Abstract
mRNA silencing and storage play an important role in gene expression under diverse circumstances, such as throughout early metazoan development and in response to many types of environmental stress. Here we demonstrate that the major mRNA-associated protein YB-1, also termed p50, is a potent cap-dependent mRNA stabilizer. YB-1 addition or overexpression dramatically increases mRNA stability in vitro and in vivo, whereas YB-1 depletion results in accelerated mRNA decay. The cold shock domain of YB-1 is responsible for the mRNA stabilizing activity, and a blocked mRNA 5′ end is required for YB-1-mediated stabilization. Significantly, exogenously added YB-1 destabilizes the interaction of the cap binding protein, eIF4E, with the mRNA cap structure. Conversely, sequestration of eIF4E from the cap increases the association of endogenous YB-1 with mRNA at or near the cap, and significantly enhances mRNA stability. These data support a model whereby down-regulation of eIF4E activity or increasing the YB-1 mRNA binding activity or concentration in cells activates a general default pathway for mRNA stabilization.
Keywords: cap structure/cold shock domain/messenger ribonucleoproteins/mRNA stability/translation
Introduction
Control of mRNA stability plays an important role in the regulation of gene expression in eukaryotes (Ross, 1995; Caponigro and Parker, 1996; Schwartz and Parker, 2000). The 3′ poly(A) tail, in association with the poly(A) binding protein (PABP), is the best studied and characterized mRNA stability factor (Bernstein and Ross, 1989; Ross, 1995; Jacobson and Peltz, 1996). PABP was first identified as a protein tightly associated with mRNA in messenger ribonucleoprotein (mRNP) particles, together with a 50 kDa protein, first termed p50, and more recently, YB-1 (Blobel, 1972; Van Venrooij et al., 1977; for review see Evdokimova and Ovchinnikov, 1999). Active polysomal mRNPs possess both YB-1 and PABP, whereas YB-1 is the predominant protein component of translationally inactive mRNPs (Van Venrooij et al., 1977; Minich et al., 1989; Spirin, 1996). A large body of evidence indicates that YB-1 is a general translational repressor; an increase in YB-1 concentration results in an inhibition of translation initiation in vitro, and YB-1 overexpression in cells causes translational repression in vivo, via a mechanism that is poorly understood (Minich et al., 1993; Davydova et al., 1997).
YB-1 belongs to the Y-box binding (YB) transcription factor protein family, so termed because of the ability of these proteins to bind to the Y-box promoter element (Wolffe et al., 1992). YB proteins are present in bacteria, plants and animals, and exhibit a wide variety of biological activities, ranging from transcriptional regulation of a large number of genes to ‘masking’ of mRNA in germinal cells. YB-1 is a nucleocytoplasmic shuttling protein (Matsumoto and Wolffe, 1998; Sommerville, 1999), and is highly conserved amongst vertebrates, including rabbit, rat, mouse, chicken, frog and human with ∼98% identity between the rabbit and human YB-1 proteins (Evdokimova et al., 1995).
YB-1 consists of an alanine/proline (AP)-rich N-terminal domain followed by a cold-shock domain (CSD) and a C-terminal segment comprised of four alternating clusters of basic and acidic amino acids (Wolffe, 1994). The most conserved region in YB proteins is the 80 amino acid CSD; YB-1 exhibits >40% identity and >60% similarity to the major Escherichia coli cold-shock protein CspA (Matsumoto and Wolffe, 1998; Sommerville, 1999). The CSD is a five-stranded β-barrel containing RNP 1 and RNP 2-like consensus motifs, which binds to duplex and single stranded DNA and RNA (Wolffe et al., 1992; Schindelin et al., 1994; Wolffe, 1994; Bouvet et al., 1995). The N-terminal AP domain of YB-1 was reported to associate with actin microfilaments, presumably contributing to mRNA localization (Ruzanov et al., 1999). The C-terminal region of YB-1 and related proteins binds DNA and RNA in a sequence-independent manner and mediates protein–protein interactions (Wolffe, 1994; Sommerville and Ladomery, 1996).
Although YB-1 has been implicated in specific stabilization of IL-2 mRNA following T-cell activation (Chen et al., 2000), its function in general mRNA stability has not been addressed. Several observations support a role for YB-1-related proteins in mRNA stabilization at early stages of metazoan development. For example, the Xenopus FRGY2 and the mouse MSY1 and MSY2 proteins are the major components of translationally inactive mRNPs, which are stored throughout gametogenesis (Richter and Smith, 1984; Murray et al., 1991; Gu et al., 1998). Consistent with a negative effect of YB proteins on protein synthesis, translational activation of stored mRNAs during embryogenesis coincides with massive degradation of ‘germinal’ YB proteins (Wolffe et al., 1992; Sommerville, 1999).
In this study we demonstrate that, coincident with translation inhibition, YB-1 dramatically stabilizes mRNA. Strikingly, the CSD of YB-1 alone engenders mRNA stabilization, but not translational repression. On the contrary, the C-terminal domain strongly inhibits mRNA translation but does not affect mRNA stability, which indicates that these activities are separated, and map to different regions of the protein. Although YB-1 stabilization of mRNA is sequence non-specific, it is strongly dependent on the presence of a blocked (m7GpppG or GpppG) mRNA 5′ end. Translation inhibitors that interfere with eIF4E cap binding function or disrupt the assembly of the eIF4F cap binding complex (such as cap analogs or the repressor protein 4E-BP1) promote an interaction between YB-1 and the mRNA 5′ cap structure, as determined by UV cross-linking experiments, and prevent mRNA degradation. Conversely, YB-1 depletion from cell extracts accelerates mRNA decay and abrogates mRNA stabilization by cap analogs. These data suggest that YB-1 is a potent, general cap-dependent mRNA stabilizer that protects mRNA against degradation when the association of eIF4E with the cap is impaired.
Results
YB-1 is a potent stabilizer of mRNAs in vitro and in vivo
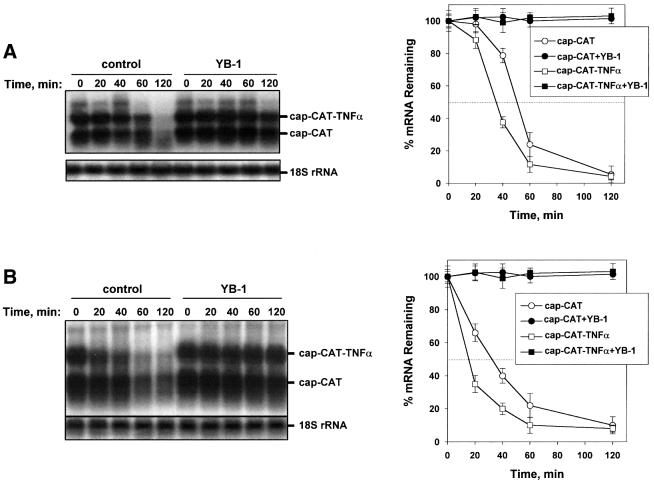
The effect of YB-1 on mRNA stability was examined in three different in vitro systems, a rabbit reticulocyte lysate, Krebs-2 ascites and HeLa cell extracts. Time-course experiments were performed to monitor the decay of a chloramphenicol acetyltransferase (CAT) mRNA, as well as the CAT mRNA fused to the 3′ UTR of the tumor necrosis factor α (TNFα) mRNA, which contains multiple destabilizing AU-rich elements (AREs). Equal amounts of total RNA were loaded in each lane, as determined by northern blot analysis of 18S rRNA (Figure 1). The half-life of CAT mRNA ranged from ∼55 min in the rabbit reticulocyte lysate (Figure 1A) to ∼35 min in the HeLa extract (Figure 1B). The half-life of CAT–3′ TNFα mRNA was somewhat shorter than that of the control CAT mRNA; ∼40 min in the reticulocyte lysate (Figure 1A) and ∼20 min in HeLa cell extract (Figure 1B). Since the difference in the half-lives of the CAT and CAT–3′ TNFα mRNAs is not large, the major decay pathway observed here is not likely to be ARE dependent. Strikingly, in the presence of YB-1 the decay rates of both the CAT and CAT–3′ TNFα mRNAs were dramatically decreased in all extracts tested, as the mRNAs remained stable even after a 2 h incubation (Figure 1). Similar results were obtained in an ascites Krebs-2 cell extract (data not shown). Taken together, these data demonstrate that YB-1 is a potent mRNA stabilizer in vitro, in several different cell extracts.
Fig. 1. YB-1 is a potent mRNA stabilizer in vitro. Capped CAT and CAT–3′ TNFα mRNAs (0.1 µg each) were incubated in a rabbit reticulocyte lysate (A) or HeLa extract (B) with buffer (control) or in the presence of YB-1 (0.5 µg). Total RNA was isolated at the times indicated, and CAT mRNA and 18S rRNA were detected by northern blot hybridization with the corresponding probes. CAT and CAT–3′ TNFα mRNAs from three independent experiments were quantified using PhosphorImaging software and normalized to 18S rRNA (right panels).
