Abstract
We have used hydroxyl radicals generated by decomposition of peroxynitrous acid to study Mg2+-dependent structure and folding of the Varkud satellite (VS) ribozyme. Protection from radical cleavage shows the existence of a solvent-inaccessible core, which includes nucleotides near two three-helix junctions, the kissing interaction between stem–loops I and V and other nucleotides, most of which have also been implicated as important for folding or activity. Kinetic folding experiments showed that the ribozyme folds very quickly, with the observed protections completely formed within 2 s of addition of MgCl2. In mutants that disrupt the kissing interaction or entirely remove stem–loop I, which contains the cleavage site, nucleotides in the three-helix junctions and a subset of those elsewhere remain protected. Unlike smaller ribozymes, the VS ribozyme retains a significant amount of structure in the absence of its substrate. Protections that depend on proper interaction between the substrate and the rest ribozyme map to a region previously proposed as the active site of the ribozyme and along both sides of helix II, identifying candidate sites of docking for the substrate helix.
Keywords: hydroxyl radical footprinting/RNA folding/RNA structure/T7 RNA polymerase/VS RNA
Introduction
Many essential biochemical functions are carried out by RNA molecules. In order to fulfill their biological roles, these molecules must adopt specific, and often complicated, three-dimensional structures. Understanding the tertiary structures that a given sequence of RNA can assume, and the pathways it may take to achieve a structure, is essential for a complete understanding of the way in which RNA-mediated biological processes occur.
Structure and folding studies on several large RNAs have provided evidence for a hierarchical assembly of smaller RNA domains to form a large, modular RNA complex (Murphy and Cech, 1993; Loria and Pan, 1996; reviewed in Brion and Westhof, 1997 and Tinoco and Bustamante, 1999). Our laboratory and others have made use of the Neurospora Varkud satellite (VS) ribozyme as a model system for the study of RNA structure–function relationships and RNA folding. This small, naturally occurring ribozyme catalyzes a site-specific self-cleavage reaction to produce 5′-OH and 2′3′-cyclic phosphate termini (Saville and Collins, 1990). Despite performing a similar chemical reaction to the other small ribozymes, the VS ribozyme possesses a unique secondary structure (Beattie et al., 1995; Sigurdsson et al., 1998). Stem–loop I of the RNA (the substrate which contains the self-cleavage site) can be physically separated from the rest of the molecule, which then acts in trans on the substrate (Guo and Collins, 1995). Of particular interest is the fact that the substrate recognized by the trans-ribozyme is a base-paired stem–loop structure, suggesting that ribozyme– substrate recognition takes place primarily, if not exclusively, through tertiary interactions.
A variety of biochemical and mutagenesis experiments have identified some features of the tertiary structure of the VS ribozyme. These include a kissing interaction between nucleotides in loops I and V (Rastogi et al., 1996), folding of the three-way junction formed by helices II, III and VI (Lafontaine et al., 2001), and a conformational change in which helix Ib shifts from an inactive (Michiels et al., 2000; Flinders and Dieckmann, 2001) to an active (Andersen and Collins, 2001) structure. In addition to these known elements of tertiary structure, a number of other positions on nucleotide bases and backbone atoms become protected from chemical modification in the presence of Mg2+ (Beattie and Collins, 1997; Sood et al., 1998), suggesting that additional tertiary interactions remain to be identified.
Hydroxyl radical footprinting analysis is a powerful technique to probe the structure of RNAs in solution (Gotte et al., 1996). In this technique, hydroxyl radicals induce cleavage of the RNA backbone by oxidation of the ribose C4′ position at backbone positions, which are accessible to the bulk solvent (Balasubramanian et al., 1998). Tertiary structure, but not secondary structure, can bring regions of RNA backbones into sufficiently close proximity that some nucleotides have reduced accessibility to solvent. Such nucleotides show decreased susceptibility to cleavage by hydroxyl radicals, allowing backbone positions on the inside of the native structure to be identified (Latham and Cech, 1989). This technique has been used to probe the structures and folding pathways of several ribozymes (Pan, 1995; Rosenstein and Been, 1996; Hampel et al., 1998) as well as other RNAs (Joseph et al., 1997; Kieft et al., 1999). Protection data have been used to model three-dimensional structure and to identify independently folding domains within larger RNA structures (Murphy and Cech, 1993, 1994; Loria and Pan, 1996). More recently determined crystallographic structures have confirmed the accuracy of this technique in identifying positions of low C4′ solvent accessibility and in predicting the RNA three-dimensional structure (Cate et al., 1996; Ferré-D’Amaré et al., 1998; Rupert and Ferré-D’Amaré, 2001).
In this work, we have identified solvent-inaccessible regions in the folded Neurospora VS ribozyme. Many of the protections fall in regions of the RNA previously identified as structurally or functionally important and all protections form very rapidly. Using a mutational approach we have identified classes of protected nucleotides that are dependent on or independent of the kissing interaction and have identified potential points of contact between the substrate (helix I) and rest of the ribozyme.
Results
Using T7 RNA polymerase to obtain RNAs with precise 3′ ends for hydroxyl radical experiments
Interpretation of data from hydroxyl radical footprinting experiments, or other experiments in which single-base resolution of cleaved, end-labeled RNAs is required, depends on being able to assign each band on the gel to cleavage of the RNA at a unique position. This, in turn, depends on the RNAs being homogeneous at the labeled end. RNAs synthesized by in vitro transcription using T7 RNA polymerase are typically a mixed population consisting of the expected sequence plus a proportion of ‘N + 1’ length RNAs that contain one or more non-templated nucleotides at the 3′ end (Milligan et al., 1987) (and sometimes the 5′ end; Pleiss et al., 1998). For synthesis of short RNAs, chemical modification of the DNA template can minimize 3′ end heterogeneity (Moran et al., 1996; Kao et al., 1999); for longer RNAs, inclusion of a cis-cleaving ribozyme or a site for cleavage by a trans-acting ribozyme is currently the only effective way of producing homogeneous 3′ ends (Grosshans and Cech, 1991). Although these approaches have been successful for many RNAs, simpler solutions to the N + 1 problem are desirable.
