Abstract
The Arabidopsis EDS1 and PAD4 genes encode lipase-like proteins that function in resistance (R) gene-mediated and basal plant disease resistance. Phenotypic analysis of eds1 and pad4 null mutants shows that EDS1 and PAD4 are required for resistance conditioned by the same spectrum of R genes but fulfil distinct roles within the defence pathway. EDS1 is essential for elaboration of the plant hypersensitive response, whereas EDS1 and PAD4 are both required for accumulation of the plant defence-potentiating molecule, salicylic acid. EDS1 is necessary for pathogen-induced PAD4 mRNA accumulation, whereas mutations in PAD4 or depletion of salicylic acid only partially compromise EDS1 expression. Yeast two-hybrid analysis reveals that EDS1 can dimerize and interact with PAD4. However, EDS1 dimerization is mediated by different domains to those involved in EDS1–PAD4 association. Co-immunoprecipitation experiments show that EDS1 and PAD4 proteins interact in healthy and pathogen-challenged plant cells. We propose two functions for EDS1. The first is required early in plant defence, independently of PAD4. The second recruits PAD4 in the amplification of defences, possibly by direct EDS1–PAD4 association.
Keywords: Arabidopsis/dimerization/EDS1/PAD4/salicylic acid
Introduction
Plants have evolved complex recognition and response mechanisms to counter attack by pathogens. Disease occurs only when the pathogen is able to avoid early detection by the plant (Feys and Parker, 2000). One of the most strongly expressed forms of plant disease resistance is conferred by resistance (R) genes whose products confer recognition of pathogen avirulence (Avr) proteins (Martin, 1999). Their highly specific interaction occurs within or on the surface of plant cells and leads to the rapid induction of plant defences (Kjemtrup et al., 2000). R gene-mediated resistance is usually, although not invariably, associated with localized plant cell necrosis, known as the hypersensitive response (HR). Accompanying the HR are a number of early cellular changes within the plant, such as an oxidative burst producing reactive oxygen intermediates (ROI), accumulation of the signaling molecules, nitric oxide (NO) and salicylic acid (SA), and the transcriptional activation of defence-related genes (McDowell and Dangl, 2000). Data suggest that cooperation between NO, ROI and SA molecules contributes to establishment of the HR and to the potentiation of defence signals in surrounding plant tissues (Shirasu et al., 1997; Delledonne et al., 1998; Klessig et al., 2000). However, the precise nature of events determining plant-pathogen recognition and downstream signaling is not known. It is also unclear how localized plant resistance induces systemic immunity (systemic acquired resistance, SAR), a broad spectrum and long lasting resistance that occurs in uninoculated parts of the plant (McDowell and Dangl, 2000).
Mutational analyses in the model plant, Arabidopsis, has led to the identification of genes required for R gene-mediated resistance or for SAR (Feys and Parker, 2000). The eds1 (enhanced disease susceptibility) mutation sup presses R gene-mediated resistance to the oomycete pathogen, Peronospora parasitica, conferred by RPP1 in accession Wassilewskija (Ws-0), RPP5 in accession Landsberg-erecta (Ler) (Parker et al., 1996), and by RPP2 and RPP4 in accession Columbia (Col-0) (Aarts et al., 1998). Mutations in EDS1 also abolish RPS4-mediated resistance present in all three accessions to the bacterial pathogen, Pseudomonas syringae expressing avrRps4 (Aarts et al., 1998). All of these R genes belong to a major R gene structural class encoding ‘TIR-NB-LRR’ proteins that have N-terminal (TIR) similarity to the intra cellular domains of human and Drosophila Toll receptors, a central nucleotide binding (NB) domain and C-terminal leucine-rich repeats (LRRs) (Parker et al., 1997; Botella et al., 1998; Gassmann et al., 1999). EDS1 is not required for resistance conferred by RPM1, RPS2 or RPS5, NB-LRR R genes that possess an N-terminal coiled coil (CC) motif and not a TIR domain (Aarts et al., 1998), pointing to the possibility that distinct resistance pathways are directed, at least in part, by particular R protein structural types. Analysis of RPS4-specified responses in wild-type and eds1 plants revealed that EDS1 operates upstream of SA-dependent defences (Falk et al., 1999). Moreover, eds1 plants are hypersusceptible to normally virulent strains of P.syringae and P.parasitica (a phenotype referred to as ‘enhanced disease susceptibility’, eds) (Parker et al., 1996; Aarts et al., 1998), suggesting defects in a basal resistance mechanism against virulent pathogens.
The screen for suppressors of RPP5 resistance in accession Ler led to the isolation of one defective allele of PAD4 (phytoalexin deficient) (pad4-2; Jirage et al., 1999). PAD4 was first identified in a mutational screen for enhanced disease susceptibility to a virulent isolate of P.syringae pv. maculicola (Glazebrook et al., 1996) and was found to be required for resistance conferred by RPP2 and RPP4 to P.parasitica in Col-0 cotyledons (Glazebrook et al., 1997). The eds phenotype of pad4 was associated with reduced accumulation of the indole phytoalexin, camalexin and the signaling molecule, SA (Glazebrook et al., 1997; Zhou et al., 1998). Neither of these responses was affected in pad4 plants responding to P.syringae expressing avrRpt2, indicating that RPS2-specified resistance does not require PAD4 (Zhou et al., 1998). PAD4 was therefore placed as an important regulator of SA accumulation in the plant response to virulent P.syringae. How ever, its position in R gene-mediated resistance responses remained unclear.
The isolation of pad4-2 as a suppressor of RPP5-mediated resistance in Ler as well as the requirement for PAD4 in RPP2 and RPP4 resistance in Col-0 shows that PAD4 participates in several EDS1-dependent responses. Both EDS1 and PAD4 encode lipase-like proteins (Falk et al., 1999; Jirage et al., 1999). Furthermore, the abundance of EDS1 and PAD4 mRNAs is upregulated by applications of SA, suggesting the operation of a positive feedback loop in the expression of both of these genes (Falk et al., 1999; Jirage et al., 1999). This raised the question of whether EDS1 and PAD4 functions are connected in plant defence.
Here we show that EDS1 and PAD4 proteins interact specifically, both in a yeast two-hybrid assay and in plant cells, suggesting that physical association between these two proteins may contribute to their activities in disease resistance. By examining, for the first time, the phenotypes of null eds1 and pad4 mutants in the same genetic background we also establish that EDS1 and PAD4 are required for resistance conditioned by an identical spectrum of R genes. We demonstrate that both EDS1 and PAD4 positively regulate SA accumulation in an EDS1/PAD4-dependent R gene-mediated response and that EDS1 is additionally required for generation of the plant HR. Furthermore, we establish that EDS1 is necessary for the upregulation of PAD4 mRNA, whereas mutations in PAD4 or depletion of SA only partially compromise enhanced EDS1 expression. Our results are consistent with placement of EDS1 and PAD4 within a defence pathway that is engaged by TIR-NB-LRR-type R genes. In this signaling mechanism, we propose two functions for EDS1. One is positioned upstream of PAD4 and triggers early plant defences. The other recruits PAD4 to potentiate plant defences through the accumulation of SA and possibly other molecules.
Results
Suppression of RPP5-mediated resistance in eds1 and pad4
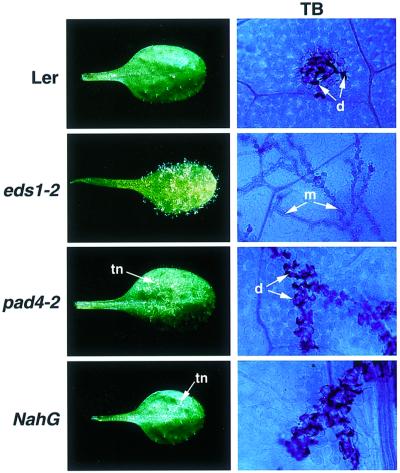
We examined the RPP5-mediated resistance phenotypes of wild-type Ler, null eds1-2 and pad4-2 mutant plants and Ler-NahG plants (expressing the SA-depleting enzyme, salicylate hydroxylase; Bowling et al., 1997) after inoculation with P.parasitica isolate Noco2. Two-week-old seedlings were sprayed with Noco2 conidia and infected leaves assessed up to 7 days after inoculation. As shown in Figure 1, Ler elaborated an HR at points of pathogen penetration that was visible microscopically after staining leaves with lactophenol Trypan Blue (TB) (Parker et al., 1993). Mycelium did not grow beyond these discrete patches of necrotic plant cells. In contrast, Noco2 colonization of eds1-2 plants was unrestricted and the mycelium rapidly ramified throughout the plant to produce abundant asexual spores on the leaf surface after 6 days (Figure 1). The phenotype of pad4-2 was strikingly different to that of eds1-2. Leaves exhibited trails of necrotic plant cells and permitted the emergence of occasional sporophores after 6–7 days. Lactophenol Trypan Blue staining revealed that pad4-2 produced an HR but the pathogen was able to grow beyond the initial infection site, giving rise to trails of dead plant cells at the plant-pathogen interface (Figure 1). Ler-NahG plants exhibited a similar trailing necrotic phenotype to pad4-2 in response to Noco2, although mycelial ingress was less extensive in leaves of NahG plants than in pad4-2 (Figure 1).
