Abstract
In the deoP2 promoter of Escherichia coli, a transcription activator, cAMP–CRP, binds at two sites, centered at –41.5 and –93.5 from the start site of transcription, while a repressor, CytR, binds to a space between the two cAMP–CRP complexes. The mechanisms for the cAMP–CRP-mediated transcription activation and CytR-mediated transcription repression were investigated in vitro using purified components. We classified the deoP2 promoter as a class II cAMP–CRP-dependent promoter, primarily by the action of cAMP–CRP at the downstream site. Interestingly, we also found that deoP2 carries an ‘UP-element’ immediately upstream of the downstream cAMP–CRP site. The UP-element overlaps with the DNA site for CytR. However, it was observed that CytR functions with the RNA polymerase devoid of the C-terminal domain of the α-subunit as well as with intact RNA polymerase. The mechanism of repression by CytR proposed in this study is that the cAMP–CRP bound at –41.5 undergoes an allosteric change upon direct interaction with CytR such that it no longer maintains a productive interaction with the N-terminal domain of α, but instead acts as a repressor to interfere with RNA polymerase acting on deoP2.
Keywords: in vitro DNA–protein interaction/prokaryotic transcription initiation/transcription activator/transcription repressor
Introduction
Bacterial transcription initiation is often modulated by the action of either the transcription activator or repressor. It has been well established that transcription activation involves a direct protein–protein interaction between RNA polymerase (RNP) and transcription activator, which binds to a DNA site in the vicinity of a promoter (Ishihama, 1993; Busby and Ebright, 1997, 1999; Ptashne and Gann, 1997; Hochschild and Dove, 1998). On the other hand, transcription repression is attributed to a mechanism of steric hindrance, in which repressor binding physically interferes with RNP binding. However, several lines of evidence support the notion that a protein–protein contact between repressors and RNP could also lead to transcription repression (Choy et al., 1995; Monsalve et al., 1996).
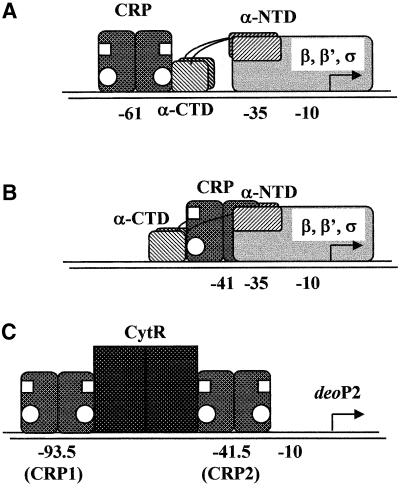
Transcription activation at CRP (cAMP-receptor protein)-dependent promoters provides a paradigm for elucidating the mechanism by which an activator makes multiple interactions with RNP to enhance transcription initiation. CRP bears two activating regions (AR1 and AR2) that recognize different determinants within the RNP α-subunit (Figure 1) (reviewed in Busby and Ebright, 1999). This subunit consists of an N-terminal domain (α-NTD) containing determinants for interaction with the remainder of RNP and with CRP, a C-terminal domain (α-CTD) containing determinants for interaction with DNA and with various activators, and an unstructured/flexible linker connecting the two subdomains (Blatter et al., 1994; Negishi et al., 1995; Jeon et al., 1997; Meng et al., 2000). At promoters that have a DNA site (approximately –60) for CRP upstream of the DNA site for RNP (class I CRP-dependent promoters), AR1 of the promoter-proximal subunit of CRP makes direct contact with α-CTD, which results in α-CTD–DNA interaction (Figure 1A) (Bell et al., 1990; Eschenlauer and Reznikoff, 1991; Zou et al., 1993a,b; Niu et al., 1994; Tang et al., 1994; Gaal et al., 1996; Murakami et al., 1996). This protein–protein interaction increases the affinity of RNP for promoter DNA (KB) and thereby increases transcription initiation (Malan et al., 1984; Dove et al., 1997; Busby and Ebright, 1999). Consistent with this model, some strong bacterial promoters with intrinsically high KB, such as rRNA promoters, carry an AT-rich sequence between –40 and –60, termed the ‘UP-element’, which is recognized and bound by α-CTD in the absence of transcription activator (Ross et al., 1993, 1998; Rao et al., 1994; Czarniecki et al., 1997; Estrem et al., 1998; Noel and Reznikoff, 1998; Tagami and Aiba, 1999; Ozoline et al., 2000). At class II CRP-dependent promoters (the DNA site for CRP is centered near or at –41, overlapping the –35 region), CRP utilizes both AR1 and AR2 to contact α-CTD and α-NTD, respectively, thereby stimulating both closed promoter complex formation (KB) and open complex formation (ki) (Figure 1B) (Bell and Busby, 1990; Williams et al., 1991, 1996; West et al., 1993; Zhou et al., 1994; Niu et al., 1996; Savery et al., 1998; Busby and Ebright, 1999). AR1 is presented as functional by the promoter-distal subunit, and AR2 is presented as functional by the promoter-proximal CRP subunit. At class II promoters, in contrast to class I promoters, the AR1 interaction does not provide direct activation but appears to overcome an inhibitory effect of α-CTD (Niu et al., 1996; Busby and Ebright, 1999). Thus, removal of α-CTD has no negative effect on class II CRP-dependent transcription and eliminates the requirement for a functional AR1 region. Finally, two properly positioned CRP molecules can work synergistically. At such compound class I/class II CRP-dependent promoters, CRP activates transcription through the set of interactions described for class I and class II CRP-dependent promoters (i.e. three sets of interactions) (Busby and Ebright, 1999).
Fig. 1. Schematic model of CRP–RNP interaction at class I (A) and class II CRP (B) dependent promoters. White circles and squares on CRP represent AR1 and AR2, respectively (see text). (C) A schematic model of two CRPs and CytR bound on deoP2.