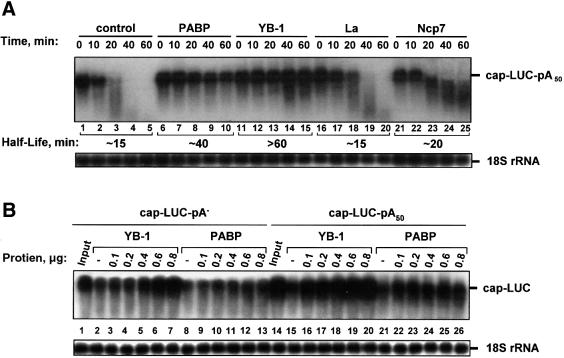
To determine whether the effect on mRNA stability is peculiar to YB-1, or is a non-specific effect elicited by any RNA binding protein, several other RNA binding proteins [La autoantigen (Gottlieb and Steitz, 1989), PABP (Görlach et al., 1994) and nucleocapsid HIV-1 protein, NCp7 (Tanchou et al., 1995)] were tested in a HeLa extract. La autoantigen and NCp7 are promiscuous high-affinity RNA binding proteins (Kd < 10 nM) (Tanchou et al., 1995; Belsham and Sonenberg, 1996), whereas PABP exhibits sequence specificity toward poly(A) (Deo et al., 1999; Görlach et al., 1994). As expected (Bernstein et al., 1989), PABP increased the stability of capped and polyadenylated luciferase (LUC) mRNA (Figure 2A, lanes 6–10), albeit to a lesser degree than YB-1 (lanes 11–15). Neither La (lanes 16–20), nor NCp7 (lanes 21–25) significantly affected LUC mRNA stability. Thus, mRNA stabilization is not a general effect elicited by all RNA binding proteins.
Fig. 2. YB-1 uniquely mediates mRNA stabilization. (A) Effect of general mRNA binding proteins on mRNA stability. Capped and polyadenylated LUC mRNA (0.1 µg) was incubated in a HeLa extract with buffer alone (–) or in the presence of 0.5 µg of the RNA binding protein indicated, and detected by northern blot hybridization. (B) Comparison of YB-1 and PABP mRNA stabilizing acitivities. LUC mRNA (0.1 µg) possessing or lacking poly(A) tail was incubated for 60 min in a HeLa extract with buffer (–) or in the presence of increasing amounts YB-1 or PABP, as indicated in the figure, and detected by northern blot hybridization. Lanes 1 and 14 (Input) show LUC mRNA isolated from the extract at 0 min of incubation.
To examine the role of the poly(A) tail in YB-1-mediated mRNA stabilization, we studied the effects of YB-1 and PABP on the stability of LUC mRNA with or without a poly(A) tail. Both non-adenylated and polyadenylated LUC mRNAs incubated in a HeLa extract in the presence of increasing amounts of YB-1 were markedly stabilized during a 60 min incubation (Figure 2B, compare lanes 2 and 15 with lanes 3–7 and 16–20, respectively). It is noteworthy, however, that more YB-1 (0.6 µg) was required for full stabilization of non-adenylated LUC mRNA in comparison with the polyadenylated counterpart (0.1 µg YB-1). As reported previously (Bernstein et al., 1989), PABP protected exclusively polyadenylated mRNA (Figure 2B, lanes 22–26), but failed to significantly stabilize non-adenylated mRNA (lanes 9–13). Therefore, in contrast to PABP, which functions primarily via the poly(A) tail, YB-1 stabilizes mRNA mainly via a non-poly(A)-dependent mechanism. However, as the polyadenylated mRNA was more effectively protected by YB-1, a role of the poly(A) tail in YB-1-mediated mRNA stabilization is evident.
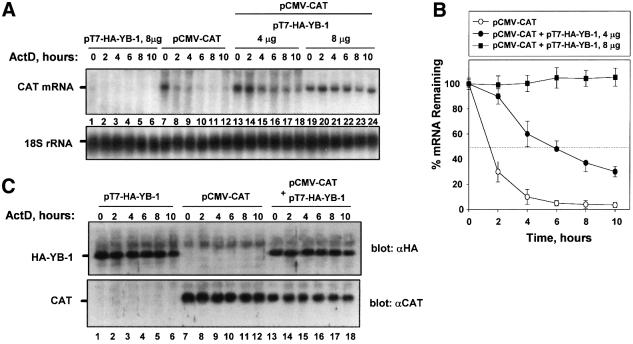
To determine whether YB-1 also stabilizes mRNA in vivo, it was overexpressed in HeLa cells using the vaccinia virus T7 RNA polymerase system (Fuerst et al., 1986) (Figure 3). The vaccinia system was used to ensure high expression of exogenous over endogenous YB-1, which is abundant in eukaryotic cells (∼0.1% of total protein, 2.6 × 106 molecules/cell; Davydova et al., 1997). Cells were infected with a recombinant vaccinia virus encoding T7 RNA polymerase, followed by transfection with an expression vector containing the CAT cDNA under the control of the human cytomegalovirus early promoter (pCMV-CAT) alone, or together with the hemagglutinin (HA)-tagged YB-1 cDNA under the control of the T7 promoter (pT7-HA-YB-1). Assuming ∼25% transfection efficiency, and considering that the signal from the exogenously expressed YB-1 in a typical transient transfection (using 8 µg of YB-1 plasmid) was about four times stronger than that from the endogenous YB-1 (data not shown), the amount of the exogenously expressed YB-1 was estimated to be ∼16-fold greater than that of the endogenous protein. To determine the effect of YB-1 overexpression on the half-life of CAT mRNA, actinomycin D (ActD) was added 20 h post-transfection to block transcription, and total RNA was isolated at various time intervals. In the absence of the overexpressed YB-1 the decay of CAT mRNA was rapid, with an apparent half-life of <2 h, as determined by northern blotting (Figure 3A, lanes 7–12). The amount of CAT protein did not change significantly during the incubation with ActD over 10 h (Figure 3C, lanes 7–12), in spite of the reduction in mRNA level, because CAT protein is very stable (half-life of ∼50 h; Thompson et al., 1991). Co-transfection of pCMV-CAT with pT7-HA-YB-1 resulted in a dramatic stabilization of CAT mRNA (Figure 3A, lanes 13–18 and 19–24). Remarkably, the level of CAT mRNA was increased up to 50-fold (at 10 h of incubation) when 8 µg of pT7-HA-YB-1 was co-transfected (Figure 3A and B). Consistent with earlier data (Davydova et al., 1997), YB-1 overproduction also resulted in translational repression of CAT mRNA; CAT protein level was decreased ∼2-fold (Figure 3C, compare lanes 7 and 13). Thus, the overproduction of YB-1 in vivo potently stabilizes CAT mRNA, while moderately inhibiting its translation.
Fig. 3. Overexpression of YB-1 in HeLa cells results in mRNA stabilization and translational repression. (A) Northern blot analysis of CAT mRNA decay in the absence or presence of overexpressed YB-1. HeLa cells infected with the recombinant vaccinia virus vT7F7-3 were co-transfected with pCMV-CAT (4 µg/plate) and with vector (pcDNA3-HA) alone or pcDNA3-HA-YB-1 (pT7-HA-YB-1; 4 or 8 µg/plate), as described in Materials and methods. Following addition of ActD for the times indicated, total cellular RNA was harvested and CAT mRNA and 18S rRNA were detected by hybridization with the corresponding probes. (B) Kinetic analysis of CAT mRNA decay. CAT mRNA from three independent experiments similar to that shown in (A) was quantified by PhosphorImaging software and normalized to 18S rRNA. (C) Western blot analysis of HA-YB-1 and CAT proteins. Five percent of HeLa extracts obtained from each time point in (A) were resolved by SDS–12% PAGE and analyzed with anti-HA or anti-CAT antibodies.
The CSD of YB-1 confers mRNA stability, but not translational repression
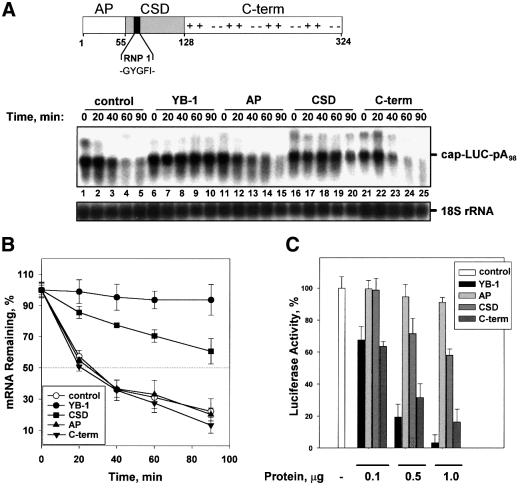
To determine which domains of YB-1 are responsible for mRNA stabilization and translational repression, the effects of different YB-1 fragments on the half-life and translation efficiency of the LUC mRNA were examined in a rabbit reticulocyte lysate. Unexpectedly, the CSD alone conferred stability on LUC mRNA almost as efficiently as the full-length YB-1 protein (Figure 4A, compare lanes 16–20 and 6–10). After 90 min of incubation, ∼100 and 60%, respectively, of LUC mRNA remained intact in the presence of the full-length YB-1 and the CSD fragment (Figure 4B). Although the CSD and the C-terminal portion of YB-1 (and related YB proteins) possess similar affinities for RNA (data not shown, see also Bouvet et al., 1995; Matsumoto et al., 1996), the C-terminal fragment failed to stabilize LUC mRNA (Figure 4A, lanes 21–25). Similarly, the AP domain, which also contributes to mRNA binding (Bouvet et al., 1995), did not affect mRNA stability (lanes 11–15). Thus, the non-specific RNA binding of YB-1 fragments is not sufficient to protect mRNA, and only the CSD exhibits stabilizing activity.