Fig. 1. RPP5 resistance phenotypes of wild-type Ler, eds1-2, pad4-2 and NahG leaves inoculated with P.parasitica isolate Noco2. Two-week-old seedlings were spray-inoculated with a suspension of P.parasitica conidia (5 × 104/ml) and incubated as described in Materials and methods. Whole leaves were photographed 6 days after inoculation. Trailing necrosis (tn) in pad4-2 and NahG is indicated by an arrow. Leaf tissue was stained with lactophenol Trypan Blue (TB) 5 days after inoculation to visualize pathogen mycelium (m) and necrotic plant cells (d). The TB-stained material, viewed under a light microscope, is shown at ×400 magnification.
We concluded from these analyses that EDS1 and PAD4 have different functions in RPP5-mediated resistance. Whereas EDS1 is an indispensable component of the HR and is associated with early plant defences, PAD4 appears to exert a resistance strengthening or potentiating activity that is downstream or independent of HR development. Similarity between the phenotypes of pad4-2 and NahG leaves suggests that a major role of PAD4 is to promote SA accumulation in the RPP5-conditioned response.
EDS1 and PAD4 interact specifically in a yeast two-hybrid assay
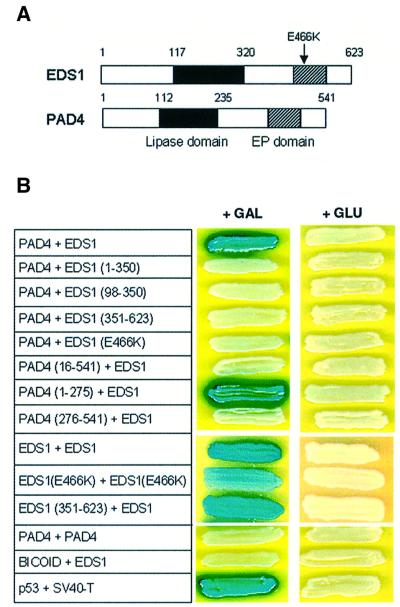
In order to identify potential protein interactors with EDS1, the full-length EDS1 cDNA was fused to the LexA DNA-binding domain in the bait vector pLexA (pLex-EDS1). A Landsberg-erecta two-hybrid cDNA library derived from pathogen-challenged plant material was constructed in the pJG4-5 activation domain (AD) vector (see Materials and methods). The pLex-EDS1 bait was verified not to auto-activate the LEU2 and LacZ reporter genes and to move to the nucleus. After transformation of the two-hybrid library in yeast strain EGY191(p8op- LacZ/pLex-EDS1), 6 000 000 primary transformants were obtained and 60 000 000 yeast clones were screened for potential EDS1 interactors. The dominant class of interactor (in 11 isolates) was identified as PAD4. Only full-length PAD4 inserts were recovered from the screen. EDS1–PAD4 interaction was also tested in the reciprocal combination with PAD4 fused to the LexA domain and was found to be stronger than the original interaction (data not shown). EDS1 did not interact with the control bait protein Bicoid (Figure 2B), making it unlikely that EDS1 is a sticky protein associating non-specifically with PAD4. In addition, a separate two-hybrid screen using full-length PAD4 yielded 50 positive interactors, 37 of which encoded full-length EDS1 (data not shown).
Fig. 2. Interaction between EDS1 and PAD4 in a yeast two-hybrid assay. (A) Schematic representation of the domain structure of the Arabidopsis EDS1 and PAD4 proteins. The lipase domain (filled box) and EP (EDS1 and PAD4-defined) domain (hatched box) are indicated. The position of the eds1-1 (E466K) mutation is shown with an arrow. The EP domain lies between residues 405 and 554 (EDS1) and residues 332 and 457 (PAD4). (B) Two-hybrid interactions between EDS1 and PAD4. Full-length proteins or defined subdomains of EDS1 and PAD4 were tested for specific interactions under inducing (+GAL) or repressing conditions (+GLU). Combinations are shown with the first protein fused to the LexA domain and the second partner fused to the AD domain. Numbers refer to amino acid positions in the full-length protein. Positive interactions are defined by activation of the LacZ (shown) and LEU2 (same pattern as LacZ; data not shown) reporter genes. The interaction between p53 and SV40-T serves as a positive control for the assay.
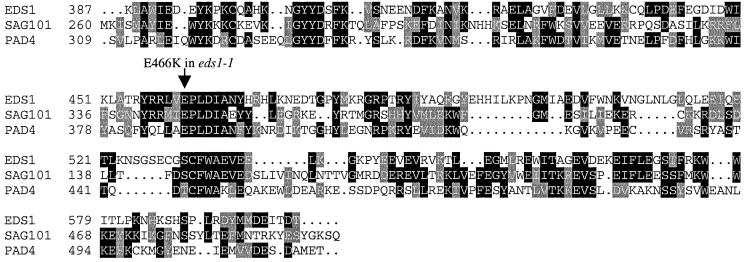
Comparison of the EDS1 and PAD4 protein sequences revealed a novel conserved domain in the C-terminus that we have named the EP domain (for EDS1 and PAD4-defined; Figure 2A), which is not present in other known proteins outside the plant kingdom. The only other Arabidopsis gene containing the EP domain is SAG101, of unknown function, which is expressed during plant senescence (He et al., 2001). Figure 3 shows a sequence alignment of the EP domain in EDS1, PAD4 and SAG101.
Fig. 3. Sequence alignment of the EP domain in EDS1, PAD4 and SAG101. Sequences of the Arabidopsis EDS1 and PAD4 proteins were aligned with SAG101, a senescence-associated gene of unknown function (He et al., 2001) across the EP domain. The position of the eds1-1 (E466K) mutation is indicated with an arrow. The alignment was generated using Clustal_W and shaded using BoxShade (see Materials and methods). Positions are relative to the full-length protein. The DDBJ/EMBL/GenBank accession No. for SAG101 is AAF78583.
In order to define regions of EDS1 and PAD4 that are required for interaction, we tested combinations of different EDS1 and PAD4 subdomains in the yeast two-hybrid assay. A schematic diagram of the designated EDS1 and PAD4 domains is shown in Figure 2A. Western blot analysis was performed on all combinations to confirm stable expression of the fusion proteins (data not shown). We found that PAD4 interacted with full-length EDS1 but not with any EDS1 subdomain tested, as shown in Figure 2B. PAD4 association with EDS1 was through its N-terminal region, comprising the predicted lipase domain. In particular, the first 15 amino acids of PAD4 were indispensable for interaction with EDS1. We next tested whether EDS1 and PAD4 were each capable of dimerization. EDS1, but not PAD4, strongly interacted with itself, suggesting that EDS1 may function in both homomeric associations as well as in heteromeric complexes with PAD4. The C-terminus of EDS1 was sufficient for interaction with full-length EDS1 (Figure 2B), suggesting that EDS1 dimerization occurs through the C-terminal end. No interaction between full-length EDS1 and its N-terminal domain (amino acids 1–350) was observed (data not shown). The absence of PAD4 dimerization in the two-hybrid system indicates a degree of specificity in interactions between members of this class of lipase-like proteins.
Previously, we isolated the eds1-1 mutant carrying a single amino acid substitution that changes a highly conserved glutamate at position 466 within the EP domain to an oppositely charged lysine residue (E466K in Figure 3). The eds1-1 mutant has a complete loss of function phenotype (Parker et al., 1996; Falk et al., 1999). Western blot analysis showed that fusions of EDS1 (E466K) to both the LexA and AD domain were stably expressed in yeast cells during two-hybrid interaction assays (data not shown). We assessed the effect of the E466K mutation on two-hybrid interactions and found that EDS1–PAD4 association was abolished, whereas it reduced, but did not abolish EDS1 dimerization (Figure 2B). Quantification of β-galactosidase activity (see Materials and methods) gave the following values for the interactions: EDS1 + EDS1, 107 ± 16 units; EDS1 (E466K) + EDS1(E466K), 17 ± 4 units. The contrasting effect of the E466K mutation on the EDS1–PAD4 and EDS1–EDS1 interactions provides further evidence that the mode of EDS1 association with PAD4 is different to that of EDS1 dimerization.