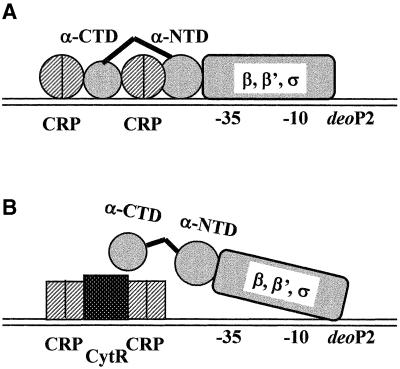
The Escherichia coli deoP2 promoter contains two DNA sites for cAMP–CRP, an upstream site centered at position –93.5 (CRP1) and a downstream site centered at position –41.5 (CRP2), and a DNA site for CytR, centered at position –65 (Figure 1C). Under conditions in which CytR is inactive (i.e. in the presence of cytidine), CRP interacts with RNP to form transcriptionally active CRP–RNP–promoter complexes, whereas under conditions in which CytR is active (in the absence of cytidine), two CRPs interact with CytR to form transcriptionally inactive (CRP)2–CytR–deoP2 complexes (Søgaard-Andersen et al., 1990a,b, 1991a,b; Søgaard-Andersen and Valentin-Hansen, 1993; Meibom et al., 1999). The formation of these complexes involves both CytR–DNA interaction and CytR–CRP interaction. The action of CytR in vivo is absolutely dependent on the presence of cAMP–CRP, and therefore CRP interacts alternatively, and mutually exclusively, with CytR or RNP. Based on these observations, it has been suggested that CytR and RNP are rivals that compete for binding with CRP to deoP2, and that CytR acts as an anti-activator, nullifying the activation by cAMP–CRP (Søgaard-Andersen et al., 1990a,b; Møllegaard et al., 1993; Kristensen et al., 1997; Meibom et al., 2000).
In this paper, we address how the formation of CytR–CRP promoter complexes at deoP2 might result in transcription repression. Recent work has shown that only one subunit of the CRP dimer, the subunit proximal to CytR, functionally interacts with CytR in (CRP)2– CytR–promoter complexes (Figure 1C), ruling out the proposal that masking of the activation region of CRP (AR2) is responsible for the transcriptional inactivity of the complexes (Meibom et al., 2000). Therefore two, not mutually exclusive, repression models seem possible: (i) CytR may interfere with interactions between AR1 of CRP and α-CTD of RNP (by occupying the DNA segment adjacent to CRP, preventing access of α-CTD to this DNA segment) (Valentin-Hansen et al., 1996); and (ii) CytR may interfere with interaction between RNP and the core promoter. A prediction of the first model is that the formation of CytR–CRP promoter complexes should not result in repression if transcription is conducted by RNP derivatives lacking α-CTD (Igarashi and Ishihama, 1991). To test this idea, we have investigated CRP and CytR regulation of transcription initiation at deoP2 with reconstituted wild-type RNP and RNP derivatives lacking α-CTD.
Results
cAMP–CRP activation of deoP2
We constructed pdeoP2 to examine the regulation of deoP2 in vitro. This plasmid is a derivative of the well characterized transcription vector pSA508 (Choy and Adhya, 1993), and carries all the deoP2 sequence information (from –116 to +25) required for cAMP–CRP activation and CytR repression, followed by a strong factor-independent transcription terminator. When supercoiled pdeoP2 DNA was used as a template in transcription assays, two small transcripts were generated: a 65 nucleotide deoP2 transcript and a 108 nucleotide RNA-I transcript originating from the plasmid native RNA-I promoter. The RNA-I transcript served as an internal control for the various transcription assays shown below.
In the first set of experiments, activation of deoP2 by cAMP–CRP was examined. Transcription was performed at increasing cAMP concentrations with constant CRP and wild-type RNP using supercoiled pdeoP2 as the DNA template (Figure 2A). Addition of cAMP had a marked effect on deoP2 transcription, but no significant effects on transcription initiation at the RNA-I promoter. The lower panel (Figure 2B) shows the quantification of deoP2 RNA as a function of cAMP concentration. The cAMP concentration needed for maximal transcription from deoP2 was ∼100 µM. At this concentration of cAMP, transcription was activated ∼4-fold.
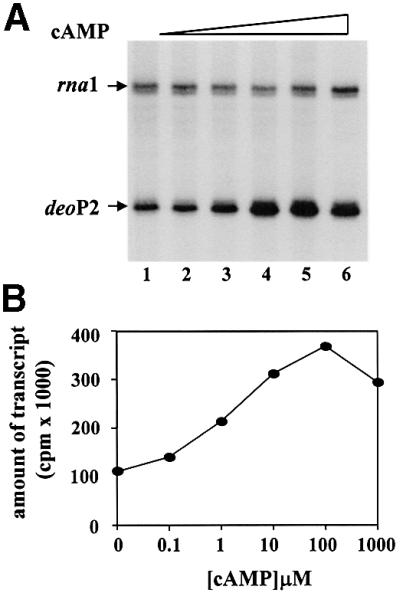
Fig. 2. cAMP was titrated in the presence of 0.1 µM CRP using deoP2 DNA template. (A) The transcription products were resolved in denaturing 8% polyacrylamide gel (see Materials and methods). The rna1 and deoP2 RNAs were labeled as such. The cAMP concentration in each lane was as follows: 0, 0.1, 1, 10, 100 and 1000 µM for lanes 1–6. (B) deoP2 RNAs were quantified with a β-scanner (PhosphorImager; Molecular Dynamics, CA) and the total c.p.m. were plotted as a function of cAMP concentration.