Fig. 4. The cold-shock domain of YB-1 confers mRNA stability, but not translational repression. (A) Top: schematic representation of YB-1 functional domains. The N-terminal AP-rich domain, the central CSD containing RNP 1 consensus motif, and the C-terminal domain with alternating basic (+) and acidic (–) amino acid clusters are indicated. Amino acids are numbered according to Evdokimova et al. (1995). (A) Bottom: effect of YB-1 fragments on mRNA stability. Capped and polyadenylated LUC mRNA (0.1 µg) was incubated in a rabbit reticulocyte lysate with buffer (control) or in the presence of 0.5 µg of YB-1 or its fragments. Total RNA was isolated at the times indicated and LUC mRNA and 18S rRNA were detected by northern blot hybridization with the corresponding probes. (B) Kinetic analysis of LUC mRNA decay. The amounts of LUC mRNA from three independent experiments similar to that shown in (A) were quantified by PhosphorImaging software and normalized to 18S rRNA. (C) Effect of YB-1 fragments on mRNA translation. Capped and polyadenylated LUC mRNA (0.1 µg) was incubated in a rabbit reticulocyte lysate with buffer alone (–) or increasing amounts of the protein indicated. Translation reactions were incubated at 30°C for 60 min and luciferase activity was monitored by luminometer. The luciferase activity in the presence of buffer alone (control) was set as 100%. Error bars denote the standard error from three independent experiments.
To establish whether mRNA stabilization correlates with translational repression, the effects of full-length YB-1 or individual YB-1 domains on luciferase synthesis were monitored. In agreement with previous data (Matsumoto et al., 1996; Izumi et al., 2001), the AP domain and the CSD affected translation only slightly (∼10 and 40%, respectively, at the highest concentrations used), while the C-terminal domain of YB-1 repressed translation to almost the same degree as the full-length YB-1 protein (Figure 4C). Thus, although the precise mechanism by which YB-1 represses translation remains to be determined (see also Discussion), these data indicate that the two activities of YB-1, mRNA stabilization and translational inhibition, are distinct and map to separate domains of the protein.
YB-1 stabilizes mRNA in a cap-dependent manner
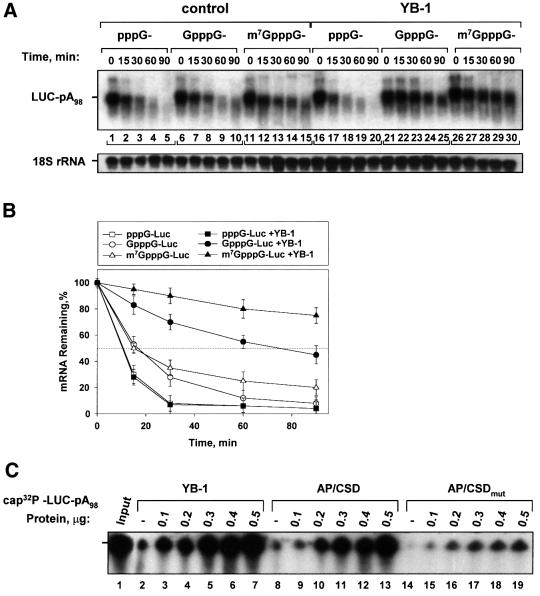
The 5′ cap structure and 3′ poly(A) tail are the primary targets for mRNA degradation pathways, and play critical roles in mRNA stability (Ross, 1995; Jacobson and Peltz, 1996; Schwartz and Parker, 2000). Because the poly(A) tail was not absolutely required for YB-1-mediated mRNA stabilization (Figure 2B), we next analyzed whether mRNA stabilization by YB-1 is dependent upon the presence of the cap structure. To assess the role of the cap in YB-1-mediated mRNA stabilization, polyadenylated LUC mRNAs possessing either a methylated (m7GpppG) or unmethylated (GpppG) cap, or lacking the cap structure altogether (pppG), were examined. Decay rates of the differentially cap-modified LUC mRNAs in the presence or absence of YB-1 in a HeLa extract are presented in Figure 5A. In agreement with previous data (e.g. Furuichi et al., 1977), LUC mRNAs possessing a blocked 5′ end (m7GpppG or GpppG) were more stable than uncapped mRNA (Figure 5A, compare lanes 1–5, 6–10 and 11–15). While YB-1 did not affect the stability of uncapped LUC mRNA, it dramatically stabilized the mRNAs harboring m7GpppG or GpppG cap structures (compare lanes 16–20 and 21–30), increasing their half-life by ∼4-fold (Figure 5B). In the presence of YB-1 a significant portion of the capped and polyadenylated LUC mRNAs remained intact even after 90 min of incubation, as compared with <10% for the uncapped mRNA counterpart.
Fig. 5. YB-1 protects mRNA against degradation in a cap-dependent manner. (A) Polyadenylated LUC mRNAs (0.05 µg) with differently modified 5′ ends were incubated in a HeLa extract in the absence or presence of YB-1 (0.2 µg). Total RNA was isolated at the times indicated and LUC mRNA and 18S rRNA were detected by northern blot hybridization with the corresponding probes. (B) Kinetic analysis of LUC mRNA decay. LUC mRNA from three independent experiments similar to that shown in (A) was quantified by PhosphorImaging software and normalized to 18S rRNA. (C) Effect of YB-1 on mRNA decapping. 32P-cap-labeled and polyadenylated LUC mRNA (0.05 µg) was incubated in a HeLa extract for 60 min with buffer alone (–) or in the presence of increasing amounts of either YB-1, AP/CSD or AP/CSD mutant, as indicated, and analyzed by 4% acrylamide–7 M urea gel electrophoresis and autoradiography. Lane 1 (Input) shows LUC mRNA isolated from the extract at 0 min of incubation.
Next, the effect of YB-1 on mRNA cap structure stability was assayed directly. LUC mRNA was 32P-labeled exclusively in the 5′ cap structure and incubated in a HeLa extract in the presence of increasing amounts of YB-1. The mRNA cap structure was almost completely removed after 60 min of incubation (Figure 5C, compare lanes 1 and 2, 8, 14), but YB-1 significantly inhibited mRNA decapping and degradation, even at the lowest amount added (0.1 µg; lane 3). Increasing the YB-1 concentration to 0.5 µg (lane 7) resulted in complete retention of the cap structure during the incubation time. To determine whether the CSD can confer cap protection as well, a YB-1 fragment (AP/CSD) and a form bearing two point mutations within the RNP 1 motif (GYGFI → GAGAI) were tested. The RNP 1 motif is a probable contact site between YB-1 and RNA; these mutations were based on previous reports showing an essential role in RNA binding for conserved aromatic amino acids in the RNP 1 motif (Bouvet et al., 1995; Schroder et al., 1995). We chose to use the two AP/CSD domains of YB-1 in this assay, instead of just the CSD, to minimize the effects of the point mutations on the general RNA binding capacity of the fragments. Strikingly, in a manner similar to the full-length protein, the AP/CSD fragment was sufficient to protect the cap-labeled mRNA, while the AP/CSD mutant exhibited much less of a protective effect (Figure 5C, compare lanes 9–13 and 15–19). The AP domain alone and the C-terminal fragment of YB-1 failed to prevent mRNA decapping and degradation, although they were stable during the incubation time (data not shown, see also Figure 4A). Therefore, the CSD and, in particular, the RNP 1 motif, appear to play an important role in protection of the cap structure and mRNA against degradation. Taken together, these data suggest that YB-1 functions as a cap-protective protein that modulates the stability of capped mRNAs.
YB-1 destabilizes the interaction of eIF4E with the cap structure
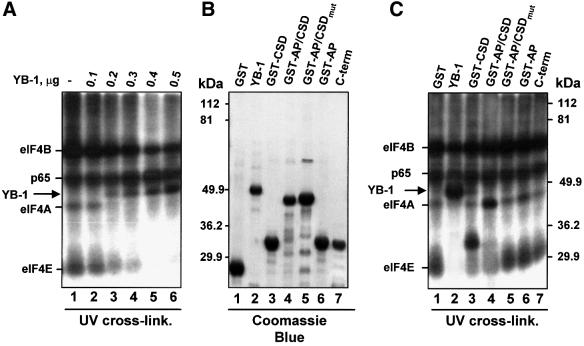
To elucidate the mechanism by which YB-1 prevents the decapping and degradation of mRNA, the effect of YB-1 on the interaction of the cap binding protein eIF4E with the mRNA cap structure was studied. LUC mRNA possessing a 32P-labeled cap was incubated in a reticulocyte lysate to allow for the assembly of RNA–protein complexes, and then subjected to UV irradiation. The translation initiation factors eIF4B (80 kDa), eIF4A (46 kDa) and eIF4E (24 kDa) were cross-linked to the capped RNA (Figure 6A, lane 1). The assignment of the cross-linked proteins is based on numerous previous reports (e.g. Pelletier and Sonenberg, 1985). Cross-linking of eIF4E to the cap structure is dependent on direct binding of eIF4E to the cap, whereas eIF4A and eIF4B interaction with the cap is a secondary event (Gingras et al., 1999). The non-specific cross-linking of a 65 kDa protein has also been reported previously (Pelletier and Sonenberg, 1985). Addition of increasing amounts of YB-1 resulted in a gradual inhibition of eIF4E, eIF4A and eIF4B cross-linking to the cap-labeled mRNA. This was accompanied by the appearance of a major cross-linked band of ∼50 kDa (Figure 6A, lanes 2–6). The ∼50 kDa cross-linked polypeptide was identified as YB-1 by immunoprecipitation (data not shown; see also Figure 7B). It is also noteworthy that YB-1 failed to decrease the non-specific cross-linking of p65. Thus, YB-1-mediated stabilization of capped mRNAs is accompanied by a specific inhibition of eIF4E (and associated initiation factors) interaction with the cap.