In planta interaction between EDS1 and PAD4
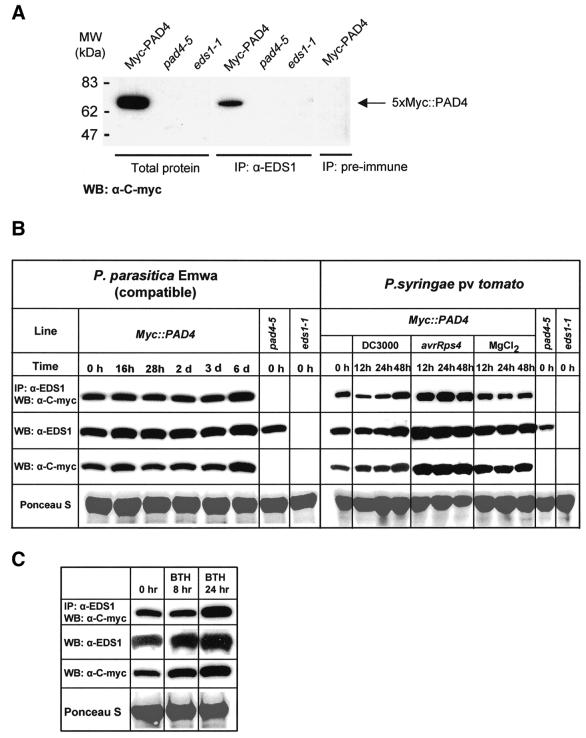
In order to study protein interactions in plant cells, a null mutant pad4 line (pad4-5; see below) was stably transformed with wild-type PAD4 containing an N-terminal c-Myc epitope tag expressed from the native PAD4 promoter (Myc::PAD4). The c-Myc epitope did not interfere with PAD4 function, as several independent transgenic lines exhibited full resistance to P.parasitica isolate Noco2 (data not shown). In addition, we generated anti-EDS1 polyclonal antiserum (see Materials and methods). Both the anti-c-Myc antibody and the anti-EDS1 antiserum detected single protein bands of the expected size (69.6 kDa for Myc::PAD4, 71.5 kDa for EDS1) in total soluble extracts of the Myc::PAD4 transgenic line, but not in protein extracts from the pad4-5 or eds1-1 mutant, respectively (Figure 4A and B).
Fig. 4. In planta protein interaction between EDS1 and PAD4. (A) Co-immunoprecipitation of EDS1 and PAD4 in total plant protein extracts. Protein extracts were prepared from the transgenic pad4-5 (5× Myc::PAD4) line, indicated as Myc-PAD4, or from the pad4-5 and eds1-1 mutants. For immunoprecipitation reactions pre-immune (as control) or EDS1 antiserum was used, followed by western blotting detection with anti-c-Myc antibody. Total protein extracts were analyzed on the same western blot to show the specificity of the anti-c-Myc antibody. (B) Analysis of EDS1 and PAD4 protein expression and co-immunoprecipitation in healthy and pathogen-challenged plants. Leaves of the 5× Myc::PAD4 epitope-tagged transgenic line were spray inoculated with P.parasitica spores (1 × 105/ml in dH20) or infiltrated with suspensions (5 × 106/ml colony forming units in 10 mM MgCl2) of DC3000, DC3000 expressing avrRps4 or 10 mM MgCl2 alone, and tissues harvested at the time points indicated. Levels of EDS1 and PAD4 protein were measured on western blots of total soluble extracts probed with anti-EDS1 and anti-c-Myc antibody, respectively. Co-immunoprecipitations were performed on the same tissue extracts, as described in (A). Equal loading of blots is indicated by Ponceau S staining of total protein. An independent experiment gave similar results. (C) Analysis of EDS1 and PAD4 protein expression and co-immunoprecipitation in leaves treated with BTH. Tissues were harvested and analyzed as described in (B). Similar results were obtained in an independent experiment.
Immunoprecipitation of EDS1 protein from extracts of the Myc::PAD4 line using the anti-EDS1 antiserum, followed by detection on western blots using the anti-c-Myc antibody, showed that the EDS1 and PAD4 proteins interact in healthy plant tissue (Figure 4A). Pre-immune serum was not able to co-immunoprecipitate the EDS1– PAD4 complex, indicating that immunoprecipitation was specific for EDS1. In addition, no signal was detected when co-immunoprecipitation was performed with protein extracts from the pad4-5 mutant line, showing that the Myc::PAD4 protein was specifically detected after immunoprecipitation (Figure 4A). Co-immunoprecipitation experiments, performed by first immunoprecipitating Myc::PAD4 with anti-c-Myc antibodies, followed by western blot detection of EDS1, also showed specific interaction between EDS1 and PAD4 (data not shown). Co-immunoprecipitation experiments using independent Myc::PAD4 transgenic lines gave identical results (data not shown). These data demonstrate that interaction between EDS1 and PAD4 in the yeast two-hybrid assay reflects their ability to associate in the plant.
We tested, by co-immunoprecipitation, whether the extent of EDS1–PAD4 association changed after pathogen inoculation of plants. EDS1 and PAD4 protein levels in total leaf extracts did not alter substantially after infection with a virulent P.parasitica isolate, Emwa1, except at day 6 when increases in the abundance of both proteins were detectable (Figure 4B). At all stages of P.parasitica infection, PAD4 co-immunoprecipitated with EDS1. At day 6, enhanced levels of both proteins correlated with increased detection of co-immunoprecipitable PAD4 (Figure 4B), suggesting that EDS1 and PAD4 expression and physical association respond to pathogen colonization. Similar results were obtained after inoculation of plants with avirulent P.parasitica isolate, Noco2, although in this interaction no increases in total or co-immunoprecipitable EDS1 and PAD4 were observed by day 6 (data not shown). Leaves of mature plants were infiltrated with the virulent P.syringae pv. tomato strain, DC3000, or with avirulent DC3000 expressing avrRps4 (DC3000/avrRps4), allowing synchronous infection of a larger area of tissue than could be achieved with P.parasitica. Enhanced levels of EDS1 and PAD4 protein were observed in tissues responding to DC3000/avrRps4 12 h after infiltration and these persisted for up to 48 h (Figure 4B). Increased EDS1 and PAD4 expression correlated with enhanced detection of co-immunoprecipitable PAD4 (Figure 4B). A slight increase in EDS1 and PAD4 expression was also observed after inoculation with virulent DC3000, although this occurred later (48 h) than in tissues inoculated with DC3000/avrRps4 (Figure 4B). In addition, leaves were sprayed with the bioactive SA analogue, benzothiadiazole (BTH). Here, elevated EDS1 and PAD4 expression corresponded with higher levels of co-immunoprecipitable PAD4 24 h after treatment (Figure 4C).
Analysis of the spectrum of Arabidopsis R genes requiring EDS1 and PAD4
Our findings that EDS1 and PAD4 interact in a two-hybrid assay and in plant cells prompted us to assess the effects of eds1 and pad4 mutants on a broader range of R gene-mediated responses within the same plant genetic background. This would establish whether EDS1 and PAD4 functions are tightly associated genetically or can be partially separated into different pathways.
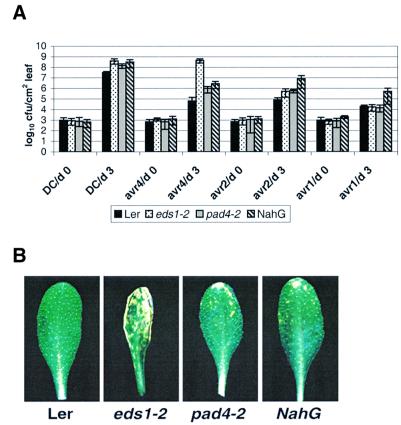
First, the effects of eds1-2 and pad4-2 on RPP genes recognizing distinct P.parasitica isolates were measured in accession Ler. As shown in Table I, resistance responses mediated by RPP5 and RPP21 that are fully EDS1 dependent (Aarts et al., 1998) were partially compromised by pad4, producing the characteristic trailing necrotic phenotype (see also Figure 1). In contrast, RPP7- and RPP8-mediated resistance operated independently of both EDS1 and PAD4 (Table I). Analysis was extended to R genes expressed in accession Ws-0 by isolating a line containing a T-DNA insertion in PAD4 (pad4-5; see Materials and methods). The pad4-5 mutant is an mRNA null mutant, since transcripts could not be detected using sensitive TaqMan analysis (see below). Here, we found that the EDS1-dependent RPP1A, 1B and 1C genes (Aarts et al., 1998) conferred partial resistance in pad4-5 with a similar phenotype to that observed for RPP5 and RPP21 in pad4-2 (Table I). We then measured the effects of eds1-2, pad4-2 and the NahG transgene on RPM1-, RPS2- and RPS4-mediated resistance in Ler to P.syringae pv. tomato DC3000 expressing, respectively, avrRpm1, avrRpt2 and avrRps4. Leaves were dipped into bacterial suspensions and bacterial growth and disease symptoms monitored over 5 days. Wild-type plants restricted growth of all the avirulent strains compared with growth of virulent DC3000 (Figure 5A). The eds1-2 mutation fully suppressed RPS4 resistance but did not affect resistance mediated by RPM1 and weakly compromised RPS2 resistance (Figure 5A). In leaves of pad4-2 and Ler-NahG, growth of DC3000/avrRps4 was intermediate between that observed in wild-type Ler and eds1-2 (Figure 5A). This correlated with a slow and sporadic appearance of chlorotic symptoms in pad4-2 and Ler-NahG, compared with eds1-2 (Figure 5B). Like eds1-2, pad4-2 caused a slight relaxation of RPS2-mediated resistance but had no effect on resistance conditioned by RPM1 (Figure 5A). Interestingly, Ler-NahG permitted significant growth of both DC3000/avrRpt2 and DC3000/avrRpm1 (Figure 5A), suggesting that SA is furnished in an EDS1- and PAD4-independent manner in these responses.