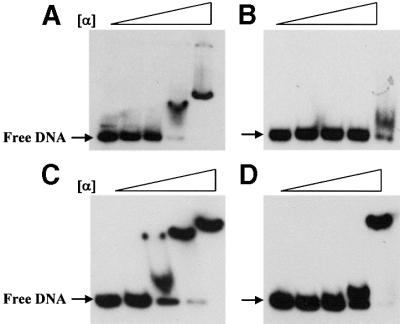
In the next set of experiments we examined activation of deoP2 transcription using RNP reconstituted from purified subunits. Three α-subunits were employed: α-wild type and two different derivatives lacking αCTD and part of the flexible linker (α-235 and α-256) (Igarashi and Ishihama, 1991). In these experiments, a pSA508 derivative carrying the lac promoter (placZP) was included in all reactions. lacZP acted as a control promoter, which is activated by cAMP–CRP only when transcribed by RNP carrying an intact α-subunit (Igarashi and Ishihama, 1991). The results of in vitro transcription experiments with the reconstituted RNPs are presented in Figure 3. The transcriptional activity of the reconstituted RNPs appeared to be very similar: a constant amount of the RNA-I transcript as well as a constant amount of factor-independent lacZP transcript were generated. As expected, only when transcribed by RNP containing the intact α-subunit (α-WT; Figure 3, lane 2) could the cAMP–CRP activate transcription at lacZP. By contrast, cAMP–CRP stimulated the transcription at deoP2 when transcribed by RNP reconstituted with truncated α-subunits as well as intact α-subunits. A similar activation pattern has been described at the E.coli galP1 promoter, the prototype class II cAMP–CRP-dependent promoter (Igarashi and Ishihama, 1991; West et al., 1993; Busby and Ebright, 1999). These results are consistent with the previous reports (Søgaard-Andersen et al., 1990a) that CRP-2 plays no role, or a very small one, in cAMP–CRP activation of transcription at deoP2. How ever, it was also noted that removal of α-CTD from RNP had a pronounced effect on the basal level deoP2 promoter strength (Figure 3, lanes 3 and 5). Therefore, the fold activation of deoP2 by cAMP–CRP when transcribed by the RNP reconstituted with the truncated α-subunit appeared to be much greater than that obtained with the RNP reconstituted with intact α, although the actual activity is less. This feature of deoP2 transcription suggested the possibility that the promoter bears an UP-element within the deoP2 promoter, which enhances transcription initiation by wild-type RNP but not by RNP derivatives lacking α-CTD. To clarify this possibility we performed a gel-shift experiment with a DNA fragment carrying an intact deoP2 promoter. The results are presented in Figure 4. Addition of purified α-subunit (>273 nM) resulted in a complete retardation of deoP2 DNA probe (Figure 4A). When a DNA probe containing a sequence within deoC was used, only a partial retardation was observed even at 820 nM α-subunit (Figure 4B). In this experiment, E.coli malT promoter DNA known to carry the UP-element was included as a control (Tagami and Aiba, 1999). The DNA probe carrying wild-type malT promoter was shifted partially with 91 nM α-subunit and completely with 273 nM α-subunit (Figure 4C). However, a complete retardation of a mutant malT promoter DNA deleted of the UP-element was observed with 820 nM α-subunit (Figure 4D). Taken together, our results show that deoP2 bears an UP-element, and that the interaction between this element and α-CTD contributes significantly to the basal and activated level of deoP2 activity.
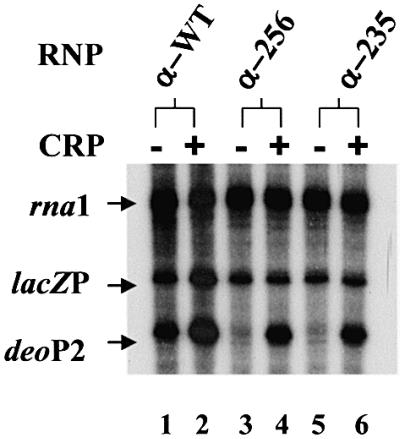
Fig. 3. cAMP–CRP activation of deoP2 with RNP carrying intact α (α-WT) (37 nM), α with 256 amino acids (α-256) (150 nM) or with 235 amino acids (α-235) (150 nM). In the same reaction tube was also added 2 nM lacZp DNA. cAMP was present at 0.1 mM in all lanes and CRP (0.1 µM) was present only in the even-numbered lanes.
Fig. 4. A mobility shift assay with purified α and a DNA fragment carrying deoP2 [–116 to +25; (A)], a structural part of deoC [+58 to +193; (B)], wild-type malT promoter [–121 to +18; (C)] and a mutant malT promoter [–121 to +18; (D)] (see text for details). The mutant malT promoter carries GCGTCGCGA instead of ATAAAAAAA between –53 and –43 of the wild-type promoter (Tagami and Aiba, 1999). The α concentration in each lane is 0, 30.3, 91, 273.8 and 820 nM from left to right.
CytR–CRP-mediated repression at deoP2
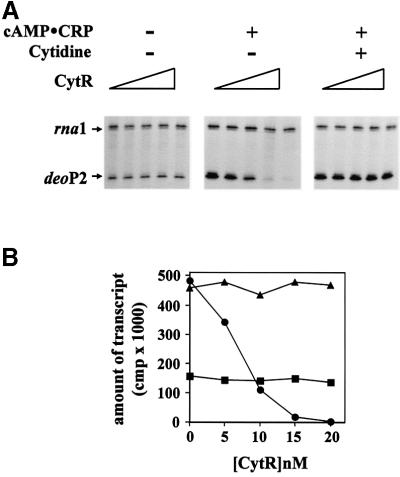
To assess CytR regulation at deoP2 in vitro, we first examined transcription with intact RNP in the presence of CytR, CytR and cAMP–CRP, and CytR, cAMP–CRP and cytidine. The results are presented in Figure 5. In the absence of cAMP–CRP, addition of CytR had no effect on deoP2 transcription (Figure 5, left panel). Addition of increasing amounts of CytR to transcription assays containing cAMP–CRP gradually decreased transcription (Figure 5, middle panel). This repressive effect of CytR was abolished by the addition of cytidine, an allosteric effector for CytR (Figure 5, right panel). The regulatory patterns were as expected. However, the finding that the formation of (CRP)2–CytR–promoter complexes resulted in a complete repression of deoP2 transcription (i.e. to a promoter activity that was considerably lower than that in the absence of cAMP–CRP) was unexpected. Thus, it appears that CytR–CRP communication at deoP2 also affected the cAMP–CRP-independent transcription.
Fig. 5. CytR titration in the absence (left panel) and presence (middle and right panels) of cAMP–CRP. The right panel is in the presence of 20 mM cytidine in addition to cAMP–CRP. The concentration of CytR in each lane for all three panels was as follows: 0, 5, 10, 15 and 20 nM from left to right. (A) The transcription products were resolved in denaturing 8% polyacrylamide gel. (B) deoP2 RNAs were quantified with a β-scanner and the total c.p.m. were plotted as a function of CytR concentration.
In an attempt to determine the role of α-CTD in CytR/CRP-mediated repression at deoP2, we next investigated transcription by the reconstituted RNP derivatives in the presence of a constant concentration of cAMP–CRP and varying amounts of CytR. The results are presented in Figure 6A, and the quantification of deoP2 transcripts is given in Figure 6B. Clearly, the formation of repression complexes at deoP2 turned off transcription by the RNP derivatives lacking α-CTD in a fashion quite similar to that seen for intact RNP. This observation is inconsistent with the previously proposed ‘anti-activation’ model, in which CytR interferes with the interaction between α-CTD of RNP and the DNA site immediately upstream of CRP1 (Valentin-Hansen et al., 1996). Rather, the in vitro results shown in Figures 5 and 6 suggest that an important component of the mechanism for CytR-mediated repression should involve interference with interactions between RNP and the core promoter.