Fig. 6. YB-1 inhibits the interaction of eIF4E with the cap. (A) Cross-linking of proteins to the mRNA cap structure. 32P-cap-labeled and polyadenylated LUC mRNA (0.1 µg) was incubated in a rabbit reticulocyte lysate for 15 min with buffer alone (–) or in the presence of increasing amounts of YB-1, as indicated. RNA–protein complexes were covalently cross-linked by UV irradiation, treated with RNase A and subjected to SDS–12% PAGE. (B) Coomassie Blue staining of YB-1 or its fragments (2 µg each). (C) Cross-linking of YB-1 fragments to the cap structure. 32P-cap-labeled and polyadenylated LUC mRNA was incubated in a rabbit reticulocyte lysate as in (A) with buffer alone (–) or in the presence of 1 µg of the indicated proteins. RNA–protein complexes were cross-linked by UV irradiation and detected as in (A).
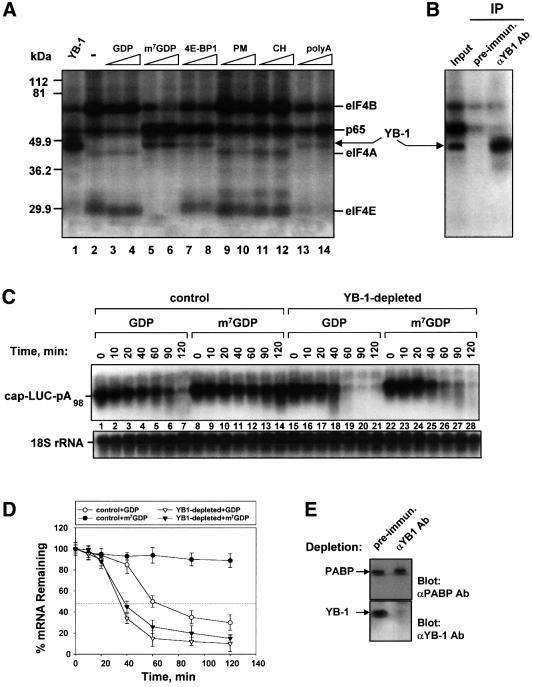
Fig. 7. YB-1 cross-links to the cap and promotes mRNA stabilization when the eIF4E interaction is reduced. (A) Effect of the translation inhibitorson YB-1 cross-linking. 32P-cap-labeled and polyadenylated LUC mRNA (0.1 µg) was incubated in a rabbit reticulocyte lysate with buffer alone (–) or in the presence of YB-1 (1 µg), GDP (0.5 and 1 mM), m7GDP (0.5 and 1 mM), 4E-BP1 (1 and 4 µg), pactamycin (PM) (2 and 10 µg/ml), cycloheximide (CH) (10 and 50 µg/ml) or poly(A) (40 and 80 µg/ml). Reaction mixtures were incubated for 15 min at 30°C, UV irradiated, treated with RNase A and subjected to SDS–12% PAGE. (B) Cross-linked proteins obtained as in (A) in the presence of m7GDP (0.5 mM) were immunoprecipitated (IP) with either pre-immune or anti-YB-1 antibodies as described in Materials and methods and subjected to SDS–12% PAGE. (C) Northern blot analysis of LUC mRNA decay in a rabbit reticulocyte lysate pre-incubated with either pre-immune (control) or anti-YB-1 (YB-1-depleted) antibodies. Capped and polyadenylated LUC mRNA (0.1 µg) was incubated in the control or YB-1-depleted rabbit reticulocyte lysate in the presence of GDP (0.5 mM) or m7GDP (0.5 mM), as indicated in the figure. Total RNA was isolated at the times indicated and LUC mRNA and 18S rRNA were detected by sequential hybridization with the corresponding probes. (D) Kinetic analysis of LUC mRNA decay. The amounts of LUC mRNA from three independent experiments identical to that shown in (C) were quantified by PhosphorImaging software and normalized to 18S rRNA. (E) A western blot showing the efficiency of YB-1 immunodepletion from the rabbit reticulocyte lysate preincubated with either pre-immune or anti-YB-1 antibodies coupled to protein A–Sepharose. Western blot analysis was performed with pre-immune or YB-1-depleted reticulocyte lysate using anti-PABP antibodies as a loading control (upper panel) or anti-YB-1 antibodies (lower panel).
Next, the various YB-1 domains (Figure 6B) were examined for their ability to cross-link to cap-labeled mRNA. Coincident with the cap-protective activity exhibited by the CSD (Figure 5B), the CSD alone, or when linked to the AP domain, cross-linked to the cap as effectively as full-length YB-1 (Figure 6C, lanes 2–4). The AP/CSD mutant (lane 5), the AP domain alone (lane 6), and the C-terminal fragment of YB-1 (lane 7) failed to cross-link to the cap-labeled mRNA, and did not affect cross-linking of the cap binding protein complex. Therefore, the CSD alone is sufficient, and the RNP 1 motif is necessary, for the cross-linking of YB-1 to the cap.
YB-1 binding at or near the cap structure results in robust mRNA stabilization
The previous data suggest that exogenous YB-1 can destabilize the eIF4E–cap interaction. It is therefore conceivable that displacement of eIF4E from the cap by other means may also increase the binding of endogenous YB-1 to the cap-proximal region of mRNA. Cap-labeled LUC mRNA was thus translated in a rabbit reticulocyte lysate in the presence of different translation inhibitors, followed by UV irradiation. The cap analog, m7GDP, was used to specifically prevent eIF4E from binding to the cap. We also tested the eIF4E binding protein 4E-BP1, which prevents the assembly of the eIF4F cap binding complex by disrupting the eIF4E–eIF4G interaction (Haghighat et al., 1995; for reviews see Gingras et al., 1999; Raught et al., 2000). In addition, we examined several translation inhibitors such as pactamycin (PM), which prevents 60S ribosomal subunit joining and stabilizes the 40S–Met–tRNA initiation complex (Fresno et al., 1976), and cycloheximide (CH), which blocks the peptidyl transferase reaction (Obrig et al., 1971). Finally, poly(A) was utilized as a competitor to determine whether PABP removal affects the binding of proteins to the cap. While GDP had only a minor effect (Figure 7A, compare lane 2 with lanes 3 and 4), m7GDP (lanes 5 and 6) significantly reduced the cross-linking of eIF4E, eIF4A and eIF4B to the cap, as expected (Lee et al., 1983; Pelletier and Sonenberg, 1985). Although 4E-BP1 does not directly inhibit eIF4E cap binding activity, addition of 4E-BP1 decreased eIF4E cross-linking (lanes 7 and 8). This most likely reflects a requirement for eIF4G in stabilizing the eIF4E interaction with capped mRNA (Haghighat and Sonenberg, 1997; Ptushkina et al., 1998). Poly(A) also inhibited the cross-linking of the initiation factors to mRNA (Figure 7A, lanes 13 and 14), possibly due to disruption of the interaction between PABP and eIF4G, which may decrease the stability of the eIF4F–cap interaction. Coincident with the decrease in eIF4E binding, the cross-linking of a ∼50 kDa polypeptide increased (lanes 5–8 and 13–14). Immunoprecipitation experiments using anti-YB-1 antibodies confirmed that the 50 kDa protein that cross-links to cap-labeled mRNA in the presence of m7GDP is YB-1 (Figure 7B). PM or CH neither inhibited the cross-linking of initiation factors to the cap, nor did they stimulate the cross-linking of YB-1 (Figure 7A, lanes 9–12). These data demonstrate that endogenous YB-1 can be cross-linked to the cap-labeled mRNA when the stability of the eIF4E–cap interaction is impaired.
If YB-1 cross-linking correlates with mRNA protection against decapping and degradation, it stands to reason that m7GDP or 4E-BP1 could inhibit mRNA decay by destabilizing the eIF4E–cap interaction and rendering the cap-proximal region more accessible to YB-1. To demonstrate that YB-1 is directly responsible for the mRNA stabilization observed under these conditions, YB-1 was specifically removed from the rabbit reticulocyte lysate using anti-YB-1 antibodies (Figure 7E). Immunodepletion with pre-immune antibodies did not affect mRNA stability, whereas removal of YB-1 significantly accelerated LUC mRNA degradation, decreasing its half-life from 60 to 30 min (Figure 7C, compare lanes 1–7 and 15–21 and Figure 7D). Upon replenishment, YB-1 fully restored mRNA stability (data not shown). More importantly, in the control lysate LUC mRNA was rendered stable over 2 h of incubation in the presence of m7GDP (Figure 7C, lanes 8–14). However, in the YB-1-depleted extract no stabilization by m7GDP was observed (Figure 7C, lanes 22–28). This result demonstrates clearly that YB-1 is required for the mRNA stabilization that is elicited by m7GDP. Taken together, these data support a model whereby YB-1 protects the cap structure, and consequently the mRNA, against degradation, when the association of eIF4E with the cap is reduced.