Table I. Suppression of RPP gene-mediated resistance to P.parasitica in leaves of eds1 and pad4 in accessions Ler and Ws-0.
| Plant R gene (P.parasitica isolate) |
||||||
| RPP5 | RPP8 | RPP4/8 | RPP21 | RPP7a | – | |
| Plant Line |
(Noco2) |
(Emco5) |
(Emwa1) |
(Maks9) |
(Hiks1) |
(Cala2) |
| Ler | R | R | R | R | R | S |
| eds1-2 | S* | R | R | S* | R | S* |
|
pad4-2 |
(S) |
R |
R |
(S) |
R |
S* |
| RPP1A,B,C | RPP1A,B | RPP1A | – | |||
| |
(Noco2) |
(Emoy2) |
(Cala2) |
(Emwa1) |
|
|
| Ws-0 | R | R | R | S | ||
| eds1-1 | S* | S* | S* | S-S* | ||
| pad4-5 | (S) | (S) | (S) | S-S* | ||
Two-week-old seedlings were scored 5 and 7 days after inoculation with P.parasitica. Ler is genetically susceptible to P.parasitica isolate Cala2 and Ws-0 is susceptible to isolate Emwa1. Phenotypes were assigned as R (fully resistant, wild-type HR), S (susceptibility of genetically compatible lines), S* (hypersusceptible, permitting more abundant sporulation than the genetically susceptible line), (S) (partially susceptible, mycelium development accompanied by trailing plant cell necrosis and occasional sporophores). Similar results were obtained in two independent experiments. While eds1-2 and pad4-2 reproducibly exhibited hypersusceptibility to Cala2, eds1-1 and pad4-5 gave variable results between experiments, as indicated.
aRPP7 in Ler is defined as an R locus cosegregating with Col-0 RPP7 in >4000 Col-0 × Ler F2 seedlings (E.Holub, personal communication).
Fig. 5. Growth and symptom development of different P.syringae strains in leaves of wild-type Ler, eds1-2, pad4-2 and NahG plants. (A) Leaves of 4-week-old short day grown plants were infiltrated with a suspension (1 × 105 colony forming units/ml) of P.syringae pv. tomato strain DC3000 containing an empty vector (DC3000) or DC3000 expressing avrRps4, avrRpt2 or avrRpm1. Bacterial titres were measured at 0 and 3 days after inoculation. The measurements and standard errors are derived from four replicates per treatment. An independent experiment gave similar results. (B) Leaves were dipped in a suspension (1 × 107 c.f.u./ml) of DC3000 expressing avrRps4 and disease symptoms observed over 6 days. As shown at day 5, Ler plants appear healthy, eds1-2 plants develop severe leaf spotting symptoms, while pad4-2 and NahG plants exhibit mild leaf spotting.
The P.parasitica and P.syringae infection studies reveal that all strongly EDS1-dependent R gene responses examined have a partial requirement for PAD4, and that EDS1-independent interactions are also independent of PAD4. These data suggest that PAD4 and EDS1 function within the same defence signaling pathway.
Pathogen-induced SA accumulation in eds1 and pad4
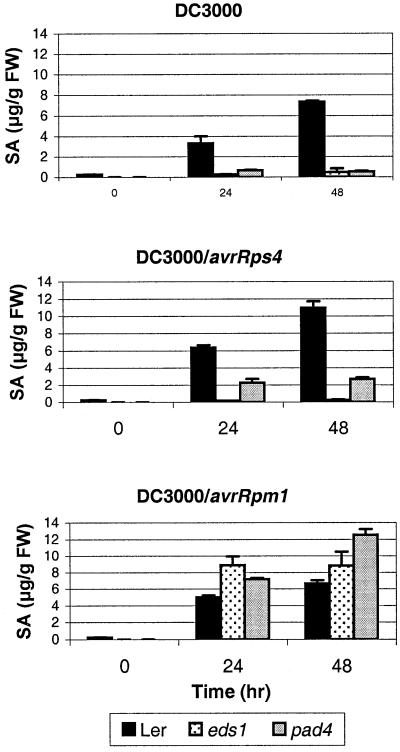
In independent studies, PAD4 and EDS1 have been implicated as regulators of SA-dependent defences (Zhou et al., 1998; Falk et al., 1999). We therefore wished to examine the relative contributions of EDS1 and PAD4 to SA accumulation in the same R gene response. Salicylic acid accumulation profiles of wild-type Ler, eds1-2 and pad4-2 leaves were analyzed after triggering an EDS1/PAD4-dependent R gene response (RPS4-mediated resistance), an EDS1/PAD4-independent response (RPM1-mediated resistance) or in a compatible interaction with P.syringae DC3000. We found that eds1-2 and pad4-2 severely depleted SA accumulation after infection with virulent DC3000 (Figure 6). Salicylic acid accumulation was also abolished in eds1-2 plants and was strongly reduced in pad4-2 after inoculation with DC3000/avrRps4 (Figure 6). In contrast, eds1-2 and pad4-2 did not compromise SA accumulation in plants responding to DC3000/avrRpm1 (Figure 6). The importance of EDS1 and PAD4 as regulators of SA levels therefore correlates with a genetic requirement for their functions in RPS4-mediated resistance. In RPM1-mediated resistance SA accumulation bypasses both EDS1 and PAD4, consistent with the bacterial growth data in NahG plants showing a requirement of RPM1 resistance for SA but not for EDS1 or PAD4 (Figure 5A).
Fig. 6. Accumulation of total salicylic acid in Ler, eds1-2 and pad4-2 plants after inoculation with virulent and avirulent P.syringae pv. tomato DC3000 strains Leaves of 4-week-old short day grown plants were dipped in a suspension (1 × 107 c.f.u./ml) of DC3000 (top panel), DC3000 expressing avrRps4 (middle panel) or avrRpm1 (bottom panel). Total salicylic acid (SA) was extracted and quantified after 0, 24 and 48 h by HPLC as described in Materials and methods. Salicylic acid measurements and standard errors are derived from three replicate samples per treatment. Salicylic acid was present in trace amounts in Ler-NahG plants at all stages of infection (data not shown).
Analysis of EDS1 and PAD4 transcripts
EDS1 (Falk et al., 1999) and PAD4 (Jirage et al., 1999) mRNAs are induced in response to pathogen inoculation or SA treatment. Here, we examined whether their mode of expression is affected by a mutation in either gene or by the presence of NahG. Leaves were infiltrated with a suspension of DC3000/avrRps4 in 10 mM MgCl2 or with 10 mM MgCl2 alone, or were sprayed with BTH. Total RNA was extracted at various time points up to 48 h after treatment, reverse transcribed into cDNA, and EDS1 and PAD4 mRNA levels measured using real-time quantitative PCR and TaqMan chemistry (Holland et al., 1991). This procedure (Wang and Brown, 1999) is particularly suitable for measuring expression levels of rare transcripts, such as EDS1 (Falk et al., 1999; Clarke et al., 2001). The Arabidopsis actin gene ACT2 was chosen as a normalization standard because of its constitutive expression in nearly all vegetative tissues in juvenile and mature plants (An et al., 1996; see Materials and methods).
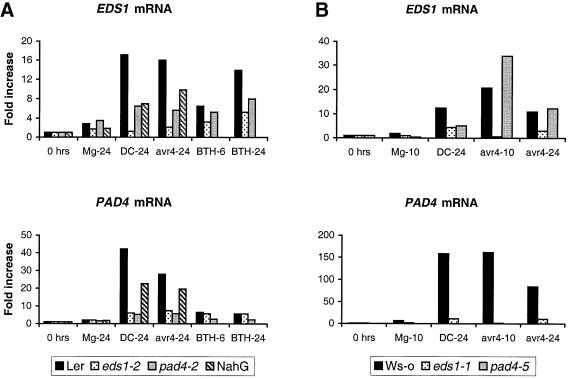
The fold induction of EDS1 and PAD4 mRNA levels in Ler wild-type and mutant lines after bacterial inoculation or BTH treatment is shown in Figure 7A. EDS1 expression was induced after inoculation with DC3000 alone or DC3000 expressing avrRps4. The presence of pad4-2 or depletion of SA by NahG partially compromised pathogen-induced EDS1 expression. Expression of PAD4 mRNA was more strongly induced than EDS1 in pathogen-treated Ler plants. This induction was severely reduced in eds1-2 plants and partially compromised by NahG. Neither mutant transcript responded strongly to pathogen inoculation, suggesting a requirement for functional protein in optimal upregulation and/or stability of their respective transcripts. As shown previously, applications of BTH induced expression of EDS1 and PAD4 mRNAs (Falk et al., 1999; Jirage et al., 1999). Mutations in either gene had little or no effect on the induction of the other gene, suggesting that BTH may lead to increased expression of EDS1 and PAD4 independently of each other.