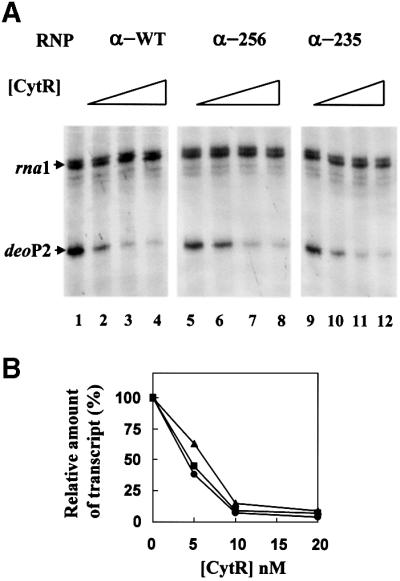
Fig. 6. Repression of deoP2 by CytR with RNP carrying intact α (α-WT) (37 nM), α with 256 amino acids (α-256) (150 nM) or α with 235 amino acids (α-235) (150 nM). The CytR concentration used in each lane was as follows: 0 for lanes 1, 5 and 9; 5 nM for lanes 2, 6 and 10; 10 nM for lanes 3, 7 and 11; 20 nM for lanes 4, 8 and 12. (A) The transcription products were resolved on denaturing 8% polyacrylamide gel. (B) deoP2 RNAs were quantified with a β-scanner and the total c.p.m. in the presence of CytR relative to that in the absence of CytR (100%) were plotted as a function of CytR concentration.
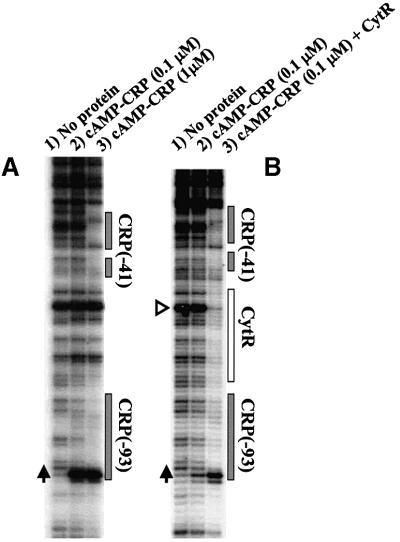
Subsequently, we examined the DNase I footprint of cAMP–CRP and CytR bound to deoP2. The DNase I footprint was obtained by a primer extension method following DNase I treatment of DNA–protein complexes since supercoiled DNA was used as a template (Figure 7). Figure 7A shows cAMP–CRP footprints where CRP was present at 0.1 µM (lane 2) or 1 µM (lane 3) in the presence of a saturating concentration of cAMP (0.1 mM). With CRP at 0.1 µM, the cAMP–CRP footprint was barely detectable only at the upstream site (centered at –93) with a hyper-reactive T (–99, vertical arrows), even though the activation by cAMP–CRP, as determined by transcription assay (see Figure 2), was maximal at this concentration. cAMP–CRP footprint at the downstream site (centered at –41) was detected only in the presence of 1 µM CRP (Figure 7A, lane 3). Figure 7B shows the cAMP–CRP footprint at 0.1 µM CRP and 0.1 mM cAMP in the absence (lane 2) or presence of CytR (20 nM) (lane 3). CytR binding shows a marked disappearance of hyper-reactive A (–65, open triangle) together with the bands extended from the downstream CRP site to the upstream CRP site. The cAMP–CRP-specific hyper-reactive T at –99 was enhanced further. Taken together, the CytR (20 nM) binding significantly enhanced the cAMP–CRP binding, particularly at the downstream site, suggesting that a protein–protein interaction between two proteins resulted in increased cAMP–CRP binding to the DNA sites.
Fig. 7. cAMP–CRP binding to deoP2 DNA as probed with DNase I. The DNase I footprint was revealed by a primer extension method since a supercoiled DNA template (pdeoP2) was used (see Materials and methods). (A) Different concentrations of CRP were reacted with deoP2 in the presence of 0.1 mM cAMP. (B) cAMP (0.1 mM) and CRP (0.1 µM) were reacted with pdeoP2 in the presence or absence of CytR (15 nM). The cAMP–CRP binding site is shown as a shaded box and CytR as an open box. Vertical arrows show the hyper-reactive T at –99, and the open triangle in (B) a hyper-reactive A (–65) in the middle of the CytR binding site (see text). The deoP2 promoter sequence (–105 to –25) is 5′-AATTATTTGAACCAGATCGCATTA CAGTGATGCAACTTGTAAGTAGATTTCCTTAATTGTGATGTG TATCGAAGTGTGTTG-3′, in which the two cAMP–CRP sites are underlined.
Discussion
Activation of deoP2
We propose here that deoP2 should be categorized in the class II CRP-dependent promoters, based on the observations that (i) a deletion of upstream CRP site (centered at –93.5) did not affect the cAMP–CRP activation of deoP2 in vivo (Søgaard-Andersen et al., 1990a) and (ii) cAMP–CRP activated deoP2 in vitro with RNP reconstituted either with truncated α at CTD or intact α equally well (Figures 2 and 3). Transcription activation at class II CRP-dependent promoters has two components: a first component involving anti-inhibition and a second component involving direct activation. Anti-inhibition is through an interaction between the AR1 of CRP and α-CTD, which overcomes an inhibitory effect of α-CTD (Figure 1); thereby, the CRP activation of class II CRP-dependent promoter is unaltered by the mutant RNP devoid of α-CTD (Attey et al., 1994; Zhou et al., 1994; Belyaeva et al., 1996). Direct activation is through an interaction of α-NTD with AR2 of cAMP–CRP (see Figure 1B) (Attey, 1994; Belyaeva et al., 1996; Murakami et al., 1997; Rhodius et al., 1997; Savery, 1998; Busby and Ebright, 1999). Therefore, in the case of deoP2, the cAMP–CRP at –41.5 should be primarily responsible for the activation. It has been shown that at some artificial CRP-dependent promoters, cAMP–CRP centered near position –103, –93 or –83 could synergistically activate transcription with a cAMP–CRP centered near position –42, a class III CRP-dependent promoter (Belyaeva et al., 1996, 1998; Murakami et al., 1997; Busby and Ebright, 1999). However, the observation with deoP2 suggests that the synergistic activation by two or more cAMP–CRP should depend on the context of the promoter.