Discussion
In this report we demonstrate that YB-1 is a potent cap-dependent mRNA stabilizer in vivo and in vitro, in several different mammalian cell extracts. mRNA stabilization facilitated by YB-1 is general and sequence non-specific, as it is exerted on CAT, CAT–3′ TNFα and LUC mRNAs. The striking finding that only those mRNAs possessing GpppG or m7GpppG cap structures are stabilized by YB-1 likely indicates an important physiological role; uncapped truncated mRNAs would not be protected and, thus, would be prevented from generating dominant-negative proteins. It is also of interest that YB-1 does not prevent endonuclease cleavage of mRNA (Evdokimova et al., 1995; and data not shown) arguing for its involvement in mRNA protection against exonucleolytic degradation rather than in the protection of mRNAs whose decay is mediated by specific endonucleases.
In Saccharomyces cerevisiae two major pathways of mRNA degradation involving an initial deadenylation, followed by decapping and either 5′→3′ or 3′→5′ exonucleolytic degradation, have been defined. A yeast decapping enzyme, Dcp1p, removes the mRNA cap structure and renders the 5′ end of mRNA accessible to the potent 5′→3′ Xrn1p exonuclease (Schwartz and Parker, 2000). Although less is known about the mechanism(s) of mRNA turnover in mammalian cells, mRNA decapping and 5′→3′ exonuclease activities have been reported (Nuss et al., 1975; Furuichi et al., 1977). More recently, a mammalian mRNA decapping activity that behaves similarly to the yeast counterpart has been identified (Gao et al., 2001). In addition, a mammalian deadenylase termed PARN (also known as DAN) interacts with the 5′ cap and facilitates the degradation of capped and polyadenylated mRNAs (Dehlin et al., 2000; Gao et al., 2000; Martinez et al., 2000). Thus, the cap structure plays a major role in determining mRNA stability.
Several findings reported here point to an important role for YB-1 in the inhibition of mRNA decapping and degradation. YB-1 protects mRNA possessing GpppG or m7GpppG blocking structures at the 5′ end mRNA (Figure 5). A striking and unexpected finding of this study is that inhibitors that reduce the eIF4E–cap interaction, such as cap analogs or the repressor protein 4E-BP1, facilitate the cross-linking of YB-1 to the cap and increase mRNA stability. In contrast, immunodepletion of YB-1 from extracts abrogates mRNA stabilization (Figure 7). The CSD alone effectively stabilizes capped mRNA and cross-links to the mRNA cap structure (Figures 4–6). The CSD together with the flanking N- and C-terminal domains are also involved in anchoring the YB-1 protein to the mRNA in a sequence-independent manner (Wolffe, 1994; Bouvet et al., 1995; Sommerville and Ladomery, 1996). Taken together, these data support a model in which YB-1 is normally bound to the mRNA body and interacts with the cap only when eIF4E function is impaired. It is also likely that different events leading to a decrease in eIF4F interaction with the mRNA, such as disruption of the eIF4E–eIF4G or eIF4G–PABP interactions (Figure 7A), result in YB-1 binding at or near the cap. Thus, association of eIF4F or YB-1 with the cap-proximal region of mRNA is mutually exclusive. Consistent with this model, we demonstrated previously that a dominant-negative eIF4A mutant that prevents eIF4F interaction with the cap structure stimulates cross-linking of a 50 kDa protein, probably YB-1, to the cap (Figure 7B; Svitkin et al., 2001).
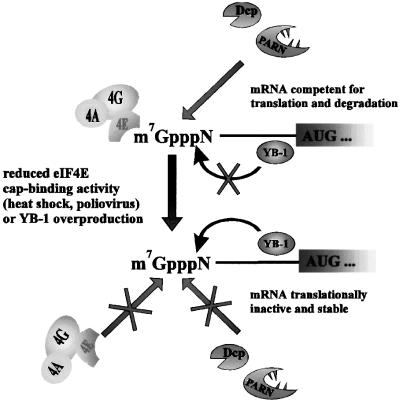
YB-1 may act as a ‘sensor’ to monitor the cellular environment and stabilize mRNA under stress conditions (Figure 8). One prediction from this idea is that inhibition of eIF4E cap binding by stress would lead to an increase in the YB-1 interaction with the cap, and mRNA stabilization. Indeed, a large number of mRNAs are stabilized during heat shock (Lindquist, 1981) and after infection with various viruses, such as poliovirus (Kaufmann et al., 1976). Heat shock elicits dephosphorylation of 4E-BP1, which results in the association of 4E-BP1 with eIF4E, and a less stable interaction of eIF4E with capped mRNAs (for review see Schneider, 2000). Infection with poliovirus inhibits cap-dependent translation by cleaving eIF4G, and indirectly destabilizing the interaction of eIF4E with the cap structure (Lee and Sonenberg, 1982; Belsham and Jackson, 2000). It is also of interest that prokaryotic YB proteins, termed cold-shock proteins, which are comprised solely of a CSD, are indispensable for survival during cold-shock adaptation and have been implicated in diverse environmental stress responses (Yamanaka et al., 1998). Given the function of the CSD of YB-1 in mRNA stabilization in mammalian cells, a role for prokaryotic proteins in the regulation of mRNA stability needs to be assessed.
Fig. 8. Model: YB-1 functions as a ‘sensor’ to monitor eIF4E and mediate mRNA stabilization. Under standard growth conditions the mRNA m7GpppN cap (where m is a methyl group, and N is any nucleotide) is accessible to the eIF4F cap binding complex as well as to degradative enzymes, such as Dcp1 and PARN. YB-1 is anchored to the mRNA body due to its general non-specific RNA binding activity, and its interaction with the cap-proximal region is prevented by eIF4F. Under certain conditions, such as during early embryogenesis or stress, an increase in YB-1 activity or sequestration of eIF4E from the cap stimulates YB-1 binding at or near the cap, leading to general mRNA stabilization.
An increase in YB-1 concentration in cells would be predicted, based on our results, to result in eIF4E displacement, and subsequent mRNA silencing and stabilization. It is likely that such a mechanism operates in germinal cells, during spermatogenesis and oogenesis, in which mRNAs are translationally masked by YB proteins (Richter and Smith, 1984; Murray et al., 1991; Kwon et al., 1993; Bouvet and Wolffe, 1994; Matsumoto and Wolffe, 1998; Sommerville, 1999).
The exact mechanism by which YB-1 represses translation remains to be determined. While the C-terminal portion of YB-1 strongly inhibits translation, the CSD is much less inhibitory (Figure 4C), despite the fact that it displaces eIF4E (Figure 6C). This is not entirely surprising, since translation in rabbit reticulocyte lysate is relatively cap independent (e.g. Svitkin et al., 1996). The ability of the C-terminal domain of YB-1, as well as the related Xenopus protein FRGY2, to repress translation was also demonstrated in earlier reports (Matsumoto et al., 1996; Izumi et al., 2001). This domain can be stably incorporated in mRNPs, while the CSD alone can not (Matsumoto et al., 1996). Perhaps the homomultimerization of YB-1 induced by the C-terminal domain is required for the inhibition of translation.
Interestingly, YB-1 overexpression in several human tumors correlates with resistance to chemotherapy and UV irradiation (Ohga et al., 1996; Bargou et al., 1997). Increased stability of c-fos, c-myc, cyclin B1 and other short-lived mRNAs in response to UV irradiation or therapeutic drugs is well documented (Laird-Offringa et al., 1990; Maity et al., 1995; Blattner et al., 2000). It would thus be of interest to determine whether YB-1 affects the stability of short-lived mRNAs encoding proto-oncogenes, growth factors and cytokines, thereby contributing to cell proliferation. It will also be important to determine whether and how the activity of YB-1 is regulated in the cell, particularly in response to extracellular stimuli or environmental stress.
Materials and methods
Plasmids
The plasmid pBluescript II-KS-CAT–3′ TNFα consisting of a 76 bp CAT 5′ UTR, CAT coding sequence and 3′ UTR TNFα was derived from pBluescript II-KS-CAT–TNFα CAT (Beutler and Brown, 1991) by excision of the KpnI–ClaI fragment (containing 2000 bp of the 5′ TNFα genomic region), blunting the ends and religation. The plasmids encoding CAT mRNA, pSP64-CAT (Parkin et al., 1988), LUC mRNAs, pT3-LUC and pT3-LUC-poly(A98) (Iizuka et al., 1994) and pGEM-18S cDNA (Chan et al., 1984) have been described previously.
To express proteins in HeLa cells, pcDNA3-HA-YB-1 was generated by subcloning the YB-1 coding region from pET-3-1-YB-1 (p50) (Evdokimova et al., 1995) into the NdeI–XhoI sites of pcDNA3-HA containing the T7 promoter (Imataka et al., 1998). The pCMV-CAT expression plasmid lacking the T7 promoter was derived from pCMV-myc P2-CAT (Nanbru et al., 1997) by excision of the XbaI–EcoRV fragment encoding the c-myc 5′ UTR. For bacterial expression and protein purification of the YB-1 fragments, PCR-amplified cDNAs encoding AP/CSD (amino acids 1–128) and CSD (amino acids 55–128) were digested with NdeI–EcoRI and BamHI–EcoRI, respectively, and ligated into pGEX2T (Pharmacia). To construct the plasmid encoding the AP/CSD mutant bearing two point mutations within the RNP 1 motif (GYGFI → GAGAI) of the CSD, pGEX2T-AP/CSD was used as a template for PCR amplification. A plasmid encoding the AP domain (amino acids 1–55) was derived from pGEX2T-AP/CSD by deletion of the CSD using two SmaI sites and religation. A cDNA encoding the C-terminal domain of YB-1 (amino acids 128–324) was PCR amplified, digested with NdeI/XhoI and ligated into pET-15b (Novagen). All constructs were confirmed by sequencing.