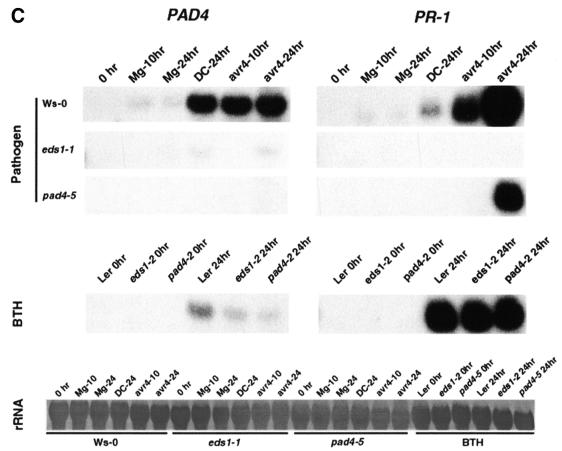
Fig. 7. Abundance of EDS1, PAD4 and PR-1 mRNAs in pathogen-inoculated and BTH-treated plants. (A) Leaves of 5-week-old Ler, eds1-2, pad4-2 and Ler-NahG plants were hand-infiltrated with 10 mM MgCl2 (Mg), 1 × 107 c.f.u./ml P.syringae DC3000 (DC) or P.syringae DC3000 expressing avrRps4 (avr4), or sprayed with 300 µM benzothiadiazole (BTH). Material was harvested after 24 h for the bacterial inoculations and 6 and 24 h for the BTH treatment. Messenger RNA abundance was determined using TaqMan chemistry (see Materials and methods). EDS1 and PAD4 mRNA levels are normalized relative to the internal control ACT2, and are calculated relative to expression at 0 h. (B) Leaves of 5-week-old Ws-0, eds1-1 and pad4-5 plants were pathogen challenged as in (A). Material was harvested 10 and 24 h after challenge. PAD4 mRNA is undetectable in the pad4-5 mutant. Relative quantification using TaqMan chemistry is as described in (A). (C) RNA gel blot analysis of PAD4 and PR-1 mRNA expression. Samples from (B) plus BTH-treated samples from (A) were analysed to verify TaqMan results for PAD4 and examine the expression of PR-1. Results for PAD4 (left) and PR-1 (right) are shown after pathogen challenge (top panel) and BTH treatment (middle panel). Control for equal loading is shown in the bottom panel (rRNA).
The Ler data show that loss of EDS1 function has a much stronger negative effect on PAD4 expression than defective PAD4 does on EDS1 expression. The results were reinforced by a similar analysis of Ws-0 wild type and corresponding eds1-1 and pad4-5 mutant lines (Figure 7B). Inoculation of Ws-0 with DC3000 or DC3000/avrRps4 induced levels of both EDS1 and PAD4 mRNAs, although the overall fold induction of PAD4 mRNA was considerably higher than that observed in Ler. Pathogen induction of PAD4 expression was almost completely abolished in eds1-1 plants. In contrast, pad4-5 did not suppress induction of EDS1 mRNA in the plant response to DC3000/avrRps4 and had a partial effect on EDS1 mRNA induction in response to DC3000.
TaqMan analysis of mRNA abundance was shown previously to accurately reflect EDS1 expression in wild-type and mutant plants (Clarke et al., 2001). We found that our estimations of PAD4 mRNA abundance by TaqMan analysis also correlated well with PAD4 mRNA levels measured on an RNA gel blot (compare Figure 7B bottom panel with Figure 7C). RNA gel blot analysis of selected samples was further used to determine the consequences of the eds1 and pad4 mutations on downstream plant defences by measuring expression levels of the SA-responsive marker gene PR1. As shown in Figure 7C, pathogen-induced expression of PR1 in wild-type Ws-0 plants was abolished by eds1-1 and strongly suppressed by pad4-5. In these tests, PR1 expression was fully rescued in eds1-2 and pad4-2 in response to BTH treatment (Figure 7C), consistent with the placement of EDS1 and PAD4 upstream of SA accumulation.
Discussion
We present genetic and molecular evidence that EDS1 and PAD4, two plant disease resistance signaling proteins, function within the same defence pathway. They are required for an identical spectrum of R genes recognizing avirulent P.parasitica and P.syringae isolates, as well as for restriction of growth of virulent isolates of these pathogens. EDS1 and PAD4 interact specifically in a yeast two-hybrid assay and co-immunoprecipitate in total plant protein extracts, suggesting that direct association may be important for their cellular roles. Also, EDS1 and PAD4 are necessary for accumulation of the plant defence signal ing molecule, SA, in response to virulent P.syringae or in resistance triggered by an EDS1/PAD4-dependent R gene, RPS4, but not in resistance conferred by an EDS1/PAD4-independent R gene, RPM1. Furthermore, upregulation of PAD4 expression after pathogen attack depends on EDS1 function.
In R gene-mediated responses that exhibited a requirement for EDS1 and PAD4, the null eds1 and pad4 mutant phenotypes were quite distinct. While eds1 plants failed to elaborate a hypersensitive response and were hypersusceptible to P.parasitica infection, pad4 plants retained the HR and exhibited intermediate susceptibility. Thus, the HR in pad4 is not sufficient to restrict pathogen coloniz ation fully. We conclude from these data that wild-type EDS1 and PAD4 have intrinsically different functions within the defence pathway. EDS1 appears to exert a critical, early role during race-specific resistance, whereas PAD4 serves to reinforce the initial resistance response. This idea is supported by the results of another study showing that EDS1 but not PAD4 is required for an HR-associated oxidative burst triggered by either RPP1- or RPS4-mediated pathogen recognition (Rustérucci et al., 2001).
Zhou et al. (1998) demonstrated that PAD4 is a regulatory component of SA accumulation in plants after inoculation with a virulent P.syringae pv. maculicola isolate. Salicylic acid is also a necessary component of systemic immunity (Dempsey et al., 1999) and can function as a resistance-potentiating signal in cooperation with ROI (Shirasu et al., 1997; Delledonne et al., 1998; Klessig et al., 2000). Here we demonstrate that PAD4 is required to furnish SA in EDS1-dependent R gene- triggered resistance (Figure 6). A major role of PAD4 in this pathway is therefore likely to positively regulate SA accumulation. This is supported by the similar levels of intermediate RPS4 resistance observed in pad4-2 and NahG plants after challenge with P.syringae carrying avrRps4 (Figure 5). Partial loss of RPP5-mediated resistance to P.parasitica Noco2 was also exhibited by NahG plants (Figure 1). However, the extent of P.parasitica growth and trailing plant cell necrosis was less in NahG than in pad4-2, suggesting that PAD4 wild-type protein has an additional defence role besides regulating SA accumulation, at least in RPP5-mediated resistance.
The eds1-2 mutation almost completely abolished SA accumulation in RPS4-mediated resistance and this correlated with the absence of detectable PR1 gene expression compared with wild-type plants (Figures 6 and 7C). In contrast, pad4-2 depleted but did not totally remove SA, tallying with a partial suppression of PR1 induction. Thus, an EDS1-dependent but PAD4-independent mechanism exists to generate residual SA, invoking a function for EDS1 that is separable from processes requiring both EDS1 and PAD4. It is possible that the low level of SA accumulation in pad4 plants is derived from an early, EDS1-regulated mechanism associated with the HR and that subsequent, enhanced SA generation depends on EDS1 and PAD4 activities.