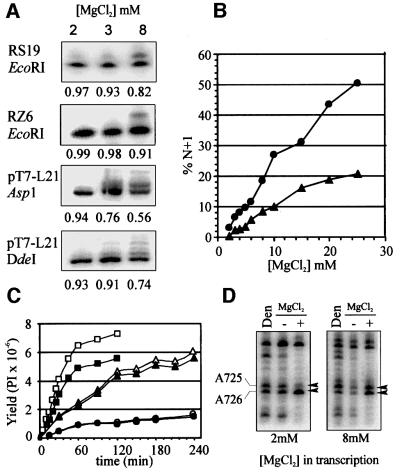
During our ongoing studies of the self-cleaving VS ribozyme, we had noticed a large variation in the proportion of N + 1 RNAs among different RNA preparations using different reagents, templates and synthesis conditions. We recently realized that the 3′ end-labeled RNAs that produced the highest quality chemical modification patterns were those that had been transcribed at reduced concentrations of MgCl2, a procedure that we have employed to minimize self-cleavage of VS precursor RNAs during transcription (Collins and Olive, 1993). We have now investigated this correlation systematically, and have found that there is indeed a substantial positive correlation between the concentration of MgCl2 in the transcription reaction mixture and the fraction of N + 1 length transcripts (Figure 1 and data not shown). This effect is independent of the template sequences examined and the restriction enzyme (blunt or 5′ overhang) used to linearize the template, and was observed with three independently purified preparations of polymerase (one of which was from a commercial supplier). RNAs that are almost free of N + 1 contaminants can be synthesized from many templates simply by lowering the concentration of MgCl2 during transcription. It should also be possible to adjust the effective Mg2+ concentration by increasing the concentration of nucleotide triphosphates (Milligan et al., 1987), although we have not directly tested this. The yield of RNA is decreased when synthesis is carried out for standard times at low MgCl2 concentrations; however, this is due primarily to a decrease in the rate of RNA synthesis, and the yield of N-length RNA can be improved by increasing the time of the transcription reaction (Figure 1C).
Fig. 1. Minimizing 3′ heterogeneity in T7 transcripts. (A) Plasmid templates (see Materials and methods) were digested with the indicated restriction enzymes and transcribed in mixtures containing 2, 3 or 8 mM MgCl2. Sample loading was adjusted to approximately equalize the amount of N-length RNA in each lane. The fraction of N-length RNA is indicated below each panel. (B) Quantification of N + 1 (and longer) RNAs as a percentage of total RNA in transcription reactions of RS19 (circles) or RZ6 (triangles) templates in reactions containing 2–25 mM MgCl2. (C) Time course of synthesis of RS19 RNA in various concentrations of MgCl2. The yields in PhosphorImager (PI) counts of total RNA (open symbols) and N-length RNA (filled symbols) from reactions containing 2, 3 and 8 mM MgCl2 are indicated by circles, triangles and squares, respectively. (D) DEPC structure probing of 3′ end-labeled G11 D RNA synthesized with 2 or 8 mM MgCl2. RNAs were partially modified with DEPC at 37°C in 200 mM HEPES pH 8.0, 50 mM KCl, ±20 mM MgCl2 or in denaturing (Den) conditions (200 mM HEPES pH 8.0, 1 mM EDTA, 90°C). Base numbers are as in Beattie et al. (1995).
The utility of this transcription protocol in interpreting data from chemical modification structure probing reactions is illustrated in Figure 1D for a region of VS RNA characterized previously (Beattie and Collins, 1997). With RNA transcribed in 2 mM MgCl2, it was easy to see that the reactivities of A725 and A726 were decreased and increased, respectively, in the presence of MgCl2. In contrast, with RNA transcribed in the presence of 8 mM MgCl2, the N + 1 band of A726, which co-migrates with A725, was so intense that accurate interpretation of the effects on A725 was impossible.
Hydroxyl radical footprinting of VS RNA
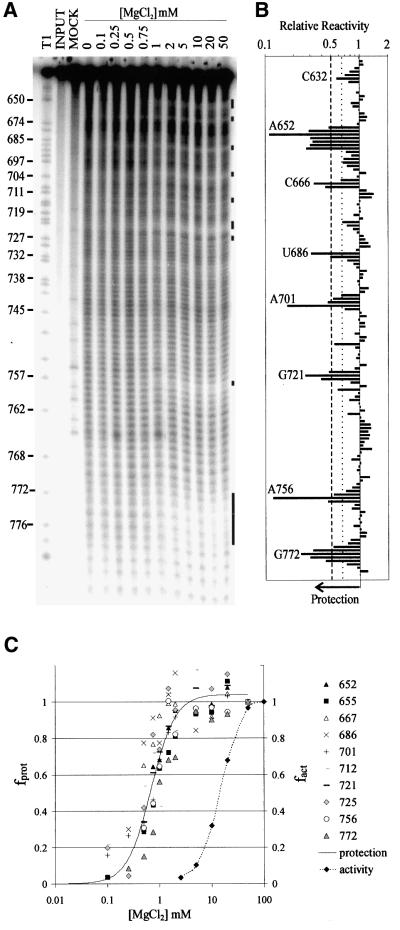
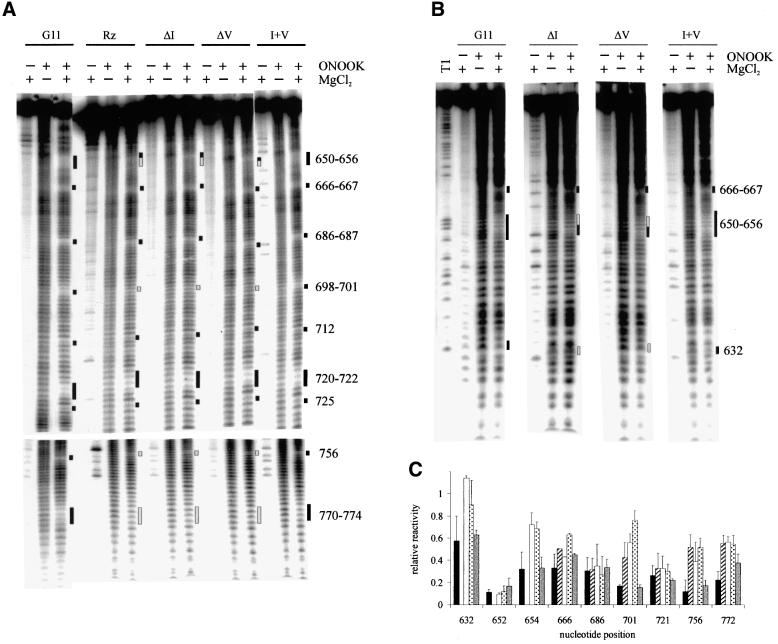
To determine whether the tertiary structure of the VS ribozyme contains regions that are inaccessible to solvent, we performed hydroxyl radical footprinting analysis on RNAs that had been pre-incubated in a range of magnesium concentrations (Figure 2A). We used the downstream cleavage product from the well characterized VS RNA deletion construct G11 (G11 D; see Figure 3) for these studies because it allowed us to look at folding of the RNA independently of the self-cleavage activity and without the mixture of precursor and product produced by self-cleavage in the higher concentrations of Mg2+. Also, G11 D differs from an active cis-cleaving construct by only one nucleotide, whose identity is unimportant, upstream of the site of self-cleavage (Guo and Collins, 1995). In the absence of Mg2+, a relatively, but not perfectly, even single-base ladder was observed, as expected if the RNA has little or no stable tertiary structure under these conditions. At increasing Mg2+ concentrations, regions of protection were observed around the two three-helix junctions (666–667, 686–687, 712, 720–722, 725), in the kissing interaction between loops I and V (632, 698–701), along both strands of helix II (650–656, 770–774) and in an internal loop in helix VI (755–757; identified by black bars to the right of Figure 2A; quantified in B; summarized in Figure 5). Most of the protected regions comprised only 2–4 nucleotides; however, up to seven nucleotides were protected along the 5′ side of helix II. The regions of strongest protection (∼8-fold) were at the bulged A652 in helix II and A756 in an internal loop in helix VI. These data provide the first physical evidence that a solvent-protected core exists in the VS ribozyme.