At least three lines of evidence support the view that there is an UP-element in deoP2, presumably between two cAMP–CRP sites: (i) a gel mobility shift assay showed purified α binding to deoP2 promoter DNA; (ii) the basal level deoP2 promoter activity is significantly impaired with the RNP carrying truncated α at CTD; (iii) there is AT-rich sequence (25/36) immediately upstream of the CRP1 site centered at –41.5 (Figures 3, 4 and 6). Previously, DNase I and uranyl footprinting of RNP complex on deoP2 revealed that there was a contact at this site in addition to those in the core promoter region (Møllegaard et al., 1993), which is presumably ascribed to the interaction between α-CTD and the UP-element. Moreover, transcription activation is sensitive to replacements in the A/T-rich sequence present upstream of the CRP1 site (Kristensen et al., 1997). Thus, all available data are consistent with the proposal that deoP2 is a class II CRP-dependent promoter which is equipped with a supplementary promoter element. It has been proposed that transcription activation of class II CRP-dependent promoters is established by positioning α-CTD to the DNA site immediately upstream of the cAMP–CRP site in addition to direct activation (Attey et al., 1994; Belyaeva et al., 1996, 1998; Niu et al., 1996; Busby and Ebright, 1999). This DNA sequence-independent interaction between the DNA adjacent to cAMP–CRP and α-CTD of RNP has been implicated as playing a role in class II cAMP–CRP-dependent activation (Savery et al., 1995, 1998; Lloyd et al., 1998). In the case of deoP2, however, there is an UP-element having a natural affinity for α-CTD. It has been shown that placement of an UP-element (from rrnB P1) immediately upstream in the E.coli melR promoter, a class II CRP-dependent promoter, increased the promoter activity without altering its dependence on CRP (Savery et al., 1995). Therefore, the contributions of α-CTD–UP-element interactions and α-CTD–CRP interactions to promoter activity were suggested to be independent and additive. Nevertheless, in the case of deoP2, if α-CTD–UP-element interactions and α-CTD–CRP interactions both recruit α-CTD to the DNA site, the activation system appears to be redundant. It is possible, however, that the above two interactions, although both with α-CTD, may affect different steps in transcription initiation. It should be noted that there is an UP-element binding protein in E.coli, methylated Ada protein, which binds to an UP-like element sequence in ada and aidB promoters, and stimulates transcription by modifying the nature of the RNP–promoter interaction (Landini and Volkert, 1995).
Repression of deoP2
Repression of deoP2 by CytR was only observed in the presence of cAMP–CRP, in agreement with the in vivo observation that CytR binding requires cAMP–CRP (Søgaard-Andersen et al., 1990a,b). It was therefore proposed that CytR acts as an anti-activator, especially since CytR binding centered at around –65 is too far from the promoter to interfere with RNP directly (Søgaard Andersen et al., 1990a,b; Møllegaard et al., 1993; Valentin-Hansen et al., 1996; Kristensen et al., 1997; Meibom et al., 2000). In this model, CytR–CRP and CytR–DNA interactions block transcription activation by CRP, by sterically blocking the functional AR1 of CRP and by sterically preventing α-CTD from interacting with the DNA segment immediately upstream of the CRP dimer at the CRP1 site. We observed in this study that CytR could block transcription in vitro to a level that is significantly lower than that observed in the absence of CRP (Figure 5). Moreover, CytR inhibits deoP2 transcription conducted by RNP reconstituted with α-subunits lacking α-CTD (Figure 6). These results suggest that CytR repression also involves interference with interactions between RNP and the core promoter (‘direct repression’). ‘Oriented heterodimer’ analysis showed that only one subunit of the CRP dimer, the subunit proximal to CytR, functionally interacts with CytR in the repression complexes (Meibom et al., 2000). We therefore envisage that CytR contact with the promoter upstream subunit of the CRP dimer at the CRP1 DNA site may induce a conformational change in the AR2 region of the promoter downstream subunit that renders it incompetent to interact properly with RNP. Consistently, it was noted that cAMP–CRP binding was greatly enhanced in the presence of CytR, especially at the CRP1 site, suggesting a protein–protein interaction between two proteins (Figure 7). Taking together the results presented in this study, we propose a simple mechanism to account for the repression by CytR: the cAMP–CRP complex at the CRP1 site, upon interaction with CytR, undergoes allosteric changes such that its binding to the DNA site is improved but its interaction with α-NTD is no longer productive, but rather destructive, leading to transcription repression (Figure 8). This is a likely possibility since cAMP–CRP activation of a CRP II-dependent promoter should require precise interaction between those amino acid determinants on CRP and α-NTD, and therefore a subtle change in protein conformation could interfere with RNP and core promoter interaction. In this context, cAMP–CRP, an activator, is converted through an interaction with CytR to a repressor that interferes with RNP acting at deoP2.
Fig. 8. Schematic model. The interaction between α-NTD and cAMP–CRP at –41 (CRP1 site) should, therefore, be important for activation of deoP2, class II CRP-dependent promoter (A). In the presence of CytR (B), the cAMP–CRP, especially that bound at the promoter-proximal site, undergoes an allosteric change such that it no longer interacts with α-NTD positively, but instead negatively. In this model, we speculate that CytR converts cAMP–CRP, especially at the promoter-proximal site, from an activator into a repressor to interfere with the RNP acting on deoP2.
Materials and methods
Plasmids
The pdeoP2 and placZP plasmids were constructed by cloning a 141 bp fragment of the deoP2 promoter (–116 to +25) and a 201 bp fragment of the lacZ promoter (–124 to +76), respectively, into the EcoRI and PstI sites of the transcriptional vector pSA508 (Choy and Adhya, 1993).
Proteins
CRP was purified from an E.coli strain carrying the crp+ gene on a multicopy plasmid, pHA5, by fast protein liquid chromatography (S.Ryu, Seoul National University, Korea). CytR was a generous gift from C.Jorgenson (Odense University, Denmark). Wild-type RNP polymerase was purchased from Pharmacia LKB. Reconstituted RNP composed of wild-type subunits or of C-truncated α-subunits was as used by Igarashi and Ishihama (1991).