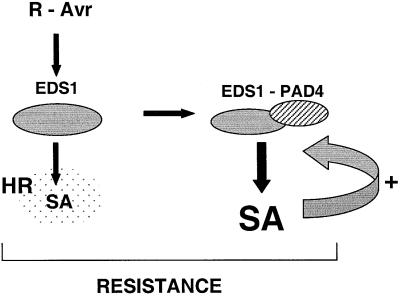
Our data point to a requirement for EDS1 upstream of PAD4 in the R gene-mediated defence pathway leading to the HR, but together with PAD4 in driving maximal SA accumulation during defence potentiation, as shown in the model in Figure 8. In this model, PAD4 activity is con tingent on the presence of EDS1. This is further supported by the dependence of PAD4 mRNA upregulation on EDS1 during pathogen challenge (Figure 7). Thus, there appear to be two distinct EDS1 activities (Figure 8). The fact that eds1 and pad4 exhibit an eds phenotype when challenged with a number of virulent pathogens (Glazebrook et al., 1996; Parker et al., 1996; Aarts et al., 1998; see also Figure 5A) reveals functions of both proteins in basal plant resistance. This low level resistance is likely to be at least partially dependent on SA accumulation since pad4-2 and eds1-2 were strongly depleted in SA after challenge with virulent P.syringae DC3000 (Figure 6). It may be that basal resistance is exerted by the combined EDS1–PAD4 ‘potentiating’ activities, consistent with the presence of a pre-existing EDS1–PAD4 protein complex in unchallenged plant tissues (Figure 4). A different attribute of EDS1 would be engaged to transduce early R-Avr protein-triggered signals leading to the HR. Other recent genetic analyses support our model. First, EDS1 and PAD4 operate at a similar position in defence pathways induced by the cpr1 and cpr6 (constitutive expressor of PR genes) mutations (Clarke et al., 2001; Jirage et al., 2001). Secondly, EDS1 and PAD4 are both necessary signaling components of runaway cell death triggered by the lsd1 mutation, in a mechanism that is separable from events associated with the plant HR (Rustérucci et al., 2001).
Fig. 8. A model for the roles of EDS1 and PAD4 in R gene-mediated resistance. Two functions are proposed for EDS1 in R-Avr protein-triggered resistance at pathogen infection foci. One lies upstream of the plant HR (indicated by the stipled area) and is required for a low level of SA accumulation. The second function recruits PAD4, possibly through direct EDS1–PAD4 interaction, and drives amplification of local defences through enhanced accumulation of SA and other molecules (indicated by the curved arrow). Complete containment of the pathogen requires both the HR and defence signal potentiation.
The yeast two-hybrid data show that EDS1 interacts with itself through the C-terminal half of at least one partner (Figure 2B). EDS1 also associates with PAD4 through the PAD4 N-terminal portion. The differential effect of the eds1 (E466K) mutation on EDS1 dimerization and EDS1–PAD4 interaction, coupled with the fact that PAD4 interacts with EDS1 through its N-terminus, whereas EDS1 homodimerization requires at least one C-terminus, suggests that the two complexes are arranged in a different way. This could result in the two associations fulfilling quite different functions during a resistance response. Western blotting analysis of the eds1-1 mutant line shows a lack of detectable mutant EDS1 protein in plant extracts (Figure 4B), in contrast with the stable expression of EDS1 (E466K) protein in yeast. The absence of detectable mutant EDS1 protein in planta may reflect a signaling failure or misfolding of the protein, leading to targeted degradation.
Our co-immunoprecipitation data show that EDS1 associates with PAD4 in healthy and pathogen-challenged plant tissues, suggesting that specific interaction may be important for their roles in defence potentiation, as depicted in Figure 8. Increased abundance of EDS1 and PAD4 proteins after pathogen inoculation or BTH treatment correlated with elevated levels of co-immunoprecipitable protein (Figure 4), indicating that at least a proportion of the increased EDS1 and PAD4 is incorporated into a complex. It is conceivable that pre-existing EDS1–PAD4 complexes play a role in basal resistance against virulent pathogens (discussed above). This raises the question of how an early, PAD4-independent function of EDS1 in TIR-NB-LRR protein-mediated pathogen recognition is exerted. Triggering of the R gene pathway does not appear to cause a dramatic alteration in EDS1–PAD4 protein association. However, we cannot discount the possibility that a sub-population of EDS1 molecules or EDS1 homodimers (as implicated by the yeast two-hybrid analysis; Figure 2) performs a critical early signaling role in R gene-mediated defence. It is interesting in this context that EDS1 protein stability does not depend on the presence of PAD4 (Figure 4). Alternatively, EDS1 may perform an additional activity within an EDS1–PAD4 complex that does not require PAD4 function.
EDS1 and PAD4 share predicted lipase catalytic motifs (Falk et al., 1999; Jirage et al., 1999), suggesting that hydrolytic activities may contribute to their signal transduction roles. Indeed, the different enzymatic specificity and/or kinetic properties of an EDS1 homodimer versus an EDS1–PAD4 heterodimer could dictate placement of the respective complexes within the resistance pathway. However, it remains to be established whether these lipase domains are enzymatically functional. There are precedents for dimerization of lipases/esterases in various systems. In mammals, hormone-sensitive lipase exists as a functional dimer with a 40-fold greater activity than the monomer (Shen et al., 2000), whereas lipoprotein lipase (LPL) exists as both an inactive monomer and an active dimer in vivo (Bergö et al., 1996). Interestingly, LPL also has a binding capacity rather than catalytic function, involved in linking triglyceride-rich lipoproteins and cholesterol-rich lipoproteins to the cell surface (Pentikäinen et al., 2000). Similarly, EDS1 and/or PAD4 could be involved in binding particular substrates, rather than enzymatically processing them. In bacteria, crystallographic analysis of the Pseudomonas fluorescens carboxylesterase, which belongs to the α/β hydrolase family of proteins that also contains lipases, reveals that this protein exists as a homodimer with the two subunits facing each other in a head-to-head fashion, thereby bringing the active sites together (Kim et al., 1997). The active LPL homodimer, on the other hand, seems to be arranged in a head-to-tail fashion (Wong et al., 1997).
Quantitation of EDS1 and PAD4 mRNA levels in wild-type, mutant, and NahG plants shows that EDS1 is essential for the upregulation of PAD4 mRNA levels in the plant–pathogen interaction, but that PAD4 contributes in a minor way to the enhanced expression of EDS1 mRNA (Figure 7A and B). Salicylic acid also appears to be a contributory factor in the expression of both genes (Figure 7A), consistent with participation of SA in a positive feedback loop (Falk et al., 1999; Jirage et al., 1999). However, other factors are likely to be involved either in the initial induction of EDS1 and PAD4 mRNAs or in the amplification of expression. For example, ROI produced during the HR (Rustérucci et al., 2001) may influence EDS1 and PAD4 expression either directly or indirectly (Levine et al., 1994; Orozco-Cárdenas et al., 2001). Indeed, the perpetuated HR and associated oxidative burst of pathogen-inoculated pad4 and NahG plants may account for some of the residual EDS1 expression observed in these backgrounds. Upregulation of EDS1 and PAD4 mRNA appears to require protein activity since the mutant eds1 and pad4 mRNAs do not respond strongly to pathogen inoculation (Figure 7), again indicative of positive feedback on expression either by the protein itself or by downstream molecules. A fuller examination of the mode of EDS1 and PAD4 protein expression, their biochemical activities and molecular associations in wild-type and mutant plants after pathogen challenge should provide important insights into their signaling roles in plant disease resistance.
Materials and methods
Plant material and pathogen strains
The isolation of eds1-1 and eds1-2 (Parker et al., 1996; Falk et al., 1999) and pad4-2 (Jirage et al., 1999) mutants has been described before. The pad4-5 mutant was identified by a reverse-genetic screen of the Feldmann T-DNA lines (Ws-0 background; Feldmann et al., 1991) for insertions into the PAD4 gene. A homozygous insertion line was identified in a subpool of 10 single insertion lines (stock CS12182 at the Arabidopsis Biological Resource Center, Columbus, OH). Sequencing of the T-DNA border–PAD4 junction revealed that the T-DNA had inserted 35 bp 5′ to the end of the unique intron in the PAD4 gene. Pseudomonas syringae strains and P.parasitica isolates used in this study have been described before (Aarts et al., 1998; McDowell et al., 2000).
Construction of c-Myc tagged PAD4 transgenic line
The PAD4 coding sequence was PCR amplified as a NcoI–BamHI fragment cloned into SLJ4C3 upstream of the NOS terminator. The 35S promoter was removed and replaced with 1 kb of PAD4 promoter amplified as an EcoRI–NcoI fragment. A NcoI cassette containing five copies of the c-Myc epitope (derived from pJR1265, R.Hampton and J.Rine, personal communication) was inserted between the PAD4 promoter and the PAD4 coding sequence to give an N-terminal fusion. The construct was transferred to the binary Basta-resistant plasmid SLJ755I5, conjugated to Agrobacterium tumefaciens GV3101, and pad4-5 mutant plants were transformed using the flower-dip method (Clough and Bent, 1998). Several independent homozygous lines with single locus T-DNA inserts were identified and confirmed to confer full resistance to P.parasitica isolate Noco2.