Fig. 2. Hydroxyl radical footprinting analysis of G11 D. (A) 3′-end-labeled G11 D RNA was incubated at 37°C in 50 mM sodium cacodylate buffer pH 6.5, 50 mM KCl, 0.1 mM EDTA and the indicated concentrations of MgCl2 followed by addition of ONOOK (see Materials and methods). RNA in the mock lane was not treated with ONOOK, but was otherwise identical to the sample lane that contained 20 mM MgCl2. An RNase T1 sequencing ladder is included to aid in assignment of nucleotides. The presence of a 5′ phosphate on the RNAs cleaved by hydroxyl radicals causes these RNAs to run slightly faster on the gel than T1 fragments of the same nucleotide length. Sites of protection from radical cleavage in the presence of MgCl2 are indicated by black bars. (B) The reactivity of each backbone position in the presence of 20 mM MgCl2 relative to the reactivity of that position in the absence of MgCl2 (relative reactivity) is plotted. Quantification of the data represents the average of 2–5 independent experiments using either 3′- or 5′-end-labeled RNA and appropriate electrophoresis times to resolve individual nucleotides [(A) and data not shown]. The variation between experiments was <20%. The most strongly protected nucleotide in each region is indicated by base number. Positions 634–639 were not quantified due to compression on the denaturing polyacrylamide gel. Positions within six nucleotides of the end of the RNA were obscured by the intense band from full-length RNA. Weak but significant protections (1.5- to 2-fold) lie between the dotted and dashed lines; strong protections (1.5- to 8.3-fold) lie to the left of the dashed line. (C) Fraction of maximal protection (fprot) for representative nucleotides in each of the protected regions identified in (B) (solid line) and the fraction of maximal self-cleavage activity (fact) of the precursor (dotted line) are plotted. For clarity a single line fit to all of the data is shown for the protection experiments. Fits of the data for individual nucleotides gave estimates of Mg2+1/2 ranging from 0.5 to 1.0 mM (mean = 0.8 ± 0.2).
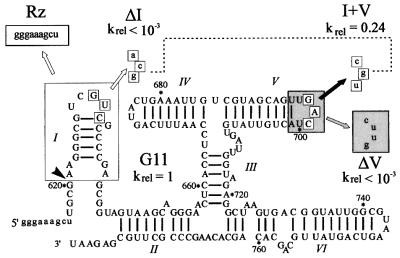
Fig. 3. Secondary structure of G11 Pre and mutant RNAs. VS nucleotides are indicated in upper case, vector or mutant nucleotides in lower case. The site of self cleavage is indicated by the arrowhead. The three-boxed nucleotides in loop I base pair with those in loop V to form a kissing interaction. Mutant ΔI disrupts the kissing interaction by substitution of the boxed nucleotides in loop I with those indicated by the unfilled arrow; mutant ΔV substitutes the nucleotides in the gray box in loop V with those indicated by the gray arrow. Mutant I + V restores the kissing interaction (dotted line) by combining the ΔI substitutions with compensatory substitutions in loop V indicated by the black arrow. Also shown is the structure of the trans-cleaving ribozyme (Rz), in which helix I (in the large unfilled box) and the nucleotides upstream of the cleavage site are replaced by the nine vector nucleotides in the small unfilled box labeled Rz (Guo and Collins, 1995). The self-cleavage rates of mutants ΔI, ΔV and I + V relative to G11 are indicated. G11 cleaves with kobs = 0.06 min–1, defined as krel = 1.
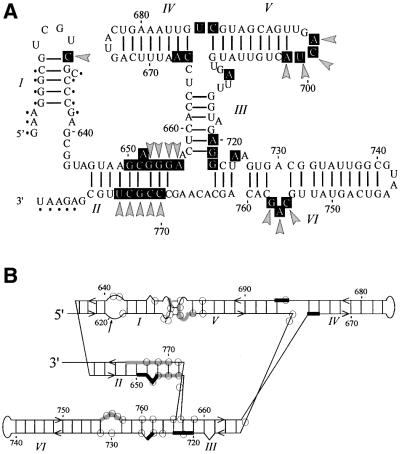
Fig. 5. Summary of protection patterns in VS RNA mapped onto secondary structure models. (A) Protected positions in G11 D (wild type) and I + V RNAs are indicated by black boxes on a standard secondary structure model. Protections that are absent or compromised in the mutant RNAs that disrupt the kissing interaction are highlighted by gray arrowheads. Positions whose reactivity was not determined because of limitations of gel resolution are indicated by small black dots. (B) Protections mapped onto an alternative representation of the secondary structure (Rastogi and Collins, 1998; Sood et al., 1998). Thick black and gray lines correspond to protections in wild-type RNA that are independent of, or dependent on, the kissing interaction, respectively. Positions whose nucleotide bases were previously identified as being functionally important based on base modification interference (Beattie and Collins, 1997) are identified by open circles.
The band intensity at each nucleotide position in the presence of each concentration of Mg2+ was quantified, normalized to the intensity in the absence of Mg2+, and fitted to a Hill equation (Hill and Chen, 1971; Hampel et al., 1998) to estimate the concentration of Mg2+ required to obtain half-maximal protection (Mg2+1/2). Figure 2C shows plots of fractional protection as a function of Mg2+ concentration for representative nucleotides from each protected region in the RNA. All of these nucleotides reached their half-maximally protected state in 0.8 ± 0.2 mM Mg2+, approximately the same value as determined from base modification protection experiments (Mg2+1/2 ≈ 1 mM; Maguire and Collins, 2001). Measure ment of the self-cleavage activity of the precursor RNA under the conditions of the hydroxyl radical protection experiment revealed that a higher concentration of Mg2+ is required for activity (Figure 2C; Mg2+1/2 = 17 mM) than for protection. The pattern of protected nucleotides that forms at low Mg2+ concentrations remains the same at Mg2+ concentrations high enough to support the maximal rate of self cleavage (Figure 2A, see also ‘Kinetic folding analysis’ below), indicating that the structure responsible for protection of specific nucleotides at low Mg2+ requires only subtle changes to adopt the active conformation.