The full-length cDNA of human PABP followed by six histidines was cloned into pET-3b (Novagen) (H.Imataka, unpublished). The pET-La expression plasmid was described previously (Chang et al., 1994).
Recombinant proteins and antibodies
Recombinant proteins were expressed in E.coli BL21(DE3). YB-1 was purified as described previously (Evdokimova et al., 1998). Briefly, bacteria were lysed by sonication in a buffer containing 2 M NaCl, 10 mM Tris–HCl pH 7.6 and 0.5 mM PMSF. Cell debris was removed by centrifugation at 10 000 g for 20 min at 4°C, and the supernatant was diluted 4-fold with 10 mM Tris–HCl pH 7.6 and passed through a heparin–Sepharose column (Pharmacia). After washing the column with loading buffer (500 mM NaCl, 10 mM Tris–HCl pH 7.6), bound YB-1 was eluted using 2 M NaCl and 10 mM Tris–HCl pH 7.6. The AP domain and CSD were purified as GST fusion proteins by glutathione–Sepharose (Pharmacia) chromatography according to the manufacturer’s recommendations. The GST tag was removed using PreScission protease (Pharmacia). The YB-1 His-tagged C-terminal domain was purified using Talon resin (Clontech) equilibrated with 500 mM NaCl, 10 mM HEPES–KOH pH 7.6 and eluted with the same buffer containing 100 mM imidazole.
To purify PABP, bacteria were lysed in 200 mM KCl, 10 mM Tris–HCl pH 7.6 and 0.5 mM phenylmethylsulfonyl fluoride (PMSF). The supernatant was clarified by centrifugation and passed through poly(A)–Sepharose (Pharmacia) equilibrated with the same buffer. After washing the column with 500 mM KCl, 20 mM Tris–HCl pH 7.6, PABP was eluted with 2 M LiCl, 50 mM Tris–HCl pH 7.6. All purified proteins were dialyzed against 100 mM KCl, 10 mM HEPES–KOH pH 7.6 and stored at –80°C.
Rabbit anti-YB-1 and anti-PABP antibodies were as described earlier (Davydova et al., 1997; Imataka et al., 1998). Mouse anti-HA and rabbit anti-CAT antibodies were purchased from Babco and Eppendorf 5′, respectively. For western blot assay, peroxidase-linked anti-mouse or anti-rabbit IgG in combination with the ECL system (Amersham) were used. For immunodepletion or immunoprecipitation, corresponding antibodies were covalently bound to protein A–Sepharose beads (Pharmacia) using 0.1% glutaraldehyde and stored in 100 mM KCl, 10 mM HEPES–KOH pH 7.6 at 4°C.
mRNA synthesis and labeling
To generate mRNAs, pSP64-CAT, pT3-LUC and pT3-LUC-poly(A98) linearized with BamHI, and pBluescript II-KS-CAT–3′ TNFα linearized with SmaI, were used as templates for in vitro transcription according to the RiboMax protocol (Promega). Capped transcripts were obtained in a reaction mixture which contained the following at a concentration of 20 µM GTP and 200 µM of either m7GpppG or GpppG. To generate 32P-cap-labeled mRNA, uncapped mRNA was capped and methylated using vaccinia virus guanylyltransferase (Gibco-BRL) in the presence of 0.4 mM S-adenosyl-l-methionine (Sigma) and 100 µCi [α-32P]GTP as described previously (Goyer et al., 1989). Labeled mRNAs were purified on CHROMA S30 spin columns (Clontech). All mRNAs used in the same experiment were synthesized in parallel.
Assay of mRNA translation and degradation
The cell-free translation system derived from mouse Krebs-2 ascites carcinoma cells, also used here for the mRNA degradation assay, was described previously (Svitkin et al., 1984). Nuclease-treated rabbit reticulocyte lysate was purchased from Promega. In vitro translation/degradation assays were performed in a final volume of 14 µl containing 10 µl of cell extract supplemented with translation ingredients and a source of energy, mRNA (50–100 ng) and buffer A (100 mM KCl, 10 mM HEPES–KOH pH 7.6). Where indicated, the reactions were supplemented with protein preparations or protein synthesis inhibitors in buffer A. Reactions were incubated at 30°C for the indicated times and luciferase activity was measured using the luciferase assay kit (Boehringer Mannheim) in a Lumat LB 9507 bioluminometer (EG&G Berthold). YB-1- or mock-depleted extracts were obtained by incubation of 200 µl of either extract with 100 µl of anti-YB-1 or preimmune antibodies immobilized on protein A–Sepharose beads for 60–90 min at 4°C with end-over-end rotation.
To prepare HeLa extract, monolayer HeLa cells were lysed in 100 mM KCl, 2 mM MgCl2, 2 mM dithiothreitol (DTT), 0.2% NP-40 and 10 mM HEPES–KOH pH 7.6. Post-mitochondrial supernatant (S10) was obtained by centrifugation at 12 000 g for 15 min. For mRNA stability assays, mRNAs (50–100 ng) were incubated with 10 µl (∼100 µg) of S10 HeLa extract in a 14 µl reaction mixture containing 100 mM KCl, 2 mM MgCl2, 10 mM HEPES–KOH pH 7.6, 2 mM DTT, 10 mM creatine phosphate, 0.5 U of creatine phosphokinase, 1 mM ATP, 0.4 mM GTP and 0.1 mM spermine.
For northern blotting analysis, reactions were incubated at 30°C for the times indicated and stopped by the addition of 50 µl of stop buffer (100 mM NaCl, 25 mM Tris–HCl pH 7.6, 0.5% SDS and 100 µg/ml yeast tRNA). Total RNA was phenol–chloroform extracted, separated on a 1% formaldehyde–agarose gel and transferred onto nitrocellulose membrane Hybond-N+ (Amersham). Blots were hybridized with randomly primed 32P-labeled cDNAs according to standard procedures (Sambrook et al., 1989). Quantifications were carried out on a Molecular Dynamics PhosphorImager using the ImageQuant software.
To analyze mRNA decay in vivo, HeLa cells growing in six-well plates were infected with vaccinia virus vTF7-3 (Fuerst et al., 1986). Four hours later, cells were co-transfected with pCMV-CAT and pcDNA3-HA-YB-1 (called pT7-HA-YB-1 in Figure 2) or vector alone (pcDNA3-HA) using Lipofectin (Gibco-BRL). Empty pcDNA3-HA vector was added to bring the total amount of co-transfected plasmids to 12 µg/plate. Two hours later the transfection mixture was changed for minimal essential medium (Life Technologies Inc., Gaithersburg, MD) supplemented with 10% fetal bovine serum. After 18–20 h of incubation, ActD was added to a final concentration of 10 µg/ml and cells were lysed at the time indicated in 400 µl of 100 mM KCl, 10 mM HEPES–KOH pH 7.8, 0.1 mM EDTA, 0.4% NP-40, 1 mM PMSF, 10 nM okadaic acid and 1 nM microcystin. Phosphatase inhibitors were added to prevent YB-1 dephosphorylation, which results in a doubling of bands on western blot. Five percent (∼20 µl) of each cell extract from each time point was used for western blotting with anti-HA or anti-CAT antibodies. The remaining extracts were phenol–chloroform deproteinized and analyzed by northern blot hybridization, as above.
UV cross-linking and immunoprecipitation
32P-cap-labeled mRNA (∼0.1 µg, 40 000 c.p.m.) was incubated in a rabbit reticulocyte lysate for 15 min at 30°C. Translation mixtures were transferred to a 96-well plate on ice and irradiated at 4°C for 20 min with a UV Stratalinker (Stratagene) at a distance of 15 cm, followed by digestion with RNase A (0.5 µg per reaction) for 15 min at 37°C. Cross-linked proteins were analyzed by SDS–12% PAGE and autoradiography, or were subjected to immunoprecipitation as follows: RIPA buffer (100 µl; 150 mM NaCl, 1% NP-40, 0.1% SDS and 50 mM Tris–HCl pH 7.6) was added to the reactions, and samples were clarified by centrifugation and incubated with 10 µl of the appropriate antibodies fixed on protein A–Sepharose beads for 2 h at 4°C with rotation. After washing with RIPA buffer, immunoprecipitates were diluted with SDS-gel loading buffer, resolved by SDS–10% PAGE and detected by autoradiography.
Acknowledgments
Acknowledgements
We thank I.Gallouzi, A.-B.Shyu and A.-C.Gingras for helpful discussions and critical reading of the paper, and C.Lister for excellent technical assistance. We also thank Y.-L.Chan, A.Sitikov and I.G.Wool for the pGEM-18S cDNA plasmid, B.Beutler for the pBluescript II-KS-CAT–TNFα plasmid, A.-C.Prats for the CAT expression plasmid and N.Iizuka for the luciferase plasmids. This work was supported by the Russian Foundation for Basic Research (RFBR-01-04-49038 and RFBR-00-15-97903) to L.P.O. and the Canadian Institute of Health Research (CIHR) to N.S., who is a Distinguished Scientist of the CIHR and a Howard Hughes Medical Institute International Scholar. V.E. was a recipient of a Human Frontier Science Program Organization long-term fellowship.