Antibody production, immunoprecipitation and western blot analysis
A mutant form of EDS1 (G125R) was highly expressed in Escherichia coli M15 (pREP4) as a fusion to a His6 tag (Qiagen), was purified on TALON metal affinity resin (Clontech) and further purified by electro-elution from a preparative SDS–PAGE gel. New Zealand white male rabbits were immunized and sera were collected.
Total protein extracts were prepared from 5-week-old leaf material after grinding in liquid nitrogen and extracting in 50 mM Tris pH 8.0, 150 mM NaCl and 1 mM EDTA, containing 1× Protease Inhibitor Cocktail (Sigma). Samples were spun at 16 000 g for 20 min and protein concentration of the soluble fraction was determined using Bradford reagent (Bio-Rad) and bovine serum albumin as a standard. For immunoprecipitation (IP) reactions, 1 mg of total protein was incubated with 5 µl of EDS1 antiserum (or pre-immune serum) in a total volume of 1 ml of IP buffer (50 mM Tris pH 8.0, 150 mM NaCl, 1 mM EDTA, 0.01% Triton X-100) and rotated end-over-end at 4°C for 90 min. Protein A/G Plus beads (35 µl, equilibrated in IP buffer; Santa Cruz Biotechnology) were added and the reactions were incubated for a further 60 min. IP reactions were washed three times with 1 ml of ice-cold IP buffer, resuspended in 40 µl of SDS–PAGE sample buffer, boiled for 5 min and 10 µl run on 7.5% SDS–PAGE gels (Bio-Rad). For standard western blotting analysis, 50 µg of total protein were loaded. Proteins were electroblotted to PVDF membranes (Amersham), blocked for 1 h at room temperature in phosphate-buffered saline (PBS)-Tween containing 5% (wt/vol) non-fat dried milk. Incubation with primary antibodies was in PBS-Tween containing 5% (wt/vol) non-fat dried milk using the following dilutions: EDS1 antiserum (1:7500); anti c-Myc 9E10 (1:5 000; Santa Cruz Biotechnology). Blots were developed using the SuperSignal West Pico Chemiluminescent kit (Pierce).
Pathogen and BTH treatments
Inoculations with P.parasitica and P.syringae, as well as calculation of growth curves were performed as described by Aarts et al. (1998). BTH wettable powder (Novartis, Basel; Lawton et al., 1996) was resuspended in water at a concentration of 300 µM active ingredient and plants were sprayed to imminent run off.
Yeast two-hybrid analysis
We used the LexA two-hybrid system (kindly provided by Roger Brent, Massachusetts General Hospital, Boston, MA; Gyuris et al., 1993). Details of the construction of the two-hybrid cDNA library are in van der Biezen et al. (2000). Two-hybrid analyses were carried out according to Golemis et al. (1998). The various EDS1 and PAD4 domains were generated by PCR, verified by sequencing on a Perkin-Elmer ABI377 sequencing machine, and subcloned into pLexA and/or pJG4-5. All pLexA constructs were tested for auto-activation of the LacZ and LEU2 reporters. Repression assays with JK101 confirmed protein fusion synthesis and nuclear localization of the LexA fusion protein. pLex-EDS1 showed slight auto-activation of the LEU2 reporter, and was therefore transformed in the less sensitive yeast strain EGY191. All other two-hybrid combinations were performed in the standard yeast strain EGY48. Western blotting analysis was performed on total yeast protein extracts, derived from galactose-induced cultures, using anti-LexA (Santa Cruz Biotechnology) and anti-HA (Roche) antibodies. The eds1-1 (E466K) mutation was introduced using site-directed mutagenesis with the QuickChange kit (Stratagene). Quantitative β-galactosidase assays were performed on a minimum of six independent yeast transformants for each combination of interactors, and the average for each interaction was calculated as described in Ausubel et al. (1996). The experiment was performed twice with similar results.
Determination of SA levels
The procedure for SA extraction and determination is described in Newman et al. (2001).
Transcript analysis
Total RNA was isolated using Tri Reagent (Sigma). For TaqMan analysis, 1–2 µg of total RNA were reverse transcribed using Expand Reverse Transcriptase (Roche) and random hexamers (Pharmacia). TaqMan reactions were carried out on an ABI 7700 Sequence Detection System (Perkin-Elmer) according to the manufacturer’s instructions. All reactions were done in triplicate.
The Arabidopsis ACT2 gene was chosen as a normalization standard for TaqMan analysis because of its high, constitutive expression in nearly all vegetative tissues in both juvenile and mature plants (An et al., 1996). ACT2 expression among the various treatments (pathogen challenge and BTH) did not vary more than 2-fold compared with rRNA levels (data not shown), and was therefore considered suitable for normalization. The ACT2 gene, including part of the 3′ UTR, from accessions Col-0, Ler and Ws-0, was PCR amplified and sequenced in order to design primers and a TaqMan probe that would work in all three accessions. The ACT2 reverse primer was derived from the 3′ UTR in order to make it specific for ACT2 mRNA. TaqMan probes for ACT2, EDS1 and PAD4 were made cDNA specific by designing them across an intron. Primers and probes were designed using Primer Express software (Perkin-Elmer) and are as follows: ACT2-F, TCGGTGGTTCCATTCTTGCT; ACT2-R, GCTTTTTAAGCCTTTGATCTTGAGAG; ACT2-probe, AGCACATTCCAGCAGATGTGGATCTCCAA; EDS1-F, CAAGAATCTTGAAGCTGTC ATTGATC; EDS1-R, TGTCCTGTGAACACTATCTGTTTTCTACT; EDS1-probe, CACAGCCATTTCCACAGAAGCTTGAAATG; PAD4-F, TGGTCGACGCTGCCATACT; PAD4-R, GGTTGAATGGCCGGTTATCA; PAD4-probe, AATTCCAATCCTTCCTTGATCTTTAACTGAAGAAAGAGT. For RNA gel blots, 10 µg of total RNA were separated on a 1.2% formaldehyde agarose gel and blotted to Hybond NX (Amersham) according to the manufacturer’s instructions. PAD4 and PR-1 32P-radiolabelled probes were generated with the Oligolabeling kit (Pharmacia).
Sequence alignment
Sequences were aligned using Clustal_W (http://www2.ebi.ac.uk/clustalw) and the alignment was shaded using BoxShade (http://www.ch.embnet.org/software/BOX_form.html).
Acknowledgments
Acknowledgements
We thank Xinnian Dong (Duke University, Durham, NC) for providing Ler-nahG seed and Eric Holub (Horticultural Research International, Wellesbourne, UK) for assessing pad4-2 cotyledon phenotypes. We also thank Eric Holub, John Rathjen and Paul Schulze-Lefert for helpful discussions, and Susheng Gan (University of Kentucky, Lexington) for communication of results prior to publication. We are grateful to the Arabidopsis Biological Resource Center for seed and DNA stocks. The Sainsbury Laboratory is funded by the Gatsby Charitable Foundation. B.J.F. and L.J.M. are also supported by grants from The Biotechnology and Biological Sciences Research Council (BBSRC).
References
- Aarts N., Metz,M., Holub,E., Staskawicz,B.J., Daniels,M.J. and Parker,J.E. (1998) Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc. Natl Acad. Sci. USA, 95, 10306–10311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An Y.Q., McDowell,J.M., Huang,S.R., McKinney,E.C., Chambliss,S. and Meagher,R.B. (1996) Strong, constitutive expression of the Arabidopsis ACT2/ACT8 actin subclass in vegetative tissues. Plant J., 10, 107–121. [DOI] [PubMed] [Google Scholar]
- Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (1996) Current Protocols in Molecular Biology. John Wiley & Sons, New York, NY.
