMBO J., 20, 1519–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson J.S. and Richardson,D.C. (1988) Amino acid preferences for specific locations at the ends of α helices. Science, 240, 1648–1652. [DOI] [PubMed] [Google Scholar]
- Robertson N.G., Skvorak,A.B., Yin,Y., Weremowicz,S., Johnson,K.R., Kovatch,K.A., Battey,J.F., Bieber,F.R. and Morton,C.C. (1997) Mapping and characterization of a novel cochlear gene in human and in mouse: a positional candidate gene is associated with late onset autosomal dominant progressive sensorineural hearing loss with vestibular defects. Genomics, 46, 345–354. [DOI] [PubMed] [Google Scholar]
- Robertson N.G. et al. (1998) Mutations in a novel cochlear gene cause DFNA9, a human nonsyndromic deafness with vestibular dysfunction. Nature Genet., 20, 299–303. [DOI] [PubMed] [Google Scholar]
- Sevilla-Sierra P., Otting,G. and Wüthrich,K. (1994) Determination of the nuclear magnetic resonance structure of the DNA-binding domain of the P22 c2 repressor (1–76) in solution and comparison with the DNA-binding domain of the 434 repressor. J. Mol. Biol., 235, 1003–1020. [DOI] [PubMed] [Google Scholar]
- Trexler M., Bányai,L. and Patthy,L. (2000) The LCCL module. Eur. J. Biochem., 267, 5751–5757. [DOI] [PubMed] [Google Scholar]