Role of the loop I–loop V kissing interaction in determining the protection pattern
Previous experiments have shown that the kissing interaction between loops I and V is essential for self-cleavage activity and for the secondary structure rearrangement that takes place in helix Ib (Rastogi et al., 1996; Andersen and Collins, 2001). To investigate the role of the kissing interaction in the formation of the solvent-inaccessible regions of the folded RNA, we performed hydroxyl radical footprinting analysis on mutants in which the interaction had been disrupted by complete deletion of helix I or more subtly by base substitutions in loops I or V (Figure 4). It has been shown previously that deletion of helix I creates a trans-acting version of the ribozyme in which helices II–VI of VS RNA can cleave a substrate RNA comprised only of stem–loop I (Guo and Collins, 1995). The Mg2+-dependent protections in the trans-ribozyme (Rz) comprise a subset of those observed in the cis-ribozyme: specifically, the same nucleotides in the two three-helix junctions are protected, as are 650–652 on the 5′ side of helix II (summarized in Figure 5). These protections must be formed by tertiary interactions within the trans-ribozyme. The nucleotides that are protected in the cis-cleaving ribozyme but not in the trans-ribozyme (indicated by gray bars in Figure 4A and B and gray arrows and boxes in Figure 5A and B, respectively) are candidates for sites of interaction of helix I with the rest of the ribozyme (see Discussion).
Fig. 4. Hydroxyl radical protection patterns in mutant VS RNAs. Reactions were carried out in the presence or absence of ONOOK and/or 20 mM MgCl2 for the trans-cleaving ribozyme (Rz) and the downstream cleavage products of each of the other RNAs indicated at the top of the figure. Positions protected from cleavage as in wild-type G11 D RNA are indicated by black bars. Positions where the protections are compromised or absent in the mutants with a disrupted kissing-interaction are indicated by gray bars. RNAs were labeled at their 3′ (A) or 5′ (B) end and electrophoresed for times appropriate to resolve regions of protection. (C) Relative reactivity (y-axis) of representative protected positions (x-axis) in the presence of 20 mM MgCl2. The solid, striped, open, dotted and gray bars represent the reactivities of G11, Rz, ΔI, ΔV and I + V RNAs, respectively. The data are the mean of two to five repeats; error bars represent the standard deviations.
To specifically address the role of the kissing interaction in the formation of protections we created two additional mutants, ΔI and ΔV, in which the nucleotides in loop I or loop V, respectively, were mutated to disrupt the interaction, and a third mutant, I + V, which restored the base-pairing interaction with complementary nucleotide changes in both loops (Figure 3). The self-cleavage activities of these RNAs were assayed and the rate constants relative to G11 (wild type) are included in Figure 3. The disruption of the kissing interaction resulted in a drop in self-cleavage activity of at least 1000-fold, while restoring the base-pairing interaction rescued the activity to within 4-fold of wild type, confirming and extending previous mutational analyses that showed the importance of this kissing interaction for self-cleavage activity. The hydroxyl radical footprinting patterns of these RNAs in the absence and presence of Mg2+ are shown in Figure 4A and B and summarized on the secondary structure model in Figure 5. The reactivity of positions representative of strongly protected regions in G11 were quantified for each mutant and are plotted in Figure 4C. The protection patterns observed in the individual loop mutants in the presence of Mg2+ are the same as in the trans-acting ribozyme, while the protection pattern observed in the compensatory mutant is qualitatively the same as in the wild-type cis-ribozyme. The protections in the vicinity of the kissing interaction in both loops I (C632) and V (A698, C699, U700 and A701) are also absent in the loop mutants and rescued by the compensatory base-pairing interaction. Interestingly, maximal protection at A756 (in helix VI) and at several nucleotides in helix II (653–656, 770–774) is also dependent on the formation of a base-pairing interaction between loops I and V. Furthermore, position G772 is protected to an intermediate degree in the compensatory mutant, suggesting that the sequence of the individual bases involved in the kissing interaction affects the magnitude of this protection. We conclude that, in addition to a role in secondary structure rearrangement in helix Ib, the kissing interaction is important for complete folding of the RNA and formation of the solvent-protected core.
Kinetic folding analysis
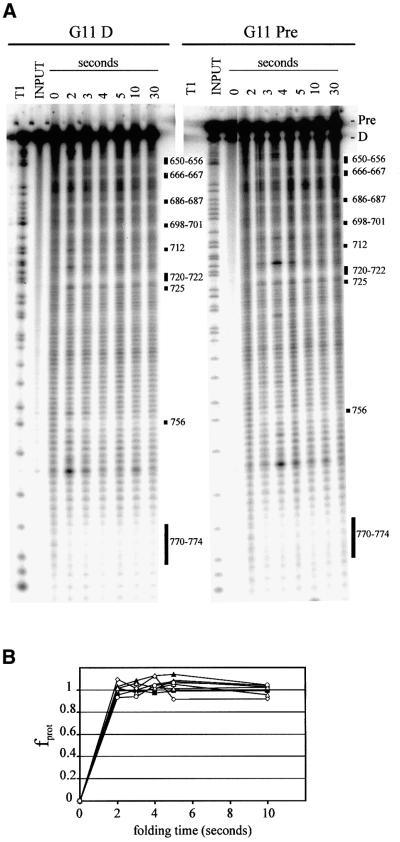
The short half-life of peroxynitrous acid at neutral pH (∼0.9 s at pH 7 or below; Beckman et al., 1990) makes this reagent a convenient source of hydroxyl radicals for monitoring the kinetics of RNA folding (Chaulk and MacMillan, 2000). To estimate the rate of folding of G11 D we monitored the protection pattern after incubation of RNA in the presence of 20 mM MgCl2 for various lengths of time. Figure 6A shows that all of the protections observed in equilibrium folding experiments (Figures 2 and 4A and B and time points up to 10 min, not shown in Figure 6A) were fully formed in 2 s, the shortest time point that could be acquired by manual pipetting (Figure 6B). Self-cleavage of the precursor RNA (G11 Pre) under these conditions is much slower (0.06 min–1; Figure 4C) than the folding of G11 D (estimated at >50 min–1 from Figure 6B), raising the possibility that D folds much faster than Pre, or that folding monitored by protection from hydroxyl radical cleavage is not the rate-limiting step in cleavage of Pre.
Fig. 6. Kinetic folding analysis. (A) Folding of 3′-end-labeled G11 D (left) or Pre (right) was initiated at time zero by the addition of MgCl2 to a final concentration of 20 mM. ONOOK was added to the RNA at the times indicated on the x-axis (and up to 300 s, data not shown) after the addition of MgCl2. Regions of protection are indicated by black bars. (B) The fraction of maximal protection versus time after addition of MgCl2 is plotted for nucleotides 686, 701, 721 and 756 using diamonds, squares, triangles and circles, respectively, for G11 Pre (filled symbols) and D (open symbols).