- Bergö M., Olivecrona,G. and Olivecrona,T. (1996) Forms of lipoprotein lipase in rat tissues: in adipose tissue the proportion of inactive lipase increases on fasting. Biochem. J., 313, 893–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botella M.A., Parker,J.E., Frost,L.N., BittnerEddy,P.D., Beynon,J.L., Daniels,M.J., Holub,E.B. and Jones,J.D.G. (1998) Three genes of the Arabidopsis RPP1 complex resistance locus recognize distinct Peronospora parasitica avirulence determinants. Plant Cell, 10, 1847–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowling S.A., Clarke,J.D., Liu,Y.D., Klessig,D.F. and Dong,X.N. (1997) The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell, 9, 1573–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke J.D., Aarts,N., Feys,B.J., Dong,X. and Parker,J.E. (2001) Constitutive disease resistance requires EDS1 in the Arabidopsis mutants cpr1 and cpr6 and is partially EDS1-dependent in cpr5. Plant J., 26, 409–420. [DOI] [PubMed] [Google Scholar]
- Clough S.J. and Bent,A.F. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J., 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Delledonne M., Xia,Y.J., Dixon,R.A. and Lamb,C. (1998) Nitric oxide functions as a signal in plant disease resistance. Nature, 394, 585–588. [DOI] [PubMed] [Google Scholar]
- Dempsey D.A., Shah,J. and Klessig,D.F. (1999) Salicylic acid and disease resistance in plants. Crit. Rev. Plant Sci., 18, 547–575. [Google Scholar]
- Falk A., Feys,B.J., Frost,L.N., Jones,J.D.G., Daniels,M.J. and Parker,J.E. (1999) EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc. Natl Acad. Sci. USA, 96, 3292–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann K.A. (1991) T-DNA insertion mutagenesis in Arabidopsis: mutational spectrum. Plant J., 1, 71–82. [Google Scholar]
- Feys B.J. and Parker,J.E. (2000) Interplay of signalling pathways in plant disease resistance. Trends Genet., 16, 449–455. [DOI] [PubMed] [Google Scholar]
- Gassmann W., Hinsch,M.E. and Staskawicz,B.J. (1999) The Arabidopsis RPS4 bacterial-resistance gene is a member of the TIR-NBS-LRR family of disease-resistance genes. Plant J., 20, 265–277. [DOI] [PubMed] [Google Scholar]
- Glazebrook J., Rogers,E.E. and Ausubel,F.M. (1996) Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics, 143, 973–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J., Zook,M., Mert,F., Kagan,I., Rogers,E.E., Crute,I.R., Holub,E.B., Hammerschmidt,R. and Ausubel,F.M. (1997) Phytoalexin- deficient mutants of Arabidopsis reveal that PAD4 encodes a regulatory factor and that four PAD genes contribute to downy mildew resistance. Genetics, 146, 381–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golemis E.A., Serebriiskii,I., Finley,R.L., Kolonin,M.G., Gyuris,J. and Brent,R. (1998) Interaction trap/two-hybrid system to identify interacting proteins. In Chanda,V.B. (ed.), Current Protocols in Protein Science. John Wiley & Sons, New York, NY, pp. 19.2.1–19.2.40.
- Gyuris J., Golemis,E., Chertkov,H. and Brent,R. (1993) Cdi1, a human G1-phase and S-phase protein phosphatase that associates with Cdk2. Cell, 75, 791–803. [DOI] [PubMed] [Google Scholar]
- He Y., Tang,W., Swain,J.D., Green,L., Jack,T.P. and Gan,S. (2001). Networking senescence-regulating pathways by using Arabidopsis enhancer trap lines. Plant Physiol., 126, 707–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland P.M., Abramson,R.D., Watson,R. and Gelfand,D.H. (1991) Detection of specific polymerase chain-reaction product by utilizing the 5′-3′ exonuclease activity of Thermus aquaticus DNA-polymerase. Proc. Natl Acad. Sci. USA, 88, 7276–7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirage D., Tootle,T.L., Reuber,T.L., Frost,L.N., Feys,B.J., Parker,J.E., Ausubel,F.M. and Glazebrook,J. (1999) Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc. Natl Acad. Sci. USA, 96, 13583–13588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirage D., Zhou,N., Cooper,B., Clarke,J.D., Dong,X. and Glazebrook,J. (2001) Constitutive salicylic acid-dependent signaling in cpr1 and cpr6 mutants requires PAD4. Plant J., 26, 395–407. [DOI] [PubMed] [Google Scholar]
- Kim K.K., Song,H.K., Shin,D.H., Hwang,K.Y., Choe,S., Yoo,O.J. and Suh,S.W. (1997) Crystal structure of carboxylesterase from Pseudomonas fluorescens, an α/β hydrolase with broad substrate specificity. Structure, 5, 1571–1584. [DOI] [PubMed] [Google Scholar]
- Kjemtrup S., Nimchuk,Z. and Dangl,J.L. (2000) Effector proteins of phytopathogenic bacteria: bifunctional signals in virulence and host recognition. Curr. Opin. Microbiol., 3, 73–78. [DOI] [PubMed] [Google Scholar]
- Klessig D.F. et al. (2000) Nitric oxide and salicylic acid signalling in plant defence. Proc. Natl Acad. Sci. USA, 97, 8849–8855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawton K.A., Friedrich,L., Hunt,M., Weymann,K., Delaney,T., Kessmann,H., Staub,T. and Ryals,J. (1996) Benzothiadiazole induces disease resistance in Arabidopsis by activation of the systemic acquired resistance signal transduction pathway. Plant J., 10, 71–82. [DOI] [PubMed] [Google Scholar]
- Levine A., Tenhaken,R., Dixon,R. and Lamb,C. (1994) H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell, 79, 583–593. [DOI] [PubMed] [Google Scholar]
- Martin G.B. (1999) Functional analysis of plant disease resistance genes and their downstream effectors. Curr. Opin. Plant Biol., 2, 273–279. [DOI] [PubMed] [Google Scholar]
- McDowell J.M. and Dangl,J.L. (2000) Signal transduction in the plant immune response. Trends Biochem. Sci., 25, 79–82. [DOI] [PubMed] [Google Scholar]
- McDowell J.M., Cuzick,A., Can,C., Beynon,J., Dangl,J.L. and Holub,E.B. (2000) Downy mildew (Peronospora parasitica) resistance genes in Arabidopsis vary in functional requirements for NDR1, EDS1, NPR1 and salicylic acid accumulation. Plant J., 22, 523–529. [DOI] [PubMed] [Google Scholar]
- Newman M.A., von Roepenack-Lahaye,E., Parr,A., Daniels,M.J. and Dow,J.M. (2001) Induction of hydroxycinnamoyl-tyramine conjugates in pepper by Xanthomonas campestris: a plant defense response activated by hrp gene-dependent and -independent mechanisms. Mol. Plant Microbe Interact., 14, 785–792. [DOI] [PubMed] [Google Scholar]
- Orozco-Cardenás M.L., Narváez-Vásquez,J. and Ryan,C.A. (2001) Hydrogen peroxide acts as a second messenger for the induction of defense genes in tomato plants in response to wounding, systemin, and methyl jasmonate. Plant Cell, 13, 179–191. [PMC free article] [PubMed] [Google Scholar]
- Parker J.E., Szabò,V., Staskawicz,B.J., Lister,C., Dean,C., Daniels,M.J. and Jones,J.D.G. (1993) Phenotypic characterization and molecular mapping of the Arabidopsis thaliana locus RPP5, determining disease resistance to Peronospora parasitica. Plant J., 4, 821–831. [Google Scholar]
- Parker J.E., Holub,E.B., Frost,L.N., Falk,A., Gunn,N.D. and Daniels,M.J. (1996) Characterization of eds1, a mutation in Arabidopsis suppressing resistance to Peronospora parasitica specified by several different RPP genes. Plant Cell, 8, 2033–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker J.E. et al. (1997) The Arabidopsis downy mildew resistance gene RPP5 shares similarity to the Toll and Interleukin-1 receptors with N and L6. Plant Cell, 9, 879–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pentikäinen M.O., Oorni,K. and Kovanen,P.T. (2000) Lipoprotein lipase (LPL) strongly links native and oxidized low density lipoprotein particles to decorin-coated collagen—roles for both dimeric and monomeric forms of LPL. J. Biol. Chem., 275, 5694–5701. [DOI] [PubMed] [Google Scholar]
- Rustérucci, C., Aviv,D.H., Holt,B.,III, Dangl,J.L. and Parker,J.E. (2001). The disease resistance signaling components EDS1 and PAD4 are essential regulators of the cell death pathways controlled by LSD1 in Arabidopsis. Plant Cell, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W.J., Patel,S., Hong,R. and Kraemer,F.B. (2000) Hormone-sensitive lipase functions as an oligomer. Biochemistry, 39, 2392–2398. [DOI] [PubMed] [Google Scholar]
- Shirasu K., Nakajima,H., Rajasekhar,V.K., Dixon,R.A. and Lamb,C. (1997) Salicylic acid potentiates an agonist-dependent gain control that amplifies pathogen signals in the activation of defense mechanisms. Plant Cell, 9, 261–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Biezen E.A., Sun,J., Coleman,M.J., Bibb,M.J. and Jones,J.D.G. (2000) Arabidopsis RelA/SpoT homologs implicate (p)ppGpp in plant signaling. Proc. Natl Acad. Sci. USA, 97, 3747–3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T. and Brown,M.J. (1999) mRNA quantification by real time TaqMan polymerase chain reaction: validation and comparison with RNase protection. Anal. Biochem., 269, 198–201. [DOI] [PubMed] [Google Scholar]
- Wong H., Yang,D., Hill,J.S., Davis,R.C., Nikazy,J. and Schotz,M.C. (1997) A molecular biology-based approach to resolve the subunit orientation of lipoprotein lipase. Proc. Natl Acad. Sci. USA, 94, 5594–5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou N., Tootle,T.L., Tsui,F., Klessig,D.F. and Glazebrook,J. (1998) PAD4 functions upstream from salicylic acid to control defense responses in Arabidopsis. Plant Cell, 10, 1021–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]