Transcription assay
Transcription reactions were carried out using the procedure described by Choy and Adhya (1993). Briefly, 2 nM DNA template, 1 mM ATP, 0.1 mM GTP, 0.1 mM CTP, 0.01 mM UTP and 10–20 µCi of [α-32P]UTP were pre-incubated in buffer [20 mM Tris–acetate pH 7.8, 10 mM magnesium acetate, 100 mM potassium glutamate, 1 mM dithiothreitol (DTT)] at 37°C for 5 min. When present, a repressor was included in the pre-incubation mix at concentrations described in the figure legends. Transcription was initiated by the addition of RNP (20 nM) in a total volume of 20 µl, and terminated after 10 min at 37°C by the addition of an equal volume (20 µl) of RNA loading buffer [80% (v/v) deionized formamide, 1× TBE (89 mM Tris, 89 mM boric acid, 2 mM EDTA), 0.025% bromophenol blue, 0.025% xylene cyanol]. The mixture was heated at 90°C for 2 min and electrophoresed in 8 M urea–8% polyacrylamide sequencing gels (40 cm × 0.4 mm). The RNA transcripts were quantified as counts per minute (c.p.m.) as determined with an Ambis β-scanner.
DNase I experiments
The experiments were conducted essentially as described by Brenowitz et al. (1986). The reaction conditions (in 20 µl) were the same as in in vitro transcription reactions, except that the nucleotides were omitted. After incubation at 37°C for 10 min, probing with DNase I was performed by adding 1 ng of the enzyme to the reaction buffer. The incubation was continued at 37°C for 5 min and stopped by addition of 20 µl of 40 mM EDTA. Modified DNA was purified using a PCR clean-up kit (Qiagen).
Primer extension analysis
Primer extension analysis used the alkaline denaturation procedure described in Rostock et al. (2000). The primer was a 33mer carrying the top strand sequence immediately upstream of multiple cloning sites in pSA508. Briefly, 1 µl of 32P-labeled primer (1 pmol) and 4 µl of 10 mM NaOH were added to 35 µl of modified DNA and incubated for 2 min at 80°C, which was followed by a quick chilling in ice (for 5 min). To the denatured DNA were added 5 µl of hybridization buffer (0.5 M Tris–HCl pH 7.2, 100 mM MgSO4, 2 mM DTT) with incubation for 3 min at 45°C, followed by a quick chilling in ice (for 5 min). Primer extension was carried out for 10 min at 50°C in the presence of dNTP (5 µl of 5 mM stock) and Klenow (0.5–1 U; Pharmacia). The reaction was stopped with 35 µl of 5 M NH4CH3COO, and the DNA was precipitated with ethanol and dissolved in formamide loading buffer. The mixture was heated for 2 min at 90°C and electrophoresed on 6% polyacrylamide DNA sequencing gels containing 8 M urea (40 cm × 0.4 mm).
Gel mobility shift assay
Gel mobility assays were performed with PCR fragments of deoP2 promoter (–116 to +25), deoC structural part (+58 to +193), wild-type malT promoter (–121 to +18) and an A/T deleted mutant malT promoter (–121 to +18) (see the legend to Figure 4 for DNA sequence alteration). For the mutant malT promoter DNA fragment, pMT203(–A/T) (Tagami and Aiba, 1999), kindly provided by H.Aiba (Nagoya University, Japan), was used as the DNA template for PCR. The PCR fragment was end-labeled with [γ-32P]ATP using T4 polynucleotide kinase (Promega). The reaction mixture containing 2 nM end-labeled deoP2 DNA fragments and different concentrations of purified α-subunit was incubated at 37°C in transcription buffer (50 mM Tris–HCl pH 7.9, 50 mM NaCl, 3 mM DTT, 5% glycerol) for 5 min. The gel running buffer was 1× Tris/borate/EDTA containing 5% glycerol, and the reaction mixtures were run on 5% acrylamide gels (acrylamide:bis-acrylamide = 50:1) containing 1× Tris/borate/EDTA and 5% glycerol.
Acknowledgments
Acknowledgements
This research was supported by a fund from a grant (HMP-99-M-04-0002) of the 1999 Good Health R and D project, Ministry of Health and Welfare, Republic of Korea. The authors are grateful to Dr H.Aiba at Nagoya University, Japan, for providing the wild-type and mutant malT promoter DNA.
References
- Attey A., Belyaeva,T., Savery,N., Hoggett,J., Fujita,N., Ishihama,A. and Busby,S. (1994) Interactions between the cyclic AMP receptor protein and the α subunit of RNA polymerase at the E.coli galP1 promoter. Nucleic Acids Res., 22, 4375–4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell A., Gaston,K., William,R., Chapman,K., Kolb,A., Buc,H., Minchin,S., Williams,J. and Busby,S. (1990) Mutations that alter the ability of Escherichia coli cyclic AMP receptor protein to activate transcription. Nucleic Acids Res., 18, 7243–7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyaeva T., Bown,J., Fujita,N., Ishihama,A. and Busby,S. (1996) Location of the C-terminal domain of the RNA polymerase α subunit in different open complexes at the E.coli galactose operon regulatory region. Nucleic Acids Res., 24, 2242–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyaeva T., Rhodius,V., Webster,C. and Busby,S. (1998) Transcription activation at promoters carrying tandem DNA sites for the Escherichia coli cyclic AMP receptor protein: organisation of the RNA polymerase α subunits. J. Mol. Biol., 277, 789–804. [DOI] [PubMed] [Google Scholar]
- Blatter E., Ross,W., Tang,H., Gourse,R. and Ebright,R. (1994) Domain organization of RNA polymerase α subunit: C-terminal 85 amino acids constitute a domain capable of dimerization and DNA binding. Cell, 78, 889–896. [DOI] [PubMed] [Google Scholar]
- Brenowitz M., Snear,D.F., Shea,M.A. and Achers,G.K. (1986) Quantitative DNase I footprint titration; A method for studying protein–DNA interactions. Method Enzymol., 130, 132–181. [DOI] [PubMed] [Google Scholar]
- Busby S. and Ebright,R.H. (1997) Transcription activation at class II CAP-dependent promoters. Mol. Microbiol., 23, 853–859. [DOI] [PubMed] [Google Scholar]
- Busby S. and Ebright,R.H. (1999) Transcription activation by catabolite activator protein (CAP). J. Mol. Biol., 293, 199–213. [DOI] [PubMed] [Google Scholar]