To distinguish these possibilities, we performed kinetic folding experiments on G11 Pre. Figure 6A and B shows that G11 Pre was also completely folded at the earliest time point, but no self-cleavage (above the 2% contaminating D present in the input RNA) was observed. This suggests that the formation of the observed protections does not reflect the rate-limiting step of the self-cleavage reaction. In the longer time points, self-cleavage was observed without any detectable change in the hydroxyl radical protection pattern, suggesting that no large-scale structural rearrangements are required to adopt the active conformation.
Discussion
We have examined the hydroxyl radical cleavage pattern of VS RNA in the presence of Mg2+ and identified several regions of the RNA backbone that become protected from radical cleavage upon tertiary folding. Protection from radical cleavage was observed in and near the two three-helix junctions, the kissing loop interaction between stem–loops I and V, and several other nucleotides in helices II and VI, most of which have also been implicated by other approaches as important for folding or activity (summarized in Figure 5). Approximately 20% of the nucleotides in the wild-type RNA (G11 D) became protected (>1.5-fold) from cleavage in the presence of Mg2+, and the extent of protection reached 8.5-fold in the most strongly protected regions. The solvent-protected core of the G11 D version of the VS ribozyme forms rapidly (estimated at >50 min–1), and at low concentrations of Mg2+ (Mg2+1/2 = 0.8 ± 0.2 mM), and does not change at higher Mg2+ concentrations or longer times. Taking advantage of the short half-life of peroxynitrous acid, we determined that the precursor RNA (G11 Pre) forms the same Mg2+-dependent protection pattern equally rapidly. In longer incubations, self cleavage was observed without any detectable change in the hydroxyl radical protection pattern, consistent with the idea that no large-scale, stable structural rearrangements are required to obtain the active conformation of the RNA.
Formation of the kissing interaction between loops I and V is important for self-cleavage (Rastogi et al., 1996), in part because this interaction is required for helix I to adopt the conformation required for activity (Andersen and Collins, 2000). More recent experiments have suggested that the kissing interaction also plays an additional role(s) in the cleavage reaction, which might include positioning or docking of helix I into the rest of the ribozyme (Andersen and Collins, 2001). The Mg2+-dependent protection of several nucleotides in loops I (632) and V (698–701) provide the first evidence that the kissing loops interact with the rest of the ribozyme: the formation of base pairs alone is not sufficient to provide protection from hydroxyl radical cleavage (Latham and Cech, 1989) and it is unlikely that the interaction of two small loops would form a local structure of adequate complexity to achieve solvent inaccessibility of the sugars (Chang and Tinoco, 1997; Kim and Tinoco, 2000). Disrupting the kissing interaction by mutations in either loop I or V eliminates or substantially decreases the degree of protection of the loop I and V nucleotides; restoration of the kissing interaction with different base pairs restores protection. These observations suggest that the kissing interaction itself, not the identities of particular base pairs nor the individual loops I or V, is the structural feature that is most effectively recognized by the rest of the ribozyme.
The nucleotides in and around the three-helix junctions are also protected in the mutants that disrupt the kissing interaction between loops I and V, indicating that these junctions fold independently of interactions with the substrate (helix I). Using FRET experiments Lilley and co-workers have shown that the II–III–VI junction folds independently of any other ribozyme sequences and that the correct folding of this junction is important for self-cleavage activity (Lafontaine et al., 2001). The hydroxyl radical protection experiments also showed that the III–IV–V junction became protected in the absence of the kissing interaction or helix I, indicating that both junctions fold independently of association with the substrate.
Adjacent to the II–III–VI junction, nucleotides 650–652 in helix II are also well protected in all constructs tested, indicating that the 5′ side of helix II interacts with the other regions of the ribozyme even in the absence of the I–V kissing interaction. Additional protections on the 5′ side of helix II (653–656) and on the 3′ side (770–774) were observed only in the presence of the I–V kissing interaction, suggesting that the kissing interaction itself or other parts of helix I interact with helix II. The alternative explanation that helix I interacts elsewhere, and induces these protections allosterically, is less likely because there are no helix I-dependent protections in the trans-ribozyme other than those in loop V, which are involved in the kissing interaction, and those in helices II and VI, which have been identified as potential sites of interaction between helix I and the trans-cleaving ribozyme (identified in gray in Figure 5), that would indicate such a hypothetical alternative site of interaction. These data provide the first physical evidence that helix II is in the core of the ribozyme.
The internal loop in helix VI that contains bases 755–757 has been previously suggested as a candidate for the active site of the ribozyme based on deletion analysis, modification interference and protection experiments and molecular modeling (Beattie and Collins, 1997; Rastogi and Collins, 1998; Sood et al., 1998). In this model, the cleavage site would dock into the internal loop, inducing a sufficiently compact structure to cause protection from hydroxyl radicals. Consistent with this hypothesis, the protections in this region (755–757) are severely compromised, although not absent, in the trans-ribozyme which lacks the cleavage site and in mutants with a disrupted kissing interaction between loops I and V (see Figure 4A–C). The observation that these protections are not completely absent in the absence of helix I suggests that a combination of interactions within the trans-cleaving ribozyme, and between helix I and the trans-cleaving ribozyme, are responsible for complete protection of the core from solvent. The extent of protection at position 756 is much greater in the presence of the substrate (8.5-fold) than in the trans-cleaving ribozyme alone (1.5-fold), indicating that contributions from interactions between helix I and the trans-cleaving ribozyme are far more significant than contributions from interactions within the trans-cleaving ribozyme in stabilizing the fully folded state of the wild-type cis-cleaving RNA. The protections in the internal loop in helix VI (755–757) are restored in the I + V mutant, which restores a kissing interaction between loops I and V (see Figure 4A–C), suggesting that in addition to being required to shift helix Ib into the active conformation (Andersen and Collins, 2001), the kissing interaction between loops I and V is necessary for correct interaction between helix I and the rest of the ribozyme.
Kinetic folding experiments revealed that both the downstream cleavage product and the precursor RNA fold very rapidly. The slowest folding step monitored by this technique occurs with a significantly faster rate constant than the self-cleavage reaction of the precursor molecule. Differences between the rate constants for cleavage and for folding measured by protection from hydroxyl radical cleavage have also been observed for the Tetrahymena intron (kcleavage = 0.03 s–1, kfolding = 0.07 s–1; Sclavi et al., 1998; Chaulk and MacMillan, 2000) RNase P RNA (kcleavage = 0.009 s–1, kfolding = 0.02 s–1; Kent et al., 2000) and the hairpin ribozyme (kcleavage = 0.5 min–1, kfolding = 2 min–1; Walter et al., 1998; Hampel and Burke, 2001). It has been speculated that one or more slow local rearrangements occur following the formation of the globally folded structure which are rate-limiting for catalysis. It is likely that a similarly subtle structural rearrangement is required to obtain the active conformation of the VS ribozyme.