References
- Bargou R.C. et al. (1997) Nuclear localization and increased levels of transcription factor YB-1 in primary breast cancers are associated with intrinsic MDR1 gene expression. Nature Med., 3, 447–450. [DOI] [PubMed] [Google Scholar]
- Belsham G.J. and Jackson,R.J. (2000) Translation initiation on picornavirus RNA. In Sonenberg,N., Hershey,J.W.B. and Mathews,M.B. (eds), Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 869–900.
- Belsham G.J. and Sonenberg,N. (1996) RNA–protein interactions in regulation of picornavirus RNA translation. Microbiol. Rev., 60, 499–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein P. and Ross,J. (1989) Poly(A), poly(A)-binding protein and the regulation of mRNA stability. Trends Biochem. Sci., 14, 373–377. [DOI] [PubMed] [Google Scholar]
- Bernstein P., Peltz,S.W. and Ross,J. (1989) The poly(A)–poly(A)-binding protein complex is a major determinant of mRNA stability in vitro.Mol. Cell. Biol., 9, 659–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beutler B. and Brown,T. (1991) A CAT reporter construct allows ultrasensitive estimation of TNF synthesis and suggests that the TNF gene has been silenced in non-macrophage cell lines. J. Clin. Invest., 87, 1336–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner C., Kannouche,P., Litfin,M., Bender,K., Rahmsdorf,H.J., Angulo,J.F. and Herrlich,P. (2000) UV-induced stabilization of c-fos and other short-lived mRNAs. Mol. Cell. Biol., 20, 3616–3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G. (1972) Proteins tightly bound to globin mRNA. Biochem. Biophys. Res. Commun., 47, 88–95. [DOI] [PubMed] [Google Scholar]
- Bouvet P. and Wolffe,A.P. (1994) A role for transcription and FRGY2 in masking maternal mRNA within Xenopus oocytes. Cell, 77, 931–941. [DOI] [PubMed] [Google Scholar]
- Bouvet P., Matsumoto,K. and Wolffe,A.P. (1995) Sequence-specific RNA recognition by the Xenopus Y-box proteins. J. Biol. Chem., 270, 28297–28303. [DOI] [PubMed] [Google Scholar]
- Caponigro G. and Parker,R. (1996) Mechanisms and control of mRNA turnover in Saccharomyces cerevisiae. Microbiol. Rev., 60, 233–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Y.-L., Gutell,R., Noller,H.F. and Wool,I.G. (1984) The nucleotide sequence of a rat 18S ribosomal ribonucleic acid gene and a proposal for the secondary structure of 18S ribosomal ribonucleic acid. J. Biol. Chem., 259, 224–230. [PubMed] [Google Scholar]
- Chang Y.-N., Kenan,D.J., Keene,J.D., Gatignol,A. and Jeang,K.-T. (1994) Direct interactions between autoantigen La and human immunodeficiency virus leader RNA. J. Virol., 68, 7008–7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.-Y., Gherzi,R., Andersen,J.S., Gaietta,G., Jurchott,K., Royer, H.-D., Mann,M. and Karin,M. (2000) Nucleolin and YB-1 are required for JNK-mediated interleukin-2 mRNA stabilization during T-cell activation. Genes Dev., 14, 1236–1248. [PMC free article] [PubMed] [Google Scholar]
- Davydova E.K., Evdokimova,V.M., Ovchinnikov,L.P. and Hershey,J.W.B. (1997) Overexpression in COS cells of p50, the major core protein associated with mRNA, results in translation inhibition. Nucleic Acids Res., 25, 2911–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehlin E., Wormington,M., Körner,C.G. and Wahle,E. (2000) Cap-dependent deadenylation of mRNA. EMBO J., 19, 1079–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deo R.C., Bonanno,J.B., Sonenberg,N. and Burley,S.K. (1999) Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell, 98, 835–845. [DOI] [PubMed] [Google Scholar]
- Evdokimova V.M. and Ovchinnikov,L.P. (1999) Translational regulation by Y-box transcription factor: involvement of the major mRNA-associated protein, p50. Int.J. Biochem. Cell Biol., 31, 139–149. [DOI] [PubMed] [Google Scholar]
- Evdokimova V.M., Sitikov,A.S., Simonenko,P.N., Lazarev,O.A., Vasilenko,K.S., Ustinov,V.A., Wei,C.-L., Hershey,J.W.B. and Ovchinnikov,L.P. (1995) The major protein of messenger ribonucleoprotein particles in somatic cells is a member of the Y-box binding transcription factor family. J. Biol. Chem., 270, 3186–3192. [DOI] [PubMed] [Google Scholar]
- Evdokimova V.M., Kovrigina,E.A., Nashchekin,D.V., Davydova,E.K., Hershey,J.W.B. and Ovchinnikov,L.P. (1998) Major core mRNP protein p50 promotes initiation of protein biosynthesis in vitro.J. Biol. Chem., 273, 3574–3581. [DOI] [PubMed] [Google Scholar]
- Fresno M., Carrasco,L. and Vazquez,D. (1976) Initiation of the polypeptide chain by reticulocyte cell-free systems. Survey of different inhibitors of translation. Eur. J. Biochem., 68, 355–364. [DOI] [PubMed] [Google Scholar]
- Fuerst T.R., Niles,E.G., Studier,F.W. and Moss,B. (1986) Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl Acad. Sci. USA, 83, 8122–8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuichi Y., LaFiandra,A. and Shatkin,A.J. (1977) 5′-terminal structure and mRNA stability. Nature, 266, 235–239. [DOI] [PubMed] [Google Scholar]
- Gao M., Fritz,D.T., Ford,L.P. and Wilusz,J. (2000) Interaction between a poly(A)-specific ribonuclease and the 5′ cap influences mRNA deadenylation rates in vitro.Mol. Cell, 5, 479–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M., Wilusz,C.J., Peltz,S.W. and Wilusz,J. (2001) A novel mRNA-decapping activity in HeLa cytoplasmic extracts is regulated by AU-rich elements. EMBO J., 20, 1134–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras A.-C., Raught,B. and Sonenberg,N. (1999) eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem., 68, 913–963. [DOI] [PubMed] [Google Scholar]
- Görlach M., Burd,C.G. and Dreyfuss,G. (1994) The mRNA poly(A)-binding protein: localization, abundance and RNA-binding specificity. Exp. Cell Res., 211, 400–407. [DOI] [PubMed] [Google Scholar]
- Gottlieb E. and Steitz,J.A. (1989) The RNA binding protein La influences both the accurancy and the efficiency of RNA polymerase III transcription in vitro. EMBO J., 8, 841–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyer C., Altmann,M., Trachsel,H. and Sonenberg,N. (1989) Identification and characterization of cap-binding proteins from yeast. J. Biol. Chem., 264, 7603–7610. [PubMed] [Google Scholar]
- Gu W., Tekur,S., Reinbold,R., Eppig,J.J., Choi,Y.-C., Zheng,J.Z., Murray,M.T. and Hecht,N.B. (1998) Mammalian male and female germ cells express a germ cell-specific Y-box protein, MSY2. Biol. Reprod., 59, 1266–1274. [DOI] [PubMed] [Google Scholar]
- Haghighat A. and Sonenberg,N. (1997) eIF4G dramatically enchances the binding of eIF4E to the mRNA 5′-cap structure. J. Biol. Chem., 272, 21677–21680. [DOI] [PubMed] [Google Scholar]
- Haghighat A., Mader,S., Pause,A. and Sonenberg,N. (1995) Repression of cap-dependent translation by 4E-binding protein 1: competition with p220 for binding to eukaryotic initiation factor-4E. EMBO J., 14, 5701–5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizuka N., Najita,L., Franzusoff,A. and Sarnow,P. (1994) Cap-dependent and cap-independent translation by internal initiation of mRNAs in cell extracts prepared from Saccharomyces cerevisiae.Mol. Cell. Biol., 14, 7322–7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imataka H., Gradi,A. and Sonenberg,N. (1998) A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J., 17, 7480–7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi H. et al. (2001) Y box-binding protein-1 binds preferentially to single-stranded nucleic acids and exhibits 3′ to 5′ exonuclease activity. Nucleic Acids Res., 29, 1200–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson A. and Peltz,S.W. (1996) Interrelationships of the pathways of mRNA decay and translation in eucaryotic cells. Annu. Rev. Biochem., 65, 693–739. [DOI] [PubMed] [Google Scholar]
- Kaufmann Y., Goldstein,E. and Penman,S. (1976) Poliovirus-induced inhibition of polypeptide initiation in vitro on native polyribosomes. Proc. Natl Acad. Sci. USA, 73, 1834–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon Y.K., Murray,M. and Hecht,N.B. (1993) Proteins homologous to the Xenopus germ cell-specific RNA-binding proteins p54/p56 are temporally expressed in mouse male germ cells. Dev. Biol., 158, 99–100. [DOI] [PubMed] [Google Scholar]