- Choy H. and Adhya,S. (1993) RNA polymerase idling and clearance in gal promoters: use of supercoiled minicircle DNA template made in vivo. Proc. Natl Acad. Sci. USA, 90, 472–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy H.E., Park,S.W., Aki,T., Parrack,P., Fujita,N., Ishihama,A. and Adhya,S. (1995) Repression and activation of transcription by Gal and Lac repressors: involvement of α subunit of RNA polymerase. EMBO J., 14, 4523–4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czarniecki D., Noel,R. and Reznikoff,W. (1997) The –45 region of the Escherichia coli lac promoter: CAP-dependent and CAP-independent transcription. J. Bacteriol., 179, 423–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dove S.L., Jung,J.K. and Hochschild,A. (1997) Activation of prokaryotic transcription through arbitrary protein–protein contacts. Nature, 386, 627–630. [DOI] [PubMed] [Google Scholar]
- Eschenlauer A. and Reznikoff,W. (1991) Escherichia coli catabolite gene activator protein mutants defective in positive control of lac operon transcription. J. Bacteriol., 173, 5024–5029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrem S.T., Gaal,T., Ross,W. and Gourse,R.L. (1998) Identification of an UP element consensus sequence for bacterial promoter. Proc. Natl Acad. Sci. USA, 95, 9761–9766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaal T., Ross,W., Blatter,E., Tang,H., Jia,X., Krishnan,V., Assa-Munt,N., Ebright,R. and Gourse,R. (1996) DNA-binding determinants of the α subunit of RNA polymerase: novel DNA-binding domain architecture. Genes Dev., 10, 16–26. [DOI] [PubMed] [Google Scholar]
- Hochschild A. and Dove,S.L. (1998) Protein–protein contacts that activate and repress prokaryotic transcription. Cell, 92, 597–600. [DOI] [PubMed] [Google Scholar]
- Igarashi K. and Ishihama,A. (1991) Bipartite functional map of the E.coli RNA polymerase α subunit: involvement of the C-terminal region in transcription activation by cAMP–CRP. Cell, 65, 1015–1022. [DOI] [PubMed] [Google Scholar]
- Ishihama A. (1993) Protein–protein communication within the transcription apparatus. J. Bacteriol., 175, 2483–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon Y.H., Yamazaki,T., Otomo,T., Ishihama,A. and Kyogoku,Y. (1997) Flexible linker in the RNA polymerase α subunit facilitates the independent motion of the C-terminal activator contact domain. J. Mol. Biol., 267, 953–962. [DOI] [PubMed] [Google Scholar]
- Kristensen H.H., Valentin-Hansen,P. and Søgaard-Andersen,L. (1997) Design of CytR regulated, cAMP–CRP dependent class II promoters in Escherichia coli: RNA polymerase–promoter interactions modulate the efficiency of CytR repression. J. Mol. Biol., 266, 866–876. [DOI] [PubMed] [Google Scholar]
- Landini P. and Volkert,M.R. (1995) RNA polymerase α subunit binding site in positively controlled promoters: a new model for RNA polymerase–promoter interaction and transcriptional activation in the Escherichia coli ada and aidB genes. EMBO J., 14, 4329–4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd G., Busby,S. and Savery,N. (1998) Spacing requirements for interactions between the C-terminal domain of the α subunit of Escherichia coli RNA polymerase and the cAMP receptor protein. Biochem. J., 330, 413–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malan R., Kolb,A., Buc,H. and McClure,W. (1984) Mechanism of CRP-cAMP activation of the lac operon: transcription initiation activation of the P1 promoter. J. Mol. Biol., 180, 881–909. [DOI] [PubMed] [Google Scholar]
- Meibom K.L., Søgaard-Andersen,L., Mironov,A.S. and Valentin-Hansen,P. (1999) Dissection of a surface-exposed portion of the cAMP–CRP complex that mediates transcription activation and repression. Mol. Microbiol., 32, 497–504. [DOI] [PubMed] [Google Scholar]
- Meibom K.L., Kalliipolitis,B.H., Ebright,R. and Valentin-Hansen,P. (2000) Identification of the subunit of cAMP receptor protein (CRP) that functionally interacts with CytR in CRP–CytR-mediated transcriptional repression. J. Biol. Chem., 275, 11951–11957. [DOI] [PubMed] [Google Scholar]
- Meng W., Savery,N.J., Busby,S.J. and Thomas,M.S. (2000) The Escherichia coli RNA polymerase α subunit linker: length requirements for transcription activation at CRP-dependent promoters. EMBO J., 19, 1555–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møllegaard N.E., Rasmussen,P.B., Valentin-Hansen,P. and Nielsen,P.E. (1993) Characterization of promoter recognition complexes formed by CRP and CytR for repression and by CRP and RNA polymerase for activation of transcription on the Escherichia coli deoP2 promoter. J. Biol. Chem., 268, 17471–17477. [PubMed] [Google Scholar]
- Monsalve M., Mencia,M., Salas,M. and Rojo,F. (1996) Protein p4 represses phage φ29 A2c promoter by interacting with the α subunit of Bacillus subtilis RNA polymerase. Proc. Natl Acad. Sci. USA, 93, 8913–8918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami K., Fujita,N. and Ishihama,A. (1996) Transcription factor recognition surface on the RNA polymerase α subunit is involved in contact with the DNA enhancer element. EMBO J., 15, 4358–4367. [PMC free article] [PubMed] [Google Scholar]
- Murakami K., Owens,J., Belyaeva,T., Meares,C., Busby,S. and Ishihama,A. (1997) Positioning of two α subunit C-terminal domains of RNA polymerase at promoters by two transcription factors. Proc. Natl Acad. Sci. USA, 94, 11274–11278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negishi, T., Fujita,N. and Ishihama,A. (1995) Structural map of the α subunit of Escherichia coli RNA polymerase: structural domains identified by proteolytic cleavage. J. Mol. Biol., 248, 723–728. [DOI] [PubMed] [Google Scholar]
- Niu W., Zhou,Y., Dong,Q., Ebright,Y. and Ebright,R. (1994) Characterization of the activating region of Escherichia coli catabolite gene activator protein (CAP). Saturation and alanine-scanning mutagenesis. J. Mol. Biol., 243, 595–602. [DOI] [PubMed] [Google Scholar]
- Niu W., Kim,Y., Tau,G., Heyduk,T. and Ebright,R. (1996) Transcription activation at class II CAP-dependent promoters: two interactions between CAP and RNA polymerase. Cell, 87, 1123–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noel R. and Reznikoff,W. (1998) CAP, the –45 region and RNA polymerase: three partners in transcription initiation at lacP1 in Escherichia coli. J. Mol. Biol., 282, 495–504. [DOI] [PubMed] [Google Scholar]