Hydroxyl radical cleavage experiments have been used extensively to study RNA folding and to aid in modeling three-dimensional structure. The accuracy of the technique in determining areas of solvent inaccessibility has been confirmed for the cases in which both models based on biochemical data (including protection from radical cleavage) and X-ray crystal structures exist (Cate et al., 1996; Ferré-D’Amaré et al., 1998; Rupert and Ferré-D’Amaré, 2001). Recently, the solvent-inaccessible core of the Group II intron was shown to include all of the known tertiary interactions, and to exclude a known peripheral folding domain (Swisher et al., 2001). These types of RNA structure studies have revealed that both long-range helical packing as well as local junction folding can cause nucleotides to become protected from hydroxyl radical cleavage (Kieft et al., 1999). The protection pattern observed in VS RNA suggests that both of these types of interactions occur in VS: longer stretches of protections have been observed, particularly along helix II and in both loops involved in the kissing interaction, as well as protections in a number of other regions, primarily around the three-helix junctions.
When the protected regions are superimposed on models of the secondary structure of the VS ribozyme, it is clear that the physical core deduced from hydroxyl radical protection (black boxes in Figure 5A; black and gray bars in Figure 5B) overlaps almost completely with the functional core of the ribozyme deduced from previous modification interference experiments (Beattie and Collins, 1997; open circles in Figure 5B) and site-directed mutagenesis (Rastogi and Collins, 1998; V.D.Sood, S.L.Hiley and R.A.Collins, unpublished data). We have combined all of this information to produce a model of the tertiary structure of VS RNA (Figure 7). In the absence of the I–V kissing interaction the tertiary structure of helices II–VI is sufficient to induce protection at the three-helix junctions and along part of the 5′ side of helix II. The kissing interaction is required for docking of helix I into the rest of the ribozyme, which causes extended protection along both sides of helix II, the internal loop in helix VI and in the kissing interaction itself. We conclude that the structure monitored by hydroxyl radical protection approximates the functionally relevant conformation of the VS ribozyme and that these data will be useful in evaluating the functional relevance of future atomic resolution structures of the VS ribozyme.
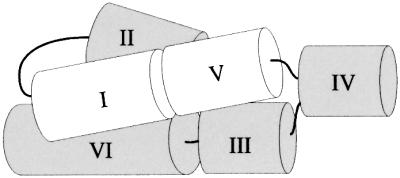
Fig. 7. Three-dimensional model of G11 Pre. A model of the tertiary structure consistent with hydroxyl radical protection and other biochemical data is shown (see text for details). Helices I–VI are represented as cylinders and their relative orientations are shown. Thin black lines represent covalent connections between helices; the connection between helices II and III is not visible in this orientation.
Materials and methods
Minimizing 3′ end heterogeneity in T7 transcripts
Clones RS19 and RZ6 are circular permutations of G11 (Andersen and Collins, 2000; R.Zamel and R.A.Collins, unpublished data) that differ slightly in sequence from each other. Plasmid DNAs were linearized with the indicated restriction enzymes according to standard procedures. Transcription reactions were carried out at 37°C for 1 h (except where specific times are specified in Figure 1C) in a 20 µl volume containing 1 µg of template DNA, 40 mM Tris–HCl pH 8.0, 25 mM NaCl, 2 mM spermidine, 1 mM each NTP, 4 mM dithiothreitol (DTT) and 2–20 mM µCi of [α-32P]GTP (Amersham; 3000 Ci/mmol), 0.3 µg of T7 polymerase at MgCl2 concentrations as indicated in the figures. All of the transcription reactions were repeated three to five times and consistent results were observed. For chemical modification structure probing with diethylpyrocarbonate (DEPC), non-radioactive RNA was transcribed for 2 h in 100 µl reactions containing proportionately more template and polymerase.
Synthesis of RNAs by in vitro transcription
Clones G11, ΔV and the trans-cleaving ribozyme (Rz; alternately referred to as clone A3 or the Ava ribozyme) have been described previously (Guo et al., 1993; Guo and Collins, 1995; Rastogi and Collins, 1998). ΔI and I + V were constructed by PCR mutagenesis in which the 5′ primer introduced the desired base substitutions into loop I. Plasmid DNAs were purified and RNAs were transcribed under standard in vitro conditions (Milligan and Uhlenbeck, 1989) in 200 µl reactions containing ∼10 µg of linearized DNA, 3 µg of T7 RNA polymerase, 40 mM Tris–HCl pH 8.0, 25 mM NaCl and 2 mM spermidine. The reactions contained only 2.5 mM MgCl2 to minimize 3′ end heterogeneity of the transcripts (see Results). The transcription reactions were incubated at 37°C for at least 3 h in order to maximize the yield of RNA with the low concentration of MgCl2. The RNAs were cleaved (ΔI and ΔV in trans by a ribozyme with complementary sequences in loop V and I, respectively, to restore a kissing interaction and permit cleavage to occur) as previously described (Collins and Olive, 1993; Guo and Collins, 1995). The RNAs were purified from a 4% polyacrylamide/8.3 M urea gel, visualized by UV shadowing, excised and eluted into 2 ml of DEPC-treated water at 65°C for 1 h. Eluted RNAs were filtered through 0.8 µm/0.2 µm acrodiscs (Gelman Sciences) and ethanol precipitated. RNAs were labeled at the 3′ end with 5′-[32P]pCp, or the 5′ end with [γ-32P]ATP, gel-purified a second time and redissolved in Milli-Q water.