- Laird-Offringa I.A., de Wit,C.L., Elfferich,P. and van der Eb,A.J. (1990) Poly(A) tail shortening is the translation-dependent step in c-myc mRNA degradation. Mol. Cell. Biol., 10, 6132–6140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.A. and Sonenberg,N. (1982) Inactivation of cap-binding proteins accompanies the shut-off of host protein synthesis by poliovirus. Proc. Natl Acad. Sci. USA, 79, 3447–3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.A.W., Guertin,D. and Sonenberg,N. (1983) mRNA secondary structure as a determinant in cap recognition and initiation complex formation. J. Biol. Chem., 258, 707–710. [PubMed] [Google Scholar]
- Lindquist S. (1981) Regulation of protein synthesis during heat shock. Nature, 293, 311–314. [DOI] [PubMed] [Google Scholar]
- Maity A., McKenna,W.G. and Muschel,R.J. (1995) Evidence for post-transriptional regulation of cyclin B1 mRNA in the cell cycle and following irradiation in HeLa cells. EMBO J., 14, 603–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J., Ren,Y.-G., Thuresson,A.-C., Hellman,U., Astrom,J. and Virtanen,A. (2000) A 54-kDa fragment of the poly(A)-specific ribonuclease is an oligomeric, processive and cap-interacting poly(A)-specific 3′ exonuclease. J. Biol. Chem., 275, 24222–24230. [DOI] [PubMed] [Google Scholar]
- Matsumoto K. and Wolffe,A. (1998) Gene regulation by Y-box proteins: coupling control of transcription and translation. Trends Cell Biol., 8, 318–323. [DOI] [PubMed] [Google Scholar]
- Matsumoto K., Meric,F. and Wolffe,A.P. (1996) Translational repression dependent on the interaction of the Xenopus Y-box protein FRGY2 with mRNA. Role of the cold shock domain, tail domain and selective RNA sequence recognition. J. Biol. Chem., 271, 22706–22712. [DOI] [PubMed] [Google Scholar]
- Minich W.B., Korneyeva,N.L. and Ovchinnikov,L.P. (1989) Translational active mRNPs from rabbit reticulocytes are qualitatively different from free mRNA in their translatability in cell-free system. FEBS Lett., 257, 257–259. [DOI] [PubMed] [Google Scholar]
- Minich W.B., Maidebura,I.P. and Ovchinnikov,L.P. (1993) Purification and characterization of the major 50 kDa repressor protein from cytoplasmic mRNP of rabbit reticulocytes. Eur. J. Biochem., 212, 633–638. [DOI] [PubMed] [Google Scholar]
- Murray M., Krohne,G. and Franke,W. (1991) Different forms of soluble cytoplasmic mRNA binding proteins and particles in Xenopus laevis oocytes and embryos. J. Cell Biol., 112, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanbru C., Lafon,I., Audigier,S., Gensac,M.C., Vagner,S., Huez,G. and Prats,A.C. (1997) Alternative translation of the proto-oncogene c-myc by an internal ribosome entry site. J. Biol. Chem., 272, 32061–32066. [DOI] [PubMed] [Google Scholar]
- Nuss D.L., Furuichi,Y., Koch,G. and Shatkin,A.J. (1975) Detection in HeLa cell extracts of a 7-methyl guanosine specific enzyme activity that cleaves m7GpppNm. Cell, 6, 21–27. [DOI] [PubMed] [Google Scholar]
- Obrig T.G., Culp,W.J., McKeehan,W.L. and Hardesty,B. (1971) The mechanism by which cycloheximide and related glutarimide antibiotics inhibit peptide synthesis on reticulocyte ribosomes. J. Biol. Chem., 246, 174–181. [PubMed] [Google Scholar]
- Ohga T., Koike,K., Ono,M., Makino,Y., Itagaki,Y., Tanimoto,M., Kuwano,M. and Kohno,K. (1996) Role of the human Y box-binding protein YB-1 in cellular sensitivity to the DNA-damaging agents cisplatin, mitomycin C and ultraviolet light. Cancer Res., 56, 4224–4228. [PubMed] [Google Scholar]
- Parkin N.T., Cohen,E.A., Darveau,A., Rosen,C., Haseltine,W. and Sonenberg,N. (1988) Mutational analysis of the 5′ non-coding region of human immunodeficiency virus type 1: effects of secondary structure on translation. EMBO J., 7, 2831–2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J. and Sonenberg,N. (1985) Photochemical cross-linking of cap binding proteins to eukaryotic mRNAs: effect of mRNA 5′ secondary structure. Mol. Cell. Biol., 5, 3222–3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptushkina M., von der Haar,T., Vasilescu,S., Frank,R., Birkenhager,R. and McCarthy,J.E.G. (1998) Cooperative modulation by eIF4G of eIF4E-binding to the mRNA 5′ cap in yeast involves a site partially shared by p20. EMBO J., 17, 4798–4808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raught B., Gingras,A.-C. and Sonenberg,N. (2000) Regulation of ribosomal recruitment in eukaryotes. In Sonenberg,N., Herhey,J.W.B. and Mathews,M.B. (eds), Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 245–293.
- Richter J.D. and Smith,L.D. (1984) Reversible inhibition of translation by Xenopus oocyte-specific proteins. Nature, 309, 378–380. [DOI] [PubMed] [Google Scholar]
- Ross J. (1995) mRNA stability in mammalian cells. Microbiol. Rev., 59, 423–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzanov P.V., Evdokimova,V.M., Korneeva,N.L., Hershey,J.W.B. and Ovchinnikov,L.P. (1999) Interaction of the universal mRNA-binding protein, p50, with actin: a possible link between mRNA and microfilaments. J. Cell Sci., 112, 3487–3496. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 7.39–7.52.
- Schindelin H., Jiang,W., Inouye,M. and Heinemann,U. (1994) Crystal structure of CspA, the major cold shock protein of Escherichia coli.Proc. Natl Acad. Sci. USA, 91, 5119–5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider R.J. (2000) Translational control during heat shock. In Sonenberg,N., Hershey,J.W.B. and Mathews,M.B. (eds), Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 581–593.
- Schroder K., Graumann,P., Schnuchel,A., Holak,T.A. and Marahiel,M.A. (1995) Mutational analysis of the putative nucleic acid-binding surface of the cold-shock domain, CspB, revealed an essential role of aromatic and basic residues in binding of single-stranded DNA containing the Y-box motif. Mol. Microbiol., 16, 699–708. [DOI] [PubMed] [Google Scholar]
- Schwartz D.C. and Parker,R. (2000) Interaction of mRNA translation and mRNA degradation in Sacharomyces cerevisiae. In Sonenberg,N., Hershey,J.W.B. and Mathews,M.B. (eds), Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 807–825.
- Sommerville J. (1999) Activities of cold-shock domain proteins in translation control. Bioessays, 21, 319–325. [DOI] [PubMed] [Google Scholar]
- Sommerville J. and Ladomery,M. (1996) Masking of mRNA by Y-box proteins. FASEB J., 10, 435–443. [DOI] [PubMed] [Google Scholar]
- Spirin A.S. (1996) Masked and translatable messenger ribonucleoproteins in higher eukaryotes. In Sonenberg,N., Hershey,J.W.B. and Mathews,M.B. (eds), Translational Control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 319–334.
- Svitkin Y.V., Lyapustin,V.N., Lashkevich,V.A. and Agol,V.I. (1984) Differences between translation products of tick-borne encephalitis virus RNA in cell-free systems from Krebs-2 cells and rabbit reticulocytes: involvement of membranes in the processing of nascent precursors of flavivirus structural proteins. Virology, 135, 536–541. [DOI] [PubMed] [Google Scholar]
- Svitkin Y.V., Ovchinnikov,L.P., Dreyfuss,G. and Sonenberg,N. (1996) General RNA binding proteins render translation cap dependent. EMBO J., 15, 7147–7155. [PMC free article] [PubMed] [Google Scholar]
- Svitkin Y.V., Pause,A., Haghighat,A., Pyronnet,S., Witherell,G., Belsham,G. and Sonenberg,N. (2001) The requirement for eukaryotic initiation factor 4A (eIF4A) in translation is in direct proportion to the degree of mRNA 5′ secondary structure. RNA, 7, 382–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanchou V., Gabus,C., Rogemond,V. and Darlix,J.-L. (1995) Formation of stable and functional HIV-1 nucleoprotein complexes in vitro. J. Mol. Biol., 252, 563–571. [DOI] [PubMed] [Google Scholar]
- Thompson J.F., Hayes,L.S. and Lloyd,D.B. (1991) Modulation of firefly luciferase stability and impact on studies of gene regulation. Gene, 103, 171–177. [DOI] [PubMed] [Google Scholar]
- Van Venrooij W.J., Van Eekelen,C.A.G., Jansen,R.T.P. and Princen, J.M.G. (1977) Specific polyA-binding protein of 76 000 molecular weight in polyribosomes is not present on polyA of free cytoplasmic mRNP. Nature, 270, 189–191. [DOI] [PubMed] [Google Scholar]
- Wolffe A.P. (1994) Structural and functional properties of the evolutionarily ancient Y-box family of nucleic acid binding proteins. BioEssays, 16, 245–251. [DOI] [PubMed] [Google Scholar]
- Wolffe A.P., Tafuri,S., Ranjan,M. and Familiari,M. (1992) The Y-box factors: a family of nucleic acid binding proteins conserved from Escherichia coli to man. New Biol., 4, 290–298. [PubMed] [Google Scholar]
- Yamanaka K., Fang,L. and Inouye,M. (1998) The CspA family in Escherichia coli: multiple gene duplication for stress adaptation. Mol. Microbiol., 27, 247–255. [DOI] [PubMed] [Google Scholar]