- Ozoline O.N., Fujita,N. and Ishihama,A. (2000) Transcription activation mediated by the C-terminal domain of the RNA polymerase α-subunit. Multipoint monitoring using a fluorescent probe. J. Biol. Chem., 275, 1119–1127. [DOI] [PubMed] [Google Scholar]
- Ptashne M. and Gann,A. (1997) Transcriptional activation by recruitment. Nature, 386, 569–577. [DOI] [PubMed] [Google Scholar]
- Rao L., Ross,W., Appleman,J., Gaal,T., Leirom,S. and Schhlax,P. (1994) Factor independent activation of rrnB P1. An ‘extended’ promoter with an upstream element that dramatically increases promoter strength. J. Mol. Biol., 235, 1421–1435. [DOI] [PubMed] [Google Scholar]
- Rhodius V., West,D., Webster,C., Busby,S. and Savery,N. (1997) Transcription activation at class II CRP-dependent promoters: the role of different activating regions. Nucleic Acids Res., 25, 326–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross W., Gosink,K., Salomon,J., Igarashi,K., Zou,C., Ishihama,A., Severinov,K. and Gourse,R. (1993) A third recognition element in bacterial promoters: DNA binding by the α subunit of RNA polymerase. Science, 262, 1407–1413. [DOI] [PubMed] [Google Scholar]
- Ross W., Aiyar,S.E., Salomon,J. and Gourse,R.L. (1998) Escherichia coli promoters with UP elements of different strengths: modular structure of bacterial promoters. J. Bacteriol., 180, 5375–5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostock N., Park,S. and Choy,H.E. (2000) Reiterative transcription initiation from galP2 promoter of Escherichia coli. Biochim. Biophys. Acta, 1491, 185–195. [DOI] [PubMed] [Google Scholar]
- Savery N., Lloyd,G., Kainz,M., Gaal,T., Ross,W., Ebright,R., Gourse,R. and Busby,S. (1998) Transcription activation at Class II CRP-dependent promoters: identification of determinants in the C-terminal domain of the RNA polymerase α subunit. EMBO J., 17, 3439–3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savery N., Rhodius,V., Wing,H. and Busby,S. (1995) Transcription activation at Escherichia coli promoters dependent on the cyclic AMP receptor protein: effects of binding sequences for the RNA polymerase α-subunit. Biochem. J., 309, 77–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Søgaard-Andersen L. and Valentin-Hansen,P. (1993) Protein–protein interactions in gene regulation: the cAMP–CRP complex sets the specificity of a second DNA-binding protein, the CytR repressor. Cell, 75, 557–566. [DOI] [PubMed] [Google Scholar]
- Søgaard-Andersen L., Martinussen,J., Mollegaard,N.E., Douthwaithe,S.R. and Valentin-Hansen,P. (1990a) The CytR repressor antagonizes cyclic AMP–cyclic AMP receptor protein activation of the deoCp2 promoter of Escherichia coli K-12. J. Bacteriol., 172, 5706–5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Søgaard-Andersen L., Møllegaard,N.E., Douthwaite,S.R. and Valentin-Hansen,P. (1990b) Tandem DNA-bound cAMP–CRP complexes are required for transcriptional repression of the deoP2 promoter by the CytR repressor in Escherichia coli. Mol. Microbiol., 4, 1595–1601. [PubMed] [Google Scholar]
- Søgaard-Andersen L., Mironov,A., Pedersen,H., Sukhodelets,V. and Valentin-Hansen,P. (1991a) Single amino acid substitutions in the cAMP receptor protein specifically abolish regulation by the CytR repressor in Escherichia coli. Proc. Natl Acad. Sci. USA, 88, 4921–4925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Søgaard-Andersen L., Pedersen,H., Holst,B. and Valentin-Hansen,P. (1991b) A novel function of the cAMP–CRP complex in Escherichia coli: cAMP–CRP functions as an adaptor for the CytR repressor in the deo operon. Mol. Microbiol., 5, 969–975. [DOI] [PubMed] [Google Scholar]
- Tagami H. and Aiba,H. (1999) An inactive open complex mediated by an UP element at Escherichia coli promoters. Proc. Natl Acad. Sci. USA, 96, 7202–7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Severinov,K., Goldfarb,A., Fenyo,D., Chait,B. and Ebright,R. (1994) Location, structure and function of the target of a transcriptional activator protein. Genes Dev., 8, 3058–3067. [DOI] [PubMed] [Google Scholar]
- Valentin-Hansen P., Søgaard-Andersen,L. and Pedersen,H. (1996) A flexible partnership: the CytR anti-activator and the cAMP–CRP activator protein, comrades in transcription control. Mol. Microbiol., 20, 461–466. [DOI] [PubMed] [Google Scholar]
- West D., Williams,R., Rhodius,V., Bell,A., Sharma,N., Zou,C., Fukita,N., Ishihama,A. and Busby,S. (1993) Interactions between the Escherichia coli cyclic AMP receptor protein and RNA polymerase at class II promoters. Mol. Microbiol., 10, 789–797. [DOI] [PubMed] [Google Scholar]
- Williams R., Bell,A., Sima,G. and Busby,S. (1991) The role of two surface exposed loops in transcription activation by the Escherichia coli CRP and FNR proteins. Nucleic Acids Res., 19, 6705–6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams R., Rhodius,V., Bell,A., Kolb,A. and Busby,S. (1996) Orientation of functional activating regions in the E.coli CRP protein during transcription activation at class II promoters. Nucleic Acids Res., 24, 1112–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Pendergrast,P., Bell,A., Williams,R., Busby,S. and Ebright,R. (1994) The functional subunit of a dimeric transcription activator protein depends on promoter architecture. EMBO J., 13, 4549–4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y., Zang,X. and Ebright,R. (1993a) Identification of the activating region of catabolite gene activator protein (CAP): isolation and characterization of mutants of CAP specifically defective in transcription activation. Proc. Natl Acad. Sci. USA, 90, 6081–6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y., Busby,S. and Ebright,R. (1993b) Identification of the functional subunit of a dimeric transcription activator protein by use of ‘oriented heterodimers’. Cell, 73, 335–370. [DOI] [PubMed] [Google Scholar]