Equilibrium hydroxyl radical footprinting reactions
Reactions were carried out in volumes of 20 µl containing 50–150 ng (125 000 c.p.m.) end-labeled RNA, 50 mM sodium cacodylate buffer (adjusted to pH 6.5 with HCl), 50 mM KCl, 0.1 mM EDTA and the concentration of MgCl2 indicated in the figures. For the analysis of the mutant VS RNA constructs, native reactions contained 20 mM MgCl2. Reactions were allowed to equilibrate at 37°C for 2–5 min after which time the entire reaction was transferred to another microfuge tube containing 1 µl of 100–125 mM potassium peroxynitrite (ONOOK; generously provided by S.Chaulk and A.MacMillan). The reactions were mixed by vigorous pipetting for 20 s, and ethanol precipitated. The RNAs were subjected to denaturing polyacrylamide electrophoresis and visualized by exposure to a PhosphorImager screen (Molecular Dynamics, Sunnyvale, CA). Sites of cleavage were identified by comparing electrophoretic mobilities of hydroxyl radical cleavage products with those of a partial T1 nuclease digest of the same RNA, accounting for the faster mobility of the hydroxyl radical products because of the presence of a 5′ phosphate (Gotte et al., 1996). Data were quantified using TotalLab v.1.11 (Nonlinear Dynamics, Newcastle upon Tyne, UK) and corrected for lane loading by normalizing to the reactivity of the average of several bands whose intensity did not change in the presence of Mg2+. The reactivity of each nucleotide at 20 mM Mg2+ divided by its reactivity in the absence of Mg2+ is plotted. Positions that displayed a decrease in reactivity of ≥1.5-fold were defined as protected. Mg2+ titration and activity data were fitted to a Hill equation f = fmax ([Mg2+]n/[Mg2+]n + Kdn), where f = fraction of maximal protection at a given [Mg2+]. The Mg2+1/2 value was obtained by calculating the [Mg2+] when f = fmax/2. Prior to performing reactions with labeled RNA, mock reactions with an equivalent amount of tRNA were carried out and the pH was measured to ensure that the buffer was able to maintain neutral pH upon addition of the highly basic ONOOK. To estimate the pseudo first-order rate constants for the self-cleavage reactions, precursor RNAs (internally labeled with [α-32P]GTP) were incubated under identical conditions to the footprinting reactions but in a volume of 60 µl. The self-cleavage reaction was initiated with the addition of MgCl2. Aliquots were removed into 2 vol. of formamide loading dye at appropriate time points to stop the reaction. The products were analyzed by denaturing gel electrophoresis, quantified using ImageQuant 3.3 and the data fitted to f = fmax – ae(–kt ) where fmax is the total fraction of RNA cleaved at t = ∞ and a is the fraction of RNA-cleaving at rate constant k.
Kinetic hydroxyl radical footprinting reactions
RNAs were allowed to equilibrate in 50 mM sodium cacodylate buffer pH 6.5, 50 mM KCl and 0.1 mM EDTA for 2 min at 37°C as described above. The reaction was transferred to another microfuge tube containing 0.1 vol. of 10× MgCl2 and mixed for discrete amounts of time before being transferred to a third microfuge tube containing the ONOOK. This method allowed for optimal mixing of the RNA with MgCl2 and the complete reaction with ONOOK in the short reaction time of these experiments. RNAs were analyzed as described above.
Acknowledgments
Acknowledgements
We are grateful to Dr A.MacMillan and S.Chaulk for providing us with potassium peroxynitrite, and to members of the Collins laboratory for helpful discussions. This work was supported by a grant from the Canadian Institutes of Health Research. S.L.H. was supported in part by an NSERC PGS B award. R.A.C. is a fellow of the Canadian Institute for Advanced Research Program in Evolutionary Biology and a Canada Research Chair.
References
- Andersen A.A. and Collins,R.A. (2000) Rearrangement of a stable RNA secondary structure during VS ribozyme catalysis. Mol. Cell, 5, 469–478. [DOI] [PubMed] [Google Scholar]
- Andersen A.A. and Collins,R.A. (2001) Intramolecular secondary structure rearrangement by the kissing interaction in the Neurospora VS ribozyme. Proc. Natl Acad. Sci. USA, 98, 7730–7735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian B., Pogozelski,W.K. and Tullius,T.D. (1998) DNA strand breaking by the hydroxyl radical is governed by the accessible surface areas of the hydrogen atoms of the DNA backbone. Proc. Natl Acad. Sci. USA, 95, 9738–9743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beattie T.L. and Collins,R.A. (1997) Identification of functional domains in the self-cleaving Neurospora VS ribozyme using damage selection. J. Mol. Biol., 267, 830–840. [DOI] [PubMed] [Google Scholar]
- Beattie T.L., Olive,J.E. and Collins,R.A. (1995) A secondary-structure model for the self-cleaving region of Neurospora VS RNA. Proc. Natl Acad. Sci. USA, 92, 4686–4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckman J.S., Beckman,T.W., Chen,J., Marshall,P.A. and Freeman,B.A. (1990) Apparent hydroxyl radical production by peroxynitrite: implications for endothelial injury from nitric oxide and superoxide. Proc. Natl Acad. Sci. USA, 87, 1620–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brion P. and Westhof,E. (1997) Hierarchy and dynamics of RNA folding. Annu. Rev. Biophys. Biomol. Struct., 26, 113–137. [DOI] [PubMed] [Google Scholar]
- Cate J.H., Gooding,A.R., Podell,E., Zhou,K., Golden,B.L., Kundrot,C.E., Cech,T.R. and Doudna,J.A. (1996) Crystal structure of a group I ribozyme domain: principles of RNA packing. Science, 273, 1678–1685. [DOI] [PubMed] [Google Scholar]
- Chang K.Y. and Tinoco,I.,Jr (1997) The structure of an RNA ‘kissing’ hairpin complex of the HIV TAR hairpin loop and its complement. J. Mol. Biol., 269, 52–66. [DOI] [PubMed] [Google Scholar]
- Chaulk S.G. and MacMillan,A.M. (2000) Characterization of the Tetrahymena ribozyme folding pathway using the kinetic foot printing reagent peroxynitrous acid. Biochemistry, 39, 2–8. [DOI] [PubMed] [Google Scholar]
- Collins R.A. and Olive,J.E. (1993) Reaction conditions and kinetics of self-cleavage of a ribozyme derived from Neurospora VS RNA. Biochemistry, 32, 2795–2799. [DOI] [PubMed] [Google Scholar]
- Ferré-D’Amaré A.R., Zhou,K. and Doudna,J.A. (1998) Crystal structure of a hepatitis delta virus ribozyme. Nature, 395, 567–574. [DOI] [PubMed] [Google Scholar]
- Flinders J. and Dieckmann,T. (2001) A pH controlled conformational switch in the cleavage site of the VS ribozyme substrate RNA. J. Mol. Biol., 308, 665–679. [DOI] [PubMed] [Google Scholar]
- Gotte M., Marquet,R., Isel,C., Anderson,V.E., Keith,G., Gross,H.J., Ehresmann,C., Ehresmann,B. and Heumann,H. (1996) Probing the higher order structure of RNA with peroxonitrous acid. FEBS Lett., 390, 226–228. [DOI] [PubMed] [Google Scholar]
- Grosshans C.A. and Cech,T.R. (1991) A hammerhead ribozyme allows synthesis of a new form of the Tetrahymena ribozyme homogeneous in length with a 3′ end blocked for transesterification. Nucleic Acids Res., 19, 3875–3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H.C. and Collins,R.A. (1995) Efficient trans-cleavage of a stem–loop RNA substrate by a ribozyme derived from Neurospora VS RNA. EMBO J., 14, 368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H.C., De Abreu,D.M., Tillier,E.R., Saville,B.J., Olive,J.E. and Collins,R.A. (1993) Nucleotide sequence requirements for self-cleavage of Neurospora VS RNA. J. Mol. Biol., 232, 351–361. [DOI] [PubMed] [Google Scholar]
- Hampel K.J. and Burke,J.M. (2001) A conformational change in the ‘loop e-like’ motif of the hairpin ribozyme is coincidental with domain docking and is essential for catalysis. Biochemistry, 40, 3723–3729. [DOI] [PubMed] [Google Scholar]
- Hampel K.J., Walter,N.G. and Burke,J.M. (1998) The solvent-protected core of the hairpin ribozyme-substrate complex. Biochemistry, 37, 14672–14682. [DOI] [PubMed] [Google Scholar]
- Hill T.L. and Chen,Y.D. (1971) Cooperative effects in models of steady-state transport across membranes. IV. One-site, two-site and multisite models. Biophys. J., 11, 685–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph S., Weiser,B. and Noller,H.F. (1997) Mapping the inside of the ribosome with an RNA helical ruler. Science, 278, 1093–1098. [DOI] [PubMed] [Google Scholar]
- Kao C., Zheng,M. and Rudisser,S. (1999) A simple and efficient method to reduce nontemplated nucleotide addition at the 3′ terminus of RNAs transcribed by T7 RNA polymerase. RNA, 5, 1268–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent O., Chaulk,S.G. and MacMillan,A.M. (2000) Kinetic analysis of the M1 RNA folding pathway. J. Mol. Biol., 304, 699–705. [DOI] [PubMed] [Google Scholar]
- Kieft J.S., Zhou,K., Jubin,R., Murray,M.G., Lau,J.Y. and Doudna,J.A. (1999) The hepatitis C virus internal ribosome entry site adopts an ion-dependent tertiary fold. J. Mol. Biol., 292, 513–529. [DOI] [PubMed] [Google Scholar]
- Kim C.H. and Tinoco,I.,Jr (2000) A retroviral RNA kissing complex containing only two G.C base pairs. Proc. Natl Acad. Sci. USA, 97, 9396–9401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine D.A., Norman,D.G. and Lilley,D.M. (2001) Structure, folding and activity of the VS ribozyme: importance of the 2-3-6 helical junction. EMBO J., 20, 1415–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latham J.A. and Cech,T.R. (1989) Defining the inside and outside of a catalytic RNA molecule. Science, 245, 276–282. [DOI] [PubMed] [Google Scholar]
- Loria A. and Pan,T. (1996) Domain structure of the ribozyme from eubacterial ribonuclease P. RNA, 2, 551–563. [PMC free article] [PubMed] [Google Scholar]
- Maguire J.L. and Collins,R.A. (2001) Effects of cobalt hexammine on folding and self-cleavage of the Neurospora VS ribozyme. J. Mol. Biol., 309, 45–56. [DOI] [PubMed] [Google Scholar]
- Michiels P.J., Schouten,C.H., Hilbers,C.W. and Heus,H.A. (2000) Structure of the ribozyme substrate hairpin of Neurospora VS RNA: a close look at the cleavage site. RNA, 6, 1821–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan J.F. and Uhlenbeck,O.C. (1989) Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol., 180, 51–62. [DOI] [PubMed] [Google Scholar]
- Milligan J.F., Groebe,D.R., Witherell,G.W. and Uhlenbeck,O.C. (1987) Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res., 15, 8783–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran S., Ren,R.X., Sheils,C.J., Rumney,S.T. and Kool,E.T. (1996) Non-hydrogen bonding ‘terminator’ nucleosides increase the 3′-end homogeneity of enzymatic RNA and DNA synthesis. Nucleic Acids Res., 24, 2044–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy F.L. and Cech,T.R. (1993) An independently folding domain of RNA tertiary structure within the Tetrahymena ribozyme. Biochemistry, 32, 5291–5300. [DOI] [PubMed] [Google Scholar]
- Murphy F.L. and Cech,T.R. (1994) GAAA tetraloop and conserved bulge stabilize tertiary structure of a group I intron domain. J. Mol. Biol., 236, 49–63. [DOI] [PubMed] [Google Scholar]
- Pan T. (1995) Higher order folding and domain analysis of the ribozyme from Bacillus subtilis ribonuclease P. Biochemistry, 34, 902–909. [DOI] [PubMed] [Google Scholar]
- Pleiss J.A., Derrick,M.L. and Uhlenbeck,O.C. (1998) T7 RNA poly merase produces 5′ end heterogeneity during in vitro transcription from certain templates. RNA, 4, 1313–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastogi T. and Collins,R.A. (1998) Smaller, faster ribozymes reveal the catalytic core of Neurospora VS RNA. J. Mol. Biol., 277, 215–224. [DOI] [PubMed] [Google Scholar]
- Rastogi T., Beattie,T.L., Olive,J.E. and Collins,R.A. (1996) A long-range pseudoknot is required for activity of the Neurospora VS ribozyme. EMBO J., 15, 2820–2825. [PMC free article] [PubMed] [Google Scholar]
- Rosenstein S.P. and Been,M.D. (1996) Hepatitis delta virus ribozymes fold to generate a solvent-inaccessible core with essential nucleotides near the cleavage site phosphate. Biochemistry, 35, 11403–11413. [DOI] [PubMed] [Google Scholar]
- Rupert P.B. and Ferré-D’Amaré,A.R. (2001) Crystal structure of a hairpin ribozyme-inhibitor complex with implications for catalysis. Nature, 410, 780–786. [DOI] [PubMed] [Google Scholar]
- Saville B.J. and Collins,R.A. (1990) A site-specific self-cleavage reaction performed by a novel RNA in Neurospora mitochondria. Cell, 61, 685–696. [DOI] [PubMed] [Google Scholar]
- Sclavi B., Sullivan,M., Chance,M.R., Brenowitz,M. and Woodson,S.A. (1998) RNA folding at millisecond intervals by synchrotron hydroxyl radical footprinting. Science, 279, 1940–1943. [DOI] [PubMed] [Google Scholar]
- Sigurdsson S.T., Thomson,J.B. and Eckstein,F. (1998) Small ribozymes. In Simons,R.W. and Grunberg-Manago,M. (eds), RNA Structure and Function. Cold Spring Harbor Laboratory Press, Plainview, NY, pp. 339–376.
- Sood V.D., Beattie,T.L. and Collins,R.A. (1998) Identification of phosphates involved in metal binding and tertiary interactions in the core of the Neurospora VS ribozyme. J. Mol. Biol., 282, 741–750. [DOI] [PubMed] [Google Scholar]
- Swisher J., Duarte,C.M., Su,L.J. and Pyle,A.M. (2001) Visualizing the solvent-inaccessible core of a group II intron ribozyme. EMBO J., 20, 2051–2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinoco I. Jr and Bustamente,C. (1999) How RNA folds. J. Mol. Biol., 293, 271–281. [DOI] [PubMed] [Google Scholar]
- Walter N.G., Hampel,K.J., Brown,K.M. and Burke,J.M. (1998) Tertiary structure formation in the hairpin ribozyme monitored by fluorescence resonance energy transfer. EMBO J., 17, 2378–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]